Abstract
The ease of performing both forward and reverse genetics in Saccharomyces cerevisiae, along with its stable haploid state and short generation times, has made this budding yeast the consummate model eukaryote for genetics. The major advantage of using budding yeast for reverse genetics is this organism’s highly efficient homology-directed repair, allowing for precise genome editing simply by introducing DNA with homology to the chromosomal target. Although plasmid- and PCR-based genome editing tools are quite efficient, they depend on rare spontaneous DNA breaks near the target sequence. Consequently, they can generate only one genomic edit at a time, and the edit must be associated with a selectable marker. However, CRISPR/Cas technology is efficient enough to permit markerless and multiplexed edits in a single step. These features have made CRISPR/Cas popular for yeast strain engineering in synthetic biology and metabolic engineering applications, but it has not been widely employed for genetic screens. In this review, we critically examine different methods to generate multi-mutant strains in systematic genetic interaction screens and discuss the potential of CRISPR/Cas to supplement or improve on these methods.
Keywords: Genetic interaction screens, Synthetic genetic array, CRISPR/Cas9, Functional genomics, dSLAM, Green Monster, Saccharomyces cerevisiae
Introduction
The budding yeast Saccharomyces cerevisiae has been a workhorse model organism for genetics for over 50 years now. One of the major advances in yeast genetics was the generation of loss-of-function (LOF)-mutant strain collections by systematically deleting all non-essential annotated open reading frames (Winzeler et al. 1999). These collections are derived from a common set of background strains and include haploid strains, homozygous diploid strains, and heterozygous diploid strains. To supplement these collections, groups have also generated collections of strains (in the same genetic background) carrying temperature-sensitive, transcriptionally repressible, or hypomorphic alleles of essential genes (Mnaimneh et al. 2004; Breslow et al. 2008; Li et al. 2011; Wang et al. 2015; Kofoed et al. 2015).
While functional analyses of single-gene perturbations are useful, by combining LOF mutations in the same cell, one can uncover suppressing or enhancing epistatic interactions that provide clues about the network structures of cellular pathways (Salminen and Novick 1987; Adams et al. 1990, 1993; Bendert and Pringle 1991; Costigan et al. 1992; Frank et al. 1992; Scidmore et al. 1993). There are two types of genetic interactions (GIs) that can occur when two or more LOF mutations are combined in the same cell–synthetic suppression or synthetic enhancement. An extreme example of synthetic suppression is synthetic rescue, when one or more of the single mutants is inviable, but the synthetic combination is viable. An extreme example of synthetic enhancement is synthetic lethality, when all single mutants are viable, but the synthetic combination is lethal.
Gain-of-function (GOF) mutations are routinely generated by gene overexpression (dosage) from a plasmid or an integrated construct using a strong promoter. Dosage screens in yeast are largely successful because there is little dosage compensation at the single-gene level (Deutschbauer et al. 2005; Springer et al. 2010; Ishikawa et al. 2017).
LOF mutations can also be combined with GOF alleles in the same cell and assessed for dosage interactions. The extreme case of dosage suppression is dosage rescue of an otherwise lethal mutation. The extreme case of dosage enhancement is dosage lethality, in which neither overexpression of gene x nor the mutation of gene y are lethal, but the combination is. One can also overexpress two genes to find double dosage lethality, but this is not a common approach (Youn et al. 2017).
The goal of these mutant combinations is to identify epistatic GIs from which one can infer pathway hierarchies (Forsburg 2001; Boone et al. 2007; Costanzo et al. 2010; Magtanong et al. 2011). For example, synthetic suppression and dosage enhancement are both consistent with a network in which the two gene products are functionally antagonistic. In contrast, synthetic enhancement and dosage suppression are both consistent with gene products that work together or in parallel in the same cellular process.
Traditionally, GIs were discovered by random mutagenesis of LOF or GOF mutant query strains, or by transforming such strains with high copy number cDNA or genomic DNA plasmid libraries, followed by a selection regimen or visual screen to find suppressing or enhancing interactions (Reed et al. 1989; Albertini and Zimmermann 1991; Costigan et al. 1992; Chowdhury et al. 1992; Flescher et al. 1993; Puziss et al. 1994; Blázquez and Gancedo 1994; Machin et al. 1995; Kroll et al. 1996; Kaytor and Livingston 1996; Akada et al. 1997; Mullen et al. 2001; Stevenson et al. 2001; Kitazono and Kron 2002; Bogomolnaya et al. 2004; Kaplan and Kupiec 2007; Carlsson et al. 2018). Although the process of screening using mutagenesis or pooled plasmid libraries is relatively quick and easy, the resulting strains require extensive characterization to ensure screen “saturation” (i.e., obtaining multiple alleles of the same genes), and to identify the genes responsible for the interactions.
Now, GI screens are routinely performed using systematic arrayed yeast strain collections or arrayed plasmid collections, which ensure comprehensive genomic coverage. There are several approaches researchers have taken to perform such screens—transformation of a systematic collection of overexpression (OE) plasmids into one or a few mutant query strains, transformation of one or a few query plasmids into a systematic collection of yeast mutant strains, or crossing one or a few query mutant strains to a systematic collection of mutant strains and obtaining haploid double mutants. Although such screens require complex high-throughput protocols up front, the back-end analysis and identification of the interacting genes is much faster and easier than in traditional GI screens.
In the following sections, we will examine and compare traditional methods to perform systematic GI screens and discuss the advantages and limitations of each approach. These approaches combine mutations either by crossing mutant strains, or by introducing a common mutation into a collection of pre-existing mutants. In both approaches, query mutations are introduced into cells by homology-directed repair (HDR) with transformed donor DNA [for a comprehensive review of how HDR works, see (Gallagher and Haber 2018)]. Chromosomal recombination with the donor DNA depends on the spontaneous generation of double-strand breaks (DSBs) at the target locus. These methods require a selectable marker in the donor DNA to select for the rare recombinants and are also too inefficient to perform more than one type of edit at a time (multiplexing). Sequence-specific endonucleases improve editing efficiency enough to allow markerless and multiplexed edits (Guha and Edgell 2017). CRISPR/Cas is the most popular method of endonuclease-mediated gene editing because of the ease of programming the target specificity. We will discuss how CRISPR/Cas has the potential to complement and streamline current methods of performing GI screens. Our hope is that this review will help yeast geneticists to make informed decisions about what approaches best suit their particular genetic screens.
Traditional genetic interaction screens
Overexpression plasmid transformation
The simplest approach to dosage interaction screens is to transform overexpression (OE) plasmids into mutant strains. These screens can be performed systematically using the genomic collections of yeast strains we mentioned earlier or genomic collections of OE plasmids.
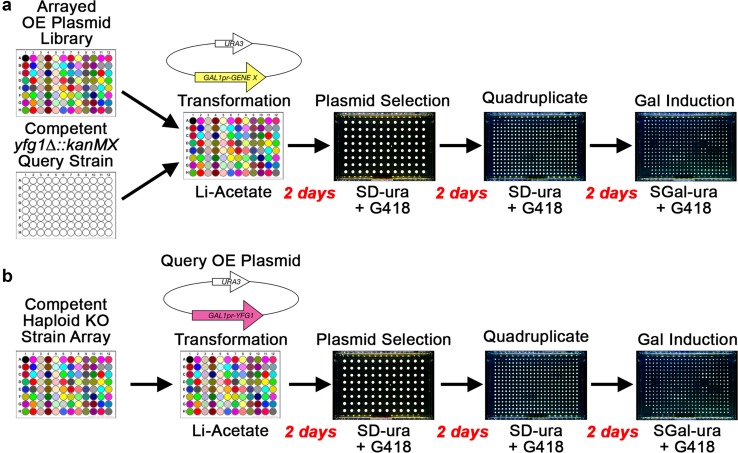
To introduce plasmids into large arrays of different yeast strains, or to introduce large arrays of different plasmids into the same yeast strain, transformations can be readily performed in 96-well plates to accommodate 4000 or more strains or plasmids (Fleming and Gitler 2011) (Fig. 1).
Fig. 1.
Dosage GI screens using OE plasmids. a The dosage interaction screen starts with two 96-well plates. One plate contains an arrayed OE plasmid library as DNA, each well overexpressing a different single ORF. Every well in the second plate contains the same query mutant strain denoted by your favorite gene yfg1∆. b The dosage interaction screen starts with a query overexpression plasmid and a 96-well plate containing an array of haploid deletion strains, each well containing a different deletion strain. Transformations are performed in liquid in 96-well plates. Transformants may undergo selection in liquid by transferring cells into fresh plates or by replica pinning to agar selection plates (shown). In this example, selection of the plasmid is maintained by growth on media lacking uracil (SD-ura), and selection of the gene deletions is maintained by adding G418 as the deletion collections carry kanMX. Transformants are assayed for growth under non-induced (SD-ura) and induced (SGal-ura) conditions. Transformants should be quadruplicated to 384 arrays for statistical measurements of growth. Plates are imaged and colony sizes are measured to determine if two genes interact. Red text indicates how many days are required for each step in the workflow. kanMX: G418-resistance marker, OE: overexpression, yfg1∆: deletion of your favorite gene, Gal: galactose; colored wells indicate that the contents of each well are different
Using plasmid transformation, systematic GI screens have been performed in two different ways. First, one could transform query mutants with systematic arrays of OE plasmids to find synthetic dosage enhancement or lethality interactions (Zimmermann et al. 2017), or to find dosage suppression or rescue interactions (Magtanong et al. 2011; Patra et al. 2017) (Fig. 1a). The alternative approach is to transform a query OE plasmid into a systematic mutant strain collection. We do not know of any examples in which this approach was performed to find synthetic dosage interactions using the entire yeast deletion collection, but it has been done with a collection of mutants (Kroll et al. 1996) (Fig. 1b). One could also transform a query OE plasmid into a subset of the haploid deletion strains that show low fitness, or into hypomorphic/conditional essential gene collections to find dosage suppression interactions.
Available systematic collections of plasmids for dosage screens have been constructed by inserting PCR-amplified ORFs behind a strongly galactose-inducible GAL1 promoter (Zhu et al. 2001; Gelperin et al. 2005; Hu et al. 2007), or by cloning the entire PCR-amplified gene, including its natural promoter and terminator, into a 2 µ plasmid (Moriya et al. 2006; Magtanong et al. 2011). 2 µ plasmids use an origin of replication that replicates independently of the host’s cell cycle. These plasmids replicate to high copy number, but because they do not have centromeres, they randomly segregate to daughter cells and are present in the population of cells at a wide range of copy numbers. Both the inducible and 2 µ plasmids are maintained as episomal circular DNA.
When performing a systematic search for negative GIs such as synthetic dosage lethality, high copy number plasmids should be avoided. The reason for this is that a lack of growth of the transformants could be due to synthetic dosage lethality or simply a technical failure of the transformation. Using an inducible overexpression system avoids this problem because transformants can be selected under uninduced conditions before exposing them to overexpression conditions.
In addition, about 20% of genes can have negative effects on cellular growth on their own (Makanae et al. 2013). One way to study synthetic dosage interactions with these genes is to control the level of their expression. When using the inducible OE plasmid collections, there is little ability to tune the level of overexpression because the GAL1 promoter used to drive gene overexpression responds to the presence of galactose in the medium in a switch-like manner (Hawkins and Smolke 2006). Some overexpression genomic DNA libraries and systematic collections use a 2 µ plasmid carrying the URA3 selection marker and a second marker consisting of a partial loss-of-function leu2-89 (LEU2-d) allele (Moriya et al. 2006; Carlsson et al. 2018). Growth of the transformants on media lacking uracil alone allows cells to maintain a relatively low average copy number, but restricting or removing leucine in the medium selects for cells with higher copy numbers to allow sufficient production of leucine from the weakened Leu2-89 enzyme. Therefore, the copy number and gene dosage can be tuned with the concentration of leucine in the medium.
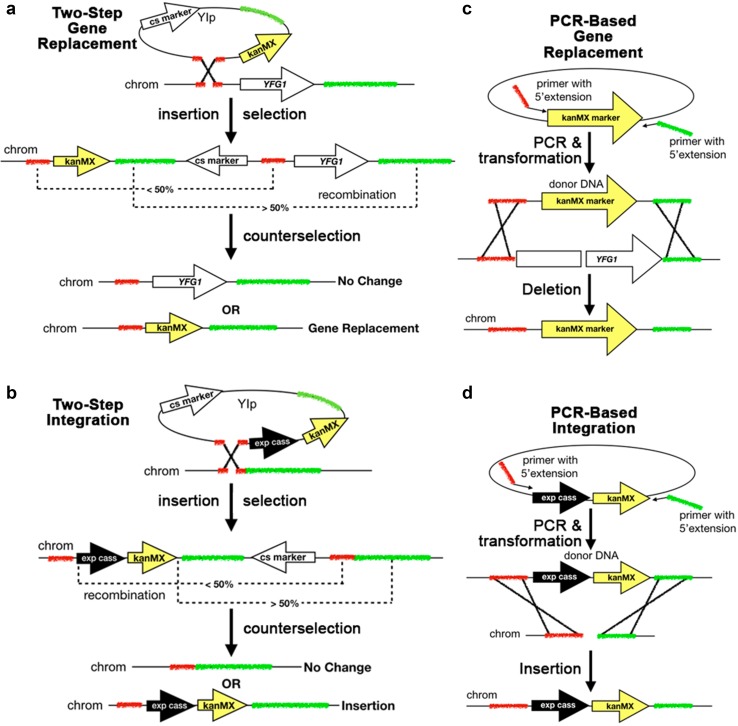
In theory, a special class of plasmids called yeast integrating plasmids (YIps) could be used in systematic GI screens (Sikorski and Hieter 1989). YIps do not carry a yeast origin of replication and, consequently, the only way they are stably inherited is by chromosomal integration. Stable integration of YIps is accomplished in two steps using yeast’s very efficient homology-directed repair (HDR) machinery for initial integration of the linearized YIp after transformation, and recombination and counterselection of the plasmid backbone (Storici et al. 2001; Nair and Zhao 2009) (Fig. 2a, b). YIps can be used to introduce gene expression cassettes or gene deletions (Rudolph et al. 1985; Alani et al. 1987; Lopes et al. 1989; Parekh et al. 1996; Voth et al. 2001; Akada et al. 2002; Sakai et al. 2004; Sadowski et al. 2007). However, YIps are only used for systematic GI screens as query plasmids because there are no systematic YIp collections.
Fig. 2.
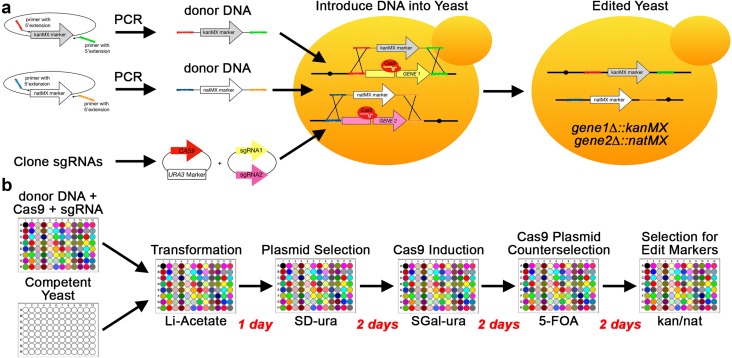
Gene editing approaches to make query mutant strains for GI screens YIp- or PCR-based methods for gene editing introduce donor DNA that serves as a template for homology-directed repair (HDR) when a spontaneous DNA double-strand break (DSB) occurs in the target locus denoted by your favorite gene yfg1∆. a, b YIp-based gene editing is performed in two steps. In the first step, the YIp linearized at a short cloned region of homology is integrated into the homologous chromosome region (red). The integrated construct generates direct repeats, which can spontaneously recombine and “pop-out” the construct. However, recombination is more likely to occur at the longer repeat region (green), resulting in stable integration of the construct. Recombinants are obtained by counterselection. a This Yip is designed to delete YFG1 and replace it with the kanR (kanMX) marker. b This YIp is designed to integrate an expression cassette (exp cass) in an intergenic chromosomal target. c, d PCR-based gene editing uses plasmid cassettes that contain a selectable marker. The target sequences are encoded in 40–60 nts of the PCR primer at 5′ overhangs (red and green). The primers also have ~ 20 nts of homology to the plasmid backbone flanking the cassettes at their 3′ ends (arrows). The same gene-specific primers can be used to insert various types of cassettes at the chromosomal target (red and green). c PCR-based gene deletion/replacement with a selectable marker. d PCR-based insertion of a marked expression cassette. kanMX: G418-resistance marker; YFG1: wild-type your favorite gene; X: homologous recombination event, chromI: chromosomal locus; exp cass: expression cassette, cs marker: counterselectable marker
Synthetic genetic arrays (SGA)
Prior to the synthetic genetic array (SGA) method, it was not practical to systematically cross mutants to test GIs. Genetic crosses were performed in small batches and the haploid segregants could only be obtained in one of two ways—tetrad dissection or random spore analysis (Sherman 2002). In tetrad dissection, the sporulation mix, which consists of unsporulated diploid cells and sporulated haploid segregants encased in asci, is mildly digested with a glycosidase to break open the asci. The mix is spread on a plate and the tetrad spores from each ascus (still clustered together) are individually dissected onto different grid positions on the plate and allowed to form clonal colonies. In random spore analysis, sporulation mixes are treated with diethyl ether to kill off all diploids while leaving a small set of resistant viable haploid spores. The treated mix is then spread onto plates to obtain colonies from each surviving spore. In both methods, colonies are assessed for mating type and the presence of each marked mutation to find the desired mutant combinations by replica plating to various types of media.
The power of the SGA approach is that it utilizes a haploid-specific reporter gene to select for haploids of one mating type. This haploid selection reporter uses a MATa-specific promoter, specifically those driving a-factor or α-receptor expression (MFA1pr or STE2pr, respectively), to drive expression of the HIS3 auxotrophic marker in his3 mutant strains. MATa haploid cells are selected on media lacking histidine. The original reporter, MFA1pr-HIS3, showed leaky expression and could recombine with the small his3∆1 deletion allele present in BY4741/2/3 strains, allowing growth of diploids and MATα strains. An improved version of the haploid reporter uses the more stringent STE2pr driving expression of the Schizosaccharomyces pombe ortholog Sphis5 to prevent recombination with his3∆1 (Daniel et al. 2006). The reporter also knocks out the CAN1 arginine permease gene for better haploid selection using the toxic arginine analog canavanine. Heterozygous diploid CAN1/can1∆::STE2pr–Sphis5 cells have an intact copy of the permease, which allows canavanine into the cells leading to death. The his + MATa haploids are can1∆ and resistant to canavanine. This additional selection prevents the growth of diploids that have undergone gene conversion to MATa/MATa, a source of false negatives (Tong and Boone 2007).
A single-query strain carries the LOF or GOF allele of interest plus the haploid reporter gene (Tong and Boone 2006). Introduction of these alleles into the SGA haploid selection strain can be done by crossing an existing mutant strain with the SGA parent strain, or by directly gene editing the SGA parent strain. Gene editing of the query strain is routinely done by YIp-mediated gene deletion/replacement or integration of an overexpression cassette (Fig. 2a, b) or by PCR-mediated gene deletion/replacement or expression cassette integration (Fig. 2c, d), but query strains may also carry episomal plasmids for gene overexpression.
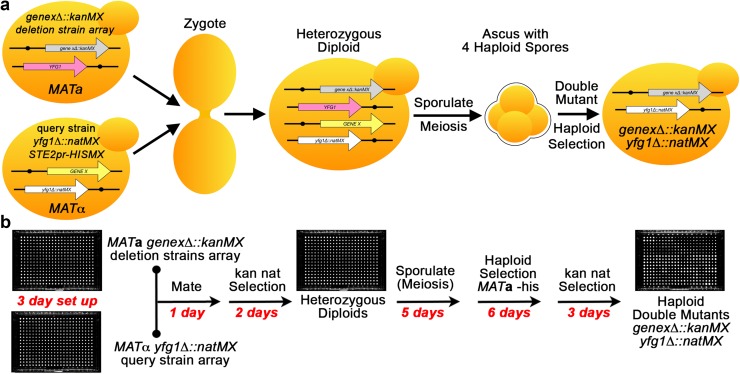
The query strain is then mated to arrayed colonies from a strain collection (Fig. 3). The query mutation must be linked to a selectable marker that is different from the marker used in the strain collection to allow selection of double mutants. Hundreds of different marked deletion strains can be arrayed on each rectangular agar plate, pinned on top of the query strain to allow each pair of strains to mate, and replica pinned onto double selection media to select for diploids. The diploids are then sporulated, and haploid cells carrying the desired mutant combinations are selected.
Fig. 3.
GI screens using SGA. a Schematic of the DNA molecules and manipulations resulting in the generation of a double mutant by SGA. b The SGA workflow starts with two agar plates of arrayed colonies. Each colony on the arrayed deletion collection plate is a different deletion strain marked with the same selection marker and in the same mating type. The query deletion strain, denoted by your favorite gene yfg1∆, is arrayed on a second plate and carries a different selection marker and has the opposite mating type. Colonies from the two parent plates are replica pinned onto the same plate to mix two mutants and allow them to mate and form a heterozygous diploid zygote. The diploids are replica pinned onto sporulation media. The resulting sporulation mix is plated on a series of selection media to obtain haploid strains carrying both parental deletions. Double mutants should be quadruplicated to 384 arrays for statistical measurements of growth. Plates are imaged and colony sizes are measured to determine if two genes interact. Red text indicates how many days are required for each step in the workflow. yfg1∆::natMX: query mutation of your favorite gene deleted and replaced with nourseothricin-resistance marker, genex∆::natMX: haploid strain collection gene deletion replaced with G418-resistance marker, STE2pr-HISMX MATa: haploid selection marker that confers growth on media lacking histidine
The explosion in yeast genetic analyses over the past decade enabled by the SGA method was due to the ability to make thousands of pairwise genetic crosses in parallel (as of this writing, a PubMed search for “synthetic genetic array” and “cerevisiae” produces 120 publications since 2001). At last count, GIs have been tested for over 75% of yeast genes, and a little under 1/3 of all possible ~ 18,000,000 pairwise combinations of annotated yeast genes have been tested (Costanzo et al. 2010).
SGAs were initially used to find synthetic lethal GIs (Tong et al. 2001, 2004; Davierwala et al. 2005; Ooi et al. 2006). Subsequent SGA screens and its variations analyzed more subtle effects of GIs on cellular fitness, including positive (suppression) as well as negative (enhancement) synthetic interactions (Schuldiner et al. 2005; Collins et al. 2010; Costanzo et al. 2010; Baryshnikova et al. 2010; Lindén et al. 2011; Piening et al. 2013; Stundon and Zakian 2015).
SGA has also been used to find synthetic dosage interactions by crossing query strains carrying integrated overexpression cassettes with the deletion collection (Youn et al. 2017), by crossing query strains transformed with OE plasmids (Measday et al. 2005; Douglas et al. 2012) to the deletion strain collection, or by crossing query strains to a collection of strains in which each carries a different OE plasmid (Sopko et al. 2006; Liu et al. 2009; Duffy et al. 2016). Usually, the resulting LOF–GOF double mutants are analyzed as arrays, but they can also be pooled to perform batch analysis of growth by quantifying unique barcodes (by microarray or sequencing) in the arrayed strain collection (Douglas et al. 2012).
Although the SGA approach allows high-throughput genetic crosses, making thousands of strains carrying just two genetic modifications remains a daunting endeavor that requires specialized robotic equipment (about one-third of SGA publications have come from the Boone lab). Although a less expensive semi-automated robotic system has been developed (Rotor HDA by Singer Instruments) to allow SGA automation within the scope of a NIH R01 grant, this tactic also reduces the throughput, adds significant cost to the consumables, and increases the manual labor required. Because of the cost of consumables and extra labor, one should perform a cost–benefit analysis of the Rotor HDA system compared to more automated systems using fewer consumables (e.g., S&P Robotics Inc. systems) using reasonable expectations of the scope and frequency of SGA screens to be performed within 5–10 years.
Another technical limitation of SGA is that it tends to produce false-negative interactions. One reason for this is that haploid deletion strains are under selective pressure to grow faster and often pick up suppressor mutations or become disomic with both the deletion allele and the wild-type gene. For example, when testing purchased haploid deletion strains upon arrival, we found a suppressing mutation in WHI5 (a transposon insertion) in the MATacln3∆ strain (YSC6273-201934719)(unpublished data), and disomy in the MATakar9∆ mutant (YSC6273-201917550), which normally has a chromosome segregation defect and is likely aneuploid (unpublished data).
Another technical problem with SGA is cross-contamination from neighboring wells in both the deletion collection and during the SGA process. For example, we found that the MATamsn5∆ strain (YSC6273-201934942) consisted mostly or entirely of a strain deleted in the adjacent ORF/plate well, MRX8 (unpublished data). The integrity of the haploid deletion strains is also an issue when transforming a query plasmid into a haploid collection for systematic dosage screens.
Finally, it may be impossible to obtain diploids from strains with strong mating defects or combinations of strains that have bilateral mating defects. However, if one images all diploid selection plates, the missing crosses can be excluded from subsequent SGA analysis of haploid mutant combinations.
Diploid-based synthetic lethality analysis on microarrays (dSLAM)
Our lab and others have avoided the problem of modifier mutations in the haploid strain collections using fresh haploid deletion strains derived from the heterozygous deletion strain collection. There is little to no selection for modifier mutations in the heterozygous diploids. However, this approach requires sporulation and dissection or random spore analysis of haploid spores to derive strain arrays for SGA and is not practical for genome-wide SGA screens. In a streamlined variation of this approach, diploid-based synthetic lethality analysis on microarrays (dSLAM) introduces the SGA haploid reporter construct into pooled strains of the heterozygous diploid deletion collection (Pan et al. 2004) (Fig. 4). The pooled diploid strains are then transformed with a YIp to delete or overexpress a gene. The pool of double mutants is selected and then sporulated. After sporulation haploid double mutant progeny are selected and the pool is analyzed by microarray (or sequencing) for the relative abundances of unique barcodes associated with each deletion in the strain collection compared to the control strains, in which YFG1 is replaced by wild-type YFG1::URA3. Because strain identification is performed by microarray or sequencing of each gene deletion’s unique barcode, strain identity cannot be misattributed as it can with potentially cross-contaminated plate arrays. In addition, the various heterozygous diploid strains largely grow at the same rate, avoiding the bias in gene representation seen in pooled haploid deletion strains.
Fig. 4.
GI screens using dSLAM. a Schematic of the DNA molecules and manipulations resulting in the generation of a double mutant by dSLAM. b The dSLAM workflow starts with a pool of various (usually the entire collection) competent diploid heterozygous deletion strains that contain the SGA MATa selection marker. The pooled cells are transformed with a query YIp that will introduce a gene edit such as a deletion (denoted by your favorite gene yfg1∆) or an overexpression cassette. The edited cells undergo selection for the edit (nat) and the gene deletions (kan), and are sporulated. After sporulation, MATa haploid cells are selected on media lacking histidine, and then double mutant haploids are selected. The barcode abundances of the various deletion strain are measured by microarray or sequencing. Depletion of barcodes indicates a negative GI with the query mutation. gene 1/2∆::kanMX: heterozygous strain collection gene deletions replaced with G418-resistance marker, YFG1: wild-type your favorite gene, yfg1∆::natMX: query mutation of your favorite gene deleted and replaced with nourseothricin-resistance marker, MFA1pr-HISMX MATa: haploid selection marker that confers growth on media lacking histidine and marked with G418-resistance marker striped regions—barcodes unique to each gene deletion
To our knowledge, dSLAM has been used only by the group that developed the method. The lack of widespread adoption of this approach is likely due to the technical complexity of barcode quantification by microarray hybridization or sequencing. Both methods require very careful bioinformatics analysis of the raw data that are beyond the capabilities of labs that do not routinely use such genomic methods and do not have a bioinformatician on staff. Nevertheless, dSLAM has been used to screen for synthetic lethal, dosage synthetic lethal, dosage suppression, and synthetic suppression interactions (Pan et al. 2004; Kim et al. 2009).
Although dSLAM seeks to avoid many of the limitations of SGA, it is very likely to lose completeness of genome coverage due to lack of transformation of random deletion strains in the pool of strains. More than 5% of genes will be excluded from any given analysis because the deletion strain did not integrate the query mutation or the marked wild-type query allele for the control (Pan et al. 2004). Moreover, some of the diploid deletion barcodes have been mutated during strain construction and do not hybridize well to microarrays, which are designed for the haploid strain barcodes (Pan et al. 2004). Using sequencing approaches to quantify the barcodes (amplifying from flanking universal primer tags) should solve this problem.
The Green Monster
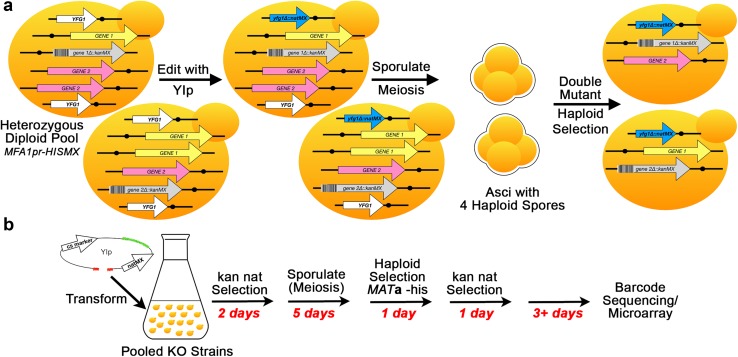
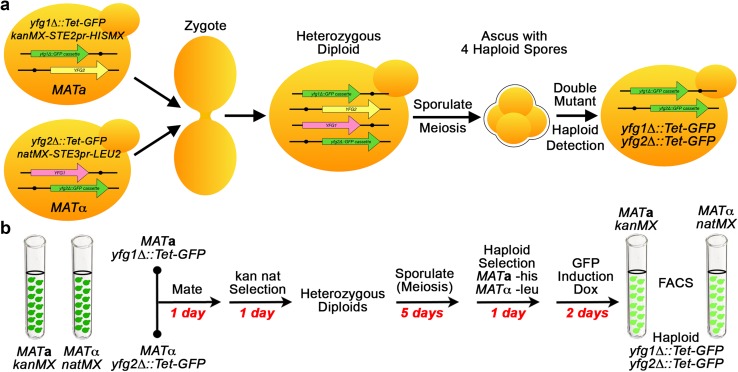
An approach to multi-mutant strain construction, called the Green Monster, is essentially SGA performed with a pooled collection of strains (Fig. 5). Different ORFs are replaced with a green fluorescent protein (GFP) expression cassette in strains carrying the SGA haploid selection markers (Suzuki et al. 2011). Using GFP, the Green Monster method bypasses the need to mark each gene deletion with a different selectable marker. The authors found a near-linear correlation between fluorescence intensity and the number of deletions. Combinations of deletions are made by mating and sporulating pools of deletion/GFP strains and then passing them through a fluorescence activated cell sorter to find progressively brighter cells. One may also perform multiple rounds of mating, sporulation, and haploid selection and only sort cells after numerous rounds to save time. Performing all steps in liquid cultures saves time and cost of media, and, because strains bearing multiple deletions from previous rounds can mate, fewer rounds of crosses are needed to obtain cells carrying all of the gene deletions in the pool compared to SGA.
Fig. 5.
GI screens using green monster. a Schematic of the DNA molecules and manipulations resulting in the generation of a double mutant by Green Monster. b The Green Monster workflow starts with various strains from MATa and MATα haploid deletion collections in which the kanMX marker is replaced with a Tet-induced GFP expression cassette (denoted by your favorite gene yfg1∆ and yfg2∆). The MATa strains also carry a GMToolkit-a (kanMX-STE2pr-HISMX) and the MATα strains carry a GMToolkit-α (natMX-STE3pr-LEU2), both of which are derived from the SGA haploid selection reporter. These strains are randomly mated in bulk, and diploids are grown under double selection conditions. The resulting heterozygous diploids are sporulated and haploids of both mating types are selected. FACS analysis sorts cells that have twice the GFP intensity of the single mutants and, therefore, carry two deletions. The entire culture can be re-mated without FACS analysis to make mutants that carry 3, or more gene deletions. Red text indicates how many days are required for each step in the workflow. yfg1/2∆::Tet-GFP: query deletions of your favorite genes deleted and replaced with Tet-inducible GFP, kanMX-STE2pr-HISMX MATa: haploid selection marker that confers growth on media lacking histidine and marked with G418-resistance marker, natMX-STE3pr-LEU2 MATα: haploid selection marker that confers growth on media lacking leucine and marked with nourseothricin-resistance marker
Suzuki et al. (2011) used the Green Monster method to make a mutant in which 16 different ABC transporter genes associated with multi-drug resistance are deleted. The mutant’s drug sensitivity spectrum was analyzed and compared to the wild-type parent and a mutant carrying deletions of eight different ABC transporters. To the best of our knowledge, this study is the only use of the Green Monster for genetic analysis.
Although this method is much faster than SGA, it still requires multiple rounds of crosses and also careful quantification of GFP by fluorescence-activated cell sorting (FACS). Furthermore, finding specific intermediate strains requires considerable screening of sorted cells and it is difficult to determine if the inability to find certain combinations of mutations is due to negative GIs or technical problems, especially when using large numbers of deletion strains. The authors obtained intermediate strains by crossing pools of subsets of all the deletion strains. The requirement for single-cell sorting and screening to find intermediate mutants and for precise fluorescence measurements by FACS has probably limited adoption of this method, although few researchers have attempted to make large numbers of strains carrying 4 or more mutant alleles.
Genetic screens in the age of CRISPR/Cas
The efficiency of gene editing by HDR with donor DNA is vastly improved when a DSB is introduced at the target locus using a sequence-specific endonuclease (Guha and Edgell 2017). Because of the ease of programming its sequence specificity, the most versatile endonuclease-based genome editing system is CRISPR/Cas (Fig. 6a). There are several excellent reviews on the discovery and mechanism of CRISPR/Cas (Lander 2016; Stovicek et al. 2017; Raschmanová et al. 2018).
Fig. 6.
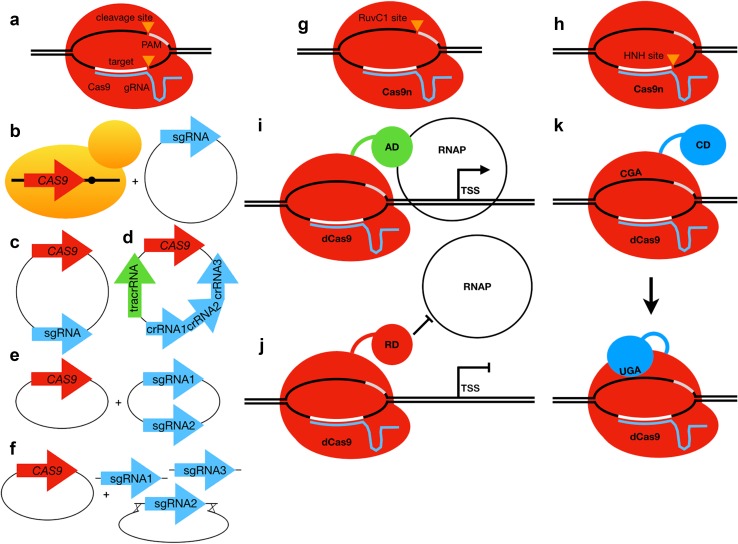
Using CRISPR/Cas for genome editing. a Schematic showing how Cas9-gRNA generate DSBs at programmed target sites. b–f Different expression systems for CRISPR/Cas9 editing in yeast. b A yeast strain expressing inducible Cas9 from an integrated cassette requires a plasmid expressing sgRNA. c A CRISPR/Cas expression system requiring only one plasmid expressing both Cas9 and sgRNA. d A multiplex CRISPR/Cas system expressing Cas9, a tracrRNA, and a CRISPR/Cas array containing different crRNAs. e A multiplex CRISPR/Cas system expressing Cas9 and two or more sgRNAs on separate plasmids. f A multiplex CRISPR/Cas system expressing Cas9 from one plasmid, and using in vivo gap repair of a selectable plasmid by linear sgRNA cassettes to generate the sgRNA expression plasmid. g Schematic showing how nCas9 (D10A mutation in the RuvC1 endonuclease active site) generates a nick on the opposite strand of the target site. h Schematic showing how nCas9 (H840A mutation in the HNH endonuclease active site) generates a nick on the same strand of the target site. i Schematic showing how catalytically inactive dCas9 can be fused to an activation domain (AD) to recruit RNA polymerase (RNAP) and promote transcription of the target gene. j Schematic showing how catalytically inactive dCas9 can be fused to a repressor domain (RD) to inhibit RNA polymerase (RNAP) and prevent transcription of the target gene. k Schematic showing how catalytically inactive dCas9 can be fused to cytidine deaminase (CD) to convert a cytosine to a uracil. A round of repair and replication either converts the U back to C, or converts the U to a T to generate a TGA STOP codon. PAM: protospacer adjacent motif, gRNA: guide RNA including the Cas-interacting stem-loop structure, tracrRNA: gRNA portion that hybridizes with crRNA and contains stem-loop, crRNA: gRNA portion that hybridizes with crRNA and contains DNA targeting sequence, sgRNA: a gRNA that combines the tracrRNA and crRNA in a single RNA molecule, AD: activation domain, RD: repressor domain, CD: cytidine deaminase, RNAP: RNA polymerase II, TSS: transcription start site
Yeast CRISPR/Cas systems
The CRISPR/Cas system was first adapted for gene editing in mammalian cells (Jinek et al. 2013; Cong et al. 2013; Mali et al. 2013), soon followed by applications in many other organisms, including budding yeast (Dicarlo et al. 2013). In the case of mammalian cells, DSBs generated by endonucleases are usually repaired by non-homologous end joining (NHEJ) even in the presence of donor DNA for HDR (Guha and Edgell 2017). Mammalian cell researchers have used a variety of methods to increase the frequency of HDR (Kooistra et al. 2004; Pöggeler and Kück 2006; Choquer et al. 2008; Fox et al. 2009; Certo et al. 2012; Delacôte et al. 2013; Verbeke et al. 2013; Ran et al. 2013; He et al. 2014; Lin et al. 2014; Schwartz et al. 2017). However, in S. cerevisiae, efforts to increase HDR over NHEJ are not necessary. In fact, NHEJ in yeast is so rare that only HDR is used for editing (although CRISPR/Cas is useful for mechanistic studies of NHEJ) (Gallagher and Haber 2018; Lemos et al. 2018).
There are over two dozen CRISPR/Cas toolkits for S. cerevisiae, most of which have been described in detail by Stovicek et al. (2017) and Raschmanová et al. (2018). Most CRISPR/Cas editing tools combine the naturally separate CRISPR RNA (crRNA) and trans-activating CRISPR RNA (tracrRNA) in a hybrid small guide RNA (sgRNA). The generic term gRNA refers to either tracrRNA-crRNA, crRNA, or sgRNA. The main differences between the various yeast CRISPR/Cas systems are their plasmid structures.
There are a few systems that integrate the Cas9 expression construct into the yeast genome (Fig. 6b) (Horwitz et al. 2015; Mans et al. 2015; Vanegas et al. 2017), but most systems express Cas9 from a plasmid (Fig. 6c–f). Integration of Cas9 has several advantages. It circumvents the requirement for transformation with a Cas9 expression plasmid (leaving more available markers for multiplexing) and subsequent counterselection of the plasmid to remove Cas9. The integrated Cas9 cassettes are also inducible. However, to remove the potential influence of the integrated Cas9 on strain phenotype (due to leaky basal expression), the Cas9 cassette should be removed by targeting it with a further gRNA and replacing the endogenous locus (Finnigan and Thorner 2016). In some yeast CRISPR/Cas systems, the Cas9 and gRNA expression cassettes are on the same plasmid, which improves the efficiency of transformation (Fig. 6c, d). However, single plasmid systems are incompatible with some of the gRNA cloning approaches. For example, because Cas9 is large and generating a Cas9-gRNA plasmid by PCR amplification can be challenging (Ryan and Cate 2014). In addition, editing efficiencies are much higher if Cas9 is expressed before introduction of the gRNA (Walter et al. 2016), making CRISPR/Cas editing inefficient when using a single Cas-gRNA plasmid. Finally, making many different mutant combinations of genes by multiplexing could be more easily accomplished by having separate expression cassettes for each gene (Fig. 6e, f).
There is significant variability in efficiency between different loci and gRNAs, probably due to differences in gRNA secondary structure that affect gRNA binding to its target and/or nucleosome occupancy at or near the target sequence (Smith et al. 2016; Horlbeck et al. 2016; Thyme et al. 2016). Because of this unpredictable variability, several gRNAs must be tested for each target gene (Ryan et al. 2014; Shalem et al. 2014; Wang et al. 2014; Koike-Yusa et al. 2014; Zhou et al. 2014; Konermann et al. 2015; Chen et al. 2017; Smith et al. 2017; Sadhu et al. 2018). It is generally presumed that as more gRNAs are tested in yeast, empirical or mechanistic design rules will improve. An additional and important consideration in designing gRNAs is the potential for off-target DNA binding due to sequence similarities. Although off-target mutations when using CRISPR to edit genes are of debatable concern in higher eukaryotes (Fu et al. 2013; Cho et al. 2014; Wang et al. 2015, 2018a, b; Iyer et al. 2015, 2018; Muhammad et al. 2016; Zhang et al. 2018), off-target mutations are considered to be unlikely in yeast due to their small genomes (making it easier to find stringent target sequences) and low error-prone NHEJ activity (Ryan et al. 2014; Jakočiunas et al. 2015a). Consistently, deep sequencing of CRISPR edited strains showed no off-target mutations (Jakočiunas et al. 2015a).
Nevertheless, off-target editing is likely to depend largely on the gRNA sequence and Cas9 activity. Reducing the likelihood of off-target mutations can be accomplished with numerous online gRNA design tools (reviewed in Stovicek et al. 2017). One can also ensure target specificity using Cas9n, a nickase in which one of the two endonuclease active sites is mutated (Fig. 6g, h), with two closely spaced gRNAs. Off-target DNA nicking by a single Cas9n will be quickly repaired, but only the combination of two close nicks on opposite strands will produce a (staggered) DSB.
CRISPR/Cas has seen only limited use for systematic functional genomics or genetic screens in yeast (Smith et al. 2016, 2017; Chen et al. 2017; Roy et al. 2018; Sadhu et al. 2018; Guo et al. 2018), largely because there were already many different genomic mutant collections when CRISPR/Cas gene-editing was developed. Nevertheless, CRISPR/Cas has been used to probe gene function in ways that current strain collections cannot, including examining ORFs that are not mutated in any strain collection. In addition, CRISPR/Cas editing can be markerless and scarless, avoiding possible phenotypic effects of selectable markers and exogenous DNA (Acton et al. 2017; Elison and Acar 2018). When making markerless/scarless edits, if the desired mutation(s) does not occur in an available gRNA target, the donor DNA must include multiple silent mutations in the PAM and/or target sequence to prevent cyclic re-cutting (Horwitz et al. 2015). Of course, in non-coding regions, neutral mutations cannot be predicted. One can avoid making silent mutations using a two-step process in which one first replaces the PAM-target sequence with heterologous “stuffer” donor sequence (Biot-Pelletier and Martin 2016; Elison and Acar 2018). The stuffer can then be replaced with donor DNA encoding only the desired point mutation by exchanging the original gRNA with a gRNA targeting the stuffer. The second donor DNA can reconstitute the original PAM-target sequence without consequence.
Yeast CRISPR/Cas libraries
Several yeast CRISPR/Cas libraries have been designed for generating LOF mutants. In the library produced by Sadhu et al. (2018), gRNAs are paired with donor DNA designed to introduce STOP codons. This library was designed to target all annotated essential genes in yeast consisting of ~ 10,000 gRNA-donor plasmids targeting different portions of ~ 1000 essential genes (Sadhu et al. 2018). The gRNAs were designed to replace the cas-binding protospacer adjacent motif (PAM) sites (NGG) in the target DNA with stop codons (TGA or TAG). This functional genomic screen showed that many essential genes could tolerate C-terminal truncations. Guo et al. (2018) similarly used a paired gRNA-donor CRISPR/Cas library to make small START codon deletions in a set of ~ 300 verified ORFs and an equal number of unverified small ORFs (smORFs) to determine which of these ORFs (which are not deleted in the KO collection) are functional (Guo et al. 2018). Roy et al. (2018) used a gRNA-donor CRISPR/Cas library to individually introduce ~ 35,000 natural sequence variants (SNPs and indels) from a wine yeast into the S288C lab strain (Roy et al. 2018). The sequence variants were selected based on whether they altered a PAM or nearby sequences that would be included in the gRNA sequence.
Rather than developing gRNA libraries, a number of groups have developed donor DNA libraries for massive editing of a handful of genomic loci with just a few gRNAs. For instance, Kuivanen et al. developed a synthetic promoter library for multiplexed promoter replacement (Kuivanen et al. 2018). Si et al. (2017) developed a cDNA library that expresses sense or anti-sense RNA depending on the orientation of the ORF in a standard expression cassette. This library included > 90% of annotated genes. They then used CRISPR/Cas9 to integrate the pooled library into multiple ∂ sites in a yeast strain that expresses RNAi machinery (Dicer, Ago2 and TRBP; S. cerevisiae does not naturally perform RNAi) (Si et al. 2017). Sense strand cassettes generated GOF alleles while anti-sense cassettes generated LOF alleles. They used this approach to perform multiplex integrations of random GOF and LOF alleles and screened the pooled transformants for improvements in several metabolic traits.
Instead of directly editing genes to analyze LOF mutations, CRISPR/Cas can also be used to modulate gene expression to find GOF or LOF phenotypes. One approach is to use dCas9, a catalytically inactive cas9 mutant, fused to a transcriptional activator or repressor domain to produce GOF or LOF conditions—CRISPRa and CRISPRi, respectively (Fig. 6i, j) (Larson et al. 2013; Perez-Pinera et al. 2013; Gilbert et al. 2013, 2014; Maeder et al. 2013; Konermann et al. 2015). Smith et al. (2016) developed a pooled plasmid library of 989 gRNAs against the 5′UTRs of 20 genes to perform CRISPRi for a chemical genetics screen (Smith et al. 2016). This group subsequently developed an arrayed strain collection in which a CRISPRi gRNA library and inducible dCas9 were chromosomally integrated to knock-down 1357 essential yeast genes in 3,832 strains (Smith et al. 2017). In contrast, Chen et al. (2017) searched for GOF interactions by developing a yeast CRISPRa pooled plasmid library consisting of ~ 107 random gRNAs, which they used in conjunction with a dCas9-VP64 synthetic transcriptional activator (Chen et al. 2017). They used this gRNA library to perform a screen for sgRNAs that suppressed lethality of human α-synuclein overexpression.
So far, the studies of Si et al. (2017) and Chen et al. (2017) are the only uses of CRISPR/Cas to perform systematic GI screens, and both employed CRISPRi/a without mutating genes. There are several limitations to adoption of CRISPR/Cas editing to perform systematic GI screens. Most important is the lack of genome-wide gRNA plasmid libraries suitable for GI screens. LOF gRNA libraries that could be used to transform query mutant strains for GI screens include the premature termination codon library of Sadhu et al. (2018) and the small deletion library of Guo et al. (2018). However, both libraries focus on ORFs not represented in the haploid deletion strain collections namely essential genes and smORFs, respectively. The only genome-wide (and random) gRNA library currently available is the GOF (CRISPRa) library of Chen et al. (2017). Second, all current systematic gRNA plasmid libraries are designed to generate genetic variation in pooled strains, requiring one to select for, or screen variants within a heterogenous population of transformants (Smith et al. 2016; Chen et al. 2017; Sadhu et al. 2018; Guo et al. 2018). Although, identifying genetic changes associated with phenotypes can be determined by sequencing the gRNA expression cassettes or associated barcodes, it is not possible to consistently find and measure the fitness of strains exhibiting subtle GIs in these pools. Finally, with the exception of the Chen et al. (2017) CRISPRa gRNA library, current gRNA plasmid libraries pair the gRNA with donor DNA on the same plasmid, locking in the type of gene edit.
Crucially, in all of these studies, transformations were performed in a manner to ensure that most cells receive only one plasmid, preventing multiplexing. Even if transformations were performed with high titers of plasmid, there is no way to select for rare multi-mutant transformants in such pools as the donor DNA is unmarked. In the Chen et al. (2017) study, multiple genes were down-regulated using a single gRNA—in essence mirroring multi-gene LOF GIs. Whether the gRNA was able to directly target each of these genes, or whether it knocked down a common transcription factor for these genes was not determined. They also found suppressed strains carrying multiple gRNAs, but did not study these further (Chen et al. 2017).
None of the CRISPR/Cas libraries discussed above have been deposited in commercial or public repositories, but some should be available by request.
Hypothetical workflow comparison of SGA and CRISPR/Cas
Regardless of the availability of CRISPR/Cas plasmid libraries, if one is interested in a few dozen genes, using CRISPR/Cas to make thousands of possible combinations of edited versions of these genes is very feasible from a cost perspective, and could be accomplished much faster than could be done by SGA (Fig. 7).
Fig. 7.
GI screens using multiplexed CRISPR/Cas gene editing. a Schematic of the DNA molecules and manipulations resulting in the generation of a double mutant using multiplexed CRISPR/Cas to generated two gene deletions replaced with two different selectable markers. b High-throughput workflow to generate multiplex edits. All CRISPR/Cas manipulations are performed in 96-well plates. In this example, all wells carry competent cells of the same strain (wild-type or mutant), but two different sgRNA-Donor plasmids are combined in each well. All wells also receive the same Cas9 expression plasmid. After selection of triply transformed cells, Cas9 expression is induced for multiple generations. Counterselection of the Cas9 plasmid shuts off gene editing, and doubly edited cells are selected (each edit has a different selectable marker). The red text indicates how many days are required for each step in the workflow. kanMX: G418-resistance marker, natMX: nourseothricin-resistance marker; colored wells indicate that the contents of each well are different
For example, if one would like to test all combinations of two to six deletions of a set of 10 genes (837 possible combinations) to find synthetic suppressor/enhancer interactions, one would have to perform 5 consecutive SGA crosses to first make double mutants, then triple mutants, etc. These iterations with single mutants are necessary because the products of SGA crosses cannot be combined, for example, to make quadruple mutants from two sets of double mutants made by SGA of single mutants (Tong and Boone 2006).
CRISPR/Cas does not require crossing strains and/or sporulation of diploids and, therefore, is much faster than SGA, dSLAM, and Green Monster approaches. To make the same mutants by CRISPR/Cas, one would start by designing, cloning, and testing 2–3 gRNAs and 10 donor DNAs, one for each of the 10 genes to determine which gRNAs are best able to perform the desired edits. Multiple gRNAs are tested for each target gene because of the aforementioned problems in predicting gRNA efficiency. Using CRISPR/Cas with validated gRNAs, one could potentially make all 837 mutant strains by performing transformations in 96-well plates with the appropriate combination of gRNA plasmids and donor DNA in each well (Fig. 7b). Although markerless edits would be possible, the screening process would not easily translate to a high-throughput workflow, especially when dealing with editing efficiencies of < 25% for 4 + multiplexed edits. Therefore, this process would benefit from the use of a different selectable marker for each gene in the desired combination.
Using CRISPR, a separate strain does not have to be made for each deletion and marker pair to facilitate crosses. Instead, each deletion is encoded in a donor DNA-gRNA pair that can be combined with other donor DNAs and gRNAs to multiplex gene deletions. These donor DNAs are easily generated using a single primer pair with homology arms specific to each target gene to PCR amplify a set of marker cassettes using common primer binding sites (Fig. 2a). Nevertheless, planning which gRNAs and donor DNAs go into each well and minimizing the number of CRISPR/Cas donor DNAs required to make the desired genetic combinations could benefit from automated workflow design (Pratapa et al. 2018).
An alternative application of CRISPR/Cas that would bypass the requirement for plasmid and donor DNA transformation employs a gene drive CRISPR/Cas cassette. In this approach, the cassette replaces an ORF in a haploid strain and consists of inducible Cas9 and sgRNA expression constructs (DiCarlo et al. 2015; Roggenkamp et al. 2017; Vaschetto 2018). The sgRNA targets the wild-type sequence of the gene deleted by the gene drive cassette. Mating the query strain to an array of collection strains such as the OE collection, produces a heterozygous diploid. Subsequent Cas9 induction will cut and convert the wild-type gene into the gene drive cassette, thus generating a homozygous diploid that can be assessed for synthetic dosage interactions. The cassette could also include a recessive allele of the target gene rather than a deletion (Roggenkamp et al. 2017). This method bypasses the need to sporulate and select for haploids, thus shaving a few weeks off the standard SGA approach, and has been used in Candida albicans to generate arrays of a few hundred double mutants targeting genes with related functions (Shapiro et al. 2018).
Discussion and future work
Despite the small size and simplicity of the S. cerevisiae genome, systematically interrogating its pairwise GI spectrum has occupied yeast geneticists for nearly two decades, and is ongoing (Costanzo et al. 2010). It is clear from this massive effort that there exists a significant level of robustness in yeast cells, partly due to network architecture resulting in compensatory feedback mechanisms and alternative pathways, and partly due to functional redundancy between genes (Gu et al. 2003; Guan et al. 2007; Bajić et al. 2014; Bauer et al. 2015; Bader et al. 2015; Cohen et al. 2016; Patra et al. 2017; Ishikawa et al. 2017; Veitia 2017). To better understand genetic robustness and genotype–phenotype relationships, it is imperative that we map multi-gene interactions. Efforts to do so are in their infancy (Haber et al. 2013; Kuzmin et al. 2018), largely because performing such systematic screens is technically demanding with current methods. So far, the multiplexing capabilities of CRISPR/Cas have been exploited mostly by metabolic engineers and synthetic biologists constructing complex genetic circuits and biosynthetic pathways in a handful of strains (Shao et al. 2009; Mikkelsen et al. 2012; Hasunuma et al. 2014; Ryan et al. 2014; Stovicek et al. 2015; Tsai et al. 2015; Jakočiunas et al. 2015b; Ronda et al. 2015; Walter et al. 2016; Jessop-Fabre et al. 2016; Shi et al. 2016; Garst et al. 2017; Kuivanen et al. 2018). However, geneticists could also take advantage of the multiplexing capabilities of CRISPR/Cas to perform systematic GI screens to generate large numbers of strains carrying three or more alleles.
We have described a variety of methods that have been utilized to perform systematic GI screens. In Table 1, we compare and contrast the main features of these approaches. Only three methods—SGA, the Green Monster, and CRISPR/Cas editing—are capable of systematically generating strains carrying three or more mutant alleles. However, it is unclear if the generation of strains carrying three or more mutations is even practical by iterative SGA. Because of the high frequency of false negatives, the SGA approach requires validation of the progeny strains before using them in further crosses. Studies that have generated triple mutants by SGA have all started with double mutant query strains (Haber et al. 2013; Kuzmin et al. 2018).
Table 1.
Comparison of GI screen methods
| Method | Available systematic strain collections |
Available systematic plasmid libraries |
Time to combine 2 allelesa | Time to combine 3 allelesa | Types of GIsb | Selection method | Plasmids required |
|---|---|---|---|---|---|---|---|
| OE plasmid | Yesc | Yesg | 3 days | N/A | DS, DE | 2 markers | Episomal |
| SGA | Yesd | Yesg | 17 days | 34 days | DS, DE, SS, SE | 2 + markers | Noi |
| dSLAM | Yesd | No | 11 days | N/A | DS, DE, SS, SE | 2 markers | YIp |
| Green Monster | Noe | N/A | 11 days | 22 days | SS, SE | GFP intensity | No |
| CRISPR/Cas | Yesf | Yesh | 7 days | 7 days | DS, DE, SS, SE | 2 + markers | Episomal |
aStarting with single-gene alleles. Not including genotyping of strains. Approximate time given in days
bDS dosage suppression, DE dosage enhancement including synthetic dosage lethality, SS synthetic suppression, SE synthetic enhancement including synthetic lethality
cSopko et al. (2006), Douglas et al. (2012), Youn et al. (2017), also deletion strain collections can be used in conjunction with a query plasmid
dGiaever et al. (2002)
eGiaever et al. (2002) the haploid KO collection is used as the base strain, but the kanMX cassettes must be converted to the Green Monster GFP cassette
fSmith et al. (2017)
gZhu et al. (2001), Gelperin et al. (2005), Moriya et al. (2006), Hu et al. (2007), Magtanong et al. (2011)
hSmith et al. (2016), Chen et al. (2017), Guo et al. (2018), Sadhu et al. (2018)
iFor dosage screens, the query strain can carry an episomal OE plasmid or an integrated OE cassette
Moreover, the reagents do not currently exist for the Green Monster to perform systematic screens, which would require the replacement of the kanMX cassette with the GFP cassette in at least a large proportion of the ~ 5000 haploid KO strains. Finally, current CRISPR/Cas gRNA libraries do not include most of the non-essential ORFs and these libraries are not well-suited to multiplexing because their donor DNAs are unmarked. To use unmarked alleles in a high-throughput genetic screen would require multiplex editing efficiencies of nearly 100% (in practice < 75% for 3 edits), particularly when one is searching for negative GIs, as cells that do not incorporate the desired edits would have growth advantages.
One might wonder what benefit genome-scale yeast CRISPR/Cas libraries could have over current yeast strain collections? Even when analyzing singly edited strains, the use of CRISPR/Cas libraries have produced more specific genetic modifications than current collections, resulting in new insights into gene structure and function (Sadhu et al. 2018; Guo et al. 2018). In addition, even simple LOF genome-scale CRISPR/Cas libraries have the potential to quickly assess highly combinatorial GI networks by editing existing systematic mutant strain collections. However, to expand their combinatorial potential, future CRISPR/Cas library designs will have to enhance multiplexing.
Although the gRNAs are the smallest component of CRISPR/Cas systems, the cost of automation and DNA reagents to make genome-scale gRNA plasmid collections is likely prohibitive for most labs. As we mentioned above, gRNA libraries must include several gRNAs for each target, making the size of genome-wide gRNA libraries very large (tens or hundreds of thousands), even for a small genome like yeast. Smith et al. (2017) addressed this issue by developing a more cost-effective method of generating genome-wide sequence-verified gRNA libraries (several thousand dollars in reagents and a few months of labor), and applied this method to the construction of a collection of CRISPRi strains allowing inducible knockdowns of most essential genes (Smith et al. 2017). In addition to the cost of gRNA oligonucleotide libraries, cloning of the gRNAs into expression vectors is a labor-intensive process, even with streamlined methods of cloning [reviewed in (Stovicek et al. 2017)].
Making genome-wide CRISPR/Cas libraries in yeast could also be simplified by using Cas9 derivatives that do not use HDR and donor DNA. We already discussed how CRISPRa/i can produce knockdown or dosage effects by directly influencing expression of the target genes without needing to also introduce a donor DNA (Gilbert et al. 2013; Smith et al. 2016, 2017; Chen et al. 2017). However, positioning of the dCas9 fusion protein is crucial to transcriptional activity. Therefore, the gRNA libraries for CRISPRi/a tend to be even larger than the knock-out libraries since each gene is targeted by more gRNAs (e.g., 10 per target gene). Another CRISPR/Cas method that does not require donor DNA uses a catalytically inactive dCas9 fused to cytidine deaminase to change targeted CAG, CGA, CAA or TGG to the STOP codons TAG, TGA or TAA, respectively, after a round of repair and replication (Fig. 6k) (Nishida et al. 2016; Komor et al. 2016; Kuscu et al. 2017; Billon et al. 2017). The mismatch repair machinery of the cell has a 50–50 chance of repairing the U instead of the G, but by making a nick in the G-containing strand using an Cas9n-cytidine deaminase fusion, one can trick the cell into saving the modified base. Unfortunately, the deaminase tends to also modify off-target cytidines, so future improvements to this method will couple the deaminase activity to PAM-target binding (Komor et al. 2016).
Because efficient gene editing in S. cerevisiae has been possible for more than 20 years, CRISPR/Cas has not been as widely adopted as in other model systems. Consequently, there are not yet many yeast CRISPR/Cas libraries, and these are pooled, requiring microarrays or sequencing to identify mutants that grow better or worse. In contrast, because yeast plasmid libraries and strain collections are arrayed, the function of each gene can be assessed for more complex phenotypes than relative growth/fitness in a population. Although they are more labor-intensive than pooled libraries, arrayed CRISPR/Cas libraries would allow one to perform more diverse phenotypic tests, including high content imaging (Henser-Brownhill et al. 2017; de Groot et al. 2018). Arrayed libraries also ensure that no genomic targets are missed, a distinct possibility when using pooled plasmid libraries that require occasional amplification. The generation of arrayed yeast CRISPR/Cas libraries is an inevitable next step.
Almost all of the current yeast strain systematic collections have been generated by gene editing using transformation with PCR products amplified from plasmids carrying cassettes of selectable markers alone or combined with protein-tagging or promoter-replacement sequences (Längle-Rouault and Jacobs 1995; Longtine et al. 1998; Baker Brachmann et al. 1998; Bähler et al. 1998; Goldstein and McCusker 1999; Gueldener 2002; Janke et al. 2004; Hentges et al. 2005; Akada et al. 2006). CRISPR/Cas could be used to modify existing genomic strain collections using a single gRNA and, in most cases, a single-donor DNA, in a manner akin to integrating a target sequence into various chromosomal loci and using a single gRNA to edit at all locations (Finnigan and Thorner 2016; Giersch and Finnigan 2017), except that the target sequences are already integrated in the yeast strain collection.
Using CRISPR/Cas to edit deletion strain collections, one could remove the kanMX marker from the deletion strains and replace it with markerless donor DNA fragments homologous to the two ends of the kanMX cassette. Although other methods have been used to make markerless deletions from the deletion collection, they also remove the deletion barcodes (Carvalho et al. 2013; Soreanu et al. 2018). Similarly, one could swap the kanMX marker from the deletion strains with other MX markers. Marker swapping methods using plasmids or PCR cassettes have been in use for over a decade (Alani et al. 1987; Longtine et al. 1998; Baker Brachmann et al. 1998; Goldstein and McCusker 1999; Voth et al. 2003). However, none have been used to systematically swap the markers of genomic collections. Marker-swapped collections would be useful for SGA, facilitating comprehensive SGA screens and also making the production of three or more edited combinations easier. One could use CRISPR/Cas to replace deletion strain markers with the GFP expression cassette used for the Green Monster method allowing for systematic construction of multi-deletion strains faster than possible using SGA. A systematic collection of gene drive deletion strains could be generated by replacing the kanMX cassette with gene drive cassettes containing an sgRNA specific to the wild-type copy of each deleted gene. This collection could be used to make double mutants using an SGA approach to cross query gene drive strains to the gene drive collection and inducing Cas9 to generate homozygous double mutants. Finally, one could introduce fiducial markers at any chromosomal location to study chromatin-chromosome dynamics. For example, using CRISPR/Cas9, Soreanu et al. (2018) replaced a natMX gene deletion with a TetO array in cells expressing TetR-tdTomato (Soreanu et al. 2018). The same approach could be used to systematically mark positions along all chromosomes using the deletion collection, or using a gRNA library paired with a dCas9-fluorescent protein (Chen and Huang 2014).
Using CRISPR/Cas to edit the GFP or TAP strain collection, one could replace the C-terminal fusion cassette with modern variants of GFP, other fluorescent protein derivatives, or other protein fusion tags. Replacement of the old cassettes could simultaneously remove the marker. Roggenkamp et al. (2017) tested a CRISPR/Cas method to do just this, but so far this approach has only been used to modify a few strains at a time (Roggenkamp et al. 2017). One could also replace the C-terminal tag cassette with a markerless MS2 or PP7 cassette for mRNA tagging. However, in this approach, the homology arms of the donor DNA would have to be specific to each ORF to assure that the mRNA tag alone is integrated between the STOP codon and 3′UTR.
Despite the versatility and ease of gene editing in yeast, the generation of genome-wide strain collections is a costly and time-consuming process usually performed by a consortium of labs. Consequently, yeast cell biologists and geneticists have had to rely on strain collections that are sometimes not optimal for their needs. Moreover, current high-throughput methods of generating multi-mutant strains rely on slow methods that all require the production of multi-mutant diploids and sporulation to obtain multi-mutant haploids. CRISPR/Cas has the potential both to rapidly enhance our current genomic strain collections and to systematically generate genome-wide 3-way or even higher dimensional GI maps.
Acknowledgements
We thank an anonymous reviewer for suggesting the application of CRISPR/Cas gene drives to genetic interaction screens. This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health (R01GM078989) and the National Science Foundation awards (1832320 and 1759900).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Acton E, Lee AH-Y, Zhao PJ, et al. Comparative functional genomic screens of three yeast deletion collections reveal unexpected effects of genotype in response to diverse stress. Open Biol. 2017;7:160330. doi: 10.1098/rsob.160330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams AE, Johnson DI, Longnecker RM, et al. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J Cell Biol. 1990;111:131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams AE, Cooper JA, Drubin DG. Unexpected combinations of null mutations in genes encoding the actin cytoskeleton are lethal in yeast. Mol Biol Cell. 1993;4:459–468. doi: 10.1091/mbc.4.5.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akada R, Yamamoto J, Yamashita I. Screening and identification of yeast sequences that cause growth inhibition when overexpressed. Mol Gen Genet. 1997;254:267–274. doi: 10.1007/s004380050415. [DOI] [PubMed] [Google Scholar]
- Akada R, Hirosawa I, Kawahata M, et al. Sets of integrating plasmids and gene disruption cassettes containing improved counter-selection markers designed for repeated use in budding yeast. Yeast. 2002;19:393–402. doi: 10.1002/yea.841. [DOI] [PubMed] [Google Scholar]
- Akada R, Kitagawa T, Kaneko S, et al. PCR-mediated seamless gene deletion and marker recycling in Saccharomyces cerevisiae. Yeast. 2006;23:399–405. doi: 10.1002/yea.1365. [DOI] [PubMed] [Google Scholar]
- Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertini S, Zimmermann FK. The detection of chemically induced chromosomal malsegregation in Saccharomyces cerevisiae D61.M: a literature survey (1984–1990) Mutat Res Genet Toxicol. 1991;258:237–258. doi: 10.1016/0165-1110(91)90011-j. [DOI] [PubMed] [Google Scholar]
- Bader DM, Wilkening S, Lin G, et al. Negative feedback buffers effects of regulatory variants. Mol Syst Biol. 2015;11:785. doi: 10.15252/msb.20145844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J, Wu J-Q, Longtine MS, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bajić D, Moreno-Fenoll C, Poyatos JF. Rewiring of genetic networks in response to modification of genetic background. Genome Biol Evol. 2014;6:3267–3280. doi: 10.1093/gbe/evu255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker Brachmann C, Davies A, Cost GJ, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Baryshnikova A, Costanzo M, Kim Y, et al. Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat Methods. 2010;7:1017–1024. doi: 10.1038/nmeth.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer CR, Li S, Siegal ML. Essential gene disruptions reveal complex relationships between phenotypic robustness, pleiotropy, and fitness. Mol Syst Biol. 2015;11:773. doi: 10.15252/msb.20145264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendert A, Pringle JR. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:1295–1305. doi: 10.1128/mcb.11.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billon P, Bryant EE, Joseph SA, et al. CRISPR-mediated base editing enables efficient disruption of eukaryotic genes through induction of STOP codons. Mol Cell. 2017;67:1068–1079.e4. doi: 10.1016/j.molcel.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biot-Pelletier D, Martin VJJ. Seamless site-directed mutagenesis of the Saccharomyces cerevisiae genome using CRISPR-Cas9. J Biol Eng. 2016;10:1–5. doi: 10.1186/s13036-016-0028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez MA, Gancedo C. Identification of extragenic suppressors of the cif1 mutation in Saccharomyces cerevisiae. Curr Genet. 1994;25:89–94. doi: 10.1007/BF00309531. [DOI] [PubMed] [Google Scholar]
- Bogomolnaya LM, Pathak R, Cham R, et al. A new enrichment approach identifies genes that alter cell cycle progression in Saccharomyces cerevisiae. Curr Genet. 2004;45:350–359. doi: 10.1007/s00294-004-0497-5. [DOI] [PubMed] [Google Scholar]
- Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nat Rev Genet. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- Breslow DK, Cameron DM, Collins SR, et al. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Methods. 2008;5:711–718. doi: 10.1038/nmeth.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson M, Hu G-Z, Ronne H. Gene dosage effects in yeast support broader roles for the LOG1, HAM1 and DUT1 genes in detoxification of nucleotide analogues. PLoS One. 2018;13:e0196840. doi: 10.1371/journal.pone.0196840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho Â, Pereira F, Johansson B. The MX4blaster cassette: Repeated and clean Saccharomyces cerevisiae genome modification using the genome-wide deletion collection. FEMS Yeast Res. 2013;13:711–719. doi: 10.1111/1567-1364.12076. [DOI] [PubMed] [Google Scholar]
- Certo MT, Gwiazda KS, Kuhar R, et al. Coupling endonucleases with DNA end–processing enzymes to drive gene disruption. Nat Methods. 2012;9:973–975. doi: 10.1038/nmeth.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Huang B. Imaging genomic elements in living cells using CRISPR/Cas9. Methods Enzymol. 2014;546:337–354. doi: 10.1016/B978-0-12-801185-0.00016-7. [DOI] [PubMed] [Google Scholar]
- Chen YC, Farzadfard F, Gharaei N, et al. Randomized CRISPR-cas transcriptional perturbation screening reveals protective genes against alpha-synuclein toxicity. Mol Cell. 2017;68:247–257.e5. doi: 10.1016/j.molcel.2017.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim Y, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquer M, Robin G, Le Pêcheur P, et al. Ku70 or Ku80 deficiencies in the fungus Botrytis cinerea facilitate targeting of genes that are hard to knock out in a wild-type context. FEMS Microbiol Lett. 2008;289:225–232. doi: 10.1111/j.1574-6968.2008.01388.x. [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Smith KW, Gustin MC. Osmotic stress and the yeast cytoskeleton: phenotype-specific suppression of an actin mutation. J Cell Biol. 1992;118:561–571. doi: 10.1083/jcb.118.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen O, Oberhardt M, Yizhak K, Ruppin E. Essential genes embody increased mutational robustness to compensate for the lack of backup genetic redundancy. PLoS One. 2016;11:e0168444. doi: 10.1371/journal.pone.0168444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Roguev A, Krogan NJ. Quantitative genetic interaction mapping using the E-MAP approach. Methods Enzymol. 2010;470:205–231. doi: 10.1016/S0076-6879(10)70009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costigan C, Gehrung S, Snyder M. A synthetic lethal screen identifies SLK1, a novel protein kinase homolog implicated in yeast cell morphogenesis and cell growth. Mol Cell Biol. 1992;12:1162–1178. doi: 10.1128/mcb.12.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JA, Yoo J, Bettinger BT, et al. Eliminating gene conversion improves high-throughput genetics in Saccharomyces cerevisiae. Genetics. 2006;172:709–711. doi: 10.1534/genetics.105.047662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davierwala AP, Haynes J, Li Z, et al. The synthetic genetic interaction spectrum of essential genes. Nat Genet. 2005;37:1147–1152. doi: 10.1038/ng1640. [DOI] [PubMed] [Google Scholar]
- de Groot R, Lüthi J, Lindsay H, et al. Large-scale image-based profiling of single-cell phenotypes in arrayed CRISPR-Cas9 gene perturbation screens. Mol Syst Biol. 2018;14:e8064. doi: 10.15252/msb.20178064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacôte F, Perez C, Guyot V, et al. High frequency targeted mutagenesis using engineered endonucleases and DNA-end processing enzymes. PLoS One. 2013;8:e53217. doi: 10.1371/journal.pone.0053217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschbauer AM, Jaramillo DF, Proctor M, et al. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics. 2005;169:1915–1925. doi: 10.1534/genetics.104.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicarlo JE, Norville JE, Mali P, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo JE, Chavez A, Dietz SL, et al. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat Biotechnol. 2015;33:1250–1255. doi: 10.1038/nbt.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AC, Smith AM, Sharifpoor S, et al. Functional analysis with a barcoder yeast gene overexpression system. G3 Genes Genom Genet. 2012;2:1279–1289. doi: 10.1534/g3.112.003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S, Fam HK, Wang YK, et al. Overexpression screens identify conserved dosage chromosome instability genes in yeast and human cancer. Proc Natl Acad Sci. 2016;113:9967–9976. doi: 10.1073/pnas.1611839113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elison GL, Acar M. Scarless genome editing: progress towards understanding genotype–phenotype relationships. Curr Genet. 2018 doi: 10.1007/s00294-018-0850-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnigan GC, Thorner J. mCAL: a new approach for versatile multiplex action of Cas9 using one sgRNA and loci flanked by a programmed target sequence. G3 Genes Genom Genet. 2016;6:2147–2156. doi: 10.1534/g3.116.029801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming MS, Gitler AD. High-throughput yeast plasmid overexpression screen. J Vis Exp. 2011 doi: 10.3791/2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flescher EG, Madden K, Snyder M. Components required for cytokinesis are important for bud site selection in yeast. J Cell Biol. 1993;122:373–386. doi: 10.1083/jcb.122.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL. The art and design of genetic screens: yeast. Nat Rev Genet. 2001;2:659–668. doi: 10.1038/35088500. [DOI] [PubMed] [Google Scholar]
- Fox BA, Ristuccia JG, Gigley JP, Bzik DJ. Efficient gene replacements in Toxoplasma gondii strains deficient for nonhomologous end joining. Eukaryot Cell. 2009;8:520–529. doi: 10.1128/EC.00357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D, Patterson B, Guthrie C. Synthetic lethal mutations suggest interactions between U5 small nuclear rna and four proteins required for the second step of splicing. Mol Cell Biol. 1992;12:5197–5205. doi: 10.1128/mcb.12.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher DN, Haber JE. Repair of a site-specific DNA cleavage: old-school lessons for Cas9-mediated gene editing. ACS Chem Biol. 2018;13:397–405. doi: 10.1021/acschembio.7b00760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst AD, Bassalo MC, Pines G, et al. Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat Biotechnol. 2017;35:48–55. doi: 10.1038/nbt.3718. [DOI] [PubMed] [Google Scholar]
- Gelperin DM, White MA, Wilkinson ML, et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection daniel. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Giersch RM, Finnigan GC. Method for multiplexing CRISPR/Cas9 in Saccharomyces cerevisiae using artificial target DNA sequences. BioProtocols. 2017;7:e2557. doi: 10.21769/BioProtoc.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LAA, Larson MHH, Morsut L, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LAA, Horlbeck MAA, Adamson B, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Gu Z, Steinmetz LM, Gu X, et al. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- Guan Y, Dunham MJ, Troyanskaya OG. Functional analysis of gene duplications in Saccharomyces cerevisiae. Genetics. 2007;175:933–943. doi: 10.1534/genetics.106.064329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueldener U. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:23e–23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha TK, Edgell DR. Applications of alternative nucleases in the age of CRISPR/Cas9. Int J Mol Sci. 2017 doi: 10.3390/ijms18122565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Chavez A, Tung A, et al. High-throughput creation and functional profiling of DNA sequence variant libraries using CrIsPr–Cas9 in yeast. Nat Biotechnol. 2018;36:540–546. doi: 10.1038/nbt.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE, Braberg H, Wu Q, et al. Systematic triple-mutant analysis uncovers functional connectivity between pathways involved in chromosome regulation. Cell Rep. 2013;3:2168–2178. doi: 10.1016/j.celrep.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasunuma T, Hori Y, Sakamoto T, et al. Development of a GIN11/FRT-based multiple-gene integration technique affording inhibitor-tolerant, hemicellulolytic, xylose-utilizing abilities to industrial Saccharomyces cerevisiae strains for ethanol production from undetoxified lignocellulosic hemicelluloses. Microb Cell Fact. 2014;13:145. doi: 10.1186/s12934-014-0145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins KM, Smolke CD. The regulatory roles of the galactose permease and kinase in the induction response of the GAL network in Saccharomyces cerevisiae. J Biol Chem. 2006;281:13485–13492. doi: 10.1074/jbc.M512317200. [DOI] [PubMed] [Google Scholar]
- He Y, Shao Y, Chen F. Efficient gene targeting in ligase IV-deficient Monascus ruber M7 by perturbing the non-homologous end joining pathway. Fungal Biol. 2014;118:846–854. doi: 10.1016/j.funbio.2014.07.003. [DOI] [PubMed] [Google Scholar]
- Henser-Brownhill T, Monserrat J, Scaffidi P. Generation of an arrayed CRISPR-Cas9 library targeting epigenetic regulators: from high-content screens to in vivo assays. Epigenetics. 2017;12:1065–1075. doi: 10.1080/15592294.2017.1395121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentges P, Van Driessche B, Tafforeau L, et al. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast. 2005;22:1013–1019. doi: 10.1002/yea.1291. [DOI] [PubMed] [Google Scholar]
- Horlbeck MA, Witkowsky LB, Guglielmi B, et al. Nucleosomes impede Cas9 access to DNA in vivo and in vitro. Elife. 2016;5:e12677. doi: 10.7554/eLife.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz AA, Walter JM, Schubert MG, et al. Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-Cas. Cell Syst. 2015;1:88–96. doi: 10.1016/j.cels.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Hu Y, Rolfs A, Bhullar B, et al. Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae. Genome Res. 2007;17:536–543. doi: 10.1101/gr.6037607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K, Makanae K, Iwasaki S, et al. Post-translational dosage compensation buffers genetic perturbations to stoichiometry of protein complexes. PLoS Genet. 2017;13:e1006554. doi: 10.1371/journal.pgen.1006554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V, Shen B, Zhang W, et al. Off-target mutations are rare in Cas9-modified mice. Nat Methods. 2018;12:479. doi: 10.1038/nmeth.3408. [DOI] [PubMed] [Google Scholar]
- Iyer V, Boroviak K, Thomas M, et al. No unexpected CRISPR-Cas9 off-target activity revealed by trio sequencing of gene-edited mice. PLoS Genet. 2018;14:e1007503. doi: 10.1371/journal.pgen.1007503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakočiunas T, Bonde I, Herrgård M, et al. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng. 2015;28:213–222. doi: 10.1016/j.ymben.2015.01.008. [DOI] [PubMed] [Google Scholar]
- Jakočiunas T, Rajkumar AS, Zhang J, et al. CasEMBLR: Cas9-facilitated multiloci genomic integration of in vivo assembled DNA parts in Saccharomyces cerevisiae. ACS Synth Biol. 2015;4:1126–1134. doi: 10.1021/acssynbio.5b00007. [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Jessop-Fabre MM, Jakočiūnas T, Stovicek V, et al. EasyClone-MarkerFree: a vector toolkit for marker-less integration of genes into Saccharomyces cerevisiae via CRISPR-Cas9. Biotechnol J. 2016;11:1110–1117. doi: 10.1002/biot.201600147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan Y, Kupiec M. A role for the yeast cell cycle/splicing factor Cdc40 in the G1/S transition. Curr Genet. 2007;51:123–140. doi: 10.1007/s00294-006-0113-y. [DOI] [PubMed] [Google Scholar]
- Kaytor MD, Livingston DM. Allele-specific suppression of temperature-sensitive mutations of the Saccharomyces cerevisiae RAD52 gene. Curr Genet. 1996;29:203–210. doi: 10.1007/BF02221549. [DOI] [PubMed] [Google Scholar]
- Kim J-H, Zhao Y, Pan X, et al. The unfolded protein response is necessary but not sufficient to compensate for defects in disulfide isomerization. J Biol Chem. 2009;284:10400–10408. doi: 10.1074/jbc.M900377200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazono AA, Kron SJ. An essential function of yeast cyclin-dependent kinase Cdc28 maintains chromosome stability. J Biol Chem. 2002;277:48627–48634. doi: 10.1074/jbc.M207247200. [DOI] [PubMed] [Google Scholar]
- Kofoed M, Milbury KL, Chiang JH, et al. An updated collection of sequence barcoded temperature-sensitive alleles of yeast essential genes. G3 Genes Genom Genet. 2015;5:1879–1887. doi: 10.1534/g3.115.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan E-PP, et al. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra R, Hooykaas PJJ, Steensma HY. Efficient gene targeting in Kluyveromyces lactis. Yeast. 2004;21:781–792. doi: 10.1002/yea.1131. [DOI] [PubMed] [Google Scholar]
- Kroll ES, Hyland KM, Hieter P, et al. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics. 1996;143:95–102. doi: 10.1093/genetics/143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuivanen J, Holmström S, Lehtinen B, et al. A high-throughput workflow for CRISPR/Cas9 mediated combinatorial promoter replacements and phenotype characterization in yeast. Biotechnol J. 2018;13:1700593. doi: 10.1002/biot.201700593. [DOI] [PubMed] [Google Scholar]
- Kuscu C, Parlak M, Tufan T, et al. CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nat Methods. 2017;14:710–712. doi: 10.1038/nmeth.4327. [DOI] [PubMed] [Google Scholar]
- Kuzmin E, VanderSluis B, Wang W, et al. Systematic analysis of complex genetic interactions. Science. 2018;360:eaao1729. doi: 10.1126/science.aao1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES. The Heroes of CRISPR. Cell. 2016;164:18–28. doi: 10.1016/j.cell.2015.12.041. [DOI] [PubMed] [Google Scholar]
- Längle-Rouault F, Jacobs E. A method for performing precise alterations in the yeast genome using a recycable selectable marker. Nucleic Acids Res. 1995;23:3079–3081. doi: 10.1093/nar/23.15.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MH, Gilbert LA, Wang X, et al. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos BR, Kaplan AC, Bae JE, et al. CRISPR/Cas9 cleavages in budding yeast reveal templated insertions and strand-specific insertion/deletion profiles. Proc Natl Acad Sci. 2018;115:201716855. doi: 10.1073/pnas.1716855115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Vizeacoumar FJ, Bahr S, et al. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat Biotechnol. 2011;29:361–367. doi: 10.1038/nbt.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindén RO, Eronen V-P, Aittokallio T. Quantitative maps of genetic interactions in yeast—comparative evaluation and integrative analysis. BMC Syst Biol. 2011;5:45. doi: 10.1186/1752-0509-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, van Dyk D, Li Y, et al. A genome-wide synthetic dosage lethality screen reveals multiple pathways that require the functioning of ubiquitin-binding proteins Rad23 and Dsk2. BMC Biol. 2009;7:75. doi: 10.1186/1741-7007-7-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M, McKenzie A, Demartini D, et al. Heterologous modules for versatile and economical PCR -based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;951:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lopes TS, Klootwijk J, Veenstra AE, et al. High-copy-number integration into the ribosomal DNA of Saccharomyces cerevisiae: a new vector for high-level expression. Gene. 1989;79:199–206. doi: 10.1016/0378-1119(89)90202-3. [DOI] [PubMed] [Google Scholar]
- Machin NA, Lee JM, Barnes G. Microtubule stability in budding yeast: characterization and dosage suppression of a benomyl-dependent tubulin mutant. Mol Biol Cell. 1995;6:1241–1259. doi: 10.1091/mbc.6.9.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Cascio VM, et al. CRISPR RNA–guided activation of endogenous human genes. Nat Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magtanong L, Ho CH, Barker SL, et al. Dosage suppression genetic interaction networks enhance functional wiring diagrams of the cell. Nat Biotechnol. 2011;29:505–511. doi: 10.1038/nbt.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makanae K, Kintaka R, Makino T, et al. Identification of dosage-sensitive genes in Saccharomyces cerevisiae using the genetic tug-of-war method. Genome Res. 2013;23:300–311. doi: 10.1101/gr.146662.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans R, van Rossum HM, Wijsman M, et al. CRISPR/Cas9: A molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 2015;15:1–15. doi: 10.1093/femsyr/fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday V, Baetz K, Guzzo J, et al. Systematic yeast synthetic lethal and synthetic dosage lethal screens identify genes required for chromosome segregation. Proc Natl Acad Sci. 2005;102:13956–13961. doi: 10.1073/pnas.0503504102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Buron LD, Salomonsen B, et al. Microbial production of indolylglucosinolate through engineering of a multi-gene pathway in a versatile yeast expression platform. Metab Eng. 2012;14:104–111. doi: 10.1016/j.ymben.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Mnaimneh S, Davierwala AP, Haynes J, et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Moriya H, Shimizu-Yoshida Y, Kitano H. In vivo robustness analysis of cell division cycle genes in Saccharomyces cerevisiae. PLoS Genet. 2006;2:e111. doi: 10.1371/journal.pgen.0020111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad J, Ahmed Khan F, Da L, et al. Keeping CRISPR/Cas on-target. Curren Issues Mol Biol. 2016;20:1–12. [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair NU, Zhao H. Mutagenic inverted repeat assisted genome engineering (MIRAGE) Nucleic Acids Res. 2009;37:5–10. doi: 10.1093/nar/gkn943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Arazoe T, Yachie N, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353:aaf8729. doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- Ooi SL, Pan X, Peyser BD, et al. Global synthetic-lethality analysis and yeast functional profiling. Trends Genet. 2006;22:56–63. doi: 10.1016/j.tig.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Pan X, Yuan DS, Xiang D, et al. A robust toolkit for functional profiling of the yeast genome. Mol Cell. 2004;16:487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Parekh RN, Shaw MR, Wittrup KD. An integrating vector for tunable, high copy, stable integration into the dispersed Ty delta sites of Saccharomyces cerevisiae. Biotechnol Prog. 1996;12:16–21. doi: 10.1021/bp9500627. [DOI] [PubMed] [Google Scholar]
- Patra B, Kon Y, Yadav G, et al. A genome wide dosage suppressor network reveals genomic robustness. Nucleic Acids Res. 2017;45:255–270. doi: 10.1093/nar/gkw1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Kocak DD, Vockley CM, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piening BD, Huang D, Paulovich AG, et al. Novel connections between DNA replication, telomere homeostasis, and the DNA damage response revealed by a genome-wide screen for TEL1/ATM interactions in Saccharomyces cerevisiae. Genetics. 2013;193:1117–1133. doi: 10.1534/genetics.113.149849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöggeler S, Kück U. Highly efficient generation of signal transduction knockout mutants using a fungal strain deficient in the mammalian ku70 ortholog. Gene. 2006;378:1–10. doi: 10.1016/j.gene.2006.03.020. [DOI] [PubMed] [Google Scholar]
- Pratapa A, Adames N, Kraikivski P, et al. CrossPlan: systematic planning of genetic crosses to validate mathematical models. Bioinform. 2018;13:2237–2244. doi: 10.1093/bioinformatics/bty072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puziss JW, Hardy TA, Johnson RB, et al. MDS1, a Dosage suppressor of an mckl mutant, encodes a putative yeast homolog of glycogen synthase Kinase 3. Mol Cell Biol. 1994;14:831–839. doi: 10.1128/mcb.14.1.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin C-Y, et al. Double nicking by RNA-Guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschmanová H, Weninger A, Glieder A, et al. Implementing CRISPR-Cas technologies in conventional and non-conventional yeasts: current state and future prospects. Biotechnol Adv. 2018;36:641–665. doi: 10.1016/j.biotechadv.2018.01.006. [DOI] [PubMed] [Google Scholar]
- Reed SI, Hadwiger JA, Richardson HE, Wittenberg C. Analysis of the Cdc28 protein kinase complex by dosage suppression. J Cell Sci Suppl. 1989;12:29–37. doi: 10.1242/jcs.1989.supplement_12.4. [DOI] [PubMed] [Google Scholar]
- Roggenkamp E, Giersch RM, Wedeman E, et al. CRISPR-UnLOCK: Multipurpose Cas9-based strategies for conversion of yeast libraries and strains. Front Microbiol. 2017;8:1–24. doi: 10.3389/fmicb.2017.01773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronda C, Maury J, Jakočiunas T, et al. CrEdit: CRISPR mediated multi-loci gene integration in Saccharomyces cerevisiae. Microb Cell Fact. 2015;14:1–11. doi: 10.1186/s12934-015-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy KR, Smith JD, Vonesch SC, et al. Multiplexed precision genome editing with trackable genomic barcodes in yeast. Nat Biotechnol. 2018;36:512–520. doi: 10.1038/nbt.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph H, Koenig-Rauseo I, Hinnen A. One-step gene replacement in yeast by cotransformation. Gene. 1985;36:87–95. doi: 10.1016/0378-1119(85)90072-1. [DOI] [PubMed] [Google Scholar]
- Ryan OW, Cate JHD. Multiplex engineering of industrial yeast genomes using CRISPRm. Methods Enzymol. 2014;546:473–489. doi: 10.1016/B978-0-12-801185-0.00023-4. [DOI] [PubMed] [Google Scholar]
- Ryan OW, Skerker JM, Maurer MJ, et al. Selection of chromosomal DNA libraries using a multiplex CRISPR system. Elife. 2014;3:1–15. doi: 10.7554/eLife.03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadhu MJ, Bloom JS, Day L, et al. Highly parallel genome variant engineering with CRISPR-Cas9. Nat Genet. 2018;50:510–514. doi: 10.1038/s41588-018-0087-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I, Su T-C, Parent J. Disintegrator vectors for single-copy yeast chromosomal integration. Yeast. 2007;24:447–455. doi: 10.1002/yea.1469. [DOI] [PubMed] [Google Scholar]
- Sakai A, Shimizu Y, Hishinuma F. Integration of heterologous genes into the chromosome of Saccharomyces cerevisiae using a delta sequence of yeast retrotransposon Ty. Appl Microbiol Biotechnol. 2004;33:302–306. doi: 10.1007/BF00164526. [DOI] [PubMed] [Google Scholar]
- Salminen A, Novick PJ. A ras-like protein is required for a post-Golgi event in yeast secretion. Cell. 1987;49:527–538. doi: 10.1016/0092-8674(87)90455-7. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Schwartz C, Frogue K, Ramesh A, et al. CRISPRi repression of nonhomologous end-joining for enhanced genome engineering via homologous recombination in Yarrowia lipolytica. Biotechnol Bioeng. 2017;114:2896–2906. doi: 10.1002/bit.26404. [DOI] [PubMed] [Google Scholar]
- Scidmore MA, Okamura HH, Rose MD. Genetic interactions between KAR2 and SEC63, encoding eukaryotic homologues of DnaK and DnaJ in the endoplasmic reticulum. Mol Biol Cell. 1993;4:1145–1159. doi: 10.1091/mbc.4.11.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Zhao H, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37:e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro RS, Chavez A, Porter CBM, et al. A CRISPR–Cas9-based gene drive platform for genetic interaction analysis in Candida albicans. Nat Microbiol. 2018;3:73–82. doi: 10.1038/s41564-017-0043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- Shi S, Liang Y, Zhang MM, et al. A highly efficient single-step, markerless strategy for multi-copy chromosomal integration of large biochemical pathways in Saccharomyces cerevisiae. Metab Eng. 2016;33:19–27. doi: 10.1016/j.ymben.2015.10.011. [DOI] [PubMed] [Google Scholar]
- Si T, Chao R, Min Y, et al. Automated multiplex genome-scale engineering in yeast. Nat Commun. 2017;8:1–12. doi: 10.1038/ncomms15187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JD, Suresh S, Schlecht U, et al. Quantitative CRISPR interference screens in yeast identify chemical-genetic interactions and new rules for guide RNA design. Genome Biol. 2016;17:1–16. doi: 10.1186/s13059-016-0900-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JD, Schlecht U, Xu W, et al. A method for high-throughput production of sequence-verified DNA libraries and strain collections. Mol Syst Biol. 2017;13:913. doi: 10.15252/msb.20167233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R, Huang D, Preston N, et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Soreanu I, Hendler A, Dahan D, et al. Marker-free genetic manipulations in yeast using CRISPR/CAS9 system. Curr Genet. 2018;0:1–11. doi: 10.1007/s00294-018-0831-y. [DOI] [PubMed] [Google Scholar]
- Springer M, Weissman JS, Kirschner MW. A general lack of compensation for gene dosage in yeast. Mol Syst Biol. 2010;6:368. doi: 10.1038/msb.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson LF, Kennedy BK, Harlow E. A large-scale overexpression screen in Saccharomyces cerevisiae identifies previously uncharacterized cell cycle genes. Proc Natl Acad Sci USA. 2001;98:3946–3951. doi: 10.1073/pnas.051013498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F, Lewis LK, Resnick MA. In vivo site-directed mutagenesis using oligonucleotides. Nat Biotechnol. 2001;19:773–776. doi: 10.1038/90837. [DOI] [PubMed] [Google Scholar]
- Stovicek V, Borodina I, Forster J. CRISPR-Cas system enables fast and simple genome editing of industrial Saccharomyces cerevisiae strains. Metab Eng Commun. 2015;2:13–22. doi: 10.1016/j.meteno.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stovicek V, Holkenbrink C, Borodina I. CRISPR/Cas system for yeast genome engineering: advances and applications. FEMS Yeast Res. 2017;17:1–16. doi: 10.1093/femsyr/fox030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stundon JL, Zakian VA. Identification of Saccharomyces cerevisiae genes whose deletion causes synthetic effects in cells with reduced levels of the nuclear Pif1 DNA helicase. Genes Genom Genet. 2015;5:2913–8. doi: 10.1534/g3.115.021139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Onge RPS, Mani R, et al. Knocking out multigene redundancies via cycles of sexual assortment and fluorescence selection. Nat Methods. 2011;8:159–164. doi: 10.1038/nmeth.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyme SB, Akhmetova L, Montague TG, et al. Internal guide RNA interactions interfere with Cas9-mediated cleavage. Nat Commun. 2016;7:11750. doi: 10.1038/ncomms11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol. 2006;313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- Tong A, Boone C. High-throughput strain construction and systematic synthetic lethal screening in Saccharomyces cerevisiae. Methods Microbiol. 2007;36:369–386. [Google Scholar]
- Tong A, Evangelista M, Parsons AB, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Tong A, Lesage G, Bader GD, et al. global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Tsai CS, Kong II, Lesmana A, et al. Rapid and marker-free refactoring of xylose-fermenting yeast strains with Cas9/CRISPR. Biotechnol Bioeng. 2015;112:2406–2411. doi: 10.1002/bit.25632. [DOI] [PubMed] [Google Scholar]
- Vanegas KG, Lehka BJ, Mortensen UH. SWITCH: a dynamic CRISPR tool for genome engineering and metabolic pathway control for cell factory construction in Saccharomyces cerevisiae. Microb Cell Fact. 2017;16:25. doi: 10.1186/s12934-017-0632-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaschetto LM. Modulating signaling networks by CRISPR/Cas9-mediated transposable element insertion. Curr Genet. 2018;64:405–412. doi: 10.1007/s00294-017-0765-9. [DOI] [PubMed] [Google Scholar]
- Veitia RA. Gene duplicates: agents of robustness or fragility? Trends Genet. 2017;33:377–379. doi: 10.1016/j.tig.2017.03.006. [DOI] [PubMed] [Google Scholar]
- Verbeke J, Beopoulos A, Nicaud J-M. Efficient homologous recombination with short length flanking fragments in Ku70 deficient Yarrowia lipolytica strains. Biotechnol Lett. 2013;35:571–576. doi: 10.1007/s10529-012-1107-0. [DOI] [PubMed] [Google Scholar]
- Voth WP, Richards JD, Shaw JM, Stillman DJ. Yeast vectors for integration at the HO locus. Nucleic Acids Res. 2001;29:e59. doi: 10.1093/nar/29.12.e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth WP, Jiang YW, Stillman DJ. New “marker swap” plasmids for converting selectable markers on budding yeast gene disruptions and plasmids. Yeast. 2003;20:985–993. doi: 10.1002/yea.1018. [DOI] [PubMed] [Google Scholar]
- Walter JM, Chandran SS, Horwitz AA. CRISPR-cas-assisted multiplexing (CAM): simple same-day multi-locus engineering in yeast. J Cell Physiol. 2016;231:2563–2569. doi: 10.1002/jcp.25375. [DOI] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–85. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang Y, Wu X, et al. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat Biotechnol. 2015;33:175–178. doi: 10.1038/nbt.3127. [DOI] [PubMed] [Google Scholar]
- Wang Q, Xiao L, Zhou L, et al. Comparison of the off-target effects among one-base to three-base mismatched targets of gRNA using a blue to white assay. J Nanosci Nanotechnol. 2018;18:1594–1598. doi: 10.1166/jnn.2018.13813. [DOI] [PubMed] [Google Scholar]
- Wang X, Liu J, Niu Y, et al. Low incidence of SNVs and indels in trio genomes of Cas9-mediated multiplex edited sheep. BMC Genom. 2018;19:397. doi: 10.1186/s12864-018-4712-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Youn J-Y, Friesen H, Nguyen Ba AN, et al. Functional analysis of kinases and transcription factors in Saccharomyces cerevisiae using an integrated overexpression library. G3 Genes Genom Genet. 2017;7:911–921. doi: 10.1534/g3.116.038471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Xing H-L, Wang Z-P, et al. Potential high-frequency off-target mutagenesis induced by CRISPR/Cas9 in Arabidopsis and its prevention. Plant Mol Biol. 2018;96:445–456. doi: 10.1007/s11103-018-0709-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- Zhu H, Bilgin M, Bangham R, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- Zimmermann C, Garcia I, Omerzu M, et al. Mapping the synthetic dosage lethality network of CDK1/CDC28. G3 Genes Genom Genet. 2017;7:1753–1766. doi: 10.1534/g3.117.042317. [DOI] [PMC free article] [PubMed] [Google Scholar]