Abstract
BACKGROUND
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors with high mortality-to-incidence ratios. Nuclear factor erythroid 2-like 3 (NFE2L3), also known as NRF3, is a member of the cap ‘n’ collar basic-region leucine zipper family of transcription factors. NFE2L3 is involved in the regulation of various biological processes, whereas its role in HCC has not been elucidated.
AIM
To explore the expression and biological function of NFE2L3 in HCC.
METHODS
We analyzed the expression of NFE2L3 in HCC tissues and its correlation with clinicopathological parameters based on The Cancer Genome Atlas (TCGA) data portal. Short hairpin RNA (shRNA) interference technology was utilized to knock down NFE2L3 in vitro. Cell apoptosis, clone formation, proliferation, migration, and invasion assays were used to identify the biological effects of NFE2L3 in BEL-7404 and SMMC-7721 cells. The expression of epithelial-mesenchymal transition (EMT) markers was examined by Western blot analysis.
RESULTS
TCGA analysis showed that NFE2L3 expression was significantly positively correlated with tumor grade, T stage, and pathologic stage. The qPCR and Western blot results showed that both the mRNA and protein levels of NFE2L3 were significantly decreased after shRNA-mediated knockdown in BEL-7404 and SMMC-7721 cells. The shRNA-mediated knockdown of NFE2L3 could induce apoptosis and inhibit the clone formation and cell proliferation of SMMC-7721 and BEL-7404 cells. NFE2L3 knockdown also significantly suppressed the migration, invasion, and EMT of the two cell lines.
CONCLUSION
Our study showed that shRNA-mediated knockdown of NFE2L3 exhibited tumor-suppressing effects in HCC cells.
Keywords: Nuclear factor erythroid 2-like 3, Hepatocellular carcinoma, The Cancer Genome Atlas, Short hairpin RNA, Epithelial-mesenchymal transition
Core tip: Nuclear factor erythroid 2-like 3 (NFE2L3), as a key regulator of the cellular stress response, is involved in the regulation of various tumors, whereas its role in hepatocellular carcinoma (HCC) has not been elucidated. Our present study identified that NFE2L3 was closely associated with the grade and stage of HCC patients based on The Cancer Genome Atlas database. Furthermore, our study showed that short hairpin RNA-mediated knockdown of NFE2L3 exhibited tumor-suppressing effects in HCC cells.
INTRODUCTION
Hepatocellular carcinoma (HCC), one of the most common malignant tumors, ranks as the third leading cause of cancer-related death worldwide, resulting in at least 700000 deaths each year[1,2]. Microvascular invasion, occult metastasis, recurrence, and resistance to chemotherapy contribute to the poor prognosis and low survival rate of HCC patients. Hepatitis B virus (HBV) and hepatitis C virus (HCV) infection, alcoholic cirrhosis, non-alcoholic steatohepatitis, and the ingestion of aflatoxin B1 are the main risk factors for HCC development[3]. Current therapeutic strategies for HCC include chemotherapy, surgical resection, liver transplantation, radiofrequency ablation, and drug therapy[4,5]. The mechanism of HCC is very complex, and more research is needed to provide more detailed clues.
Nuclear factor erythroid 2-like 3 (NFE2L3), also known as NRF3, was first identified and reported in 1999 and is a member of the cap ‘n’ collar basic-region leucine zipper family of transcription factors[6]. The human NFE2L3 transcript encodes a 694-amino acid protein that can bind to antioxidant response elements by heterodimerizing with small musculoaponeurotic fibrosarcoma factors[6,7]. NFE2L3, as a key regulator of the cellular stress response, is expressed in a wide variety of tissues such as the heart, brain, lung, liver, kidney, and pancreas, particularly abundant in the placenta[8,9]. Accumulating evidence suggests that NFE2L3 is involved in the regulation of various biological processes, such as transcription, signal transduction, cell cycle, growth, development, differentiation, and inflammation. In addition to these functions, Chevillard et al[10] showed that the absence of NFE2L3 predisposes mice to lymphoma development, suggesting a protective role for this transcription factor in hematopoietic malignancies. Wang et al[11] reported that NFE2L3 is necessary for RCAN1-4-mediated enhanced growth and invasion, and its overexpression independently enhances these effects in thyroid cancer cells. Chowdhury et al[12] demonstrated that NFE2L3 promotes colon cancer cell proliferation by activating UHMK1 gene expression. These findings suggest that NFE2L3 may be a crucial regulator of cancer progression. However, the roles of NFE2L3 in HCC and their underlying molecular mechanisms have yet to be elucidated.
In the present study, we analyzed the expression of NFE2L3 in HCC and its correlation with clinicopathological parameters based on the Cancer Genome Atlas (TCGA) data portal. In addition, we utilized short hairpin RNA (shRNA) interference technology to knock down NFE2L3 in vitro and systematically evaluated the biological function of NFE2L3 in HCC cells.
MATERIALS AND METHODS
HCC patient data in the Cancer Genome Atlas
A total of 344 HCC patients with clinicopathological information and RNA-Seq expression data were obtained from TCGA database (https://cancergenome.nih.gov/). The gene expression data were normalized using the RNA normalization method described in TCGA National Cancer Institute (NCI) Wiki.
Cell lines and cell culture
Human HCC cell lines (SMMC-7721 and BEL-7404) were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and identified by short tandem repeat analysis. SMMC-7721 and BEL-7404 cells were cultured in RPMI 1640 medium (Gibco, NY, United States) supplemented with 10% fetal bovine serum (FBS) (Gibco, Sydney, Australia), 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco, NY, United States) and incubated at 37 °C in a humidified atmosphere with 5% CO2.
Short hairpin RNA lentivirus infection
An NFE2L3 shRNA interference lentiviral vector was constructed and synthesized by GeneChem Co., Ltd (Shanghai, China). The NFE2L3 shRNA interference target sequence was 5’-AGTCAATCCCAACCACTAT-3’ (shNFE2L3), and a scramble sequence 5’-TTCTCCGAACGTGTCACGT-3’ was used as a negative control (shCtrl). The lentiviral vectors were transfected into SMMC-7721 and BEL-7404 cells according to the manufacturer’s instructions. The cells were seeded (2 × 105 cells/mL) onto 6-well plates and incubated for 24 h to reach 50% confluence, and then replaced with infection medium containing lentiviral vectors at a multiplicity of infection of 10 plaque-forming units/cell. Successfully infected cells were green fluorescent protein positive and observed under a fluorescence microscope after 72 h, and the interference efficiency of NFE2L3 shRNA was determined using quantitative real-time PCR (qPCR) and Western blot.
RNA extraction and real-time PCR
Total RNA was extracted with TRIzol reagent (Pufei Biotechnology, Shanghai, China). The RNA concentration and purity were assessed using the OD260 and OD260/OD280 ratio, respectively, and cDNA was synthesized with M-MLV RT (Promega, United States) according to the manufacturers’ instructions. qPCR was performed using a SYBR Green master mix (Takara Biotechnology, Dalian, China) on the Stratagene Mx3000P (Agilent Technologies, United States). The sequences of the primers are as follows: NFE2L3, forward: 5’-ACACTTACCACTTACAGCCAACT-3’, reverse: 5’-CTTCGTCTGATGTCACGGAT-3’; GAPDH, forward: 5’-TGACTTCAACAGCGACACCCA-3’, reverse: 5’-CACCCTGTTGCTGTAGCCAAA-3’. Relative mRNA levels were calculated by the comparative threshold cycle method (2-ΔCt)[13] using GAPDH as the internal control.
Flow cytometry assay
The cells were seeded (2 × 105 cells/mL) onto 6-well plates at 72 h posttransfection and incubated to reach approximately 85% confluence. Both supernatant and adherent cells were harvested, centrifuged, washed with D-Hanks solution, and re-suspended at a density of 1 × 106 cells/mL in 1 × binding buffer solution. The cells were stained with Annexin V-APC for 15 min at room temperature using the Annexin V Apoptosis Detection Kit APC (eBioscience, San Diego, CA, United States) following the manufacturer’s instructions. Flow cytometry was performed on a Guava easyCyte HT system (Millipore, Billerica, MA, United States) and analyzed using Guava InCyte software (Millipore).
Clone-forming assay
The cells were seeded (8 × 102 cells/well) onto 6-well plates at 72 h posttransfection and cultured for 9 d with a medium change every 3 d. The cell clones were photographed using a fluorescence microscope (Olympus, Tokyo, Japan) before the termination of the culture. The cells were fixed with 4% paraformaldehyde for 30 min and washed once with phosphate-buffered saline (PBS), followed by staining with Giemsa (Sigma-Aldrich, United States). After washing with distilled deionized water and drying completely, the cell clones were photographed with a digital camera and then counted. Each experimental group was performed in triplicate.
Cell proliferation assay
The cells were seeded onto 96-well plates at a density of 2 × 103 cells/well and cultured at 37 °C in 5% CO2 for 24 h. Direct counting of cells in the 96-well plates was scanned and analyzed using a Celigo cytometer (Nexcelom, Manchester, United Kingdom) from the next day of plating for a continuous 5 d. By adjusting the input parameters of the analysis settings, the number of cells with green fluorescence was accurately calculated and statistically analyzed. Cell count-fold represents the cell count at each time point relative to the average on day 1, indicating changes in cell proliferation. Cell growth curves were plotted based on the cell count-fold value at different time points.
The cells were seeded onto a 96-well plate at a density of 5 × 103 cells/well and incubated for 24 h. Then, 20 μL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT; Genview, Houston, TX, United States; 5 mg/mL) in PBS was added to each well, and the cells were incubated for an additional 4 h. After termination of incubation, the supernatants were discarded and replaced with 100 μL dimethyl sulfoxide to dissolve the formazan crystals. The optical density values were measured at 490 nm to estimate viable cells. OD490-fold represents the OD values at each time point relative to the average of day 1, indicating changes in cell proliferation.
Cell migration assay
The cell density was adjusted to 9 × 104 cells/well with basic medium. The apical chamber was loaded with 100 μL of cell suspension, and the basolateral chamber was supplied with 600 μL of the culture medium containing 30% FBS. After incubation for 48 h, the medium was removed from the upper chamber, and the cells were scraped off with a cotton swab. The cells that invaded to the other side of the membrane were fixed with 4% paraformaldehyde for 30 min, followed by staining with Giemsa (Sigma-Aldrich, United States). Migratory cells were counted under an inverted microscope (200 ×) in at least three randomly selected fields.
Cell invasion assay
The cell density was adjusted to 5 × 104 cells/well with basic medium. Before seeding, 24-well transwell chambers with 8 μm pores (Corning, United States) were coated with Matrigel (BD Biosciences, San Jose, CA, United States), followed by the hydration of the Matrigel matrix layer. The apical chamber was loaded with 500 μL of cell suspension, and the basolateral chamber was supplied with 750 μL of the culture medium containing 30% FBS. After incubation for 60 h, the medium was removed from the upper chamber, and the cells were scraped off with a cotton swab. The cells that invaded to the other side of the membrane were stained with Giemsa. The invaded cells were counted under an inverted microscope in at least three randomly selected fields.
Western blot analysis
The cells were harvested and washed twice with ice-cold PBS and then lysed using protein RIPA lysis buffer (Beyotime, Shanghai, China). The protein concentration was determined using a BCA protein assay kit (Beyotime, Shanghai, China). The protein samples were mixed with 5× SDS-PAGE loading buffer (Beyotime, Shanghai, China) and boiled for 5 min. A total of 30 μg of protein was separated using 10% SDS-PAGE and then transferred onto polyvinylidene fluoride membranes (Merck Millipore, Billerica, MA, United States). Subsequently, the membranes were blocked in Tris-buffered saline/Tween 20 (TBST) solution containing 5% skim milk at room temperature for 1 h and then incubated separately overnight at 4 °C with the following primary antibodies: Anti-NFE2L3 (NBP2-30870, 1:200, Novus), anti-N-Cadherin (13116, 1:1000, Cell Signaling Technology), anti-Vimentin (ab92547, 1:2500, Abcam), anti-Snail1 (3879, 1:1000, Cell Signaling Technology), anti-Snail2 (9585, 1:1000, Cell Signaling Technology), and anti-GAPDH (sc-32233, 1:2000, Santa Cruz). GAPDH was used as the internal control to normalize protein loading. After washing four times with TBST, the membranes were incubated for 1.5 h at room temperature with horseradish peroxidase-conjugated AffiniPure goat anti-mouse IgG (sc-2005, Santa Cruz) or anti-rabbit IgG (sc-2004, Santa Cruz). The protein bands were visualized with the Pierce ECL Western Blotting Substrate kit (Thermo Scientific, Rockford, IL, United States) after washing with TBST.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 6.0 software (GraphPad Prism 6.0 Software Inc., San Diego, CA, United States). We used a chi-square test to analyze the differences between groups and the Spearman rank correlation test to examine the correlations. Student’s t test (two-tailed) was used to determine significant differences between two groups. One-way analysis of variance (ANOVA), followed by Dunn’s multiple comparison test, was used for multiple comparisons. The data are presented as the mean ± SD. Differences with P-values < 0.05 were considered statistically significant.
RESULTS
Correlation between NFE2L3 expression and clinicopathological features
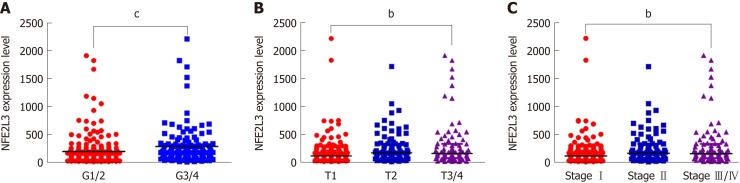
To evaluate NFE2L3 expression in HCC, we analyzed 344 HCC patients with detailed clinicopathological information, and RNA-Seq expression data were obtained from the TCGA database. For the statistical analysis, the patients were divided into two groups based on the median expression level of NFE2L3: A low expression group (n = 172) and a high expression group (n = 172). The association between NFE2L3 expression and clinicopathological features are listed in Table 1, which revealed that NFE2L3 expression was related to tumor grade (P = 0.0006), T stage (P = 0.007), and pathologic stage (P = 0.0138). As shown in Figure 1A, the expression level of NFE2L3 in G3/4 HCC patients was significantly higher than that in G1/2 grade patients. Figure 1B and C shows that NFE2L3 expression gradually increased with the advancement of T stage and pathologic stage, and the difference was significant in T3/4 patients compared to T1 patients (P < 0.01) and was also significant in stage III/IV patients compared to stage I patients (P < 0.01). In addition, the Spearman rank correlation analysis demonstrated that there was a significant correlation between NFE2L3 expression and tumor grade (P < 0.0001), T stage (P = 0.0004), and pathologic stage (P = 0.0007), but there was no significant correlation between different clinicopathological features (Table 2).
Table 1.
Association of nuclear factor erythroid 2-like 3 expression with clinicopathological features of hepatocellular carcinoma patients in The Cancer Genome Atlas dataset n (%)
| Variable |
NFE2L3 expression |
Total cases | P-value | |
| Low (n = 172) | High (n = 172) | |||
| Tumor grade | 0.0006 | |||
| G1/2 | 122 (70.9) | 91 (52.9) | 213 | |
| G3/4 | 50 (29.1) | 81 (47.1) | 131 | |
| T stage | 0.0070 | |||
| T1 | 100 (58.1) | 71 (41.2) | 171 | |
| T2 | 35 (20.3) | 52 (30.2) | 87 | |
| T3/4 | 37 (21.5) | 49 (28.5) | 86 | |
| Pathologic stage | 0.0138 | |||
| I | 98 (57.0) | 71 (41.2) | 169 | |
| II | 35 (20.3) | 50 (29.1) | 85 | |
| III/IV | 39 (22.7) | 51 (29.7) | 90 | |
NFE2L3: Nuclear factor erythroid 2-like 3.
Figure 1.
Association of nuclear factor erythroid 2-like 3 expression with clinicopathological features of hepatocellular carcinoma patients in The Cancer Genome Atlas dataset. A: Nuclear factor erythroid 2-like 3 (NFE2L3) expression in G1/2 (n = 213) and G3/4 (n = 131) hepatocellular carcinoma (HCC) patients; B: NFE2L3 expression in T1 (n = 171), T2 (n = 87) and T3/4 (n = 86) HCC patients; C: NFE2L3 expression in stage I (n = 169), stage II (n = 85), and stage III/IV (n = 90) HCC patients. bP < 0.01, cP < 0.001. NFE2L3: Nuclear factor erythroid 2-like 3; HCC: Hepatocellular carcinoma.
Table 2.
Pearson correlation coefficients for the relationship of nuclear factor erythroid 2-like 3 expression and clinicopathological features
|
NFE2L3 expression |
Tumor grade |
T stage |
Pathologic stage |
|||||
| r | P-value | r | P-value | r | P-value | r | P-value | |
| NFE2L3 expression | 1.000 | 0.2354 | < 0.0001 | 0.1913 | 0.0004 | 0.1811 | 0.0007 | |
| Tumor grade | 0.2354 | < 0.0001 | 1.000 | -0.0032 | 0.9522 | -0.0069 | 0.8985 | |
| T stage | 0.1913 | 0.0004 | -0.0032 | 0.9522 | 1.000 | 0.9814 | < 0.0001 | |
| Pathologic stage | 0.1811 | 0.0007 | -0.0069 | 0.8985 | 0.9814 | < 0.0001 | 1.000 | |
NFE2L3: Nuclear factor erythroid 2-like 3.
Expression of NFE2L3 mRNA and protein in HCC cell lines
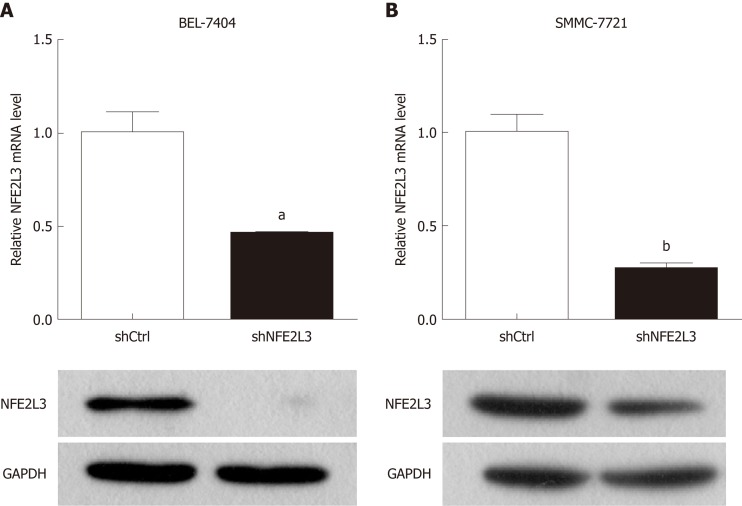
To investigate the potential role of NFE2L3, we performed shRNA interference to knock down NFE2L3 in BEL-7404 and SMMC-7721 cells. The interference efficiency of NFE2L3 shRNA was measured by qPCR and Western blot. As shown in Figure 2A and B, NFE2L3 mRNA levels were significantly reduced by more than 50% (P < 0.05) in the two cell lines infected with NFE2L3 shRNA (shNFE2L3) compared with the negative control cells (shCtrl). Additionally, the Western blot results indicated that the protein level of NFE2L3 was markedly decreased after shRNA-mediated knockdown in BEL-7404 and SMMC-7721 cells.
Figure 2.
Expression of nuclear factor erythroid 2-like 3 mRNA and protein in hepatocellular carcinoma cell lines. A: Nuclear factor erythroid 2-like 3 (NFE2L3) mRNA levels in BEL-7404 and SMMC-7721 cells infected with shNFE2L3 (NFE2L3 short hairpin RNA) were measured by qPCR; B: NFE2L3 protein levels in BEL-7404 and SMMC-7721 cells infected with shNFE2L3 were measured by Western blot. Data shown are the mean ± SEM, aP < 0.05, bP < 0.01 vs shCtrl. NFE2L3: Nuclear factor erythroid 2-like 3; shCtrl: Negative control cells; shNFE2L3: Nuclear factor erythroid 2-like 3 short hairpin RNA.
Role of NFE2L3 in the apoptosis, clone formation, and proliferation of HCC cells
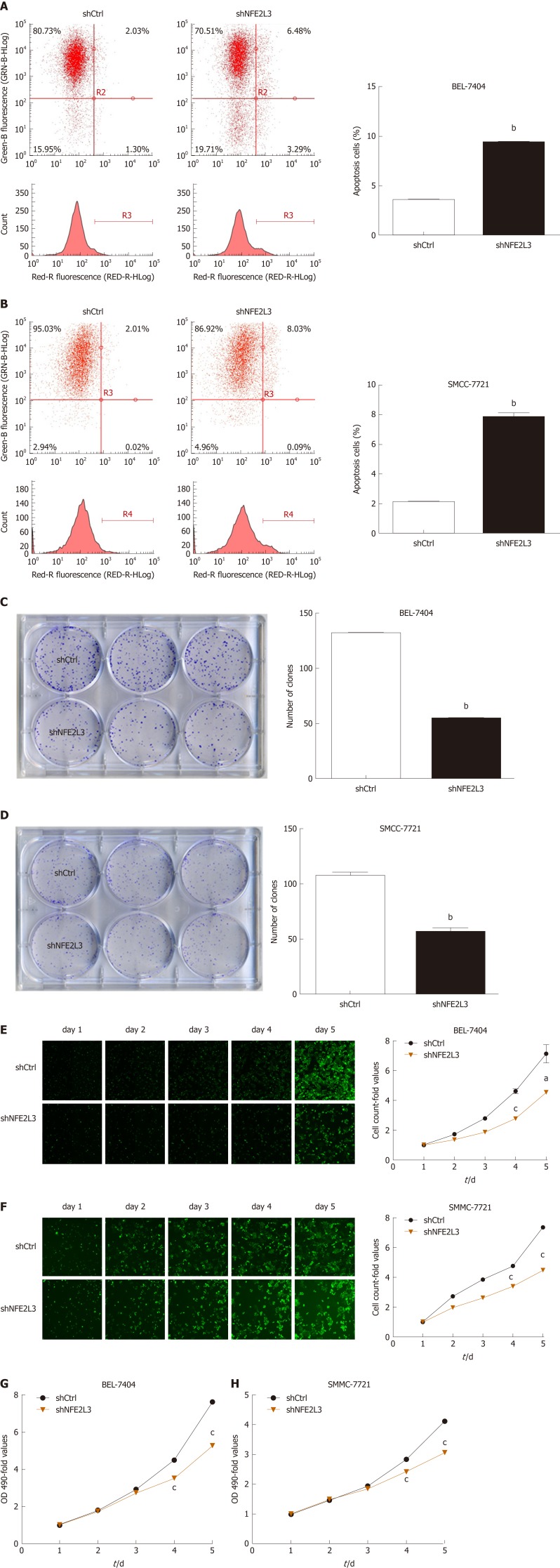
Flow cytometry results demonstrated that the apoptosis rate of the shNFE2L3 group was significantly higher than that of the shCtrl group in BEL-7404 and SMMC-7721 cells (P < 0.05, Figure 3A and B). The clone formation assay of BEL-7404 and SMMC-7721 cells showed that NFE2L3 shRNA reduced the number of clones by approximately 50% compared with those in the shCtrl group (Figure 3C and D). After 5 d of continuous detection by Celigo cytometry, the number of cells with green fluorescence was accurately calculated and statistically analyzed. The cell count-fold values of the shNFE2L3 group were significantly lower than those of the shCtrl group in BEL-7404 and SMMC-7721 cells, especially on day 4 and day 5 (Figure 3E and F). Subsequently, the MTT results revealed that the OD 490-fold values of the shNFE2L3 group in BEL-7404 and SMMC-7721 cells were significantly lower than those of the shCtrl group on days 4 and 5 (Figure 3G and H), which are consistent with the results detected by the Celigo cytometer.
Figure 3.
Role of nuclear factor erythroid 2-like 3 in apoptosis, clone formation, and proliferation of hepatocellular carcinoma cells. A and B: Apoptotic cells were stained with Annexin V-APC and measured using flow cytometry. The abscissa represents red fluorescence (RED-R-HLog), and the ordinate represents green fluorescence (GRN-B-HLog) and cell count; C and D: Cell clones were stained with Giemsa and photographed with a digital camera. The number of clones was accurately calculated and statistically analyzed; E and F: Successfully infected cells were green fluorescent protein positive and cell images were taken with a Celigo cytometer for a continuous 5 d. Cell count-fold represents cell count at each time point relative to the average of day 1; G and H: OD values were measured at 490 nm after the treatment of MTT. Cell growth curves were plotted based on OD 490-fold at different time point. Data shown are the mean ± SEM. Student’s t test was used to analyze significant differences, aP < 0.05, bP < 0.01, cP < 0.001 vs shCtrl. shCtrl: Negative control cells; shNFE2L3: Nuclear factor erythroid 2-like 3 short hairpin RNA.
ShRNA-mediated knockdown of NFE2L3 suppresses the migration, invasion, and epithelial-mesenchymal transition (EMT) of HCC cells
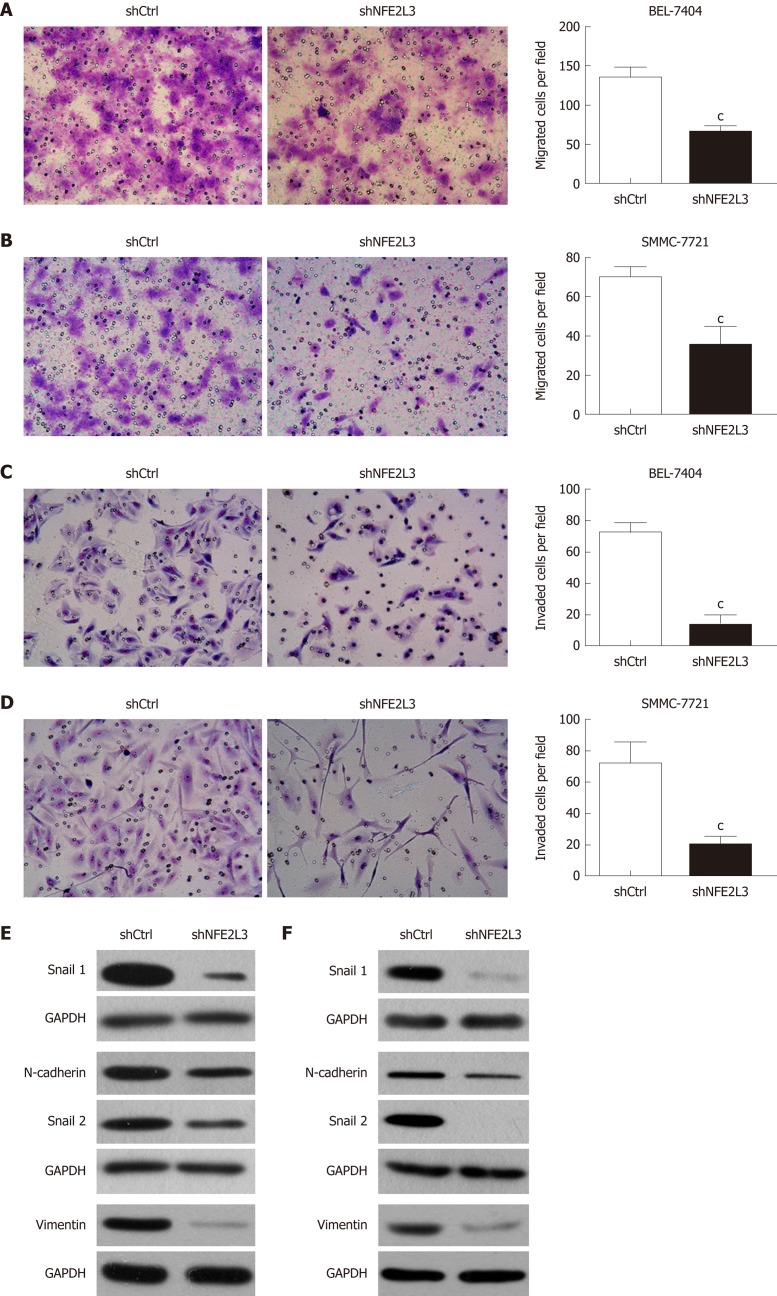
We further explored the effect of NFE2L3 knockdown on the migration, invasion, and EMT of HCC cells. The cell migration assay showed that compared with the shCtrl group, shRNA-mediated knockdown of NFE2L3 in BEL-7404 and SMMC-7721 cells resulted in markedly decreased migration ability, as indicated by a significant reduction in the average number of migrated cells (Figure 4A and B). Furthermore, the transwell invasion assay showed that NFE2L3 knockdown significantly weakened the invasion capacity of BEL-7404 and SMMC-7721 cells in vitro, as evidenced by a significant decrease in the mean number of cells that invaded through the Matrigel-coated membrane (Figure 4C and D). As shown in Figure 4E and F, shRNA-mediated knockdown of NFE2L3 dramatically decreased the levels of mesenchymal markers (N-cadherin and Vimentin) and EMT transcription regulators (Snail1 and Snail2) in BEL-7404 and SMMC-7721 cells.
Figure 4.
Short hairpin RNA-mediated knockdown of nuclear factor erythroid 2-like 3 suppresses migration, invasion, and epithelial-mesenchymal transition of hepatocellular carcinoma cells. A and B: Cell migration assay showed that migrated cells of the shNFE2L3 group were less than that of the shCtrl group in BEL-7404 and SMMC-7721 cells; C and D: Cell invasion assay showed that invaded cells of the shNFE2L3 group were less than that of the shCtrl group in BEL-7404 and SMMC-7721 cells. Both cell migration and invasion were measured with transwell assays. Migrated and invaded cells were stained with Giemsa and imaged and counted under a microscope. Scare bar = 150 μm; E and F: Snail 1, N-cadherin, Snail 2, and Vimentin protein levels were analyzed using Western blot. GAPDH was a loading control. Data shown are the mean ± SEM. Student’s t test was used to analyze significant differences, cP < 0.001 vs shCtrl. shCtrl: Negative control cells; shNFE2L3: Nuclear factor erythroid 2-like 3 short hairpin RNA.
DISCUSSION
HCC is the result of many variable etiological factors, such as HBV, HCV, alcohol, aflatoxin, congenital and acquired metabolic diseases[14,15]. Thus, HCC caused by different risk factors may have a different molecular basis involving the loss of cell cycle control and senescence control, the dysregulation of apoptosis, liver inflammation, and hepatocarcinogenesis[16]. As a potent transcriptional regulator, NFE2L3 can regulate carcinogenesis, inflammation, and stem cell differentiation by activating or inhibiting the transcription of its target genes depending on the cellular context[12,17]. Additionally, DNA methylation is associated with metabolic diseases[18], which are the main risk factors of HCC. Thus, the methylation level of NFE2L3 gene may be involved in hepatocarcinogenesis. However, the role of NFE2L3 in HCC and its underlying molecular mechanisms have not been investigated.
In the present study, we first reported the expression and biological function of NFE2L3 in HCC. TCGA analysis showed that NFE2L3 expression was significantly positively correlated with tumor grade, T stage, and pathologic stage. Subsequently, we selected SMMC-7721 and BEL-7404 for shRNA interference and functional studies. Knockdown of NFE2L3 could induce apoptosis and inhibit the clone formation and cell proliferation of HCC cells. NFE2L3 knockdown also significantly suppressed the migration and invasion of HCC cells. Additionally, our results showed that shRNA-mediated knockdown of NFE2L3 dramatically decreased the levels of mesenchymal markers (N-cadherin and Vimentin) and EMT transcription regulators (Snail1 and Snail2) in BEL-7404 and SMMC-7721 cells. EMT is essential for tumor metastasis and involves a cellular reprogramming process in which polarized, immotile epithelial cells lose adherent and tight junctions, exhibit increased motility, and acquire a mesenchymal phenotype[19,20]. N-cadherin, a mesenchymal cadherin associated with EMT, has been widely studied in various tumor types. Vimentin, as a type 3 intermediate filament protein, is also a critical EMT mesenchymal cell marker. Several transcription factors, including the Snail/Slug family, Twist, and ZEB1, function as molecular switches of the EMT program[21]. As a critical regulator of multiple signaling pathways leading to EMT, the expression of Snail is closely associated with cancer metastasis. Slug, another member of the Snail family of transcription factors, has been characterized as a strong E-cadherin repressor and a major EMT inducer and is correlated with distant metastasis[22]. Thus, the decrease in the levels of N-cadherin, Vimentin, Snail1, and Snail2 represents a block in the EMT process. These observations will help us to further investigate the molecular mechanisms of NFE2L3 in HCC, which will be reported in the near future.
Based on the published literature, we speculated on the possible mechanism underlying the role of NFE2L3 in HCC. Chenais et al[7] found that NRF3 (NFE2L3) mRNA and protein expression is upregulated by the proinflammatory cytokine tumor necrosis factor-α (TNF-α). Huang et al[23] reported that TNF-α could enhance migration and inhibit the apoptosis of the HCC cell line HepG2 by upregulating heat shock protein 70 (HSP70). Jing et al[24] indicated that TNF-α promoted HCC carcinogenesis through the activation of hepatic progenitor cells. Therefore, we hypothesized that the cancer-promoting role of NFE2L3 in HCC might be regulated by TNF-α. Moreover, Xiao et al[25] indicated that NFE2L3 could directly activate the phospholipase A2, group 7 (Pla2g7) gene at the transcriptional level. Pla2g7 was upregulated in more than half of the tumors in hepatitis C virus-associated HCC[26]. Pla2g7, associated with aggressive prostate cancer, promoted prostate cancer cell migration and invasion and was inhibited by statins[27]. Low et al[28] found that Pla2g7 is important in regulating tumor cell migration and acts as a novel tumor-promoting factor in nasopharyngeal carcinoma. It could be speculated that NFE2L3 might promote HCC cell migration and invasion via directly activating Pla2g7 transcription. Additionally, NFE2L3 was identified as a negative regulator of the peroxiredoxin 6 (PRDX6) promoter in human pulmonary A549 cells by reporter gene assay[9,29]. PRDX6, as a bifunctional enzyme with both peroxidase and calcium-independent phospholipase A2 (iPLA2) activity, induces S phase arrest and promotes apoptosis in HCC cells, and inhibits HCC tumorigenicity in mice injected with cancer cells[30]. From these data, we hypothesized that shRNA-mediated knockdown of NFE2L3 might induce apoptosis and inhibit hepatocarcinogenesis through negative regulation of PRDX6. The discovery and identification of NFE2L3 target genes are essential for understanding the molecular mechanisms underlying their roles. Although several potential target genes of NFE2L3 have been reported, further experiments are required to confirm whether the genes mentioned above are genuine targets of NFE2L3 in HCC.
While our current research provides important and instructive findings, some limitations do exist. Due to partial loss of follow-up, it was not possible to perform survival analyses for some clinical cases. In future studies, we will expand the sample size, strengthen follow-up, and conduct detailed analyses. In addition, we have investigated the function of NFE2L3 at the cellular level in vitro, but no animal experiments have been performed in vivo. We need to further validate the role of NFE2L3 through in vivo experiments and explore the molecular mechanisms underlying its roles in HCC.
In conclusion, we identified that NFE2L3 was closely associated with the grade and stage of HCC patients based on the TCGA database. Furthermore, our study showed that shRNA-mediated knockdown of NFE2L3 exhibited tumor-suppressing effects in HCC cells.
ARTICLE HIGHLIGHTS
Research background
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors. Many factors can induce HCC, such as viral infection, alcoholic cirrhosis, and aflatoxin, while the underlying mechanism remains unclear. Nuclear factor erythroid 2-like 3 (NFE2L3) is a member of the cap ‘n’ collar basic-region leucine zipper family of transcription factors. Studies have shown that NFE2L3 acts as a crucial regulator in a variety of cancer progression involving proliferation, migration, and invasion of tumor cells.
Research motivation
The roles of NFE2L3 in HCC and their underlying molecular mechanisms have yet to be elucidated.
Research objectives
Our study aimed to examine NFE2L3 expression in HCC and to analyze its association with clinicopathological features, and to systematically investigate the biological effects of NFE2L3 in HCC cell lines.
Research methods
Based on The Cancer Genome Atlas data portal, we analyzed NFE2L3 expression in 344 HCC patients and its correlation with clinicopathological features. Short hairpin RNA (shRNA) interference technology was utilized to knock down NFE2L3 in SMMC-7721 and BEL-7404 cells. NFE2L3 mRNA levels were quantified using qPCR. Flow cytometry, clone-forming, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, and transwell assays were performed to evaluate the apoptosis, clone formation, proliferation, migration, and invasion of HCC cells. The protein levels of NFE2L3 and epithelial-mesenchymal transition (EMT) markers were examined by Western blot.
Research results
Our results revealed that NFE2L3 expression in G3/4 HCC patients was significantly higher than that in G1/2 grade patients, and its expression gradually increased with the advancement of T stage and pathologic stage. The Spearman rank correlation analysis showed that NFE2L3 expression was significantly correlated with tumor grade, T stage, and pathologic stage. The qPCR and Western blot results indicated that both mRNA and protein levels of NFE2L3 were markedly decreased after shRNA-mediated knockdown in BEL-7404 and SMMC-7721 cells. Flow cytometry results demonstrated that shRNA-mediated knockdown of NFE2L3 could promote the apoptosis of HCC cells. Meanwhile, NFE2L3 knockdown significantly suppressed the clone formation, cell proliferation, migration, and invasion of HCC cells. Additionally, our results showed that NFE2L3 knockdown dramatically decreased the levels of mesenchymal markers (N-cadherin and Vimentin) and EMT transcription regulators (Snail1 and Snail2) in HCC cells.
Research conclusions
The present study identified that NFE2L3 was closely associated with the grade and stage of HCC patients, and shRNA-mediated knockdown of NFE2L3 exhibited tumor-suppressing effects in HCC cells.
Research perspectives
Our study preliminarily explored the possible role of NFE2L3 in HCC. Future studies should focus on the following aspects. First, we will expand the sample size, strengthen follow-up, and conduct survival analyses. Second, animal experiments will be performed to further validate the role of NFE2L3 in vivo. Finally, we need to further explore the molecular mechanisms underlying its roles in HCC.
Footnotes
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Germany
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
Institutional review board statement: The clinicopathological features involved in this study are all from the TCGA database, thus, no institutional review board document is provided.
Conflict-of-interest statement: None declared.
Data sharing statement: No additional data are available.
ARRIVE guidelines statement: The authors have read the ARRIVE guidelines and prepared the manuscript accordingly.
Peer-review started: January 4, 2019
First decision: January 23, 2019
Article in press: February 16, 2019
P- Reviewer: Gazouli M, Tarantino G S- Editor: Yan JP L- Editor: Wang TQ E- Editor: Yin SY
Contributor Information
Miao-Mei Yu, Comprehensive Laboratory, the Third Affiliated Hospital of Soochow University, Changzhou 213003, Jiangsu Province, China.
Yue-Hua Feng, Comprehensive Laboratory, the Third Affiliated Hospital of Soochow University, Changzhou 213003, Jiangsu Province, China.
Lu Zheng, Comprehensive Laboratory, the Third Affiliated Hospital of Soochow University, Changzhou 213003, Jiangsu Province, China.
Jun Zhang, Comprehensive Laboratory, the Third Affiliated Hospital of Soochow University, Changzhou 213003, Jiangsu Province, China.
Guang-Hua Luo, Comprehensive Laboratory, the Third Affiliated Hospital of Soochow University, Changzhou 213003, Jiangsu Province, China. shineroar@163.com.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Waller LP, Deshpande V, Pyrsopoulos N. Hepatocellular carcinoma: A comprehensive review. World J Hepatol. 2015;7:2648–2663. doi: 10.4254/wjh.v7.i26.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, Gores G. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 4.Tunissiolli NM, Castanhole-Nunes MMU, Biselli-Chicote PM, Pavarino EC, da Silva RF, da Silva RC, Goloni-Bertollo EM. Hepatocellular Carcinoma: A Comprehensive Review of Biomarkers, Clinical Aspects, and Therapy. Asian Pac J Cancer Prev. 2017;18:863–872. doi: 10.22034/APJCP.2017.18.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medavaram S, Zhang Y. Emerging therapies in advanced hepatocellular carcinoma. Exp Hematol Oncol. 2018;7:17. doi: 10.1186/s40164-018-0109-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi A, Ito E, Toki T, Kogame K, Takahashi S, Igarashi K, Hayashi N, Yamamoto M. Molecular cloning and functional characterization of a new Cap'n' collar family transcription factor Nrf3. J Biol Chem. 1999;274:6443–6452. doi: 10.1074/jbc.274.10.6443. [DOI] [PubMed] [Google Scholar]
- 7.Chénais B, Derjuga A, Massrieh W, Red-Horse K, Bellingard V, Fisher SJ, Blank V. Functional and placental expression analysis of the human NRF3 transcription factor. Mol Endocrinol. 2005;19:125–137. doi: 10.1210/me.2003-0379. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Kobayashi A, Yamamoto M, Hayes JD. The Nrf3 transcription factor is a membrane-bound glycoprotein targeted to the endoplasmic reticulum through its N-terminal homology box 1 sequence. J Biol Chem. 2009;284:3195–3210. doi: 10.1074/jbc.M805337200. [DOI] [PubMed] [Google Scholar]
- 9.Chevillard G, Blank V. NFE2L3 (NRF3): The Cinderella of the Cap'n'Collar transcription factors. Cell Mol Life Sci. 2011;68:3337–3348. doi: 10.1007/s00018-011-0747-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chevillard G, Paquet M, Blank V. Nfe2l3 (Nrf3) deficiency predisposes mice to T-cell lymphoblastic lymphoma. Blood. 2011;117:2005–2008. doi: 10.1182/blood-2010-02-271460. [DOI] [PubMed] [Google Scholar]
- 11.Wang C, Saji M, Justiniano SE, Yusof AM, Zhang X, Yu L, Fernández S, Wakely P, Jr, La Perle K, Nakanishi H, Pohlman N, Ringel MD. RCAN1-4 is a thyroid cancer growth and metastasis suppressor. JCI Insight. 2017;2:e90651. doi: 10.1172/jci.insight.90651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chowdhury AMMA, Katoh H, Hatanaka A, Iwanari H, Nakamura N, Hamakubo T, Natsume T, Waku T, Kobayashi A. Multiple regulatory mechanisms of the biological function of NRF3 (NFE2L3) control cancer cell proliferation. Sci Rep. 2017;7:12494. doi: 10.1038/s41598-017-12675-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C (T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 14.Sengupta S, Parikh ND. Biomarker development for hepatocellular carcinoma early detection: Current and future perspectives. Hepat Oncol. 2017;4:111–122. doi: 10.2217/hep-2017-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scalera A, Tarantino G. Could metabolic syndrome lead to hepatocarcinoma via non-alcoholic fatty liver disease? World J Gastroenterol. 2014;20:9217–9228. doi: 10.3748/wjg.v20.i28.9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Stefano F, Chacon E, Turcios L, Marti F, Gedaly R. Novel biomarkers in hepatocellular carcinoma. Dig Liver Dis. 2018;50:1115–1123. doi: 10.1016/j.dld.2018.08.019. [DOI] [PubMed] [Google Scholar]
- 17.Siegenthaler B, Defila C, Muzumdar S, Beer HD, Meyer M, Tanner S, Bloch W, Blank V, Schäfer M, Werner S. Nrf3 promotes UV-induced keratinocyte apoptosis through suppression of cell adhesion. Cell Death Differ. 2018;25:1749–1765. doi: 10.1038/s41418-018-0074-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwenk RW, Vogel H, Schürmann A. Genetic and epigenetic control of metabolic health. Mol Metab. 2013;2:337–347. doi: 10.1016/j.molmet.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 20.Prieto-García E, Díaz-García CV, García-Ruiz I, Agulló-Ortuño MT. Epithelial-to-mesenchymal transition in tumor progression. Med Oncol. 2017;34:122. doi: 10.1007/s12032-017-0980-8. [DOI] [PubMed] [Google Scholar]
- 21.Gheldof A, Berx G. Cadherins and epithelial-to-mesenchymal transition. Prog Mol Biol Transl Sci. 2013;116:317–336. doi: 10.1016/B978-0-12-394311-8.00014-5. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Shi J, Chai K, Ying X, Zhou BP. The Role of Snail in EMT and Tumorigenesis. Curr Cancer Drug Targets. 2013;13:963–972. doi: 10.2174/15680096113136660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang BP, Lin CS, Wang CJ, Kao SH. Upregulation of heat shock protein 70 and the differential protein expression induced by tumor necrosis factor-alpha enhances migration and inhibits apoptosis of hepatocellular carcinoma cell HepG2. Int J Med Sci. 2017;14:284–293. doi: 10.7150/ijms.17861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jing Y, Sun K, Liu W, Sheng D, Zhao S, Gao L, Wei L. Tumor necrosis factor-α promotes hepatocellular carcinogenesis through the activation of hepatic progenitor cells. Cancer Lett. 2018;434:22–32. doi: 10.1016/j.canlet.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Xiao Q, Pepe AE, Wang G, Luo Z, Zhang L, Zeng L, Zhang Z, Hu Y, Ye S, Xu Q. Nrf3-Pla2g7 interaction plays an essential role in smooth muscle differentiation from stem cells. Arterioscler Thromb Vasc Biol. 2012;32:730–744. doi: 10.1161/ATVBAHA.111.243188. [DOI] [PubMed] [Google Scholar]
- 26.Smith MW, Yue ZN, Geiss GK, Sadovnikova NY, Carter VS, Boix L, Lazaro CA, Rosenberg GB, Bumgarner RE, Fausto N, Bruix J, Katze MG. Identification of novel tumor markers in hepatitis C virus-associated hepatocellular carcinoma. Cancer Res. 2003;63:859–864. [PubMed] [Google Scholar]
- 27.Vainio P, Lehtinen L, Mirtti T, Hilvo M, Seppänen-Laakso T, Virtanen J, Sankila A, Nordling S, Lundin J, Rannikko A, Orešič M, Kallioniemi O, Iljin K. Phospholipase PLA2G7, associated with aggressive prostate cancer, promotes prostate cancer cell migration and invasion and is inhibited by statins. Oncotarget. 2011;2:1176–1190. doi: 10.18632/oncotarget.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Low HB, Png CW, Li C, Wang Y, Wong SB, Zhang Y. Monocyte-derived factors including PLA2G7 induced by macrophage-nasopharyngeal carcinoma cell interaction promote tumor cell invasiveness. Oncotarget. 2016;7:55473–55490. doi: 10.18632/oncotarget.10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chowdhury I, Mo Y, Gao L, Kazi A, Fisher AB, Feinstein SI. Oxidant stress stimulates expression of the human peroxiredoxin 6 gene by a transcriptional mechanism involving an antioxidant response element. Free Radic Biol Med. 2009;46:146–153. doi: 10.1016/reeradbiomed.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu X, Lu D, Zhuang R, Wei X, Xie H, Wang C, Zhu Y, Wang J, Zhong C, Zhang X, Wei Q, He Z, Zhou L, Zheng S. The phospholipase A2 activity of peroxiredoxin 6 promotes cancer cell death induced by tumor necrosis factor alpha in hepatocellular carcinoma. Mol Carcinog. 2016;55:1299–1308. doi: 10.1002/mc.22371. [DOI] [PubMed] [Google Scholar]