Abstract
Alternative polyadenylation (APA) produces from the same gene multiple mature RNAs with varying 3’ ends. Although APA is commonly believed to generate beneficial functional diversity and be adaptive, we hypothesize that most genes have one optimal polyadenylation site and that APA is caused largely by deleterious polyadenylation errors. The error hypothesis, but not the adaptive hypothesis, predicts that, as the expression level of a gene rises, its polyadenylation diversity declines, relative use of the major (presumably optimal) polyadenylation site increases, and that of each minor (presumably nonoptimal) site decreases. It further predicts that the number of polyadenylation signals per gene is smaller than the random expectation and that polyadenylation signals for major but not minor sites are under purifying selection. All of these predictions are confirmed in mammals, suggesting that numerous defective RNAs are produced in normal cells, many phenotypic variations at the molecular level are nonadaptive, and that cellular life is noisier than is appreciated.
Keywords: molecular error, expression level, natural selection, nonadaptive, polyadenylation signal, posttranscriptional modification
INTRODUCTION
Upon transcription, a typical eukaryotic messenger RNA (mRNA) undergoes polyadenylation in the nucleus, which is a two-step process consisting of an endonucleolytic cleavage followed by the addition of a poly(A) tail (Edmonds, 2002; Zhao et al., 1999). This process is orchestrated by a complex enzymatic system of up to 85 proteins that recognizes a polyadenylation signal (PAS), binds to downstream sequence elements, and eventually achieves the cleavage and polyadenylation (Edmonds, 2002; Shi et al., 2009; Tian and Graber, 2012; Zheng and Tian, 2014). The added poly(A) tail plays important roles in mRNA nuclear export, stability, and translation (Edmonds, 2002).
Polyadenylation may occur at one of several sites in an mRNA molecule, a phenomenon known as alternative polyadenylation (APA) (Shen et al., 2008; Tian et al., 2005). Recent transcriptomic surveys revealed a high abundance of APA across multiple species (Derti et al., 2012; Graber et al., 2013; Jan et al., 2011; Li et al., 2012; Mangone et al., 2010; Wu et al., 2011). For instance, ~70% of human genes show APA and ~50% have three or more polyadenylation sites (Derti et al., 2012). APA allows the production from a single gene multiple mature mRNAs that differ in their 3’ ends, including the untranslated region (UTR) and sometimes even the coding region (Di Giammartino et al., 2011; Lutz, 2008). Variation in the coding sequence can alter protein functions and variation in the 3’ UTR may impact the subcellular localization (Jansen, 2001), stability (Barrett et al., 2012), and translation (de Moor et al., 2005) of an mRNA. Thus, APA can be functionally important. Indeed, the mouse immunoglobulin heavy constant mu (Ighm) gene expresses a secreted form using a proximal polyadenylation site and the membrane-bound form using a distal site (Peterson, 2007). The mouse transcription factor gene Bzw1 has three polyadenylation sites, allowing making mature mRNAs with different translation efficiencies (Yu et al., 2006). Furthermore, some APA choices vary among cell types, developmental stages, and physiological/pathological states (Elkon et al., 2012; Fu et al., 2011; Hoque et al., 2013; Ji et al., 2009; Lianoglou et al., 2013; Mayr and Bartel, 2009; Miura et al., 2013; Sandberg et al., 2008; Ulitsky et al., 2012). These observations led to the prevailing view that APA is a beneficial and widely used mechanism of post-transcriptional regulation (Mayr, 2016). For instance, it is often suggested that APA expands the transcriptome diversity such that one gene can encode several mature mRNAs with distinct functions or regulations that may be used in different tissues or at different times (Di Giammartino et al., 2011; Elkon et al., 2013; Mayr, 2016).
Nonetheless, recent genome-wide studies failed to detect a clear relationship between APA and mRNA stability, mRNA concentration, translational efficiency, or protein concentration at the global scale (Gruber et al., 2014; Spies et al., 2013). For example, the global 3’ UTR shortening caused by APA in proliferating T cells of humans and mice was found to have a limited effect on mRNA and protein concentrations (Gruber et al., 2014). Another study concluded that APA has surprisingly small impacts on the stability and translational efficiency of most mRNAs in mouse fibroblasts (Spies et al., 2013). It is possible that APA plays global regulatory roles that are currently undetected owing to the limited numbers of cell types, species, or aspects of regulation studied or methodological limitations. It is also possible that the existence of APA largely reflects molecular errors caused by imprecise polyadenylation rather than adaptation. Because all biochemical processes, including polyadenylation, are stochastic in nature, error is inevitable. While the error rate may have been reduced by natural selection, it may not be zero, either due to the limited power of natural selection (Lynch, 2011) or because reducing the error rate beyond a certain level could be more costly than the error itself. Analyzing high-throughput mRNA 3’ end sequencing data from multiple tissues of five mammals, we here offer congruent evidence supporting the latter hypothesis, which we refer to as the error hypothesis.
RESULTS
Polyadenylation diversity decreases with gene expression
In a given tissue at a given developmental stage, if a gene has one optimal polyadenylation site and APA results from imprecise polyadenylation, we would expect APA to be deleterious because it may (i) reduce the fraction of functional mRNA molecules, (ii) diminish the mean functionality of mRNA molecules, (iii) waste materials and energy in the synthesis of defective mRNAs and possibly defective proteins, (iv) waste energy and other resources in the degradation of defective mRNAs and possibly defective proteins, and/or (v) result in toxic mRNA or protein products. Given the polyadenylation error rate per mRNA molecule, the harms associated with (i) and (ii) are independent of the expression level of the gene concerned, while those originating from (iii) to (v) rise with the expression level. Thus, the total harm of APA in a gene is expected to increase with the expression level of the gene. Consequently, natural selection against APA should intensify and the resultant rate of APA should decrease, as gene expression increases. By contrast, no general trend is predicted if APA is beneficial and adaptive, because, under this hypothesis, the ideal APA rate of a gene depends on the specific function and regulation of the gene.
To distinguish between the error hypothesis and the adaptive hypothesis of APA, we analyzed polyadenylation sites in a total of 24 tissue samples from human, macaque, mouse, rat, and dog inferred by PolyA-seq, a strand-specific and quantitative method for high-throughput sequencing of 3’ ends of polyadenylated transcripts (Derti et al., 2012). We aligned the filtered polyadenylation sites in each tissue sample to protein-coding genes and then counted the PolyA-seq reads for each site. We used two indices to measure the polyadenylation diversity for each protein-coding gene in each tissue sample. The first is Simpson’s index of diversity (Simpson, 1949), which is commonly used in ecology to measure the probability that two randomly picked individuals from a sample belong to different species. The second is Shannon diversity index (Shannon, 1948), which was developed in information science and is popularly applied to biodiversity research. Both Simpson and Shannon indices take into account the number of polyadenylation sites present in a gene as well as the relative uses of different sites (see STAR Methods).
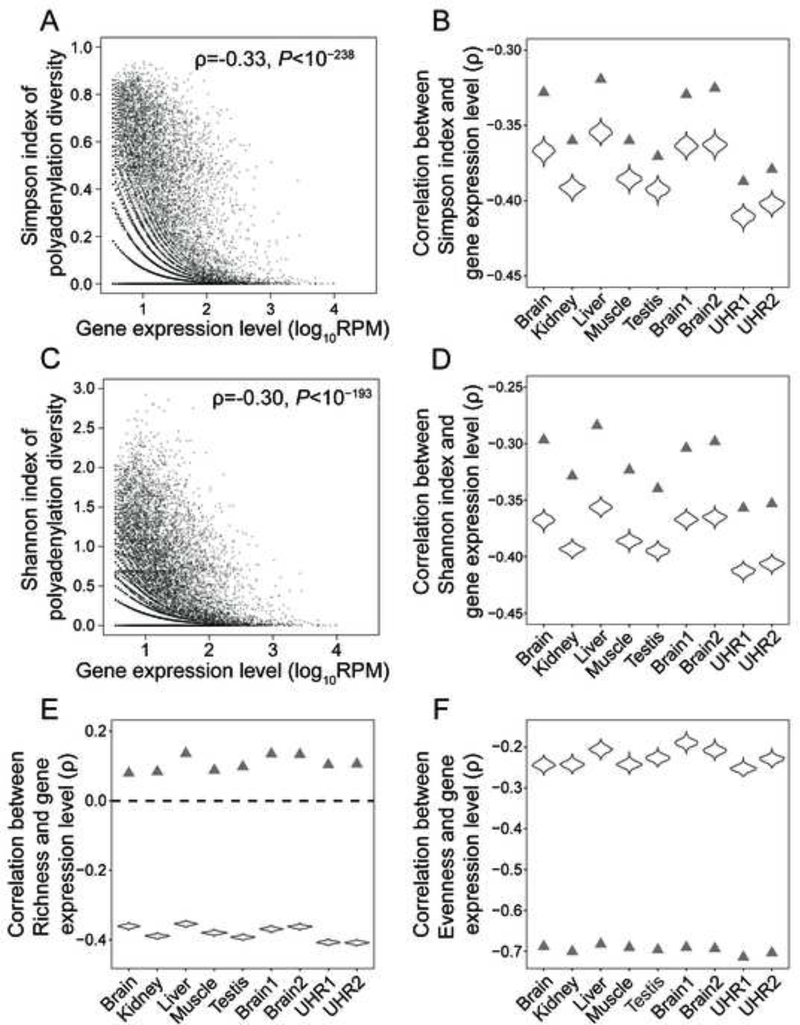
Using the human brain as an example, we first studied the relationship between the Simpson index of polyadenylation diversity for a gene and the expression level of the gene, which is measured by the number of PolyA-seq reads mapped to the gene per million reads mapped in the entire sample (RPM). Because of poor estimation of polyadenylation diversity when the number of reads mapped to a gene is too small, we restricted the analysis to genes with at least 10 reads in the tissue concerned. Consistent with the prediction of the error hypothesis, the rank correlation (ρ) between the expression level of a gene and its Simpson index of polyadenylation diversity is significantly negative, and this negative correlation is apparent across the entire expression range (Figure 1A). This trend remains when only polyadenylation sites downstream of the last stop codon of all annotated transcripts of each gene are considered (ρ = −0.13, P < 10−35) or when only those sites with relative usages ≥ 5% are considered (ρ = −0.40, P < 10−300). As a negative control, we simulated PolyA-seq reads under the assumption of no correlation between gene expression level and Simpson index, and indeed detected no correlation by our analysis. Because sequencing depth and the precision of APA survey for a gene rise with its expression level, it is important to confirm that the correlation in Figure 1A is not an artifact of unequal APA surveys of different genes. To this end, we down-sampled our data by randomly picking 10 PolyA-seq reads per gene for all genes with at least 10 reads and then re-estimated the Simpson index. The correlation (ρ’) between the gene expression level and re-estimated Simpson index becomes even more negative (Figure 1B). Because the down-sampling of reads is stochastic, we repeated this process 1000 times. The frequency distribution of ρ’ shows that the above result is robust to the stochasticity of read down-sampling (Figure 1B). Examining the other eight human tissue samples in the dataset reveals similar patterns (Figure 1B). The human tissues examined are brain, kidney, liver, muscle, testis, MAQC-Brain1, MAQC-Brain2, MAQC-UHR1, and MAQC-UHR2. The last four samples were from a MicroArray Quality Control (MAQC) study, where MAQC-Brain1 and MAQC-Brain2 are replicates of a human brain reference RNA from Ambion (Shi et al., 2006), whereas MAQC-UHR1 and MAQC-UHR2 are replicates of a universal human reference RNA from Stratagene. We further confirmed our results using gene expression levels measured by an independent mRNA-sequencing experiment (see STAR Methods).
Figure 1. Polyadenylation diversity declines as gene expression increases in humans.
(A) Relationship between the expression level of a gene in human brain and its Simpson index of polyadenylation diversity. In (A) and (C), each dot represents one gene. Spearman’s rank correlation coefficient (ρ) and associated P-values are presented. RPM, number of PolyA-seq reads mapped to a given gene per million reads mapped to all genes in the sample.
(B) Spearman’s correlation between gene expression level and Simpson index in each of nine human tissue samples. In (B), (D), (E), and (F), triangles show the ρ on the basis of the original data, whereas the violin plots show the frequency distributions of ρ on the basis of 1000 down-sampled data in which 10 PolyA-seq reads are randomly sampled per gene. P < 10−37 for all correlations in all panels.
(C) Relationship between the expression level of a gene in human brain and its Shannon index of polyadenylation diversity.
(D) Spearman’s correlation between gene expression level and Shannon index in each of nine human tissue samples.
(E) Spearman’s correlation between gene expression level and polyadenylation site Richness in each of nine human tissue samples.
(F) Spearman’s correlation between gene expression level and polyadenylation site use Evenness in each of nine human tissue samples.
See Methods for the definitions of Simpson index, Shannon index, Richness, and Evenness, and main text for descriptions of tissue samples. See also Figures S1-S3.
Using the Shannon index to measure polyadenylation diversity similarly yielded a negative correlation between gene expression level and polyadenylation diversity, as shown in Figure 1C for the human brain and Figure 1D for all nine human tissue samples. Even stronger negative correlations were obtained upon down-sampling of PolyA-seq reads to 10 per gene (Figure 1D). To examine the robustness of our results from the down-sampled data, we down-sampled PolyA-seq reads to as few as 5 and as many as 80 reads per gene from genes with at least that many reads and found our results to remain qualitatively unchanged for both Simpson and Shannon indices (Figures S1A and S1B). Although using raw and down-sampled data yielded qualitatively similar results in all of the above analyses, results from down-sampled data are more reliable due to equal surveys of polyadenylation among genes.
A reduction in the polyadenylation diversity of a gene may be caused by a decrease in the number of polyadenylation sites (i.e., Richness), a decrease in the evenness of the relative usages of different polyadenylation sites (i.e., Evenness), or both (see STAR Methods). While a positive correlation between gene expression level and Richness is observed in each human tissue sample (Figure 1E), this trend could be an artifact of higher sequencing depths and thus deeper APA surveys of more highly expressed genes. Indeed, the correlation becomes significantly negative for each tissue sample after we down-sampled the data to 10 reads per gene (Figure 1E). Down-sampling 5, 20, 40, or 80 reads yielded similar results (Figure S1C). As mentioned, because results from down-sampled data are more reliable than those from raw data and because they are qualitatively different here, down-sampling is necessary for fairly comparing polyadenylation site Richness among genes. We found the correlation between Evenness and gene expression level to be significantly negative in both the original and down-sampled data for all tissue samples (Figures 1F and S1D). Thus, both the polyadenylation site Richness and usage Evenness decrease as gene expression increases.
All of the above analyses compared a heterogeneous set of genes that vary in many properties. To minimize the impacts of potential confounding factors, we repeated these analyses by comparing paralogous genes of different expression levels, because paralogous genes are generated by gene duplication and are similar in gene structure, DNA sequence, regulation, and function (Zhang, 2013). For a pair of paralogs to be included in our analysis, we required that the expression level of the relatively highly expressed paralog must at least double that of the relatively lowly expressed one to allow sufficient statistical power. Consistent with the results from all genes (Figures 1A-1D), there is a significant trend for Simpson and Shannon indices to be lower in the relatively highly expressed paralog than in the relatively lowly expressed one, and these observations generally hold after down-sampling PolyA-seq reads to 10 per gene (Figure S2). Thus, the trends in Figure 1 are not attributable to the potential variation in polyadenylation diversity among genes of different functions.
Employing the method used for comparing all human genes, we analyzed the other four mammals (macaque, mouse, rat, and dog) in the dataset and found the results to be highly similar to those in humans (Figure S3), supporting the error hypothesis of APA in a diverse set of mammals.
Relative uses of all polyadenylation sites except the major one decrease with gene expression level
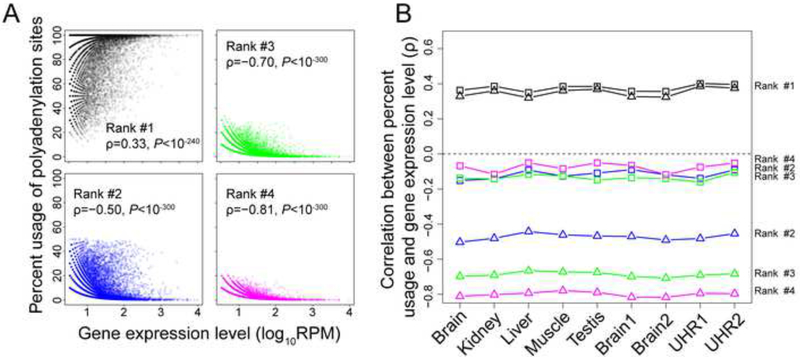
While the above observations support the hypothesis that a large fraction of APA in a tissue results from harmful molecular error, it does not tell us exactly how much. For example, in a gene with four polyadenylation sites, it is possible that the use of only one of the sites is optimal and desired in a given tissue, while the use of all other sites reflects error. It is also possible that the uses of two or even three of the four sites are desired. To address this question, we calculated and ranked the relative usages of all polyadenylation sites in each gene that has at least 10 PolyA-seq reads. The relative usage of a polyadenylation site is the number of PolyA-seq reads mapped to the site divided by the total number of PolyA-seq reads mapped to all polyadenylation sites of the gene. For a given gene, the polyadenylation site with the highest relative usage (i.e., ranked #1) will be referred to as the major site while all others will be referred to as minor sites. Given the importance of the poly(A) tail, at least one polyadenylation site should be functional and desired in a gene. Intuitively, this site should have the highest relative usage in most genes. Because natural selection against polyadenylation error intensifies with gene expression level, the relative usage of each desired polyadenylation site should increase while that of each undesired site should reduce as the gene expression level increases. We first tested this prediction in the human brain. Indeed, the relative usage of the major site in a gene increases with gene expression (upper-left plot in Figure 2A). Although this result dictates that the total use of all minor sites must decrease with gene expression, this trend does not have to apply to every minor site rank. Notwithstanding, each minor site rank examined has a reduced use as gene expression rises, suggesting that no rank of minor sites is desired. For example, among all genes with at least two polyadenylation sites, the relative usage of the second most frequently used site in a gene decreases with gene expression level (lower-left plot in Figure 2A). A similar negative correlation is observed for the third most frequently used sites among genes with at least three polyadenylation sites (upper-right plot in Figure 2A) and for the fourth most frequently used sites among genes with at least four polyadenylation sites (lower-right plot in Figure 2A). These trends remain unchanged when only polyadenylation sites downstream of the last stop codon of all annotated transcripts of each gene are considered (P < 10−187). We also observed a negative correlation when the analysis in Figure 2A is extended to the 5th, 6th, …, and 10th most frequently used polyadenylation sites among genes with at least 5, 6, …, and 10 polyadenylation sites, respectively. We confirmed the results presented in Figure 2A by down-sampling the original data to 10 PolyA-seq reads per gene and re-ranking polyadenylation sites using the down-sampled data (Figure 2B). The other human tissue samples show similar patterns (Figure 2B), which were also confirmed using gene expression levels measured by an independent mRNA-sequencing experiment (see STAR Methods). We further verified that the statistical trends in Figure 2 generally hold even when we limited the analysis to the common set of genes with at least four polyadenylation sites (Figure S4). Analysis of the other four mammals in our data yielded similar results (Figure S5). These observations strongly suggest that, for most genes in any tissue of any species surveyed, only the major polyadenylation site is desired while all other sites are undesired and reflect polyadenylation error.
Figure 2. Increased use of the most frequently used polyadenylation site of a gene and reduced uses of all other sites as gene expression level rises in humans.
(A) Spearman’s correlation between the expression level of a gene and the relative use of a polyadenylation site in the gene in human brain. Polyadenylation sites are ranked on the basis of their relative uses in the tissue concerned, with rank #1 being the most frequently used site. Each dot represents a gene.
(B) Spearman’s rank correlation between gene expression level and the relative use of each polyadenylation site in each of the nine human tissue samples examined. P < 0.02 in all cases. Triangles and squares indicate the correlations on the basis of the original data and down-sampled data, respectively. In both panels, the correlation for polyadenylation sites with a particular rank is calculated using the genes that have at least that particular number of polyadenylation sites. See also Figures S4-S6.
We also validated the above human results using paralogous genes, which should be more comparable as aforementioned. For the human brain, in 62% of the 490 pairs of paralogous genes analyzed, the major polyadenylation site is used more often in the relatively highly expressed paralog than in the relatively lowly expressed one, significantly more than the random expectation of 50% (P < 10−6, binomial test; Figure S6A). By contrast, for the second, third, and fourth most frequently used polyadenylation sites, respectively, 74%, 85%, and 91% of gene pairs show lower usages in the relatively highly expressed gene than in the relatively lowly expressed one (Figure S6A). Other tissues show similar patterns (Figure S6B). These trends generally hold in down-sampled data (Figure S6B).
Using proximal minor polyadenylation sites is more harmful than using distal minor sites
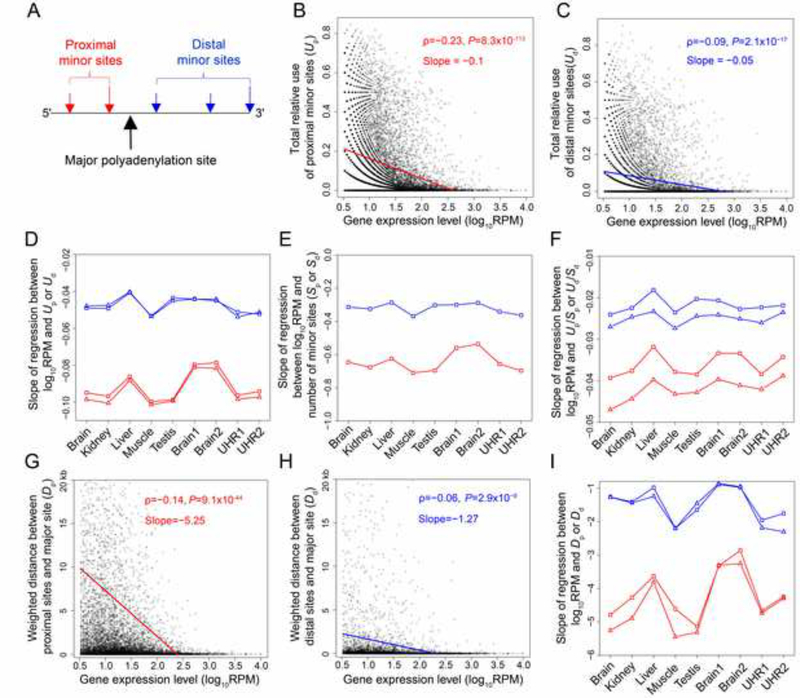
The 3’ UTR of a gene plays important roles in post-transcriptional regulations, and hence the 3’ UTR often contains regulatory sequences such as those that are bound by microRNAs or RNA-binding proteins (Mignone et al., 2002; Zhao et al., 1999). Because the major polyadenylation site of a gene is likely the optimal site, the regulatory sequences in the 3’ UTR should be located upstream of the major site. Therefore, the error hypothesis of APA predicts that using minor polyadenylation sites that are upstream of the major site (i.e., proximal to the stop codon) is more harmful than using minor sites that are downstream of the major site (i.e., distal to the stop codon), because the former is more likely than the latter to disrupt regulatory sequences in the 3’ UTR (Figure 3A). To verify this prediction, we respectively calculated the total relative use of all proximal minor sites (Up) and that of all distal minor sites (Ud) of a gene in a tissue. Using the human brain as an example, we found that both Up and Ud decrease with gene expression level, but the correlation between Up and gene expression level (Figure 3B) is much stronger than that between Ud and gene expression level (Figure 3C). Furthermore, the slope of the linear regression between Up and expression level (Figure 3B) is about twice that of the linear regression between Ud and expression level (Figure 3C) and their difference is statistically significant (P < 10−16). This pattern is consistently observed in the original and down-sampled data of all human tissue samples (Figure 3D).
Figure 3. Uses of proximal and distal minor polyadenylation sites in humans.
(A) Proximal and distal minor sites are defined on the basis of whether they are upstream or downstream of the major site.
(B) Total relative use of proximal sites in a gene decreases with the expression level of the gene. In (B) and (C), each dot represents one gene, and the solid line is the linear least-square regression.
(C) Total relative use of distal sites in a gene decreases with the expression level of the gene.
(D) The slope of the linear regression between the total relative use of proximal (red) or distal (blue) sites in a gene and the gene expression level in each of nine human tissue samples. In (D)-(F) and (I), triangles and squares denote results from the original and down-sampled data, respectively. Results from the original data are not presented in (E) due to the known bias caused by variable sequencing depths of different genes.
(E) The slope of the linear regression between the number of proximal (red) or distal (blue) sites in a gene and the gene expression level.
(F) The slope of the linear regression between the mean relative usage per proximal (red) or distal (blue) site in a gene and the gene expression level.
(G) Weighted distance between the proximal sites and major site (Dp) in a gene decreases with the expression level of the gene. In (G) and (H), each dot represents one gene. The solid line is the linear least-square regression. Only genes with weighted distance smaller than 20 kb are shown, but the correlations and regressions are based on all genes.
(H) Weighted distance between the distal sites and major site (Dd) in a gene decreases with the expression level of the gene.
(I) The slope of the linear regression between gene expression level and Dp (red) or Dd (blue) in each of nine human tissue samples.
We further examined the number of proximal minor polyadenylation sites (Sp) and that of distal minor sites (Sd). Because the number of polyadenylation sites observed is seriously influenced by sequencing depth, we analyzed the down-sampled data only. We found both Sp and Sd to decrease with gene expression level, but Sp decreases faster than Sd and this pattern is consistent among all human tissue samples examined (Figure 3E). We also investigated the mean relative usage per proximal site (Up/Sp) and that per distal site (Ud/Sd). Again, Up/Sp decreases faster than Ud/Sd as gene expression level increases (Figure 3F). Thus, both the number of proximal minor sites and the relative use of each proximal minor site decrease, relative to the corresponding values of distal minor sites, as gene expression level increases.
The harm of using a proximal minor site should decrease as the site gets closer to the major site, because the probability of disrupting regulatory sequences in 3’ UTR becomes smaller. Similarly, the harm of using a distal minor site should decrease as the site gets closer to the major site, because the probability of acquiring a deleterious regulatory sequence becomes lower. Therefore, the error hypothesis of APA predicts that, as the expression level of a gene rises, selection against erroneous polyadenylation intensifies and consequently both the weighted mean distance between the proximal minor sites and major site (Dp) and that between the distal minor sites and major site (Dd) decrease, where the weights are the relative usages of individual minor sites. Furthermore, it predicts that Dp decreases faster than Dd, because the probability of disrupting regulatory sequences per kb of 3’ UTR shortened is expected to exceed that of acquiring deleterious sequences per kb of 3’ UTR added. In the human brain data analyzed, we indeed observed that both Dp and Dd decrease with gene expression level (Figures 3G and 3H). Furthermore, the correlation between Dp and gene expression level is much stronger than that between Dd and expression level, and the slope of the linear regression between Dp and expression quadruples that of the linear regression between Dd and expression level (P < 10−16; Figures 3G and 3H). These patterns are consistently observed in the original and down-sampled data of all human tissue samples analyzed (Figure 3I). Taken together, our results from the analyses of the relative positions and uses of minor sites strongly support the error hypothesis of APA. Note that our results are not inconsistent with a previous report that short 3’ UTRs are more abundant in highly expressed genes than in lowly expressed genes (Ji et al., 2011), because we measured the positions of minor sites relative to the major site, while the previous study measured them relative to the most proximal site and because the position of the major site relative to the most proximal site differs among genes with different expression levels. In fact, we were able to replicate previously reported trends (Ji et al., 2011) using our data.
Across-tissue and among-species comparisons support the error hypothesis
APA varies across tissues (Hoffman et al., 2016; Lianoglou et al., 2013; Miura et al., 2013; Sandberg et al., 2008). Our analyses suggest that these variations are generally explainable by the error hypothesis and that only a small fraction of genes have different desired polyadenylation sites in different tissues (Figures S7 and S8; see STAR Methods). Recent genome-wide APA studies reported that APA varies among species (Derti et al., 2012; Wodniok et al., 2007). We found that these variations are consistent with predictions of the error hypothesis (Figures S9 and S10; see STAR Methods).
Natural selection on polyadenylation signals
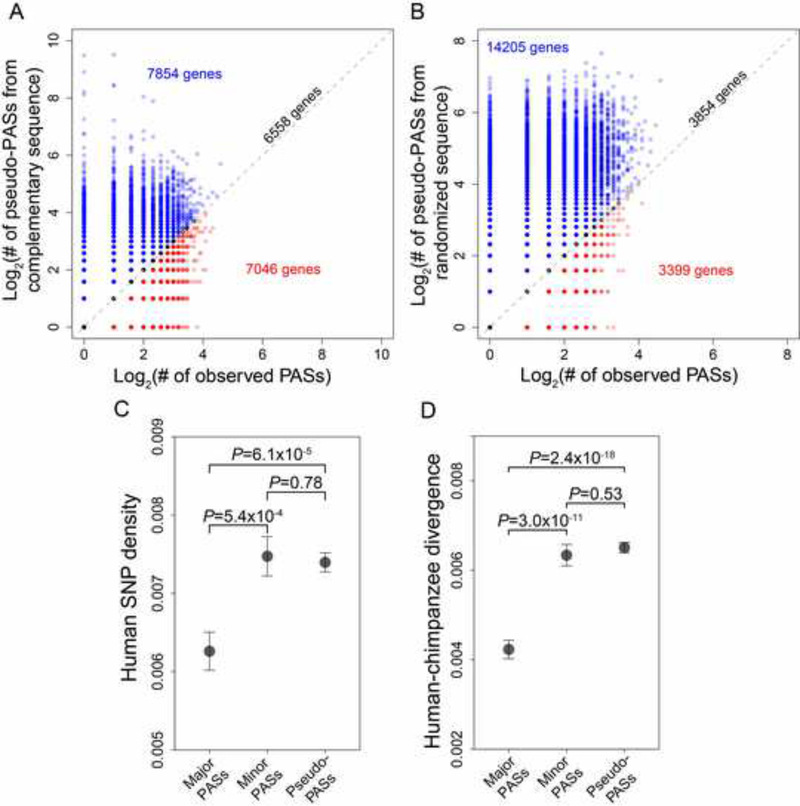
Polyadenylation signals (PASs) are sequence motifs recognized by the RNA cleavage complex as signals for polyadenylation (Beaudoing et al., 2000; Tian et al., 2005). In mammals, they are thought to be the AATAAA hexamer and 12 variants, typically located within 40 nucleotides upstream of the cleavage site (Lee et al., 2007). If APA is generally deleterious, the number of PASs per gene should be smaller than the random expectation under no selection. By contrast, if APA is adaptive, the opposite may be true. We used the number of pseudo-PASs as a proxy for the expected number of PASs under no selection, where pseudo-PASs are PAS hexamers identified from the complementary sequence of the region of mRNA where PASs are searched. We were able to test these predictions in humans, thanks to a recent study that computationally identified all PASs and pseudo-PASs in human genes (Kainov et al., 2016). The mean number of PASs per gene in the 21,458 human genes examined is 1.87, significantly lower than the mean number of pseudo-PASs per gene (3.85; P < 10−125, paired t-test). Furthermore, genes having fewer PASs than pseudo-PASs significantly outnumber genes having more PASs than pesudo-PASs (Figure 4A). These results strongly suggest that a substantial fraction of PASs have been removed by natural selection due to their deleterious effects, consistent with the error hypothesis of APA. The significant deficiency of PASs relative to the random expeactation is observed for each quartile of the data (P < 10−17) when genes are bined by expression level, demonstrating that the negative correlation between polyadenylation diversity and gene expression level (Figure 1) is not due to adaptive APA of weakly expressed genes. To exclude the possiblity that the pattern in Figure 4A is due to any potential strand bias in nucleotide composition, for each 3’ UTR, we further identified pseudo-PASs from a control 3’ UTR, which is a random sequence with the same length and nucletide composition as the real 3’ UTR. The mean number of pseudo-PASs per gene becomes 9.67, significantly exceeding the actual number of 1.87 (P < 10−324, pair t-test). The number of genes with fewer PASs than pseudo-PASs is 4.2 times the number of genes with more PASs than pseudo-PASs (Figure 4B).
Figure 4. Natural selection acting on human polyadenylation signals (PASs).
(A) Number of PASs and that of pseudo-PASs in the complementary 3’ UTR sequence in each gene. Each dot represents a gene. Dots above, on, and below the diagonal are colored in blue, black, and red, respectively, with their numbers indicated in the corresponding color. Blue dots significantly outnumber red dots (P < 10−10, binomial test).
(B) Number of PASs and that of pseudo-PASs in a randomized 3’ UTR sequence of each gene. Blue dots significantly outnumber red dots (P < 10−324).
(C) Single nucleotide polymorphism (SNP) density at PASs of global major polyadenylation sites, PASs of global minor sites, and pseudo-PASs (from complementary sequences).
(D) Number of substitutions per site between humans and chimpanzees at PASs of global major polyadenylation sites, PASs of global minor sites, and pseudo-PASs (from complementary sequences). Error bars show one standard error, and P-values are from Fisher’s exact test. See also Table S1.
Our finding that, for most human genes, only one polyadenylation site is desired per gene even when multiple tissues are considered predicts that different polyadenylation sites are under different selective constraints. The PAS corresponding to the desired site should be under purifying selection because sequence variation at this PAS is expected to be harmful. By contrast, the PASs corresponding to undesired sites should not be under purifying selection because sequence variation should be neutral or even beneficial if it removes a deleterious site. To test these predictions, we merged all PolyA-seq reads from the five human tissues and identified the global major polyadenylation site in each gene. We found that, compared with pseudo-PASs (in complementary sequences), which are presumably neutrally evolving, PASs for major polyadenylation sites have a significantly lower single nucleotide polymorphism (SNP) density in humans (Figure 4C) and a significantly lower divergence (number of substitutions per site) between humans and chimpanzees (Figure 4D). Notably, no significant difference is observed in either SNP density (Figure 4C) or divergence (Figure 4D) between pseudo-PASs and PASs for minor polyadenylation sites. To confirm that the above results are not simply due to any mutation rate difference between the three groups of sites, we computed the ratio of the numbers of substitutions and SNPs for each group of sites, which becomes independent of mutation rate. This ratio is significantly lower for PASs of major polyadenylation sites than for pseudo-PASs, but is not significantly different between PASs of minor polyadenylation sites and pseudo-PASs (Table S1). These results, showing purifying selection on PASs for major but not minor polyadenylation sites, support the error hypothesis of APA and contradict the adaptive hypothesis.
Note that the above analyses were based on the assumptions that each of the 13 hexamers considered can function as a PAS and that no other PAS motif exists. These assumptions may not be correct for all genes. Consequently, some of the computationally identified PASs may not be real while some real PASs may be missed. These errors add noise to the above analyses and reduce their statistical power. Therefore, our conclusions are most likely conservative. Nevertheless, because the same 13 hexamers were used in identifying PASs and pseudo-PASs, their comparison is unbiased.
DISCUSSION
Next-generation sequencing revealed huge polyadenylation diversities that include variations of polyadenylation among mRNA molecules of the same gene, among tissues or developmental stages for the same gene, and among species for orthologous genes. But the biological significance of these diversities has been elusive, despite the common belief that they are beneficial and adaptive (Di Giammartino et al., 2011; Elkon et al., 2013; Mayr, 2016). Prompted by the report of limited functional effects of APA in a few human and mouse cell types examined (Gruber et al., 2014; Spies et al., 2013), we proposed that APA largely reflects deleterious imprecise polyadenylation and tested this hypothesis by a series of comparative analysis of polyadenylation data in a total of 24 tissues samples from five mammals. We found strong and consistent evidence supporting the error hypothesis and refuting the adaptive hypothesis. That APA is generally harmful does not preclude its occasional use for adaptation, as has been found in some cases (Berkovits and Mayr, 2015; Di Giammartino et al., 2011; Elkon et al., 2013; Mayr, 2016). But the general pattern revealed in this study argues that APA should be considered slightly deleterious and nonadaptive unless proven otherwise.
If APA is generally harmful as our results strongly suggest, one cannot help but wonder why APA is still present and not removed completely by natural selection. The simple answer is that the most deleterious polyadenylation sites have been eliminated by natural selection; the deleterious effects of the remaining polyadenylation errors may be too small to be effectively removed by natural selection. That polyadenylation diversity is lower in highly expressed genes than in lowly expressed genes is consistent with this explanation, because natural selection against deleterious polyadenylation in a gene intensifies with the expression level of the gene. This explanation also solves the puzzle of why functional effects of most observed APA is experimentally undetectable. The efficacy of natural selection against deleterious mutations is higher in species with larger effective population sizes. Thus, we predict that, everything else being equal, polyadenylation error rate and polyadenylation diversity in a species should be negatively correlated with the effective population size of the species. This prediction is worth testing in the future when APA data become available from species with drastically different population sizes.
Some may wonder why polyadenylation cannot be more precise so that it does not make any noticeable error. The fact that a polyadenylation site is primarily (albeit not completely) determined by a PAS (Tian et al., 2005) means that, for polyadenylation to be precise, the PAS has to be sufficiently specific. Under no nucleotide frequency bias, the probability for a random hexamer to be a PAS is 13×0.256 = 0.0032 in mammals. Thus, approximately every 300 nucleotides contain a potential PAS. Transcription is hard to stop and often extends to the downstream gene (Proudfoot, 2016). Given the very long distance between the end of the coding region in a transcript and the end of the transcript (Proudfoot, 2016), many potential PASs are expected in a transcript, creating imprecise polyadenylation. Why cannot the sequence motif for PAS be longer so that PASs can be more specific? There are several non-mutually exclusive possibilities. First, there may be a mechanistic constraint that limits the length of an RNA sequence motif that can be accurately recognized by the polyadenylation complex. Second, it is possible that the cost for a more precise polyadenylation system is greater than the harm caused by imprecise polyadenylation. Third, it is possible that the selective pressure for a more precise polyadenylation system is simply not strong, especially because the cost of imprecise polyadenylation is lowered after the selective removal of many spurious PASs.
Polyadenylation is but one of a large array of post-transcriptional modifications, which also include 5’ capping, splicing, circularization (Salzman et al., 2012), and more than 100 different forms of nucleotide modifications such as pseudouridylation and N6-adenosine methylation (m6A) (Gilbert et al., 2016). The present finding on polyadenylation, along with the reports that adenosine-to-inosine (A-to-I) editing (Xu and Zhang, 2014) and cytidine-to-uridine (C-to-U) editing (Liu and Zhang, 2017a) of human coding RNAs are largely owing to imprecise targeting by promiscuous enzymes and are nonadaptive, that most m6A modifications in coding sequences are unconserved and likely nonfunctional (Liu and Zhang, 2017b), and that a sizable proportion of alternative splicing is due to splicing error (Saudemont et al., 2017), suggests the intriguing possibility that a large fraction of the reported post-transcriptional modification events are manifestations of molecular errors rather than adaptations. Future studies are required to test this hypothesis. Regardless, our findings, in conjunction with the other findings mentioned above, suggest that numerous defective RNAs are made in normal cells, highlighting the currently underappreciated fact that the cellular life is full of noise and far from an orderly and harmonious picture that is commonly portrayed.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jianzhi Zhang (jianzhi@umich.edu).
METHOD DETAILS
Polyadenylation sites
The PolyA-seq data used (Derti et al., 2012) were downloaded from NCBI Gene Expression Omnibus (GSE30198). The polyadenylation sites identified by the original authors through rigorous filtering (Derti et al., 2012) were used in our analysis. Briefly, Derti et al. aligned the raw PolyA-seq reads to the respective genome and retained only uniquely aligned reads. They then filtered internal priming events to acquire genuine polyadenylation sites. They reported relatively low rates of false positive error (internal priming events treated as genuine polyadenylation sites, 2.5%) and false negative error (genuine polyadenylation sites treated as internal priming events, 14.4%). More importantly, because these errors are random, they do not bias our analysis.
Considering the heterogeneity of the cleavage site directed by a given PAS, Derti et al. clustered all polyadenylation site peaks within 30 nucleotides on the same strand and summed read counts within each cluster; the highest peak within each cluster was retained to represent the polyadenylation site. When comparing different tissue samples from the same species, we similarly considered polyadenylation sites within 30 nucleotides as the same site. Briefly, we merged polyadenylation sites of different tissues and clustered cleavage positions that are within 30 nucleotides. If a cluster is wider than 30 nucleotides, we followed Hoque et al. (2013) to first identify the cleavage site with the greatest number of PolyA-seq reads and then re-cluster reads located >30 nucleotides from the position. This process was repeated until all sites in the cluster were defined.
Human (hg19) and mouse (mm9) genomic annotations were downloaded from UCSC (https://genome.ucsc.edu/), while macaque (rheMac2), rat (rn4), and dog (canFam2) genome annotations were obtained from Ensembl (http://useast.ensembl.org/index.html). We focused our analysis on protein-coding genes only. For each gene, the region between the 5’ most and 3’ most coordinates among all annotated transcripts was considered to belong to the gene. Our data contained 21,458 human genes, 21,023 macaque genes, 21,089 mouse genes, 22,937 rat genes, and 19,305 dog genes. Polyadenylation sites mapped to a gene or within 1 kb downstream of the most distal polyadenylation site previously annotated for the gene were considered to belong to the gene (Derti et al., 2012). Sites mapped to more than one gene were removed. When combining polyadenylation sites from multiple tissues, we regarded sites within 30 nucleotides on the same strand as the same site (Derti et al., 2012). In total, human, macaque, mouse, rat, and dog had 198,189, 77,216, 65,105, 66,167, and 74,149 polyadenylation sites, respectively.
Measures of polyadenylation diversity and gene expression level
The Simpson and Shannon indices of polyadenylation diversity for a gene are respectively defined by and , where S, also known as Richness, is the number of polyadenylation sites in the gene and pi is the proportion of RNA molecules of the gene that use the ith site. Evenness is a measure of the evenness of pi values and is defined by the Shannon index divided by the largest possible Shannon index given S.
Because PolyA-seq only sequences the 3’ end of an mRNA, the expression level of a gene is proportional to the number of PolyA-seq reads mapped to the gene (Derti et al., 2012). The expression level of a gene is measured by the total number of PolyA-seq reads mapped to the polyadenylation sites of the gene multiplied by 106 and then divided by the total number of reads mapped to all polyadenylation sites of all genes in the sample. This is referred to as reads per million reads mapped (RPM).
PolyA-seq is an established method for gene expression profiling (Derti et al., 2012; Fu et al., 2011; Hoque et al., 2013), and is as reliable as the more wildly used method of mRNA-sequencing (mRNA-seq) when benchmarked against the gold standard of quantitative reverse transcription polymerase chain reaction (qRT-PCR) for both expression ratios between tissues and absolute expression levels in one tissue (Derti et al., 2012). Nevertheless, to examine the robustness of our findings, we verified key results using mRNA-seq-based gene expression measures. Specifically, Bullard et al. (2010) used mRNA-seq to study gene expression levels (SRA: SRX016359 and SRX016367) in two of the samples we analyzed: MAQC-brain (corresponding to the two replicates labeled Brain1 and Brain2 in our Figures 1B and 2B) and MAQC-UHR (corresponding to the two replicates of UHR1 and UHR2 in our Figures 1B and 2B). For MAQC-brain, the correlation between the Simpson index of polyadenylation diversity and gene expression level measured by mRNA-seq is −0.28 (P = 1.4×10−184), while the corresponding value based on PolyA-seq is −0.33 (Figure 1B). The correlation between the percent usage of the rank #1 polyadenylation site and gene expression level measured by mRNA-seq is 0.27 (P = 2.5×10−177), while the corresponding value based on PolyA-seq is 0.33 (Figure 2B). For MAQC-UHR, the correlation between the Simpson index of polyadenylation diversity and gene expression level measured by PolyA-seq is −0.32 (P = 6.6×10−261), while the corresponding value based on PolyA-seq is −0.39 (Figure 1B). The correlation between the percent usage of rank #1 polyadenylation site and gene expression level measured by mRNA-seq is 0.32 (P = 9.2×10−253), while the corresponding value based on PolyA-seq is 0.38 (Figure 2B).
Down-sampling
To remove the potential influence of unequal sequencing depths of different genes in a sample on our analysis, we conducted down-sampling analyses. Briefly, we randomly picked the same number of PolyA-seq reads from all genes. Unless otherwise noted, we randomly picked 10 PolyA-seq reads per gene for all genes with at least 10 reads.
Major and minor polyadenylation sites
The major polyadenylation site of a gene in a tissue is the most frequently used polyadenylation site of the gene in the tissue, while all other polyadenylation sites of the gene are considered minor sites. A proximal minor site of a gene is a minor site that is upstream of the major site, while a distal minor site is a minor site that is downstream of the major site. For genes with no proximal or distal polyadenylation sites, the corresponding usage was set at 0.
Paralogs and orthologs
Paralogous genes in humans as well as one-to-one orthologous genes among human, macaque, mouse, rat, and dog were downloaded from Ensembl (release 75; Feb 2014) using BioMart (http://useast.ensembl.org/biomart/martview/). We obtained 4,423,592 human paralogous gene pairs that belong to 3594 families, of which 1625 families are mapped with at least two UCSC knownGene IDs that we used. When comparing between paralogs, from each family we randomly selected only one paralogous pair that has at least a two-fold expression difference. Because of the expression level variation among tissues, the included paralogous pairs in our analysis vary by tissue. The number of orthologs between human and the other four mammals is 16,694, 15,087, 14,735 and 15,563, respectively. From these data, we obtained 12,243 one-to-one orthologs among the five mammals. To identify orthologous polyadenylation sites between species, we used the UCSC liftOver tool (https://genome.ucsc.edu/util.html) to align polyadenylation sites of other mammals with the human genome. If a polyadenylation site of a non-human species is within 30 nucleotides of a human polyadenylation site and if they belong to orthologous genes, we regarded them as orthologous.
Across-tissue comparisons of APA
Under the error hypothesis, among-tissue variations in APA are due to the stochastic nature of polyadenylation errors. Thus, it predicts that the between-tissue difference in the relative uses of various polyadenylation sites of a gene decreases as the expression level of the gene rises, because the sensitivity of fitness to a change in the relative uses of polyadenylation sites increases with gene expression level. By contrast, no such prediction is made by the adaptive hypothesis, because the between-tissue difference in APA would depend on the specific tissues and genes. To verify the prediction of the error hypothesis, we measured the distance in the relative uses of polyadenylation sites of a gene between two tissues. Specifically, to measure the difference in polyadenylation for a gene between PolyA-seq samples A and B, we used a net correlational distance defined by dAB - 0.5dA - 0.5dB. Here dAB equals 1 minus Pearson’s correlation coefficient between samples A and B in the relative uses of all polyadenylation sites of the gene, dA is the same as dAB except that the two samples used are two PolyA-seq bootstrap samples derived from sample A, and dB is the same as dAB except that the two samples used are two PolyA-seq bootstrap samples derived from sample B. We considered the five human tissues (brain, kidney, liver, muscle, and testis) but excluded the four MAQC samples due to their overlaps with the five tissues. To ensure accuracy in measuring polyadenylation diversity in each tissue, we analyzed only those genes that have at least 10 PolyA-seq reads in each of the five tissues. We correlated the polyadenylation distance with the gene’s mean expression level in the two tissues compared. To avoid the influence by different sequencing depths of different genes, we also sampled 10 PolyA-seq reads per gene from each tissue for all genes that have at least 10 reads in each of the five human tissues. We found that in all 10 pairs of tissues compared, the correlation is significantly negative regardless of whether the original or down-sampled data are used (Figure S7A), supporting the error hypothesis. As a negative control, we generated two bootstrapped PolyA-seq datasets from either the original or down-sampled PolyA-seq data of one tissue and treated them as samples from a pseudo pair of tissues. Indeed, for such pseudo tissue pairs, no negative correlation was observed between the gene expression level and polyadenylation distance.
Because a given gene can have different expression levels in different tissues, the error hypothesis further predicts a negative correlation between its polyadenylation diversity and its expression level across tissues. For each gene, we calculated the correlation between its expression level and its Simpson or Shannon index of polyadenylation diversity across the five human tissues. Indeed, significantly more genes exhibit negative correlations than expected by chance regardless of the specific measure of polyadenylation diversity (Simpson or Shannon index) and data (original or down-sampled) used (Figures S7B and S7C). Furthermore, based on the down-sampled data, which are uninfluenced by different sequencing depths of different genes, for both polyadenylation site Richness (Figure S7D) and site use Evenness (Figure S7E), significantly more genes than expected by chance exhibit a negative correlation with expression level, indicating that the reduction in polyadenylation diversity in a tissue where the gene expression is elevated is achieved by reducing both the number of polyadenylation sites used and the evenness of the relative uses of these sites.
We then investigated the relative use of each polyadenylation site of a gene in the five human tissues. We defined the major and minor sites in each tissue separately, and it is possible that the major site in one tissue differs from that in another tissue. For each gene, we computed the across-tissue rank correlation between its expression level in a tissue and the relative use of the polyadenylation site of a certain rank in that tissue. For rank #1, 57% of genes show a positive correlation, significantly more than the random expectation of 50% (Figure S7F). By contrast, for each of ranks #2, #3, and #4, only 23–32% of genes show a positive correlation, significantly less than the random expectation (Figures S7G-S7I). These patterns are also evident in down-sampled data (Figures S7F-S7I). Together, the among-tissue variation in polyadenylation diversity and that in relative uses of polyadenylation sites support the hypothesis that, for most genes, only the use of the major polyadenylation site in a tissue is desired for that tissue and the uses of all minor sites in the tissue reflect errors.
Although the above analyses established that, for most genes, only one polyadenylation site is preferred per tissue, it remains possible that the preferred site is different in different tissues. Under this scenario, differential polyadenylation among tissues would be adaptive. To assess this hypothesis, we first repeated the analysis in Figures S7F-S7I by defining the global major and minor polyadenylation sites of each gene using the combined PolyA-seq reads from all five human tissues. For the global major site, 58% of genes show a positive correlation between the relative usage in a tissue and the expression level in the tissue, significantly more than the random expectation (Figure S8A). By contrast, for all global minor sites examined, only 41–43% of genes show a positive correlation, significantly less than the random expectation (Figures S8B-S8D). Comparing these results with those in Figures S7F-S7I suggests the possibility that only a small fraction of genes have different desired polyadenylation sites in different tissues.
To estimate this fraction, we first counted the number of different major polyadenylation sites observed in each gene across the five human tissues (N; Figure S8E), because if all five tissues share the same major site, it is most likely that they all share the same preferred site. We found that, of 6936 genes examined, 5590 (or 80.6%) have the same major site in all tissues (i.e., N = 1). We also examined the maximum possible number of different major sites in the five tissues (M) for each gene, which would be the smaller of 5 and the total number of polyadenylation sites observed in the five tissues for the gene (Figure S8E). When M ≥ 2, 99.8% of genes show N < M, suggesting that tissue-specific optimization of polyadenylation is far less than what APA could potentially offer, consistent with the hypothesis that among-tissue variations in APA are largely nonadaptive.
Even when more than one major polyadenylation site is found for a gene, it is still possible that all tissues actually have the same preferred site and that the observation of different major sites in different tissues is due to sampling error caused by limited sequencing depth. To examine this possibility, for each gene, we randomly shuffled its PolyA-seq reads among the five tissues without altering the number of reads in each tissue. We then used the shuffled data to count the number of different major sites for the gene in the five tissues. We repeated this process 10,000 times and estimated the mean number of major sites for the gene in the shuffled data (n) (Figure S8F) and the fraction of times (f) when the number of major sites observed in the shuffled data equals to or exceeds that in the actual data. Here, f is an estimate of the one-tailed P-value in testing the null hypothesis that all tissues share the same major site. We converted the P-values to Q-values to control for multiple testing, and found that 181 genes have Q < 0.05 (red dots in Figure S8F). Thus, approximately 2.6% of genes examined may have different preferred APA sites in different tissues.
In the above, we assumed that, after the exclusion of sampling error, the major polyadenylation site of a gene in a tissue is the preferred polyadenylation site in the tissue. This may not always be the case, because using the preferred site more than any other site in every tissue for very gene could be difficult, due to the limited power of polyadenylation regulation. Thus, it is possible that, even after the exclusion of sampling error, the observed major site is still not the optimal site for some genes in some tissues. This kind of high usage of suboptimal sites should have a fitness cost that rises with the gene expression level. Consequently, such phenomenon should have been reduced by natural selection to lower occurrences in more highly expressed genes. However, this is not an easy-to-test hypothesis, because of the confounding factor of sequencing depth, which also depends on gene expression level. To this end, for each gene, we sampled 10 PolyA-seq reads from each tissue and re-estimated N and n as was done using the original data. We then divided all genes into two groups: those with N > n (regardless of statistical significance of this inequality), and the rest. The expression level is significantly lower for the former group of 884 genes than the latter group of 6052 genes (Figure S8G). This observation supports the idea that a substantial fraction of genes whose N exceeds n do not necessarily have different preferred sites in different tissues; rather, they cannot use the single preferred polyadenylation site in all tissues to the highest level. In other words, the fraction of genes with adaptive differential APA among tissues is likely lower than the above estimate of 2.6%.
Among-species comparisons of APA
Precise polyadenylation is under stronger selective pressures in highly expressed genes than in lowly expressed genes. Thus, under the neutral evolution hypothesis, inter-specific difference in relative usages of various polyadenylation sites of a gene should decrease as the expression level of the gene rises. No such prediction is made by the adaptive hypothesis, because, under the adaptive hypothesis, different genes should show different patterns according to the specific functions of the genes concerned. To distinguish between the two hypotheses, we measured the distance in the relative uses of polyadenylation sites of a gene in the same tissue between two species (see STAR Methods). Because only the brain and testis were surveyed in all five mammals in our dataset, we studied these two tissues separately. We first identified one-to-one orthologous genes among the five mammals. For each tissue, we correlated this distance with the gene’s mean expression level in a pair of species compared. In the analysis of a tissue (brain or testis), only genes with at least 10 PolyA-seq reads in the tissue of each of the five species were analyzed. To avoid the influence by different sequencing depths of different genes, we also sampled 10 PolyA-seq reads per gene from the tissue of each species. When the original brain data were used, the correlation was significantly negative for each of the 10 species pairs (Figure S9A). When the down-sampled brain data were used, nine of the 10 species pairs showed negative correlations and six of these negative correlations were significant, whereas the sole positive correlation was not significant (Figure S9A). These observations are consistent with the prediction of the neutral evolution hypothesis. Qualitatively identical results were obtained for the testis (Figure S10A).
Because a gene can have different expression levels in a given tissue of different species, the error hypothesis predicts a negative correlation between its polyadenylation diversity and its expression level across species. Indeed, more genes exhibit negative correlations than expected by chance regardless of whether the Simpson (Figures S9B and S10B) or Shannon (Figures S9C and S10C) index is used. Randomly sampling 10 reads per species for each gene does not qualitatively alter this finding (Figures S9B, S9C, S10B, and S10C). The reduction in polyadenylation diversity with gene expression level among species is attributable to decreases in polyadenylation site Richness (Figures S9D and S10D) and site use Evenness (Figures S9E and S10E).
We also conducted an among-species comparison of the relative uses of different polyadenylation sites in the brain (Figure S9) and testis (Figure S10), respectively. For most genes, the relative usage of the major polyadenylation site identified in a species increases with the expression level of the gene in the species (Figures S9F and S10F). But for minor sites, the opposite is true (Figures S9G, S9H, S10G, and S10H). Thus, between-species variations in APA are consistent with the error hypothesis and are generally nonadaptive.
Polyadenylation signals and single nucleotide polymorphisms
The PAS hexamers and pseudo-PASs in human genes, as well as human SNP densities, were acquired from Kainov et al. (2016). Briefly, Kainov et al. downloaded the PAS hexamers from PAS.db2 (Lee et al., 2007) and converted the genomic coordinates of PASs from hg17 to hg19 using liftOver from UCSC. Human SNP data were downloaded from Interim Phase 1 of the 1000 Genomes Project (Genomes Project et al., 2012). Nucleotide substitutions at PASs were based on a comparison between human (hg19) and chimpanzee (PanTro4) genomes through liftOver from UCSC.
Because PAS hexamers are generally located within the 40-nucleotide segment upstream of polyadenylation sites (Lee et al., 2007), we assigned PAS hexamers to corresponding polyadenylation sites observed in the five independent tissues (Derti et al., 2012). SNP density is the number of SNPs within the hexamers considered divided by the total length of the hexamers. Divergence is the number of nucleotide differences between humans and chimpanzees in the hexamers considered divided by the total homologous sites of all the hexamers considered.
DATA AND SOFTWARE AVAILABILITY
All data used have been published and all programs used are available upon request.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited Data | ||
| PolyA-seq data | Derti et al., 2012 | https://www.ncbi.nlm.nih.gov/geo/; GSE: GSE30198 |
| RNA-seq data | Bullard et al., 2010 | https://trace.ddbj.nig.ac.jp/DRASearch/; SRA: SRX016359 and SRX016367 |
| SNP and PASs | Kainov et al., 2016 | N/A |
| Software and Algorithms | ||
| R | The R Foundation | https://www.r-project.org/ |
| Perl | The Perl Foundation | https://www.perl.org/ |
| LiftOver | UCSC | https://genome.ucsc.edu/util.html |
| BioMart | ENSEMBL | http://useast.ensembl.org/biomart/martview/ |
ACKNOWLEDGEMENTS
We thank the authors of Kainov et al. (2016) for sharing the human PAS hexamer data, Tamir Tuller for sharing yeast transcription elongation speed data, and members of the Zhang lab for valuable comments. This work was supported in part by research grant R01GM120093 from the U.S. National Institutes of Health to J.Z. C.X. was supported by China Scholarship Council.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Barrett LW, Fletcher S, and Wilton SD (2012). Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell. Mol. Life Sci 69, 3613–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoing E, Freier S, Wyatt JR, Claverie JM, and Gautheret D (2000). Patterns of variant polyadenylation signal usage in human genes. Genome Res. 10, 1001–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovits BD, and Mayr C (2015). Alternative 3’ UTRs act as scaffolds to regulate membrane protein localization. Nature 522, 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard JH, Purdom E, Hansen KD, and Dudoit S (2010). Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moor CH, Meijer H, and Lissenden S (2005). Mechanisms of translational control by the 3’ UTR in development and differentiation. Semin. Cell Dev. Biol 16, 49–58. [DOI] [PubMed] [Google Scholar]
- Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, and Babak T (2012). A quantitative atlas of polyadenylation in five mammals. Genome Res. 22, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino DC, Nishida K, and Manley JL (2011). Mechanisms and consequences of alternative polyadenylation. Mol. Cell 43, 853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds M (2002). A history of poly A sequences: from formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol 71, 285–389. [DOI] [PubMed] [Google Scholar]
- Elkon R, Drost J, van Haaften G, Jenal M, Schrier M, Oude Vrielink JA, and Agami R (2012). E2F mediates enhanced alternative polyadenylation in proliferation. Genome biol. 13, R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Ugalde AP, and Agami R (2013). Alternative cleavage and polyadenylation: extent, regulation and function. Nat. Rev. Genet 14, 496–506. [DOI] [PubMed] [Google Scholar]
- Fu Y, Sun Y, Li Y, Li J, Rao X, Chen C, and Xu A (2011). Differential genome-wide profiling of tandem 3’ UTRs among human breast cancer and normal cells by highthroughput sequencing. Genome Res. 21, 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, and McVean GA (2012). An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert WV, Bell TA, and Schaening C (2016). Messenger RNA modifications: Form, distribution, and function. Science 352, 1408–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber JH, Nazeer FI, Yeh PC, Kuehner JN, Borikar S, Hoskinson D, and Moore CL (2013). DNA damage induces targeted, genome-wide variation of poly(A) sites in budding yeast. Genome Res. 23, 1690–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Martin G, Muller P, Schmidt A, Gruber AJ, Gumienny R, Mittal N, Jayachandran R, Pieters J, Keller W, et al. (2014). Global 3’ UTR shortening has a limited effect on protein abundance in proliferating T cells. Nat. Commun 5, 5465. [DOI] [PubMed] [Google Scholar]
- Hoffman Y, Bublik DR, A PU, Elkon R, Biniashvili T, Agami R, Oren M, and Pilpel Y. (2016). 3’UTR shortening potentiates microRNA-based repression of pro-differentiation genes in proliferating human cells. PLoS Genet. 12, e1005879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G, and Tian B (2013). Analysis of alternative cleavage and polyadenylation by 3’ region extraction and deep sequencing. Nat. Methods 10, 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Friedman RC, Ruby JG, and Bartel DP (2011). Formation, regulation and evolution of Caenorhabditis elegans 3’UTRs. Nature 469, 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RP (2001). mRNA localization: Message on the move. Nat. Rev. Mol. Cell Bio 2, 247256. [DOI] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, and Tian B (2009). Progressive lengthening of 3’ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc. Natl. Acad. Sci. USA 106, 7028–7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Luo W, Li W, Hoque M, Pan Z, Zhao Y, and Tian B (2011). Transcriptional activity regulates alternative cleavage and polyadenylation. Mol. Syst. Biol 7, 534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainov YA, Aushev VN, Naumenko SA, Tchevkina EM, and Bazykin GA (2016). Complex selection on human polyadenylation signals revealed by polymorphism and divergence data. Genome Biol. Evol 8, 1971–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Yeh I, Park JY, and Tian B (2007). PolyA_DB 2: mRNA polyadenylation sites in vertebrate genes. Nucleic Acids Res. 35, D165–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sun Y, Fu Y, Li M, Huang G, Zhang C, Liang J, Huang S, Shen G, Yuan S, et al. (2012). Dynamic landscape of tandem 3’ UTRs during zebrafish development. Genome Res. 22, 1899–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lianoglou S, Garg V, Yang JL, Leslie CS, and Mayr C (2013). Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 27, 2380–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, and Zhang J (2017a). Human C-to-U coding RNA editing is largely nonadaptive. Mol. Biol. Evol 35, 963–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, and Zhang J (2017b). Most m6A RNA modifications in protein-coding regions are evolutionarily unconserved and likely nonfunctional. Mol. Biol. Evol 35, 666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CS (2008). Alternative polyadenylation: a twist on mRNA 3’ end formation. ACS Chem Biol 3, 609–617. [DOI] [PubMed] [Google Scholar]
- Lynch M (2011). The lower bound to the evolution of mutation rates. Genome Biol. Evol 3, 1107–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, et al. (2010). The landscape of C. elegans 3’UTRs. Science 329, 432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C (2016). Evolution and biological roles of alternative 3’UTRs. Trends Cell Biol. 26, 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, and Bartel DP (2009). Widespread shortening of 3’UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138, 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignone F, Gissi C, Liuni S, and Pesole G (2002). Untranslated regions of mRNAs. Genome biol. 3, REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura P, Shenker S, Andreu-Agullo C, Westholm JO, and Lai EC (2013). Widespread and extensive lengthening of 3’ UTRs in the mammalian brain. Genome Res. 23, 812–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson ML (2007). Mechanisms controlling production of membrane and secreted immunoglobulin during B cell development. Immunol. Res 37, 33–46. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ (2016). Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science 352, aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Gawad C, Wang PL, Lacayo N, and Brown PO (2012). Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7, e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, and Burge CB (2008). Proliferating cells express mRNAs with shortened 3’ untranslated regions and fewer microRNA target sites. Science 320, 1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudemont B, Popa A, Parmley JL, Rocher V, Blugeon C, Necsulea A, Meyer E, and Duret L (2017). The fitness cost of mis-splicing is the main determinant of alternative splicing patterns. Genome biol. 18, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon CE (1948). A mathematical theory of communication. Bell Syst. Tech. J 27, 379–423 and 623–656. [Google Scholar]
- Shen Y, Ji G, Haas BJ, Wu X, Zheng J, Reese GJ, and Li QQ (2008). Genome level analysis of rice mRNA 3’-end processing signals and alternative polyadenylation. Nucleic Acids Res. 36, 3150–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, et al. (2006). The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 24, 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YS, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, Frank J, and Manley JL (2009). Molecular architecture of the human pre-mRNA 3 ‘ processing complex. Mol. Cell 33, 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson EH (1949). Measurement of diversity. Nature 163, 688. [Google Scholar]
- Spies N, Burge CB, and Bartel DP (2013). 3’ UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome Res. 23, 2078–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, and Graber JH (2012). Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip. Rev. RNA 3, 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, and Lutz CS (2005). A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 33, 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Subtelny AO, Koppstein D, Bell GW, Sive H, and Bartel DP (2012). Extensive alternative polyadenylation during zebrafish development. Genome Res. 22, 2054–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodniok S, Simon A, Glockner G, and Becker B (2007). Gain and loss of polyadenylation signals during evolution of green algae. BMC Evol. Biol 7, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Liu M, Downie B, Liang C, Ji G, Li QQ, and Hunt AG (2011). Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc. Natl. Acad. Sci. USA 108, 12533–12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, and Zhang J (2014). Human coding RNA editing is generally nonadaptive. Proc. Natl. Acad. Sci. USA 111, 3769–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Sha H, Gao Y, Zeng H, Zhu M, and Gao X (2006). Alternative 3’ UTR polyadenylation of Bzw1 transcripts display differential translation efficiency and tissue-specific expression. Biochem. Biophys. Res. Commun 345, 479–485. [DOI] [PubMed] [Google Scholar]
- Zhang J (2013). Gene duplication. In The Princeton Guide to Evolution, Losos J, ed. (Princeton, New Jersey: Princeton University Press; ), pp. 397–405. [Google Scholar]
- Zhao J, Hyman L, and Moore C (1999). Formation of mRNA 3’ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, and Tian B (2014). RNA-binding proteins in regulation of alternative cleavage and polyadenylation. Adv. Exp. Med. Biol 825, 97–127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used have been published and all programs used are available upon request.