Abstract
Interferon is known as a pleiotropic factor in innate immunity, cancer immunity and therapy. Despite an objective short-term response of interferon (IFN) therapy in renal cell carcinoma (RCC) patients, the potential adverse effect of IFN on RCC cells is not fully understood. In this study, we demonstrate that IFNs can enhance RCC invasion via a new mechanism of IFIT5-mediated tumor suppressor microRNA (miRNA) degradation resulted in the elevation of Slug and ZEB1 and epithelial-to-mesenchymal transition (EMT). Clinically, a significant upregulation of IFNγ signaling pathway (such as IFNGR1, IFNGR2, STAT1 and STAT2) is observed in RCC patients with metastatic disease. Overall, this study provides a new mechanism of action of IFN-elicited canonical pathway in regulating suppressor miRNAs. Most importantly, it highlights the potential pro-metastatic effect of IFNs, which could undermine the clinical applicability of IFNs for treating RCC patients.
Keywords: Interferon (IFN), interferon-induced tetratricopeptide repeat 5 (IFIT5), epithelial-to-mesenchymal transition (EMT)
Introduction
Renal cell carcinoma (RCC) is by far the most lethal urologic malignancy of cancer-specific mortality of 40% compared with 25% of overall mortality of prostate and bladder cancers because it is resistant to chemotherapeutics and radiotherapy [1]. Pathologically, RCC is a heterogenous disease consisting three major types: clear cell RCC (ccRCC) [2], papillary RCC [3] and chromophobe RCC [4]. As for the therapeutic strategy for RCC patients, usually localized primary tumor can be managed successfully with radical nephrectomy. On the contrary, despite the prognosis of metastatic RCC (mRCC) patients has improved with targeted therapies, 20-25% of the patients are refractory to chemotherapy at the first response assessment and acquire drug resistance during the treatment [5].
Accumulating studies have demonstrated that tumor-associated immune cells play a critical role in cancer development. The presence of tumor-associated macrophage (TAM) is able to increase tumor cell proliferation and dissemination in different cancer types [6-12]. Action of these tumor-infiltrating lymphocytes (TILs) is mainly mediated through secretion of cytokines such as interleukins (IL-1, IL-6, IL-8 and IL-10), tumor necrosis factors (TNF), and interferons (IFNs). Many studies have shown that these cytokines could stimulate tumor cell proliferation, protect tumor cells from apoptosis, or promote angiogenesis and metastasis. Interleukin-6 (IL-6) is known to act as pro-tumorigenic cytokine by facilitating cell growth and anti-apoptosis in multiple myeloma [13,14]. High concentration of interleukin-1 (IL-1) is associated with more malignant tumor phenotype [15-18]. Both IL-1α and IL-β are implied to aggravate tumor angiogenesis and invasiveness via induction of vascular endothelial cell growth factor (VEGF) and tumor necrosis factor (TNF) [18]. On the other hand, the role of IFNs in cancer development remains controversial. For example, both IFN and IFNγ can exhibit anti-tumor activities [19-25]. IFNγ is also responsible for antigen-specific tumor immunity [26-29]. In contrast, IFNγ was reported to facilitate lung metastasis in melanoma 30 and peritoneal dissemination of ovarian cancer [31]. In prostate cancer, our recent study also indicates that IFNγ can promote epithelial-to-mesenchymal transition (EMT) via a new mechanism of IFIT5 in specific microRNA (miRNA) turnover [32].
Clinically, a significant elevation of several key effectors (such as IFNγ receptor 1 and 2 [IFNGR1, IFNGR2], STAT1, STAT2) in IFNγ signaling pathway is associated with metastatic RCC tumor [33]. Nevertheless, the role of IFNs in RCC development is not fully characterized. In this study, we demonstrate that IFN-elicited RCC invasion is mediated by IFIT5-XRN1 complex responsible for specific microRNA (such as miR-363) turnover; loss of miR-363 expression has been detected in RCC specimens [34]. Taken together, IFN is a potent tumor promoter that is mediated by a new mechanism of action of IFIT5 in degrading tumor suppressor miRNA.
Materials and methods
Cell lines
ACHN, 768O and 769P cell lines were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS). 293 cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% FBS. Stable IFIT5-shRNA knockdown (shIFIT5) and control (shCon) RCC cell lines were established from ACHN, 786O and 769P cell lines using pLK0.1-shCon or pLK0.1-shIFIT5 plasmids provided from Academia Sinica, Taiwan. All RCC cell lines were authenticated using the short tandem repeat (STR) profiling by Genomic Core in UT Southwestern Medical Center (UTSW). Mycoplasma testing was performed using MycoAlert® kit (Lonza Walkersville, Inc. Walkersville, MD) every quarterly to ensure Mycoplasma-free condition.
Invasion assay
RCC cells cultured in the serum-free RPMI-1640 medium for 18 hrs were plated onto the upper chamber of Transwell (8-um (i.e., 8-micrometer) pore size, Corning) pre-coated with 2.5% Matrigel, and bottom chamber contained RPMI-1640 medium with 10% FBS. After 24 hrs, cells from the bottom side of chamber were fixed by 4% paraformaldehyde, stained with 0.4% Crystal Violet and observed under microscope (Keyance). The crystal-violet-stained cells from each field were quantified using BZ-X Analyzer software. Relative invaded cells were normalized to control group of each experiment. Each experiment was performed in triplicates.
Construction of SSMut and DSMut pre-miRNA-expression plasmid
miR-363 and miR-128 expression plasmid was initially purchased from Origene and Genecopoeia, respectively. Native miR-363 or miR-128 expression plasmid was engineered to generate mutant pre-miR-363 or mutant miR-128 with 5’-end six nucleotides single stranded overhang (pre-SS6Mut-miR-363 or pre-SS6Mut-miR-128) or double-stranded blunt end (pre-DSMut-miR-363 or pre-DSMut-miR-128) constructs using QuickChange II site-directed mutagenesis kit (Agilent Technologies) and the mutated sequence was confirmed by DNA sequencing from Genomic Core in UTSW.
RNA purification
RCC cells were pelleted by centrifuge at 5000 rpm for 2 mins then snap-frozen in liquid nitrogen before RNA extraction. Chilled 1-Thioglycerol/Homogenization Solution (200 µl) was added to re-suspend the pellet, followed by additional of 200 µl Lysis Buffer with 15 µl Proteinase K solution. Samples were vortexed and incubated at room temperature for 10 mins before loading into Maxwell® cartridge. RNA sample was purified in the Maxwell® RSC Instrument using the microRNA Tissue Kit and eluted in 40-60 µl Nuclease-Free Water.
Quantitative real-time RT-PCR (qRT-PCR)
For the quantification of miRNA expression level, total RNA 2.5 ug (i.e., 2.5 microgram) was subjected to miScript II RT kit (QIAGEN) then 2.0 µl cDNA was applied to a 25-μL reaction volume using miScript SYBR® Green PCR kit (QIAGEN) in CFX96 Touch Real-Time PCR detection system (BioRad). Primer assays for each miRNA species were purchased from QIAGEN. The relative expression levels of matured miRNAs from each sample were determined by normalizing to SNORD95 small RNA. For the quantification of mRNA expression level, total RNA (2 g) was subjected to iScript advanced cDNA synthesis kit (BioRad) then 2.0 μl cDNA was applied to 25-μl qRT-PCR reaction volume using iTaq Universal SYBR® Green supermix (BioRad) in CFX384 Touch Real-Time PCR detection system (BioRad). The relative expression levels of IFIT5, Slug, ZEB1, E-cadherin, and Vimentin mRNA from each sample were determined by normalizing to 18S mRNA. All quantitative data were analyzed using ΔCt (Ct value normalized to internal SNORD95 miRNA or 18S RNA) and the fold change (ΔΔCt) was obtained after normalizing with the control group.
Western blot analysis
RCC Cells were lysed in lysis buffer [50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% Triton X-100, 1 mM sodium orthovanadate, 1 mM sodium fluoride, 1 mM sodium pyrophosphate, 10 mg/mL, aprotinin, 10 mg/mL leupeptin, 2 mM phenylmethylsulfonyl fluoride, and 1 mM EDTA] for 30 mins on ice. Cell lysates were spin down at 20,000×g for 20 mins at 4°C. Protein extracts were subjected to SDS-PAGE using Bolt 4-12% Bis-Tris Plus gel (Invitrogen), and transferred to nitrocellulose membrane using Trans-Blot Turbo Transfer system (BIORAD). Membranes were incubated with primary antibodies against IFIT5 (ProteinTech), Slug (Cell Signaling Technology), ZEB1 (Cell Signaling Technology), E-Cadherin (BD Transduction Laboratory), Vimentin (Sigma-Aldrich), STAT1 (Santa Cruz Biotechnology), GAPDH (Santa Cruz Biotechnology) or HRP-conjugated Flag (Sigma-Aldrich) antibodies at 4°C for 16-18 hrs, followed by incubation with horseradish peroxidase-conjugated secondary antibodies at room temperature for 1.5 hrs. Results were visualized with ECL chemiluminescent detection system (Thermo Scientific Pierce) using Alphaimager instrument. The relative protein expression level in each sample was normalized by GAPDH levels.
Luciferase reporter assay
Established stable clones of control (shCon) or STAT1-knockdown (shSTAT1) 786O cells (8×104) were seeded onto 12-well plates at 75% confluence before transfection with IFIT5 promoter-luciferase reporter plasmid. Cells were harvested and lysed with Passive Lysis buffer (Promega) at 48 hrs after transfection. Luciferase activity was measured using the Firefly luciferase reporter assay (Promega) on the Veritas Microplate Luminometer (Turner Biosystems). Relative luciferase activity was determined by normalizing with control. Each experiment was performed in triplicates.
Statistics analysis
Statistics analyses were performed using GraphPad Prism software. Statistical significance was evaluated using Student t-test. P values of P<0.05, P<0.001 and P<0.00001 were considered significant difference between compared groups and marked with asterisks.
Results
IFN promotes RCC invasion via induction of IFIT5
Prior to targeted therapy for RCC patients, IFNα has demonstrated a short-term efficacy as a single agent [35-39] and improved the overall survival by combining with other agents such as cyclooxygenase-2 inhibitor (celecoxib) [40], interleukin-2 [41], capecitabine [42] or sorafenib [43,39,44]. On the other hand, IFNγ therapy resulted in minimal anti-tumor activity among mRCC patients [45,46]. Thus, we decided to study potential adverse effect of IFNγ and observed that either IFNα or IFNγ was able to facilitate cell invasion of ACHN and 786O cell lines using Transwell invasion assay (Figure 1A). Since IFNγ appeared more potent than IFNα, we decided to focus IFNγ to unveil its mechanism of action. Indeed, IFNγ treatment is able to activate the canonical pathway of STAT1 phosphorylation to increase IFIT5 expression (Figures 1B, S1A and S1B), a bona fide IFN-induced gene [47,48] that is capable of promoting EMT in prostate cancer [32]. To elucidate the role of IFIT5 in IFNγ-induced RCC cell invasion, we knocked down IFIT5 in ACHN, 786O and 769P cell lines and demonstrated that IFNγ-induced cell invasion is diminished in IFIT5-knockdown (KD) cells (Figures 1C, S1C and S1D). Indeed, IFNγ is able to increase both Slug and ZEB1 gene expression leading to EMT in ACHN cells (Figure 1D and 1E). However, in IFIT5-knockdown (KD) cells, IFNγ failed to induce Slug and ZEB1 gene expression as well as EMT change based on E-Cadherin and Vimentin expression (Figures 1D, 1E and S1E). Taken together, these data support the notion that IFIT5 is the key mediator in IFNγ-induced RCC invasion.
Figure 1.
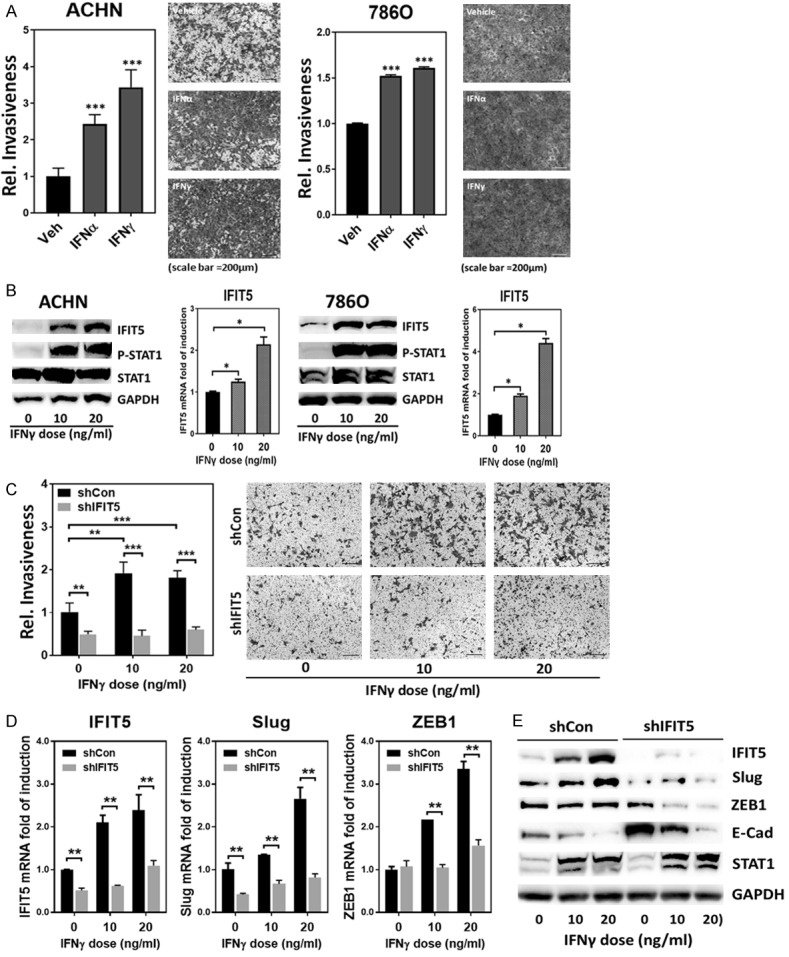
IFNγ promotes renal cancer invasion via induction of IFIT5. A. Increased invasiveness of ACHN or 786O cells after IFNα or IFNγ treatment (20 ng/ml) for 48 hrs, compared to vehicle control. (***P<0.00001). B. Dose-dependent elevation of IFIT5 protein and mRNA level in renal cancer cell lines (ACHN and 786O) treated with IFNγ for 48 hrs, compared to vehicle control. (*P<0.05). C. The impact of IFIT5 loss (shIFIT5) on the IFNγ-enhanced aggravation of invasiveness in ACHN cells, compared to shCon. (**P<0.001, ***P<0.00001). D. The impact of IFIT5 loss (shIFIT5) on the IFNγ-induced elevation of Slug and ZEB1 mRNA level in ACHN cells, compared to shCon (**P<0.001). E. The impact of IFIT5 loss (shIFIT5) on the IFNγ-induced alteration of Slug, ZEB1 and E-Cadherin (E-Cad) protein level in ACHN cells, compared to shCon.
IFIT5 complex regulates miR-363 turnover
IFIT5 has been characterized to function as a binding protein for various RNA species (such as viral RNA, tRNA) [47,49,50]. Our recent data [32] demonstrate that a new function of IFIT5 is to recruit XRN1 exoribonuclease to degrade miRNA by recognizing the unique 5’-structure of precursor miRNA. In addition, loss of miR-363 is detected in RCC specimens [34]. Thus, we further validated whether IFIT5 with a similar functional role could contribute the loss of miR-363 in RCC cells. Noticeably, miR-363 belongs to the miR-106a-363 cluster containing miR-106a, miR-18b, miR-20b, miR-19b, miR-92a-2 and miR-363 [51-54]. As shown in Figure 2A and 2B, only mature miR-363 but no other miRNA species from this cluster was elevated in IFIT5-KD cells.
Figure 2.
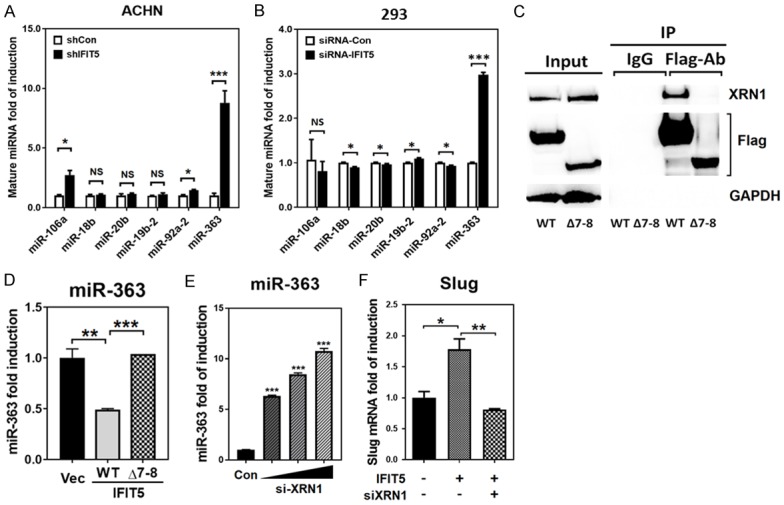
Recruitment of XRN1 is required for the machinery of IFIT5-mediated miR-363 degradation. A, B. The impact of IFIT5 shRNA knockdown (shIFIT5) on the expression level of miRNAs derived from the miR-106a-363 cluster (miR-106a, miR-18b, miR-20b, miR-19b-2, miR-92a-2 and miR-363) in ACHN and 293T cells, compared to control shRNA (shCon). (*P<0.05, ***P<0.00001). C. Co-Immunoprecipitation using flag antibody to pulled down flag-tagged WT or mutant (Δ7-8 TPR deletion) IFIT5 protein overexpressed in 293 cells, and immunoblotted with or and flag antibody. D. Expression level of miR-363 in cells overexpressed with WT or mutant (Δ7-8 TPR deletion) IFIT5, compared to vector control (**P<0.001, ***P<0.0001). E. Dose-dependent expression level of miR-363 in IFIT5-positive 293 cells transfected with siRNA-knockdown of XRN1 (***P<0.0001). F. Expression level of Slug in IFIT5-overexpressed 786O cells transfected with siRNA-XRN1, compared to vector control (*P<0.05, **P<0.001).
Since XRN1 is required for the activity of IFIT5 complex, we further determine the binding domain in IFIT5 for XRN1 and found 7-8 TPR domain (Δ7-8) as a key binding domain (Figure 2C). This truncated IFIT5 was incapable of degradation of miR-363 (Figure 2D). Furthermore, our data demonstrate that XRN1 is required for IFIT5-mediated miRNA degradation (Figure 2E). Consistently, the restoration of miR-363 level by siRNA-knockdown of XRN1 resulted in the suppression of Slug mRNA expression in 786O cells (Figure 2F). Taken together, we believe that the presence of IFIT5-XRN1 contributes to the loss of miR-363 in RCC cells.
miR-363 functions as a potent suppressor in IFNγ-induced cell invasion
Knowing IFIT5 as a potent regulator for miR-363 turnover (Figure 2A and 2B), we further demonstrated that IFNγ exhibited the same specific degradation of miR-363 from miR-106a-363 cluster in both ACHN and 786O cells (Figure 3A and 3B). Also, IFNγ-induced downregulation of miR-363 can be reversed in IFIT5 KD cells (Figure 3C), supporting the mechanism of IFNγ on miRNA turnover is mediated by IFIT5 complex.
Figure 3.
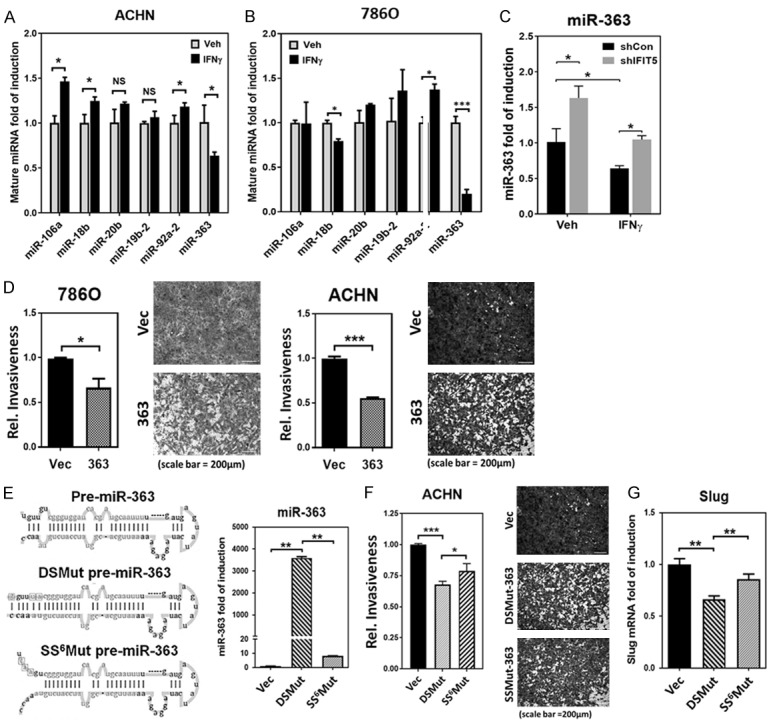
IFIT5 regulates miR-363 turnover via recognition of precursor miRNA 5’end. A, B. The impact of IFNγ on the expression level of miRNAs derived from the miR-106a-363 cluster (miR-106a, miR-18b, miR-20b, miR-19b-2, miR-92a-2 and miR-363) in ACHN and 786O cells, compared to vehicle control (*P<0.05, ***P<0.0001). C. The impact of IFIT5 loss (shIFIT5) on the IFNγ-induced downregulation of miR-363 level in ACHN cells. (*P<0.05). D. Attenuated cancer invasiveness by miR-363 overexpression in 786O or ACHN cells, compared to vector control (*P<0.05, ***P<0.00001). E. Left: Mutation of nucleotides (box) for generating 5’-end 6 nucleotides single stranded pre-miR-363 (SS6Mut pre-miR-363) and blunt 5’-end double stranded pre-miR-363 (DSMut pre-miR-363). Both mature miR-363 and miR-363* sequence are shown in lighter gray. Right: Expression level of mature miR-363 in 293 cells transfected with plasmids carrying mutant SS6Mut or DSMut pre-miR-363 sequence. (**P<0.001). F. The impact of mutant SS6Mut or DSMut pre-miR-363 on suppression of cell invasion in ACHN cells (*P<0.05, ***P<0.00001). G. Expression level of Slug mRNA in 293 cells transfected with plasmids carrying mutant SS6Mut or DSMut pre-miR-363 sequence. (**P<0.001).
By increasing miR-363 expression in either ACHN and 786O cells, a significant reduction of IFNγ-elicited cell invasion was observed (Figure 3D). Recently, we have demonstrated that IFIT5 preferentially bind to 5’-end single-stranded overhang sequence of precursor miRNAs [32]. Thus, the blunt-end (i.e., DSMut) of pre-miR-363 appears more resistant to IFIT5-mediated degradation than open-end (i.e., SS6Mut) of pre-miR-363 (Figure 3E). Functionally, the DSMut of pre-miR-363 is more potent in suppressing cell invasion of several RCC cells (Figures 3F and S2A) as well as Slug gene expression (Figure 3G). Overall, these data provide some understanding of induction of Slug by IFNγ in RCC cells.
IFIT5 regulates the turnover of miR-128 that can target ZEB1
In addition, ZEB1 induction was detected in IFNγ-treated RCC cells (Figure 1E). We hypothesized that this regulation could be mediated by the same mechanism. Thus, by searching IFIT5-binding miRNA candidates, we identified pre-miR-128 with as similar single-stranded overhang structure at its 5’-end (Figure 4A left panel) and targeting ZEB1. By modify its native structure, the DSMut of pre-miR-128 became more resistant than the SS6Mut of pre-miR-128 (Figure 4A right panel). Similar to miR-363, the DSMut of pre-miR-128 is more potent in suppressing cell invasion of several RCC cells (Figures 4B and S2B) as well as ZEB1 gene expression (Figure 4C).
Figure 4.
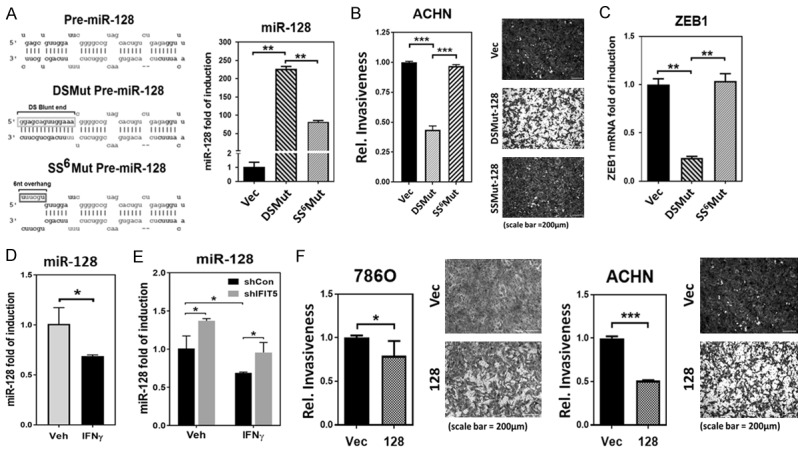
IFIT5 regulates miR-128 turnover via recognition of 5’-end of precursor miRNA. A. Left: Mutation of nucleotides (box) for generating 5’-end 6 nucleotides single stranded pre-miR-128 (SS6Mut pre-miR-128) and blunt 5’-end double stranded pre-miR-128 (DSMut pre-miR-128). Both mature miR-128 and miR-128* sequence are shown in lighter gray. Right: Expression level of mature in 293 cells transfected with plasmids carrying mutant SS6Mut or DSMut pre-miR-128 sequence. (**P<0.001). B. The impact of mutant DSMut or SS6Mut pre-miR-128 on suppression of cell invasion in ACHN cells (***P<0.00001). C. Expression level of ZEB1 mRNA in 293 cells transfected with plasmids carrying mutant SS6Mut or DSMut pre-miR-128 sequence. (**P<0.001). D. The impact of IFNγ and IFIT5 knockdown on the expression level of miR-128, compared to vehicle and control shRNA, respectively. (*P<0.05). E. The impact of IFIT5 loss (shIFIT5) on the IFNγ-induced downregulation of miR-128 level in ACHN cells. (*P<0.05). F. Attenuated cancer invasiveness by miR-128 overexpression in 786O or ACHN cells, compared to vector control (*P<0.05, ***P<0.00001).
Consistent with the down-regulation of miR-363 by IFNγ (Figure 3A), we also observed the reduction of miR-128 level in IFNγ-treated ACHN cells (Figure 4D). Moreover, IFNγ-induced down-regulation of miR-128 can be rescued in the IFIT5 KD cells (Figure 4E). On the other hand, by increasing miR-128 expression in either ACHN and 786O cells, a significant reduction of IFNγ-elicited cell invasion was observed (Figure 4F). Taken together, we believe that decreased miR-128 regulated by IFIT5 complex underlies IFNγ-induced ZEB1 contributing to RCC cell invasion.
IFNγ-signaling pathway is correlated with RCC progression
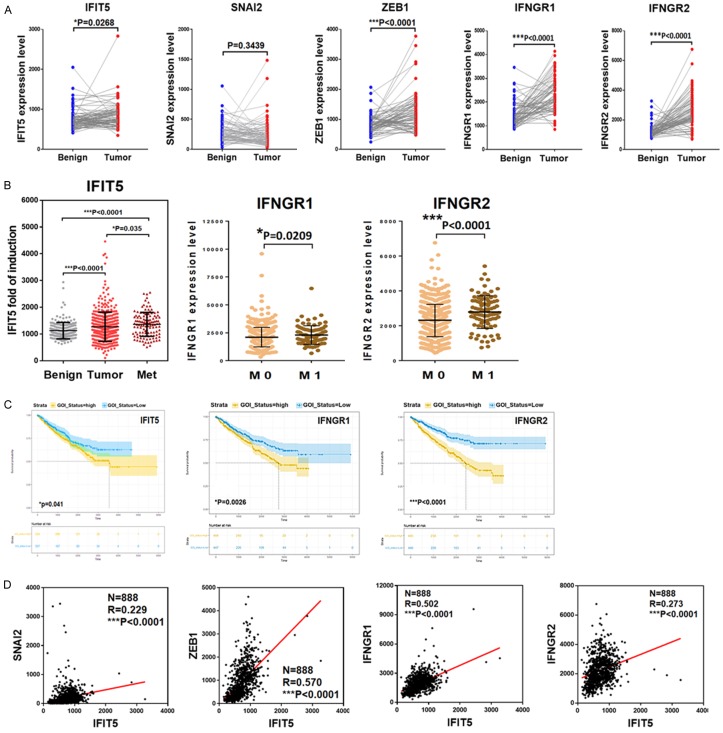
To strengthen our observation from in vitro model, we further analyzed the IFNγ-signaling pathway from 75 paired adjacent benign and tumor tissues of ccRCC patients and the results (Figure 5A) indicated a significant elevation of IFNγ receptor 1 or 2 (IFNGR1, IFNGR2) and IFIT5 in tumor specimens. In addition, the expression levels of IFNGR1, IFNGR2, and IFIT5 in mRCC patients are significantly higher than primary tumor or benign tissue (Figure 5B). Moreover, patients with higher level of elevated IFNGR1, IFNGR2, and IFIT5 predicts poor overall survival of RCC patients based on the Kaplan-Meier curves analyses (Figure 5C). As expected, a positive correlation between IFIT5 and Slug, ZEB1, IFNGR1 or IFNGR2 expression level is also observed after analyzing TCGA RCC dataset (Figure 5D). Overall, these clinical data support the pro-tumorigenic role of IFNγ-signaling axis in RCC progression.
Figure 5.
Machinery of IFIT5-mediated miRNA degradation is enhanced in the metastatic progression of RCC. A. Expression level of IFIT5, SNAI2/Slug, ZEB1, IFNGR1 and IFNGR2 in paired adjacent benign and tumor specimens derived from 75 RCC patients. B. Expression of IFIT5 in the patient-derived xenografts from normal benign (N=191), primary (N=294) and metastatic tumors (N=86) of RCC patients. Expression level of IFNGR1 and IFNGR2 genes in tumor specimens derived from RCC patients with non-metastasis (M0, N=550) and metastasis (M1, N=90) status. C. Kaplan-Meier overall survival curves estimate patients with RCC including types of clear cell RCC, papillary RCC and Chromophobe RCC, stratified in low and high expression of IFIT5, IFNGR1 or IFNGR2 genes. D. Clinical correlation between IFIT5 and SNAI2(Slug), ZEB1, IFNGR1 or IFNGR2 in TCGA dataset derived from 888 RCC patient tumor specimens.
Discussion
The incidence of RCC has been steadily rising by 2-4% each year; a 5-fold increase in the incidence and a two-fold increase in mortality compared to 1971 [55]. RCC is by far the most lethal urologic malignancy of cancer-specific mortality of 40% compared with 25% of overall mortality of prostate and bladder cancers once it becomes metastatic. The median OS of un-treated metastatic disease is 5 months with 1-year survival of only 29%. Although, a recent study elegantly demonstrated the predisposition of different genetic mutation in RCC patients to various organ metastases [56,57], the influence of tumor microenvironment and its regulatory network are not fully characterized.
At the initial stage of cancer metastasis, tumor cells could undergo a phenotypic change from epithelial type to mesenchymal type (i.e., EMT), leading to loss of cell-cell adhesion, increased cell motility and invasion into vascular circulation [58]. Loss of E-Cadherin (epithelial marker) and gain of Vimentin (mesenchymal marker) is associated with RCC metastasis [59] and the presence of a sarcomatoid component is often found in metastatic lesion of RCC and associated with high mortality. Slug (Snail2), an EMT transcription repressor of E-Cadherin, has been demonstrated to induce EMT in RCC cell lines [60-62]. Slug expression is substantially increased in high-grade ccRCC tissues and sarcomatoid carcinoma, which predicts the worse prognosis for patients with RCC [63]. Tumor surrounding microenvironment is known to play an important role in EMT [64-67] induction by producing several cytokines such as TNF-α [68], TGF-β [69] and IL-10 [67].
In RCC, a number of cytokines have shown anti-tumor activity, the most consistent results have been reported with IFNα [70]. IL-2 and IFNγ have also been given to patients with metastatic RCC with limited success [70]. Although the mechanism of action of these cytokines is poorly understood, anti-tumor effects in murine models have been linked to the direct killing of tumor cells by activated T cells and natural killer cells, as well as to anti-angiogenic effects [71-73]. However, in this study, we demonstrated the pro-tumorigenic role of IFNγ in promoting RCC cancer invasion. IFNγ signaling pathway is highly elevated in primary as well metastatic RCC and significantly correlated with OS RCC patients, suggesting the oncogenic role of IFNγ-IFNGR1/IFNGR2-IFIT5 signal axis in the progression of RCC. Noticeably, this study unveils a new mechanism of action of IFIT5 in regulating turnover of tumor suppressor miRNAs (miR-363 and miR-128), leading to the upregulation of EMT drivers (Slug and ZEB1). Moreover, IFNγ promoting cancer metastasis is not only limited to RCC; it has been recently reported in other malignancies such as prostate [32] and gastric cancer [74]. In summary, we demonstrate an adverse effect of IFNγ on RCC progression via a unique regulation of miRNA turnover. Therefore, targeting IFNγ signaling pathway using inhibitory RNA strategy may provide a new avenue for the clinical management of RCC progression.
Acknowledgements
We thank Dr. Samarpita Sengupta for editorial assistance. This work was in part supported by grants from the United States Army (W81XWH-11-1-0491 and W81XWH-16-1-0474 to JTH) and (W81XWH-14-1-0249 to UGL), and China Scholarship Council (201508440279 to JB).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, Heng DY, Larkin J, Ficarra V. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009. doi: 10.1038/nrdp.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–9. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Linehan WM, Spellman PT, Ricketts CJ, Creighton CJ, Fei SS, Davis C, Wheeler DA, Murray BA, Schmidt L, Vocke CD, Peto M, Al Mamun AA, Shinbrot E, Sethi A, Brooks S, Rathmell WK, Brooks AN, Hoadley KA, Robertson AG, Brooks D, Bowlby R, Sadeghi S, Shen H, Weisenberger DJ, Bootwalla M, Baylin SB, Laird PW, Cherniack AD, Saksena G, Haake S, Li J, Liang H, Lu Y, Mills GB, Akbani R, Leiserson MD, Raphael BJ, Anur P, Bottaro D, Albiges L, Barnabas N, Choueiri TK, Czerniak B, Godwin AK, Hakimi AA, Ho TH, Hsieh J, Ittmann M, Kim WY, Krishnan B, Merino MJ, Mills Shaw KR, Reuter VE, Reznik E, Shelley CS, Shuch B, Signoretti S, Srinivasan R, Tamboli P, Thomas G, Tickoo S, Burnett K, Crain D, Gardner J, Lau K, Mallery D, Morris S, Paulauskis JD, Penny RJ, Shelton C, Shelton WT, Sherman M, Thompson E, Yena P, Avedon MT, Bowen J, Gastier-Foster JM, Gerken M, Leraas KM, Lichtenberg TM, Ramirez NC, Santos T, Wise L, Zmuda E, Demchok JA, Felau I, Hutter CM, Sheth M, Sofia HJ, Tarnuzzer R, Wang Z, Yang L, Zenklusen JC, Zhang J, Ayala B, Baboud J, Chudamani S, Liu J, Lolla L, Naresh R, Pihl T, Sun Q, Wan Y, Wu Y, Ally A, Balasundaram M, Balu S, Beroukhim R, Bodenheimer T, Buhay C, Butterfield YS, Carlsen R, Carter SL, Chao H, Chuah E, Clarke A, Covington KR, Dahdouli M, Dewal N, Dhalla N, Doddapaneni HV, Drummond JA, Gabriel SB, Gibbs RA, Guin R, Hale W, Hawes A, Hayes DN, Holt RA, Hoyle AP, Jefferys SR, Jones SJ, Jones CD, Kalra D, Kovar C, Lewis L, Li J, Ma Y, Marra MA, Mayo M, Meng S, Meyerson M, Mieczkowski PA, Moore RA, Morton D, Mose LE, Mungall AJ, Muzny D, Parker JS, Perou CM, Roach J, Schein JE, Schumacher SE, Shi Y, Simons JV, Sipahimalani P, Skelly T, Soloway MG, Sougnez C, Tam A, Tan D, Thiessen N, Veluvolu U, Wang M, Wilkerson MD, Wong T, Wu J, Xi L, Zhou J, Bedford J, Chen F, Fu Y, Gerstein M, Haussler D, Kasaian K, Lai P, Ling S, Radenbaugh A, Van Den Berg D, Weinstein JN, Zhu J, Albert M, Alexopoulou I, Andersen JJ, Auman JT, Bartlett J, Bastacky S, Bergsten J, Blute ML, Boice L, Bollag RJ, Boyd J, Castle E, Chen YB, Cheville JC, Curley E, Davies B, DeVolk A, Dhir R, Dike L, Eckman J, Engel J, Harr J, Hrebinko R, Huang M, Huelsenbeck-Dill L, Iacocca M, Jacobs B, Lobis M, Maranchie JK, McMeekin S, Myers J, Nelson J, Parfitt J, Parwani A, Petrelli N, Rabeno B, Roy S, Salner AL, Slaton J, Stanton M, Thompson RH, Thorne L, Tucker K, Weinberger PM, Winemiller C, Zach LA, Zuna R. Comprehensive molecular characterization of papillary renal-cell carcinoma. N Engl J Med. 2016;374:135–45. doi: 10.1056/NEJMoa1505917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis CF, Ricketts CJ, Wang M, Yang L, Cherniack AD, Shen H, Buhay C, Kang H, Kim SC, Fahey CC, Hacker KE, Bhanot G, Gordenin DA, Chu A, Gunaratne PH, Biehl M, Seth S, Kaipparettu BA, Bristow CA, Donehower LA, Wallen EM, Smith AB, Tickoo SK, Tamboli P, Reuter V, Schmidt LS, Hsieh JJ, Choueiri TK, Hakimi AA The Cancer Genome Atlas Research Network. Chin L, Meyerson M, Kucherlapati R, Park WY, Robertson AG, Laird PW, Henske EP, Kwiatkowski DJ, Park PJ, Morgan M, Shuch B, Muzny D, Wheeler DA, Linehan WM, Gibbs RA, Rathmell WK, Creighton CJ. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014;26:319–330. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buti S, Bersanelli M, Sikokis A, Maines F, Facchinetti F, Bria E, Ardizzoni A, Tortora G, Massari F. Chemotherapy in metastatic renal cell carcinoma today? A systematic review. Anticancer Drugs. 2013;24:535–54. doi: 10.1097/CAD.0b013e3283609ec1. [DOI] [PubMed] [Google Scholar]
- 6.Zhang C, Gao L, Cai Y, Liu H, Gao D, Lai J, Jia B, Wang F, Liu Z. Inhibition of tumor growth and metastasis by photoimmunotherapy targeting tumor-associated macrophage in a sorafenib-resistant tumor model. Biomaterials. 2016;84:1–12. doi: 10.1016/j.biomaterials.2016.01.027. [DOI] [PubMed] [Google Scholar]
- 7.Grossman JG, Nywening TM, Belt BA, Panni RZ, Krasnick BA, DeNardo DG, Hawkins WG, Goedegebuure SP, Linehan DC, Fields RC. Recruitment of CCR2(+) tumor associated macrophage to sites of liver metastasis confers a poor prognosis in human colorectal cancer. Oncoimmunology. 2018;7:e1470729. doi: 10.1080/2162402X.2018.1470729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura Y, Sumiyoshi M. Resveratrol prevents tumor growth and metastasis by inhibiting lymphangiogenesis and M2 macrophage activation and differentiation in tumor-associated macrophages. Nutr Cancer. 2016;68:667–78. doi: 10.1080/01635581.2016.1158295. [DOI] [PubMed] [Google Scholar]
- 9.Liu B, Jia Y, Ma J, Wu S, Jiang H, Cao Y, Sun X, Yin X, Yan S, Shang M, Mao A. Tumor-associated macrophage-derived CCL20 enhances the growth and metastasis of pancreatic cancer. Acta Biochim Biophys Sin (Shanghai) 2016;48:1067–1074. doi: 10.1093/abbs/gmw101. [DOI] [PubMed] [Google Scholar]
- 10.Behnes CL, Bremmer F, Hemmerlein B, Strauss A, Ströbel P, Radzun HJ. Tumor-associated macrophages are involved in tumor progression in papillary renal cell carcinoma. Virchows Arch. 2014;464:191–6. doi: 10.1007/s00428-013-1523-0. [DOI] [PubMed] [Google Scholar]
- 11.Buddingh EP, Kuijjer ML, Duim RA, Bürger H, Agelopoulos K, Myklebost O, Serra M, Mertens F, Hogendoorn PC, Lankester AC, Cleton-Jansen AM. Tumor-infiltrating macrophages are associated with metastasis suppression in high-grade osteosarcoma: a rationale for treatment with macrophage activating agents. Clin Cancer Res. 2011;17:2110–9. doi: 10.1158/1078-0432.CCR-10-2047. [DOI] [PubMed] [Google Scholar]
- 12.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4:71–8. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 13.Shen JK, Dong LH, Qi H, Zhang GS. Clinical significance of serum vascular endothelial growth factor and interleukin-6 in multiple myeloma. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2005;30:68–71. [PubMed] [Google Scholar]
- 14.Harmer D, Falank C, Reagan MR. Interleukin-6 interweaves the bone marrow microenvironment, bone loss, and multiple myeloma. Front Endocrinol (Lausanne) 2019;9:788. doi: 10.3389/fendo.2018.00788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bani MR, Garofalo A, Scanziani E, Giavazzi R. Effect of interleukin-1-beta on metastasis formation in different tumor systems. J Natl Cancer Inst. 1991;83:119–23. doi: 10.1093/jnci/83.2.119. [DOI] [PubMed] [Google Scholar]
- 16.Jagielska J, Kapopara PR, Salguero G, Scherr M, Schütt H, Grote K, Schieffer B, Bavendiek U. Interleukin-1 assembles a proangiogenic signaling module consisting of caveolin-1, tumor necrosis factor receptor-associated factor 6, p38-mitogen-activated protein kinase (MAPK), and MAPK-activated protein kinase 2 in endothelial cells. Arterioscler Thromb Vasc Biol. 2012;32:1280–8. doi: 10.1161/ATVBAHA.111.243477. [DOI] [PubMed] [Google Scholar]
- 17.Watari K, Shibata T, Kawahara A, Sata K, Nabeshima H, Shinoda A, Abe H, Azuma K, Murakami Y, Izumi H, Takahashi T, Kage M, Kuwano M, Ono M. Tumor-derived interleukin-1 promotes lymphangiogenesis and lymph node metastasis through M2-type macrophages. PLoS One. 2014;9:e99568. doi: 10.1371/journal.pone.0099568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voronov E, Shouval DS, Krelin Y, Cagnano E, Benharroch D, Iwakura Y, Dinarello CA, Apte RN. IL-1 is required for tumor invasiveness and angiogenesis. Proc Natl Acad Sci U S A. 2003;100:2645–50. doi: 10.1073/pnas.0437939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gresser I. The antitumor effects of interferon: a personal history. Biochimie. 2007;89:723–8. doi: 10.1016/j.biochi.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 20.Ferrantini M, Proietti E, Santodonato L, Gabriele L, Peretti M, Plavec I, Meyer F, Kaido T, Gresser I, Belardelli F. Alpha 1-interferon gene transfer into metastatic friend leukemia cells abrogated tumorigenicity in immunocompetent mice: antitumor therapy by means of interferon-producing cells. Cancer Res. 1993;53:1107–12. [PubMed] [Google Scholar]
- 21.Gabriele L, Kaido T, Woodrow D, Moss J, Ferrantini M, Proletti E, Santodonato L, Rozera C, Maury C, Belardelli F, et al. Local and systemic response of mice to interferon-alpha 1-transfected Friend leukemia cells. Am J Pathol. 1995;147:445–460. [PMC free article] [PubMed] [Google Scholar]
- 22.Shin EC, Shin WC, Choi Y, Kim H, Park JH, Kim SJ. Effect of interferon-gamma on the susceptibility to Fas (CD95/APO-1)-mediated cell death in human hepatoma cells. Cancer Immunol Immunother. 2001;50:23–30. doi: 10.1007/s002620000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Egwuagu CE, Li W, Yu CR, Che Mei Lin M, Chan CC, Nakamura T, Chepelinsky AB. Interferon-gamma induces regression of epithelial cell carcinoma: critical roles of IRF-1 and ICSBP transcription factors. Oncogene. 2006;25:3670–9. doi: 10.1038/sj.onc.1209402. [DOI] [PubMed] [Google Scholar]
- 24.Xu Z, Hurchla MA, Deng H, Uluçkan O, Bu F, Berdy A, Eagleton MC, Heller EA, Floyd DH, Dirksen WP, Shu S, Tanaka Y, Fernandez SA, Rosol TJ, Weilbaecher KN. Interferon-gamma targets cancer cells and osteoclasts to prevent tumor-associated bone loss and bone metastases. J Biol Chem. 2009;284:4658–66. doi: 10.1074/jbc.M804812200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aulitzky W, Gastl G, Aulitzky WE, Herold M, Kemmler J, Mull B, Frick J, Huber C. Successful treatment of metastatic renal cell carcinoma with a biologically active dose of recombinant interferon-gamma. J. Clin. Oncol. 1989;7:1875–84. doi: 10.1200/JCO.1989.7.12.1875. [DOI] [PubMed] [Google Scholar]
- 26.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, Schreiber RD. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–11. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 27.Panelli MC, Wang E, Shen S, Schluter SF, Bernstein RM, Hersh EM, Stopeck A, Gangavalli R, Barber J, Jolly D, Akporiaye ET. Interferon gamma (IFNgamma) gene transfer of an EMT6 tumor that is poorly responsive to IFNgamma stimulation: increase in tumor immunogenicity is accompanied by induction of a mouse class II transactivator and class II MHC. Cancer Immunol Immunother. 1996;42:99–107. doi: 10.1007/s002620050258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phan-Lai V, Kievit FM, Florczyk SJ, Wang K, Disis ML, Zhang M. CCL21 and IFNgamma recruit and activate tumor specific T cells in 3D scaffold model of breast cancer. Anticancer Agents Med Chem. 2014;14:204–10. doi: 10.2174/18715206113136660375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou F. Molecular mechanisms of IFN-gamma to up-regulate MHC class I antigen processing and presentation. Int Rev Immunol. 2009;28:239–260. doi: 10.1080/08830180902978120. [DOI] [PubMed] [Google Scholar]
- 30.Taniguchi K, Petersson M, Höglund P, Kiessling R, Klein G, Kärre K. Interferon gamma induces lung colonization by intravenously inoculated B16 melanoma cells in parallel with enhanced expression of class I major histocompatibility complex antigens. Proc Natl Acad Sci U S A. 1987;84:3405–9. doi: 10.1073/pnas.84.10.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abiko K, Matsumura N, Hamanishi J, Horikawa N, Murakami R, Yamaguchi K, Yoshioka Y, Baba T, Konishi I, Mandai M. IFN-gamma from lymphocytes induces PD-L1 expression and promotes progression of ovarian cancer. Br J Cancer. 2015;112:1501–9. doi: 10.1038/bjc.2015.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lo UG, Pong RC, Yang D, Gandee L, Hernandez E, Dang A, Lin CJ, Santoyo J, Ma S, Sonavane R, Huang J, Tseng SF, Moro L, Arbini AA, Kapur P, Raj GV, He D, Lai CH, Lin H, Hsieh JT. IFN-r-induced IFIT5 promotes epithelial-to-mesenchymal transition in prostate cancer via microRNA processing. Cancer Res. 2018 doi: 10.1158/0008-5472.CAN-18-2207. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 33.Au-Yeung N, Mandhana R, Horvath CM. Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAKSTAT. 2013;2:e23931. doi: 10.4161/jkst.23931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung M, Mollenkopf HJ, Grimm C, Wagner I, Albrecht M, Waller T, Pilarsky C, Johannsen M, Stephan C, Lehrach H, Nietfeld W, Rudel T, Jung K, Kristiansen G. MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. J Cell Mol Med. 2009;13:3918–28. doi: 10.1111/j.1582-4934.2009.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hofmockel G, Wirth MP, Heimbach D, Frohmüller HG. Results of low dosage cyclic interferon-gamma therapy of metastatic renal cell carcinoma. Urologe A. 1993;32:290–4. [PubMed] [Google Scholar]
- 36.Nishisaka N, Yoshihara H, Nakatani T, Sugimura K, Maekawa T, Asakawa M, Yasumoto R, Umeda M, Sakamoto W, Kawakita J, et al. Clinical study of immunotherapy with interferon alpha and gamma in metastatic renal cell carcinoma. Nihon Hinyokika Gakkai Zasshi. 1993;84:1987–93. doi: 10.5980/jpnjurol1989.84.1987. [DOI] [PubMed] [Google Scholar]
- 37.Ellerhorst JA, Kilbourn RG, Amato RJ, Zukiwski AA, Jones E, Logothetis CJ. Phase II trial of low dose gamma-interferon in metastatic renal cell carcinoma. J Urol. 1994;152:841–5. doi: 10.1016/s0022-5347(17)32587-9. [DOI] [PubMed] [Google Scholar]
- 38.Farace F, Pallardy M, Angevin E, Hercend T, Escudier B, Triebel F. Metastatic renal-cell carcinoma patients treated with interleukin 2 or interleukin 2 plus interferon gamma: immunological monitoring. Int J Cancer. 1994;57:814–21. doi: 10.1002/ijc.2910570609. [DOI] [PubMed] [Google Scholar]
- 39.Fosså SD. Interferon in metastatic renal cell carcinoma. Semin Oncol. 2000;27:187–93. [PubMed] [Google Scholar]
- 40.Onishi T, Suzuki H, Igarashi T. A case of favorable response after combination treatment with interferon-alpha and cyclooxygenase-2 inhibitor against metastatic renal cell carcinoma. Hinyokika Kiyo. 2012;58:25–9. [PubMed] [Google Scholar]
- 41.Donskov F, Jensen NV, Smidt-Hansen T, Brøndum L, Geertsen P. A randomized phase II trial of interleukin-2 and interferon-alpha plus bevacizumab versus interleukin-2 and interferon-alpha in metastatic renal-cell carcinoma (mRCC): results from the Danish renal cancer group (DaRenCa) study-1. Acta Oncol. 2018;57:589–594. doi: 10.1080/0284186X.2018.1433324. [DOI] [PubMed] [Google Scholar]
- 42.Amato RJ, Mohammad T. Interferon-alpha plus capecitabine and thalidomide in patients with metastatic renal cell cancer. J Exp Ther Oncol. 2008;7:41–7. [PubMed] [Google Scholar]
- 43.Eto M, Kawano Y, Hirao Y, Mita K, Arai Y, Tsukamoto T, Hashine K, Matsubara A, Fujioka T, Kimura G, Shinohara N, Tatsugami K, Hinotsu S, Naito S Japan RCC Trialist Collaborative Group (JRTCG) investigators. Phase II clinical trial of sorafenib plus interferon-alpha treatment for patients with metastatic renal cell carcinoma in Japan. BMC Cancer. 2015;15:667. doi: 10.1186/s12885-015-1675-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawano Y, Takahashi W, Eto M, Kamba T, Miyake H, Fujisawa M, Kamai T, Uemura H, Tsukamoto T, Azuma H, Matsubara A, Nishimura K, Nakamura T, Ogawa O, Naito S. Prognosis of metastatic renal cell carcinoma with first-line interferon-alpha therapy in the era of molecular-targeted therapy. Cancer Sci. 2016;107:1013–7. doi: 10.1111/cas.12951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aulitzky WE, Lerche J, Thews A, Lüttichau I, Jacobi N, Herold M, Aulitzky W, Peschel C, Stöckle M, Steinbach F, et al. Low-dose gamma-interferon therapy is ineffective in renal cell carcinoma patients with large tumour burden. Eur J Cancer. 1994;30A:940–5. doi: 10.1016/0959-8049(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 46.Otto F, Mackensen A, Mertelsmann R, Engelhardt R. Complete response of metastatic renal cell carcinoma to low-dose interferon-gamma treatment. Cancer Immunol Immunother. 1995;40:115–8. doi: 10.1007/BF01520293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katibah GE, Qin Y, Sidote DJ, Yao J, Lambowitz AM, Collins K. Broad and adaptable RNA structure recognition by the human interferon-induced tetratricopeptide repeat protein IFIT5. Proc Natl Acad Sci U S A. 2014;111:12025–30. doi: 10.1073/pnas.1412842111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santhakumar D, Rohaim MAMS, Hussein HA, Hawes P, Ferreira HL, Behboudi S, Iqbal M, Nair V, Arns CW, Munir M. Chicken interferon-induced protein with tetratricopeptide repeats 5 antagonizes replication of RNA viruses. Sci Rep. 2018;8:6794. doi: 10.1038/s41598-018-24905-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katibah GE, Lee HJ, Huizar JP, Vogan JM, Alber T, Collins K. tRNA binding, structure, and localization of the human interferon-induced protein IFIT5. Mol Cell. 2013;49:743–50. doi: 10.1016/j.molcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang B, Liu X, Chen W, Chen L. IFIT5 potentiates anti-viral response through enhancing innate immune signaling pathways. Acta Biochim Biophys Sin (Shanghai) 2013;45:867–74. doi: 10.1093/abbs/gmt088. [DOI] [PubMed] [Google Scholar]
- 51.Dylla L, Jedlicka P. Growth-promoting role of the miR-106a~363 cluster in Ewing sarcoma. PLoS One. 2013;8:e63032. doi: 10.1371/journal.pone.0063032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khuu C, Sehic A, Eide L, Osmundsen H. Anti-proliferative properties of miR-20b and miR-363 from the miR-106a-363 cluster on human carcinoma cells. Microrna. 2016;5:19–35. doi: 10.2174/2211536605666160322151813. [DOI] [PubMed] [Google Scholar]
- 53.Khuu C, Utheim TP, Sehic A. The three paralogous microRNA clusters in development and disease, miR-17-92, miR-106a-363, and miR-106b-25. Scientifica (Cairo) 2016;2016:1379643. doi: 10.1155/2016/1379643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gruszka R, Zakrzewska M. The oncogenic relevance of miR-17-92 cluster and its paralogous miR-106b-25 and miR-106a-363 clusters in brain tumors. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19030879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 56.Turajlic S, Xu H, Litchfield K, Rowan A, Horswell S, Chambers T, O’Brien T, Lopez JI, Watkins TBK, Nicol D, Stares M, Challacombe B, Hazell S, Chandra A, Mitchell TJ, Au L, Eichler-Jonsson C, Jabbar F, Soultati A, Chowdhury S, Rudman S, Lynch J, Fernando A, Stamp G, Nye E, Stewart A, Xing W, Smith JC, Escudero M, Huffman A, Matthews N, Elgar G, Phillimore B, Costa M, Begum S, Ward S, Salm M, Boeing S, Fisher R, Spain L, Navas C, Grönroos E, Hobor S, Sharma S, Aurangzeb I, Lall S, Polson A, Varia M, Horsfield C, Fotiadis N, Pickering L, Schwarz RF, Silva B, Herrero J, Luscombe NM, Jamal-Hanjani M, Rosenthal R, Birkbak NJ, Wilson GA, Pipek O, Ribli D, Krzystanek M, Csabai I, Szallasi Z, Gore M, McGranahan N, Van Loo P, Campbell P, Larkin J, Swanton C TRACERx Renal Consortium. Deterministic evolutionary trajectories influence primary tumor growth: TRACERx renal. Cell. 2018;173:595–610. e11. doi: 10.1016/j.cell.2018.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mitchell TJ, Turajlic S, Rowan A, Nicol D, Farmery JHR, O’Brien T, Martincorena I, Tarpey P, Angelopoulos N, Yates LR, Butler AP, Raine K, Stewart GD, Challacombe B, Fernando A, Lopez JI, Hazell S, Chandra A, Chowdhury S, Rudman S, Soultati A, Stamp G, Fotiadis N, Pickering L, Au L, Spain L, Lynch J, Stares M, Teague J, Maura F, Wedge DC, Horswell S, Chambers T, Litchfield K, Xu H, Stewart A, Elaidi R, Oudard S, McGranahan N, Csabai I, Gore M, Futreal PA, Larkin J, Lynch AG, Szallasi Z, Swanton C, Campbell PJ TRACERx Renal Consortium. Timing the landmark events in the evolution of clear cell renal cell cancer: TRACERx renal. Cell. 2018;173:611–623. e17. doi: 10.1016/j.cell.2018.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Piva F, Giulietti M, Santoni M, Occhipinti G, Scarpelli M, Lopez-Beltran A, Cheng L, Principato G, Montironi R. Epithelial to mesenchymal transition in renal cell carcinoma: implications for cancer therapy. Mol Diagn Ther. 2016;20:111–7. doi: 10.1007/s40291-016-0192-5. [DOI] [PubMed] [Google Scholar]
- 59.Katagiri A, Watanabe R, Tomita Y. E-cadherin expression in renal cell cancer and its significance in metastasis and survival. Br J Cancer. 1995;71:376–9. doi: 10.1038/bjc.1995.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harten SK, Shukla D, Barod R, Hergovich A, Balda MS, Matter K, Esteban MA, Maxwell PH. Regulation of renal epithelial tight junctions by the von Hippel-Lindau tumor suppressor gene involves occludin and claudin 1 and is independent of E-cadherin. Mol Biol Cell. 2009;20:1089–101. doi: 10.1091/mbc.E08-06-0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Evans AJ, Russell RC, Roche O, Burry TN, Fish JE, Chow VW, Kim WY, Saravanan A, Maynard MA, Gervais ML, Sufan RI, Roberts AM, Wilson LA, Betten M, Vandewalle C, Berx G, Marsden PA, Irwin MS, Teh BT, Jewett MA, Ohh M. VHL promotes E2 box-dependent E-cadherin transcription by HIF-mediated regulation of SIP1 and snail. Mol Cell Biol. 2007;27:157–69. doi: 10.1128/MCB.00892-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Esteban MA, Tran MG, Harten SK, Hill P, Castellanos MC, Chandra A, Raval R, O’brien TS, Maxwell PH. Regulation of E-cadherin expression by VHL and hypoxia-inducible factor. Cancer Res. 2006;66:3567–75. doi: 10.1158/0008-5472.CAN-05-2670. [DOI] [PubMed] [Google Scholar]
- 63.Mikami S, Katsube K, Oya M, Ishida M, Kosaka T, Mizuno R, Mukai M, Okada Y. Expression of Snail and Slug in renal cell carcinoma: E-cadherin repressor Snail is associated with cancer invasion and prognosis. Lab Invest. 2011;91:1443–58. doi: 10.1038/labinvest.2011.111. [DOI] [PubMed] [Google Scholar]
- 64.Li S, Xu F, Zhang J, Wang L, Zheng Y, Wu X, Wang J, Huang Q, Lai M. Tumor-associated macrophages remodeling EMT and predicting survival in colorectal carcinoma. Oncoimmunology. 2017;7:e1380765. doi: 10.1080/2162402X.2017.1380765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Su S, Liu Q, Chen J, Chen J, Chen F, He C, Huang D, Wu W, Lin L, Huang W, Zhang J, Cui X, Zheng F, Li H, Yao H, Su F, Song E. A positive feedback loop between mesenchymal-like cancer cells and macrophages is essential to breast cancer metastasis. Cancer Cell. 2014;25:605–20. doi: 10.1016/j.ccr.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 66.Yuan ZY, Luo RZ, Peng RJ, Wang SS, Xue C. High infiltration of tumor-associated macrophages in triple-negative breast cancer is associated with a higher risk of distant metastasis. Onco Targets Ther. 2014;7:1475–80. doi: 10.2147/OTT.S61838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu CY, Xu JY, Shi XY, Huang W, Ruan TY, Xie P, Ding JL. M2-polarized tumor-associated macrophages promoted epithelial-mesenchymal transition in pancreatic cancer cells, partially through TLR4/IL-10 signaling pathway. Lab Invest. 2013;93:844–54. doi: 10.1038/labinvest.2013.69. [DOI] [PubMed] [Google Scholar]
- 68.Hagemann T, Robinson SC, Schulz M, Trümper L, Balkwill FR, Binder C. Enhanced invasiveness of breast cancer cell lines upon co-cultivation with macrophages is due to TNF-alpha dependent up-regulation of matrix metalloproteases. Carcinogenesis. 2004;25:1543–9. doi: 10.1093/carcin/bgh146. [DOI] [PubMed] [Google Scholar]
- 69.Zhang S, Che D, Yang F, Chi C, Meng H, Shen J, Qi L, Liu F, Lv L, Li Y, Meng Q, Liu J, Shang L, Yu Y. Tumor-associated macrophages promote tumor metastasis via the TGF-beta/SOX9 axis in non-small cell lung cancer. Oncotarget. 2017;8:99801–99815. doi: 10.18632/oncotarget.21068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McDermott DF, Atkins MB. Application of IL-2 and other cytokines in renal cancer. Expert Opin Biol Ther. 2004;4:455–68. doi: 10.1517/14712598.4.4.455. [DOI] [PubMed] [Google Scholar]
- 71.Rosenberg SA, Mulé JJ, Spiess PJ, Reichert CM, Schwarz SL. Regression of established pulmonary metastases and subcutaneous tumor mediated by the systemic administration of high-dose recombinant interleukin 2. J Exp Med. 1985;161:1169–88. doi: 10.1084/jem.161.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mulé JJ, Yang JC, Afreniere RL, Shu SY, Rosenberg SA. Identification of cellular mechanisms operational in vivo during the regression of established pulmonary metastases by the systemic administration of high-dose recombinant interleukin 2. J Immunol. 1987;139:285–94. [PubMed] [Google Scholar]
- 73.Neidhart JA. Interferon therapy for the treatment of renal cancer. Cancer. 1986;57:1696–1699. doi: 10.1002/1097-0142(19860415)57:8+<1696::aid-cncr2820571312>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 74.Xu YH, Li ZL, Qiu SF. IFN-gamma induces gastric cancer cell proliferation and metastasis through upregulation of integrin beta3-mediated NF-kappaB signaling. Transl Oncol. 2018;11:182–192. doi: 10.1016/j.tranon.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.