Abstract
An induced stringent response, which is established by an increased level of (p)ppGpp, is required for the expression of β-lactam resistance in methicillin-resistant Staphylococcus aureus (MRSA). However, it is not clear whether RSH (enzyme mediating stringent response to amino acid starvation) or small alarmone synthetases (SASs) are involved in the maintenance of (p)ppGpp level in response to β-lactams. Since the S. aureus genome encodes two active SASs (RelP and RelQ), their contribution to the expression of β-lactam resistance in MRSA was investigated. It was determined that relQ deletion renders community-associated MRSA (CA-MRSA) sensitive to β-lactams by negatively affecting the expression of mecA, and induction of (p)ppGpp synthesis by mupirocin bypasses the requirement of relQ for the expression of high-level β-lactam resistance. Surprisingly, relP deletion increased the level of β-lactam resistance. Such contradictory observations could be attributed to the fact that relQ promoter is ~5-fold stronger than the relP and is induced by oxacillin as well as deletion of either of the SASs, while relP promoter responds only to oxacillin. The stronger promoter activity of relQ, coupled with the inducibility of the relQ promoter in response to the lack of relP, results in efficient expression of relQ in the relP-deleted background. This positively affects mecA expression and renders the ΔrelP strain highly resistant. These findings indicate an important role for RelQ in the expression of high-level β-lactam resistance in MRSA.
Keywords: RelP, RelQ, MRSA, MecA, Stringent Response
Introduction
Staphylococcus aureus is a Gram-positive cocci often found on human and animal skins and mucous membranes. It is commonly associated with opportunistic infections in hospitals and the community. Methicillin-resistant S. aureus (MRSA) is intrinsically resistant to most of the β-lactams due to the presence of the mecA gene encoding an altered penicillin-binding protein (PBP) known as PBP-2a (Llarrull et al., 2009). In methicillin-susceptible S. aureus (MSSA), the transpeptidase activity of PBPs is lost due to irreversible acylation of an active site serine by the β-lactam antibiotics (Williamson and Tomasz, 1985). However, the PBP-2a of MRSA is resistant to β-lactam acylation, and successfully catalyzes the DD-transpeptidation reaction, leading to a methicillin-resistant phenotype (Fuda et al., 2004). Although mecA is essential for methicillin resistance, it in itself is not sufficient, since the native PBP2 is also required for MRSA (Pinho et al., 2001) and the level of resistance expressed could be altered by varying temperature, pH, and salt concentration (Chambers and Hackbarth, 1987).
The stringent response (SR) is a highly conserved regulatory mechanism induced by amino acid starvation and various environmental stresses and is established by (p)ppGpp (nutritional alarmone) synthesis. This response adjusts the cell's biosynthetic machinery according to the availability of required precursors and energy. In Escherichia coli, (p)ppGpp is synthesized by two cytoplasmic enzymes, a (p)ppGpp synthetase (RelA), and a (p)ppGpp synthetase/hydrolase (SpoT) (Xiao et al., 1991). Both these enzymes are composed of N-terminal enzymatic (synthetase and/or hydrolase) and C-terminal regulatory domains (Mechold et al., 2002). Staphylococcus aureus harbors a Rel/SpoT homolog (RSH) and two additional small alarmone synthetases (SASs) encoded by relP and relQ (Eymann et al., 2002; Nanamiya et al., 2008; Geiger et al., 2010, 2014). The RSH has N-terminal enzymatic (synthetase and hydrolase) and C-terminal regulatory domains and performs both (p)ppGpp synthesis and degradation activities. Each SAS (RelP or RelQ) has only a synthetase domain; both the hydrolase and the C-terminal regulatory domains are missing.
The role and regulation of RelP and RelQ has been studied in Bacillus subtilis (Nanamiya et al., 2008), Streptococcus mutans (Lemos et al., 2007), and Enterococcus faecalis (Abranches et al., 2009). These studies showed that relP and relQ encode active (p)ppGpp synthatases, which are dispensable in optimal growth conditions. In MSSA, it has been shown that relP and relQ encode active (p)ppGpp synthatases, which are induced in response to cell wall-active antibiotics to mitigate such conditions (Geiger et al., 2014). Recently, ethanol-mediated induction of relP in MRSA was reported (Pando et al., 2017), but involvement of these SASs in the expression of β-lactam resistance in MRSA is still unclear. Here, we explore the involvement of RelP and RelQ in the expression of β-lactam resistance in a community-associated MRSA (CA-MRSA) strain by deleting relP and relQ and characterizing the mutants. This study reveals the importance of RelQ for full expression of β-lactam resistance in MRSA and confirms that relQ deletion negatively affects the mecA expression. The transcript and promoter activity analyses showed that the apparent opposite effect of relP (positive) and relQ (negative) deletions on the level of β-lactam resistance is a consequence of enhanced relQ induction in the relP-deleted background.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study are described in Table 1. Mueller-Hinton (MH) medium was used for drug susceptibility tests. Tryptic Soya Broth (TSB) and Tryptic Soya Agar (TSA) were used for mutagenesis work. Plasmids were propagated and maintained in E. coli DH5α (Taylor et al., 1993) and S. aureus RN4220 (Kreiswirth et al., 1983) using 100 μg/ml ampicillin (Amp) and 10 μg/ml chloramphenicol (Cm), respectively. The nucleotide sequence of the primers used in this study are listed in Table 2.
Table 1.
Bacterial strains and plasmids used in this study.
| Strains or plasmids | Relevant properties | References or sources |
|---|---|---|
| E. coli STRAINS | ||
| DH5α | ΔlacU169 hsdR17 recA1 endA1 gyrA96 thiL relA1 | Taylor et al., 1993 |
| S. aureus STRAINS | ||
| RN4220 | Restriction-deficient S. aureus strain | Kreiswirth et al., 1983 |
| JE2 | Wild-type; developed from a CA-MRSA, USA300-FPR3757 | Fey et al., 2012 |
| ΔrelP | relP deletion mutant of JE2 | Pando et al., 2017 |
| ΔrelQ | relQ deletion mutant of JE2 | This work |
| ΔrelPQ | relPQ deletion mutant of JE2 | This work |
| PLASMIDS | ||
| pLI50 | E. coli-S. aureus shuttle plasmid; AmpR (E. coli); CmR (S. aureus) | Lee et al., 1991 |
| pKOR1 | ATc-inducible suicide mutagenesis vector | Bae and Schneewind, 2006 |
| pALC2073 | E. coli-S. aureus shuttle plasmid with xyl/tetO promoter; | Bateman et al., 2001 |
| pKOR1-ΔrelQ | relQ deletion plasmid | This work |
| pMN12 | relP ORF with its native promoter cloned in KpnI/XbaI site of pLI50 | This work |
| pMN13 | relQ ORF with its native promoter cloned in KpnI/SalI site of pLI50 | This work |
| pMN14 | relP promoter fused with relQ ORF and cloned in KpnI/SalI site of pLI50 | This work |
| pMN15 | relQ promoter fused with relP ORF and cloned in KpnI/XbaI site of pLI50 | This work |
| pMN18 | E. coli lacZ engineered ORF cloned in XbaI/HindIII sites of pLI50 with Bacillus subtilis spoVG ribosomal binding site. | This work |
| pMN19 | relP promoter region cloned in KpnI/XbaI sites of pMN18 | This work |
| pMN20 | SAUSA300_0906 promoter region cloned in KpnI/XbaI sites of pMN18 | This work |
| pMN21 | relQ upstream region (including SAUSA300_0906 promoter and ORF) cloned in KpnI/XbaI sites of pMN18 | This work |
| pMN25 | relQ ORF cloned in EcoRI sites of pALC2073 | This work |
| pMN26 | mecA ORF cloned in EcoRI sites of pALC2073 | This work |
AmpR, ampicillin resistance; CmR, chloramphenicol resistance.
Table 2.
Primers used in this study (additional nucleotides not specific for S. aureus are shown in bold; restriction sites are underlined; and nucleotides used to produce regions for overlap PCR are italicized).
| Primer | Sequence (5′ to 3′) |
|---|---|
| attB1-relQ-us:F | GGGGACAAGTTTGTACAAAAAAGCAGGCTCAATGATGTCATATGGTGTTGTTG |
| relQ-us:R:BamHI | CGGGATCCCCATTGATTCATAGTGCTTCACC |
| relQ-ds:F:BamHI | CGGGATCCGATTAACGAGGTGTTATAAATCATG |
| attB2-relQ-ds:R | GGGGACCACTTTGTACAAGAAAGCTGGGTGTTGTATTACGATCTAGACGCGTAAC |
| relP-up:F | GATTGGTATCGAGCGTTATCG |
| relP:R | CACACCTACTAAACATCTACTC |
| relQ-up:F | GTCGTTAATGCACCAAGTATTG |
| relQ:R | AAGGCATTAGACTTGGAGTCAC |
| rpoB:F | GTGACGCTACTTATGCTGCAC |
| rpoB:R | CGAACGTACCTGTATCAGTC |
| pbp2:RT:F | AGCGTATGGACCTGCCATTG |
| pbp2:RT:R | GTACCGTGACTCTTCGTATC |
| mecA:RT: F | CACCTTCATATGACGTCTATC |
| mecA:RT:R | GAACCTGGTGAAGTTGTAATC |
| PrelP:F:KpnI | CGGGGTACCAGAGAACCGCTTATGGATGGTCCAC |
| relP:R:SalI | ACGCGTCGACCACACCTACTAAACATCTACTC |
| P0906:F:KpnI | CGGGGTACCGCAATTTATTATAGATTGATGCAGTTATC |
| relQ:R:SalI | ACGCGTCGACAGGCATTAGACTTGGAGTCAC |
| PrelP-(Q):R | CTGATCCCATTGATTCATTTTTATACTAACCTC |
| relQ-ORF(P):F | GAGGTTAGTATAAAAATGAATCAATGGGATCAG |
| P0906-(P):R | GTTTTCGATCTACATACATGCTTAATCCTCCTCTTATTC |
| relP-ORF(Q):F | GAATAAGAGGAGGATTAAGCATGTATGTAGATCGAAAAC |
| lacZ:SVF:XbaI | TGCTCTAGAGGGAAAAGGTGGTGAACTACTGTGGAAGTTACTG |
| lacZ:SVF | AACTACTGTGGAAGTTACTGACGTAAGATTACGGGTCGACTGGGAAAACCCTGGCGTTAC |
| lacZ:R:BglII | GAAGATCTCTGCCCGGTTATTATTATTTTTGACACCAGACCAACTG |
| TF:BglII | GAAGATCTGCGATGGCTGTTTTGGCGGATGAGAG |
| TR:HindIII | CCCAAGCTTGTTTGTAGAAACGCAAAAAGGCCATC |
| PrelP:R:XbaI | GCTCTAGACGATATATAATCATCTTTATTGTACC |
| P0906:R:XbaI | GCTCTAGATCCATAACATTTTAACACAATTCAATAATAC |
| PrelQ:R:XbaI | GCTCTAGACTTTATTCAATGTCGAATGTTTCTTC |
| relQ:F:EcoRI | GGAATTCATAAAGCGGGGTGAAGCACTATG |
| relQ:R:EcoRI | GGAATTCTGATTTATAACACCTCGTTAATC |
| mecA:F:EcoRI | GGAATTCGTCTTATATAAGGAGTATATTGATG |
| mecA:R:EcoRI | GGAATTCTTATTCATCTATATCGTATTTTTTATTAC |
| relP-SP1:R | CCACATATCCATACCTATC |
| relP-SP2:R | CACACGTCGCTCCATATGATG |
| relP-SP3:R | GTGCTTATTTCCTTTAGTGCTGAC |
| relQ-SP1:R | CAATAATGACATGATACGAG |
| relQ-SP2:R | CTTAAACCAGCGATATCGTAC |
| relQ-SP3:R | CACCAACTTCATATTGTTTGCGCATG |
Mutant Construction
The markerless and in-frame relQ and relPQ deletion mutants were constructed using Gateway cloning technology-based ATc-inducible suicide mutagenesis vector pKOR1 (Bae and Schneewind, 2006), following the protocol that was used earlier for construction of ΔrelP (Pando et al., 2017). Briefly, the upstream and downstream regions of relQ ORFs were amplified using attB1-relQ-us:F/relQ-us:R:BamHI and relQ-ds:F:BamHI/attB2-relQ-ds:R primer sets (Table 2), respectively, and ligated after BamHI digestion. Afterwards, the ligated fragments were inserted in pKOR1 using the BP reaction of Gateway Technology (Invitrogen) to construct relQ deletion plasmid designated as pKOR1-ΔrelQ. The deletion plasmid was first mobilized into S. aureus RN4220, isolated from RN4220 and then transferred into JE2 (Fey et al., 2012) and ΔrelP by electroporation. Deletion plasmid was integrated (single cross-over) into JE2 and ΔrelP chromosome by growing the transformants at 43°C (non-permissive condition) in TSB supplemented with 7.5 μg/ml chloramphenicol. Integrated plasmid was excised (double cross-over) to replace the wild-type allele by growing at 30°C (permissive condition) in TSB supplemented with 10 μg/ml chloramphenicol, and plasmid-free cells were selected by growing at 30°C temperature on TSA plates supplemented with 1 μg/ml ATc.
Antibiotic Susceptibility Tests
Drug sensitivity tests were performed by the Kirby–Bauer method as described earlier (Bauer et al., 1959), and the minimal inhibitory concentration (MIC) of oxacillin was determined using the agar double-dilution method as described by Wiegand et al. (2008). The MICs were determined using oxacillin concentrations ranging from 0.25 to 1,024 μg/ml in the absence and presence of mupirocin (0.03 μg/ml), and repeated >7 times to confirm the differences in antibiotic susceptibilities.
Construction of relP and relQ Expression Plasmids
The relP and relQ genes were amplified with their native promoters using PrelP:F:KpnI/relP:R:SalI and P0906:F:KpnI/relQ:R:SalI primer sets (Table 2), respectively. The PCR products were cloned into the KpnI/SalI site of pLI50 (Lee et al., 1991) to construct pMN12 (pLI50-relP) and pMN13 (pLI50-relQ). To fuse relQ promoter with relP ORF, P0906:F:KpnI/P0906-(P):R and relP-ORF(Q):F/relP:R:SalI primer sets (Table 2) were used to amplify relQ promoter and relP ORF, respectively. P0906-(P):R and relP-ORF(Q):F primers are engineered to incorporate overlapping nucleotides in the amplified products to facilitate the overlap PCR. The overlap PCR was performed using PCR products as template with the P0906:F:KpnI/relP:R:SalI primer set. Similarly, relP promoter was fused with relQ ORF using PrelP:F:KpnI/PrelP-(Q):R and relQ-ORF(P):F/relQ:R:SalI primer sets (Table 2). Finally, fused PCR products, PrelQ-relP and PrelP-relQ, were cloned into KpnI/SalI sites of pLI50 to construct pMN14 (pLI50- PrelP-relQ) and pMN15 (pLI50- PrelQ-relP). Recombinant plasmids were first mobilized into S. aureus RN4220 by electroporation. Plasmids were isolated from RN4220 and then mobilized into parent and mutant strains by electroporation.
Construction of xyl/tetO Promoter-Driven Expression Plasmids
The relQ and mecA ORFs were amplified with their native ribosomal binding sites (RBS) using relQ:F:EcoRI/relQ:R:EcoRI and mecA:F:EcoRI/mecA:R:EcoRI primer sets, respectively (Table 2). The PCR products were cloned into pALC2073 (Bateman et al., 2001) using the EcoRI site. Recombinant plasmids were confirmed by restriction digestion and sequencing, and designated as pMN25 (pALC2073-relQ) and pMN26 (pALC2073-mecA). These plasmids were mobilized in parent and mutant strains via S. aureus RN4220.
RNA Extraction and Real-Time PCR
Cells were harvested from 1 ml of mid-log phase cultures (0.5–0.9 OD600nm) by centrifugation at 6,000 rpm for 5 min at 4°C, resuspended in 100 μl TE (30 mM Tris-Cl and 1 mM EDTA, pH 8.0), and lysed with 100 μg/ml lysostaphin (Sigma, USA) by incubating at 30°C for 5 min. Afterwards, TRIzol reagent (Invitrogen, Germany) was used to extract total RNA. RNA samples were treated with RNase-free DNase I (New England Biolabs) at a final concentration of 1 U/50 μl sample for 30 min at 37°C. RNA samples were used as template with Taq DNA polymerase to check DNA contaminations. For relative quantification, 2 μg total RNA was reverse transcribed using a cDNA Synthesis Kit (Fermentas, USA), and 1 μl of 10x diluted cDNA was used as template in 20 μl reaction volume. Real-time quantitative PCR (qPCR) was performed using SYBR Green Master mix (ROX; Fermentas) and the 7500 Real-Time PCR System (Applied Biosystems, CA, USA). An amplicon of rpoB was used as endogenous control for relative quantification by the method. Primer efficiencies were calculated by generating the standard curves for each primer pair. Efficiencies of the primer pairs used for RT-PCR were found to be 92–99%. ANOVA (analysis of variance) followed by Tukey's post-hoc test was performed using SPSS 17.0 software for data analysis, and p-values < 0.05 were considered as significantly different in relative expression level.
Construction of lacZ:Reporter Vector and Promoter-lacZ Transcriptional Fusions
Escherichia coli genomic DNA was used as template with lacZ:SV:F/lacZ:R:BglII (Table 2) primers to amplify lacZ ORF. This amplified product was used as template with lacZ:SVF:XbaI/lacZ:R:BglII (Table 2) primers for the second round PCR amplification to add B. subtilis spoVG RBS and modify N-terminal of lacZ ORF as present in pMUTIN2 (Vagner et al., 1998). Primer lacZ:SVF:XbaI and lacZ:SV:F were designed with additional/engineered nucleotides to add B. subtilis spoVG ribosome-binding site and modified N-terminal of lacZ ORF as present in pMUTIN2. The nucleotide sequence of pMUTIN2 (GenBank accession No. AF072806) was followed for this manipulation. A 428 bp fragment encompassing t1 t2 terminators of the E. coli rrnB operon known to be active in B. subtilis (Peschke et al., 1985) was amplified using pMMB206 as template (Morales et al., 1991) with TF:BglII/TR:HindIII primers (Table 2). The lacZ ORF and the terminator were cloned into XbaI/HindIII restriction sites of pLI50 using three fragment ligation to construct pMN18. The upstream regions of relP (485 bp), SAUSA300_0906 (231 bp), and relQ (599 bp) were amplified using PrelP:F:KpnI/PrelP:R:XbaI, P0906:F:KpnI/0906:R:XbaI and P0906:F:KpnI/relQ-Pr:R:XbaI primers, respectively (Table 2), and cloned into KpnI/XbaI sites of pMN18 to construct relP (pMN19), SAUSA300_0906 (pMN20), and relQ (pMN21) promoter:lacZ fusions.
β-Galactosidase Assay
Staphylococcus aureus strains harboring promoter:lacZ transcriptional fusions or the promoterless vector (pMN18) were grown in MHB up to mid-log phase (0.5–0.6 OD600nm). A culture of each strain was then divided into two parts. One part of each strain was supplemented with 4 μg/ml of oxacillin and incubated at 37°C for 60 min, while the other part was kept as control. The cell number in the treated and control cultures of different strains were equalized by adjusting OD600nm = 0.5 using MHB with or without oxacillin. β-galactosidase assays were performed with 1 ml of adjusted culture as described earlier (Miller, 1972). ANOVA (analysis of variance) followed by Tukey's post-hoc test was performed using SPSS 17.0 software for data analysis, and p-values < 0.05 were considered as significantly different in β-galactosidase activities.
5′ RACE
The relP and relQ transcription start sites (TSS) were identified using a 3′/5′ RACE kit, 2nd Generation (Roche, Germany), following the manufacturer's protocol. Briefly, relP and relQ transcripts were reverse transcribed from total RNA into cDNA using relP- or relQ-SP1:R (Table 2). cDNAs were purified and 3′-poly(dA) tailed and then used as template in two PCRs designed to amplify the 5′ ends of relP and relQ using oligo(dT)-anchor/relP-SP2:R or oligo(dT)-anchor/relQ-SP2:R primers, respectively. First PCR products were separately used as template in second PCR using anchor/relP-SP3:R and anchor/relQ-SP3:R primer sets. PCR products were ligated into pGEM-T Easy (Promega, USA), and the clones were sequenced.
Results
Deletion of relQ Results in β-Lactam Sensitivity
The role of (p)ppGpp in increased/homogenous expression of β-lactam resistance has been demonstrated in S. aureus (Mwangi et al., 2013). However, contribution of RelP, RelQ, and RSH to maintain the (p)ppGpp level required for the expression of β-lactam resistance is not clear. To examine the contribution of RelP and RelQ to the expression of drug resistance, relQ was deleted in JE2 (a CA-MRSA, wild-type strain) and ΔrelP following a previously described protocol (Pando et al., 2017). The in-frame deletion mutants, ΔrelQ and ΔrelPQ, were constructed by deleting 618 bp of relQ (636 bp) in JE2 and ΔrelP, respectively, and confirmed by PCR amplification of the relQ locus, and sequencing of amplicons obtained from JE2 and mutants. As expected, the relP-up:F/relP:R (Table 2) primer set produced 1,209 bp fragment with JE2 or ΔrelQ genomic DNA while a smaller amplicon of 532 bp was produced with that of ΔrelP or ΔrelPQ (Supplementary Figure 1). Similarly, the relQ-up:F/relQ:R (Table 2) primer set produced ~1,600 bp amplicon with JE2 or ΔrelP genomic DNA, and a 998 bp amplicon with that of ΔrelQ or ΔrelPQ (Supplementary Figure 1). PCR amplification of expected amplicons and their sequencing confirmed the in-frame deletion of relQ in the mutant strains (data not shown). The relQ and relPQ deletion mutants were designated as ΔrelQ and ΔrelPQ, respectively.
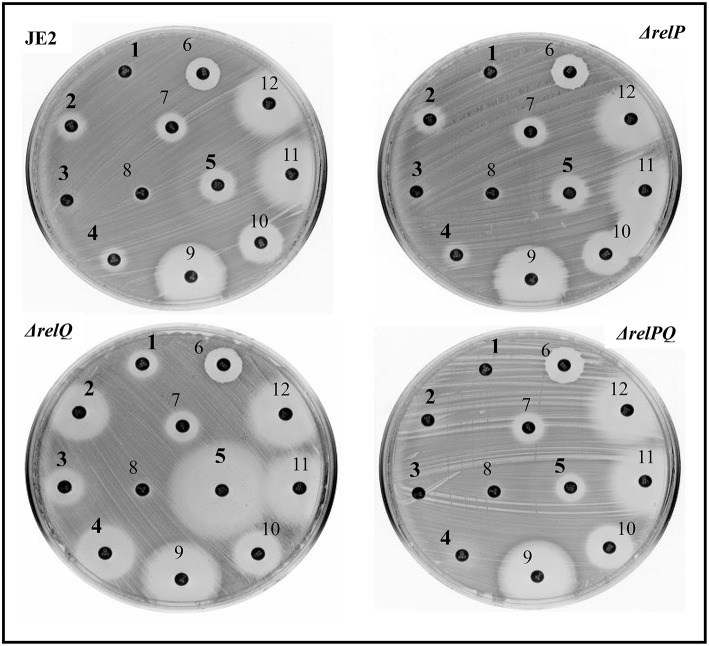
Disc-susceptibility testing revealed that non-β-lactam antibiotic (vancomycin, streptomycin, ciprofloxacin, erythromycin, amikacin, linezolid, and spectinomycin) disks produced either no or an equal-size zone of inhibition with parent and mutant strains. However, β-lactam antibiotic (ceftazidime, cefepime, oxacillin, ceftriaxone, and tazobactam/piperacillin) disks produced a larger zone of inhibition with ΔrelQ compared to the parent (Figure 1). Intriguingly, this test also revealed that the zone of inhibitions produced by β-lactams in the case of ΔrelP and ΔrelPQ were equal to that of the parent (JE2). Since ΔrelPQ was less sensitive than ΔrelQ, relP was again deleted in the ΔrelQ strain, and the disc susceptibility test was repeated. This revealed that deletion of relP from the ΔrelQ genome renders the strain ΔrelPQ less sensitive compared to that of ΔrelQ (data not shown). To validate these observations, MIC testing was performed, which revealed that relQ deletion decreased oxacillin MIC 16-fold while relP deletion increased it 4-fold as compared to JE2. Interestingly, ΔrelPQ was only 4-fold more sensitive than JE2 but 4-fold more resistant than the ΔrelQ strain (Table 3). These results indicate an important role of relQ in β-lactam resistance expressed by MRSA, and also that deletion of relP and relQ affect the β-lactam resistance in an apparently opposite manner.
Figure 1.
Disc-susceptibility test of JE2, ΔrelP, ΔrelQ, and ΔrelPQ using Kirby-Bauer antibiotic testing. The numbers indicate different antibiotic disks; ceftazidime (1), cefepime (2), oxacillin (3), ceftriazone (4), tezobactam/piperacillin (5), vancomycin (6), streptomycin (7), ciprofloxacin (8), erythromycin (9), amikacin (10), linozelid (11), and spectinomycin (12). Numbers in bold indicate the disks for which differences in the sensitivity were observed.
Table 3.
Effect of relP and relQ deletion on oxacillin MIC (determined by agar double-dilution method).
| Strains/mutants | Oxacillin MICs (μg/ml) | |
|---|---|---|
| (–) Mupirocin | (+) Mupirocin (0.03 μg/ml) | |
| JE2 | 128 | 256 |
| ΔrelP | 512 | 512 |
| ΔrelQ | 8 | 256 |
| ΔrelPQ | 32 | 256 |
Mupirocin Restores the β-Lactam Resistance in ΔrelQ and ΔrelPQ
Since relQ is an active (p)ppGpp synthetase (Geiger et al., 2014), and since a higher level of this alarmone is required for the MRSA phenotype (Kim et al., 2013; Mwangi et al., 2013), it was speculated that (i) the sensitivity of ΔrelQ might be a consequence of a relQ deletion-mediated decrease in (p)ppGpp level and, if so, the requirement of relQ for expression of β-lactam resistance should be bypassed by induction of (p)ppGpp synthesis in ΔrelQ; and (ii) higher resistance of ΔrelPQ than ΔrelQ might be a consequence of (p)ppGpp synthesis by the RSH in the absence of RelP and RelQ and, if so, ΔrelPQ resistance level should further increase by induction of (p)ppGpp synthesis via RSH. To test this, oxacillin MICs were determined in the presence of mupirocin, an isoleucine homolog capable of inducing the (p)ppGpp synthesis in Staphylococcus via RSH by inhibiting isoleucyl-tRNA synthetase (Cassels et al., 1995). The results indicated that mupirocin restores the β-lactam resistance in ΔrelQ and ΔrelPQ to the parental level (Table 3). These results suggest that relQ plays an important role in maintaining a higher level of (p)ppGpp required for the expression of β-lactam resistance in MRSA.
Deletion of relQ Reduces the mecA Expression Level
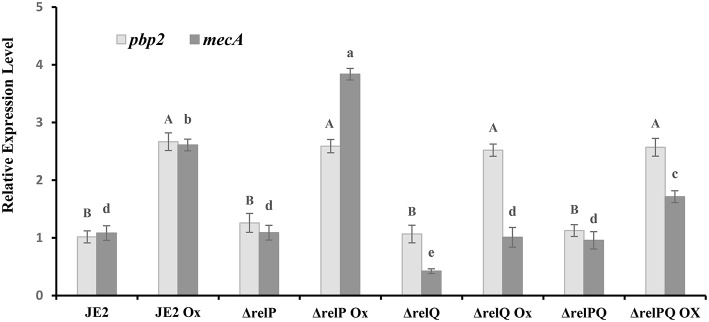
The β-lactam sensitivity of ΔrelQ prompted us to investigate whether lack of relQ affected the mecA expression level. To examine this, relative quantification of mecA transcript was performed to compare its level in parent and mutants grown in the presence or absence of oxacillin. The pbp2, which is known to induce in response to oxacillin (Boyle-Vavra et al., 2003), was used as a positive control for this analysis and was found to follow the reported oxacillin-inducible pattern in every strain (Figure 2). Transcript analysis revealed that in JE2, ΔrelP, and ΔrelPQ have almost equal basal levels of mecA transcript; however, it is >2-fold repressed in ΔrelQ. It is evident from Figure 2 that although oxacillin induces the mecA in all the strains, its level was the lowest in ΔrelQ. It was also noted that relP deletion does not affect the basal level of mecA expression (Figure 2), but it positively affects mecA inducibility by oxacillin (~4-fold), providing an explanation for higher levels of β-lactam resistance in ΔrelP than the parent. Interestingly, deletion of both the SASs restored the mecA basal level in ΔrelPQ almost equal to the parent, but the oxacillin-induced level was lower than that of the parent (Figure 2). Comparison between ΔrelQ and ΔrelPQ revealed that the oxacillin-induced mecA level in ΔrelPQ is almost 2-fold higher than that of ΔrelQ. These results explain why the double mutant is more resistant than the ΔrelQ mutant.
Figure 2.
Expression analysis of mecA in JE2 and mutant strains. RNA was isolated from the exponentially grown (0.5–1.2 OD600nm) cultures treated with or without oxicillin (Ox) 4 μg/ml for 60 min in MHB. cDNA was prepared from RNA samples and used as template with appropriate primer sets, and relative expression levels were calculated as mentioned in the Materials and Methods section. Each bar shows the mean and standard deviation of values obtained from three replicates. The effect of mutation and oxacillin on expression of pbp2 and mecA was analyzed by performing multiple pairwise comparisons for pbp2 (uppercase letters) and mecA (lowercase letters) using ANOVA followed by Tukey's post-hoc test, and p-values < 0.05 were considered as significantly different. Different letters show statistically significant differences.
Expression of relP or relQ Complements ΔrelQ and ΔrelPQ Mutants
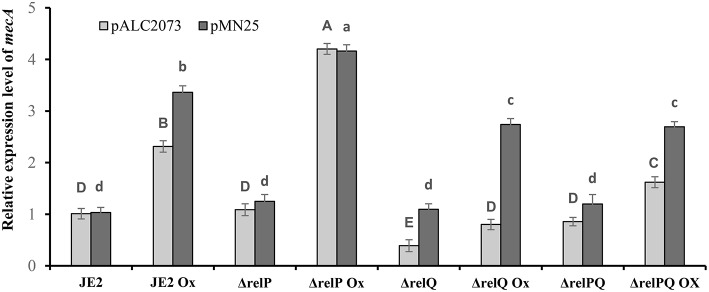
Since both relP and relQ synthesize the same alarmone (Geiger et al., 2014), we examined whether expression of either of these can complement ΔrelQ and ΔrelPQ. For this, relP and relQ were cloned with their native promoters in pLI50 to construct pMN12 and pMN13, respectively (Table 1). When these plasmids were mobilized in RN4220, transformants were found for pMN12 while no transformants could be obtained for pMN13, even after several electroporations and longer incubations. We anticipated that either plasmid-borne over-expression of relQ or SAUSA300_0906 ORF, which encodes a hypothetical protein and is located between the promoter and relQ ORF (Figure 5B), might be a reason for such toxicity. To examine further, relP promoter was fused with relQ ORF (pMN14) and relQ promoter (without SAUSA300_0906 ORF) with relP ORF (pMN15). Transformants were obtained for pMN14 (pLI50-PrelP-relQ), while none could be obtained for pMN15 (pLI50-PrelQ-relP), suggesting that relQ promoter-driven plasmid-borne over-expression of either of the SASs is toxic to the cells. MIC determination revealed that relP promoter-driven plasmid-borne expression of relP (via pMN12) or relQ (via pMN14) restored resistance in ΔrelQ and ΔrelPQ (Table 4). To validate the relQ ORF-mediated complementation, pMN25 was constructed by cloning the relQ ORF with its native RBS into pLAC2073 to express this gene in a tetracycline-inducible manner. The MIC test revealed that pMN25-borne relQ expression restored the resistance in ΔrelQ and ΔrelPQ (Table 4). For further validation, it was examined whether relQ expression could restore the mecA expression level in the ΔrelQ and ΔrelPQ strains. To examine this, mecA transcript level was compared in parent and mutants harboring pALC2073 or pMN25 and was grown in the presence or absence of oxacillin. Figure 3 clearly shows that the expression of relQ restores the mecA level in the ΔrelQ and ΔrelPQ strains almost equal to the parent. Complementation of ΔrelQ and ΔrelPQ by plasmid-borne expression of relP or relQ validated their involvement in the expression of β-lactam resistance and ruled out the possibility of secondary mutations or polar effects in ΔrelQ and ΔrelPQ strains.
Table 4.
Effect of expression of relP, relQ, and mecA on oxacillin MIC (determined by agar double-dilution method).
| Plasmids present in S. aureus strains/mutants | Oxacillin MICs (μg/ml) of S. aureus strains | |||
|---|---|---|---|---|
| JE2 | ΔrelP | ΔrelQ | ΔrelPQ | |
| pLI50 (Empty plasmid) | 128 | 512 | 8 | 32 |
| pMN12 (pLI50-PrelP-relP) | 256 | 512 | 128 | 256 |
| pMN14 (pLI50-PrelP-relQ) | 256 | 512 | 128 | 256 |
| pALC2073 (Empty plasmid) | 128 | 512 | 8 | 32 |
| pMN25 (pALC2073-relQ) | 256 | 512 | 128 | 128 |
| pMN26 (pALC2073-mecA) | 512 | 512 | 256 | 256 |
Figure 3.
Effect of relQ expression on mecA transcription in parent and mutant strains. Transcript level was monitored by RT-PCR. cDNA was prepared by reverse-transcription of the RNA samples isolated from exponentially grown cultures treated with or without oxicillin (Ox) 4 μg/ml for 60 min in MHB. The relative expression levels were calculated using the method. Each bar shows the mean and standard deviation of values obtained from three replicates. The effect of relQ expression on mecA transcription level (lowercase letters) in different strains was analyzed by performing multiple pairwise comparisons using ANOVA followed by Tukey's post-hoc test, and p-values < 0.05 were considered to represent significant difference. Tukey's post-hoc test was also performed to analyze the effect of empty plasmid on the relative transcription level of mecA (uppercase letters) in different strains. Different letters show statistically significant differences.
Expression of mecA Restores β-Lactam Resistance in ΔrelQ and ΔrelPQ
The relQ deletion-mediated oxacillin sensitivity and reduced expression/induction of mecA prompted us to examine whether plasmid-borne expression by a known promoter, which is free from SR-mediated regulation, could restore oxacillin resistance in ΔrelQ and ΔrelPQ. For this, mecA ORF with its RBS was cloned into pALC2073. Comparison of MICs in the presence of 0.2 μg/ml tetracycline revealed that the ΔrelQ and ΔrelPQ harboring pMN26 (pALC2073-relQ) became oxacillin-resistant (MIC 256 μg/ml) while the presence of pALC2073 did not make any difference (Table 4). This confirmed that relQ deletion affects oxacillin sensitivity mainly by negatively affecting the mecA expression.
relQ Promoter Is Stronger and Responds to Lack of relP and relQ
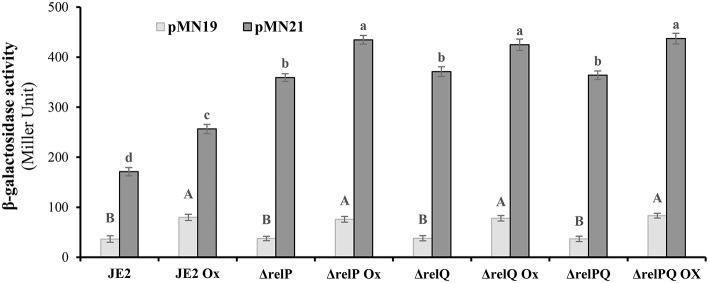
Since an induced level of this alarmone is required for β-lactam resistance (Mwangi et al., 2013), it was hypothesized that positive effects of relP deletion on β-lactam resistance might be a consequence of relQ induction in the relP-deleted background. To examine this, relP/Q promoter activities were monitored in response to their deletions and oxacillin. For this, E. coli lacZ ORF was engineered and cloned with a terminator into the XbaI-HinDIII site of pLI50 to construct reporter vector pMN18 (as described in the Materials and Methods section). Using pMN18, the upstream regions of relP (485 bp), SAUSA300_0906 (231 bp), and relQ (599 bp, including SAUSA300_0906 ORF and the 231 bp upstream region), without their RBS, were transcriptionally fused with lacZ to construct pMN19, pMN20, and pMN21, respectively. These constructs were mobilized into parent and mutant strains via S. aureus RN4220. Isolation of transformant for pMN21 (SAUSA300_0906 with its upstream (relQ) promoter region) indicated that plasmid-borne over-expression of SAUSA300_0906 was not toxic to the cells; this confirmed that relQ promoter-driven plasmid-borne over-expression of the relQ was the reason for the toxicity observed during transformation of pMN13. β-galactosidase assays revealed that the empty vector (pMN18) resulted in zero activity, and pMN20 and pMN21 resulted in equal activity (data not shown), suggesting that relQ is transcribed from the promoter located upstream of SAUSA300_0906. Results showed that, (i) relP promoter activity was equal in all the strains, and oxacillin induces it ~2-fold in every strain; (ii) relQ promoter is ~5-fold stronger than relP and induced (~1.5-fold) by oxacillin; and interestingly (iii) its activity is 2-fold higher in relP- and/or relQ- deleted backgrounds compared to that of the parent (Figure 4). These results suggest that the relP promoter responds only to oxacillin while the relQ promoter induces in response to oxacillin as well as the lack of relP and/or relQ, which enables it to express relQ efficiently in the ΔrelP strain.
Figure 4.
β-galactosidase activity of JE2 and its mutants harboring lacZ transcriptionally fused to relP (pMN19) and relQ (pMN21) promoter. β-galactosidase activity was performed in triplicate at three different occasions with cells harvested from exponentially grown (0.6–1.2 OD600nm) cultures treated with or without oxicillin (Ox) 4 μg/ml for 60 min in MHB. Data for empty plasmid (pMN18) are omitted because of its undetectable β-galactosidase activity. Data for pMN20 (SAUSA300_0906 promoter:lacZ fusion) are not included, as it gives the activity equal to the pMN21. Each bar shows a mean and standard deviation of values obtained from three replicates. The effect of mutation and oxacillin on the promoter activity of relP and relQ was analyzed by performing multiple pairwise comparisons of β-galactosidase activities in different strains due to relP (uppercase letters) or relQ (lowercase letters) promoter using ANOVA followed by Tukey's post-hoc test, and p-values < 0.05 were considered to represent significant difference. Different letters show statistically significant differences.
Identification of relP and relQ TSSs
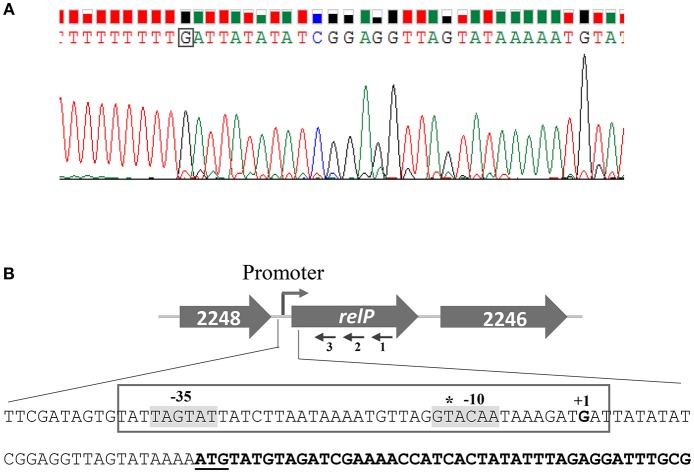
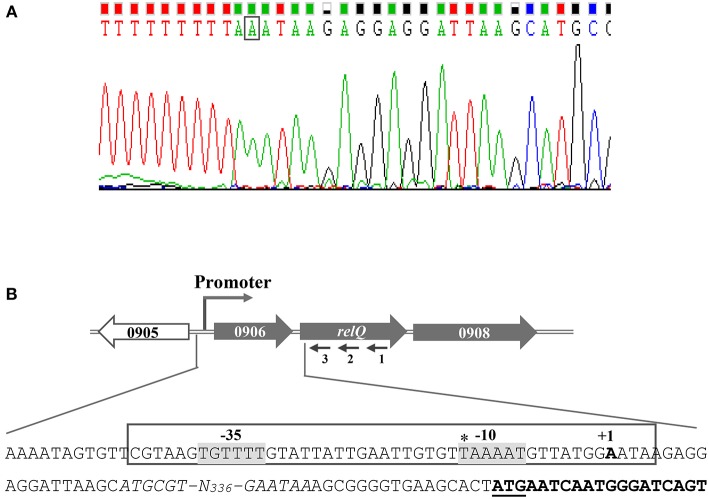
Difference in the promoter activity/inducibility of relP/Q prompted us to examine the differences in their promoter elements. Although relP/Q TSSs have been mapped and primary sigma factor (SigA) binding motifs have been predicted in their upstream regions using transcriptomic approaches (Mader et al., 2016), the mapped TSSs have been shown to have an upshift tendency, which creates problems in identification of actual TSSs and promoter motifs. 5′ RACE was performed to identify the actual TSSs, which allowed the identification of their possible −35 and −10 elements. The identified TSSs are 27 and 19 bp upstream of relP and SAUSA300_0906 start codons, respectively (Figures 5, 6). These are 13 and 14 bp downstream of the earlier reported TSSs of relP and relQ, respectively. Sequence analysis revealed that relP had TAGTAT (−35) and GTACAA (−10), and relQ had TGTTTT (−35) and TAAAAT (−10) promoter elements. This indicates a significant difference in their −35 and −10 elements that might be the reason for difference in their promoter activities.
Figure 5.
Determination of the relP transcription start site (TSS) by 5′ RACE. (A) Electropherogram, showing TSS (boxed nucleotide), is representative of results from sequencing of several distinct clones obtained after 5′ RACE experiments. (B) Schematic representation of S. aureus relP chromosomal region. Large and filled arrows represent the relative size, location, and transcriptional orientation of ORFs. Small and thin arrows represent the regions used to design primers for 5′RACE experiment; relP-SP1:R (1), relP-SP2:R (2), and relP-SP3:R (3). Nucleotide sequences from −78 to +44 of relP start codon (underlined) showing TSS (indicated as +1) and possible −35 and −10 elements (gray background). relP ORF is indicated by bold nucleotides. The earlier predicted TSS is shown by asterisk.
Figure 6.
Determination of the transcription start site (TSS) of relQ by 5′ RACE. (A) Electropherogram showing TSS (boxed nucleotide) is representative of results from sequencing of several distinct clones obtained after 5′ RACE experiment. (B) Schematic representation of S. aureus relQ chromosomal region. Large and filled arrows represent the relative size, location, and transcriptional orientation of ORFs. Small and thin arrows represent the regions used to design primers for the 5′RACE experiment; relQ-SP1:R (1), relQ-SP2:R (2), and relQ-SP3:R (3). Nucleotide sequences from −436 to +19 of relQ start codon (underlined) showing TSS (indicated as +1) and possible −35 and −10 elements (gray background). SAUSA300_0906 ORFs are shown by italicized nucleotides, and the portion of relQ ORF is shown in bold. The earlier predicted TSS is shown with an asterisk.
Discussion
Several studies have shown that SR, which is established by an increased level of (p)ppGpp, is involved in bacterial stress adaptation, drug resistance, pathogenesis and persistence. Recently, involvement of (p)ppGpp in the expression of β-lactam resistance was reported by showing that laboratory or clinical hetero-resistant MRSA strains required an induced level of (p)ppGpp (either by exposure to mupirocin or truncation of RSH) for the expression of homogeneous and increased levels of oxacillin resistance (Kim et al., 2013; Mwangi et al., 2013). Because every MRSA isolate does not carry a truncated RSH or need exposure of mupirocin-like SR-inducers, how (p)ppGpp synthesis is induced in response to β-lactams in MRSA is still not clear. Since exposure of β-lactams is not supposed to pose any nutrient starvation, direct involvement of RSH in β-lactam-induced (p)ppGpp synthesis does not seem logical. With these considerations, we initiated this study based on a hypothesis that RelP/Q might be involved in (p)ppGpp synthesis in response to β-lactams because these SASs are active (p)ppGpp synthetases and induced in response to cell-wall stresses in S. aureus (Geiger et al., 2014).
Although recent reports show that RelP/Q are induced in response to cell-wall stresses to mitigate such conditions in MSSA (Geiger et al., 2014), their contribution to the expression of β-lactam resistance in MRSA is still unclear. To examine their roles, we deleted relP and relQ in a CA-MRSA and kept RSH intact to emphasize their function. Since the (p)ppGpp synthetic activity of these SASs has been reported in vivo and in vitro (Geiger et al., 2014), we did not feel the need to measure the (p)ppGpp level in the SAS-deleted strains, though lack of (p)ppGpp estimation facility was another reason. Our observation that deletion of relQ renders CA-MRSA sensitive to several β-lactams suggests an important role of RelQ in the expression of β-lactam resistance. This finding corroborates an earlier observation that a laboratory-generated highly oxacillin-resistant S. aureus strain reverted to a reduced-resistance strain due to a point mutation in the synthetase domain of RelQ (Mwangi et al., 2013). Since RelQ synthesizes (p)ppGpp, it was examined whether mupirocin-induced (p)ppGpp synthesis could bypass the requirement of RelQ for β-lactam resistance. The results showed that mupirocin fully restored the resistance in ΔrelQ, suggesting that relQ deletion increases the sensitivity mainly by negatively affecting the (p)ppGpp level.
Our observations that relQ deletion reduces the basal level expression and oxacillin inducibility of mecA provides an explanation for β-lactam sensitivity of ΔrelQ. Complementation of ΔrelQ by a xyl/tetO promoter-driven mecA expression further validates that relQ deletion increases sensitivity by reducing mecA expression. Interestingly, we observed that deletion of both the SASs renders ΔrelPQ more resistant than ΔrelQ by restoring the mecA expression level almost equal to the parent. Since requirement of (p)ppGpp for mecA expression has been shown (Kim et al., 2013; Mwangi et al., 2013), it seems logical to hypothesize that the reduced sensitivity of ΔrelPQ might be a consequence of RSH-mediated synthesis of (p)ppGpp to maintain the basal level of this alarmone in the absence of both the SASs. Our data also show that although the basal level of mecA is restored in ΔrelPQ, its oxacillin-induced level was lower than the parent, indicating the importance of RelP/Q for efficient induction of mecA in response to β-lactams.
Lack of polar effects or secondary mutations is supported by our genetic complementation data, which shows that xyl/tetO promoter-driven expression of relQ fully complements the mutant. It was also observed that multi-copy plasmid-borne relP promoter-driven expression of relP or relQ ORF complements the mutants while relQ promoter-driven plasmid-borne expression of either of these results in lethality. It appears that relP promoter activity is insufficient for the expression of relP to compensate the effect of relQ deletion when it is present as a single copy on the ΔrelQ chromosome, but its presence on a multi-copy plasmid amplified the promoter activity and complemented the mutant. Based on promoter activity data, which showed that relQ promoter is >5-fold stronger than the relP, it can be inferred that the toxicity associated with relQ promoter-driven plasmid-borne expression of either of the SASs might be a consequence of a high level of (p)ppGpp due to further amplification of the relQ promoter activity by the plasmid copy-number. However, relQ promoter-driven expression of relQ ORF from a single chromosomal copy is not toxic to S. aureus.
In addition to revealing the importance of relQ, the results also indicate a positive effect of relP deletion on β-lactam resistance. Expression analysis provides an explanation by showing that relP deletion increases the oxacillin inducibility of mecA. Our data shows that relQ promoter is induced in response to oxacillin as well as deletion of either of the SASs while the relP promoter responds only to oxacillin. These observations suggest that relQ is efficiently expressed in the relP-deleted background, which enhances the mecA expression and renders the ΔrelP strain highly resistant. Inducibility of relQ promoter in relP- and/or relQ-deleted backgrounds indicates toward its ability to induce in response to low (p)ppGpp levels. Although SigA binding motifs have been predicted in relP/Q upstream regions, their −35 and −10 elements have not been identified because the mapped TSSs have an upshift tendency (Mader et al., 2016). By promoter mapping, we have identified the TSSs and the most probable −35 and −10 elements. The results revealed that the identified −35 elements of relP (TAGTAT) and relQ (TGTTTT) showed similarity with the reported SigA −35 (TGATAA and TTTATT) consensus elements (Deora and Misra, 1996), which supports their SigA-dependence predicted earlier (Geiger et al., 2014; Mader et al., 2016). However, differences in their −35 and −10 elements provide an insight into difference in their promoter activities.
Recently, it was shown that although RelQ deletion does not affect the level of β-lactam resistance in N315 and Mu3, its overexpression increases the resistance level several-fold (Matsuo et al., 2019). These studies suggest that RelQ plays an important role in the expression of β-lactam resistance, but the requirement of its expression level may depend on the strains. Based on our findings, we propose that possibly a certain level of (p)ppGpp is required for the expression of mecA in JE2, and both the SASs are involved in maintaining this level, which might be affected by deletion of either of these. However, an enhanced oxacillin-inducibility of relQ in the ΔrelP compensates for the effect of relP deletion, while the effect of relQ deletion cannot be compensated due to the lack of relP inducibility in response to relQ deletion. This difference in regulation of relP and relQ results in sensitivity in ΔrelQ and resistance in ΔrelP. Our findings show that RelQ mediates the expression of mecA in response to β-lactams in MRSA, but how these SASs are regulated differently is a matter of further research.
Author Contributions
AB performed most of the experiments and compiled the data. PP, AD, and AZ helped in promoter mapping and promoter activity analysis. GN helped in data analysis and discussion. MM designed the study and experiments, interpreted the results, and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by DST-INSPIRE Faculty awarded (IFA13-LSBM-94) to MM. The fellowship to AB by DST, New Delhi, is gratefully acknowledged. The authors are thankful to Dr. John Gustafson, BMB, NRC, OSU, Stillwater, OK, USA for providing the strains and facility to construct mutants in his Lab. We are thankful to Dr. Ambrose L. Cheung, Dartmouth Medical School, Hanover, NH, USA for providing pALC2073 plasmid. We thank Dr. Sanjiv Kumar, IISR, Lucknow, for carefully reading and improving the English, and Dr. Ashutosh Awasthi, Department of Microbial Technology, CSIR- CIMAP, Lucknow, for his help in statistical analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00339/full#supplementary-material
References
- Abranches J., Martinez A. R., Kajfasz J. K., Chávez V., Garsin D. A., Lemos J. A. (2009). The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J. Bacteriol. 191, 2248–2256. 10.1128/JB.01726-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae T., Schneewind O. (2006). Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55, 58–63. 10.1016/j.plasmid.2005.05.005 [DOI] [PubMed] [Google Scholar]
- Bateman B. T., Donegan N. P., Jarry T. M., Palma M., Cheung A. L. (2001). Evaluation of a tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect. Immun. 69, 7851–7857. 10.1128/IAI.69.12.7851-7857.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A. W., Perry D. M., Kirby W. N. M. (1959). Single disc antibiotic sensitivity testing of staphylococci. Arch. Int. Med. 104, 208–216. 10.1001/archinte.1959.00270080034004 [DOI] [PubMed] [Google Scholar]
- Boyle-Vavra S., Yin S., Challapalli M., Daum R. S. (2003). Transcriptional induction of the penicillin-binding protein 2 gene in Staphylococcus aureus by cell wall-active antibiotics oxacillin and vancomycin. Antimicrob. Agents Chemother. 47, 1028–1036. 10.1128/AAC.47.3.1028-1036.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassels R., Oliva B., Knowles D. (1995). Occurrence of the regulatory nucleotides ppGpp and pppGpp following induction of the stringent response in staphylococci. J. Bacteriol. 177, 5161–5165. 10.1128/jb.177.17.5161-5165.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers H. F., Hackbarth C. J. (1987). Effect of NaCl and nafcillin on penicillin-binding protein 2a and heterogeneous expression of methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 31, 1982–1988. 10.1128/AAC.31.12.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deora R., Misra T. K. (1996). Characterization of the primary σ factor of Staphylococcus aureus. J. Bio. Chem. 271, 21828–21834. 10.1074/jbc.271.36.21828 [DOI] [PubMed] [Google Scholar]
- Eymann C., Homuth G., Scharf C., Hecker M. (2002). Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J. Bacteriol. 184, 2500–2520. 10.1128/JB.184.9.2500-2520.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey P. D., Endres J. L., Yajjala V. K., Widhelm T. J., Boissy R. J., Bose J. L., et al. (2012). A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537–e00512. 10.1128/mBio.00537-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuda C., Suvorov M., Vakulenko S. B., Mobashery S. (2004). The basis for resistance to β-lactam antibiotics by penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. J. Bio. Chem. 279, 40802–40806. 10.1074/jbc.M403589200 [DOI] [PubMed] [Google Scholar]
- Geiger T., Goerke C., Fritz M., Schafer T., Ohlsen K., Liebeke M., et al. (2010). Role of the (p)ppGpp synthetase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus. Infect. Immun. 78, 1873–1883. 10.1128/IAI.01439-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T., Kästle B., Gratani F. L., Goerke C., Wolz C. (2014). Two small (p)ppGpp synthetases in Staphylococcus aureus mediate tolerance against cell envelope stress conditions. J. Bacteriol. 196, 894–902. 10.1128/JB.01201-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C., Mwangi M., Chung M., Milheirço C., Lencastre H., Tomasz A. (2013). The mechanism of heterogeneous β-lactam resistance in MRSA: key role of the stringent stress response. PLoS ONE 8:e82814. 10.1371/journal.pone.0082814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiswirth B. N., Löfdahl S., Betley M. J., O'Reilly M., Schlievert P. M., Bergdoll M. S., et al. (1983). The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305, 709–712. [DOI] [PubMed] [Google Scholar]
- Lee C. Y., Buranen S. L., Ye Z. H. (1991). Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103, 101–105. 10.1016/0378-1119(91)90399-V [DOI] [PubMed] [Google Scholar]
- Lemos J. A., Lin V. K., Nascimento M. M., Abranches J., Burne R. A. (2007). Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol. Microbiol. 65, 1568–1581. 10.1111/j.1365-2958.2007.05897.x [DOI] [PubMed] [Google Scholar]
- Llarrull L. I., Fisher J. F., Mobashery S. (2009). Molecular basis and phenotype of methicillin resistance in Staphylococcus aureus and insights into new β-lactams that meet the challenge. Antimicrob. Agents Chemother. 53, 4051–4063. 10.1128/AAC.00084-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader U., Nicolas P., Depke M., Pané-Farré J., Debarbouille M., van der Kooi-Pol M. M., et al. (2016). Staphylococcus aureus transcriptome architecture: from laboratory to infection-mimicking conditions. PLoS Genet. 12:e1005962. 10.1371/journal.pgen.1005962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo M., Yamamoto N., Hishinuma T., Hiramatsu K. (2019). Identification of a novel gene associated with high-level β-lactam resistance in heterogeneous vancomycin-intermediate Staphylococcus aureus strain Mu3 and methicillin-resistant S. aureus strain N315. Antimicrob. Agents Chemother. 10.1128/AAC.00712-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechold U., Murphy H., Brown L., Cashel M. (2002). Intramolecular regulation of the opposing (p)ppGpp catalytic activities of RelSeq, the Rel/Spo enzyme from Streptococcus equisimilis. J. Bacteriol. 184, 2878–2888. 10.1128/JB.184.11.2878-2888.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 352–355. [Google Scholar]
- Morales V. M., Backman A., Bagdasarian M. (1991). A series of wide-host-range low copy-number vectors that allow direct screening for recombinants. Gene 97, 39–47. [DOI] [PubMed] [Google Scholar]
- Mwangi M. M., Kim C., Chung M., Tsai J., Vijayadamodar G., Benitez M., et al. (2013). Whole-genome sequencing reveals a link between beta-lactam resistance and synthetases of the alarmone (p)ppGpp in Staphylococcus aureus. Microbial Drug Resist. 19, 153–159. 10.1089/mdr.2013.0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanamiya H., Kasai K., Nozawa A., Yun C. S., Narisawa T., Murakami K., et al. (2008). Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol. Microbiol. 67, 291–304. 10.1111/j.1365-2958.2007.06018.x [DOI] [PubMed] [Google Scholar]
- Pando J. M., Pfeltz R. F., Cuaron J. A., Nagarajan V., Mishra M. N., Torres N. J., et al. (2017). Ethanol-induced stress response of Staphylococcus aureus. Can J. Microbiol. 63, 1–13. 10.1139/cjm-2017-0221 [DOI] [PubMed] [Google Scholar]
- Peschke U., Beuck V., Bujard H., Gentz R., Le Grice S. (1985). Efficient utilization of Escherichia coli transcriptional signals in Bacillus subtilis. J. Mol. Biol. 186, 175–182. 10.1016/0022-2836(85)90129-9 [DOI] [PubMed] [Google Scholar]
- Pinho M. G., de Lencastre H., Tomasz A. (2001). An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc. Natl. Acad. Sci. U.S.A. 98, 10886–10891. 10.1073/pnas.191260798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R. G., Walker D. C., Mclnnes R. R. (1993). E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Res. 21, 1677–1678. 10.1093/nar/21.7.1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner V., Dervyn E., Ehrlich S. D. (1998). A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144, 3097–3104. 10.1099/00221287-144-11-3097 [DOI] [PubMed] [Google Scholar]
- Wiegand I., Hilpert K., Hancock R. E. W. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protocol. 2, 163–175. 10.1038/nprot.2007.521 [DOI] [PubMed] [Google Scholar]
- Williamson R., Tomasz A. (1985). Inhibition of cell wall synthesis and acylation of the penicillin binding proteins during prolonged exposure of growing Streptococcus pneumoniae to benzylpenicillin. Ear. J. Biochem. 151, 475–483. 10.1111/j.1432-1033.1985.tb09126.x [DOI] [PubMed] [Google Scholar]
- Xiao H., Kalman M., Ikehara K., Zemel S., Glaser G., Cashel M. (1991). Residual guanosine 3′, 5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266, 5980–5990. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.