Abstract
Glioblastomas (GBM) are deadly brain tumors that currently do not have long-term patient treatments available. GBM overexpress the angiogenesis factor VEGF and its receptor VEGFR2. ETLD1 (epidermal growth factor, latrophilin and seven transmembrane domain-containing protein 1), a G-protein coupled receptor (GPCR) protein, we discovered as a biomarker for high-grade gliomas, is also a novel regulator of angiogenesis. Since it was established that VEGF regulates ELTD1, we wanted to establish if VEGFR2 is also associated with ELTD1, by targeted antibody inhibition. G55 glioma-bearing mice were treated with either anti-ELTD1 or anti-VEGFR2 antibodies. With the use of MRI molecular imaging probes, we were able to detect in vivo levels of either ELTD1 (anti-ELTD1 probe) or VEGFR2 (anti-VEGFR2 probe). Protein expressions were obtained for ELTD1, VEGF or VEGFR2 via immunohistochemistry (IHC). VEGFR2 levels were significantly decreased (P < 0.05) with anti-ELTD1 antibody treatment, and ELTD1 levels were significantly decreased (P < 0.05) with anti-VEGFR2 antibody treatment, both compared to untreated tumors. IHC from mouse tumor tissues established that VEGFR2 and ELTD1 were co-localized. The mouse anti-ELTD1 antibody treatment study indicated that anti-VEGFR2 antibody treatment does not significantly increase survival, decrease tumor volumes, or alter tumor perfusion (measured as relative cerebral blood flow or rCBF). Conversely, anti-ELTD1 antibody treatment was able to significantly increase animal survival (P < 0.01), decrease tumor volumes (P < 0.05), and reduce change in rCBF (P < 0.001), when compared to untreated or IgG-treated tumor bearing mice. Anti-ELTD1 antibody therapy could be beneficial in targeting ELTD1, as well as simultaneously affecting VEGFR2, as a possible GBM treatment.
Keywords: Glioblastoma (GBM), ELTD1, VEGFR2, orthotopic G55 xenograft glioma model, in vivo, MRI, rCBF, molecular-targeted MRI (mt-MRI)
Introduction
Gliomas are classified as primary or secondary tumors, and categorized from WHO grades I to IV based on their growth pattern, behaviors and genetic driver mutations [1]. The most aggressive, grade IV Glioblastomas (GBM), are extremely fatal diffuse and invasive tumors [1,2]. Known GBM markers include IDH1/2 mutation status, 1p/19q co-deletion, and mutations in EGFR, Cyclin D1/3, MDM2, and PTEN [3-6]. These and many other genetic and epigenetic alterations allow glioma cells to evade regulatory processes that normal cells go through, allowing them to thrive and alternatively result in mutated or harmful cells going through apoptosis [5]. These mutations or upregulated proteins are used as biomarkers to characterize gliomas [7]. Biomarkers are important components of gliomas that facilitate not only more accurate classification of malignancy, but also a better way to direct treatment options specific to patients [6].
The ability of glioma cells to rapidly migrate and aggressively infiltrate throughout the brain make it impossible for successful total surgical resection [8,9]. Although FDA-approved Bevacizumab targeting VEGFA has had some success in reducing angiogenesis in patients, this treatment overall does not increase patient survival even following radiotherapy and/or chemotherapy [7,10,11]. Furthermore, treatment with Bevacizumab has been linked to drug resistance in patients [7,10,11]. GBM, which accounts for the majority of all gliomas, are the most common of all malignant central nervous system (CNS) tumors [2]. GBM have the highest incidence rate (3.2 per 100,000 population) and the highest number of cases of all malignant tumors with 12,760 cases projected in 2018 [2]. Sadly, the five-year survival rate is 5.5% for glioblastomas [2].
High-grade gliomas are highly vascular tumors, as angiogenesis, the formation of new blood vessels, is essential for tumor growth to supply oxygen and nutrients delivered to tumor tissue [3,6]. Angiogenesis in gliomas is the result of the upregulation of microvascular proliferation factors such as VEGF, PDGF (platelet-derived growth factor), and bFGF (basic fibroblast growth factor) [8]. The idea that the interruption of angiogenesis will lead to tumor regression led to the development of drugs targeting VEGF/VEGFR2 signaling pathways, such as Bevacizumab, a monoclonal antibody against VEGF and other tyrosine kinase inhibitors [8]. VEGF is a main regulator of angiogenesis, as it binds to VEGFR2 found on endothelial cells. The binding of VEGF-A to VEGFR2 induces a cascade of different signaling pathways [8,12]. The dimerization of the receptor and the following autophosphorylation of the intracellular TK (tyrosine kinase) domains lead to the simultaneous activation of PLC-γ-Raf kinase-MEK-MAP kinase and PI3K-AKT pathways, causing cellular proliferation and endothelial-cell survival [12].
Low success of VEGFA as a therapeutic target can be attributed to the complex processes and many factors involved in tumor angiogenesis such as HIF-1α (hypoxia inducing factor 1α), PDGF, bFGF, IL-8 (interleukin 8), thrombospondin1/2, endostatin, and interferons that are up or down regulated in gliomas due to genetic mutations [4]. Knowledge of biomarkers or pathways including the Rb pathway, the p53 pathway, mitogenic signaling pathways including PI3K and MAPK, PI3K/PTEN/AKTK, EGFR, PDGFR give researchers the ability to develop molecular targeted therapies that can act not only as prognostic markers, but also as therapeutic targets [8,13]. Current clinical trials focus on molecular targeted therapies, as well as combined with radiotherapy and chemotherapy, such as temozolomide (TMZ) [13,14]. The need for new therapeutic targets is of crucial importance as there is no effective treatment for patients diagnosed with gliomas.
ELTD1 (Epidermal growth factor, latrophilin and seven transmembrane domain-containing protein 1), or alternatively named ADGRL4 (Adhesion G Protein-Coupled Receptor L4), is a regulator of angiogenesis [15]. Initially discovered in developing cardiomyocytes, ELTD1, has been found to be highly expressed at both mRNA and protein levels in glioblastoma blood vessels, and its expression is highly associated with tumor grade [16-20]. ELTD1 plays a key role in angiogenesis both in vitro and in vivo by regulating the tip cell activity required for the sprouting of blood vessels [17,21]. Furthermore, in vivo siRNA data indicate that ELTD1 is also important in promoting tumor growth and metastasis [17]. In vivo experiments with zebrafish showed that ELTD1 is important in vascular development, specifically in the formation of smaller vessels [17].
Seeking a highly expressed glioma biomarker that could potentially be a therapeutic target, our lab identified that the angiogenic marker, ELTD1, could be a promising therapeutic target for high-grade gliomas [20]. Our group initially found that ELTD1 is highly associated with high-grade gliomas, and is expressed both on endothelial and tumor cells [19,22]. We found ELTD1 was also highly expressed in rat F98 gliomas from preclinical studies [19].
Chemotherapy, radiotherapy and Bevacizumab are the standard-of-care for patients diagnosed with gliomas [10,23]. These treatments have not been effective overall in prolonging patient survival. Research to find effective treatments is essential [11]. We hypothesized that because ELTD1 is expressed both on endothelial and glioma tumor cells, inhibiting it might have a significant therapeutic effect against high-grade gliomas [22].
Our preclinical studies focused on determining if inhibiting ELTD1 by using a polyclonal antibody would have a therapeutic effect in a mouse model [22]. We used C57BL/6 mice injected orthotopically with GL261 cells, as well as a human G55 GBM cells injected orthotopically in nude mice, as high-grade glioma models [22]. Monitoring tumor growth via MRI, we treated the mice with either anti-VEGF, anti-c-Met, anti-ELTD1 or IgG antibodies, once we identified tumors that were 10-15 mm3 in volumes within the mouse brains [22]. Briefly, we found that by inhibiting ELTD1, percent survival significantly increased while tumor volumes significantly decreased compared to untreated tumor-bearing mice [22]. Anti-ELTD1 polyclonal Ab treatments also had a significant effect on tumor perfusion, angiogenesis, and microvessel density, which are all drastically altered in gliomas [22]. We concluded that targeting ELTD1 as a therapy in gliomas would be advantageous not only because of its angiogenic properties, but because we found significantly less hemorrhaging in the anti-ELTD1 Ab treated group, compared to the anti-VEGF Ab group [22].
Although information about ELTD1 is limited, published reports indicate that endothelial cell ELTD1 expression is induced by VEGF, bFGF, and TGFβ2, but reduced by DLL4-Notch signaling as shown in in vitro and in vivo [17,18]. This information might give us an insight into how ELTD1 is stimulated. Characterizing ELTD1 will expand our knowledge on vascular growth, angiogenesis, and our overall understanding regarding glioma malignancy. Additionally, it might lead to the use of anti-ELTD1 therapy in other types of cancer, as it has been found to be expressed in colorectal cancer, breast cancer, head and neck squamous cell carcinoma and others [17].
The objective of the current study was to determine whether targeting ELTD1 or VEGFR2 with antibodies alters animal survival, tumor volumes cell growth, in vivo expression levels via assessment with molecular-targeted MRI, and ex vivo expression via immunohistochemistry (IHC), in order to establish if there is a relationship between ELTD1 and VEGFR2, the receptor for VEGF.
Materials and methods
Intracerebral glioma cell implantation and treatments
All animal studies were conducted with the approval (protocol 13-10) of the Oklahoma Medical Research Foundation (OMRF) Institutional Animal Care Use Committee (IACUC) policies, which follow NIH guidelines. Two-month-old male nude mice (Hsd:Athymic Nude-Foxn1nu mice; Harlan Inc., Indianapolis, IN) were implanted intracerebrally with human G55 xenograft cells (1 × 106) per mL suspended in 4 μL in cell culture media of 1% agarose solution. The heads of anesthetized mice were immobilized (stereotaxic unit; Kopf Instruments, Tujunga, CA), and with aseptic techniques, a 1 mm burr hole was drilled in the skull 1 mm anterior and 2 mm lateral to the bregma on the left side. A 20 μL gas-tight Hamilton syringe was used to inject G55 cells into the left frontal lobe at a depth of 1.5 mm relative to the dural surface in a stereotaxic unit. The cell lines were maintained and expanded immediately prior to inoculation, and not used for more than 20 passages. Following injection, the skin was closed with surgical sutures. Once tumors reached 10-15 mm3 (determined via MRI), mice were treated with 2 mg/kg of anti-ELTD1 (Bioss), anti-VEGFR2 (Santa Cruz) or anti-VEGF (bevacizumab; Avastin, Genentech) antibodies every 2-3 days until the tumor reached 130-150 mm3, or were left untreated. All mice were euthanized when tumors reached ≥ 150 mm3 prior to tumor-induced death. Nonspecific mouse immunoglobulin IgG (Alpha Diagnostics) was also administered at 2 mg/kg in sterile saline via tail vein injections every 2-3 days.
In vivo magnetic resonance (MR) techniques
Morphological imaging
Nude mice were anesthetized and positioned in a stereotaxic cradle. A 30-cm horizontal bore Bruker Biospin magnet operating at 7 Tesla (T; Bruker BioSpin GmbH, Karlsruhe, Germany), was used with a S116 gradient set to perform all MRI experiments. A mouse head coil was used for signal detection and a 72 mm quadrature volume coil for transmission. Multiple-slice, multiple echo (MSME) imaging (FOV = 2.50 × 2.50 cm2, TR = 2000 ms, TE = 17.5 or 58.2 ms, matrix = 192, averages = 2, slices = 16, slice thickness = 1 mm) was used to calculate tumor volumes and to inspect tumor morphology. Starting ten days after the G55 tumor cells inoculation, each mouse brain was imaged in vivo every 2-3 days until the end of the study.
Perfusion imaging
In order to assess microvascular alterations associated with tumor capillaries, the perfusion imaging method, arterial spin labeling, was used as previously described [23]. Briefly, perfusion maps were obtained on a single axial slice of the brain located on the point of the rostro-caudal axis where the tumor had the largest cross section. The imaging geometry was a 3.5 × 3.5 mm2 slice, of 1.5 mm in thickness, with a single shot echo-planar encoding over a 64 × 64 matrix. An echo time of 20 ms and a repetition time of 18 s were used. To obtain perfusion contrast, the flow alternating inversion recovery scheme was used, where inversion recovery images were acquired using selective and nonselective slices. For each type of inversion, 8 images were acquired with inversion times evenly spaced at 20-2820 ms. For perfusion data, the recovery curves obtained from each pixel for nonselective or selective inversion images were numerically fitted to derive pixelwise T 1 and T 1* values, respectively, and longitudinal recovery rates were then used to calculate the cerebral blood flow, CBF (mL/(100 g·min)), from: CBF = λ[(1/T*1)-(1/T1)]. The partition coefficient, λ, was scaled by the value of 0.9 mL/g [25,26]. To calculate differences in relative (r)CBF values, tumor rCBF values were obtained at early and late stage tumor progression and were normalized to rCBF values in the contralateral brain region of corresponding animals.
Molecular-targeted MR imaging (mt-MRI): The contrast agent, biotin-BSA (bovine serum albumin)-Gd (gadolinium)-DTPA, was prepared as previously described by our group [26], based on the modification of the method developed by Dafni et al. [27]. Anti-VEGFR-2 Ab (Santa Cruz Biotech, Inc., CA, USA) or anti-ELTD1 Ab (Abcam, Cambridge MA) was conjugated to the albumin moiety through a sulfo-NHS-EDC link according to the protocol of Hermanson [28]. Molecular MRI was performed when the tumor volumes were close to their maximum tumor volumes (120-180 mm3). Molecular probes with a biotin-albumin-Gd-DTPA construct bound to either anti-VEGFR2 or anti-ELTD1 antibodies were injected via a tail vein catheter in mice. A non-specific IgG was used with the biotin-albumin-Gd-DTPA construct as a negative control. A variable-TR RARE sequence (rapid acquisition with refocused echoes), with multiple TRs of 200, 400, 800, 1200 and 1600 ms, TE of 15 ms, FOV of 3.5 × 3.5 cm2, matrix size of 256 × 256 and a spatial resolution of 0.137 mm) was used to obtain T1-weighted images before and after administration of probe or control contrast agents. Relative probe (contrast agent) concentrations were calculated to assess the levels of VEGFR-2 or ELTD1, as well as the non-specific IgG contrast agent, in each animal. A contrast difference image was created from the pre- and (120 minutes) post-contrast datasets for the slice of interest, by computing the difference in T1 relaxation times between the post-contrast and the pre-contrast image on a pixel basis. From difference images ten regions of interest (ROI), of equal size (0.05 cm2), were drawn within areas with the highest T1 relaxation at the TR 800 ms, in the tumor parenchyma and contralateral side of the brains of each animal, after either anti-VEGFR-2 or anti-ETLD1 probe injections. T1 relaxation times are affected by the presence of the Gd-containing molecular imaging probes. T1 values obtained from the ROIs in the tumor regions were normalized to the corresponding contralateral sides. The T1 relaxation values of the specified ROIs were computed from all pixels in the ROIs, by the following equation [29] (processed by ParaVision 5.0, Bruker): S (TR) = S0 (1 - e-TR/T1), where TR is the repetition time, S0 is the signal intensity (integer machine units) at TR, T1 and TE = 0, and T1 is the constant of the longitudinal relaxation time. Overlays of contrast difference images and T1-weighted images were generated using the 3D Analysis Software for Life Sciences Amira® (Fei, Hillsboro, Oregon).
Immunohistochemistry
All mice were euthanized after the last MRI examination. The brain of each animal was removed, preserved in 10% neutral buffered formalin, and processed routinely. Paraffin-embedded tissues were sectioned in 5 μm sections, mounted on super frost plus glass slides, stained with hematoxylin and eosin (H&E), and examined by light microscopy. IHC was done to establish ELTD1, VEGF, and VEGFR2 levels by staining tissue samples with anti-ELTD1 (rabbit anti-ELT, 10 µg/mL; #ab150489, Abcam), anti-VEGF (Rabbit anti VEGFA antibody; ab 46154; 1:100 = 10 µg/mL), or anti-VEGFR2 (Rabbit anti VEGF Receptor 2 antibody; ab2349; 1:50 = 4 µg/mL) antibodies. Samples were additionally stained for Streptavidin/Biotin (Vector labs, SP-2002). For the co-localization assay, the tissue was washed with PBS and incubated with 15% sucrose before embedding in an Optimal Cutting Temperature (O.C.T.) compound and freezing in liquid nitrogen. Frozen tissue blocks were sectioned, mounted on slides, and stained for VEGFR2 (Rabbit anti VEGF Receptor 2 antibody; ab2349; 1:50 = 4 µg/mL) and ELTD1 (rabbit anti-ELT, 10 µg/mL; #ab150489, Abcam). Fluorescein (FITC) or Cy3 dye were used to tag secondary antibodies. Slides were then observed under a fluorescence microscope. Only areas containing tumor tissue were analyzed for IHC expression. Areas without tumor tissue and areas with necrosis or significant artifacts (e.g. tissue folding) were deselected and excluded from analysis. The number of positive pixels was divided by the total number of pixels (negative and positive) in the analyzed area. Three regions of interest (ROI) in each group were identified, and measurements were captured digitally for each selected ROI. They were then analyzed as well as imaged using Aperio ImageScope (Leica Biosystems, Buffalo Grove, IL).
Bioinformatics
Bioinformatics methods previously described [19,20,30-32] pair coexpression data with literature-based analysis of protein commonalities. Global Microarray Meta-Analysis (GAMMA) is a predictive algorithm that uses public microarray and RNAseq datasets from NCBI’s Gene Expression Omnibus (GEO) to identify groups of transcripts that are highly correlated with each other, and then identify shared biological associations within MEDLINE. Using GAMMA, we identified 40 most correlated transcripts for eltd1 and vegfr2.
Statistical analysis
Survival curves were analyzed using Kaplan-Meier curves. Tumor volumes, changes in normalized rCBF, and immunohistochemistry protein levels, were analyzed and compared by two-way ANOVA with multiple comparisons. Molecular-targeted MRI data were analyzed using a student t-test. Data were represented as mean ± SD, and p-values of either *0.05, **0.01, ***0.001, and ****0.0001 were considered statistically significant.
Results
Mice treated with anti-VEGFR2 antibodies did not significantly increase percent survival in tumor-bearing mice compared to IgG Ab treated mice, while anti-ELTD1 Ab (P < 0.0001) treatments significantly increased percent survival in the G55 human xenograft model. Anti-VEGFR2 antibody treatment also did not significantly decrease tumor volumes compared to the IgG treated groups on day 21 of tumor detection, while anti-ELTD1 (P < 0.01) antibody treatments significantly reduced tumor volumes (Figure 1).
Figure 1.
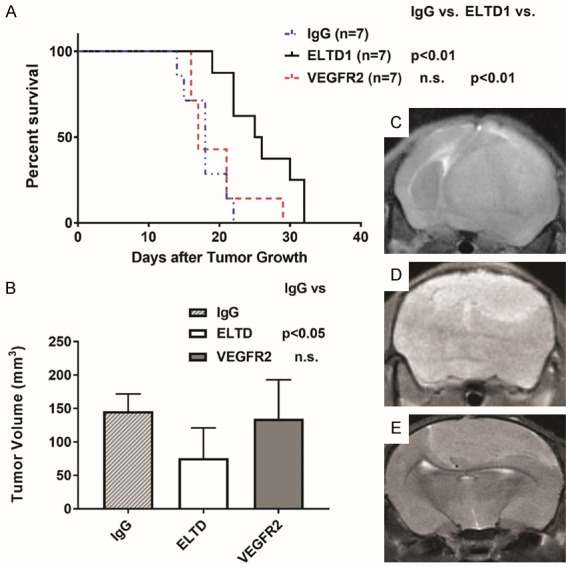
Anti-ELTD1 Ab treatment is more effective than anti-VEGFR2 Ab treatment in increasing animal survival and decreasing tumor volumes in a G55 human xenograft model. (A) Percent survival curve for IgG- (n = 7), anti-ELTD1- (n = 7), and anti-VEGFR2 Ab (n = 7) treated mice. Anti-ELTD1 Ab treatment significantly increased animal survival (P < 0.01), compared to either IgG-treated or anti-VEGFR2 Ab-treated mice. IgG was a non-specific IgG used as a control. (B) Tumor volumes of treatment groups (mean ± SD). Anti-ELTD1 Ab treatment significantly decreased tumor volumes at 21 days post-tumor detection (P < 0.05), compared to IgG-treated mice. Morphological representative MR images for IgG- (C), anti-ELTD1 Ab- (D) and anti-VEGFR2 Ab-treated (E) mice at 21 days following initial tumor detection.
Additionally, comparing early to late tumor perfusion via MRI ASL (arterial spin labeling), we found that anti-VEGFR2 antibody treatments did not alter perfusion in tumor-bearing mice compared to IgG treated mice, while anti-ELTD1 antibody treatment (P < 0.001) decreased tumor perfusion significantly less than IgG treated mice (Figure 2).
Figure 2.
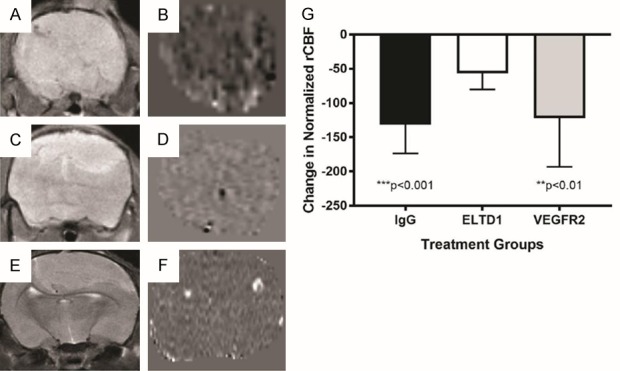
Anti-ELTD1 Ab treatment is more effective in restoring tumor-associated vascularity to normal, compared to anti-VEGFR2 Ab treatment. Representative T2-weighted MR images from G55 glioma-bearing mice either IgG- (A), anti-ELTD1 Ab- (C), or anti-VEGFR2 Ab-treated (E). Representative MR perfusion maps from G55 tumors either IgG- (B), anti-ELTD1-Ab- (D), or anti-VEGFR2-Ab-treated (F). (G) Normalized (against muscle tissue) tumor rCBF differences (late to early time periods) for tumor growth measured from MR perfusion images obtained from treated G55 tumors. n = 5 for IgG, n = 12 for anri-ELTD1 Ab, and n = 7 for anti-VEGFR2-Ab-treated mice. Anti-ELTD1 Ab treatement was found to significantly reduce rCBF differences when compared to IgG (P < 0.001) or anti-VEGFR2 Ab treatment (P < 0.01). Mean ± SD.
Fluorescence imaging detected both VEGFR2 and ELTD1 expression in an untreated mouse glioma showing that they are co-localized in blood vessels as well as in glioma cells (Figure 3).
Figure 3.
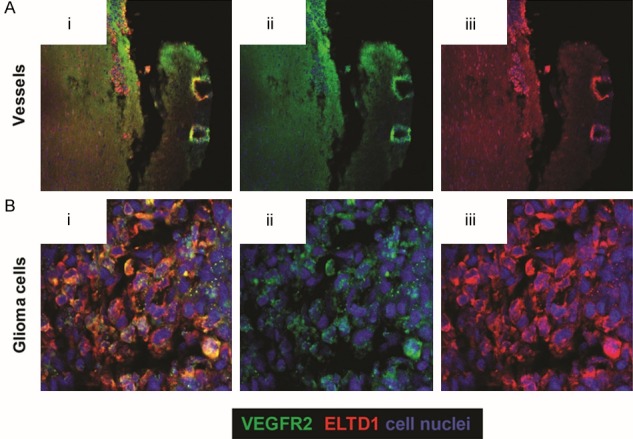
ELTD1 and VEGFR2 are co-localized within blood vessels and glioma cells in G55 glioma tumors. Blood vessels are depicted in (A), and glioma cells are shown in (B). Ex-vivo fluorescence co-localization of VEGFR2 (ii: green) and ELTD1 (iii: red) demonstrates co-localization (i). Cell nuclei are stained blue with DAPI.
Molecular-targeted MR imaging was used to assess in vivo levels of both ELTD1 and VEGFR2 in untreated and treated G55 xenografts. From changes in T1 relaxation, expression of ELTD1 (measured by the presence of the anti-ELTD1 probe), as well as the control IgG contrast agent, were significantly reduced in mice treated with anti-VEGFR2 antibody (P < 0.05), compared to untreated mice, as determined by a difference in T1 relaxation times (Post-Pre/Pre) (Figure 4). Inversely, expression of VEGFR2, and the control IgG contrast agent, were significantly reduced in mice treated with anti-ELTD1 antibody (P < 0.005) compared to untreated mice, as established by a difference in T1 relaxation times (Post-Pre/Pre) (Figure 5).
Figure 4.
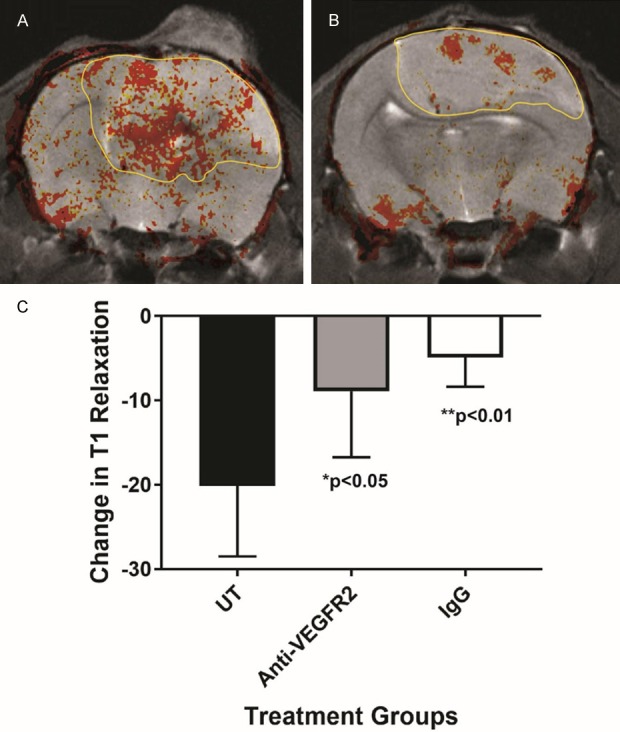
ELTD1 expression in vivo significantly decreases in G55 glioma-bearing mice treated with anti-VEGFR2 Ab treatment. Differences in T1 relaxation (indicated presence of an ELTD1-targeting molecular MR imaging probe) were significantly reduced for anti-VEGFR2 Ab-treated mice, compared to untreated animals (n = 5 for each group) (P < 0.05). IgG-treated mice (n = 5) also had significantly reduced differences in T1 relaxation, when compared to UT mice (P < 0.01). Mean ± SD.
Figure 5.
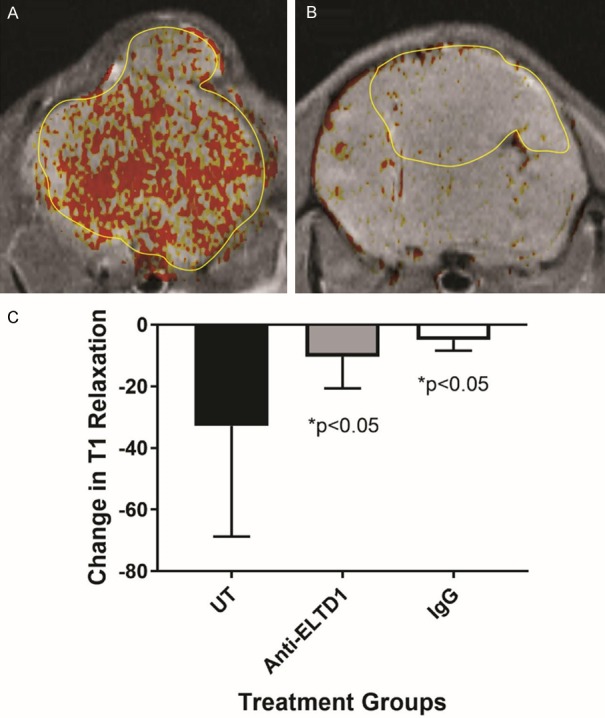
VEGFR2 expression in vivo significantly decreases in G55 glioma-bearing mice treated with anti-ELTD1 Ab treatment. Differences in T1 relaxation (indicated presence of a VEGFR2-targeting molecular MR imaging probe) were significantly reduced for anti-ELTD1 Ab-treated mice, compared to untreated animals (n = 5 for each group) (P < 0.05). IgG-treated mice (n = 5) also had significantly reduced differences in T1 relaxation, when compared to UT mice (P < 0.05). Mean ± SD.
Mouse glioma tissues were stained with streptavidin-horseradish peroxidase (HRP), which binds to the biotin in our molecular probe, detecting the levels of ELTD1 or VEGFR2 probes within our mouse glioma samples. We found that the ELTD1-targeted probe was significantly decreased (P < 0.01) with anti-VEGFR2 Ab treatment compared to our untreated mouse glioma sample (Figure 6). We found the similar results with our anti-ELTD1 Ab treated group as levels of the VEGFR2-targeted probe were significantly reduced (P < 0.05) compared to untreated tissue (Figure 7). Expression of ELTD1, VEGF, and VEGFR2 in mouse glioma tissues were also measured from our mouse studies using Immunohistochemistry. Data showed that both ELTD1 and VEGF are significantly expressed more than VEGFR2 in mouse glioma tissue (P < 0.0001 for both) (Figure 8).
Figure 6.
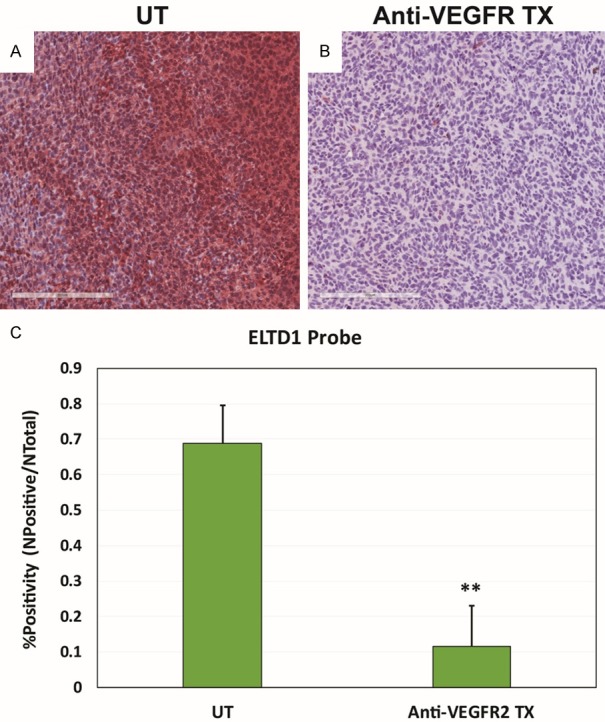
ELTD1 expression ex vivo significantly decreased in G55 glioma tissue in anti-VEGFR2 Ab treated mice. SA-HRP staining of the ELTD1 probe (via the biotin moiety) in untreated G55 tumors (A), compared to anti-VEGFR2 Ab treated glioma tissue (B). (C) Percent positivity for the ELTD1 probe was significantly reduced in anti-VEGFR2 Ab-treated gliomas, compared to untreated tumors (P < 0.01). Mean ± SD, n = 5.
Figure 7.
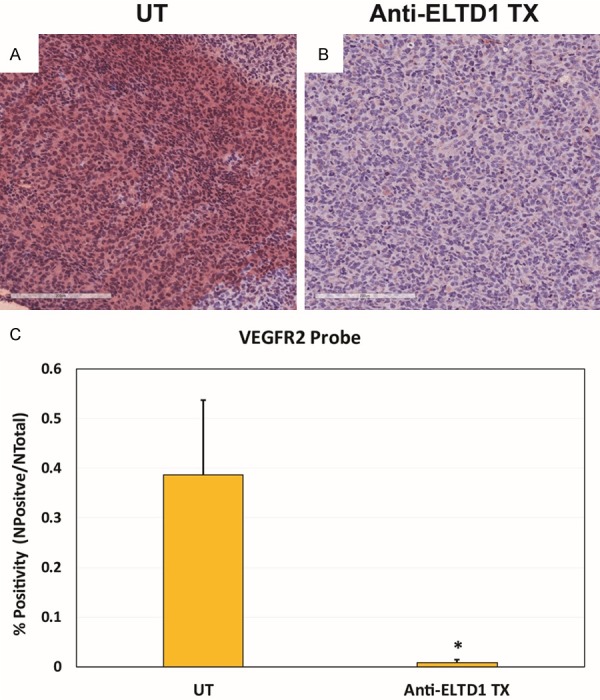
VEGFR2 expression ex vivo significantly decreased in G55 glioma tissue in anti-ELTD1 Ab treated mice. SA-HRP staining of the VEGFR2 probe (via the biotin moiety) in untreated G55 tumors (A), compared to anti-ELTD1 Ab treated glioma tissue (B). (C) Percent positivity for the VEGFR2 probe was significantly reduced in anti-VEGFR2 Ab-treated gliomas, compared to untreated tumors (P < 0.05). Mean ± SD, n = 4.
Figure 8.
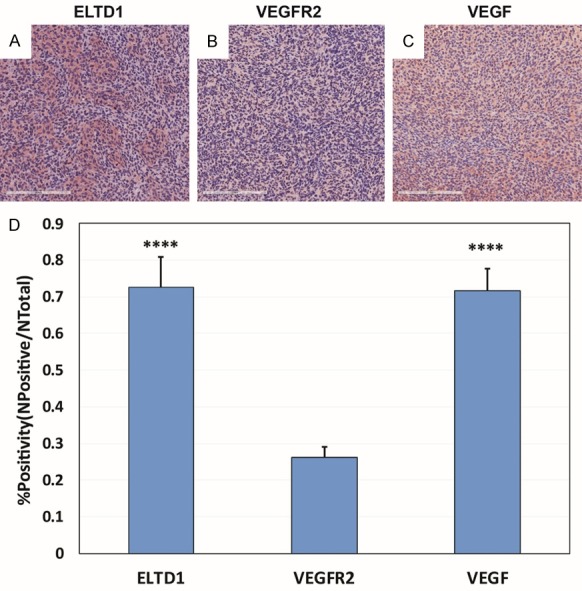
ELTD1 and VEGF are expressed significantly higher than VEGFR2 in a G55 glioma model. Staining for ELTD1 (A), VEGFR2 (B), and VEGF (C) showed that ELTD1 (P < 0.0001) and VEGF (P < 0.0001) were expressed significantly higher than VEGF in G55 tumors (as calculated by percent (%) positivity (D). Mean ± SD, n = 6.
To further study the relationship between ELTD1 and VEGF or VEGFR2, we stained mouse glioma tissue from different treatment groups for either ELTD1, VEGF, or VEGFR2 using IHC. Expression of both VEGFR2 and ELTD1 was significantly reduced (P < 0.05, P < 0.01 respectively) in mice treated with anti-VEGFR2 Ab compared to untreated mice (Figure 9). Similarly, expression of both ELTD1 and VEGFR2 was significantly reduced (both P < 0.05) in mice treated with anti-ELTD1 Ab compared to untreated mice (Figure 10). Alternatively, our anti-VEGF Ab treated group showed a significant decrease in ELTD1 and VEGF expression (both P < 0.05), but had no significant effect on VEGFR2 expression (Figure 11).
Figure 9.
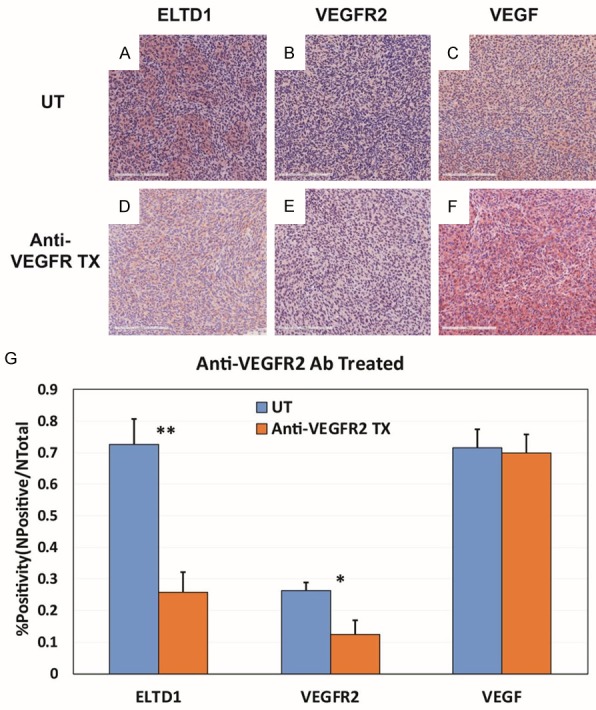
Anti-VEGFR2 Ab treatment significantly reduced the expressions of ELTD1 and VEGFR2. Representative images of ELTD1 (A), VEGFR2 (B) and VEGF (C) expression levels in untreated G55 tumors, compared to anti-VEGFR2 Ab treated tissues (D-F, respectively). (G) Percent positivities for either ELTD1, VEGFR2 or VEGF, show a significant decrease in ELTD1 (P < 0.01) and VEGFR2 (P < 0.05) expressions, but did not show a significant decrease in VEGF expression. Mean ± SD, n = 6.
Figure 10.
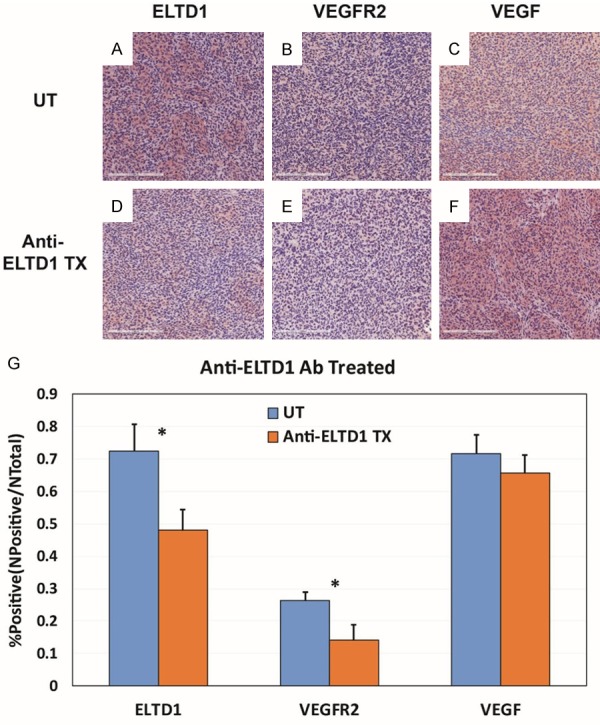
Anti-ELTD1 Ab treatment significantly reduced the expressions of ELTD1 and VEGFR2. Representative images of ELTD1 (A), VEGFR2 (B) and VEGF (C) expression levels in untreated G55 tumors, compared to anti-ELTD1 Ab treated tissues (D-F, respectively). (G) Percent positivities for either ELTD1, VEGFR2 or VEGF, show a significant decrease in ELTD1 (P < 0.05) and VEGFR2 (P < 0.05) expressions, but did not show a significant decrease in VEGF expression. Mean ± SD, n = 6.
Figure 11.
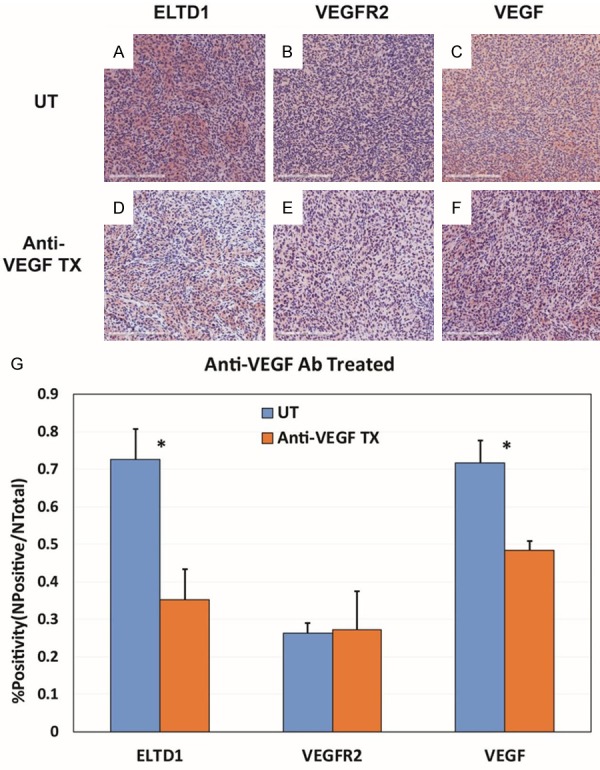
Anti-VEGF Ab treatment significantly reduced the expressions of ELTD1 and VEGF. Representative images of ELTD1 (A), VEGFR2 (B) and VEGF (C) expression levels in untreated G55 tumors, compared to anti-ELTD1 Ab treated tissues (D-F, respectively). (G) Percent positivities for either ELTD1, VEGFR2 or VEGF, show a significant decrease in ELTD1 (P < 0.05) and VEGF (P < 0.05) expressions, but did not show a significant decrease in VEGFR2 expression. Mean ± SD, n = 6.
To further assess if there is an association between ELTD1 and VEGFR2, public microarray datasets were analyzed to identify the genes most correlated in their expression levels with both eltd1 and vegfr2 (separately) and then predict what genes are most relevant to their genetic network using an approach previously described [30,33]. Then, we narrowed the list to include only the genes with protein-protein interactions (PPIs) by cross-referencing the predictions with STRING v10 [34]. The goal was to identify a list of candidate interactions relevant to both ELTD1 and VEGFR2. The genes on the resulting list (see Table 1) have been found in other studies to be relevant to angiogenesis and, given that we have found molecular links between the two, could be reasonable candidates to further explore how the two may be related.
Table 1.
vegfr2 (KDR) is highly associated with eltd1. GAMMA-predicted association analysis identifies the top 8 genes associated with eltd1 based on GAMMA scores
| Gene | Function | Ref. |
|---|---|---|
| FLT4 (Vascular Endothelial Growth Factor Receptor 3) | Involved in lymphangiogenesis and maintenance of the lymphatic endothelium. Promotes proliferation, survival and migration of endothelial cells, and regulates angiogenic sprouting. | [49-53] |
| KDR (Vascular Endothelial Growth Factor Receptor 2) | Main mediator of VEGF-induced endothelial proliferation, survival, migration, tubular morphogenesis and sprouting. | [54-58] |
| ANGPT2 (Angiopoietin 2) | Regulates vascular remodeling by promoting EC survival, proliferation, and migration and destabilizing the interaction between EC and perivascular cells. | [47,59-61] |
| MMP1 (Matrix Metallopeptidase 1) | Involved in the breakdown of extracellular matrix in normal physiological processes, such as embryonic development, reproduction, and tissue remodeling, as well as in disease processes, such as arthritis and metastasis. | [62-66] |
| CDH5 (Cadherin 5) | Plays an important role in endothelial cell biology through control of the cohesion and organization of the intercellular junctions. Acts in concert with KRIT1 to establish and maintain correct endothelial cell polarity and vascular lumen. | [67-71] |
| PTK2 (Protein Tyrosine Kinase 2) | Plays an essential role in regulating cell migration, adhesion, spreading, reorganization of the actin cytoskeleton, formation and disassembly of focal adhesions and cell protrusions, cell cycle progression, cell proliferation and apoptosis | [72-76] |
| FLT1 (Vascular Endothelial Growth Factor Receptor 1) | Acts as a cell-surface receptor for VEGFA, VEGFB and PGF, and plays an essential role in the development of embryonic vasculature, the regulation of angiogenesis, cell survival, cell migration, macrophage function, chemotaxis, and cancer cell invasion | [77-81] |
| PDGFRB (Platelet Derived Growth Factor Receptor Beta) | Plays an essential role in the regulation of embryonic development, cell proliferation, survival, differentiation, chemotaxis and migration. | [82-86] |
Discussion
While our early results on the effect of anti-ELTD1 antibodies on tumor growth and vascularization seemed promising, there is still very little known about ELTD1 and its mechanisms within gliomas [35]. Our objective was to further characterize ELTD1 in terms of its effect on gliomas, and establish if targeting ELTD1 will also affect VEGFR2.
Although using inhibitors against tyrosine kinase receptors (RTKs) has had little overall success in treating patients diagnosed with glioblastomas in the past, current drugs and small molecules in clinical trials specifically targeting VEGFR2 might have more promising results [14,36-38]. Our goal was to compare anti-VEGFR2 to anti-ELTD1 Ab treatments in a mouse model as both are regulators of angiogenesis [39]. Considering that gliomas are highly heterogeneous, treatments against VEGFR2 might not be effective in some patients, whereas anti-ELTD1 drug treatments may target different factors associated with tumor growth and angiogenesis. Some patients could also overexpress ELTD1, supporting research that compare anti-ELTD1 and anti-VEGFR2 treatment results with one another.
An important tool used by clinicians and researchers is MRI [14]. It is used not only for predictive diagnostic purposes, but to monitor patient responses to different treatments using imaging techniques such as diffusion-weighed MRI, contrast-enhanced perfusion, and MR spectroscopy [14]. With the help of MRI to monitor tumor growth post glioma cell injection, we found that inhibiting VEGFR2 via antibodies did not have a significant effect on mouse survival nor on tumor volumes compared to the untreated group as well as the anti-ELTD1 Ab treated group. Furthermore, VEGFR2 inhibition had no significant effect on normalizing tumor perfusion. Perfusion, measured as relative cerebral blood flow (rCBF) can be used to assess microvascular alterations associated with tumor angiogenesis and anti-angiogenic therapies [40,41]. Perfusion typically decreases within tumor regions, as vasculature loses its hierarchy due to uncontrolled angiogenesis [40,41]. Our results indicate that inhibiting VEGFR2 did not have an effect on microvascular changes related to tumor angiogenic growth. VEGFR2 may not be as involved in microvascular changes as ELTD1.
It raises the question is why inhibiting VEGF using Bevacizumab has a much better outcome in mice and human studies, while targeting VEGFR2 has had poor or little effect on increasing survival or decreasing angiogenesis? VEGFR2 has been shown to promote angiogenesis, cell viability and invasion in gliomas, but studies such as those by Lu et al. found that blocking VEGFR2 may stimulate invasion by activating the c-MET pathway [42,43]. Previous preclinical studies show that inhibiting VEGFR2 can have a significant therapeutic effect in some glioma models, but not in others [44]. The reason for this variance might be the overall heterogeneity of glioblastomas. Responses to VEGFR pathway stimulation or inhibitions may vary depending on the differential expression of VEGF family ligands and receptors [44].
Our IHC also shows that VEGFR2 is significantly less expressed in xenograft G55 mice glioma model compared to VEGF and ELTD1, and although this might account for the lack of therapeutic effect, different antibodies (monoclonal versus polyclonal) may have different affinities for their respective receptors. Additionally, factors such as intracellular VEGFR phosphorylation, tumoral versus endothelial expression, tumor heterogeneity, microenvironment, and alternatively spliced VEGFR variants may be affecting the efficacy of treatment using antibodies [45].
Internalization and dimerization of VEGFR2 are important steps in VEGF-induced angiogenesis signaling transduction activity by phosphorylation [46]. Recent studies show that VEGFR2 might be more effective intracellularly [46]. This indicates that using an antibody against VEGFR2 might not be as effective as it is designed to bind only to the extracellular region of the receptor, suggesting that intracellular expression may be ignored. Ultimately, the changes occurring downstream of VEGFA-induced binding of VEGFR2 might be a possible reason for the different outcomes between the two targets.
We found that ELTD1 was a much better therapeutic target in our mouse GBM model compared to VEGFR2. Serban et al. found that VEGF regulates ELTD1 both in vitro and in vivo [15]. As discussed earlier, there are many pathways and factors induced by VEGF, making it difficult to predict exactly how VEGF is able to regulate ELTD1. To further elucidate one aspect of this relationship, we examined whether there was an association between ELTD1 and VEGF’s receptor, VEGFR2.
From the bioinformatics analysis, we found that vegfr2 is highly associated with eltd1, along with other vascularization promoting proteins such as angiopoietin 2 that promotes endothelial cell survival by regulating vascular remodeling (Table 1) [47,48]. Furthermore, by using mouse glioma tissue, we were able to do fluorescence IHC microscopic imaging and found both ELTD1 and VEGFR2 are co-expressed both in glioma and endothelial cells. This finding along with the bioinformatics data suggests that there may be a possible correlation between ELTD1 and VEGFR2.
Using molecular probes made from constructs consisting of biotin-albumin-Gd-DTPA bound to either anti-VEGFR2 or anti-ELTD1 antibodies, we were able to determine that levels of ELTD1 were significantly reduced in vivo in mice treated with anti-VEGFR2 Ab compared to untreated mice and vice versa. Our SA-HRP staining also confirmed these results, as expression of ELTD1 significantly lowered for those treated with anti-VEGFR2 antibodies in glioma mouse tissue and vice versa. These data tell us not only the efficacy of using antibodies in our in vivo model, particularly for anti-ELTD1 Ab treatment, but that they are able to cross the blood brain barrier, as the molecular probes were detected bound to their respective targets.
Finally, IHC analysis on mouse glioma tissue for all treatment groups showed significantly less expression of both VEGFR2 and ELTD1 for both the anti-VEGFR2 and anti-ELTD1 Ab treated groups. These IHC data supports both our in vivo and ex vivo molecular targeting data. Anti-VEGF Ab treated tissue showed a significant decrease in both VEGF and ELTD1 expression levels, while it did not significantly reduce VEGFR2. Interestingly, VEGF expression levels did not decrease with either anti-VEGFR2 or anti-ELTD1 Ab treated groups. This suggests that neither ELTD1 nor VEGFR2 is regulating VEGF in our mouse model. As previously mentioned, there are many factors that can influence the expression of VEGF [87].
Our goal was to determine if targeting ELTD1 was a better therapeutic approach than VEGFR2, considering current data suggests that drugs targeting RTKs, such as VEGFR2 are overall ineffective [37]. Additionally, we wanted to elucidate the possible relationship between VEGFR2 and ELTD1, as both are important factors of angiogenesis. Glioma therapies have shifted dramatically to utilizing dual therapies by combining anti-angiogenic drugs with cytotoxic agents such as TMZ. Research such as this might be helpful for scientists to understand angiogenesis better, as it a process highly associated with tumor malignancy in many cancers. This growing understanding of vascular regulators may help us find more therapeutic targets or combinations of drugs that might target novel biomarkers, such as ELTD1. Future studies will examine both RNA and protein expressions in anti-ELTD1 antibody treated vs. non-treated G55 gliomas using RNA-seq and ELISA, respectively.
Conclusions
We conclude that targeting ELTD1 is a better therapeutic approach as it significantly increases survival, decrease tumor volumes, and alters perfusion rates in our model compared to our untreated group much better than by targeting VEGFR2. Additionally, we conclude that targeting ELTD1 may also elicit an effect on VEGFR2.
Acknowledgements
Research reported in this publication was supported in part by the Oklahoma Medical Research Foundation (RAT), and the National Cancer Institute Cancer Center Support Grant P30CA225520 awarded to the University of Oklahoma Stephenson Cancer Center regarding use of the Tissue Pathology Share Resource. Funding for JDW was supported in part by the NIH grant 2P20GM103636. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Disclosure of conflict of interest
None.
References
- 1.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Ostrom QT, Gittleman H, Liao P, Vecchione-Koval T, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2010-2014. Neuro Oncol. 2017;19:v1–v88. doi: 10.1093/neuonc/nox158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 4.Dunn GP, Rinne ML, Wykosky J, Genovese G, Quayle SN, Dunn IF, Agarwalla PK, Chheda MG, Campos B, Wang A, Brennan C, Ligon KL, Furnari F, Cavenee WK, Dephino RA, Chin L, Hahn WC. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26:756–784. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parker NR, Khong P, Parkinson JF, Howell VM, Wheeler HR. Molecular heterogeneity in glioblastoma: potential clinical implications. Front Oncol. 2015;5:55. doi: 10.3389/fonc.2015.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bastien JI, McNeill KA, Fine HA. Molecular characterizations of glioblastoma, targeted therapy, and clinical results to date. Cancer. 2015;121:502–516. doi: 10.1002/cncr.28968. [DOI] [PubMed] [Google Scholar]
- 7.Narayana A, Kelly P, Golfinos J, Parker E, Johnson G, Knopp E, Zagzag D, Fischer I, Raza S, Medabalmi P, Eagan P, Gruber ML. Antiangiogenic therapy using bevacizumab in recurrent high-grade glioma: impact on local control and patient survival. J Neurosurg. 2009;110:173–180. doi: 10.3171/2008.4.17492. [DOI] [PubMed] [Google Scholar]
- 8.Olar A, Aldape KD. Using the molecular classification of glioblastoma to inform personalized treatment. J Pathol. 2014;232:165–177. doi: 10.1002/path.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sunit D, Marsden PA. Angiogenesis in glioblastoma. N Engl J Med. 2013;369:1561–1563. doi: 10.1056/NEJMcibr1309402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai A, Tran A, Nghiemphu PL, Pope WB, Solis OE, Selch M, Filka E, Yong WH, Mischel PS, Liau LM, Phuphanich S, Black K, Peak S, Green RM, Sier CE, Kolevska T, Polikoff J, Fehrenbacher L, Elashoff R, Cloughesy T. Phase II study of bevacizumab plus temozolomide during and after radiation therapy for patients with newly diagnosed glioblastoma multiforme. J. Clin. Oncol. 2011;29:142–148. doi: 10.1200/JCO.2010.30.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chamberlain MC. Bevacizumab for recurrent malignant gliomas: efficacy, toxicity, and patterns of recurrence. Neurology. 2009;72:772–773. doi: 10.1212/01.wnl.0000339387.03225.0a. author reply 773-774. [DOI] [PubMed] [Google Scholar]
- 12.Stiver SI, Tan X, Brown LF, Hedley-Whyte ET, Dvorak HF. VEGF-A angiogenesis induces a stable neovasculature in adult murine brain. J Neuropathol Exp Neurol. 2004;63:841–855. doi: 10.1093/jnen/63.8.841. [DOI] [PubMed] [Google Scholar]
- 13.Reifenberger G, Wirsching HG, Knobbe-Thomsen CB, Weller M. Advances in the molecular genetics of gliomas - implications for classification and therapy. Nat Rev Clin Oncol. 2017;14:434–452. doi: 10.1038/nrclinonc.2016.204. [DOI] [PubMed] [Google Scholar]
- 14.Szopa W, Burley TA, Kramer-Marek G, Kaspera W. Diagnostic and therapeutic biomarkers in glioblastoma: current status and future perspectives. Biomed Res Int. 2017;2017:8013575. doi: 10.1155/2017/8013575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serban F, Artene SA, Georgescu AM, Purcaru SO, Tache DE, Alexandru O, Dricu A. Epidermal growth factor, latrophilin, and seven transmembrane domain-containing protein 1 marker, a novel angiogenesis marker. Onco Targets Ther. 2015;8:3767–3774. doi: 10.2147/OTT.S93843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nechiporuk T, Urness LD, Keating MT. ETL, a novel seven-transmembrane receptor that is developmentally regulated in the heart. ETL is a member of the secretin family and belongs to the epidermal growth factor-seven-transmembrane subfamily. J Biol Chem. 2001;276:4150–4157. doi: 10.1074/jbc.M004814200. [DOI] [PubMed] [Google Scholar]
- 17.Masiero M, Simões FC, Han HD, Snell C, Peterkin T, Bridges E, Mangala LS, Wu SY, Pradeep S, Li D, Han C, Dlaton H, Lopez-Berestein G, Tuynman JB, Mortensen N, Li JL, Patient R, Sood AK, Banham AH, Harris AL, Buffa FM. A core human primary tumor angiogenesis signature identifies the endothelial orphan receptor ELTD1 as a key regulator of angiogenesis. Cancer Cell. 2013;24:229–241. doi: 10.1016/j.ccr.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dieterich LC, Mellberg S, Langenkamp E, Zhang L, Zieba A, Salomäki H, Teichert M, Huang H, Edqvist PH, Kraus T, Augustin HG, Olofsson T, Larsson E, Söderberg O, Molema G, Pontén F, Georgii-Hemming P, Alafuzoff I, Dimberg A. Transcriptional profiling of human glioblastoma vessels indicates a key role of VEGF-A and TGFβ2 in vascular abnormalization. J Pathol. 2012;228:378–390. doi: 10.1002/path.4072. [DOI] [PubMed] [Google Scholar]
- 19.Towner RA, Jensen RL, Colman H, Vaillant B, Smith N, Casteel R, Saunders D, Gillespie DL, Silasi-Mansat R, Lupu F, Giles CB, Wren JD. ELTD1, a potential new biomarker for gliomas. Neurosurgery. 2013;72:77–90. doi: 10.1227/NEU.0b013e318276b29d. discussion 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Towner RA, Jensen RL, Vaillant B, Colman H, Saunders D, Giles CB, Wren JD. Experimental validation of 5 in-silico predicted glioma biomarkers. Neuro Oncol. 2013;15:1625–1634. doi: 10.1093/neuonc/not124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serban F, Daianu O, Tataranu LG, Artene SA, Emani G, Georgescu AM, Alexandru O, Purcaru SO, Tache DE, Daniciulescu MM, Sfredel V, Dricu A. Silencing of epidermal growth factor, latrophilin and seven transmembrane domain-containing protein 1 (ELTD1) via siRNA-induced cell death in glioblastoma. J Immunoassay Immunochem. 2017;38:21–33. doi: 10.1080/15321819.2016.1209217. [DOI] [PubMed] [Google Scholar]
- 22.Ziegler J, Pody R, Coutinho de Souza P, Evans B, Saunders D, Smith N, Mallory S, Njoku C, Dong Y, Chen H, Dong J, Lerner M, Mian O, Tummala S, Battiste J, Fung KM, Wren JD, Towner RA. ELTD1, an effective anti-angiogenic target for gliomas: preclinical assessment in mouse GL261 and human G55 xenograft glioma models. Neuro Oncol. 2017;19:175–185. doi: 10.1093/neuonc/now147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu W, Kato Y, Artemov D. Heterogeneity of tumor vasculature and antiangiogenic intervention: insights from MR angiography and DCE-MRI. PLoS One. 2014;9:e86583. doi: 10.1371/journal.pone.0086583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garteiser P, Doblas S, Watanabe Y, Saunders D, Hoyle J, Lerner M, He T, Floyd RA, Towner RA. Multiparametric assessment of the anti-glioma properties of OKN007 by magnetic resonance imaging. J Magn Reson Imaging. 2010;31:796–806. doi: 10.1002/jmri.22106. [DOI] [PubMed] [Google Scholar]
- 25.Herscovitch P, Raichle ME. What is the correct value for the brain--blood partition coefficient for water? J Cereb Blood Flow Metab. 1985;5:65–69. doi: 10.1038/jcbfm.1985.9. [DOI] [PubMed] [Google Scholar]
- 26.Towner RA, Smith N, Doblas S, Garteiser P, Watanabe Y, He T, Saunders D, Herlea O, Silasi-Mansat R, Lupu F. In vivo detection of inducible nitric oxide synthase in rodent gliomas. Free Radic Biol Med. 2010;48:691–703. doi: 10.1016/j.freeradbiomed.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 27.Dafni H, Landsman L, Schechter B, Kohen F, Neeman M. MRI and fluorescence microscopy of the acute vascular response to VEGF165: vasodilation, hyper-permeability and lymphatic uptake, followed by rapid inactivation of the growth factor. NMR Biomed. 2002;15:120–131. doi: 10.1002/nbm.724. [DOI] [PubMed] [Google Scholar]
- 28.Hermanson GT. Bioconjugate techniques. New York: Academic Press; 1996. p. 176. [Google Scholar]
- 29.Haacke EM. Magnetic resonance imaging: physical principles and sequence design. New York: Wiley-Liss; 1999. [Google Scholar]
- 30.Wren JD. A global meta-analysis of microarray expression data to predict unknown gene functions and estimate the literature-data divide. Bioinformatics. 2009;25:1694–1701. doi: 10.1093/bioinformatics/btp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wren JD, Garner HR. Shared relationship analysis: ranking set cohesion and commonalities within a literature-derived relationship network. Bioinformatics. 2004;20:191–198. doi: 10.1093/bioinformatics/btg390. [DOI] [PubMed] [Google Scholar]
- 32.Wren JD. Extending the mutual information measure to rank inferred literature relationships. BMC Bioinformatics. 2004;5:145. doi: 10.1186/1471-2105-5-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dozmorox MG, Giles CB, Wren JD. Predicting gene ontology from a global meta-analysis of 1-color microarray experiments. BMC Bioinformatics. 2011;12(Suppl 10):S14. doi: 10.1186/1471-2105-12-S10-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–52. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Favara DM, Banham AH, Harris AL. A review of ELTD1, a pro-angiogenic adhesion GPCR. Biochem Soc Trans. 2014;42:1658–1664. doi: 10.1042/BST20140216. [DOI] [PubMed] [Google Scholar]
- 36.Reardon DA, Vredenburgh JJ, Desjardins A, Peters K, Gururangan S, Sampson JH, Marcello J, Herndon JE 2nd, McLendon RE, Janney D, Friedman AH, Bigner DD, Friedman HS. Effect of CYP3A-inducing anti-epileptics on sorafenib exposure: results of a phase II study of sorafenib plus daily temozolomide in adults with recurrent glioblastoma. J Neurooncol. 2011;101:57–66. doi: 10.1007/s11060-010-0217-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galanis E, Anderson SK, Lafky JM, Uhm JH, Giannini C, Kumar SK, Kimlinger TK, Northfelt DW, Flynn PJ, Jaeckle KA, Kaufmann TJ, Buckner JC. Phase II study of bevacizumab in combination with sorafenib in recurrent glioblastoma (N0776): a north central cancer treatment group trial. Clin Cancer Res. 2013;19:4816–4823. doi: 10.1158/1078-0432.CCR-13-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kreisl TN, Smith P, Sul J, Salgado C, Iwamoto FM, Shih JH, Fine HA. Continuous daily sunitinib for recurrent glioblastoma. J Neurooncol. 2013;111:41–48. doi: 10.1007/s11060-012-0988-z. [DOI] [PubMed] [Google Scholar]
- 39.Reardon DA, Turner S, Peters KB, Desjardins A, Gururangan S, Sampson JH, McLendon RE, Herndon JE 2nd, Jones LW, Kirkpatrick JP, Friedman AH, Vredenburgh JJ, Bigner DD, Fiedman HS. A review of VEGF/VEGFR-targeted therapeutics for recurrent glioblastoma. J Natl Compr Canc Netw. 2011;9:414–427. doi: 10.6004/jnccn.2011.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SJ, Kim JH, Kim YM, Lee GK, Lee EJ, Park IS, Jung JM, Kang KH, Shin T. Perfusion MR imaging in gliomas: comparison with histologic tumor grade. Korean J Radiol. 2001;2:1–7. doi: 10.3348/kjr.2001.2.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Law M. Advanced imaging techniques in brain tumors. Cancer Imaging. 2009;9 Spec No A:S4–9. doi: 10.1102/1470-7330.2009.9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu KV, Chang JP, Parachoniak CA, Pandika MM, Aghi MK, Meyronet D, Isachenko N, Fouse SD, Philips JJ, Cheresh DA, Park M, Bergers G. VEGF inhibits tumor cell invasion and mesenchymal transition through a MET/VEGFR2 complex. Cancer Cell. 2012;22:21–35. doi: 10.1016/j.ccr.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamerlik P, Lathia JD, Rasmussen R, Wu Q, Bartkova J, Lee M, Moudry P, Bartek J Jr, Fischer W, Lukas J, Rich JN, Bartek J. Autocrine VEGF-VEGFR2-Neuropilin-1 signaling promotes glioma stem-like cell viability and tumor growth. J Exp Med. 2012;209:507–520. doi: 10.1084/jem.20111424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szabo E, Schneider H, Seystahl K, Rushing EJ, Herting F, Weidner KM, Weller M. Autocrine VEGFR1 and VEGFR2 signaling promotes survival in human glioblastoma models in vitro and in vivo. Neuro Oncol. 2016;18:1242–1252. doi: 10.1093/neuonc/now043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abhinand CS, Raju R, Soumya SJ, Arya PS, Sudhakaran PR. VEGF-A/VEGFR2 signaling network in endothelial cells relevant to angiogenesis. J Cell Commun Signal. 2016;10:347–354. doi: 10.1007/s12079-016-0352-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lampugnani MG, Orsenigo F, Gagliani MC, Tacchetti C, Dejana E. Vascular endothelial cadherin controls VEGFR-2 internalization and signaling from intracellular compartments. J Cell Biol. 2006;174:593–604. doi: 10.1083/jcb.200602080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan HT, Khankin EV, Karumanchi SA, Parikh SM. Angiopoietin 2 is a partial agonist/antagonist of Tie2 signaling in the endothelium. Mol Cell Biol. 2009;29:2011–2022. doi: 10.1128/MCB.01472-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim DG, Kim KH, Seo YJ, Yang H, Marcusson EG, Son E, Lee K, Sa JK, Lee HW, Nam DH. Anti-miR delivery strategies to bypass the blood-brain barrier in glioblastoma therapy. Oncotarget. 2016;7:29400–29411. doi: 10.18632/oncotarget.8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galland F, Karamysheva A, Mattei MG, Rosnet O, Marchetto S, Birnbaum D. Chromosomal localization of FLT4, a novel receptor-type tyrosine kinase gene. Genomics. 1992;13:475–478. doi: 10.1016/0888-7543(92)90277-y. [DOI] [PubMed] [Google Scholar]
- 50.Ghalamkarpour A, Holnthoner W, Saharinen P, Boon LM, Mulliken JB, Alitalo K, Vikkula M. Recessive primary congenital lymphoedema caused by a VEGFR3 mutation. J Med Genet. 2009;46:399–404. doi: 10.1136/jmg.2008.064469. [DOI] [PubMed] [Google Scholar]
- 51.Matsuura M, Onimaru M, Yonemitsu Y, Suzuki H, Nakano T, Ishibashi H, Shirasuna K, Sueishi K. Autocrine loop between vascular endothelial growth factor (VEGF)-C and VEGF receptor-3 positively regulates tumor-associated lymphangiogenesis in oral squamoid cancer cells. Am J Pathol. 2009;175:1709–1721. doi: 10.2353/ajpath.2009.081139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Connell FC, Ostergaard P, Carver C, Brice G, Williams N, Masour S, Mortimer PS, Jeffery S Lymphoedema Consortium. Analysis of the coding regions of VEGFR3 and VEGFC in Milroy disease and other primary lymphoedemas. Hum Genet. 2009;124:625–631. doi: 10.1007/s00439-008-0586-5. [DOI] [PubMed] [Google Scholar]
- 53.Butler MG, Dagenais SL, Rockson SG, Glover TW. A novel VEGFR3 mutation causes Milroy disease. Am J Med Genet A. 2007;143a:1212–1217. doi: 10.1002/ajmg.a.31703. [DOI] [PubMed] [Google Scholar]
- 54.Terman BI, Dougher-Vermazen M, Carrion ME, Dimitrov D, Armellino DC, Gospodarowicz D, Böhlen P. Identification of the KDR tyrosine kinase as a receptor for vascular endothelial cell growth factor. Biochem Biophys Res Commun. 1992;187:1579–1586. doi: 10.1016/0006-291x(92)90483-2. [DOI] [PubMed] [Google Scholar]
- 55.Yang Y, Xie P, Opatowsky Y, Schlessinger J. Direct contacts between extracellular membrane-proximal domains are required for VEGF receptor activation and cell signaling. Proc Natl Acad Sci U S A. 2010;107:1906–1911. doi: 10.1073/pnas.0914052107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim DH, Xu W, Kamel-Reid S, Liu X, Jung CW, Kim S, Lipton JH. Clinical relevance of vascular endothelial growth factor (VEGFA) and VEGF receptor (VEGFR2) gene polymorphism on the treatment outcome following imatinib therapy. Ann Oncol. 2010;21:1179–1188. doi: 10.1093/annonc/mdp452. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Z, Neiva KG, Lingen MW, Ellis LM, Nör JE. VEGF-dependent tumor angiogenesis requires inverse and reciprocal regulation of VEGFR1 and VEGFR2. Cell Death Differ. 2010;17:499–512. doi: 10.1038/cdd.2009.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.An SJ, Chen ZH, Lin QX, Su J, Chen HJ, Lin JY, Wu YL. The -271 G>A polymorphism of kinase insert domain-containing receptor gene regulates its transcription level in patients with non-small cell lung cancer. BMC Cancer. 2009;9:144. doi: 10.1186/1471-2407-9-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coffelt SB, Chen YY, Muthana M, Welford AF, Tal AO, Scholz A, Plate KH, Reiss Y, Murdoch C, De Palma M, Lewis CE. Angiopoietin 2 stimulates TIE2-expressing monocytes to suppress T cell activation and to promote regulatory T cell expansion. J Immunol. 2011;186:4183–4190. doi: 10.4049/jimmunol.1002802. [DOI] [PubMed] [Google Scholar]
- 60.Daly C, Eichten A, Castanaro C, Pasnikowski E, Adler A, Lalani AS, Papadopoulos N, Kyle AH, Minchiton AI, Yancopoulos GD, Thurston G. Angiopoietin-2 functions as a Tie2 agonist in tumor models, where it limits the effects of VEGF inhibition. Cancer Res. 2013;73:108–118. doi: 10.1158/0008-5472.CAN-12-2064. [DOI] [PubMed] [Google Scholar]
- 61.Maisonpierre PC, Suri C, Jones PF, Bartunkova S, Wiegand SJ, Radziejewski C, Compton D, McClain J, Aldrich TH, Papadopoulos N, Daly TJ. Angiopoietin-2, a natural antagonist for Tie2 that disrupts in vivo angiogenesis. Science. 1997;277:55–60. doi: 10.1126/science.277.5322.55. [DOI] [PubMed] [Google Scholar]
- 62.Vairaktaris E, Serefoglou Z, Avgoustidis D, Yapijakis C, Critselis E, Vylliotis A, Spyridonidou S, Dekra S, Vassillou S, Nkenke E, Patsouris E. Gene polymorphisms related to angiogenesis, inflammation and thrombosis that influence risk for oral cancer. Oral Oncol. 2009;45:247–253. doi: 10.1016/j.oraloncology.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 63.Cheng YC, Kao WH, Mitchell BD, O’Connell JR, Shen H, McArdie PF, Gibson Q, Ryan KA, Shuldiner AR, Pollin TI. Genome-wide association scan identifies variants near Matrix Metalloproteinase (MMP) genes on chromosome 11q21-22 strongly associated with serum MMP-1 levels. Circ Cardiovasc Genet. 2009;2:329–337. doi: 10.1161/CIRCGENETICS.108.834986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ricketts C, Zeegers MP, Lubinski J, Maher ER. Analysis of germline variants in CDH1, IGFBP3, MMP1, MMP3, STK15 and VEGF in familial and sporadic renal cell carcinoma. PLoS One. 2009;4:e6037. doi: 10.1371/journal.pone.0006037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Song YQ, Ho DW, Karppinen J, Kao PY, Fan BJ, Luk KD, Yip SP, Leong JC, Cheah KS, Sham P, Chan D, Cheung KM. Association between promoter -1607 polymorphism of MMP1 and lumbar disc disease in Southern Chinese. BMC Med Genet. 2008;9:38. doi: 10.1186/1471-2350-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsuchiya N, Narita S, Kumazawa T, Inoue T, Ma Z, Tsuruta H, Saito M, Horikawa Y, Yuasa T, Satoh S, Ogawa O, Habuchi T. Clinical significance of a single nucleotide polymorphism and allelic imbalance of matrix metalloproteinase-1 promoter region in prostate cancer. Oncol Rep. 2009;22:493–499. doi: 10.3892/or_00000462. [DOI] [PubMed] [Google Scholar]
- 67.Ali J, Liao F, Martens E, Muller WA. Vascular endothelial cadherin (VE-cadherin): cloning and role in endothelial cell-cell adhesion. Microcirculation. 1997;4:267–277. doi: 10.3109/10739689709146790. [DOI] [PubMed] [Google Scholar]
- 68.Breviario F, Caveda L, Corada M, Martin-Padura I, Navarro P, Golay J, Introna M, Gulino D, Lampugnani MG, Dejana E. Functional properties of human vascular endothelial cadherin (7B4/cadherin-5), an endothelium-specific cadherin. Arterioscler Thromb Vasc Biol. 1995;15:1229–1239. doi: 10.1161/01.atv.15.8.1229. [DOI] [PubMed] [Google Scholar]
- 69.Suzuki S, Sano K, Tanihara H. Diversity of the cadherin family: evidence for eight new cadherins in nervous tissue. Cell Regul. 1991;2:261–270. doi: 10.1091/mbc.2.4.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bailey SD, Xie C, Do R, Montpetit A, Diaz R, Mohan V, Keavney B, Yusuf S, Gerstein HC, Engert JC, Anand S DREAM investigators. Variation at the NFATC2 locus increases the risk of thiazolidinedione-induced edema in the Diabetes REduction Assessment with ramipril and rosiglitazone Medication (DREAM) study. Diabetes Care. 2010;33:2250–2253. doi: 10.2337/dc10-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Graziani A, Poteser M, Heupel WM, Schleifer H, Krenn M, Drenckhahn D, Romanin C, Baumgartner W, Groschner K. Cell-cell contact formation governs Ca2+ signaling by TRPC4 in the vascular endothelium: evidence for a regulatory TRPC4-beta-catenin interaction. J Biol Chem. 2010;285:4213–4223. doi: 10.1074/jbc.M109.060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lim ST, Chen XL, Lim Y, Hanson DA, Vo TT, Howerton K, Larocque N, Fisher SJ, Schlaepfer DD, Ilic D. Nuclear FAK promotes cell proliferation and survival through FERM-enhanced p53 degradation. Mol Cell. 2008;29:9–22. doi: 10.1016/j.molcel.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salasznyk RM, Klees RF, Boskey A, Plopper GE. Activation of FAK is necessary for the osteogenic differentiation of human mesenchymal stem cells on laminin-5. J Cell Biochem. 2007;100:499–514. doi: 10.1002/jcb.21074. [DOI] [PubMed] [Google Scholar]
- 74.Ezratty EJ, Partridge MA, Gundersen GG. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat Cell Biol. 2005;7:581–590. doi: 10.1038/ncb1262. [DOI] [PubMed] [Google Scholar]
- 75.Golubovskaya VM, Finch R, Cance WG. Direct interaction of the N-terminal domain of focal adhesion kinase with the N-terminal transactivation domain of p53. J Biol Chem. 2005;280:25008–25021. doi: 10.1074/jbc.M414172200. [DOI] [PubMed] [Google Scholar]
- 76.Xia H, Nho RS, Kahm J, Kleidon J, Henke CA. Focal adhesion kinase is upstream of phosphatidylinositol 3-kinase/Akt in regulating fibroblast survival in response to contraction of type I collagen matrices via a beta 1 integrin viability signaling pathway. J Biol Chem. 2004;279:33024–33034. doi: 10.1074/jbc.M313265200. [DOI] [PubMed] [Google Scholar]
- 77.Shibuya M, Yamaguchi S, Yamane A, Ikeda T, Tojo A, Matsushime H, Sato M. Nucleotide sequence and expression of a novel human receptor-type tyrosine kinase gene (flt) closely related to the fms family. Oncogene. 1990;5:519–524. [PubMed] [Google Scholar]
- 78.Srinivas SK, Morrison AC, Andrela CM, Elovitz MA. Allelic variations in angiogenic pathway genes are associated with preeclampsia. Am J Obstet Gynecol. 2010;202:445, e441–411. doi: 10.1016/j.ajog.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 79.Muehlenbachs A, Fried M, Lachowitzer J, Mutabingwa TK, Duffy PE. Natural selection of FLT1 alleles and their association with malaria resistance in utero. Proc Natl Acad Sci U S A. 2008;105:14488–14491. doi: 10.1073/pnas.0803657105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim SY, Lim JH, Yang JH, Kim MY, Han JY, Ahn HK, Choi JS, Park SY, Kim MJ, Ryu HM. Dinucleotide repeat polymorphism in Fms-like tyrosine kinase-1 (Flt-1) gene is not associated with preeclampsia. BMC Med Genet. 2008;9:68. doi: 10.1186/1471-2350-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fan F, Wey JS, McCarty MF, Belcheva A, Liu W, Bauer TW, Somcio RJ, Wu Y, Hooper A, Hicklin DJ, Ellis LM. Expression and function of vascular endothelial growth factor receptor-1 on human colorectal cancer cells. Oncogene. 2005;24:2647–2653. doi: 10.1038/sj.onc.1208246. [DOI] [PubMed] [Google Scholar]
- 82.Jin P, Zhang J, Sumariwalla PF, Ni I, Jorgensen B, Crawford D, Phillips S, Feldman M, Shepard HM, Paleolog EM. Novel splice variants derived from the receptor tyrosine kinase superfamily are potential therapeutics for rheumatoid arthritis. Arthritis Res Ther. 2008;10:R73. doi: 10.1186/ar2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reddi AL, Ying G, Duan L, Chen G, Dimri M, Douillard P, Druker BJ, Naramura M, Band V, Band H. Binding of Cbl to a phospholipase Cgamma1-docking site on platelet-derived growth factor receptor beta provides a dual mechanism of negative regulation. J Biol Chem. 2007;282:29336–29347. doi: 10.1074/jbc.M701797200. [DOI] [PubMed] [Google Scholar]
- 84.Brück P, Wassmann B, Lopez ER, Hoelzer D, Ottmann OG. Development of hygromas or severe edema during treatment with the tyrosine kinase inhibitor STI571 is not associated with platelet-derived growth factor receptor (PDGFR) gene polymorphisms. Leuk Res. 2004;28:1153–1157. doi: 10.1016/j.leukres.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 85.Kovalenko M, Denner K, Sandström J, Persson C, Gross S, Jandt E, Vilella R, Böhmer F, Ostman A. Site-selective dephosphorylation of the platelet-derived growth factor beta-receptor by the receptor-like protein-tyrosine phosphatase DEP-1. J Biol Chem. 2000;275:16219–16226. doi: 10.1074/jbc.275.21.16219. [DOI] [PubMed] [Google Scholar]
- 86.Abe A, Emi N, Tanimoto M, Terasaki H, Marunouchi T, Saito H. Fusion of the platelet-derived growth factor receptor beta to a novel gene CEV14 in acute myelogenous leukemia after clonal evolution. Blood. 1997;90:4271–4277. [PubMed] [Google Scholar]
- 87.Xu H, Rahimpour S, Nesvick CL, Zhang X, Ma J, Zhang M, Zhang G, Wang L, Yang C, Hong CS, Germanwala AV, Elder JB, Ray-Chaudhury A, Yao Y, Gilbert MR, Lonser RR, Heiss JD, Brady RO, Mao Y, Qin J, Zhuang Z. Activation of hypoxia signaling induces phenotypic transformation of glioma cells: implications for bevacizumab antiangiogenic therapy. Oncotarget. 2015;6:11882–11893. doi: 10.18632/oncotarget.3592. [DOI] [PMC free article] [PubMed] [Google Scholar]


