Abstract
Purpose:
To identify molecular factors that determine duration of response to EGFR tyrosine kinase inhibitors and to identify novel mechanisms of drug resistance, we molecularly profiled EGFR mutant tumors prior to treatment and after progression on EGFR TKI using targeted next-generation sequencing.
Experimental Design:
Targeted next-generation sequencing was performed on 374 consecutive patients with metastatic EGFR mutant lung cancer. Clinical data were collected and correlated with somatic mutation data. Erlotinib resistance due to acquired MTOR mutation was functionally evaluated by in vivo and in vitro studies.
Results:
In 200 EGFR-mutant pre-treatment samples, the most frequent concurrent alterations were mutations in TP53, PIK3CA, CTNNB1 and RB1 and focal amplifications in EGFR, TTF1, MDM2, CDK4, and FOXA1. Shorter time to progression on EGFR TKI was associated with amplification of ERBB2 (HR=2.4, p=0.015) or MET (HR 3.7, p=0.019), or mutation in TP53 (HR 1.7, p=0.006). In the 136 post-treatment samples, we identified known mechanisms of acquired resistance: EGFR T790M (51%), MET (7%) and ERBB2 amplifications (5%). In the 38 paired samples, novel acquired alterations representing putative resistance mechanisms included BRAF fusion, FGFR3 fusion, YES1 amplification, KEAP1 loss, and an MTOR E2419K mutation. Functional studies confirmed the contribution of the latter to reduced sensitivity to EGFR TKI in vitro and in vivo.
Conclusions:
EGFR-mutant lung cancers harbor a spectrum of concurrent alterations that have prognostic and predictive significance. By utilizing paired samples, we identified several novel acquired alterations that may be relevant in mediating resistance, including an activating mutation in MTOR further validated functionally.
Introduction
EGFR-mutant lung cancers represent a distinct molecular subset of lung cancers, with the majority of such patients benefitting from treatment with EGFR tyrosine kinase inhibitor (TKI) therapy [1, 2]. Although the majority of patients respond to EGFR TKI, there is a significant heterogeneity in the clinical course of patients with EGFR-mutant lung cancers. This variability may be partially attributable to differences among EGFR mutation subtypes with exon 19 deletions associated with longer overall survival compared to the L858R missense substitution. In contrast, tumors with EGFR exon 20 insertions are typically intrinsically resistant to EGFR inhibitor therapy, with rare exceptions [3, 4] and lung cancers with certain EGFR mutations such as exon 18 alterations may be more sensitive to a particular EGFR TKI, such as afatinib, over others [5]. Among patients with the common sensitizing EGFR exon 19 deletions and EGFR L858R, there are no established molecular features that predict duration of benefit from EGFR TKI.
As comprehensive tumor genomic profiling becomes standard of care, clinicians are often informed in real time about concurrent genetic alterations that co-occur with clinically actionable driver oncogenes and which may affect prognosis and response to treatment.
For example, within KRAS-mutant lung cancers, concurrent LKB1 (STK11) and TP53 mutations may be associated with shorter survival [6–9] and the presence of the former may also confer a greater sensitivity to mTOR or MEK inhibition [10, 11]. Similarly, in EGFR-mutant lung cancers, several reports have suggested that concurrent TP53 alterations are associated with a lower likelihood of response to EGFR TKI and shorter overall survival [12, 13]. Because expression of BIM is required for induction of apoptosis by EGFR TKIs, low levels of pre-treatment BIM within the tumor are associated with a lower response rate and shorter progression-free survival on EGFR TKI [14–17]. EGFR T790M, which is most commonly seen as an acquired mutation, can sometimes occur pre-treatment, and when present, baseline EGFR T790M mutations result in a lower response rate, shorter progression-free survival on EGFR TKI and shorter overall survival in patients with metastatic EGFR-mutant lung cancers[18–20]
Immunotherapy has emerged as an invaluable treatment option for many patients with lung cancer. Significant effort has gone into identifying predictive biomarkers that could be used to prospectively select patients that are most likely to benefit from immunomodulation, with PD-L1 expression as well as tumor mutation burden having emerged as potential biomarkers of immunotherapy response [21, 22]. EGFR-mutant lung cancers have a relatively lower tumor mutation burden and less frequently express PDL1, features that may underlie the lack of response to immunotherapy largely seen in patients with EGFR-mutant lung cancers [23]. As responses to immunotherapy are limited in this population, defining the ideal sequence of treatment options remains critical.
Acquisition of EGFR T790M is the most frequently identified resistance mechanism to first and second generation EGFR TKI therapy. Less frequently identified but well established acquired alterations include MET and ERBB2 amplification, PIK3CA and BRAF mutations, and small cell histologic transformation [24, 25]. In approximately 30% of patients, the molecular basis of clinical progression is not defined, which may be attributable to the limited scope of the molecular profiling assays historically employed. Comprehensive next-generation sequencing performed on tumor samples before and after treatment provides an opportunity to discover relevant molecular alterations not previously appreciated that may suggest candidates for combination therapy to improve responses to EGFR TKI. In order to understand the impact of co-mutations and to identify novel acquired alterations that emerge on treatment, we integrated genomic data, treatment histories and clinical outcomes in patients with EGFR-mutant lung cancers who underwent clinical next generation sequencing analysis before and/or after treatment with EGFR TKI, and report functional studies of one of the novel candidate resistance mechanisms thereby identified.
Methods
We identified all patients with EGFR mutant metastatic lung cancers who had targeted hybrid capture, next-generation sequencing (NGS) performed at Memorial Sloan Kettering from March 2014 to February 2017. Genomic analysis was performed using the MSK-IMPACT assay, a clinical test designed to detect mutations, copy-number alterations, and select fusions involving 341 (version 1), 410 (version 2) or 468 (version 3) cancer-associated genes [26, 27]. All genetic alterations discussed within the manuscript involved genes present in all three versions of MSK-IMPACT. Paired analysis of tumor and matched-normal samples was performed to allow for definitive identification of somatic mutations. Cancer cell fractions were calculated as per previously published methods[28]. Patients were divided into three mutually exclusive cohorts: patients with tumor samples collected prior to EGFR TKI therapy, patients with tumor sampled collected after disease progression and patients for whom paired samples (pre-EGFR TKI and post-EGFR TKI) were available within the prespecified time frame. Data collection was approved by the MSK Institutional Review Board/Privacy Board.
We collected clinical characteristics and detailed treatment histories for all patients including sites of disease, time to progression on EGFR tyrosine kinase inhibitor (TKI) and overall survival (OS) from start of EGFR TKI. Time to progression on EGFR TKI was defined as time from start of EGFR TKI to time of radiographic RECIST progression. Fisher’s exact and log-rank tests were utilized to identify potential associations between EGFR mutation subtype, frequent co-mutations, clinical factors, and patient outcomes. Time to progression and overall survival were estimated using Kaplan-Meier methodology. Patients were followed until death; patients alive were censored at the time of last available follow up.
Please see supplemental material for detailed methods regarding the in vivo and in vitro studies.
Results
Patient and Tumor Characteristics
From March 2014 to February 2017, 374 patients with activating kinase-domain mutations in EGFR underwent clinical tumor genomic profiling. Two hundred patients had tumor for molecular analysis collected prior to initiation of EGFR TKI; 136 patients had tumor molecular profiling from a sample obtained after disease progression on EGFR TKI and 38 patients had tumors samples from both before and after treatment with an EGFR TKI (Table 1). Three post-treatment specimens (3/136, 2%) had neuroendocrine differentiation, two as small cell carcinoma and one as large cell neuroendocrine carcinoma. No component of small cell histology was seen in any of the pre-treatment specimens.
Table 1.
Characteristics of patients who had MSK-IMPACT testing prior to EGFR TKI therapy (n=200) or after EGFR TKI therapy (n=136) or both (n=38)
| Clinical characteristics | Pre EGFR-TKI N (%) |
Post EGFR-TKI N (%) |
Paired samples N (%) |
||
|---|---|---|---|---|---|
| Total | 200 | 136 | 38 | ||
| Median (range) | 63 (23–89) | 61 (29–86) | 62 (34–84) | ||
| Female | 134 (67) | 41 (30) | 27 (71) | ||
| Former smoker | 82 (41) | 57 (42) | 16 (42) | ||
| Median pack-yr (range) | 6 (1–125) | 0 (0–110) | 0 (0–30) | ||
| Large cell neuroendocrine | 0 (0) | 1 (0.7) | 0 (0) | 0 (0) | |
| EGFR mutation | |||||
| EGFR T790M | 0 (0) | 70 (52) | 0 (0) | 16 (42) | |
| Exon 19 deletion | 90 (45) | 78 (57) | 17 (45) | ||
| L858R | 72 (36) | 43 (32) | 15 (39) | ||
| Exon 20 insertions | 7 (3.5) | 1 (0.7) | 0 (0) | ||
| Exon 18 deletion | 5 (2.5) | 4 (3) | 3 (8) | ||
| Exon 19 insertion | 3 (1.5) | 1 (0.7) | 1 (3) | ||
| L861Q | 6 (3) | 2 (1.5) | 0 (0) | ||
| G719A | 3 (1.5) | 2 (1.5) | 0 (0) | ||
| L747P | 3 (1.5) | 0 (0) | 0 (0) | ||
| E709X+G719X | 4 (2) | 0 (0) | 0 (0) | ||
| G719X+S768I | 2 (1) | 3 (2) | 0 (0) | ||
| G719X+L861Q | 3 (1.5) | 0 (0) | 0 (0) | ||
| EGFR-KDD | 2 (1) | 2 (1.5) | 2 (5) | ||
| Prior treatment | |||||
| Erlotinib | 0 (0) | 128 (94) | 0 (0) | 36 (95) | |
| Afatinib | 0 (0) | 6 (4) | 0 (0) | 2 (5) | |
| Dacomitinib | 0 (0) | 2 (1) | 0 (0) | 0 (0) | |
Concurrent Genomic Alterations and Outcomes in patients whose tumors were profiled before treatment with EGFR TKI
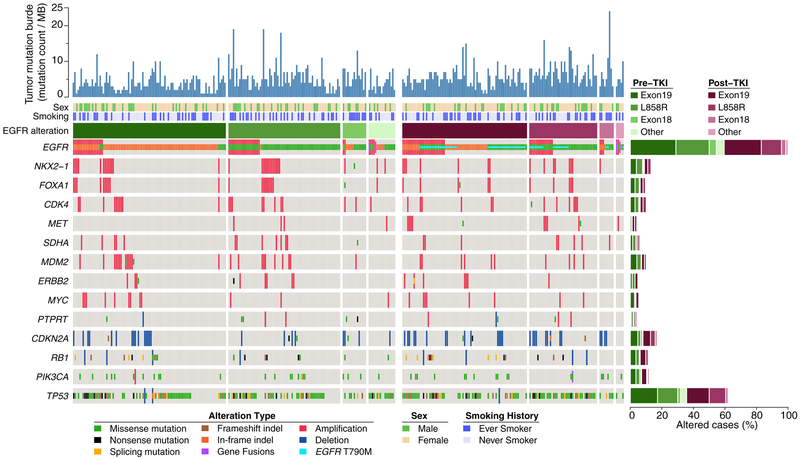
The most frequent concurrent mutations and amplifications in samples obtained prior to EGFR TKI treatment are presented in Figure 1. The median number of co-mutations was 5 (range 0–19). The most frequent co-occurring mutations in the baseline samples were TP53 (60%, n=119), PIK3CA (12%, n=23), CTNNB1 (9%, n=18) and RB1 (10%, n=19). The most frequent concurrent amplifications were EGFR (22%, n=45), NKX2.1/TTF-1 (15%, n=29), MDM2 (12%, n=23), CDK4 (10%, n=21), and FOXA1 (10%, n=20). ERBB2 amplification was seen in 4% (n=8) and MET amplification in 2% (n=4) of baseline samples. No patients had evidence of EGFR T790M prior to treatment with EGFR TKI. There were no concurrent ALK, ROS1, RET, BRAF or MET mutations in any of the pre-treatment samples. There was one sample with an EGFR L858R mutation and concurrent subclonal KRAS Q61H and KRAS Q22K mutations, described in more detail in a previous report [29]. The number of co-mutations did not correlate with specific EGFR mutation subtype, age, sex or smoking status.
Figure 1.
Oncoprint of alterations identified in tumor samples from patients with EGFR-mutant lung cancer pre-treatment (green) and after treatment (pink) with an EGFR TKI. The frequency is noted on the right. The type of genetic alteration (missense, inframe, truncated, amplification, deletion, fusion) is described in the legend, and the comutations present in ≥5% of cases were included in the figure. HER2 and MET amplification were included due to their relevance in acquired resistance.
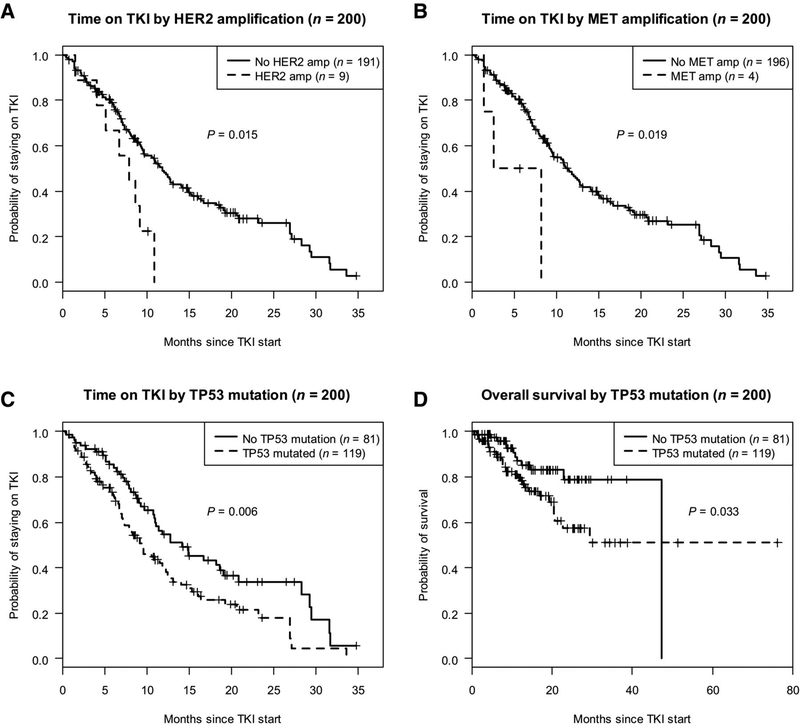
In the cohort of patients with pre-treatment tumor samples used for molecular profiling, the median time to progression (TTP) on EGFR TKI was 11 months (95% CI 9–13 months). The presence of ERBB2 amplification (median TTP 8 months, 95% CI 5-NR months; HR 2.42, p=0.018), MET amplification (median TTP 5 months, 95% CI 7–12 months; HR 3.65, p=0.029) and TP53 mutation (median TTP 6 months, range 1 to 33; HR 1.68, p=0.006) in pre-treatment samples were all associated with shorter time on EGFR TKI (Figure 2). The presence of TP53 alterations was associated with shorter overall survival from start of EGFR TKI (NR vs 47 mo, HR 2.04, p=0.036) (Figure 2). No other co-mutations had a significant association with overall survival.
Figure 2:
Progression-free survival on EGFR TKI and overall survival from start of EGFR TKI stratified by concurrent alteration present in pre-treatment tumor sample in patients with EGFR-mutant lung cancers. (A) Time on EGFR TKI stratified by presence/absence of HER2 amplification (B) Time on EGFR TKI stratified by presence/absence of MET amplification (C) Time on EGFR TKI stratified by presence/absence of p53 mutation (D) Overall survival stratified by presence/absence of p53 mutation.
Concurrent Genomic Alterations in Patients whose tumors were profiled after disease progression on EGFR TKI
The most frequent concurrent mutations and amplifications in the 136 samples obtained after EGFR TKI treatment are presented in Figure 1. EGFR T790M was identified in 52% (70/136). The most frequent co-mutations were TP53 (65%, n=89), PIK3CA (13%, n=17) and RB1 (8%, n=11), and the most frequent amplifications were EGFR (35%, n=47), TERT (11%, n=15), and NKX2.1/TTF-1 (11%, n=15). ERBB2 amplifications were seen in 4% (n=7) and MET amplification in 7% (n=9). One BRAF D594V mutation (0.7%) and two BRAF fusions were identified (2.2%). There were no concurrent ALK, ROS1, RET, KRAS, or MET alterations in any of the samples.
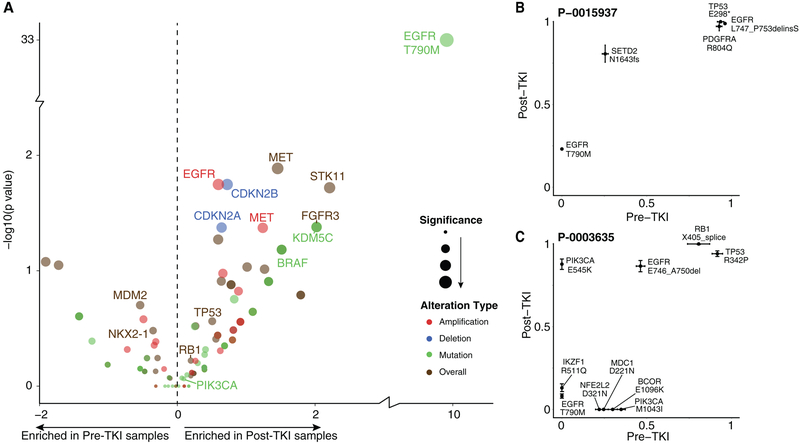
When comparing the pre-treatment and post-treatment samples, several somatic alterations were more frequently seen in the post-treatment samples (Figure 3A). EGFR T790M (52% vs 0%), BRAF alterations (5.1% vs 1%; OR=5.3, p=0.034), CDKN2A loss (21.3% vs 12.5%; OR 1.9, HR 0.035), CDKN2B loss (19.9% vs 10.5%; OR 2.1, p=0.018), EGFR amplification (35% vs 23%; OR 1.82, p=0.018), FGFR3 alterations (3.7% vs 0.5%; OR=7.55, p=0.042) and MET amplification (6.6% vs 2%; OR=3.46, p=0.042) were all more frequently seen in the post-treatment samples compared to the pre-treatment samples. The analyses performed suggest that post-treatment samples are enriched for these alterations, but we cannot confirm which alterations were acquired since these pre- and post-treatment samples were not paired samples.
Figure 3:
(A) Enrichment of genomic alterations in tumors from patients with EGFR-mutant lung cancers prior to EGFR TKI versus after progression on EGFR TKI. The level of enrichment is represented as different in frequency between the two states (X axis) and its significance (P value, Y axis). The type of alterations is represented by color. (B, C) Cancer cell fractions of mutations based on FACETS analysis comparing pre-treatment (X axis) and post-treatment tumor samples (Y axis) in two representative patients.
Analysis of Paired Pre-treatment and Acquired Resistance Specimens
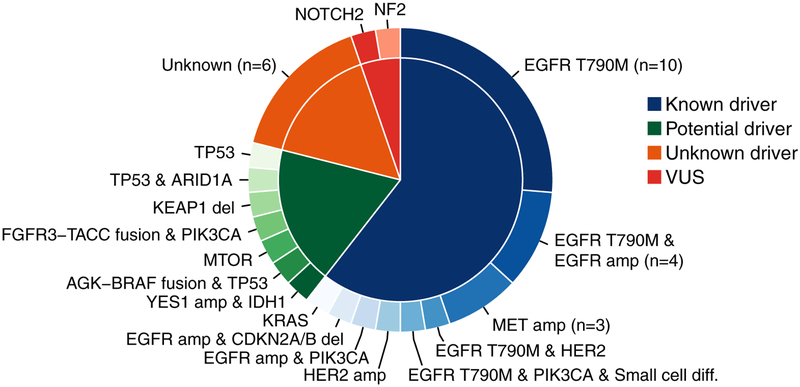
To identify candidate novel mechanisms of acquired resistance to EGFR TKI and to clarify which mutations identified in the broader analysis of resistance specimens are more likely to be acquired during therapy, we analyzed paired pre and post- EGFR TKI treatment samples from an additional 38 patients. All had adenocarcinoma histology prior to treatment; one patient had small cell histology upon rebiopsy after progression on erlotinib (2.6%, 1/38). The paired pre-treatment and post-treatment samples maintained the same sensitizing EGFR mutation in all cases. Comprehensive information regarding co-mutations present at baseline, at acquired resistance, and site of biopsy is provided in Supplemental Tables 1 and 2. Forty-two percent (16/38) had an acquired EGFR T790M mutation identified in the post-treatment sample. In these 16 patients who acquired EGFR T790M, we visually examined sequencing reads to see if low level pre-treatment EGFR T790M was present in the baseline sample but below the bioinformatics cutoff for calling an alteration, but no evidence of EGFR T790M was present in any of the baseline paired samples, indicating that small pre-existing EGFR T790M-positive clones, if present, must occur well below the limit of detection of our assay (1–2%). Acquired MET amplification was identified in 8% (3/38), acquired ERBB2 amplification in 5% (2/38) and acquired amplification of the EGFR allele with the sensitizing mutation in 16% (6/38). Notable acquired alterations that represent the presumed mechanisms of resistance to EGFR TKI for each sample are noted in Figure 4.
Figure 4:
Mechanisms of resistance in the paired samples. These mutations are acquired alterations when comparing pre-treatment samples to samples obtained after clinical progression on an EGFR TKI.
The presence of multiple concurrent alterations that could mediate resistance was common in these rebiopsy specimens, with EGFR T790M frequently arising concurrently with other ostensible mechanisms of resistance. For example, EGFR T790M was detected in two patients with EGFR amplification and in one patient with an acquired ERBB2 amplification. However, acquired EGFR T790M mutations did not occur in any patient with acquired MET amplification. In one patient’s post-treatment sample, EGFR T790M, an acquired PIK3CA mutation and small cell transformation were all identified; RB1 and TP53 loss were present in both the pre-treatment and post-treatment samples. Acquired PIK3CA E545K mutations were seen in 3 patients; however, in all instances, the acquired PIK3CA mutation was seen concurrently with other known mechanisms of resistance.
We also inferred the fraction of cancer cells that contained specific mutations where sufficient tumor content was available. Two representative cases are illustrated in Figure 3B, 3C. As expected, the EGFR sensitizing mutations were present in a very high fraction of cells in each case. Acquired mutations associated with drug resistance, in contrast, often appear to be subclonal and present in a lower fraction of cancer cells. EGFR T90M mutations when identified in a tumor, were present in an average 34% of cells (median = 26%, range = 5.7%−84%). Interestingly, RB1 and TP53 mutations appear to be truncal mutations present in an essentially clonal fashion both pre-treatment and after-treatment with EGFR TKI.
TP53 mutations were present in average 87% of cells (median = 93%, range (6–100%)) and RB1 mutations were present in average 95% of cells (median = 94%, range (93–100%)).
Upon review of the paired samples, several novel acquired alterations were identified as plausible candidate mediators of clinical resistance to EGFR TKIs as these alterations have functional significance pre-clinically or in other clinical settings (Figure 3). One patient had an acquired AGK-BRAF fusion as well as a known pathogenic TP53 mutation, G334W. Two others had acquired loss of function TP53 alterations, one in combination with a not previously reported ARID1A mutation, E1032K. Another patient had evidence of an acquired FGFR3-TACC3 fusion in their resistance sample along with an acquired PIK3CA E545K mutation. One patient had loss of KEAP1 in their resistance sample and no other acquired alterations associated with EGFR TKI resistance. One patient acquired YES1 amplification in their post-treatment sample along with an acquired IDH1 R132G mutation. One patient had an acquired mTOR mutation, E2419, in their post-treatment sample. We have further characterized two of these novel candidate EGFR TKI resistance mechanisms: functional data on the effect of MTOR E2419K on response to EGFR TKI are provided below and validation of YES1 amplification as a TKI resistance mechanism is described separately (Fan PD et al., submitted).
Tumor mutation burden
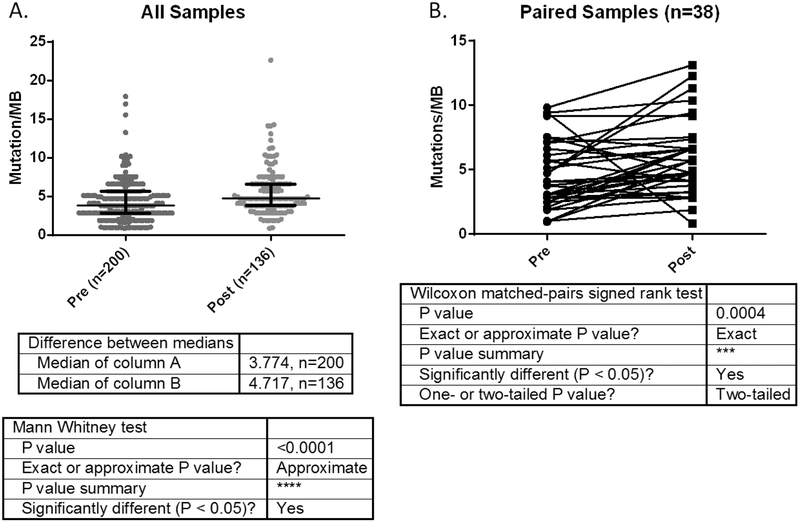
We also examined difference in tumor mutation burden (TMB, number of nonsynonymous variants, normalized per megabase covered to account for MSK-IMPACT panel versions of varying size) for all tumor samples. TMB was slightly but significantly higher in the post-EGFR TKI samples (n=136) when compared with the pre-treatment samples (n=200), 4.7 vs 3.8 mutations/MB, p<0.0001 (Figure 5A). In the 38 paired samples, the tumor mutation burden was also higher in the tumor samples after treatment with EGFR TKI, p=0.0015 (Figure 5B).
Figure 5.
A. TMB (tumor mutation burden) normalized per MB in pre-TKI cohort compared to post-TKI cohort.
B: Number of concurrent mutations in pre-TKI sample and paired post-TKI sample for 38 patients with paired samples.
Functional Analysis of mTOR E2419K mutation as a novel resistance mechanism to EGFR TKI
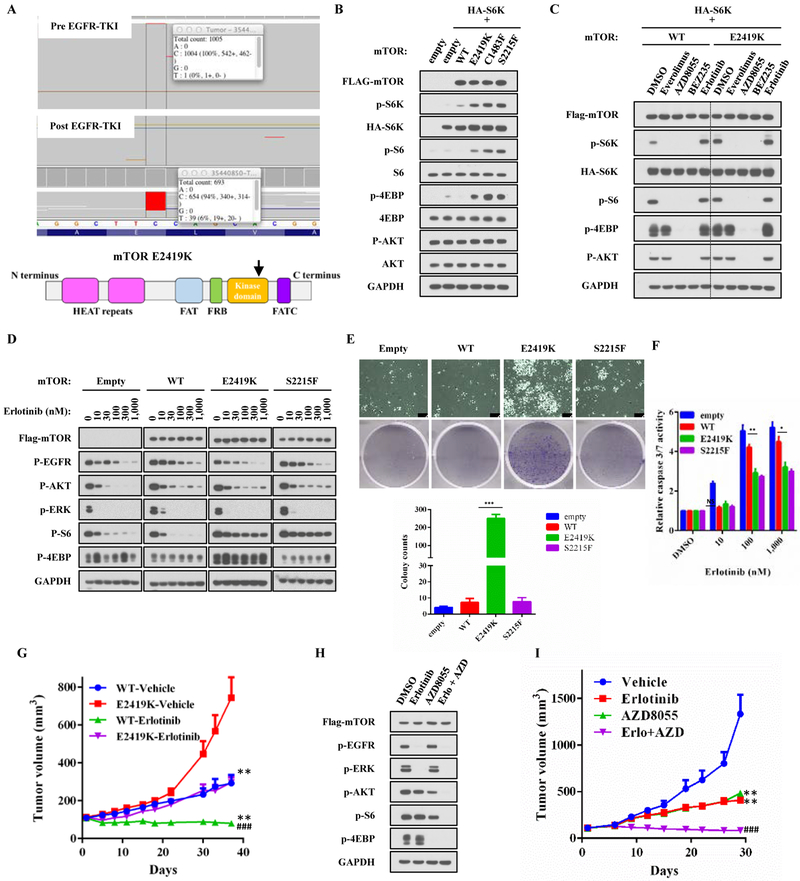
Tumor samples from one patient with paired pre-treatment and post-treatment samples demonstrated an acquired E2419K mutation in exon 53 of mTOR in 5.6% (39/693) reads, with EGFR L858R identified in 17% (112/645) reads, suggesting that approximately a third of the tumor cells in the post-treatment biopsy harbored this MTOR mutation, in the absence of other known resistance mechanisms (Figure 6A). We performed functional studies of this mutation and other common activating mTOR mutations (C1483F and S2215F) [30, 31]. Cells transiently expressing mTOR E2419K showed stronger phosphorylation of S6K, S6, and 4E-BP, which are downstream signaling pathways of mTORC1, but with little effect on phosphorylation of AKT, which is downstream from mTORC2 (Figure 6B and Supplemental Figure 1) suggesting that these mTOR mutations exclusively activate mTORC1, consistent with previous data [30]. Cells harboring the mTOR E2419K mutations displayed inhibition of downstream signaling upon treatment with mTOR inhibitors (everolimus, AZD8055, BEZ235), but not with EGFR inhibitors (erlotinib) (Figure 6C).
Figure 6: Functional analysis of acquired mTOR E2419K mutation in elrotinib-resistant EGFR mutant non-small cell lung cancer.
A. MSK-IMPACT analysis of paired samples before and after EGFR-TKI resistance revealed an acquired mTOR E2419K mutation. B. 293T cells were transiently transfected with pcDNA3 Flag-mTOR (WT, E2419K, S1483F, S2215F), vector control, or HA-S6K1. Thirty-six hours after the transfection, cells were serum starved overnight and subsequently nutrition starved in PBS for 1 hour. Lysates were subjected to immunoblotting. Band intensities were quantified using ImageJ software, and data are representative of two independent experiments (mean ± SE). ***p<0.001, compared to the respective WT+S6K group. C. 293T cells were transiently transfected with pcDNA3 mTOR (wild-type, E2419K), vector control, or HA-S6K1. Forty-eight hours after the transfection, cells were treated with everolimus (100nM), AZD8055 (500nM), BEZ235 (500nM), or erlotinib (1μM) for 3 hours and then subjected to immunoblotting. D. Isogenic stable PC9-mTOR lines were treated with increasing concentrations of erlotinib for 3 hours without serum and lysates were subjected to immunoblotting. E. A total 1.5 × 104 of cells were plated in 6-well plates, and treated with 1μM erlotinib for 14 days. The number of colonies was analyzed using ImageJ. Each experiment was assayed in duplicate determinations and data are representative of three independent experiments (mean ± SE). F. Caspase 3/7 activity was analyzed in stable PC9-mTOR lines that were treated with increasing concentrations of erlotinib for 48 hours. Each experiment was assayed in duplicate determinations and data are representative of three independent experiments (mean ± SE). *p<0.05, **p<0.01 compared to PC9-mTOR WT group. G. PC9-mTOR WT and E2419K cells were implanted into a subcutaneous flank of athymic nude mice. When tumors reached approximately 100 mm3, mice were treated with vehicle or 25 mg/kg erlotinib daily. Tumor volume was determined on the indicated days after the onset of treatment. Data represent mean ± SE (n = 5). *p<0.05, compared to the respective vehicle-treated group. #p<0.05, compared to erlotinib-treated PC9-mTOR WT group. H. PC9-mTOR E2419K cells were treated with erlotinib (1μM), AZD8055 (500nM), or a combination of erlotinib (1μM) and AZD8055 (500nM) for 3 hours. Lysates were then subjected to immunoblotting. I. PC9-mTOR E2419K cells were implanted subcutaneously into the flank of athymic nude mice. Once tumors reached approximately 100 mm3, mice were treated with vehicle, 25 mg/kg erlotinib, 20 mg/kg AZD8055, or a combination of 25 mg/kg erlotinib and 20 mg/kg AZD8055 daily. Tumor volume was determined on the indicated days after the onset of treatment. Data represent mean ± SE (n = 5). *p<0.05, compared to the vehicle treated group. #p<0.05, compared to erlotinib only treated group.
To investigate the role of mTOR mutations in mediating EGFR TKI resistance, we generated an isogenic cell line model using PC9 cells transduced with retroviral vectors driving expression of either mTOR WT or mutant. Phosphorylation of EGFR, AKT, and ERK was inhibited to a similar extent by increasing concentrations of erlotinib in both mTOR WT or mutant cells, but phosphorylation of S6 was largely insensitive to erlotinib in mTOR mutant cells (Figure 6D). To determine the sensitivity to erlotinib treatment in vitro, cell viability (measured by AlamarBlue assay), survival and proliferation (measured by clonogenic assay), and caspase-3/7 activity (measured by luminescent assay) were examined. Cells expressing mTOR E2419K formed larger and more numerous colonies and showed less caspase 3/7 activity (Figure 6E and F), whereas no differences were observed in cell viability (Supplemental Figure 2), suggesting that the mTOR E2419K mutation enhances clonogenicity and survival of EGFR mutant lung cancer cells in the presence of erlotinib rather than proliferation, cellular effects consistent with activation of the PI3K/AKT/MTOR signaling pathway rather than the MAPK pathway..
Next, we generated xenograft models of the isogenic PC9-mTOR mutant cell lines to assess response to erlotinib treatment in vivo. PC9-mTOR E2419K xenografts exhibited faster growth than PC9-mTOR WT xenografts and tumor growth of PC9-mTOR WT xenografts was almost completely inhibited with erlotinib, whereas mTOR E2419K mutant xenografts continued to grow despite erlotinib exposure, suggesting that mTOR E2419K mediates resistance to erlotinib in vivo (Figure 6G). We then assessed whether acquired resistance to erlotinib mediated by mTOR activation could be targeted with combination mTOR and EGFR inhibition. In PC9-mTOR E2419K cells, neither treatment with erlotinib or AZD8055 alone fully abrogated EGFR downstream signaling, but combination treatment with both drugs fully inhibited downstream EGFR signaling (Figure 6H). The IC50 of PC9-mTOR lines for AZD8055 ranged from 26.9 nM to 69.7 nM, and those of PC9 cells with mTOR activating mutations were slightly higher compared to empty vector control or mTOR WT (Supplemental Figure 3). These differences might reflect increases in mTOR catalytic activity associated with activating mutations. In vivo, mice bearing PC9-mTOR E2419K xenografts were treated with either erlotinib alone (25 mg/kg), AZD8055 alone (20mg/kg), or combination treatment. mTOR E2419K xenografts had some growth retardation with single agent erlotinib or AZD8055, but only combination treatment induced tumor shrinkage (Figure 6I) without a significant reduction in mouse weight (Supplemental Figure 4). Taken together, these findings suggest that the acquired mTOR E2419K mutation confers resistance to erlotinib in EGFR mutant NSCLC, and that combination therapy with EGFR and mTOR inhibitors may reverse such resistance.
Discussion
In this analysis, we have characterized the landscape of concurrent genetic alterations in a series of patients with EGFR-mutant lung cancers. We identify pre-existent ERBB2 and MET amplification as well as TP53 alterations as determinants of shorter time on EGFR TKI. Through analysis of tumors collected after disease progression on EGFR TKI, including 38 paired samples, we identified a number of molecular alterations as potential mechanisms of acquired resistance. These data support the exploration of combinatorial approaches that can overcome established drug resistance or delay the emergence of drug resistant clones. Since our analyses were exploratory, we opted not to adjust for multiple testing, and our results require validation in future studies.
We found that co-existing TP53 mutations was by far the most common co-mutation in EGFR mutant lung cancers and associated with a markedly shorter time to progression on initial EGFR TKI as well as shorter overall survival. This frequency of TP53 mutations was somewhat higher than reported in lung adenocarcinomas in prior studies (ranging from 24–55%) [32–35], but the predictive and prognostic effects seen in our report are consistent with several smaller reports [12, 13, 36]. As we reported all TP53 alterations identified (missense, nonsense, frameshift), we may be over-estimating the frequency of pathogenic alterations but chose to do so since functional data to determine whether an alteration is pathogenic is not available for all alterations reported. Lung cancers with loss of TP53 and resultant impaired cell cycle control may be more sensitive to growth factor simulation leading to a survival advantage in preclinical models for lung cancers with concurrent TP53 and EGFR alterations. The high frequency of TP53 alterations and their association with inferior outcomes suggest that this subset of patients may benefit from combination treatment strategies as initial treatment in an attempt to improve survival and response to EGFR TKI. Interestingly, we found TP53 and RB1 alterations to be essentially clonal in the majority of paired samples, suggesting this truncal mutation occurred early in oncogenesis.
Concurrent ERBB2 (HER2) and MET amplification were less common (4% and 2%, respectively), but were also independent predictors of shorter time to progression on EGFR TKI, and may be particularly clinically actionable given the clinical availability of MET and HER2 inhibitors. These copy number alterations of ERBB2 and MET were also enriched in specimens obtained after EGFR TKI resistance. Concurrent activation of ERBB2 or MET, both representing parallel or “bypass” signaling pathways that share downstream effectors with EGFR such as the RAS/RAF/MEK and PI3K/AKT pathways, have been shown in laboratory studies to make tumor cells less dependent on EGFR for activation of these downstream effectors, thereby reducing the anti-tumor efficacy of EGFR TKIs [37, 38]. Such patients clearly represent a subset that may benefit from dual EGFR/HER2 or EGFR/MET combination treatment as initial therapy in order to fully inhibit downstream signaling and thus prevent or delay the emergence of drug resistance clones.
Transformation from lung adenocarcinoma to small cell lung cancer in patients with EGFR mutant lung cancer is an uncommon but clinically important event. As expected, in all patients with small cell transformation, there was loss of TP53 and RB1 [39, 40]. There were TP53 and RB1 mutations in the pre-treatment adenocarcinoma sample of the one patient with small cell transformation, consistent with earlier reports that RB1 and TP53 inactivation is an early event in patients with eventual small cell transformation[40]. As we further identified RB1 loss in 11% of patient samples prior to EGFR TKI therapy and concurrent RB1 and TP53 loss in 9% (n=18) of baseline tumor samples, these results suggest that co-mutations in these genes are likely required but insufficient to induce a small cell phenotype. This may be a subset of patients where upfront treatment strategies that address a potential pre-existing small cell clone and/or a predisposition to small cell transformation could be important.
In the tumor samples collected after progression on EGFR TKI, EGFR T790M was frequently identified. All of the major previously reported mechanisms of EGFR TKI resistance were seen in our cohort including small cell transformation, MET and ERBB2 amplification, and BRAF and PIK3CA mutations. When comparing the post-treatment samples to the unmatched pre-treatment samples, EGFR T790M, BRAF alterations, CDKN2A/B alterations, FGFR3 alterations and EGFR and MET amplification were all enriched in the post-treatment samples suggesting they can contribute to acquired resistance to EGFR TKI therapy. In addition to the known resistance alterations, CDKN2A loss has been associated with under expression of DUSP4 which plays a central role in negative feedback regulation of EGFR signaling [41] providing a biologic rationale for its role in acquired resistance. FGFR3 alterations have also been implicated in mediating EGFR TKI resistance, potentially though epithelial to mesenchymal transition [42–44].
Thirty-eight patients had paired tumor samples collected before and after treatment with an EGFR TKI. In addition to the established mechanisms of resistance to EGFR TKI, several novel putative mechanisms of drug resistance were observed in the samples obtained upon disease progression including an AGK-BRAF fusion. BRAF fusions have been reported as oncogenic drivers in multiple solid tumors with case reports demonstrating responses to MEK inhibition [45]. BRAF fusions were recently found to be enriched in patients with concurrent EGFR mutations [46], and this is the first report of an acquired BRAF fusion as a candidate mechanism of resistance to EGFR TKI therapy. Putative loss-of-function KEAP1 alterations are frequently seen in non-small cell lung cancer [47] and pre-clinical data implicate loss of KEAP1 and resultant increased NRF2 expression as mechanisms of resistance to RTK/MAPK pathway inhibition [48]. This is the first report of a patient with an acquired KEAP1 mutation in the setting of treatment with an EGFR TKI. SFK/FAK (Src family kinases and focal adhesion kinase) signaling can sustain downstream AKT and MAPK signaling in the setting of EGFR inhibition, and the SFK family member YES1 was amplified in experimentally derived osimertinib-resistant EGFR-mutant cells [49], and separately, we have also identified YES1 amplification in a transposon mutagenesis screen as a potential mechanism of resistance to afatinib [50]. Here, for the first time, we identified an acquired YES1 amplification in a patient after treatment with erlotinib, and this has potential therapeutic relevance as SFK inhibitors such as dasatinib could be utilized in this clinical situation. FGFR fusions have been reported in non-small cell lung cancers [51] and there is converging evidence that FGFR activation may induce resistance to EGFR inhibitors [42–44, 52]. This is the first clinical report of an acquired FGFR3-TACC3 fusion after treatment with an EGFR TKI.
We demonstrate that mTOR E2419K mutation can drive resistance to EGFR TKI treatment, much like mutations of other downstream RTK effectors. [25, 53–55]. This is the first report of an acquired mTOR E2419K in a patient with EGFR-mutant lung cancer after treatment with erlotinib. We found that c ombination EGFR and mTOR inhibition controls tumor growth in vivo in EGFR TKI-resistant, mTOR E2419K mutant NSCLC. Coincidentally, this point mutation was reported in an urothelial carcinoma patient who achieved a complete response to everolimus and pazopanib therapy demonstrating that mTOR E2419K may be clinically actionable [56].
Tumor mutation burden is emerging as a predictive biomarker of response to immunotherapy [21]. Most EGFR-mutant lung cancers are largely resistant to immune checkpoint inhibitors with only limited responses to immmunotherapies seen in this subset of patients [23]. Nonetheless, there is continued interest in identifying subsets of EGFR-mutant lung cancer patients more likely to respond to immunotherapy, and also to identify the optimal timing of immunotherapy in this population. Notably, we observed a small, but significant increase in tumor mutation burden after treatment with EGFR TKIs. Available treatment algorithms do not currently incorporate temporal changes in tumor mutation burden to guide the timing of potential immunotherapy treatment. In a population that is largely resistant to immunotherapy [23], such temporal changes in tumor mutation burden may suggest that immunotherapy is better attempted after TKI therapy. However, there are also substantial preclinical data to suggest that targeted therapies can modulate immune responses by affecting T cell priming, activation, and differentiation and the tumor microenvironment[57], such that there may be multiple factors at play, and thus more data are needed to determine the optimal sequence of targeted therapies and immunotherapies in patients with EGFR mutant lung cancers.
In summary, our analysis further defines the genomic diversity present among lung cancers with an established oncogenic driver and makes clearer the impact of concurrent alterations on patient outcomes. A recent paper has reported on genetic alterations seen in cell-free DNA (cfDNA) from a large cohort of patients with EGFR-mutant lung cancer and identified alterations in CDK4/6, CTNNB1 and PIK3CA as relevant to EGFR TKI response[35]. As we learn more about the prognostic and predictive significance of concurrent genetic alterations, we may be able to use this information to improve outcomes for our patients.
Concurrent alterations that portend poorer outcomes in patients with EGFR-mutant lung cancers could be used in future studies to guide the selection of combination treatment strategies implemented at treatment onset for certain higher-risk patients. We need to continue to collect data regarding concurrent alterations from both tumor tissue and tumor cfDNA to better clarify predictive and prognostic markers of response as well as to highlight relevant acquired alterations that emerge as mechanisms of resistance to EGFR TKIs. Moving forward, the use of next-generation sequencing to molecularly profile tumors will, and should, become increasingly routine and we will need to develop strategies to nimbly assess and initiate appropriate combination therapies based on an individual patient’s sequencing results.
Supplementary Material
Statement of translational relevance.
There is heterogeneity in the clinical course of patients with EGFR-mutant lung cancers that may be attributable to concurrent molecular alterations. Concurrent HER2 amplification, MET amplification and TP53 mutations in pre-treatment tumor samples were all associated with shorter time to progression on EGFR TKI and concurrent TP53 mutations were also associated with shorter overall survival. Identification of these concurrent mutations may help us tailor the treatment of these patients by utilizing new strategies upfront to overcome primary resistance. In addition, several acquired mutations were identified at the time of clinical progression that may mediate resistance to EGFR TKIs including a BRAF fusion, FGFR3 fusion, YES1 amplification, KEAP1 loss, and an MTOR E2419K mutation that represent potential new therapeutic targets. Comprehensive genomic profiling allows us to understand clinical heterogeneity within EGFR-mutant lung cancers and may allow us to personalize targeted therapy for individual patients.
Acknowledgements:
Supported, in part, by NIH P01 CA129243 (M. Kris/M. Ladanyi), and NIH P30 CA008748 (MSKCC).
Footnotes
Conflict of Interests: Dr. Helena Yu has consulted for AstraZeneca and Lilly. Dr. Mark Kris has consulted for AstraZeneca. Dr. Bob Li has consulted for Genentech, ThermoFisher Scientific, and Guardant Health. All other authors do not report any relevant conflicts of interest.
Presented in part at the Annual Meeting of the American Society of Clinical Oncology, Chicago, IL, June 3–7, 2016.
References:
- 1.Sequist LV, et al. , Phase III Study of Afatinib or Cisplatin Plus Pemetrexed in Patients With Metastatic Lung Adenocarcinoma With EGFR Mutations. J Clin Oncol, 2013. [DOI] [PubMed] [Google Scholar]
- 2.Rosell R, et al. , Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol, 2012. 13(3): p. 239–46. [DOI] [PubMed] [Google Scholar]
- 3.Kosaka T, et al. , Response Heterogeneity of EGFR and HER2 Exon 20 Insertions to Covalent EGFR and HER2 Inhibitors. Cancer Res, 2017. 77(10): p. 2712–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naidoo J, et al. , Epidermal growth factor receptor exon 20 insertions in advanced lung adenocarcinomas: Clinical outcomes and response to erlotinib. Cancer, 2015. 121(18): p. 3212–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi Y, et al. , EGFR Exon 18 Mutations in Lung Cancer: Molecular Predictors of Augmented Sensitivity to Afatinib or Neratinib as Compared with First- or Third-Generation TKIs. Clin Cancer Res, 2015. 21(23): p. 5305–13. [DOI] [PubMed] [Google Scholar]
- 6.Huang CL, et al. , Mutations of p53 and K-ras genes as prognostic factors for non-small cell lung cancer. Int J Oncol, 1998. 12(3): p. 553–63. [DOI] [PubMed] [Google Scholar]
- 7.Fukuyama Y, et al. , K-ras and p53 mutations are an independent unfavourable prognostic indicator in patients with non-small-cell lung cancer. Br J Cancer, 1997. 75(8): p. 1125–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koivunen JP, et al. , Mutations in the LKB1 tumour suppressor are frequently detected in tumours from Caucasian but not Asian lung cancer patients. Br J Cancer, 2008. 99(2): p. 245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji H, et al. , LKB1 modulates lung cancer differentiation and metastasis. Nature, 2007. 448(7155): p. 807–10. [DOI] [PubMed] [Google Scholar]
- 10.Chen Z, et al. , A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature, 2012. 483(7391): p. 613–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahoney CL, et al. , LKB1/KRAS mutant lung cancers constitute a genetic subset of NSCLC with increased sensitivity to MAPK and mTOR signalling inhibition. Br J Cancer, 2009. 100(2): p. 370–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Canale M, et al. , Impact of TP53 Mutations on Outcome in EGFR-Mutated Patients Treated with First-Line Tyrosine Kinase Inhibitors. Clin Cancer Res, 2017. 23(9): p. 2195–2202. [DOI] [PubMed] [Google Scholar]
- 13.VanderLaan PA, et al. , Mutations in TP53, PIK3CA, PTEN and other genes in EGFR mutated lung cancers: Correlation with clinical outcomes. Lung Cancer, 2017. 106: p. 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faber AC, et al. , BIM expression in treatment-naive cancers predicts responsiveness to kinase inhibitors. Cancer Discov, 2011. 1(4): p. 352–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costa DB, et al. , BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med, 2007. 4(10): p. 1669–79; discussion 1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong Y, et al. , Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med, 2007. 4(10): p. e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng KP, et al. , A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med, 2012. 18(4): p. 521–8. [DOI] [PubMed] [Google Scholar]
- 18.Yu HA, et al. , Poor response to erlotinib in patients with tumors containing baseline EGFR T790M mutations found by routine clinical molecular testing. Ann Oncol, 2014. 25(2): p. 423–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su KY, et al. , Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J Clin Oncol, 2012. 30(4): p. 433–40. [DOI] [PubMed] [Google Scholar]
- 20.Rosell R, et al. , Pretreatment EGFR T790M mutation and BRCA1 mRNA expression in erlotinib-treated advanced non-small-cell lung cancer patients with EGFR mutations. Clin Cancer Res, 2011. 17(5): p. 1160–8. [DOI] [PubMed] [Google Scholar]
- 21.Rizvi NA, et al. , Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science, 2015. 348(6230): p. 124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel SP and Kurzrock R, PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol Cancer Ther, 2015. 14(4): p. 847–56. [DOI] [PubMed] [Google Scholar]
- 23.Gainor JF, et al. , EGFR Mutations and ALK Rearrangements Are Associated with Low Response Rates to PD-1 Pathway Blockade in Non-Small Cell Lung Cancer: A Retrospective Analysis. Clin Cancer Res, 2016. 22(18): p. 4585–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu HA, et al. , Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res, 2013. 19(8): p. 2240–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sequist LV, et al. , Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med, 2011. 3(75): p. 75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng DT, et al. , Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn, 2015. 17(3): p. 251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zehir A, et al. , Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med, 2017. 23(6): p. 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenman CD, et al. , Estimation of rearrangement phylogeny for cancer genomes. Genome Res, 2012. 22(2): p. 346–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jordan EJ, et al. , Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov, 2017. 7(6): p. 596–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J, et al. , Mechanistically distinct cancer-associated mTOR activation clusters predict sensitivity to rapamycin. J Clin Invest, 2016. 126(9):3526–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grabiner BC, et al. , A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov, 2014. 4(5): p. 554–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma X, et al. , Prognostic and Predictive Effect of TP53 Mutations in Patients with Non-Small Cell Lung Cancer from Adjuvant Cisplatin-Based Therapy Randomized Trials: A LACE-Bio Pooled Analysis. J Thorac Oncol, 2016. 11(6): p. 850–61. [DOI] [PubMed] [Google Scholar]
- 33.Chiba I, et al. , Mutations in the p53 gene are frequent in primary, resected non-small cell lung cancer. Lung Cancer Study Group. Oncogene, 1990. 5(10): p. 1603–10. [PubMed] [Google Scholar]
- 34.Schwaederle M, et al. , VEGF-A Expression Correlates with TP53 Mutations in Non-Small Cell Lung Cancer: Implications for Antiangiogenesis Therapy. Cancer Res, 2015. 75(7): p. 1187–90. [DOI] [PubMed] [Google Scholar]
- 35.Blakely CM, et al. , Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet, 2017. 49(12): p. 1693–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim SM, et al. , Targeted sequencing identifies genetic alterations that confer primary resistance to EGFR tyrosine kinase inhibitor (Korean Lung Cancer Consortium). Oncotarget, 2016. 7(24): p. 36311–36320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Engelman JA, et al. , MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science, 2007. 316(5827): p. 1039–43. [DOI] [PubMed] [Google Scholar]
- 38.Takezawa K, et al. , HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR mutant lung cancers that lack the second-site EGFR T790M mutation. Cancer Discov, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peifer M, et al. , Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet, 2012. 44(10): p. 1104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JK, et al. , Clonal History and Genetic Predictors of Transformation Into Small-Cell Carcinomas From Lung Adenocarcinomas. J Clin Oncol, 2017. 35(26): p. 3065–3074. [DOI] [PubMed] [Google Scholar]
- 41.Chitale D, et al. , An integrated genomic analysis of lung cancer reveals loss of DUSP4 in EGFR-mutant tumors. Oncogene, 2009. 28(31): p. 2773–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ware KE, et al. , Rapidly acquired resistance to EGFR tyrosine kinase inhibitors in NSCLC cell lines through de-repression of FGFR2 and FGFR3 expression. PLoS One, 2010. 5(11): p. e14117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomson S, et al. , Kinase switching in mesenchymal-like non-small cell lung cancer lines contributes to EGFR inhibitor resistance through pathway redundancy. Clin Exp Metastasis, 2008. 25(8): p. 843–54. [DOI] [PubMed] [Google Scholar]
- 44.Azuma K, et al. , FGFR1 activation is an escape mechanism in human lung cancer cells resistant to afatinib, a pan-EGFR family kinase inhibitor. Oncotarget, 2014. 5(15): p. 5908–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ross JS, et al. , The distribution of BRAF gene fusions in solid tumors and response to targeted therapy. Int J Cancer, 2016. 138(4): p. 881–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reddy VP GL, Elvin JA, Vergilio J, Suh J, Ramkissoon S, et al. BRAF fusions in clinically advanced non-small cell lung cancer: An emerging target for anti-BRAF therapies. in ASCO. 2017. Chicago, Illinois. [Google Scholar]
- 47.Singh A, et al. , Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med, 2006. 3(10): p. e420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krall EB, et al. , KEAP1 loss modulates sensitivity to kinase targeted therapy in lung cancer. Elife, 2017. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ichihara E, et al. , SFK/FAK Signaling Attenuates Osimertinib Efficacy in Both Drug-Sensitive and Drug-Resistant Models of EGFR-Mutant Lung Cancer. Cancer Res, 2017. 77(11): p. 2990–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan PD NG, Jayaprakash A, Venturini E, Robine N, Smibert P, et al. YES1 amplification as a mechanism of acquired resistance (AR) to EGFR tyrosine kinase inhibitors (TKIs) identified by a transposon mutagenesis screen and clinical genomic testing.. in ASCO. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang R, et al. , FGFR1/3 tyrosine kinase fusions define a unique molecular subtype of non-small cell lung cancer. Clin Cancer Res, 2014. 20(15): p. 4107–14. [DOI] [PubMed] [Google Scholar]
- 52.Ware KE, et al. , A mechanism of resistance to gefitinib mediated by cellular reprogramming and the acquisition of an FGF2-FGFR1 autocrine growth loop. Oncogenesis, 2013. 2: p. e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Misale S, et al. , Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature, 2012. 486(7404): p. 532–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohashi K, et al. , Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci U S A, 2012. 109(31): p. E2127–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Engelman JA, et al. , Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest, 2006. 116(10): p. 2695–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wagle N, et al. , Activating mTOR mutations in a patient with an extraordinary response on a phase I trial of everolimus and pazopanib. Cancer Discov, 2014. 4(5): p. 546–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vanneman M and Dranoff G, Combining immunotherapy and targeted therapies in cancer treatment. Nat Rev Cancer, 2012. 12(4): p. 237–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.