Abstract
The apoptosis repressor with caspase recruitment domain (ARC) protein is a strong independent adverse prognostic marker in acute myeloid leukemia (AML). We previously reported that ARC regulates leukemia-microenvironment interactions through the NFkB/IL1β signaling network. Malignant cells have been reported to release IL1β, which induces PGE2 synthesis in mesenchymal stromal cells (MSC), in turn activating β-catenin signaling and inducing the cancer stem cell phenotype. Although Cox-2 and its enzymatic product PGE2 play major roles in inflammation and cancer, the regulation and role of PGE2 in AML are largely unknown. Here we report that AML-MSC co-cultures greatly increase Cox-2 expression in MSC and PGE2 production in an ARC/IL1β-dependent manner. PGE2 induced the expression of β-catenin, which regulated ARC and augmented chemoresistance in AML cells; inhibition of β-catenin decreased ARC and sensitized AML cells to chemotherapy. NOD/SCIDIL2RγNull-3/GM/SF mice transplanted with ARC-knockdown AML cells had significantly lower leukemia burden, lower serum levels of IL1β/PGE2, and lower tissue human ARC and β-catenin levels, prolonged survival, and increased sensitivity to chemotherapy than controls. Collectively, we present a new mechanism of action of anti-apoptotic ARC by which ARC regulates PGE2 production in the tumor microenvironment and microenvironment-mediated chemoresistance in AML.
Keywords: ARC, IL1β, PGE2, β-catenin, AML microenvironment
Introduction
Acute myeloid leukemia (AML) is a highly heterogeneous hematological malignancy with poor overall survival. Intrinsic characteristics of leukemia cells such as overexpression of anti-apoptotic proteins and activation of survival signaling pathways, extrinsic microenvironmental factors such as alteration of growth factors/cytokines, and leukemia-niche interactions that further augment the changes in leukemia cells and microenvironments all support leukemia cell and leukemia stem/progenitor cell growth, survival, and resistance to therapy. Thus, targeting the survival and resistance mechanisms that are common in AML should benefit many patients with this disease. We previously reported that the apoptosis repressor with caspase recruitment domain (ARC) protein is a strong independent, poor prognosis factor in AML. ARC protects leukemia cells from apoptosis induced by multiple agents (1, 2). We subsequently found that ARC induces IL1β expression in AML cells, increases chemokine CCL2, CCL4, and CXCL12 expression in mesenchymal stromal cells (MSCs) in an ARC/IL1β-dependent manner, and facilitates leukemia-microenvironment interactions through a NFκB/IL1β signaling network (3).
Bone marrow (BM)-derived MSCs are an essential structural and functional component of the BM microenvironment, and they are critical for hematopoiesis (4). Within the context of leukemia, MSCs play an essential role in protecting leukemia cells from chemotherapeutic agents (5). IL1β was previously shown to promote apoptosis-resistance in AML blasts (6), and the IL-1 receptor accessory protein (IL1RAP) is reportedly overexpressed in AML stem/progenitor cells (7). Furthermore, targeting IL1RAP with a neutralizing antibody selectively killed AML stem cells (8). These data support the critical role of BM IL1β signaling in AML cell survival. Furthermore, a recent study demonstrated that cancer cells release IL1β that strongly induces Cox-2/PGE2 in MSCs, which in turn activates β-catenin signaling and induces a stem cell phenotype in epithelial cancer cells (9).
The role of Cox-2 and its enzymatic product PGE2 in inflammation and cancer are well documented. Cox-2-catalyzed prostaglandin synthesis is regulated by NFκB/IL1β signaling (10–12), which is frequently upregulated in cancer cells. PGE2 signals through EP1-EP4 receptors that regulate multiple signaling cascades including β-catenin (13). Cox-2 and PGE2 have been reported to enhance hematopoietic stem cell survival, self-renewal, engraftment, and homing (14). Treatment with the PGE2 analog dimethyl-PGE2 (dmPGE2) was found to upregulate CXCR4 through increasing HIF1α and enhance hematopoietic stem/progenitor cell homing and engraftment (15, 16).
Although PGE2 is known to possess potent tumor-stimulating activity (17), its role in leukemia is largely unknown. AML cell line HL-60 was reported to expresses high levels of microsomal PGE synthase 1 (mPGES-1), which is frequently induced concomitantly with Cox-2 by various pro-inflammatory stimuli and responsible for increased PGE2 production (18). mPGES-1 was undetectable in normal blood mononuclear cells and its inhibition reduced PGE2 production and viability of HL-60 cells (19). PGE2 from MSCs was shown to activate PKA signaling and antagonize p53-induced cell death (20). Furthermore, Wnt/β-catenin signaling is constitutively active in AML (21) and is required for the development of AML stem cells (22). Expression of β-catenin by AML cells portends enhanced clonogenic capacities and poor disease prognosis (23).
We therefore hypothesized that ARC exerts its role in leukemia-stromal interactions by modulating PGE2 levels. In this study, we investigated Cox-2 expression in MSCs co-cultured with AML cells in which ARC expression was genetically modified. We also investigated PGE2 levels in vitro in an AML-MSC co-culture system, in fresh BM samples from AML patients and normal controls, and in vivo in immuno-deficient mice xenografted with ARC knockdown (ARC KD) AML cells. We demonstrate that both Cox-2 expression and PGE2 generation are ARC/IL1β dependent and that ARC, regulated by β-catenin, is an integral component of an IL1β/PGE2/β-catenin circuit. Cox-2/PGE2, regulated by ARC and induced by AML-MSC co-culture contributes to MSC-mediated chemoprotection in AML.
Materials and Methods
Cells, cell culture, and cell treatments
OCI-AML3 cells, provided by Dr. M. Minden (Ontario Cancer Institute, Toronto, ON, Canada) were validated by STR DNA fingerprinting using the AmpF_STR Identifier Kit according to manufacturer’s instructions (Cat#4322288, Applied Biosystems; Foster City, CA). The STR profiles were compared to known ATCC fingerprints, and to the Cell Line Integrated Molecular Authentication database (CLIMA) version 0.1.200808 (http://bioinformatics.istge.it/clima/) (24). The STR profile was identified as unique. Mycoplasma testing was performed using the PCR Mycoplasma Detection Kit from Applied Biological Materials (Cat#G238; Richmond, BC, Canada) per manufacturer’s instructions. Authenticated and mycoplasma-free cells are stored under liquid nitrogen and are never kept in culture for >4 months. Primary samples were acquired from AML patients or normal controls after informed written consent following the institution approved protocol in accordance with Declaration of Helsinki. Patient characteristics are shown in Table 1. Mononuclear cells were isolated from primary samples by density-gradient centrifugation using Lymphocyte Separation Medium (Cat#25–072-CV, Corning; Manassas, VA). Human MSCs were isolated from BM samples obtained from healthy subjects as described previously (25). Cell lines were cultured in RPMI-1640 medium and cells from primary samples and MSCs in α-MEM medium, both supplemented with 10% heat-inactivated fetal calf serum, 2 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were kept at 37°C in a humidified atmosphere of 5% CO2. ARC KD AML cells (2) and MSCs (26) were generated as previously described. For co-culture experiments, leukemia cells were added to MSCs (4:1) that were plated the night before and cultured in α-MEM medium with supplements. AML cells, MSCs, or the co-cultured cells were treated with IL1β (Cat#200–01B) with or without IL1βRA (Cat#200–01RA) (PeproTech; Rocky Hill, NJ), dmPGE2 (16,16-dimethyl-PGE2, a PGE2 analog; Cat#14750, Cayman Chemical; Ann Arbor, MI), Ara-C with or without Cox-2 inhibitor Celecoxib (Cat#C-1502, LC Laboratories; Woburn, MA), or β-catenin inhibitor C-82 (27, 28) (provided by PRISM Pharma) with or without Ara-C.
Table 1.
Patient characteristics
| No. | % blast | source | treatments and responses | cytogenetics | mutations |
|---|---|---|---|---|---|
| 1 | 76 | PB | Untreated | 46,XY[20] | NPM1, NRAS1, TP53, STAG2, DNMT3A, FLT3-ITD |
| 2 | 58 | PB | Untreated | complex | ASXL1, ETNK1, RUNX1, SETBP1, WT1, FLT3-ITD |
| 3 | 85 | BM | Treated with multiple agents | complex | FLT3-ITD/D835, NA for other mutations |
| 4 | 90 | PB | Refractory/resistance AML | 46,XY[20] | RUNX1, WT1, FLT3-ITD/D835 |
| 5 | 63 | PB | AML, progressed from MPN. Treated with decitabine | complex | NOTCH1, JAK, TP53 |
| 6 | 54 | PB | Refractory AML | complex | RUNX1, TP53 |
| 7 | 73 | PB | Relapsed AML. Resistance to multiple agents | complex | NRAS, TP53, FLT3-D835 |
| 8 | 74 | PB | Relapsed AML | NA | NA |
| 9 | 52 | BM | Untreated | 46,XX[20] | IDH1, WT1, DMNT3A |
| 10 | 55 | BM | Treated with 2016-1084 BAY and Hydroxyurea | 47,XY,+8[8]/46,XY[12] | ASXL1, CREBBP, DNMT3A, IDH1, JAK2, U2AF1, WT1 |
| 11 | 88 | BM | Treated with chemotherapy | complex | PTPN11, CALR, CBL, FLT3-ITD/D835, WT1 STAT5A |
| 12 | 83 | BM | Treated with Hydroxyurea | 46,XY[20] | TET2, NRAS, DNMT3A, U2AF |
| 13 | 68 | BM | Treated with chemotherapy | complex | FLT3-D835, SH2B3, STAG2 |
| 14 | 81 | BM | Treated with Hydroxyurea and Cytarabine | complex | CREBBP |
| 15 | 78 | BM | Treated with Hydroxyurea | complex | ASXL1, BRAF, DNMT3A, FLT3-D835, NRAS, RUNX1, TET2 |
| 16 | 61 | BM | Treated with Hydroxyurea | 46,XX[20] | IDH2, NPM1 FLT3-ITD |
| 17 | 70 | BM | Newly diagnosed | complex | JAK2, MPL, WT1 |
| 18 | 81 | BM | Transformed from MDS to AML. Resistance to multiple therapies | complex | FLT3-ITD |
| 19 | 40 | BM | Newly diagnosed | complex | IDH1, TP53 |
| 20 | 85 | BM | Relapsed AML | 48,XY,+13,+13[20] | RUNX1, ASXL1 |
| 21 | 96 | PB | Transformed from MDS to AML. Resistance to multiple therapies | 46,XX,del(7)(q22q34)[20] | TET2, WT1 |
| 22 | 87 | PB | Relapsed/refractory AML | 46,XY[20] | NPM1, MPL, NRAS, IDH2 FLT3-ITD |
| 23 | 88 | PB | Refractory AML | complex | NA |
| 24 | 86 | PB | Newly diagnosed | Pseudodiploid clone 46,XY,t(9;11)(p22;q23)[20] | NA |
| 25 | 93 | PB | Relapsed/refractory AML | complex | FLT3-ITD, NA for other mutations |
| 26 | 85 | PB | Relapsed/refractory AML | complex | NA |
| 27 | 94 | PB | Newly diagnosed | 46,XX[20] | FLT3-D835, NA for other mutations |
NA, not available; WT, wild type.
Determination of PGE2 and human IL1β
PGE2 in the supernatant of cultured cells, mouse serum, and human BM samples was determined by PGE2 ELISA (Cat#KGE004B), and human IL1β in mouse serum and human BM samples was determined by ELISA specific for human IL1β (Cat#DLB50) (both from R&D Systems; Minneapolis, MN) following the manufacturer’s instructions.
Silencing of β-catenin expression
OCI-AML3 cells were electroporated with a scramble control or the SMARTpool ON-TARGET plus CTNNB1 siRNAs (750 nM) (Cat#L-003482–00-0005, Dharmacon; Lafayette, CO) using an Amaxa apparatus (Solution T, Cat#VCA-1002; program X-001) (Lonza; Walkersville, MD) following the manufacturer’s instructions as previously described (28).
RNA isolation and RT-PCR
RNA isolation and Taqman RT-PCR were carried out as previously described (29). Primers sets used are: β-catenin (Hs00355049_m1), ARC (Hs00358724_g1), and ABL (Hs01104728_m1) (all from ThermoFisher Scientific; Waltham, MA). The abundance of each transcript relative to that of ABL was calculated using the 2−ΔCt method, where ΔCt is the mean Ct of the transcript of interest minus the mean Ct of the transcript for ABL housekeeping gene.
ARC promoter-GFP reporter constructs and lentiviral transduction
The promoter region of the ARC coding gene NOL3, identified using the Eukaryotic Primer Database (https://epd.vital-it.ch/index.php) (residues 67173446–67174045, RefSeq NC_000016.10, human chromosome 16, GRCh38.p7) has two LEF1/TCF4E motifs (CTTTGTGC and GGCCAAAG) by searching PROMO 3.0 (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3). We amplified by PCR and inserted this region between the Cla I and Nhe I sites in the lentivector pCDH-CMV-MSC-EF1a-Puro (System Biosciences; Palo Alto, CA), to replace the CMV promoter. We then inserted the open reading for copGFP between the Nhe I and BamH1 sites such that the start codon for the GFP moiety was in frame with the NOL3 start codon. We mutated the LEF1/TCF4E motifs using a QuikChange II XL site-directed mutagenesis kit as directed by the manufacturer (Agilent; Santa Clara, CA), except that we used NEBStable cells (New England Biolabs; Ipswich, MA) in lieu of XL10-Gold cells. Mutants were identified by the presence of both Pml I and Sal I sites. Primers used for these constructions are listed in supplemental Table 1. Lentivirus was prepared by transfecting HEK293T cells (ATCC; Manassas, VA) with an equimolar mix of reporter vector and packaging plasmids psPAX2 and pMD2.G (gifts of Didier Trono, Addgene, Cambridge, MA) using JetPrime transfection reagent as directed by the manufacturer (Polyplus, Illkirch, France). OCI-AML3 cells were transduced with the lentivirus as described (2).
Western blot analysis
Protein levels were determined by western blot as described previously (3) using the Odyssey Infrared Imaging System for signal detection and Odyssey software version 3.0 for quantification (LI-COR Biosciences; Lincoln, NE). Cytoplasmic and nuclear fractions were prepared as previously described (30). Antibodies against β-catenin (Cat#8480) and ARC (Cat#NBP2–41753) were purchased from Cell Signaling Technology (Danvers, MA) and Novus (Littleton, CO), respectively. Histone H3 was used as loading control for nuclear fraction, α-tubulin for cytoplasm, and β-actin for total lysate.
Protein determination by flow cytometry
After staining with Ghost Dye™ Violet 510 (Cat#13–0870-T500, Tonbo Biosciences; San Diego, CA), cells were washed and fixed with 4% paraformaldehyde and permeabilized with 100% methanol, and then stained with Fc-blocker (Cat#130–059-901, Miltenyi Biotec; San Diego, CA), followed with Cox-2-PE (Cat#12282, 1:50, Cell Signaling Technology), CD90-PerCP/CyC5.5 (Cat#328118), and CD45-Pacific Blue (Cat#304029) (Biolegend; San Diego, CA) in 5% BSA/PBS. The stained cells were analyzed using a Gallios flow cytometer (Beckman Coulter Life Sciences; Indianapolis, IN) and quantified using FlowJo analytic platform (BD Biosciences; San Jose, CA). Cox-2 levels in MSCs or AML cells were expressed as the geometric mean fluorescent intensity (MFI) difference of cells stained with Cox-2 antibody or with IgG in CD90+ or CD45+ cells, respectively.
CyTOF analysis
AML patient samples were stained with a panel of metal-tagged antibodies for cell surface markers and intracellular proteins (Supplemental Table 2) and subjected to CyTOF analysis as previously described (3, 28, 31). Viable (cisplatin low) single cells were gated with FlowJo software (v10, FlowJo LLC) and exported as flow cytometry standard (FCS) data for subsequent analysis in Cytofkit (32). RPhenoGraph was used for unsupervised subset detection based on cell surface markers. t-SNE embedded FCS files were further analyzed in FlowJo and cell populations identified by RPhenoGraph were mimicked and gated on the t-SNE map for quantitation of intracellular marker expression. The expression level of each protein in the desired cell compartments is expressed as ArcSinh-transformed data.
Apoptosis assay
Apoptosis was estimated in CD45+ cells by flow cytometry after annexin V–Cy5.5 staining in the presence of 7-amino-actinomycin D (7-AAD) using LSRII flow cytometry (BD Biosciences). Apoptosis in primary samples was assessed in CD45+ and CD34+CD38− cells after the cells were incubated with CD34-PE, CD38-PECy7, and CD45-APCH7 antibodies (BD Biosciences) and expressed as specific apoptosis:
In vivo study
NOD/SCIDIL2RγNull-3/GM/SF (NSGS) mice (male, 3 months old) were injected with either vector control or ARC KD OCI-AML3 cells (1×106/mouse). A week later, control or ARC KD cell-injected mice (10/group) were either untreated or treated with Ara-C (100 mg/kg) via intraperitoneal injection, 3x/week for approximately 5 and half weeks. Leukemia burden was assessed by flow cytometric measurement of human CD45+ (huCD45+) cells in mouse periphery blood (PB), BM, and spleen. Serum IL1β and PGE2 levels were determined. Tissue sizes were measured and protein expression in mouse tissues was determined by Opal/TSA (tyramide signal amplification) multispectral immunohistochemistry (IHC) and Vectra imaging (see below). Mouse survival was followed. All the mouse experiments were carried out following protocols approved by the Institution Animal Care and Use Committee at MD Anderson Cancer Center.
Protein determination by Opal/TSA multiplex IHC and Vectra multispectral imaging
Paraffin-embedded mouse tissue slides were deparaffinized and rehydrated followed by antigen retrieval with Biogenex Laboratories Antigen Citra RTUSE (Cat#NC0359148, Fisher Scientific; Hampton, NH) at 95°C for 15 min and blocked with 3% BSA in TBS-T for additional 15 min. Tissues were incubated with a primary antibody followed by HRP conjugated IgG (broad spectrum) provided in SuperPicture™ polymer detection kit (Cat#878963, ThermoFisher Scientific) for 15 min at room temperature and then labeled with TSA plus reagents following the manufacturer’s protocols (Perkin Elmer; Waltham, MA). The above steps were repeated sequentially for staining each of the following proteins: ARC, CD45 and β-catenin all specific for human. For ARC detection, tissues were stained overnight at 4°C with an anti-ARC antibody (1:4000, Cat#38916S, Cell Signaling Technology) and for β-catenin detection, they were incubated 1 h at room temperature with an anti-β-catenin antibody (1:1000; Cat#LS-B4123, LifeSpan BioSciences Inc.; Seattle, WA). For huCD45 detection, tissues were stained 2 h at room temperature with an anti-huCD45 antibody (1:2000; Cat#M0701, Agilent Dako; Santa Clara, CA). Opal fluorescein reagents used for TSA were Opal 540 (Cat#FP1494001KT), Opal 570 (Cat#FP1488001KT), Opal 650 (Cat#FP1296001KT) (Perkin Elmer) for CD45, ARC, and β-catenin; respectively. Opal fluorescent reagent was diluted with 1× plus amplification diluent (1:150) (Cat#FP1498, Perkin Elmer). Each issue was incubated with 200 μl diluted Opal reagent at room temperature for 5 min and washed with TBS-T twice, 3 min each. Finally, the tissue slides were counterstained with 2.5 µg/ml DAPI (Cat#D21490, ThermoFisher Scientific) at room temperature for 5 min, sealed by anti-fade fluorescent mounting medium (Cat#S3023, Dako), and stored at 4°C.
Multispectral images were acquired using Vectra 3.0 automated quantitative pathology imaging system (CLS142338, Perkin Elmer) with ×20 objective and software version 3.012 (Perkin Elmer). Images were analyzed by InForm version 2.3 software (Perkin Elmer). Briefly, a fluorophore library was created by mono-spectral labelled tissue images to define spectral curves for each fluorophore and counterstain to adjust background noise and positive staining of biomarkers. InForm software provides objective counting of cell population and biomarkers, allows tissue separation by machine training and increases the accuracy of the statistical analysis. Algorithms were created for separating tumor tissues and normal tissues and for evaluating various protein levels via specific antibodies. For protein quantification, huCD45+ cells were selected from multiple fields and multiple cells in each field. The scan fields for spleens were selected from the central region of the tissue section to maximally cover the whole spleen avoiding the margins, and the fields for BM were selected from the whole marrow compartments away from bone fragments. The expression level is expressed as mean optical density (OD)/huCD45+ cell in each field.
Statistical analyses
Experiments were conducted in triplicate unless otherwise indicated. One way ANOVA or Student’s t test was performed to compare the differences between groups. Results were expressed as the mean±SEM. Correlation coefficient was determined by Pearson correlation analysis. The combination index (CI), expressed as the mean of CI values at the 50%, 75%, and 90% effective doses was determined by the Chou-Talalay method (33). CI<1.0 was considered synergistic; =1.0 additive; and >1.0 antagonistic. Mouse survival was estimated by the Kaplan-Meier method, and analyzed using log-rank statistics. Statistical significance was set at P < 0.05.
Results
AML-MSC co-cultures increase Cox-2 expression in MSCs in an IL1β- and ARC-dependent manner
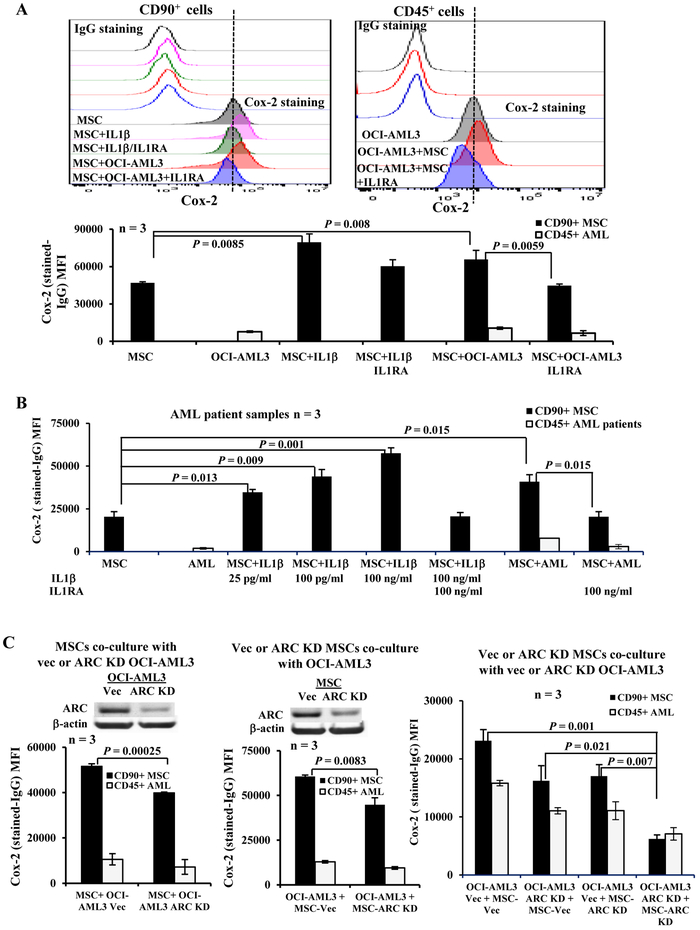
MSCs were treated with IL1β or co-cultured with OCI-AML3 cells in the absence or presence of IL1R antagonist IL1RA (48 h). Cox-2 protein levels were determined by flow cytometry in both MSCs (CD90+) and OCI-AML3 cells (CD45+). As expected, IL1β greatly induced the expression of Cox-2 in MSCs, which was suppressed by IL1RA. Co-cultures of MSCs with OCI-AML3 cells also significantly increased Cox-2 expression in MSCs, and similarly this induction was inhibited by co-treatment of cells with IL1RA (Figure 1A) suggesting that increased Cox-2 expression in MSCs co-cultured with AML cells is mediated through IL1β. By comparison, OCI-AML3 cells expressed lower levels of Cox-2 than MSCs, and co-culture of AML cells with MSCs also increased Cox-2 expression in AML cells in an IL1β-dependent manner, but to a lesser degree (Figure 1A). Importantly, similar results were obtained when primary AML patient samples (Table 1, samples 1–3) were used (Figure 1B) and IL1β, even at 25 pg/ml level, was sufficient to significantly increase Cox-2 in MSCs (P = 0.013).
Figure 1.
AML-MSC co-cultures increase Cox-2 in MSCs in an IL1β- and ARC-dependent manner. A. Cox-2 expression in MSCs (CD90+) and OCI-AML3 (CD45+) cells cultured alone or co-cultures, without or with IL1β (100 ng/ml) and/or ILIRA (100 ng/ml) treatments for 48 h. The upper panel is the histogram of one representative experiment and the lower panel is the results of 3 independent experiments with 3 different MSCs. B. Cox-2 expression in MSCs (CD90+) and primary patient samples (CD45+) (n = 3, Table 1, samples 1-3) cultured alone or co-cultures, without or with IL1β (25 pg/ml, 100 pg/ml, or 100 ng/ml) and/or ILIRA (100 ng/ml) treatments for 48 h. The same MSC was used for all three co-cultures. C. Cox-2 expression in MSC and OCI-AML3 cells by flow cytometry under MSCs co-cultured with control or ARC KD OCI-AML3 (left), control or ARC KD MSCs co-cultured with OCI-AML3 (center), or control or ARC KD MSCs co-cultured with control or ARC KD OCI-AML3 (right) for 48 h. Vec, vector control. Three independent experiments were performed.
We next co-cultured MSCs with vector control or ARC KD OCI-AML3 cells, vector control or ARC KD MSCs with OCI-AML3 cells, or vector control or ARC KD MSCs with vector control or ARC KD OCI-AML3 cells (48 h). We found that significantly less Cox-2 protein was induced in MSCs when ARC was knocked down either in AML cells (P = 0.00025) or MSCs (P = 0.0083) compared to the respective controls and that ARC KD in both MSCs and AML cells resulted in the lowest level of Cox-2 expression (P = 0.007, 0.021, or 0.001, vs ARC KD in MSCs, ARC KD in AML cells, or vector control; respectively) (Figure 1C) indicating that Cox-2 expression depends on ARC levels, which is in agreement with our previous finding that ARC regulates NFκB/IL1β signaling (3).
AML-MSC co-cultures induce PGE2 in an IL1β- and ARC-dependent manner
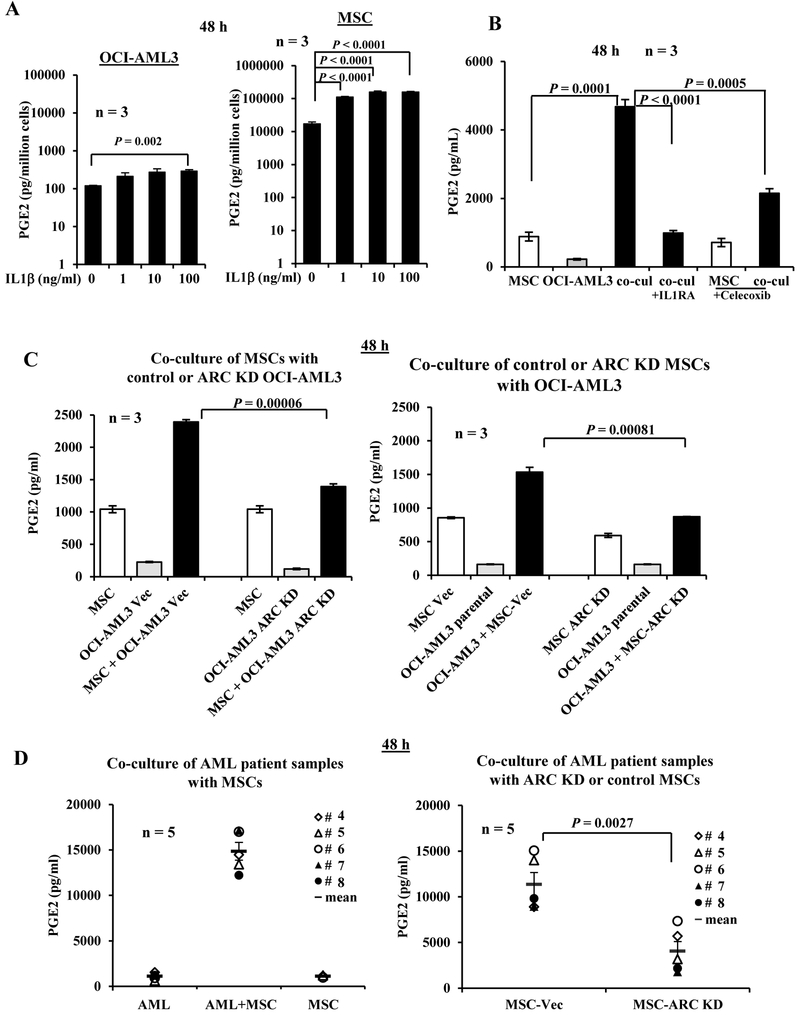
As shown in Figure 1, although Cox-2 is expressed in OCI-AML3 cells and primary patient samples, AML cells express relatively lower Cox-2 than MSCs. We next determined PGE2 levels in the culture media of OCI-AML3 and MSCs in the absence or presence of IL1β (48 h) and found that compared to OCI-AML3 cells, MSCs secreted >100-fold PGE2/cell. IL1β, at 1 to 100 ng/ml, significantly induced PGE2 production in MSCs (6.5- to 9.2-fold; P < 0.0001), while at only 100 ng/ml, IL1β significantly induced PGE2 production in OCI-AML3 cells (2.5-fold; P = 0.002) (Figure 2A) indicating that, consistent with Cox-2 expression, MSCs are the major source of PGE2.
Figure 2.
AML-MSC co-cultures increase secreted PGE2 in an IL1β- and ARC-dependent manner. A. OCI-AML3 cells or MSCs were cultured in the absence or presence of IL1β (1 to 100 ng/ml) for 48 h. B. OCI-AML3 and MSCs were cultured alone or co-cultured (co-cul) without or with IL1RA (100 ng/ml) or Celecoxib (200 nM) for 48 h. C. MSCs were co-cultured with vector control or ARC KD OCI-AML3 (left) and vector control or ARC KD MSC were co-cultured with OCI-AML3 cells (right) for 48 h. D. Cells from AML patient samples (n = 5, Table 1, samples 4-8) were co-cultured with MSCs (left) and with ARC KD or control MSCs (right) for 48 h. PGE2 levels in the supernatant were determined by ELISA. Vec, vector. Cell line experiments were done in triplicates. Three MSCs were used for A and B.
We then co-cultured MSCs and OCI-AML3 cells for 48 h and found markedly increased PGE2 levels in the medium of co-cultured cells compared to MSCs or OCI-AML3 cultured alone. This increase was suppressed by IL1 receptor antagonist IL1RA or Cox-2 inhibitor Celecoxib (Figure 2B). We next co-cultured MSCs with vector control or ARC KD OCI-AML3 cells for 48 h and found that knocking down ARC in OCI-AML3 cells significantly decreased the secreted PGE2 in the co-culture system (P < 0.0001) (Figure 2C, left). In addition, we found that in co-cultured vector control or ARC KD MSCs with OCI-AML3 cells, knock-down of ARC in MSCs significantly reduced PGE2 production (P = 0.00081) (Figure 2C, right). Our data suggest that, as with Cox-2 expression, AML-MSC co-culture induces the production of PGE2 in an ARC- and IL1β-dependent manner. Finally, co-culture cells from PB samples of primary AML patients (n = 5, Table 1, samples 4–8) with MSCs (48 h) markedly increased secreted PGE2, while PGE2 levels were significantly lower when patient cells were co-cultured with ARC KD MSCs (P = 0.0027) (Figure 2D).
PGE2 treatment or co-culture with MSCs induces β-catenin and ARC and augments chemoresistance in AML cells
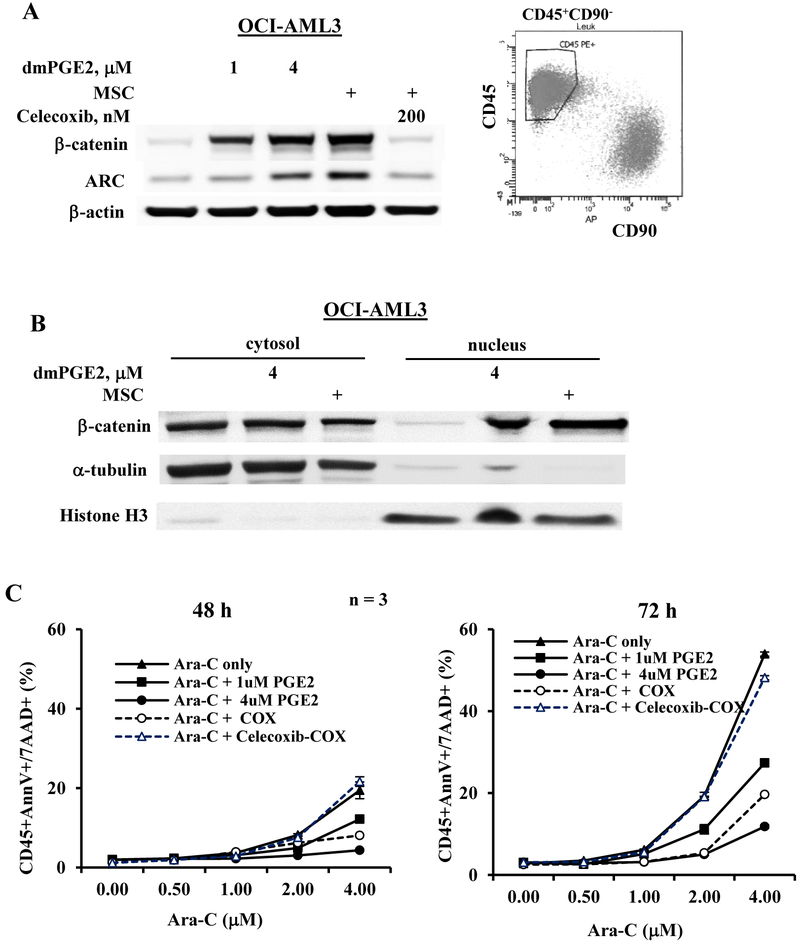
We previously showed that co-culture of AML cells with MSCs increases β-catenin (27) and ARC expression in AML (2). To demonstrate that this effect is partially mediated through PGE2 signaling, OCI-AML3 cells were treated with a PGE2 analog, dmPGE2 or co-cultured with MSCs without or with Cox-2 inhibitor Celecoxib. dmPGE2 induced protein levels of β-catenin and ARC (Figure 3A). As expected, co-culture with MSCs increased β-catenin and ARC in FACS-sorted OCI-AML3 (CD45+CD90−) cells after co-culture (Fig 3A) and this increase was completely suppressed by Celecoxib (Figure 3A). Importantly, both PGE2 and co-cultures primarily increased nuclear β-catenin in AML cells (Figure 3B). To examine the biological relevance of increased PGE2 production in the leukemia-MSC system, we treated OCI-AML3 cells with Ara-C, the commonly used chemotherapeutic agent for AML therapy with or without dmPGE2 or MSCs. As shown in Figure 3C, Ara-C induced time and dose-dependent apoptosis in OCI-AML3 cells, which was significantly suppressed by MSC co-culture and by dmPGE2 co-treatment in a dose-dependent manner. Blocking PGE2 production with Celecoxib (200 nM) significantly reduced the protective effect of MSCs. Our data indicate that increased PGE2 production in leukemia-MSC co-culture system activates β-catenin, increases ARC expression, and contributes to MSC-mediated chemoprotection.
Figure 3.
AML cells treated dmPGE2 or co-cultured with MSCs express increased β-catenin and ARC and are more resistance to chemotherapy. A. OCI-AML3 cells were treated with dmPGE2 (1 or 4 μM) or co-cultured with MSCs without or with Cox-2 inhibitor Celecoxib (200 nM) for 48 h. β-catenin and ARC protein levels were determined by western blot. For co-culture experiments, OCI-AML3 cells were FACS-sorted after co-culture and west blot was done using the lysate from the sorted CD45+CD90− cells as shown in the histogram. B. OCI-AML3 cells were treated with dmPGE2 (4 μM) or co-cultured with MSCs for 48 h. Cytosolic and nuclear β-catenin levels were determined by western blot. C. OCI-AML cells were treated with Ara-C with or without dmPGE2 (1 or 4 μM) or with MSC co-culture in the absence or presence of Cox-2 inhibitor Celecoxib (200 nM) for 48 and 72 h. Apoptosis was assessed by flow cytometry. COX, co-culture.
ARC expression in AML cells is regulated by β-catenin
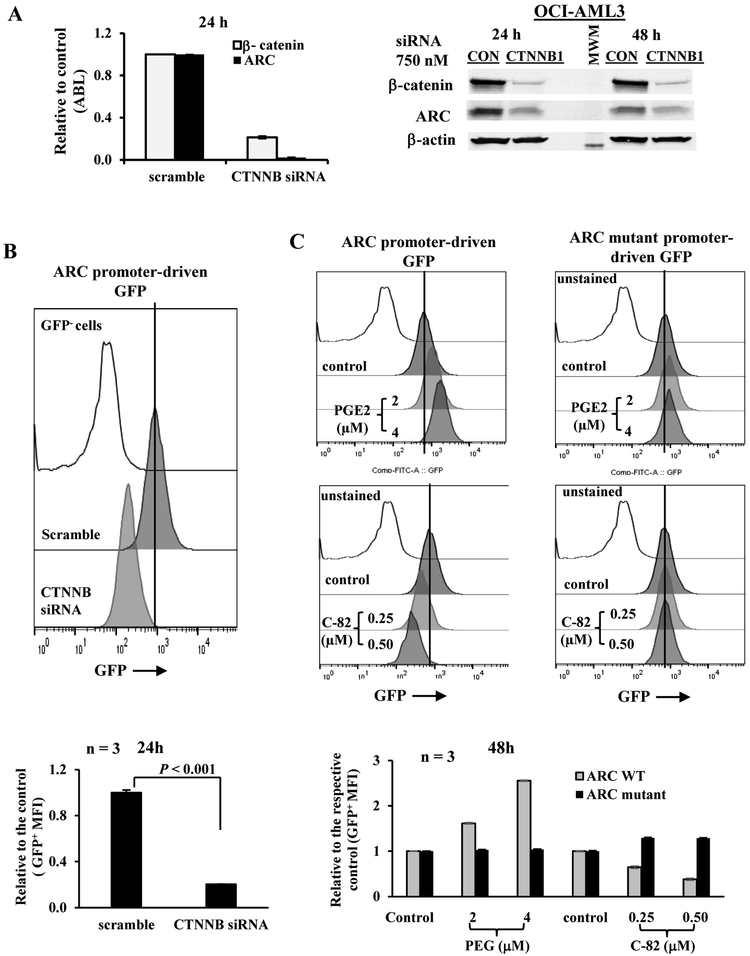
We have demonstrated that in AML-MSC co-cultures, ARC-regulates IL1β in AML cells, induces Cox-2 in MSCs, and increases secreted PGE2, which in turn induces the expression of β-catenin in AML cells. Interestingly, in addition to β-catenin, PGE2 also increased ARC level. Once activated, β-catenin is translocated to the nucleus where it interacts with DNA binding proteins T-cell factors (TCFs)/lymphoid enhancer factors (LEFs) that recognize and bind to specific sequence motifs in promoters and enhancers of target genes (34). Promoter analysis demonstrated that the ARC promoter region contains two binding sites for TCF4 (CTTTGTG and GCCAAAG)/LEF1 (CTTTGTGC and GGCCAAAG), suggesting that ARC may be regulated by β-catenin. To test that, we silenced β-catenin in AML cells by siRNAs. As shown in Figure 4A, inhibition of β-catenin suppressed ARC expression both at the RNA and protein levels, suggesting that β-catenin regulates ARC expression transcriptionally, and that the PGE2-induced ARC increase is mediated through PGE2/β-catenin signaling.
Figure 4.
β-catenin regulates ARC expression in AML cells. A. OCI-AML3 cells were transfected with a control siRNA (scramble) or smart pool siRNAs against β-catenin gene (CTNNB1 siRNA) (750 nM) by Amaxa electroporation. The RNA (24 h) and protein (24 and 48 h) levels of β-catenin and ARC were determined by RT-PCR and western blot, respectively, after transfection. CON, scramble control. MWM, molecular weight marker. B. OCI-AML3 cells transduced with ARC promoter-driven GFP reporter gene were transfected with scramble control or β-catenin siRNAs and GFP levels were determined 24 h after transfection. C. OCI-AML3 cells transduced with ARC promoter (WT)- or ARC mutant promoter-driven GFP reporter gene were treated with PGE2 or C-82 for 48 h. For B and C, histograms (top panels) are representative results from one experiment and bar grafts (lower panels) are results of triplicates. GFP levels were determined by flow cytometry. Untransduced OCI-AML3 cells were used as a negative control for GFP.
We next generated an ARC promoter-driven GFP reporter and transduced the construct into OCI-AML3 cells. Silencing β-catenin decreased the level of GFP reporter in OCI-AML3 cells (Figure 4B). We also mutated LEF1/TCF4E motifs in the ARC promoter region to generate an ARC mutant promoter-driven GFP reporter. Figure 4C shows that PGE2 markedly increased, and inhibition of β-catenin with a specific inhibitor C-82 decreased GFP levels in OCI-AML3 cells transduced with ARC promoter-driven GFP. PGE2 or C-82 had only minimal effects on GFP levels in cells transduced with ARC mutant promoter-driven GFP, further supporting that PGE2/β-catenin signaling transcriptionally regulates ARC expression.
Inhibition of ARC in AML decreases IL1β and PGE2, exhibits anti-leukemia activity, and sensitizes AML cells to chemotherapy in vivo in a human xenograft NSGS model
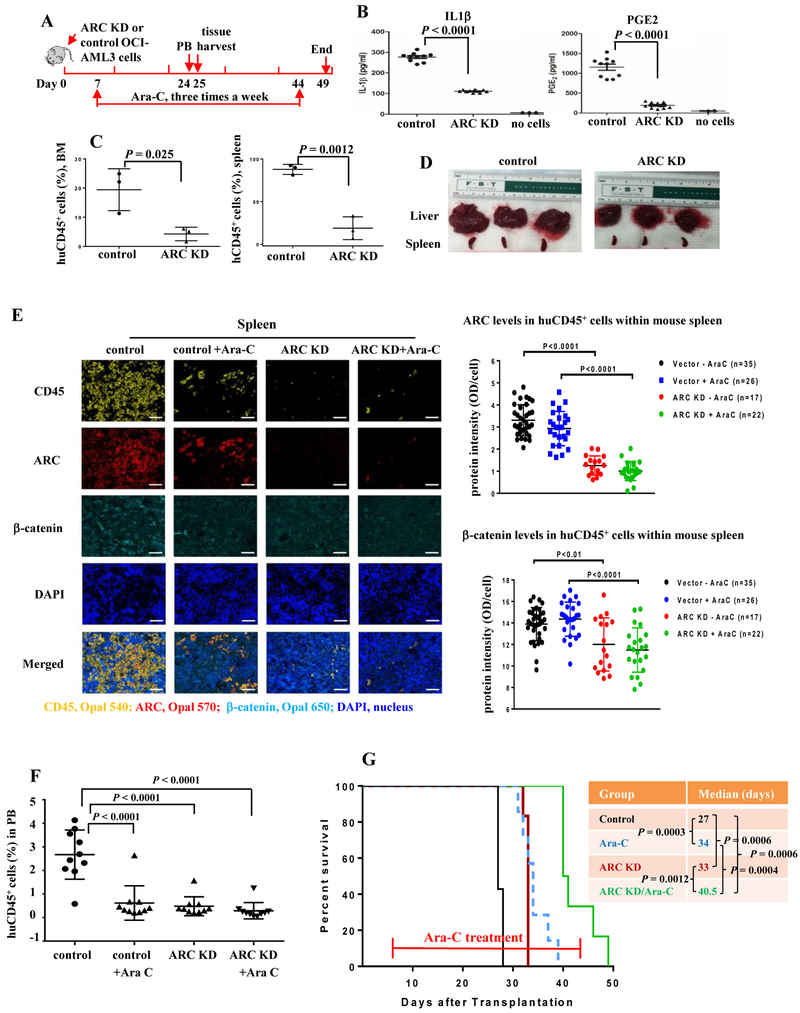
To assess the biological relevance of ARC/IL1β/PGE2/β-catenin signaling in AML in vivo, we injected NSGS mice with vector control or ARC KD OCI-AML3 cells. Mice were left untreated or treated with Ara-C and PB and tissues were collected 24 or 25 d after leukemia cell injection (Figure 5A). We first determined the levels of IL1β and PGE2 in mouse serum by ELISA and found a statistically significant decrease in serum IL1β and PGE2 in mice transplanted with ARC KD compared to control AML cells (Figure 5B). In addition, mice with ARC KD AML cells had lower leukemia burden in all tissues examined. Specifically, significantly less huCD45+ cells were found in mouse BMs and spleens (Figure 5C), along with markedly reduced liver and spleen size (Figure 5D) compared to mice injected with control cells.
Figure 5.
Inhibition of ARC in AML cells reduces serum IL1β and PGE2 levels, decreases leukemia burden in various tissues, prolongs survivals, and sensitizes to Ara-C in NSGS mice. A. In vivo experiment scheme. B. IL1β and PGE2 levels in serum of mice harboring control or ARC KD OCI-AML3 cells determined by ELISA. C. huCD45 positivity in BM and spleen of mice harboring control or ARC KD OCI-AML3 cells determined by flow cytometry. D. Liver and spleen from mice harboring control or ARC KD OCI-AML3 cells. E. Expression of ARC and β-catenin in spleen huCD45+ cells of mice harboring control or ARC KD OCI-AML3 cells with or without Ara-C treatment, determined by Opal/TSA multiplex IHC staining and Vectra multispectral imaging analysis. Left, huCD45 (yellow), ARC (red), β-catenin (cyan), DAPI (blue), and merged imaging (objective lens ×20, scale bar 50 µm). Fluorophores for CD45, ARC, and β-catenin are Opal 540, Opal 570 and Opal 650, respectively. Right, the quantitation of ARC and β-catenin expression in huCD45+ cells of mouse spleen: 35 fields in control cell-injected mice (7,164 to 10,873 cells/field, total 319,431 cells), 26 fields in control cell-injected mice with Ara-C treatment (2,635 to 4,777 cells/field, total 99,727 cells), 17 fields in ARC KD cell-injected mice (610 to 5,021 cells/fields, total 37,206 cells), and 22 fields in ARC KD cell-injected mice with Ara-C treatment (248 to 3,927 cells/field, total 45,431 cells); respectively. F. huCD45 positivity in PB of mice harboring control or ARC KD OCI-AML3 cells with or without Ara-C treatment by flow cytometry. G. Survival of mice injected with control or ARC KD OCI-AML3 cells without or with Ara-C treatment.
We next determined the expression levels of ARC and β-catenin in huCD45+ cells in mouse spleen and BM using Opal/TSA IHC and Vectra imaging that allows simultaneous measurement of multiple markers within a single tissue section. Figure 5E shows the individual or mixed fluorescent images of mouse spleen stained with huCD45, ARC, β-catenin, and DAPI (left) and quantification of ARC and β-catenin protein levels (right) in the spleens of all four experimental groups. We analyzed 35 fields in control cell-injected mice (7,164 to 10,873 cells/field, total 319,431 cells), 26 fields in control cell-injected mice with Ara-C treatment (2,635 to 4,777 cells/field, total 99,727 cells), 17 fields in ARC KD cell-injected mice (610 to 5,021 cells/fields, total 37,206 cells), and 22 fields in ARC KD cell-injected mice with Ara-C treatment (248 to 3,927 cells/field, total 45,431 cells); respectively. As expected, mice injected with ARC KD OCI-AML3 cells with or without Ara-C treatment expressed drastically lower levels of ARC protein in human spleen CD45+ cells (P < 0.0001) compared to those injected with control cells with or without treatment. Interestingly, their spleen huCD45+ cells also had significantly lower levels of β-catenin (P < 0.0001 with Ara-C treatment, P < 0.01 without Ara-C treatment), supporting the existence of an ARC/IL1β/PGE/β-catenin/ARC regulatory loop. Similarly, mice injected with ARC KD OCI-AML3 cells with or without Ara-C treatment expressed significantly lower levels of ARC and β-catenin in BM huCD45+ cells compared to those injected with control cells without treatment. However, in BM, Ara-C treated groups had lower ARC and β-catenin levels compared to their respective untreated controls, especially for BM cells obtained from control cell-injected mice (supplemental Figure 1). The reason for this difference is unclear. We did observe that in BM samples, cells from Ara-C treated mice did not attach to slides well compared to cells from untreated mice, which was not observed in spleen samples. Lower leukemia burden in mice with ARC KD AML cells was further demonstrated by Opal/TSA staining of huCD45 in spleen and BM and Vectra imaging (Figure 5E and supplemental Figure 1).
Furthermore, both ARC inhibition, and the chemotherapeutic drug Ara-C, significantly lowered leukemia burden in PB assessed by flow cytometry of huCD45+ cells (Figure 5F) and prolonged survival (Figure 5G). Mice with ARC KD AML cells lived longer (median survival 33 d, P = 0.0006), similar to mice with control cells treated with Ara-C (34 d, P = 0.0003) compared to those with control cells not receiving treatment (27 d) (Figure 5G). Importantly, inhibition of ARC sensitized AML to Ara-C: mice injected with ARC KD cells treated with Ara-C showed the longest median survival time (40.5 d) (P = 0.0006, 0.0004, and 0.0012 vs. control, control + Ara-C, and ARC KD; respectively) (Figure 5G).
IL1β/PGE2/β-catenin/ARC cascade and targeting in primary AML samples
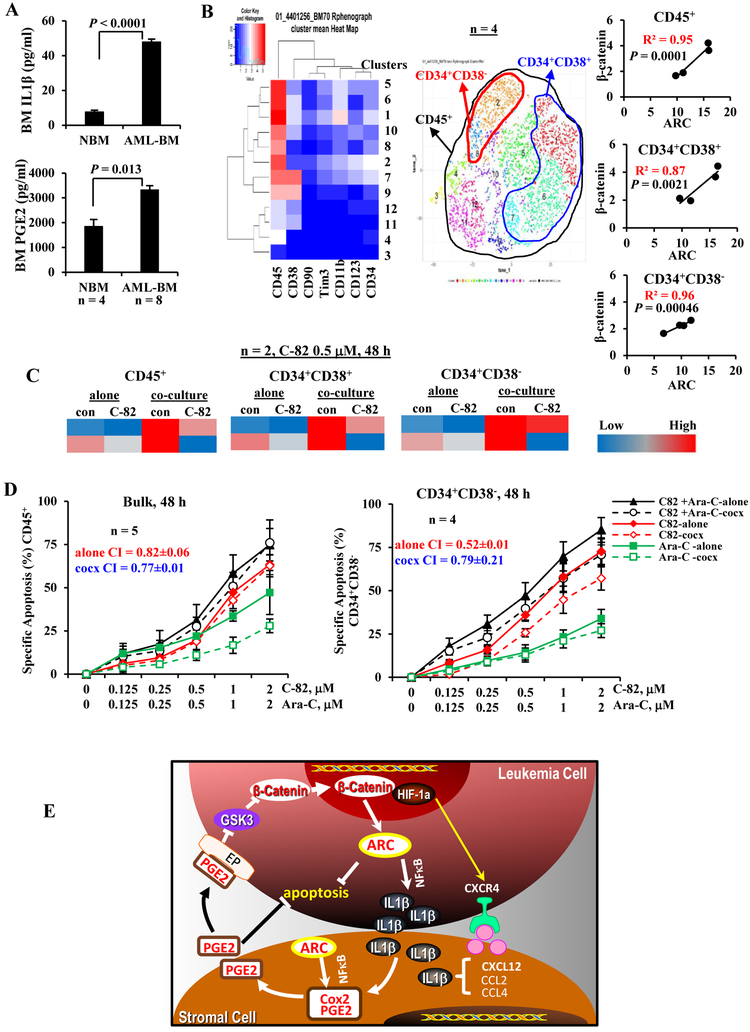
To assess potential roles of IL1β and PGE2 in AML and the AML BM microenvironment, we collected BM from AML patients (n = 8, Table 1, samples 9–16) and normal controls (n = 4) and determined IL1β and PGE2 levels: both IL1β and PGE2 levels were significantly higher in BM from AML patients compared to normal controls (Figure 6A). We then determined β-catenin and ARC protein levels in AML patient BM cells (Table 1, samples 17–20) by CyTOF mass cytometry in various cell compartments and observed strong correlations of β-catenin and ARC levels, in bulk as well as in CD34+CD38+ and CD34+CD38− stem/progenitor cells (Figure 6B). Furthermore, inhibition of β-catenin by C-82 decreased ARC and MSC co-culture-induced ARC expression in CD45+ blasts and CD34+CD38+ and CD34+CD38− stem/progenitor cells from primary samples (Figure 6C) (n = 2, Table 1, samples 21, 22). To demonstrate if targeting the IL1β/PGE2/β-catenin/ARC cascade sensitizes to chemotherapy, we treated primary AML cells with C-82 and Ara-C (Table 1, samples 23–27): C-82 and Ara-C synergistically induced apoptosis in bulk (n = 5) and CD34+CD38− cells (n = 4) even when cells were co-cultured with MSCs (CI < 1) (Figure 6D).
Figures 6.
PGE2/β-catenin/ARC cascade and targeting in primary AML samples and the proposed mechanism of ARC action. A. IL1β and PGE2 levels in BM samples from AML patients (n = 8) (Table 1, samples 9-16) and normal controls (NBM, n = 4) by ELISA. B. Correlation of ARC and β-catenin protein levels, determined by CyTOF in various BM cell populations of AML patients (n = 4) (Table 1, samples 17-20). C. Levels of ARC protein, determined by CyTOF in AML patient samples (n = 2) (Table 1, samples 21 and 22) treated with β-catenin inhibitor C-82 (0.5 μM) without or with MSC co-culture for 48 h. Protein levels determined by CyTOF are expressed as Arcsinh-transformed counts. D. AML patient samples were treated with C-82, Ara-C, or both for 48 h. Apoptosis was determined in blasts (n = 5) (Table 1, samples 23-27) and CD34+CD38− (n = 4) (Table 1, samples 23-26) cells. cocx, co-culture. E. Proposed mechanism of action: ARC, regulated by β-catenin, mediates leukemia stromal interaction through ARC-IL1β/Cox-2/PGE2/β-catenin circuit.
Discussion
ARC was first identified as an anti-apoptotic protein. We reported that ARC is a strong adverse prognostic marker in AML and protects leukemia cells from therapeutic agent-induced cell death (1, 2) consistent with its anti-apoptotic property. Interestingly, we also observed that ARC plays a role in leukemia-MSC interactions. Based on our previous finding that ARC modulates leukemia-microenvironment interactions by regulating NFκB/ILβ signaling (3), we here demonstrate that ARC regulates IL1β in AML which then induces Cox-2 expression in MSCs and PGE2 production, which in turn activates β-catenin and increases ARC in AML, while ARC itself is regulated by β-catenin. The data support the concept that in addition to directly suppressing apoptosis, ARC, regulated by β-catenin, mediates leukemia-stromal interactions through the ARC-IL1β/Cox-2/PGE2/β-catenin circuit. Furthermore, ARC in MSCs also affects Cox-2 levels and PGE2 production in AML-MSC co-culture. We previously reported that ARC in MSCs induces nuclear and phosphor-NFκB (3). NFκB is known to regulate Cox-2-catalyzed PGE2 synthesis (10–12). Therefore, ARC is an integral component of and a regulator of leukemia-microenvironment interactions (Figure 6E).
PGE2 was reported to suppress apoptosis in multiple cell types through various mechanisms such as PI3K/AKT signaling or regulating survivin expression (35–37). In this study, we found that PGE2 protects AML cells from chemotherapy and strongly induces the expression of anti-apoptotic ARC protein in AML cells, a new mechanism of PGE2-mediated anti-apoptotic effect. Furthermore, we demonstrated that MSCs confer AML drug resistance in a PGE2-dependent manner. Our data reveal that AML cells, microenvironmental factors, and leukemia-stromal interactions cooperatively support leukemia cells through the novel ARC/IL1β/Cox-2/PGE2/β-catenin circuit reported here.
We discovered that ARC is transcriptionally regulated by β-catenin. PGE2 treatment or co-culture with MSCs induces β-catenin in AML in agreement with our previous finding that leukemia cells in the marrow had higher β-catenin levels than paired leukemia cells in circulation (27), suggesting that the BM microenvironment induces β-catenin levels in AML cells at least in part through stromal cell secreted PGE2. Furthermore, using a xenograft model of human AML in NSGS mice, we demonstrated that inhibition of ARC by shRNA significantly decreased leukemia burden and serum IL1β/PGE2 levels, prolonged survival, and sensitized to chemotherapy. Interestingly, in addition to decreased ARC levels, low β-catenin levels in huCD45+ cells were also detected in mouse tissues injected with ARC KD AML cells compared to with controls, supporting the ARC/IL1β/Cox/PGE2/β-catenin regulatory circuit in vivo.
We demonstrate that both IL1β and PGE2 levels are significantly higher in BM from AML patients compared to normal controls and that the levels of BM β-catenin and ARC strongly correlate in AML bulk and stem/progenitor cells, supporting the regulatory cascade in primary AML. It is technically challenging to establish ARC knockdown or overexpression in primary AML cells and currently no specific ARC inhibitor is available. However, targeting the components of the regulatory circuit can be explored. We previously reported that inhibition of β-catenin by its selective inhibitor PRI-724 (C-82 prodrug) has anti-leukemia activity in vivo in immunodeficient mice engrafted either with an AML cell line or AML PDX cells and sensitized to tyrosine kinase inhibitor in FLT3 mutated AML (27). We here show that C-82 decreases ARC and sensitizes to Ara-C in bulk and CD34+CD38− AML cells even when they were co-cultured with MSCs.
Evasion of immunity in hematological malignancies has been increasingly recognized in recent years (38). Wnt/β-catenin and Cox-2/PGE2 signaling are both implicated in immunosuppression (39). As ARC is a component of the IL1β/Cox-2/PGE2/β-catenin cascade that connects β-catenin and IL1β and given our finding that high ARC expression is a strong adverse predictor of survival in AML, we envision that higher ARC levels in AML may also contribute to a more inflammatory and immunosuppressive microenvironment, a hypothesis that is currently under investigation.
In conclusion, we demonstrate that ARC is an integral component of a novel IL1β/Cox-2/PGE2/β-catenin circuit, plays a critical role in leukemia growth and the leukemia microenvironment, and may serve as a potential novel therapeutic target in AML. We are currently trying to develop inhibitors targeting ARC. The finding that ARC functions as a survival modulator through multiple mechanisms explains its strong adverse effect in AML. This sets ARC apart from other anti-apoptotic proteins.
Supplementary Material
Significance: The anti-apoptotic protein ARC promotes AML aggressiveness by enabling detrimental cross-talk with bone marrow mesenchymal stromal cells.
Acknowledgements
We thank Drs. Numsen Hail for editorial support and Ivo Veletic for technical assistance.
Funding: This work was supported in part by grants from the University Cancer Foundation via the Institutional Research Grant program at MD Anderson to B. Z. Carter and from the National Institutes of Health (P01CA055164), Cancer Prevention Research Institute of Texas (CPRIT, RP121010), and by the Paul and Mary Haas Chair in Genetics to M. Andreeff and MD Anderson’s Cancer Center Support Grant CA016672 (Flow Cytometry and Cellular Image Facility and Characterized Cell Line core).
Footnotes
Conflicts of interests: None
References
- 1.Carter BZ, Qiu YH, Zhang N, Coombes KR, Mak DH, Thomas DA, et al. Expression of ARC (apoptosis repressor with caspase recruitment domain), an antiapoptotic protein, is strongly prognostic in AML. Blood. 2011;117:780–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mak PY, Mak DH, Mu H, Shi Y, Ruvolo P, Ruvolo V, et al. Apoptosis repressor with caspase recruitment domain is regulated by MAPK/PI3K and confers drug resistance and survival advantage to AML. Apoptosis. 2014;19:698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter BZ, Mak PY, Chen Y, Mak DH, Mu H, Jacamo R, et al. Anti-apoptotic ARC protein confers chemoresistance by controlling leukemia-microenvironment interactions through a NFkappaB/IL1beta signaling network. Oncotarget. 2016;7(15):20054–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konopleva M, Tabe Y, Zeng Z, Andreeff M. Therapeutic targeting of microenvironmental interactions in leukemia: mechanisms and approaches. Drug Resist Updat. 2009;12:103–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turzanski J, Grundy M, Russell NH, Pallis M. Interleukin-1beta maintains an apoptosis-resistant phenotype in the blast cells of acute myeloid leukaemia via multiple pathways. Leukemia. 2004;18:1662–70. [DOI] [PubMed] [Google Scholar]
- 7.Barreyro L, Will B, Bartholdy B, Zhou L, Todorova TI, Stanley RF, et al. Overexpression of IL-1 receptor accessory protein in stem and progenitor cells and outcome correlation in AML and MDS. Blood. 2012;120:1290–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Askmyr M, Agerstam H, Hansen N, Gordon S, Arvanitakis A, Rissler M, et al. Selective killing of candidate AML stem cells by antibody targeting of IL1RAP. Blood. 2013;121:3709–13. [DOI] [PubMed] [Google Scholar]
- 9.Li HJ, Reinhardt F, Herschman HR, Weinberg RA. Cancer-stimulated mesenchymal stem cells create a carcinoma stem cell niche via prostaglandin E2 signaling. Cancer Discov. 2012;2:840–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ackerman WEt Summerfield TL, Vandre DD Robinson JM, Kniss DA Nuclear factor-kappa B regulates inducible prostaglandin E synthase expression in human amnion mesenchymal cells. Biol Reprod. 2008;78:68–76. [DOI] [PubMed] [Google Scholar]
- 11.Kaltschmidt B, Linker RA, Deng J, Kaltschmidt C. Cyclooxygenase-2 is a neuronal target gene of NF-kappaB. BMC Mol Biol. 2002;3:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto K, Arakawa T, Ueda N, Yamamoto S. Transcriptional roles of nuclear factor kappa B and nuclear factor-interleukin-6 in the tumor necrosis factor alpha-dependent induction of cyclooxygenase-2 in MC3T3-E1 cells. J Biol Chem. 1995;270:31315–20. [DOI] [PubMed] [Google Scholar]
- 13.Kalinski P Regulation of immune responses by prostaglandin E2. J Immunol. 2012;188:21–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pelus LM, Hoggatt J. Pleiotropic effects of prostaglandin E2 in hematopoiesis; prostaglandin E2 and other eicosanoids regulate hematopoietic stem and progenitor cell function. Prostaglandins Other Lipid Mediat. 2011;96:3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broxmeyer HE, Pelus LM. Inhibition of DPP4/CD26 and dmPGE(2) treatment enhances engraftment of mouse bone marrow hematopoietic stem cells. Blood Cells Mol Dis. 2014;53:34–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Speth JM, Hoggatt J, Singh P, Pelus LM. Pharmacologic increase in HIF1alpha enhances hematopoietic stem and progenitor homing and engraftment. Blood. 2014;123:203–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakanishi M, Rosenberg DW. Multifaceted roles of PGE2 in inflammation and cancer. Semin Immunopathol. 2013;35:123–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murakami M, Naraba H, Tanioka T, Semmyo N, Nakatani Y, Kojima F, et al. Regulation of prostaglandin E2 biosynthesis by inducible membrane-associated prostaglandin E2 synthase that acts in concert with cyclooxygenase-2. J Biol Chem. 2000;275:32783–92. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Yin S, Nie D, Xie S, Ma L, Wang X, et al. MK886 inhibits the proliferation of HL-60 leukemia cells by suppressing the expression of mPGES-1 and reducing prostaglandin E2 synthesis. Int J Hematol. 2011;94:472–8. [DOI] [PubMed] [Google Scholar]
- 20.Naderi EH, Skah S, Ugland H, Myklebost O, Sandnes DL, Torgersen ML, et al. Bone marrow stroma-derived PGE2 protects BCP-ALL cells from DNA damage-induced p53 accumulation and cell death. Mol Cancer. 2015;14:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon M, Grandage VL, Linch DC, Khwaja A. Constitutive activation of the Wnt/beta-catenin signalling pathway in acute myeloid leukaemia. Oncogene. 2005;24:2410–20. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ysebaert L, Chicanne G, Demur C, De Toni F, Prade-Houdellier N, Ruidavets JB, et al. Expression of beta-catenin by acute myeloid leukemia cells predicts enhanced clonogenic capacities and poor prognosis. Leukemia. 2006;20:1211–6. [DOI] [PubMed] [Google Scholar]
- 24.Romano P, Manniello A, Aresu O, Armento M, Cesaro M, Parodi B. Cell Line Data Base: structure and recent improvements towards molecular authentication of human cell lines. Nucleic Acids Res. 2009;37:D925–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Studeny M, Marini FC, Champlin RE, Zompetta C, Fidler IJ, Andreeff M. Bone marrow-derived mesenchymal stem cells as vehicles for interferon-beta delivery into tumors. Cancer Res. 2002;62:3603–8. [PubMed] [Google Scholar]
- 26.Mak PY, Mak DH, Ruvolo V, Jacamo R, Kornblau SM, Kantarjian H, et al. Apoptosis repressor with caspase recruitment domain modulates second mitochondrial-derived activator of caspases mimetic-induced cell death through BIRC2/MAP3K14 signalling in acute myeloid leukaemia. Br J Haematol. 2014;167:376–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang X, Mak PY, Mu H, Tao W, Mak D, Kornblau S, et al. Disruption of Wnt/beta-catenin exerts anti-leukemia activity and synergizes with FLT3 inhibition in FLT3-mutant acute myeloid leukemia. Clin Cancer Res. 2018;24(10):2417–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou H, Mak PY, Mu H, Mak DH, Zeng Z, Cortes J, et al. Combined inhibition of beta-catenin and Bcr-Abl synergistically targets tyrosine kinase inhibitor-resistant blast crisis chronic myeloid leukemia blasts and progenitors in vitro and in vivo. Leukemia. 2017;31:2065–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter BZ, Mak PY, Mu H, Zhou H, Mak DH, Schober W, et al. Combined targeting of BCL-2 and BCR-ABL tyrosine kinase eradicates chronic myeloid leukemia stem cells. Sci Transl Med. 2016;8:355ra117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang B, Li M, McDonald T, Holyoake TL, Moon RT, Campana D, et al. Microenvironmental protection of CML stem and progenitor cells from tyrosine kinase inhibitors through N-cadherin and Wnt-beta-catenin signaling. Blood. 2013;121:1824–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han L, Qiu P, Zeng Z, Jorgensen JL, Mak DH, Burks JK, et al. Single-cell mass cytometry reveals intracellular survival/proliferative signaling in FLT3-ITD-mutated AML stem/progenitor cells. Cytometry A. 2015;87:346–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen H, Lau MC, Wong MT, Newell EW, Poidinger M, Chen J. Cytofkit: A Bioconductor Package for an Integrated Mass Cytometry Data Analysis Pipeline. PLoS Comput Biol. 2016;12:e1005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. [DOI] [PubMed] [Google Scholar]
- 34.Cadigan KM, Waterman ML. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol. 2012;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baratelli F, Krysan K, Heuze-Vourc’h N, Zhu L, Escuadro B, Sharma S, et al. PGE2 confers survivin-dependent apoptosis resistance in human monocyte-derived dendritic cells. J Leukoc Biol. 2005;78:555–64. [DOI] [PubMed] [Google Scholar]
- 36.Fernandez-Martinez A, Molla B, Mayoral R, Bosca L, Casado M, Martin-Sanz P. Cyclo-oxygenase 2 expression impairs serum-withdrawal-induced apoptosis in liver cells. Biochem J. 2006;398:371–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.George RJ, Sturmoski MA, Anant S, Houchen CW. EP4 mediates PGE2 dependent cell survival through the PI3 kinase/AKT pathway. Prostaglandins Other Lipid Mediat. 2007;83:112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andersen MH. The targeting of immunosuppressive mechanisms in hematological malignancies. Leukemia. 2014;28:1784–92. [DOI] [PubMed] [Google Scholar]
- 39.Prima V, Kaliberova LN, Kaliberov S, Curiel DT, Kusmartsev S. COX2/mPGES1/PGE2 pathway regulates PD-L1 expression in tumor-associated macrophages and myeloid-derived suppressor cells. Proc Natl Acad Sci U S A. 2017;114:1117–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.