Abstract
The molecular chaperones Hsp70 and Hsp90 bind and fold a significant proportion of the proteome. They are responsible for the activity and stability of many disease related proteins including those in cancer. Substantial effort has been devoted to developing a range of chaperone inhibitors for clinical use. Recent studies have identified the oncogenic ribonucleotide reductase (RNR) complex as an interactor of chaperones. While several generations of RNR inhibitor have been developed for use in cancer patients, many of these produce severe side effects such as nausea, vomiting and hair loss. Development of more potent, less patient-toxic anti-RNR strategies would be highly desirable. Inhibition of chaperones and associated co-chaperone molecules in both cancer and model organisms such as budding yeast result in the destabilization of RNR subunits and a corresponding sensitization to RNR inhibitors. Going forward, this may form part of a novel strategy to target cancer cells that are resistant to standard RNR inhibitors.
Keywords: Ribonucleotide reductase, molecular chaperones, Hsp70, Hsp90, Ydj1, Hdj2, DNA damage response
Introduction
Heat shock proteins (Hsps) are a set of well-conserved, highly expressed molecular chaperones that are vital for cellular function (Freilich et al. 2018; Kim et al. 2013). Originally identified as proteins induced by thermal stress, it is now understood that Hsps play important roles in protecting cells against a range of cell stresses that include DNA damage, oxidative stress, metabolic challenges and aging (Freilich et al. 2018; Kim et al. 2013; Rodina et al. 2016; Tai et al. 2016). Heat shock protein 70 (Hsp70) and Heat shock protein 90 (Hsp90) are both imperative for the folding and stabilization of the majority of the proteome (Kim et al. 2013; Li et al. 2017). Hsp70 is responsible for many housekeeping tasks such as de novo folding, transportation across cellular compartments, folding of denatured proteins and, if needed, protein degradation of “client” proteins (Hubscher et al. 2017; Kim et al. 2013). Hsp90 is required for the stability, function and degradation of many proteins from a variety of cellular pathways (Gopinath and Leu 2017; McClellan et al. 2007; Pennisi et al. 2015; Sato and Torigoe 1998; Trepel et al. 2010). While Hsp70 and Hsp90 both contribute to protein folding in the cell, their roles differ. Hsp70 can bind unfolded polypeptides and direct them to a partially or near-native structure. After transfer of partially folded clients from Hsp70 to Hsp90, Hsp90 potentiates final structural re-arrangements that allow full client activity (Wegele et al. 2004).
Hsp70 and Hsp90 in cancer
Because of their cytoprotective qualities, overexpression of Hsp70 and Hsp90 are implicated in multiple diseases including cancer (Calderwood and Gong 2016; Calderwood and Neckers 2016; Cesa et al. 2018; Lianos et al. 2015; Sherman and Gabai 2015). High levels of Hsps are associated with poor prognosis and a resistance to standard cancer therapeutics. Many of the mutations that are present in oncoproteins render them intrinsically unstable and consequently these proteins require extra chaperone function to maintain stability and activity. Oncoproteins that Hsp90 and Hsp70 stabilize include CDK4, p53, p21, Cyclin D, ERK5, SRC and the Androgen receptor (Bachman et al. 2018; Dong et al. 2018; Hallett et al. 2017; Joshi et al. 2018; Truman et al. 2012; Truman et al. 2006).
Proteomic analyses of chaperone networks are useful for identifying novel anticancer strategies
Chaperone inhibitors trigger loss of client activity and client degradation. This knowledge can be combined with powerful proteomic technologies to identify new clients and potentially new anticancer strategies. A recent study analyzed global proteome changes in bladder cancer cells upon treatment with five established Hsp90 inhibitors AUY922, ganetespib, SNX2112, AT13387, and CUDC305 (Li et al. 2017). Substantial decreases in protein abundance were observed for proteins involved in oxidative phosphorylation, DNA synthesis and replication, cell cycle and glutathione metabolism (Li et al. 2017). While many of these proteins have been previously established as chaperone clients, several were novel interactors (Li et al. 2017). It will be interesting to see if future studies confirm these as bona fide clients.
While many interactions of chaperones have been identified on a single client basis, several studies have utilized large-scale genetic or proteomic screens to understand the Hsp70 and Hsp90 interactome (Freilich et al. 2018; Joshi et al. 2018; Mollapour and Neckers 2011; Taipale et al. 2012; Taldone et al. 2014; Trepel et al. 2010; Truman et al. 2012; Truman et al. 2015b). Chaperone-client interactions are dynamic, altering upon cell condition. Recent proteomic chaperone studies have attempted to probe this, by analyzing global changes in chaperone interaction upon chaperone phosphorylation or upon expression of various oncoproteins (Dunn et al. 2015; Mollapour et al. 2011; Truman et al. 2012; Woodford et al. 2016).
In an effort to understand the global roles of Hsp70 and Hsp90 in genome integrity, Truman et al., purified Hsp70 and Hsp90 complexes from yeast in untreated and MMS-treated (DNA damaging agent) cells were analyzed by mass spectrometry. 256 chaperone interacting proteins were identified, with 142 new interactions seen. 1.5% of Hsp70-client interactions and 2% of Hsp90-client interactions were increased upon DNA damage. Only one protein (Rnr4) was observed to increase interaction with both Hsp70 and Hsp90 upon DNA damage stress, suggesting a previously undiscovered role for chaperones in regulation of ribonucleotide reductase (Truman et al. 2015a; Truman et al. 2015b).
RNR, an important player in DNA replication
Maintenance of genome integrity is critical for cell survival and proliferation (Mikolaskova et al. 2018). Ribonucleotide reductase (RNR) is an important enzyme that is involved in the production of deoxyribonucleotides (dNTPs) which are subsequently used in DNA synthesis and repair (Cerqueira et al. 2007a; Cerqueira et al. 2005; Chabes et al. 2000; Nordlund and Reichard 2006; Wang et al. 1997). RNR is comprised of two diverse subunits, the large subunit R1 (R1 in vertebrates, Rnr1/Rnr3 in yeast) which contains the allosteric regulatory sites (Maicher and Kupiec 2018) and the small subunit R2 (R2/R2B in vertebrates, Rnr2/Rnr4 in yeast) which consists of a cell cycle regulated binuclear iron center and a tyrosyl free radical (Cerqueira et al. 2007a; Cerqueira et al. 2005; Chabes et al. 2000; Nordlund and Reichard 2006; Wang et al. 1997). Throughout the cell cycle, the expression levels of the subunits will increase or decrease depending on the cellular need for dNTPs. In S phase, the total pool of dNTPs peaks in order to permit DNA replication. Conversely, the number of dNTPs dramatically drops 10-fold in G0/G1 phase when DNA repair and mitochondrial DNA synthesis demands the bulk of dNTPs (Cerqueira et al. 2005; Chabes et al. 2000; Perlstein et al. 2005; Truman et al. 2015b). Tight regulation of RNR is key to ensure correct levels of all four dNTPs at all times. Historically, researchers focused on the allosteric regulation of this enzyme, however, recently it has been discovered that RNR control is extremely complex and may require other forms of regulation as well. This includes regulation of expression of RNR genes, proteolysis of RNR subunits, control of the cellular localization of the small RNR subunit, and regulation of RNR activity by small protein inhibitors. RNR is critical for both original DNA synthesis as well as DNA repair, making it a classical target for cancer therapeutics. Further understand of the regulatory mechanisms of RNR is vital to developing novel treatments for cancer and other diseases (Mulder et al. 2005; Nordlund and Reichard 2006).
RNR inhibitors as anticancer agents
Because RNR is an attractive target in cancer, many RNR inhibitors have been developed as cancer therapeutics (Cerqueira et al. 2007a; Mulder et al. 2005; Nordlund and Reichard 2006). One of the first RNR inhibitors developed was Hydroxyurea (HU). The anticancer properties of HU were first observed in the 1960s and since then it has been widely used to treat both cancer and sickle-cell anemia (Singh and Xu 2016; Yarbro 1992). HU works by reducing the diferric-tyrosyl radical center in the small R2 subunit via one-electron transfer from the drug complex. HU was developed for human use but works similarly in yeast for studying mechanisms (Singh and Xu 2016; Yarbro 1992). Another potent RNR inhibitor used as an anti-cancer drug is Gemcitabine (dFdC) (Cerqueira et al. 2007b; Plunkett et al. 1995). It has been approved for medical use since 1995, and has since been approved for additional uses clinically. Gemcitabine is an analog of deoxycytidine and once it enters cells is phosphorylated by deoxycytidine kinase to its active form (Cerqueira et al. 2007b; Plunkett et al. 1995). Gemcitabine is incorporated into DNA through DNA synthesis and blocks further DNA polymerase processing. This results in “masked termination” and eventually cell death (Cerqueira et al. 2007b; Plunkett et al. 1995).
RNR activity is dependent on Hsp70 and Hsp90
Interestingly, the R2 subunits in both yeast and mammalian cells are clients of both Hsp70 and Hsp90. Rnr2 and Rnr4 interact with Ssa1 and Hsp82 and inhibition of chaperone activity with VER-155008 or 17-AAG promotes Rnr2/4 degradation. Likewise Hsp70 and Hsp90 bind human R2 and treatment of MCF-7 cells with VER-155008 or 17-AAG triggers R2 degradation (Truman et al. 2015a; Truman et al. 2015b).
Given that RNR is reliant on chaperone activity, it stands to reason that chaperone inhibition would sensitize cells to RNR inhibitors. Several studies have confirmed this, with 17-AAG being utilized to sensitize both breast and lung cancer cells to gemcitabine (Arlander et al. 2003; Ghadban et al. 2017; Pedersen et al. 2015; Truman et al. 2015b).
Inhibition of co-chaperones as an anti-RNR strategy
Hsp70 and Hsp90 require the assistance of helper co-chaperone molecules in order to properly function. Co-chaperones are non-client proteins bind chaperones and assist in protein folding/stabilization activity. Co-chaperones have multiple functions-some bind and partially fold client proteins while others act as regulatory units to their chaperone counterpart (Amin-Wetzel et al. 2017; Caplan 2003; Connell et al. 2001; Craig and Marszalek 2017; Joshi et al. 2018; Kampinga and Craig 2011). Regulatory functions of co-chaperones include accelerating Hsp70/Hsp90 nucleotide binding and hydrolysis or acting as a linker between two chaperone molecules. It is assumed that co-chaperones are highly specialized in the manner in which they choose a client to present to Hsp70 or Hsp90 and facilitate binding and releasing (Caplan 2003). The most well-characterized class of Hsp70 co-chaperones are the J-domain proteins after their highly conserved chaperone binding region. These J-domain proteins are typically associated with Hsp70 and protein folding functions (Amin-Wetzel et al. 2017; Aron et al. 2007; Caplan 2003; Connell et al. 2001; Craig and Marszalek 2017; Joshi et al. 2018; Kampinga and Craig 2011; Lu and Cyr 1998).
Recently there has been interest in co-chaperones as anticancer targets, especially as co-chaperone expression in altered in a variety of cancers (Calderwood 2013; He et al. 2015; Parrales et al. 2016; Sopha et al. 2017; Tracz-Gaszewska et al. 2017; Weeks and Miller 2008). In some cases, co-chaperones themselves have been shown to play an important role in cell death related pathways such as apoptosis. While inhibitors of Hsp70 and Hsp90 are effective in killing cells, the essential nature of chaperone function makes many of these compounds highly toxic to patients (Calderwood 2013; Erlichman 2009; Weeks and Miller 2008). Targeting co-chaperones, which specifically fine-tune chaperone function may form part of a novel anticancer strategy. Identification of a novel small molecule inhibitor of mammalian HDJ2 (C86) that destabilizes the androgen receptor and inhibits the growth of castration-resistant prostate cancer furthers the idea that targeting HDJ2 and related co-chaperones may offer an alternative therapeutic avenue (Moses et al. 2018).
A recent screen of yeast co-chaperone knockout mutants for sensitivity to HU identified Ydj1 as a master regulator of the RNR complex. Ydj1 binds Rnr2 and cells lacking Ydj1 display reduced RNR subunit expression and stability (Sluder et al. 2018). Demonstrating broad conservation from yeast to human, Hdj2 and R2B (mammalian counterparts of Rnr2) interact, promoting RNR complex stability and activity in human cells. Mammalian cells that lacked Hdj2 were found to be more sensitive to RNR inhibiting drugs such as hydroxyurea, gemcitabine and triapine (Sluder et al. 2018). Although few direct inhibitors of co-chaperones have been identified, one that has interesting therapeutic potential is 116-9e. 116-9e is a small molecule inhibitor that blocks Hsp40 binding to Hsp70 through the J-domain. Treatment of 116-9e causes disruption of Rnr2-Ydj1 (yeast cells) or R2-Hdj2 (mammalian cells) and ultimately compromises RNR function, sensitizing cells to HU and triapine ((Sluder et al. 2018) and summarized in Fig. 1). In contrast to the direct action of C86, 116-9e holds promise as a low toxicity drug capable of decreasing cellular resistance to a wide variety of anticancer agents (unpublished work). Several studies have demonstrated that both Hsp90 and Hsp70 interactions with their co-chaperones can be substantially altered by post-translational modifications (PTMs) such as phosphorylation (Bachman et al. 2018; Mollapour et al. 2010; Muller et al. 2013; Nitika and Truman 2017; Truman et al. 2012; Vaughan et al. 2008). Given the promising data describing the role of co-chaperones in cancer, therapies that target the PTMs of co-chaperones may provide a future therapeutic strategy.
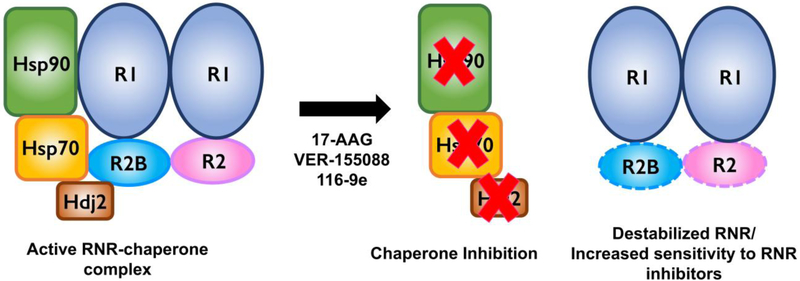
Figure 1. The RNR complex requires chaperones to function.
Hsp90, Hsp70 and the co-chaperone Hdj2 bind and stabilize the RNR complex. Treatment of cells with small molecules that target chaperone function (17-AAG or VER-155008) or co-chaperone function (116-e) result in both decreased transcription and protein stability of RNR subunits. In turn, this sensitizes cancer cells to a wide variety of RNR inhibitors that include hydroxyurea, gemicitabine and triapine.
Conclusion and perspectives
RNR has been a longstanding anticancer drug target because of its prominent role in DNA synthesis and repair (Cerqueira et al. 2007a; Mulder et al. 2005; Nordlund and Reichard 2006). Likewise, because of Hsp70 and Hsp90’s cytoprotective qualities, researchers have developed several chaperone inhibitors that show success in cancer cell studies (Assimon et al. 2013; Calderwood 2013; Fan et al. 2003; Galluzzi et al. 2009; McClellan et al. 2007; Weeks and Miller 2008). However, both RNR and chaperone inhibitors are highly toxic to patients which makes dosing and treatment difficult. Combination therapies that simultaneously target both chaperones and RNR appear to be highly synergistic, although whether this is fully translatable to patients remains to be seen. While this combination strategy seems promising, targeting the co-chaperones that regulate specific onco-clients may provide additional synergy and decreased toxicity to patients going forward.
Acknowledgements
This work was supported by NCI R15CA208773 (AWT) and NSF REU 1359271 (LED). We would like to thank Nitika for critical reading.
References
- Amin-Wetzel N, Saunders RA, Kamphuis MJ, Rato C, Preissler S, Harding HP, Ron D (2017) A J-Protein Co-chaperone Recruits BiP to Monomerize IRE1 and Repress the Unfolded Protein Response Cell 171:1625–1637 e1613 doi: 10.1016/j.cell.2017.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlander SJ, Eapen AK, Vroman BT, McDonald RJ, Toft DO, Karnitz LM (2003) Hsp90 inhibition depletes Chk1 and sensitizes tumor cells to replication stress J Biol Chem 278:52572–52577 doi: 10.1074/jbc.M309054200 [DOI] [PubMed] [Google Scholar]
- Aron R, Higurashi T, Sahi C, Craig EA (2007) J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation EMBO J 26:3794–3803 doi:7601811 [pii] 10.1038/sj.emboj.7601811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assimon VA, Gillies AT, Rauch JN, Gestwicki JE (2013) Hsp70 protein complexes as drug targets Curr Pharm Des 19:404–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman AB et al. (2018) Phosphorylation induced cochaperone unfolding promotes kinase recruitment and client class-specific Hsp90 phosphorylation Nat Commun 9:265 doi: 10.1038/s41467-017-02711-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood SK (2013) Molecular cochaperones: tumor growth and cancer treatment Scientifica (Cairo) 2013:217513 doi: 10.1155/2013/217513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood SK, Gong J (2016) Heat Shock Proteins Promote Cancer: It's a Protection Racket Trends Biochem Sci 41:311–323 doi: 10.1016/j.tibs.2016.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood SK, Neckers L (2016) Hsp90 in Cancer: Transcriptional Roles in the Nucleus Adv Cancer Res 129:89–106 doi: 10.1016/bs.acr.2015.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan AJ (2003) What is a co-chaperone? Cell Stress Chaperones 8:105–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerqueira NM, Fernandes PA, Ramos MJ (2007a) Ribonucleotide reductase: a critical enzyme for cancer chemotherapy and antiviral agents Recent Pat Anticancer Drug Discov 2:11–29 [DOI] [PubMed] [Google Scholar]
- Cerqueira NM, Fernandes PA, Ramos MJ (2007b) Understanding ribonucleotide reductase inactivation by gemcitabine Chemistry 13:8507–8515 doi: 10.1002/chem.200700260 [DOI] [PubMed] [Google Scholar]
- Cerqueira NM, Pereira S, Fernandes PA, Ramos MJ (2005) Overview of ribonucleotide reductase inhibitors: an appealing target in anti-tumour therapy Current medicinal chemistry 12:1283–1294 [DOI] [PubMed] [Google Scholar]
- Cesa LC et al. (2018) X-linked inhibitor of apoptosis protein (XIAP) is a client of heat shock protein 70 (Hsp70) and a biomarker of its inhibition J Biol Chem 293:2370–2380 doi: 10.1074/jbc.RA117.000634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabes A, Domkin V, Larsson G, Liu A, Graslund A, Wijmenga S, Thelander L (2000) Yeast ribonucleotide reductase has a heterodimeric iron-radical-containing subunit Proc Natl Acad Sci U S A 97:2474–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, Patterson C (2001) The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins Nat Cell Biol 3:93–96 doi: 10.1038/35050618 [DOI] [PubMed] [Google Scholar]
- Craig EA, Marszalek J (2017) How Do J-Proteins Get Hsp70 to Do So Many Different Things? Trends Biochem Sci 42:355–368 doi: 10.1016/j.tibs.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Wu Z, Wang D, Pascal LE, Nelson JB, Wipf P, Wang Z (2018) Hsp70 binds to the androgen receptor N-terminal domain and modulates the receptor function in prostate cancer cells Mol Cancer Ther doi: 10.1158/1535-7163.MCT-18-0432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn DM et al. (2015) c-Abl Mediated Tyrosine Phosphorylation of Aha1 Activates Its Co-chaperone Function in Cancer Cells Cell Rep 12:1006–1018 doi: 10.1016/j.celrep.2015.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlichman C (2009) Tanespimycin: the opportunities and challenges of targeting heat shock protein 90 Expert Opin Investig Drugs 18:861–868 doi: 10.1517/13543780902953699 [DOI] [PubMed] [Google Scholar]
- Fan CY, Lee S, Cyr DM (2003) Mechanisms for regulation of Hsp70 function by Hsp40 Cell Stress Chaperones 8:309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freilich R, Arhar T, Abrams JL, Gestwicki JE (2018) Protein-Protein Interactions in the Molecular Chaperone Network Acc Chem Res 51:940–949 doi: 10.1021/acs.accounts.8b00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Giordanetto F, Kroemer G (2009) Targeting HSP70 for cancer therapy Mol Cell 36:176–177 doi:S1097-2765(09)00738-2 [pii] 10.1016/j.molcel.2009.10.003 [DOI] [PubMed] [Google Scholar]
- Ghadban T et al. (2017) HSP90 is a promising target in gemcitabine and 5-fluorouracil resistant pancreatic cancer Apoptosis 22:369–380 doi: 10.1007/s10495-016-1332-4 [DOI] [PubMed] [Google Scholar]
- Gopinath RK, Leu JY (2017) Hsp90 mediates the crosstalk between galactose metabolism and cell morphology pathways in yeast Curr Genet 63:23–27 doi: 10.1007/s00294-016-0614-2 [DOI] [PubMed] [Google Scholar]
- Hallett ST et al. (2017) Differential Regulation of G1 CDK Complexes by the Hsp90-Cdc37 Chaperone System Cell Rep 21:1386–1398 doi: 10.1016/j.celrep.2017.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HL et al. (2015) Overexpression of DNAJC12 predicts poor response to neoadjuvant concurrent chemoradiotherapy in patients with rectal cancer Exp Mol Pathol 98:338–345 doi: 10.1016/j.yexmp.2015.03.029 [DOI] [PubMed] [Google Scholar]
- Hubscher V, Mudholkar K, Rospert S (2017) The yeast Hsp70 homolog Ssb: a chaperone for general de novo protein folding and a nanny for specific intrinsically disordered protein domains Curr Genet 63:9–13 doi: 10.1007/s00294-016-0610-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S, Wang T, Araujo TLS, Sharma S, Brodsky JL, Chiosis G (2018) Adapting to stress - chaperome networks in cancer Nature reviews Cancer 18:562–575 doi: 10.1038/s41568-018-0020-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA (2011) The HSP70 chaperone machinery: J proteins as drivers of functional specificity Nat Rev Mol Cell Biol 11:579–592 doi:nrm2941 [pii] 10.1038/nrm2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU (2013) Molecular chaperone functions in protein folding and proteostasis Annual review of biochemistry 82:323–355 doi: 10.1146/annurev-biochem-060208-092442 [DOI] [PubMed] [Google Scholar]
- Li QQ et al. (2017) Proteomic analysis of proteome and histone post-translational modifications in heat shock protein 90 inhibition-mediated bladder cancer therapeutics Sci Rep 7:201 doi: 10.1038/s41598-017-00143-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lianos GD et al. (2015) The role of heat shock proteins in cancer Cancer letters 360:114–118 doi: 10.1016/j.canlet.2015.02.026 [DOI] [PubMed] [Google Scholar]
- Lu Z, Cyr DM (1998) Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1 J Biol Chem 273:27824–27830 [DOI] [PubMed] [Google Scholar]
- Maicher A, Kupiec M (2018) Rnr1's role in telomere elongation cannot be replaced by Rnr3: a role beyond dNTPs? Curr Genet 64:547–550 doi: 10.1007/s00294-017-0779-3 [DOI] [PubMed] [Google Scholar]
- McClellan AJ, Xia Y, Deutschbauer AM, Davis RW, Gerstein M, Frydman J (2007) Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches Cell 131:121–135 doi: 10.1016/j.cell.2007.07.036 [DOI] [PubMed] [Google Scholar]
- Mikolaskova B, Jurcik M, Cipakova I, Kretova M, Chovanec M, Cipak L (2018) Maintenance of genome stability: the unifying role of interconnections between the DNA damage response and RNA-processing pathways Curr Genet 64:971–983 doi: 10.1007/s00294-018-0819-7 [DOI] [PubMed] [Google Scholar]
- Mollapour M, Neckers L (2011) Detecting HSP90 phosphorylation Methods Mol Biol 787:67–74 doi: 10.1007/978-1-61779-295-3_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour M, Tsutsumi S, Neckers L (2010) Hsp90 phosphorylation, Wee1 and the cell cycle Cell Cycle 9:2310–2316 doi:12054 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour M et al. (2011) Threonine 22 phosphorylation attenuates Hsp90 interaction with cochaperones and affects its chaperone activity Mol Cell 41:672–681 doi:S1097-2765(11)00094-3 [pii] 10.1016/j.molcel.2011.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses MA et al. (2018) Targeting the Hsp40/Hsp70 Chaperone Axis as a Novel Strategy to Treat Castration-Resistant Prostate Cancer Cancer Res 78:4022–4035 doi: 10.1158/0008-5472. CAN-17-3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder KW, Winkler GS, Timmers HT (2005) DNA damage and replication stress induced transcription of RNR genes is dependent on the Ccr4-Not complex Nucleic Acids Res 33:6384–6392 doi: 10.1093/nar/gki938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P, Ruckova E, Halada P, Coates PJ, Hrstka R, Lane DP, Vojtesek B (2013) C-terminal phosphorylation of Hsp70 and Hsp90 regulates alternate binding to co-chaperones CHIP and HOP to determine cellular protein folding/degradation balances Oncogene 32:3101–3110 doi: 10.1038/onc.2012.314 [DOI] [PubMed] [Google Scholar]
- Nitika Truman AW (2017) Cracking the Chaperone Code: Cellular Roles for Hsp70 Phosphorylation Trends Biochem Sci 42:932–935 doi: 10.1016/j.tibs.2017.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund P, Reichard P (2006) Ribonucleotide reductases Annual review of biochemistry 75:681–706 doi: 10.1146/annurev.biochem.75.103004.142443 [DOI] [PubMed] [Google Scholar]
- Parrales A, Ranjan A, Iyer SV, Padhye S, Weir SJ, Roy A, Iwakuma T (2016) DNAJA1 controls the fate of misfolded mutant p53 through the mevalonate pathway Nat Cell Biol 18:1233–1243 doi: 10.1038/ncb3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen KS, Kim GP, Foster NR, Wang-Gillam A, Erlichman C, McWilliams RR (2015) Phase II trial of gemcitabine and tanespimycin (17AAG) in metastatic pancreatic cancer: a Mayo Clinic Phase II Consortium study Invest New Drugs 33:963–968 doi: 10.1007/s10637-015-0246-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi R, Ascenzi P, di Masi A (2015) Hsp90: A New Player in DNA Repair? Biomolecules 5:2589–2618 doi: 10.3390/biom5042589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlstein DL, Ge J, Ortigosa AD, Robblee JH, Zhang Z, Huang M, Stubbe J (2005) The active form of the Saccharomyces cerevisiae ribonucleotide reductase small subunit is a heterodimer in vitro and in vivo Biochemistry 44:15366–15377 doi: 10.1021/bi051616+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plunkett W, Huang P, Xu YZ, Heinemann V, Grunewald R, Gandhi V (1995) Gemcitabine: metabolism, mechanisms of action, and self-potentiation Seminars in oncology 22:3–10 [PubMed] [Google Scholar]
- Rodina A et al. (2016) The epichaperome is an integrated chaperome network that facilitates tumour survival Nature 538:397–401 doi: 10.1038/nature19807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Torigoe T (1998) The molecular chaperones in cell cycle control Ann N Y Acad Sci 851:61–66 [DOI] [PubMed] [Google Scholar]
- Sherman MY, Gabai VL (2015) Hsp70 in cancer: back to the future Oncogene 34:4153–4161 doi: 10.1038/onc.2014.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Xu YJ (2016) The Cell Killing Mechanisms of Hydroxyurea Genes (Basel) 7 doi: 10.3390/genes7110099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder IT, Nitika, Knighton LE, Truman AW (2018) The Hsp70 co-chaperone Ydj1/HDJ2 regulates ribonucleotide reductase activity PLoS Genet 14:e1007462 doi: 10.1371/journal.pgen.1007462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopha P, Ren HY, Grove DE, Cyr DM (2017) Endoplasmic reticulum stress-induced degradation of DNAJB12 stimulates BOK accumulation and primes cancer cells for apoptosis J Biol Chem 292:11792–11803 doi: 10.1074/jbc.M117.785113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W, Guzman ML, Chiosis G (2016) The epichaperome: the power of many as the power of one Oncoscience 3:266–267 doi: 10.18632/oncoscience.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Krykbaeva I, Koeva M, Kayatekin C, Westover KD, Karras GI, Lindquist S (2012) Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition Cell 150:987–1001 doi: 10.1016/j.cell.2012.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taldone T, Ochiana SO, Patel PD, Chiosis G (2014) Selective targeting of the stress chaperome as a therapeutic strategy Trends Pharmacol Sci 35:592–603 doi: 10.1016/j.tips.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracz-Gaszewska Z et al. (2017) Molecular chaperones in the acquisition of cancer cell chemoresistance with mutated TP53 and MDM2 up-regulation Oncotarget 8:82123–82143 doi: 10.18632/oncotarget.18899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trepel J, Mollapour M, Giaccone G, Neckers L (2010) Targeting the dynamic HSP90 complex in cancer Nature reviews Cancer 10:537–549 doi: 10.1038/nrc2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman AW et al. (2012) CDK-dependent Hsp70 Phosphorylation controls G1 cyclin abundance and cell-cycle progression Cell 151:1308–1318 doi: 10.1016/j.cell.2012.10.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman AW et al. (2015a) The quantitative changes in the yeast Hsp70 and Hsp90 interactomes upon DNA damage Data Brief 2:12–15 doi: 10.1016/j.dib.2014.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman AW et al. (2015b) Quantitative proteomics of the yeast Hsp70/Hsp90 interactomes during DNA damage reveal chaperone-dependent regulation of ribonucleotide reductase Journal of proteomics 112:285–300 doi: 10.1016/j.jprot.2014.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman AW et al. (2006) Expressed in the yeast Saccharomyces cerevisiae, human ERK5 is a client of the Hsp90 chaperone that complements loss of the Slt2p (Mpk1p) cell integrity stress-activated protein kinase Eukaryot Cell 5:1914–1924 doi:EC.00263-06 [pii] 10.1128/EC.00263-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan CK et al. (2008) Hsp90-dependent activation of protein kinases is regulated by chaperone-targeted dephosphorylation of Cdc37 Mol Cell 31:886–895 doi:S1097-2765(08)00541-8 [pii] 10.1016/j.molcel.2008.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PJ, Chabes A, Casagrande R, Tian XC, Thelander L, Huffaker TC (1997) Rnr4p, a novel ribonucleotide reductase small-subunit protein Mol Cell Biol 17:6114–6121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks SA, Miller DJ (2008) The heat shock protein 70 cochaperone YDJ1 is required for efficient membrane-specific flock house virus RNA replication complex assembly and function in Saccharomyces cerevisiae J Virol 82:2004–2012 doi:JVI.02017-07 [pii] 10.1128/JVI.02017-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegele H, Muller L, Buchner J (2004) Hsp70 and Hsp90--a relay team for protein folding Reviews of physiology, biochemistry and pharmacology 151:1–44 doi: 10.1007/s10254-003-0021-1 [DOI] [PubMed] [Google Scholar]
- Woodford MR et al. (2016) Mps1 Mediated Phosphorylation of Hsp90 Confers Renal Cell Carcinoma Sensitivity and Selectivity to Hsp90 Inhibitors Cell Rep 14:872–884 doi: 10.1016/j.celrep.2015.12.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarbro JW (1992) Mechanism of action of hydroxyurea Seminars in oncology 19:1–10 [PubMed] [Google Scholar]