Abstract
Purpose:
To design and evaluate a small engineered protein binder targeting human programmed death-1 ligand (hPD-L1) in vivo for PET imaging in four mouse tumor models, and in situ in human cancer specimens.
Experimental Design:
The hPD-L1 protein binder, FN3hPD-L1, was engineered using a 12 kDa human fibronectin type-3 domain (FN3) scaffold. The binder’s affinity was assayed in CT26 mouse colon carcinoma cells stably expressing hPD-L1 (CT26/hPD-L1). 64Cu-FN3hPD-L1 was assayed for purity, specific activity, and immunoreactivity. Four groups of NSG mice (n=3–5/group) were imaged with 64Cu-FN3hPD-L1 PET imaging (1–24 hours post-injection of 3.7 MBq/7 μg of Do-FN3 in 200 μL PBS): Nod SCID Gamma (NSG) mice bearing (1) syngeneic CT26/hPD-L1tumors, (2) CT26/hPD-L1 tumors blocked (blk) by pre-injected non-radioactive FN3hPD-L1 binder, (3) hPD-L1-negative Raji xenografts, and (4) MDA-MB-231 xenografts. The FN3hPD-L1 binder staining was evaluated against validated hPD-L1 antibodies by immunostaining in human cancer specimens.
Results:
FN3hPD-L1 bound hPD-L1 with 1.4±0.3 nM affinity in CT26/hPD-L1 cells. 64Cu-FN3hPD-L1 radiotracer showed >70% yield and >95% purity. 64Cu-FN3hPD-L1 PET imaging of mice bearing CT26/hPD-L1 tumors showed tumor-to-muscle ratios of 5.6±0.9 and 13.1±2.3 at 1 and 4 hours post-injection, respectively. The FN3hPD-L1 binder detected hPD-L1 expression in human tissues with known hPD-L1 expression status based on two validated antibodies.
Conclusions:
The 64Cu-FN3hPD-L1 radiotracer represents a novel, small, and high-affinity binder for imaging hPD-L1 in tumors. Our data support further exploration and clinical translation of this binder for non-invasive identification of cancer patients who may respond to immune checkpoint blockade therapies.
Keywords: fibronectin domain 3 (FN3) binder, immune checkpoint, positron emission tomography (PET), immunoPET, programmed death ligand 1 (PD-L1)
INTRODUCTION
Despite multiple mechanisms employed by cancer cells to evade and inhibit the host tissue’s immunological surveillance (1), many treatment strategies based on antagonizing co-inhibitory, or activating co-stimulatory, pathways of immune cells have emerged (2). Clinical studies based on targeting these pathways clearly indicate prolonged patient survival by 6–12 months (3,4). In particular, the targeting of immune checkpoints (ICs) such as programmed death receptor-1 (PD-1, CD279) and its ligand PD-L1 (B7-H1, CD274), and anti-CTLA-4 therapy is rapidly advancing the field of cancer immunotherapy.
In the case of PD-1/PD-L1 immune checkpoint inhibitor therapy (5), PD-1 is expressed on activated T, B, and natural killer lymphocytes whereas PD-L1 is expressed in a wide variety of tumors including melanoma, non-small cell lung cancer, Merkel cell carcinoma, breast cancer, and squamous cell carcinoma (4,6). PD-L1 is also expressed on the surface of macrophages, endothelial cells, and other nonmalignant tissues (e.g. pancreatic), which aids in the prevention of autoimmune disease (7,8).
In the tumor microenvironment, PD-L1 expression is mainly promoted by the adaptive immune response mechanisms to interferon-γ (IFN-γ) secreted by antigen-bound tumor-infiltrating T cells (8,9). KRAS and PTEN gene mutations (9p24.1) in certain tumors also promote PD-L1 expression (10,11). The overall role of PD-L1 expression by tumors is to inhibit activation of infiltrating PD-1-expressing cytotoxic T cells, thereby evading the host immune system and suppressing its response (12).
In current clinical practice, the decision to proceed with IC inhibitor therapy with anti-PD-L1 immunotherapy is based on anti-PD-L1 immunohistochemistry (IHC) staining in biopsied tumor tissue sections (13,14). The amount of PD-L1 expression is also used as a surrogate for determining whether a patient might respond to anti-PD-1 immunotherapy (15). Although IHC is a well-established technique that undergoes rigorous validation prior to clinical utilization, numerous factors may contribute to inconsistent results: a) under- or mis-sampling of the tumor region with the highest PD-L1 expression at the time of biopsy, b) difficulty in obtaining adequate specimens in patients with metastatic disease, and c) variability in baseline biomarker expression due to previous anti-cancer therapies. Further, IHC lacks the spatial and temporal resolution of the biomarker expression profile within the entire tumor. On the other hand, non-invasive PET imaging allows repetitive visualization of PD-L1-expressing tumors throughout the body that enables improved lesion localization and characterization (16,17).
Many pre-clinical studies have shown that PD-L1 expression in tumors can be assessed by PET imaging (18–20). However, antibody-based PD-L1 tracer imaging studies suffer from slow clearance of the antibodies from the non-target tissues and limit the use of shorter half-life radioisotopes for labeling. For diagnostic imaging, tracers with faster clearance profiles from non-target tissues would enable quantitation of the signals from the target tissues at earlier time points (e.g. 1–4 hours post injection [p.i]). This can be achieved using small proteins (~25–50 kDa) or nanobodies (~10 kDa), a class of molecules that can be imaged in target tissue within 4 hours of administration. Such an approach would be more patient-friendly (for example the patient would not have to return the day after radiotracer injection for PET imaging) and expedite the initiation of the appropriate IC therapy. We have previously developed a class of protein scaffolds, e.g. the tenth type III domain of human fibronectin (FN3, ~10 kDa) as a platform that can provide faster clearance (<12 hours vs. ~72 hours in the case of antibodies) and demonstrates a specific in vivo targeting ability to yield excellent tumor-to-background contrast (24).
FN3 has been engineered for many targets (22,23) with picomolar to nanomolar affinity, including molecular imaging of cancer using PET in murine models (24) and in phase II clinical studies in therapeutic oncology (25). FN3 demonstrates a high stability scaffold, contains three solvent-exposed loops that can be mutated to introduce new high-affinity variants, and a single lysine that provides rapid amine conjugation of chelators (23,26,27). Similar kinds of molecules that have already been validated for clinical use include affibodies (21,28), knottins (29), nanobodies (30,31), peptides (32,33), and antibody fragments (34–36). These small molecules are designed to enhance vascular extravasation (37) and tissue penetration for delivery into solid tumors (38,39).
Anti-hPD-L1 antibody (atezolizumab)-based imaging studies revealed that hPD-L1 can be detected in the tumor microenvironment using various isotopes (111In, 64Cu, and 89Zr) in mouse tumor xenograft models (18). In this report, we present the development of a novel small protein molecule PET tracer, 64Cu-FN3hPD-L1, for imaging of hPD-L1 expression in an hPD-L1-expressing tumor mouse model at time points as early as 1-hour p.i. In human cancer tissue specimens, we further show a similarity between our FN3hPD-L1 binder and two validated hPD-L1 antibodies in regard to detecting hPD-L1 expression status.
MATERIALS and METHODS
Reagents and radiochemicals
All reagents were obtained from Sigma-Aldrich (St. Louis, MO) unless otherwise stated. N-succinimidyl-DOTA (NHS-DOTA) was purchased from Macrocyclics (Plano, TX). The CT26 mouse colon carcinoma cells engineered to express human PD-L1 were authenticated by short tandem repeat profiling in Dr. Irving Weissman’s lab, and kindly provided for this study. Quantitative analysis of indirect immunofluorescence staining in flow cytometry was used to calculate the number of hPD-L1 molecules per CT26/hPD-L1 cell (QIFKIT®, Code K0078, Dako, Denmark). The Raji human Burkitt’s lymphoma hPD-L1-negative cells were obtained from American Type Culture Collection (ATCC, catalog number CCL-86™, Manassas, VA). The MDA-MB-231 human triple negative breast adenocarcinoma (TNBC) cells naturally expressing hPD-L1 were purchased (ATCC, catalog number HTB-26™, Manassas, VA). All three cell lines were maintained according to standard techniques and used within three passages. They were maintained in DMEM supplemented with 10% fetal calf serum, 2 mmol/L glutamine, 100 units/mL penicillin, 100 μg/mL streptomycin, and 0.25 μg/mL fungizone. Media and supplements were obtained from Life Technologies (Carlsbad, CA).
High-performance liquid chromatography (HPLC) was performed on HPLC-Ultimate (ThermoFisher, Waltham, MA) with an ultraviolet detector and an inline radioactivity detector. The system used a SEC-2000 LC column (300 × 7.8 mm) with 5 μm hydrophilic bonded silica support and 400 Å pore size (Phenomenex, Torrance, CA). Matrix-assisted laser desorption ionization (MALDI) mass spectrometry was performed with a TOF/TOF™ 5800 (SCIEX, Concord, Canada) operated in linear mode with sinapinic acid as matrix.
Engineering of FN3hPD-L1
The human PD-L1 (hPD-L1, or B7-H1, CD274) protein was purchased (catalogue # 10084-H02H-100, Sino Biologics, Beijing, P.R. China), biotinylated, characterized by MALDI, and immobilized on streptavidin magnetic beads for screening of FN3 binders. The yeast surface displayed FN3 G4 library with diversified loops and was sorted and matured as previously described (26,40). Briefly, yeast displaying 2.5×108 FN3 mutants were sorted for binding to magnetic beads with immobilized hPD-L1 protein, followed by fluorescence-activated cell sorting (FACS) for full-length proteins using the C-terminal c-myc epitope. Plasmid DNA from selected clones was recovered, mutated by error-prone PCR of either the entire FN3 gene or the paratope loops, and reintroduced into yeast by electroporation with homologous recombination. As binder enrichment progressed in later evolutionary cycles, FACS for binding to soluble hPD-L1 protein was also used. Five cycles of selection and mutation were performed. Plasmid DNA was recovered, transformed into bacteria, and individual clones were sequenced by standard DNA sequencing methods. The best binder (FN3hPD-L1, Fig. 1A) was expressed in bacterial culture with a His6 tag, purified by nickel column chromatography, and reversed-phase HPLC (41).
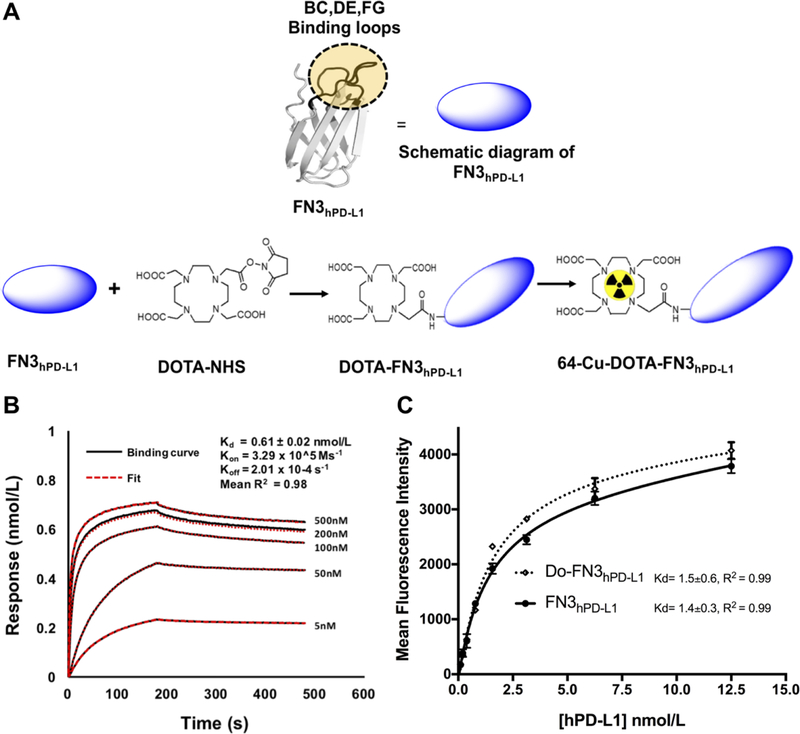
Figure 1: Determination of binding affinity of FN3hPD-L1 to hPD-L1.
A: Pictorial diagram of FN3hPD-L1 and DOTA-FN3hPD-L1. B: Biotinylated FN3hPD-L1 bound hPD-L1 protein with nmol/L affinity. Analyses were performed between the biotinylated FN3hPD-L1 binder (50 nmol/L) coated on the streptavidin-coated sensor, and various concentrations of hPD-L1. The surface changes due to binding of hPD-L1 onto the FN3hPD-L1 binder was measured as response units (nmol/L). A 1:1 binding curve was generated, yielding a Kd value of 0.6±0.02 nmol/L. Data are shown in black, with the corresponding curve fits in red. The R2 value of 0.98 represents the average value of those from the five curves. C: Comparison of binding activities of FN3hPD-L1 and Do-FN3hPD-L1 in intact cells expressing hPD-L1. hPD-L1-expressing CT26 cells were incubated with the indicated concentrations (0.5–12.5 nmol/L) of anti-6x-Histag-APC-FN3hPD-L1 or anti-6x-istag-APC-Do-FN3hPD-L1. The data are expressed as the mean±SD of 3 independent experiments.
Preparation of FN3hPD-L1
Bacterial expression plasmids were constructed to express FN3hPD-L1. The plasmids also encoded for a C-terminal His6 epitope tag for purification. Plasmids were transformed into BL21 (DE3) Escherichia coli. Cells were grown in 1 L of lysogeny broth medium for 4 hours and induced with 0.5 mmol/L isopropyl β-d-1-thiogalactopyranoside for 1 hour. Cells were pelleted, resuspended in 10 mL of lysis buffer (50 mmol/L sodium phosphate, pH 8.0, 500 mmol/L sodium chloride, 5% glycerol, 5 mmol/L CHAPS detergent, 25 mmol/L imidazole, and complete ethylenediaminetetraacetic acid–free protease inhibitor cocktail), frozen and thawed, and sonicated. The sample was centrifuged at 12,000×g for 10 minutes. Fibronectin was purified from the soluble fraction by immobilized metal affinity chromatography and reversed-phase HPLC with a C18 column (Phenomenex, San Diego, CA). Protein mass was verified by mass spectrometry.
Determination of binding affinity by FACS and Octet® biosensor
Octet experiments were conducted at 25°C in a buffer of PBS pH 7.4, 0.01% (v/v) Tween-20 and 1% bovine serum albumin (BSA), and sample plates were agitated at 1000 rpm. Biotin-FN3hPD-L1 was coupled onto streptavidin tips (Thermo Fisher Scientific, Waltham, MA). hPD-L1 protein was titrated into 5–500 nmol/L binding sites using a 1.2-fold dilution series. The FN3hPD-L1 binder was immobilized on a streptavidin coated-tip. These mixtures were allowed to bind the sensor tip-coupled FN3 binder for 10 minutes. We confirmed that neither hPD-L1 protein nor binder bound non-specifically to the unmodified tips. Samples were analyzed on duplicate tips to verify that the assay was reproducible between tips. Octet data were exported into Scrubber v.2.0a (BioLogic Software Pvt Ltd, Australia) for data processing and analysis. hPD-L1 biomarker protein was analyzed in 10 mmol/L HEPES, pH 7.4, 150 mmol/L NaCl, 0.005% (v/v) Tween-20 at five concentrations between 0 and 500 nmol/L. Assays were performed in duplicate and the response from an empty flow cell and from buffer injections was subtracted from each dataset. The data were analyzed using with a global fitting to the 1:1 binding model.
Intact cell binding flow cytometry assay
Cells were incubated at 37°C in humidified air with 5% CO2. For affinity measurement, 1×105 CT26/hPD-L1 or Raji (hPD-L1-negative) cells were washed with 0.1% BSA (w/v) in PBS and incubated with various concentrations of FN3hPD-L1. Cells were pelleted, washed with 0.1% BSA (w/v) in PBS, and incubated with 100 μL of 0.05 μg/μL Alexa Fluor® 488-conjugated mouse anti-His6 antibody (clone AD1.1.10, BIO-RAD, Hercules, CA) in 0.1% BSA (w/v) in PBS. Cells were washed and analyzed using flow cytometry. The minimum and maximum fluorescence and the affinity values were determined by minimizing the sum of squared errors assuming a 1:1 binding interaction. Experiments were performed in triplicate.
Preparation of Do-FN3hPD-L1
The DOTA-NHS ligand has already shown good biological performance when used in protein conjugation of various radionuclides such as 68Ga and 64Cu (19, 42). DOTA-FN3hPD-L1 (Do-FN3hPD-L1) tracer was prepared by conjugating DOTA-NHS to FN3hPD-L1 according to a published procedure (41). Briefly, lyophilized FN3hPD-L1 protein was resuspended in dimethylformamide with 2% triethylamine and reacted at room temperature for 1 hour with 20 equivalents of DOTA-NHS. DOTA-FN3hPD-L1 was purified by HPLC and lyophilized for 64Cu labeling. The number of DOTA chelators conjugated to each FN3hPD-L1 molecule was calculated by mass spectrometry by comparing the mass of FN3hPD-L1 and Do-FN3hPD-L1, see Fig. 1A (41).
Radiolabeling of Do-FN3hPD-L1
64CuCl2 was received from the University of Wisconsin (Madison, WI) and its specific activity at the time of shipment (24 hours prior to labeling) was 1.6±0.2 Ci/μmol. Radiolabeling of Do-FN3hPD-L1 was performed using 64CuCl2 as follows: Do-FN3hPD-L1, 25–50 μg in 100 μL of 0.25 mol/L ammonium acetate buffer (pH 5.5) was reacted with 92.5–185 MBq of neutralized 64CuCl2 solution at 37°C for 1 hour. After incubation, 0.1 M diethylenetriamine pentaacetic acid (pH 7.0) was added to a final concentration of 5 mmol/L and incubated at room temperature for 15 minutes to scavenge unchelated 64CuCl2 in the reaction mixture. Purification of the 64Cu-FN3hPD-L1 was achieved using SEC-2000 HPLC with a flow rate of 1.0 mL/minute in PBS [0.1 mol/L NaCl, 0.05 mol/L sodium phosphate (pH 7.4)]. The final radioconjugate of 64Cu-FN3hPD-L1 was filtered through a 0.2 μm filter into a sterile vial.
Radiotracer immunoreactivity assay
Immunoreactivity of the 64Cu-FN3hPD-L1 tracer was tested by cell-binding assays as previously described (41). Two hundred microliters of CT26/hPD-L1 cells were suspended in microcentrifuge tubes at concentrations of 5.0, 0.6, 0.3, 0.16 and 0.08 × 106 cells/mL in PBS (pH 7.4) with 1% bovine serum albumin (PBSA). Thereafter, each tube received aliquots of 50 μL of 64Cu-FN3hPD-L1 (from a stock solution of 10 μCi in 10 mL PBSA). The triplicate tubes containing the tracer (n=15; final volume 250 μL each) were gently vortexed and incubated at 37°C. Two hours later, the solutions were centrifuged (300×g for 3 minutes), resuspended, and washed twice with ice-cold PBS before removing the supernatant. 64Cu-activity associated with the cell pellet was measured with a gamma counter (1470 WIZARD Automatic Gamma Counter; Perkin Elmer, Waltham, MA). Competition assays were also performed by the same procedure but with the Raji cells. Linear regression analysis of a plot of total/bound activity versus 1/(normalized cell concentration) was performed, and the immunoreactive fraction was calculated as 1/y-intercept.
Small animal PET/CT imaging of hPD-L1 expression in NSG mice bearing syngeneic and xenograft tumors
Animal studies were approved by the Administrative Panel on Laboratory Animal Care (APLAC) at Stanford University. NSG (NOD.Cg-PrkdcscidIl2rgtm1wjl/SzJ) mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and maintained in-house in an AAALAC-accredited facility. The average weight of the NSG mice was 23.0±2.0 g. Six- to eight-week-old NSG mice were implanted subcutaneously with 5×106 CT26/hPD-L1 (left shoulder), 5×106 Raji (hPD-L1-negative, right shoulder) cells, or 2×106 MDA-MB-231 (naturally hPD-L1-expressing, left axilla) cells in 50 μL of PBS mixed with 50 μL of Matrigel Matrix (catalog # 356234, Corning, Inc., Corning, NY). Mice with tumors ranging in size from 350–450 mm3 (for CT26/hPD-L1 and Raji tumors) or 200–250 mm3 (for MDA-MB-231 tumors) were chosen for the studies. Four groups of NSG mice (n = 3–5/group) received 64Cu-FN3hPD-L1: (1) CT26/hPD-L1-non-blocking (nblk, no blocking of hPD-L1 with nonradioactive FN3hPD-L1 prior to radiotracer injection), (2) CT26/hPD-L1-blocking (blk), (3) Raji (hPD-L1-negative)-nblk, and (4) MDA-MB-231-nblk. Each mouse received 200 μL of 64Cu-FN3hPD-L1 diluted in PBS, corresponding to 3.7±0.4 MBq (8–10 μg of FN3hPD-L1), via tail vein injection. The blk group received a blocking dose (100-fold excess) of nonradioactive FN3hPD-L1 in 200 μL of PBS 2 hours before radiotracer injection. After radiotracer administration, the animals were imaged under 2% isofluorane delivered with 100% oxygen at 0.5, 1, 4, 18, and 24 hours p.i. using a Siemens Inveon small animal multimodality PET/CT system (Preclinical Solutions, Siemens Healthcare Molecular Imaging, Malvern, PA). Results are reported as percent injected dose per gram of tissue (%ID/g). Image files were assessed with region of interest (ROI)-based analyses using an Inveon Research Workspace (IRW, Siemens Healthcare Molecular Imaging, Malvern, PA). For each small animal PET scan, three-dimensional ROIs were drawn around the tumor, heart, liver, kidneys, and muscles on decay-corrected whole-body images. The tumor ROI analysis was performed in the same manner as that of the other organs (i.e., based on the anatomical information of the CT image in which organs and the tumor are well-delineated to enable drawing of the ROIs). The average radioactivity concentration in the ROI was obtained from the mean pixel values within the ROI volume. These data were converted to counts/mL/minute by using a predetermined conversion factor (41). The results were then divided by the injected dose to obtain an image ROI-derived %ID/g.
Cancer patient tissue specimens
De-identified tissue sections from biopsies and corresponding subsequent resections of six cancer patients were obtained through a Stanford Institutional Review Board (IRB)-approved protocol (IRB-44051). The cases consisted of primary lung adenocarcinoma (n=2), metastatic lung adenocarcinoma to brain (n=3), and oncocytic thyroid carcinoma (n=1). Supplementary Table S1 online summarizes the clinical characteristics of the patients from who the tissue sections were obtained.
Histology and immunofluorescence staining of hPD-L1
The human cancer tissues and implanted mouse xenografts (MDA-MB-231 and Raji) were obtained in paraffin-embedded blocks and sectioned into 4–5 μm-thick sections for immunohistochemistry (IHC) and hematoxylin and eosin (H&E) staining, and 10 μm-thick sections for immunofluorescence (IF). All tissue sections were de-paraffinized prior to staining, which was performed in a blinded manner. A pathologist (SRL) reviewed the IHC and IF staining results to determine the hPD-L1 expression in the H&E-confirmed tumor areas after reviewing multiple fields (4–20×) in each specimen. During scoring, the pathologist focused on areas of tumor and excluded areas of necrosis or tumor-infiltrating inflammatory cells such as lymphocytes, which are known to stain for hPD-L1.
For IHC, two rabbit monoclonal antibodies against human-PD-L1 that had been validated in placental tissue were used: clone E1L3N (catalog no. 13684, Cell Signaling Technology, Cambridge, MA) and clone SP263 (catalog no. 790–4905, Ventana Medical Systems, Inc., Tucson, AZ). The IHC methods were performed according to the manufacturers’ protocols. Briefly, clone E1L3N was diluted 1:500 on the Bond™ Polymer Refine Detection platform (catalog no. DS9800, Leica Biosystems, Buffalo Grove, IL) and clone SP263 was pre-diluted (1.61 μg/mL) and applied with the BenchMark ULTRA system (Ventana Medical Systems, Inc.) using the OptiView DAB IHC Detection Kit (catalog no. 760–700, Ventana Medical Systems, Inc.). After validation, the two hPD-L1 antibody clones were separately applied to the human cancer resection tissues from the six cases in this study (there was insufficient corresponding prior biopsy tissue on which to study the validated antibodies) and the implanted mouse xenografts (MDA-MB-231 and Raji). The slides were scanned on an UltraFast Scanner Digital (Philips, Amsterdam, The Netherlands) digital pathology slide scanner.
For IF, the microscope slides were rinsed twice with Dulbecco’s phosphate-buffered saline (DPBS) at room temperature. A PAP pen was used to outline a hydrophobic barrier around each tissue section. The sections were permeabilized with 0.1% Triton X-100 (v/v) in DPBS for 5 minutes at room temperature, followed by 3 washes (5 minutes each) with DPBS. The tissues were blocked for 1 hour in 10% (v/v) fetal bovine serum in DPBS. The blocking solution was then removed, and the sections were washed with wash buffer (0.1% bovine serum albumin [BSA] (w/v) in DPBS) twice (5 minutes each). The FN3hPD-L1 binder was diluted 1:200 in 0.01% Tween-20 (v/v) in 1% BSA (w/v) in DBPS, from the stock concentration of 5 mg/mL in DMSO to a final concentration of 25 μg/mL FN3hPD-L1. The diluted binder was then applied to the tissue sections overnight at 4oC. Afterwards, the binder was removed with three DPBS washes (5 minutes each). The DyLight® 650-conjugated anti-6X His tag® secondary antibody (ab117504, Abcam, Cambridge, MA) was diluted 1:800 in 1% BSA (w/v) in DPBS and applied to the tissues in the dark for 1 hour at room temperature. The secondary antibody was removed with three DPBS washes (5 minutes each). Cover slips were mounted and their edges sealed with clear nail polish. Images were acquired using a NanoZoomer 2.0-RS whole slide imager (Hamamatsu, Hamamatsu City, Japan) and saved as TIFF files using the NanoZoomer Digital Pathology (NDP) Scan version 2.5 software.
Statistical Analyses
Data were analyzed and plotted using Prism version 5.01 (GraphPad Software Inc., La Jolla, CA). Data were tested for normality using the Shapiro-Wilk test. An unpaired two-tailed Student’s t test was used to compare the means of groups of normally-distributed data. For non-normally distributed data, the Wilcoxon rank-sum analysis of medians was performed. A p-value less than 0.05 was considered to be statistically significant. Bonferroni correction was used to adjust alpha for multiple comparisons. Data are presented as mean±standard deviation (SD).
RESULTS
Screening, selection, and binding affinity of FN3hPD-L1
Based on the hPD-L1 cell binding study, the best unique clone against hPD-L1 protein was selected and called FN3hPD-L1. We performed the sequence analyses for confirmation of the full framework and uniqueness of the loop region. The soluble protein yield was >5mg/L with >95% purity by HPLC and demonstrated 11,826 Da molecular weight when measured by mass spectrometry (expected 11,824 Da). This soluble FN3hPD-L1 binder was biotinylated to determine the binding affinity (one biotin per FN3hPD-L1 molecule, as determined by MALDI spectrometry). The purified soluble FN3hPD-L1 binder displayed the dissociation constant (Kd) of 0.61±0.02 nmol/L for purified hPD-L1 (Fig. 1B). A flow cytometry assay with live CT26/hPD-L1 cells indicated the binding affinity of FN3hPD-L1 for cell surface hPD-L1 was 1.4±0.3 nmol/L (Fig. 1C).
Synthesis and quality control of the 64Cu-FN3hPD-L1 radiotracer
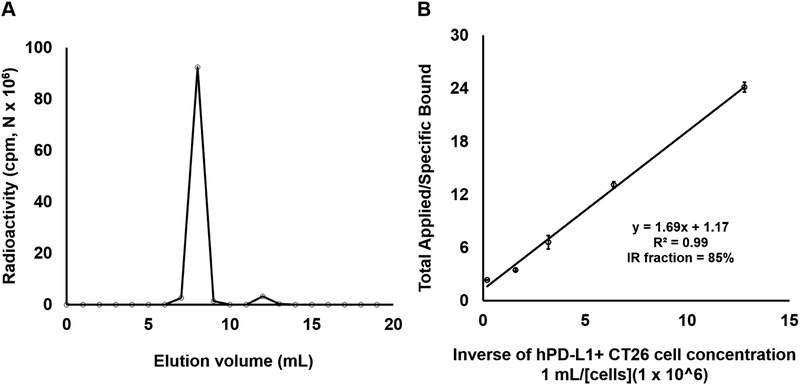
The characterization of 64Cu-FN3hPD-L1 radiotracer is summarized in Table 1. DOTA-NHS (Do) was conjugated to lysine groups of FN3hPD-L1 and yielded Do-FN3hPD-L1 with 1–2 chelates per binder, as confirmed by MALDI-TOF mass spectrometry (Supplementary Fig. S2 online). This DOTA-conjugated FN3hPD-L1 binder (Do-FN3hPD-L1) was further purified using SEC2000-HPLC column and radiolabeled with 64Cu. The final yield and purity of 64Cu-FN3hPD-L1 were greater than 70% and 95%, respectively (Fig. 2a). SEC2000-HPLC showed that the specific activity of FN3hPD-L1 was 5.3±0.5 GBq/μmol (Table 1). Immunoreactivity of the 64Cu-FN3hPD-L1 was tested against the CT26/hPD-L1 cells (mean ± SD = 3×105 ± 1.5×104 hPD-L1 molecules per CT26/hPD-L1 cell) and hPD-L1-negative Raji cells and found to be 83.6±8.8% and 4.9±0.3% (p=0.0001), respectively (Fig. 2b, results from Raji cells were not shown due to low immunoreactivity). Thus, the 64Cu-FN3hPD-L1 tracer was specific for the hPD-L1 antigen.
Table 1: Characterization of 64Cu-FN3hPD-L1 radiotracer.
DOTA = 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid, FACS = fluorescence-activated cell sorting, FN3 = fibronectin 3, HPLC = high performance liquid chromatography, TLC = thin-layer chromatography.
| Parameter | Results |
|---|---|
| FN3hPD-L1 protein production | >5mg/L |
| Binding affinity (by FACS) | 1–2 nM |
| Chemical purity (by HPLC) | >95% |
| DOTA chelates/protein | 1–2 |
| pH | 7±0.5 |
| Radiochemical yield | >70% |
| Specific activity (GBq/μmol) | 5.3±0.5 |
| 64Cu-FN3hPD-L1 purity (by TLC and HPLC) | >95% |
| Immunoreactivity fraction | 85% |
Figure 2: Characterization of 64Cu-FN3hPD-L1 radiotracer syntheses.
A: Radio- high-performance liquid chromatography (HPLC) trace showed >95% purity (area under the curve) of 64Cu-FN3hPD-L1. B: Binding assay showing that the immunoreactivity of the 64Cu-FN3hPD-L1 for CT26/hPD-L1 cells was 85±2.4%.
64Cu-FN3hPD-L1 radiotracer imaging of mouse models bearing syngeneic and xenograft tumors
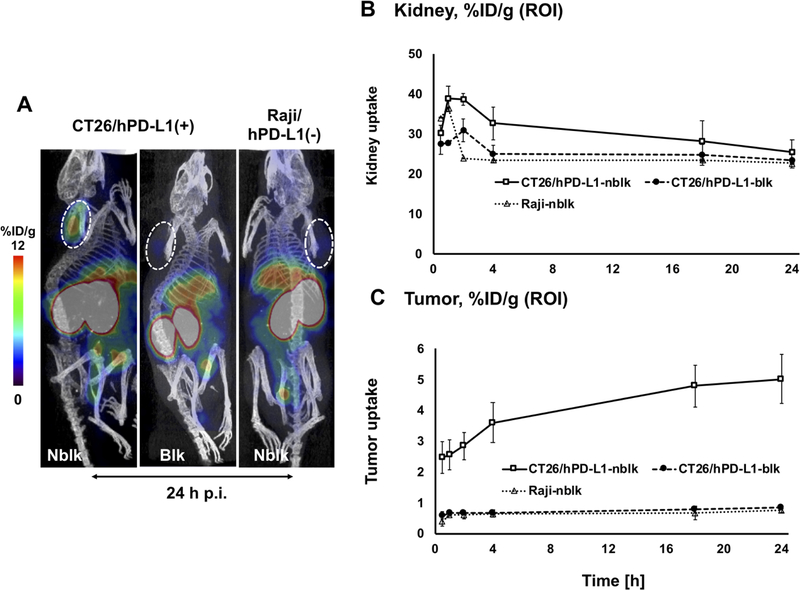
Figure 3a shows representative coronal 64Cu-FN3hPD-L1 small animal PET/CT images (at 24 hours p.i.) of NSG mice bearing the CT26/hPD-L1 tumors in the left shoulder (nblk and blk) and NSG mice bearing the hPD-L1-negative Raji tumors in the right shoulder (Raji-nblk). These PET images clearly show that the 64Cu-FN3hPD-L1 tracer was able to detect CT26 tumors that express hPD-L1. Figure 3b shows the uptake of the 64Cu-FN3hPD-L1 radiotracer in kidney at various time points from 0.5 to 24 hours p.i. (mean %ID/g±SD). The ROI analyses of kidney uptake in the three different NSG mouse groups (CT26/hPD-L1-nblk, CT26/hPD-L1-blk, and Raji-nblk) showed the radiotracer clearance pattern (mean±SD %ID/g) to be comparable at 24 hours p.i. (25.4±3.1, 23.4±0.6, and 22.7±0.6, respectively, at 24 hours p.i.). Figure 3c shows higher tumor uptake in the CT26/hPD-L1-nblk group compared to the CT26/hPD-L1-blk and Raji-nblk groups at each time point. In particular, in mice bearing CT26/hPD-L1 tumors, the difference between nblk and blk cohorts was observed as early as 1-hour p.i. (2.6±0.5 vs. 0.7±0.02 %ID/g, p=0.004) and was maintained at 4 hours p.i. (3.6±0.7 vs. 0.7±0.1 %ID/g, p=0.006). The 24-hour p.i. uptake of 64Cu-FN3hPD-L1 (mean±SD %ID/g in the CT26/hPD-L1-nblk group) was 1.2±0.18 and 4.9±0.36 in the heart and liver, respectively, compared to 5.0±0.8 in the tumor. Small animal PET/CT 3D visualization of 64Cu-FN3hPD-L1 radiotracer uptake at 1-hour p.i. in the CT26/hPD-L1-blk and CT26/hPD-L1-nblk groups is shown in Supplementary Video S3.
Figure 3: Small animal PET/CT imaging of hPD-L1 expression in syngeneic and xenograft tumors.
A: Representative small animal PET/CT coronal images showing the three mouse groups (n=3), which either received 10 mg/kg of non-radioactive FN3hPD-L1 2 hours prior to radiotracer injection for blocking (blk) or did not (non-blocking, nblk). All groups received 64Cu-FN3hPD-L1 tracer (3.7 MBq) via tail vein injections and the PET images shown were acquired 24 hours p.i. The CT26 mouse colon carcinoma cells (left shoulder implantation) were engineered to express hPD-L1, whereas the Raji human Burkitt’s lymphoma cells (right shoulder implantation) were naturally hPD-L1-negative. Liver and kidney are indicated by the yellow fonts of L and K, respectively. Tumors are outlined with a dotted white circle. B-C: Region of interest quantification of PET signals from (B) tumor and (C) kidney was performed over the post-radiotracer injection period (0.5–24 hours, x-axis). CT26/hPD-L1-blk (∙), CT26/hPD-L1-nblk (□), Raji-nblk (Δ). Data are reported as mean±SD %ID/g.
PET/CT imaging and biodistribution of 64Cu-FN3hPD-L1 in a mouse model bearing a hPD-L1-expressing MDA-MB-231 human TNBC xenograft
Supplementary Figure S5A shows representative coronal 64Cu-FN3hPD-L1 small animal PET/CT images (at 24 hours p.i.) of NSG mice bearing the MDA-MB-231 xenograft in the left shoulder (nblk). These images clearly show that the 64Cu-FN3hPD-L1 tracer was able to detect hPD-L1 protein, which is naturally expressed in this cell line. Supplementary Figure S5B shows higher 64Cu-FN3hPD-L1 ROI-based tumor uptake (mean±SD %ID/g) in the MDA-MB-231-nblk group compared to the MDA-MB-231-blk group at 4 hours (1.8±0.1 vs. 1.2±0.1 %ID/g, respectively, p=0.004) and 24 hours (3.6±0.4 vs. 1.6±0.2 %ID/g, respectively, p=0.004) p.i. The difference in tracer uptake between the nblk and blk groups was apparent as early as 1 hour p.i. (1.4±0.09 vs. 0.8±0.2 %ID/g, respectively, p=0.012). The radiotracer clearance from kidney at 24 hours p.i. (mean±SD %ID/g) in the two different MDA-MB-231 xenograft cohorts (nblk and blk) was 17.2±0.7 and 12.6±1.4, respectively (p=0.02).
Tumor-to-muscle ratio and ex vivo biodistribution of 64Cu-FN3hPD-L1 radiotracer in mice bearing syngeneic and xenograft tumors
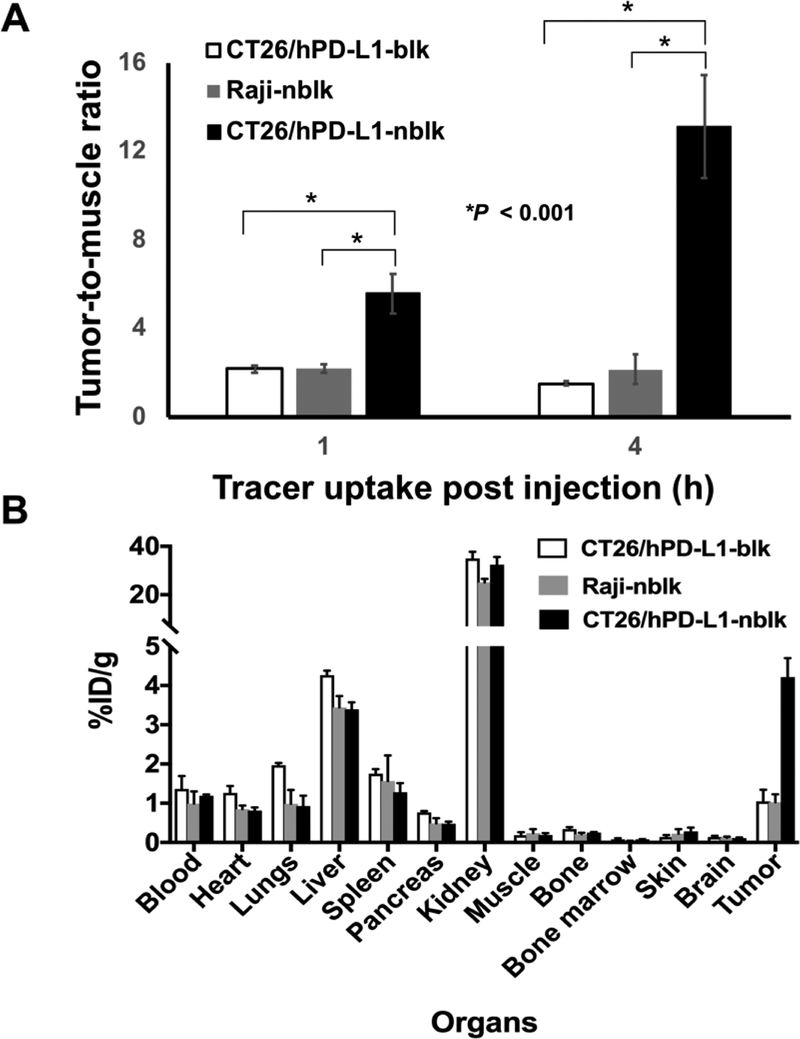
We further calculated the tumor-to-muscle ratios of the PET signal at 1 and 4 hours p.i. in the CT26/hPD-L1-nblk, CT26/hPD-L1-blk, and Raji-nblk groups as %ID/g, after decay-correction (Figure 4a). The tumor-to-muscle ratio of 64Cu-FN3hPD-L1 was significantly greater in the CT26/hPD-L1-nblk group compared to the corresponding blk group at both 1 hour p.i. (5.6±0.9 vs. 2.1±0.2, p=0.0014) and 4 hours p.i. (13.1±2.3 vs. 1.5±0.1, p=0.0005). Figure 4b shows the tracer uptake at 24 hours p.i. in a panel of ex vivo organs from the three groups of mice. Ex vivo tumor uptake in the CT26/hPD-L1-nblk, CT26/hPD-L1-blk, and Raji-nblk groups was 4.2±0.5, 1.0±0.3, and 1.0±0.2 %ID/g, respectively (p=0.001 for CT26/hPD-L1 nblk vs. blk, and p=0.003 for CT26/hPD-L1-nblk vs Raji-nblk groups, two separate t-tests). Tumor-to-muscle ratio from ex vivo data in the respective groups was 23.8±4.8, 6.3±2.9, and 4.7±1.4 (p=0.01 for CT26/hPD-L1-nblk compared to each of the other groups, two separate t-tests). Ex vivo kidney uptake (%ID/g) in the CT26/hPD-L1-nblk and -blk mice was 32.4±3.1 and 34.9±2.7, respectively (p=0.04). Ex vivo liver uptake (%ID/g) in the CT26/hPD-L1-nblk and -blk mice was 3.4±0.2 and 4.3±0.1, respectively (p=0.003). Tumor radiotracer uptake (%ID/g) in the CT26/hPD-L1-nblk group at 24 hours p.i. as measured by the ROI from the imaging study was 5.0±0.8, compared to 4.2±0.5 based on the ex vivo tissue uptake cpm count results (p=0.15).
Figure 4: Analyses of 64Cu-FN3hPD-L1 tumor-to-muscle ratios and ex vivo biodistribution.
A: ROI analyses of in vivo radiotracer uptake (tumor-to-muscle ratio, mean±SD) at two early time points (1 and 4 hours post-injection) in the CT26/hPD-L1-nblk, CT26/hPD-L1-blk, and Raji-nblk mouse groups (n=3–5/group). The 64Cu-FN3hPD-L1 tracer uptake value (expressed as %ID/g) in the CT26/hPD-L1-nblk group (5.6±0.9) was significantly greater than in the CT26/hPD-L1-blk group (2.1±0.2, p<0.001) and in the Raji-nblk group (2.2±0.2, p<0.001) at 1 hour. B: Histogram of 64Cu-FN3hPD-L1 tracer biodistribution in the three groups (n=3/group). All mice were injected with 3.7 MBq of 64Cu-FN3hPD-L1 tracer via tail vein and euthanized at 24 hours post-injection. Organs were then isolated, and uptake of tracer dose was measured by gamma counter (decay-corrected mean %ID/g ±SD,). Pre-blocked mice (blk) received 10 mg/kg of non-radioactive FN3hPD-L1 via tail vein 2 hours before 64Cu-FN3hPD-L1 radiotracer injection whereas non-blocked mice (nblk) did not.
In the mice bearing the MDA-MB-231 xenografts, the ROI-based 64Cu-FN3hPD-L1 tumor-to-muscle ratio was greater in the nblk group compared to the corresponding blk group at both 1 hour (2.2±0.2 vs. 1.3±0.1, respectively, p=0.012) and 4 hours (3.4±0.4 vs. 2.0±0.5, respectively, p=0.022) p.i. The ex vivo tumor uptake at 24 hours p.i. in the nblk and blk groups was 3.6±0.5 and 1.8±0.3 %ID/g, respectively (p=0.013). Tumor uptake (%ID/g mean±SD) in the nblk group at 24 hours p.i. was 3.6±0.4 (ROI-based) compared to 3.6±0.5 (ex vivo), p=0.97.
hPD-L1 expression status in human cancer tissues based on validated naturally hPD-L1 antibodies and the FN3hPD-L1 binder
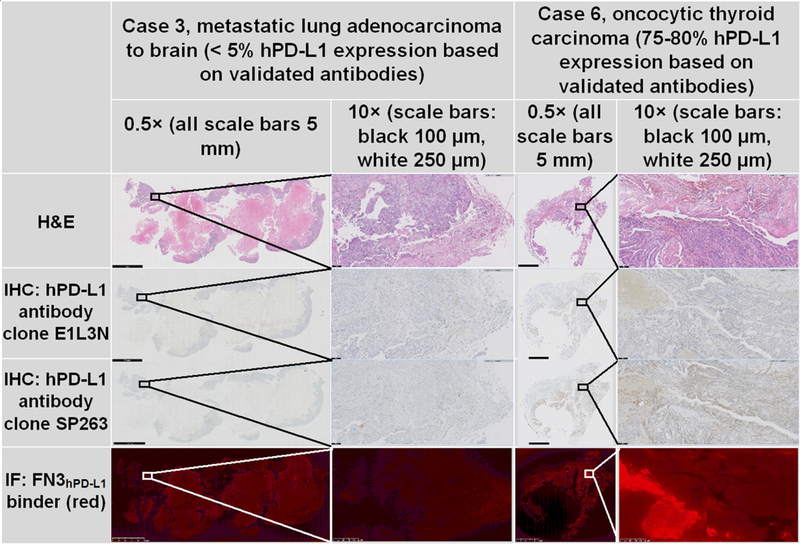
To assess the translational potential of the FN3 hPD-L1 scaffold’s ability to correctly identify the presence of hPD-L1 expression in human tissue, FN3hPD-L1 binder immunofluorescence was compared to immunohistochemistry of the two validated hPD-L1 antibodies in tissue sections from six human cancer cases (Table S1). Supplementary Figure S4 displays the hPD-L1 expression status at the time of biopsy (based on the results from an FDA-approved send-out lab) and subsequent resection (using the two validated hPD-L1 antibodies and the FN3hPD-L1 binder). Supplementary Figure S4 shows the heterogeneity of hPD-L1 expression not only from one patient to another, but also from biopsy to resection within the same patient. In the very low (<5%) hPD-L1 expressing cases (Cases 1–3, the two lung primary tumors and one of the metastatic lung to brain tumors) at the time of resection, there was 100% agreement among the two validated hPD-L1 antibodies and the FN3hPD-L1 binder. In the remaining three cases with moderate-high (30–80%) hPD-L1 expression (Cases 4–6), the two validated antibodies resulted in comparable (i.e. within 5–15% of each other) hPD-L1 expression results. In these same three cases, the magnitude of hPD-L1 expression based on the FN3hPD-L1 binder was lower but still ≥10% (Fig. S4). Fig. 5 shows H&E, immunohistochemistry (using two validated hPD-L1 antibodies), and immunofluorescence (using the FN3hPD-L1 binder) in representative low and high hPD-L1-expressing human cancer tissues (Cases 3 and 6, respectively). At these extrema, the FN3hPD-L1 staining pattern demonstrated a similar trend compared with that of the two validated hPD-L1 antibodies.
Figure 5: Low and high hPD-L1-expressing human cancer tissue specimens.
Comparison of hematoxylin and eosin (H&E), validated hPD-L1 immunohistochemistry (IHC; clones E1L3N and SP263), and immunofluorescence (IF; FN3hPD-L1 binder). Two cases with differing hPD-L1 expression in the tumor tissue were evaluated: metastatic lung adenocarcinoma to the brain (Case 3, 5% hPD-L1 expression) and oncocytic thyroid carcinoma (Case 6, 75–80% hPD-L1 expression). FN3hPD-L1 = hPD-L1 binder (red). The black and white boxes in the low-magnification images represent the area displayed in the corresponding higher-magnification images. All scale bars at magnification level 0.5× = 5 mm. At magnification level 10×, black scale bars = 100 μm and white scale bars = 250 μm.
hPD-L1 expression status in mice bearing xenografts of human tumors with different hPD-L1 levels (MDA-MB-231 and Raji), based on validated hPD-L1 antibodies
In the MDA-MB-231 xenograft that received 64Cu-FN3hPD-L1 (100 μCi / 10 μg of FN3hPD-L1 binder), hPD-L1 staining with the validated antibodies was scored as 55% using clone E1L3N and 85% using clone SP263 (Supplementary Figure S5C). We also performed validated hPD-L1 antibody staining in MDA-MB-231 (hPD-L1 expressing) and Raji (hPD-L1 non-expressing) xenografts that were not exposed to 64Cu-FN3hPD-L1 (Supplementary Figure S6). The MDA-MB-231 and Raji hPD-L1 expression was scored as 60% and 0%, respectively (clone E1L3N), and 90% and 5%, respectively (clone SP263).
DISCUSSION
In this report, we present the development of a novel radiotracer that targets hPD-L1, an immune checkpoint protein expressed in most tumors. The resulting FN3-based small protein binder of hPD-L1 (FN3hPD-L1), which was approximately one-tenth the size of an antibody, bound purified hPD-L1 protein and cells engineered to express hPD-L1 (19,24,41). Whereas antibody-based tracers can take a few days to attain optimal tumor uptake and then be cleared from the background tissues (43), small protein binders can do so within 24 hours (29). We made three molecular components (FN3hPD-L1 binder, Do-FN3hPD-L1 conjugate, and 64Cu-FN3hPD-L1 radiotracer), each of which was carefully assayed for its binding property against purified hPD-L1 protein (Figs. 1–2).
The pre-clinical in vivo imaging study revealed that the 64Cu-FN3hPD-L1 radiotracer rapidly targeted the tumors expressing hPD-L1 (Fig. 3). In fact, this tracer provided a tumor-to-muscle ratio in the CT26/hPD-L1-nblk group that was 3- and 9-times greater than that in the group pre-injected with non-radioactive FN3hPD-L1 (blk), as early as 1 and 4 hours post-injection, respectively (Figs. 3–4). Further, we evaluated the specificity of the tracer in vivo under pre-blocking with nonradioactive FN3hPD-L1 (blk) and pre-non-blocking (nblk) conditions in mice bearing hPD-L1-positive (CT26/hPD-L1) and hPD-L1-negative (Raji) tumors. The results indicated that the PET signals at 4 hours p.i. from the CT26/hPD-L1 tumors were substantially lower in the blk group compared to the nblk group by 6-fold (Figs. 3).
Previously, we developed the hPD-L1-targeting 64Cu-DOTA-HAC tracer and tested it in a NSG mouse model bearing CT26/hPD-L1 cells engineered to expresses hPD-L1 protein, which resulted in a moderate uptake of ~2 %ID/g at 1 hour p.i. (19). At 1 hour p.i., the current 64Cu-FN3hPD-L1 tracer and the 64Cu-DOTA-HAC tracer that used a different scaffold (19) each had a tumor-to-muscle ratio of ~6. A protein binder’s binding affinity is influenced by its binding domain, not its size (24). As reported in (24,40,44) and the current study, the smaller the protein binder, the faster its clearance. For example, based on ROI analysis in a study by Olafsen et al., the in vivo tumor-specific targeting of their 64Cu-anti-CD20 minibody with two binding domains in CD-20-positive tumors vs. CD-20-negative tumors was 2.3-fold higher at 4 hours p.i. and 1.9-fold higher at 19 hours p.i. (44). In contrast, based on ROI analysis the in vivo tumor-specific targeting of our single-binding domain 64Cu-FN3hPD-L1 radiotracer in hPD-L1-positive vs. hPD-L1-negative tumors was 5.6-fold higher at 4 hours p.i. and 8.1-fold higher at 24 hours p.i. The apparent increased tumor retention of our small protein binder, both within a few hours p.i as well as over time, is promising. It may be a good candidate tracer for non-invasive imaging-based determination of hPD-L1 expression status in tumors at early time points post-injection, which could help to determine likelihood of patient response to immune checkpoint blockade therapies.
In order to confirm the applicability of our tracer to human tumors with heterogeneous expression levels of hPD-L1, we performed 64Cu-FN3hPD-L1 PET imaging in a mouse model of CT26/hPD-L1 murine colon carcinoma cell line and hPD-L1-negative Raji Burkitt’s lymphoma cell line. We found that at 2 hours post-injection, the 64Cu-FN3hPD-L1 uptake in the CT26/hPD-L1 tumors was 4.7-fold higher compared to in the Raji tumors. Recently, Chatterjee and colleagues reported a peptide-based radiotracer, [64Cu]WL12, targeting hPD-L1 in mouse models of Chinese hamster ovary (CHO) tumors and CHO tumors engineered to stably express hPD-L1 (33). They reported a 3.1-fold higher uptake of their tracer in the CHO-hPD-L1 group compared to the CHO group at 2 hours p.i. (33). Donnelly et al. reported the results of 2-hour dynamic PET imaging (23). At 2 hours post-injection, the ratio of 18F-BMS-986192 uptake in the higher-hPD-L1 expressing L2987 to the lower-hPD-L1 expressing HT-29 tumors was 3.4 (23).
Although the 64Cu-FN3hPD-L1 tracer showed excellent tumor uptake at early time points and rapid clearance from most non-target tissues, its elevated signals in the liver and kidney at 24 hours p.i. were of less concerning for two reasons. The liver activity may be due to dissociation of 64Cu from DOTA (45) or charge effects of the engineered protein and the DOTA chelator. Kidney retention is a common problem for small proteins (44) because they pass through the glomerulus and can be reabsorbed in the renal tubules. Although we observed increased uptake of 64Cu-FN3hPD-L1 in the kidneys compared to that in the tumors, such uptake is similar to that of many other comparable-sized 64Cu-DOTA-labeled molecules (e.g. 64Cu-anti-CD20 minibody) (44). Over the first 4 hours after administration, the tracer uptake increased 38% in the tumor whereas it decreased by 16% in the kidney, suggesting renal clearance of the radiotracer (Fig. 3). On the other hand, in our previous study of a different class of binder derived from hPD-1 protein (high-affinity consensus [HAC]) (19), the 64Cu-DOTA-HAC radiotracer uptake decreased in both the tumor and kidney over the first four hours after administration, by 14% and 27%, respectively. Over the course of hPD-L1 tracer development in our laboratory, the 64Cu-FN3hPD-L1 radiotracer appears to have achieved improved tumor retention by 1.3-fold over the other binder class, HAC (19).
The results of our study and others (18,44,46) demonstrate that smaller binders can provide improved imaging results compared to high molecular weight antibodies both in terms of tumor-to-background ratio and absolute tumor uptake at early time points after tracer injection (e.g., 1–4 hours). For example, antibody-based tracers can take 3–10 days to attain optimal tumor uptake and be cleared from the background tissues (47,48). An ex vivo biodistribution study by Lesniak and colleagues found that the antibody-based radiotracer [64Cu]atezolizumab had a 1.3-fold higher uptake in mice bearing MDA-MB-231 TNBC xenografts compared to those bearing hPD-L1-low-expressing SUM149 TNBC xenografts 24 hours p.i (49). At the same post-injection time point, we found a 3.5-fold higher uptake of 64Cu-FN3hPD-L1 in mice bearing MDA-MB-231 xenografts compared to those bearing non-hPD-L1-expressing Raji Burkitt’s lymphoma xenografts. Hence, the FN3-based small protein binder strategy has considerable advantages as an imaging agent at the earlier post-tracer injection time points of in vivo tumor imaging in delineating the tumor-to-background tissue (e.g., muscle and blood). From the patient perspective, the development of a PET tracer to visualize hPD-L1-expressing tumor cells at early time points would enable tracer injection and PET imaging on the same day, facilitating earlier treatment decisions.
This novel tracer might be used to take advantage of the sensitivity of PET in cases of tumors with moderate to high hPD-L1 expression, as demonstrated by the results of the comparative histology analysis we performed in multiple human cancer tissue specimens (Fig. 5 and Supplemental Fig. S4). It should be noted that hPD-L1 is also expressed on lymphocytes, macrophages, and histiocytes, as well as in areas of tumor necrosis (50). In addition to the cell membrane, hPD-L1 is also expressed variably within cancer cells. The two hPD-L1 clones evaluated by validated antibodies used in this study resulted in comparable overall hPD-L1 expression in adjacent tissue sections despite not overlapping completely. This confirms the previously known heterogeneity of hPD-L1 expression patterns based on the antibody used and the respective cut-offs for positivity (13,14,16). Our FN3hPD-L1 binder identified similar hPD-L1 expression to that of the two validated hPD-L1 antibodies in the very low (<5%) hPD-L1-expressing tumors, suggesting high specificity of the FN3hPD-L1 binder for correctly identifying tumors that will likely not be responsive to IC blockade therapy, according to current clinical standards. In the remaining moderate-high (30–80%) hPD-L1-expressing tumors based on the validated antibodies, the FN3hPD-L1 binder staining was also elevated, but its magnitude was not as high (10–20%). Taken together, these results indicate that a FN3hPD-L1 binder-based value of ≤5% could serve as the cut-off to rule-out the patient subgroup of low-hPD-L1 expressing tumors. Again, based on the FN3hPD-L1 binder immunofluorescence results, a cut-off value of ≥10% may identify the moderate- to high- hPD-L1 expressing tumors. It should be reiterated that these FN3hPD-L1 binder-based cut-off values differ from the antibody-based cut-off values for low- and moderate-/high-hPD-L1 expression. Given the heterogeneity of staining patterns within and between various classes of hPD-L1 binders (e.g. antibodies, fibronectin-based), a one-to-one correlation between hPD-L1 antibody staining pattern vs. FN3hPD-L1 binder staining pattern at the cellular level was not anticipated. However, Figure 5 shows that at the tissue level (10× magnification), there was overlap between the areas stained by the hPD-L1 antibodies and by the FN3hPD-L1 binder. The immunofluorescence staining confirmed one of the goals of this study, that FN3hPD-L1 bound hPD-L1 in human cancer tissues.
Overall, PET imaging with 64Cu-FN3hPD-L1 in the CT26/hPD-L1 syngeneic tumor model indicated favorable hPD-L1-expressing tumor visualization at the earliest time points (1–4 hours p.i., Supplementary Video S3), and the PET signal was specific for hPD-L1 when compared against the pre-blocked CT26/hPD-L1 (hPD-L1-positive) group and the non-blocked Raji (hPD-L1-negative) group. Further, 64Cu-FN3hPD-L1 uptake in the tumors increased while in the background tissues it decreased at 24 hours p.i. Furthermore, we established that FN3hPD-L1 bound hPD-L1 in human cancer tissues. Based on these findings, the 64Cu-FN3hPD-L1 radiotracer has the potential for human translation to prospectively identify likely responders to immune checkpoint blockade therapy.
Conclusions
In conclusion, we developed and radiolabeled a novel 12 kDa FN3-based anti-hPD-L1 tracer (64Cu-FN3hPD-L1) for PET imaging of human PD-L1 expressed in a mouse colorectal carcinoma syngeneic tumor model. In addition, FN3hPD-L1 was confirmed to bind hPD-L1 in human cancer tissue specimens with known hPD-L1 expression status based on validated hPD-L1 antibodies. This indicates the potential for clinical translation of this radiotracer.
Supplementary Material
Translational relevance.
There is a pressing need for in vivo diagnostic imaging techniques that non-invasively measure expression of the immune checkpoint protein hPD-L1 in tumors. Such tracers could identify prospective patient responders to immune checkpoint blockade therapy at an earlier stage, thereby potentially improving treatment outcomes. In this article we present the development of a novel 12-kDa small protein scaffold of a fibronectin type 3 domain (FN3) binder that targets hPD-L1, which is expressed in most human tumors. We radiolabeled the binder, as 64Cu-FN3hPD-L1, for PET imaging. The radiotracer was evaluated in three groups of NSG mice, bearing hPD-L1-positive (with/without blocking) syngeneic or hPD-L1-negative xenograft tumors. Early assessment of tracer uptake revealed increased tumor-to-muscle ratios of 5.6±0.9 and 13.1±2.3 at 1 and 4 hours post-injection, respectively. In addition, FN3hPD-L1 bound hPD-L1 in human cancer tissues that were also evaluated with validated hPD-L1 antibodies, indicating the potential for clinical translation of this radiotracer.
Acknowledgements
We would like to thank the Ben and Catherine Ivy Foundation (SSG), National Cancer Institute (P50CA114747, SSG), Stanford Cancer Institute Fellowship for Cancer Research (CBP), and American Brain Tumor Association Basic Research Fellowship supported by the Ryan J. Hanrahan Memorial (CBP) for their support in funding this research. We acknowledge the support of Dr. Timothy Doyle for the small animal PET/CT imaging performed at the Stanford Center for Innovative In Vivo Imaging (SCi3), Ms. Pauline Chu for tissue processing and H&E staining, Ms. Ellen Gomulia for immunohistochemistry, and Ms. Karen Eliahu for protein purification.
Financial support
We would like to thank the Ben and Catherine Ivy Foundation (SSG), National Cancer Institute (P50CA114747, SSG), Stanford Cancer Institute Fellowship for Cancer Research (CBP), and American Brain Tumor Association Basic Research Fellowship supported by the Ryan J. Hanrahan Memorial (CBP) for their support in funding this research.
Footnotes
Conflict of interest
The authors declare no potential conflicts of interest with respect to the work presented in this manuscript.
REFERENCES
- 1.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013;39(1):1–10. [DOI] [PubMed] [Google Scholar]
- 2.Kuchroo VK, Das MP, Brown JA, Ranger AM, Zamvil SS, Sobel RA, et al. B7–1 and B7–2 costimulatory molecules activate differentially the Th1/Th2 developmental pathways: application to autoimmune disease therapy. Cell 1995;80(5):707–18. [DOI] [PubMed] [Google Scholar]
- 3.Alsaab HO, Sau S, Alzhrani R, Tatiparti K, Bhise K, Kashaw SK, et al. PD-1 and PD-L1 Checkpoint Signaling Inhibition for Cancer Immunotherapy: Mechanism, Combinations, and Clinical Outcome. Front Pharmacol 2017;8:561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014;515(7528):563–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swaika A, Hammond WA, Joseph RW. Current state of anti-PD-L1 and anti-PD-1 agents in cancer therapy. Mol Immunol 2015;67(2 Pt A):4–17. [DOI] [PubMed] [Google Scholar]
- 6.Straub M, Drecoll E, Pfarr N, Weichert W, Langer R, Hapfelmeier A, et al. CD274/PD-L1 gene amplification and PD-L1 protein expression are common events in squamous cell carcinoma of the oral cavity. Oncotarget 2016;7(11):12024–34 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cimino-Mathews A, Thompson E, Taube JM, Ye X, Lu Y, Meeker A, et al. PD-L1 (B7-H1) expression and the immune tumor microenvironment in primary and metastatic breast carcinomas. Hum Pathol 2016;47(1):52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyford-Pike S, Peng S, Young GD, Taube JM, Westra WH, Akpeng B, et al. Evidence for a role of the PD-1:PD-L1 pathway in immune resistance of HPV-associated head and neck squamous cell carcinoma. Cancer Res 2013;73(6):1733–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med 2012;4(127):127ra37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green MR, Monti S, Rodig SJ, Juszczynski P, Currie T, O’Donnell E, et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood 2010;116(17):3268–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 2015;5(8):860–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med 2002;8(8):793–800. [DOI] [PubMed] [Google Scholar]
- 13.Sakane T, Murase T, Okuda K, Takino H, Masaki A, Oda R, et al. A comparative study of PD-L1 immunohistochemical assays with four reliable antibodies in thymic carcinoma. Oncotarget 2018;9(6):6993–7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ancevski Hunter K, Socinski MA, Villaruz LC. PD-L1 Testing in Guiding Patient Selection for PD-1/PD-L1 Inhibitor Therapy in Lung Cancer. Mol Diagn Ther 2018;22(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Passiglia F, Bronte G, Bazan V, Natoli C, Rizzo S, Galvano A, et al. PD-L1 expression as predictive biomarker in patients with NSCLC: a pooled analysis. Oncotarget 2016;7(15):19738–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel SP, Kurzrock R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol Cancer Ther 2015;14(4):847–56. [DOI] [PubMed] [Google Scholar]
- 17.Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer 2016;16(5):275–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chatterjee S, Lesniak WG, Gabrielson M, Lisok A, Wharram B, Sysa-Shah P, et al. A humanized antibody for imaging immune checkpoint ligand PD-L1 expression in tumors. Oncotarget 2016;7(9):10215–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maute RL, Gordon SR, Mayer AT, McCracken MN, Natarajan A, Ring NG, et al. Engineering high-affinity PD-1 variants for optimized immunotherapy and immuno-PET imaging. Proc Natl Acad Sci U S A 2015;112(47):E6506–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heskamp S, Hobo W, Molkenboer-Kuenen JD, Olive D, Oyen WJ, Dolstra H, et al. Noninvasive Imaging of Tumor PD-L1 Expression Using Radiolabeled Anti-PD-L1 Antibodies. Cancer Res 2015;75(14):2928–36. [DOI] [PubMed] [Google Scholar]
- 21.Miao Z, Ren G, Liu H, Jiang L, Cheng Z. Small-animal PET imaging of human epidermal growth factor receptor positive tumor with a 64Cu labeled affibody protein. Bioconjug Chem 2010;21(5):947–54. [DOI] [PubMed] [Google Scholar]
- 22.Koide A, Gilbreth RN, Esaki K, Tereshko V, Koide S. High-affinity single-domain binding proteins with a binary-code interface. Proc Natl Acad Sci U S A 2007;104(16):6632–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donnelly DJ, Smith RA, Morin P, Lipovsek D, Gokemeijer J, Cohen D, et al. Synthesis and Biologic Evaluation of a Novel (18)F-Labeled Adnectin as a PET Radioligand for Imaging PD-L1 Expression. J Nucl Med 2018;59(3):529–35. [DOI] [PubMed] [Google Scholar]
- 24.Hackel BJ, Kimura RH, Gambhir SS. Use of (64)Cu-labeled fibronectin domain with EGFR-overexpressing tumor xenograft: molecular imaging. Radiology 2012;263(1):179–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tolcher AW, Sweeney CJ, Papadopoulos K, Patnaik A, Chiorean EG, Mita AC, et al. Phase I and pharmacokinetic study of CT-322 (BMS-844203), a targeted Adnectin inhibitor of VEGFR-2 based on a domain of human fibronectin. Clin Cancer Res 2011;17(2):363–71. [DOI] [PubMed] [Google Scholar]
- 26.Hackel BJ, Ackerman ME, Howland SW, Wittrup KD. Stability and CDR composition biases enrich binder functionality landscapes. J Mol Biol 2010;401(1):84–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lipovsek D Adnectins: engineered target-binding protein therapeutics. Protein Eng Des Sel 2011;24(1–2):3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tolmachev V, Rosik D, Wallberg H, Sjoberg A, Sandstrom M, Hansson M, et al. Imaging of EGFR expression in murine xenografts using site-specifically labelled anti-EGFR 111In-DOTA-Z EGFR:2377 Affibody molecule: aspect of the injected tracer amount. Eur J Nucl Med Mol Imaging 2010;37(3):613–22. [DOI] [PubMed] [Google Scholar]
- 29.Kimura RH, Cheng Z, Gambhir SS, Cochran JR. Engineered knottin peptides: a new class of agents for imaging integrin expression in living subjects. Cancer Res 2009;69(6):2435–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gainkam LO, Huang L, Caveliers V, Keyaerts M, Hernot S, Vaneycken I, et al. Comparison of the biodistribution and tumor targeting of two 99mTc-labeled anti-EGFR nanobodies in mice, using pinhole SPECT/micro-CT. J Nucl Med 2008;49(5):788–95. [DOI] [PubMed] [Google Scholar]
- 31.Huang L, Gainkam LO, Caveliers V, Vanhove C, Keyaerts M, De Baetselier P, et al. SPECT imaging with 99mTc-labeled EGFR-specific nanobody for in vivo monitoring of EGFR expression. Mol Imaging Biol 2008;10(3):167–75. [DOI] [PubMed] [Google Scholar]
- 32.Chen X, Hou Y, Tohme M, Park R, Khankaldyyan V, Gonzales-Gomez I, et al. Pegylated Arg-Gly-Asp peptide: 64Cu labeling and PET imaging of brain tumor alphavbeta3-integrin expression. J Nucl Med 2004;45(10):1776–83. [PubMed] [Google Scholar]
- 33.Chatterjee S, Lesniak WG, Miller MS, Lisok A, Sikorska E, Wharram B, et al. Rapid PD-L1 detection in tumors with PET using a highly specific peptide. Biochem Biophys Res Commun 2017;483(1):258–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xenaki KT, Oliveira S, van Bergen En Henegouwen PMP. Antibody or Antibody Fragments: Implications for Molecular Imaging and Targeted Therapy of Solid Tumors. Front Immunol 2017;8:1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D’Huyvetter M, De Vos J, Xavier C, Pruszynski M, Sterckx YGJ, Massa S, et al. 131I-labeled Anti-HER2 Camelid sdAb as a Theranostic Tool in Cancer Treatment. Clin Cancer Res 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vu NX, Pruksametanan N, Srila W, Yuttavanichakul W, Teamtisong K, Teaumroong N, et al. Generation of a rabbit single-chain fragment variable (scFv) antibody for specific detection of Bradyrhizobium sp. DOA9 in both free-living and bacteroid forms. PLoS One 2017;12(6):e0179983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan F, Dellian M, Fukumura D, Leunig M, Berk DA, Torchilin VP, et al. Vascular permeability in a human tumor xenograft: molecular size dependence and cutoff size. Cancer Res 1995;55(17):3752–6. [PubMed] [Google Scholar]
- 38.Schmidt MM, Wittrup KD. A modeling analysis of the effects of molecular size and binding affinity on tumor targeting. Mol Cancer Ther 2009;8(10):2861–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zahnd C, Kawe M, Stumpp MT, de Pasquale C, Tamaskovic R, Nagy-Davidescu G, et al. Efficient tumor targeting with high-affinity designed ankyrin repeat proteins: effects of affinity and molecular size. Cancer Res 2010;70(4):1595–605. [DOI] [PubMed] [Google Scholar]
- 40.Hackel BJ, Kapila A, Wittrup KD. Picomolar affinity fibronectin domains engineered utilizing loop length diversity, recursive mutagenesis, and loop shuffling. J Mol Biol 2008;381(5):1238–52 doi 10.1016/j.jmb.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Natarajan A, Hackel BJ, Gambhir SS. A novel engineered anti-CD20 tracer enables early time PET imaging in a humanized transgenic mouse model of B-cell non-Hodgkins lymphoma. Clin Cancer Res 2013;19(24):6820–9. [DOI] [PubMed] [Google Scholar]
- 42.Natarajan A, Gowrishankar G, Nielsen CH, Wang S, Iagaru A, Goris ML, et al. Positron emission tomography of 64Cu-DOTA-Rituximab in a transgenic mouse model expressing human CD20 for clinical translation to image NHL. Mol Imaging Biol 2012;14(5):608–16. [DOI] [PubMed] [Google Scholar]
- 43.Warram JM, de Boer E, Sorace AG, Chung TK, Kim H, Pleijhuis RG, et al. Antibody-based imaging strategies for cancer. Cancer Metastasis Rev 2014;33(2–3):809–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olafsen T, Betting D, Kenanova VE, Salazar FB, Clarke P, Said J, et al. Recombinant anti-CD20 antibody fragments for small-animal PET imaging of B-cell lymphomas. J Nucl Med 2009;50(9):1500–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wadas TJ, Wong EH, Weisman GR, Anderson CJ. Copper chelation chemistry and its role in copper radiopharmaceuticals. Curr Pharm Des 2007;13(1):3–16. [DOI] [PubMed] [Google Scholar]
- 46.Goux M, Becker G, Gorre H, Dammicco S, Desselle A, Egrise D, et al. Nanofitin as a New Molecular-Imaging Agent for the Diagnosis of Epidermal Growth Factor Receptor Over-Expressing Tumors. Bioconjug Chem 2017;28(9):2361–71. [DOI] [PubMed] [Google Scholar]
- 47.Jekunen A, Kairemo K, Karnani P. In vivo modulators of antibody kinetics. Acta Oncol 1996;35(3):267–71. [DOI] [PubMed] [Google Scholar]
- 48.Bakir MA, Eccles S, Babich JW, Aftab N, Styles J, Dean CJ, et al. c-erbB2 protein overexpression in breast cancer as a target for PET using iodine-124-labeled monoclonal antibodies. J Nucl Med 1992;33(12):2154–60. [PubMed] [Google Scholar]
- 49.Lesniak WG, Chatterjee S, Gabrielson M, Lisok A, Wharram B, Pomper MG, et al. PD-L1 Detection in Tumors Using [(64)Cu]Atezolizumab with PET. Bioconjug Chem 2016;27(9):2103–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu H, Boyle TA, Zhou C, Rimm DL, Hirsch FR. PD-L1 Expression in Lung Cancer. J Thorac Oncol 2016;11(7):964–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.