Abstract
Genetic studies have revealed a critical role of Distal-homeobox (Dlx) genes in bone formation, and our previous study showed that Dlx2 overexpressing in neural crest cells leads to profound abnormalities of the craniofacial tissues. The aim of this study was to investigate the role and the underlying molecular mechanisms of Dlx2 in osteogenic differentiation of mouse bone marrow stromal cells (BMSCs) and pre-osteoblast MC3T3-E1 cells. Initially, we observed upregulation of Dlx2 during the early osteogenesis in BMSCs and MC3T3-E1 cells. Moreover, Dlx2 overexpression enhanced alkaline phosphatase (ALP) activity and extracellular matrix mineralization in BMSCs and MC3T3-E1 cell line. In addition, micro-CT of implanted tissues in nude mice confirmed that Dlx2 overexpression in BMSCs promoted bone formation in vivo. Unexpectedly, Dlx2 overexpression had little impact on the expression level of the pivotal osteogenic transcription factors Runx2, Dlx5, Msx2, and Osterix, but led to upregulation of Alp and Osteocalcin (OCN), both of which play critical roles in promoting osteoblast maturation. Importantly, luciferase analysis showed that Dlx2 overexpression stimulated both OCN and Alp promoter activity. Through chromatin-immunoprecipitation assay and site-directed mutagenesis analysis, we provide molecular evidence that Dlx2 transactivates OCN and Alp expression by directly binding to the Dlx2-response cis-acting elements in the promoter of the two genes. Based on these findings, we demonstrate that Dlx2 overexpression enhances osteogenic differentiation in vitro and accelerates bone formation in vivo via direct upregulation of the OCN and Alp gene, suggesting that Dlx2 plays a crucial role in osteogenic differentiation and bone formation.
Subject terms: Mechanisms of disease, Bone development, Mechanisms of disease
Osteogenesis: Dlx2 gene plays key role in bone development
The distal-less homeobox (Dlx) gene family is related to various features of bone development, and the Dlx2 member of that family has been found to play a crucial role in bone formation. A team headed by Steve Guofang Shen at the Shanghai Jiao Tong University School of Medicine in China investigated the function of Dlx2 in osteogenic (bone development) differentiation of mouse bone marrow stromal cells (BMSCs) and MC3T3-E1 cells (precursors of osteoblasts, the major cellular component of bone). The team found that overexpression of Dlx2 promotes osteogenic differentiation in vitro and accelerates bone formation in vivo by enhancing Osteocalcin and Alp genes (both of which play critical roles in promoting osteoblast maturation). The authors conclude that their results suggest a promising future strategy for treating bone defects where BMSCs overexpress Dlx2.
Introduction
The distal-less homeobox (Dlx) gene family consists of six members (Dlx1, Dlx2, Dlx3, Dlx5, Dlx6, and Dlx7); these members are expressed in a complex pattern in the first and second branchial arch region.1 Dlx1/2 regulates the development of the upper jaw, while Dlx5/6 regulates lower jaw development.2 Together with other homeobox proteins, the Dlx family regulates osteoblast differentiation. As one of the key transcription factors regulating osteogenic differentiation, Dlx5 stimulates two other key transcription factors, Runx2 and Osterix (Osx), which sequentially induce expression of bone markers such as Osteocalcin (OCN) and Alkaline Phosphatase (ALP). The expression of Dlx5 is induced by bone morphogenetic protein-2 (BMP-2).3 Msx2, another homeobox gene and a key regulator of osteogenic differentiation, represses the expression of Alp by directly binding to its promoter, while Dlx5 activates its expression by interfering with the ability of Msx2.4 Thus, Dlx5 coordinates with Msx2 to regulate osteogenic differentiation due to their reciprocal ability to compete with each other.
Sharing strong sequence similarity with Dlx5, Dlx2 has been shown to play a crucial role in craniofacial skeletal development.5 Dlx2 is upregulated in the central area of the first branchial arch during days 9.5 and 10.5 of embryonic development in mice. This upregulation of Dlx2 is important for the differentiation and development of the primordium, as it leads to the development of the maxillofacial skeletal pattern.6 Given that Dlx5 controls osteogenic differentiation,7 it is reasonable to speculate that Dlx2 might be involved in this process. So far, only a few studies have reported that Dlx2 overexpression increases the osteogenic differentiation potential of pre-osteoblast cells.8 However, how Dlx2 regulates osteogenic differentiation and the underlying cellular and molecular mechanisms remain unknown.
In a previous study, we found that elevated Dlx2 expression led to midfacial development defects, nasal deformities, premaxillary bony deficiency, and spine deformities.9 Thus, it is crucial to examine how Dlx2 overexpression leads to abnormal bone formation both in vitro and in vivo. To investigate the role of Dlx2 during osteogenic differentiation both in vitro and in vivo, we used mouse bone marrow stromal cells (BMSCs) in our study, as the ability of BMSCs to differentiate toward adipogenic, chondrogenic, and osteogenic cell lineages has been characterized extensively in vivo and in vitro by various researchers.10 Osteogenic differentiation of BMSCs can be assayed in vitro by ALP and Alizarin red staining and in vivo by transplantation assays.11,12 Therefore, mouse BMSCs are suitable for investigating the effect of Dlx2 overexpression on osteogenesis both in vitro and in vivo. Murine osteoblastic cell line MC3T3-E1 cells were also chosen to verify the effect of Dlx2 overexpression on osteogenesis in vitro.
Initially, we observed the upregulation of Dlx2 in both mouse BMSCs and MC3T3-E1 cells during osteogenic differentiation. Moreover, forced overexpression of Dlx2 led to enhanced osteogenic differentiation potential of both BMSCs and MC3T3-E1 cells in vitro, and accelerated bone formation in vivo. These findings prompted us to explore the underlying mechanisms. To our surprise, we found that Dlx2 overexpression had no significant effect on the expression levels of Dlx5, Msx2, Runx2, and Osx, but led to upregulation of Alp and OCN in BMSCs and MC3T3-E1 cells. Considering the fact that Alp promotes the early stage of osteogenic differentiation and OCN accelerates the late stage, we next analyzed the promoter of OCN and Alp through luciferase-reporter assay and chromatin-immunoprecipitation (ChIP) analysis, and found that Dlx2 transcriptionally regulated OCN and Alp expression by directly binding to their promoters. Taken together, our data demonstrates for the first time that Dlx2 overexpression enhances the early stage of osteogenic differentiation via direct upregulation of Alp, and promotes the late stage of osteogenic differentiation via direct upregulation of OCN.
Results
Endogenous Dlx2 expression in BMSCs and MC3T3-E1 cells during osteogenesis
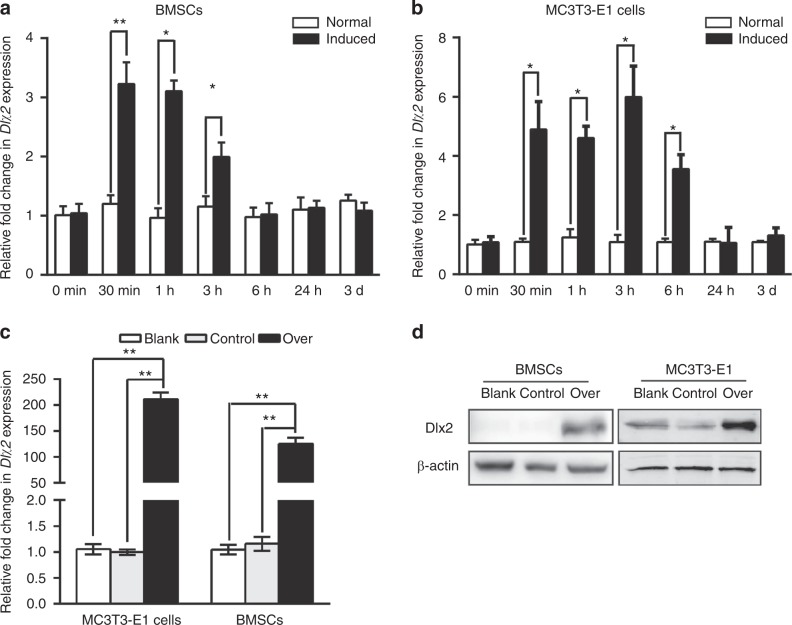
First, we examined the levels of Dlx2 expression upon osteogenic induction in mouse BMSCs and MC3T3-E1 cells. Quantitative reverse transcription polymerase chain reaction (RT-qPCR) results showed that when BMSCs were exposed to osteogenic-inducing medium (OIM), Dlx2 expression was upregulated within 0.5 and 3 h after induction (Fig. 1a). However, after 7- or 14-day culture in OIM, these cells express similar mRNA level of Dlx2 with the cells cultured in normal culture medium (data not shown). In addition, western blot analysis with an anti-Dlx2 antibody detected only a very weak signal of Dlx2 protein in BMSCs cultured both in normal medium and OIM for 3 h. This could be explained by the low protein level of endogenous Dlx2 in BMSCs. Supporting this notion is the finding that the endogenous protein level of other Dlx proteins, such as Dlx5, is also quite low in BMSCs.13
Fig. 1.
Analysis of Dlx2 expression by RT-qPCR in BMSCs and MC3T3-E1 cells upon osteogenic induction. Endogenous expression of Dlx2 in BMSCs (a) and MC3T3-E1 cells (b) at different time points upon osteogenic induction. Relative transcript levels of Dlx2 at each time point were quantified by RT-qPCR and normalized with a house-keeping gene Gapdh. Student’s t tests were used to determine statistical significance; n = 3. Error bars represent SDs. *P < 0.05; **P < 0.01. c Dlx2 expression in BMSCs and MC3T3-E1 cells was evaluated with RT-qPCR. Gene expression was normalized with Gapdh, and statistical significance was determined as described in a. d Protein levels of Dlx2 in BMSCs and MC3T3-E1 cells were measured by Western blot analysis. Blank, BMSCs/MC3T3-E1 cells; control, Lenti-CTRL transduced BMSCs/MC3T3-E1 cells; over, Lenti-DLX2 OE transduced BMSCs/MC3T3-E1 cells. β-Actin was used as an internal control
The expression pattern of Dlx2 in MC3T3-E1 cells was similar with that in BMSCs. The mRNA level of Dlx2 in MC3T3-E1 cells was upregulated within and the first 6 h after osteogenic induction (Fig. 1b), but showed no differences with that in the cells cultured in normal culture medium after culture for 7 or 14 days (data not shown). Consistently, previous study in stem cells from apical papilla (SCAP) also showed a similar expression pattern that Dlx2 was upregulated within the first few hours upon osteogenic induction, and its expression then showed no significant difference with that in cells grown in normal cell culture medium. This reduction of Dlx2 expression in the induced group could be explained by the regulation of mRNA stability by microRNAs (miRNAs). Latest study reveals that Dlx2 is a target gene of the miRNA, miR-185-5p, and its expression is negatively regulated by Dlx2.14 The initial upregulation of Dlx2 may stimulate the expression of downstream target genes, which induces miRNA expression and sequentially exert a feedback on Dlx2 expression. In addition, western blot analysis detects only weak expression of Dlx2 in MC3T3-E1 cells treated either with OIM or normal culture medium, consistent with the endogenous protein level of Dlx2 in BMSCs. Together, these results demonstrate that Dlx2 was upregulated during early osteogenesis in mouse BMSCs and MC3T3-E1 cells.
Forced overexpression of Dlx2 in BMSCs and MC3T3-E1 cells
Next, to investigate the effect of Dlx2 on osteogenesis, we established Dlx2-overexpressing BMSCs and MC3T3-E1 cells. Cultured BMSCs or MC3T3-E1 cells were transduced with Lenti-DLX2 OE lentivirus to allow stable Dlx2 overexpression. As a control, parallel BMSCs or MC3T3-E1 cells were transduced with Lenti-CTRL lentivirus as mock control. Dlx2 expression was then evaluated by RT-qPCR and western blot analysis. Lenti-DLX2 OE-transduced BMSCs (over) displayed much more Dlx2 transcripts (Fig. 1c) and correspondingly increased protein production (Fig. 1d) than wildtype or Lenti-CTRL-transduced BMSCs (control) did. Similarly, Lenti-DLX2 OE transduction in MC3T3-E1 cells also led to increased mRNA and protein levels of Dlx2 (Fig. 1c, d). These observations indicate that Dlx2 was successfully overexpressed in both cell lines.
Dlx2 overexpression enhances the osteogenic differentiation potential of BMSCs and MC3T3-E1 cells
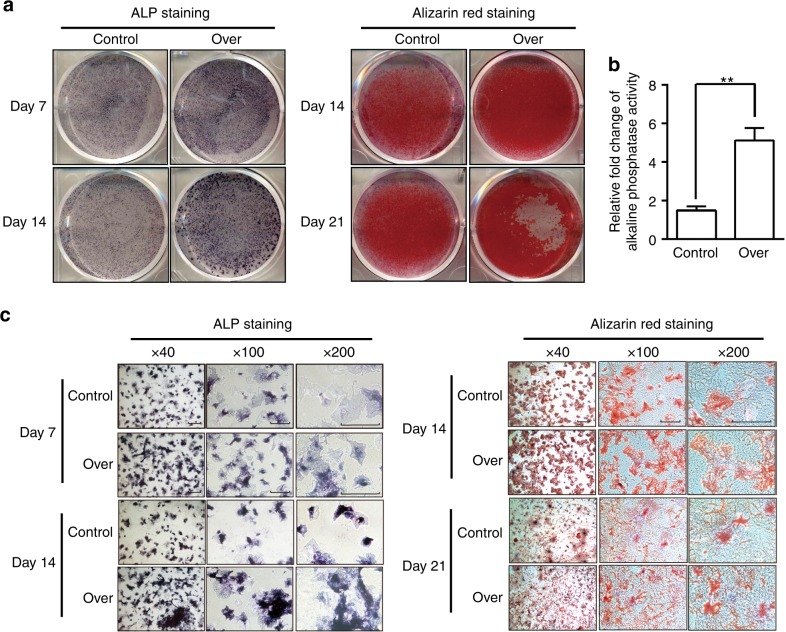
To investigate whether the overexpression of Dlx2 effects on osteogenic differentiation in vitro, we carried out ALP staining and Alizarin staining assays. Lentivirus-transduced BMSCs or MC3T3-E1 cells were cultured in OIM for 7, 14, or 21 days to assess their osteogenic differentiation potential.14 Interestingly, ALP staining was significantly enhanced at day 7 and 14 in the Dlx2-overexpressing BMSCs compared with that in control BMSCs (Fig. 2a, c). Similarly, higher ALP activity levels were observed in Dlx2-overexpressing BMSCs at day 14 after osteogenic induction (Fig. 2b). Furthermore, Alizarin red staining revealed that mineralization was markedly enhanced in Dlx2-overexpressing BMSCs during the entire culture period, especially at day 21 (Fig. 2a, c). Consistently, we also observed enhanced ALP and Alizarin red staining in Dlx2-overexpressing MC3T3-E1 cells (Fig. S1). Given that ALP activity is involved in the early stage of osteogenic differentiation while mineralization is involved in the late stage, we therefore proposed that Dlx2 overexpression accelerates the early stage of osteogenesis by increasing ALP activity and the late stage of osteogenesis by enhancing mineralization.
Fig. 2.
Forced overexpression of Dlx2 enhanced osteogenesis of BMSCs in vitro. a ALP staining was performed on days 7 or 14 after osteogenic induction. Alizarin red staining was carried out after cells were cultured in OIM for 14 or 21 day. b Semi-quantitative analysis of ALP activity in Dlx2-overexpressing BMSCs (over) and control BMSCs (control) after 14-day culture in OIM. c Magnified views of ALP staining and Alizarin red staining in a. Scale bar = 50 μm in all the panels. Left panel, 40-fold magnified image; middle panel, 100-fold magnified image; right panel, 200-fold magnified image
Forced overexpression of Dlx2 in BMSCs accelerated bone formation in vivo
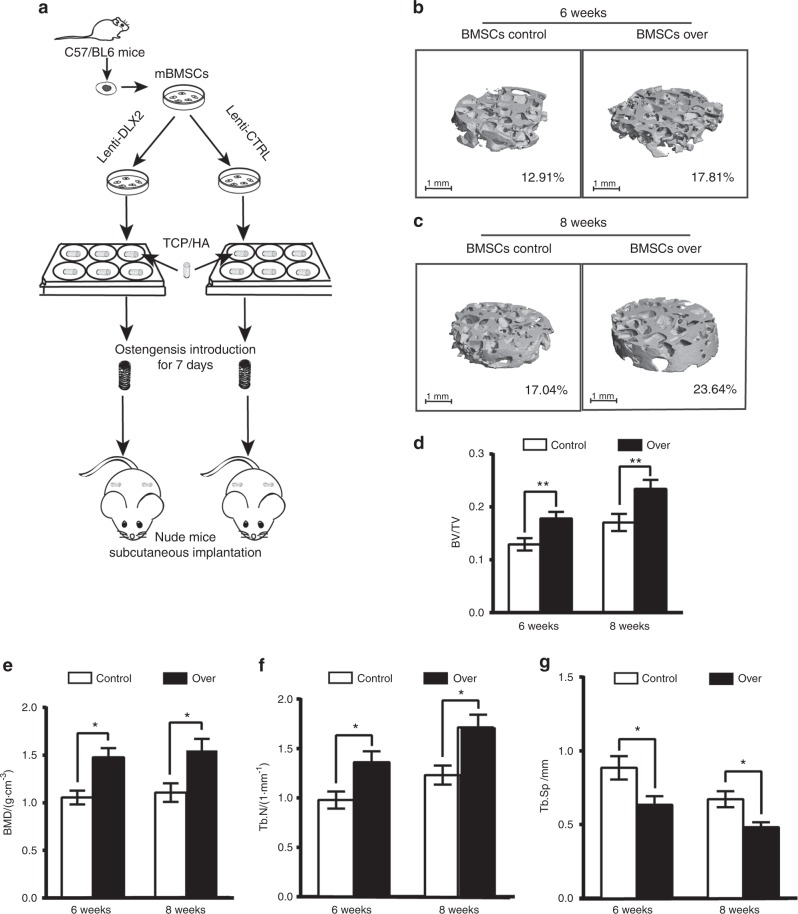
To investigate whether Dlx2 overexpression could affect osteogenesis in vivo, we subcutaneously implanted the BMSCs/β-tricalcium phosphate (β-TCP) constructs into nude mice. The whole implanted tissues were then analyzed with microscopic computed tomography (micro-CT) examination 6- or 8-weeks after implantation (Fig. 3a). Micro-CT revealed new bone formation in both control and Dlx2 overexpression groups (Fig. 3b, c). However, quantitative morphometric analysis showed that bone volume/tissue volume (BV/TV) in the Dlx2-overexpressing group (17.81% ± 1.23% and 23.64% ± 1.71% at week 6 and week 8, respectively) was significantly higher than that in the control group (12.91% ± 1.16% and 17.04% ± 1.62% at week 6 and week 8, respectively) (P < 0.05) (Fig. 3d). Similarly, the Dlx2-overexpressing group showed higher bone mineral density (BMD) of the newly formed bone ((1.477 ± 0.097) g· cm−3 and (1.550 ± 0.121) g· cm−3 at week 6 and week 8, respectively) than the control group ((1.055 ± 0.072) g· cm−3 and (1.107 ± 0.098) g· cm−3 at week 6 and week 8, respectively) (Fig. 3e). Besides, trabecular number (Tb.N) in the Dlx2-overexpressing group ((1.362 ± 0.110) g· cm−3 and (1.713 ± 0.129) g· cm−3 at week 6 and 8, respectively) was significantly higher than that in the control group ((0.979 ± 0.086) g· cm−3 and (1.232 ± 0.097) g· cm−3 at week 6 and 8, respectively) (Fig. 3f). Additionally, the Dlx2-overexpressing group exhibited decreased trabecular space (Tb.Sp) ((0.634 ± 0.059) mm and (0.482 ± 0.033) mm at week 6 and 8, respectively) when compared to the control group ((0.885 ± 0.079) mm and (0.672 ± 0.044) mm at week 6 and 8, respectively) (Fig. 3g). Taken together, these results demonstrate that overexpression of Dlx2 in BMSCs substantially triggers osteogenic differentiation and improves bone formation in vivo, defining a pivotal role of Dlx2 in osteogenic differentiation.
Fig. 3.
Dlx2-overexpressing BMSCs accelerated bone formation in vivo. a Schematic diagram of implantation experiments. The whole BMSCs/β-TCP constructs were obtained for micro-CT analysis 6 or 8 weeks after implantation. b, c Representative 3D reconstructed micro-CT results of the BMSCs/β-TCP constructs at weeks 6 and 8 after subcutaneous implantation. Implants were then harvested after 6 (b) or 8 (c) weeks, and were scanned by micro-CT. Scale bar = 1 μm. Average BV/TV is indicated below. Over, BMSCs transduced with Lenti-DLX2 OE; control, BMSCs transduced with Lenti-CTRL. d–g Analysis of the bone volume/tissue volume (BV/TV), bone mineral density (BMD), trabecular number (Tb.N), and trabecular spacing (Tb.Sp) in the respective groups. Statistical significance was determined as described in Fig. 1; n = 6, *P < 0.05, **P < 0.01
Effect of forced overexpression of Dlx2 on the expression of osteogenic genes in BMSCs
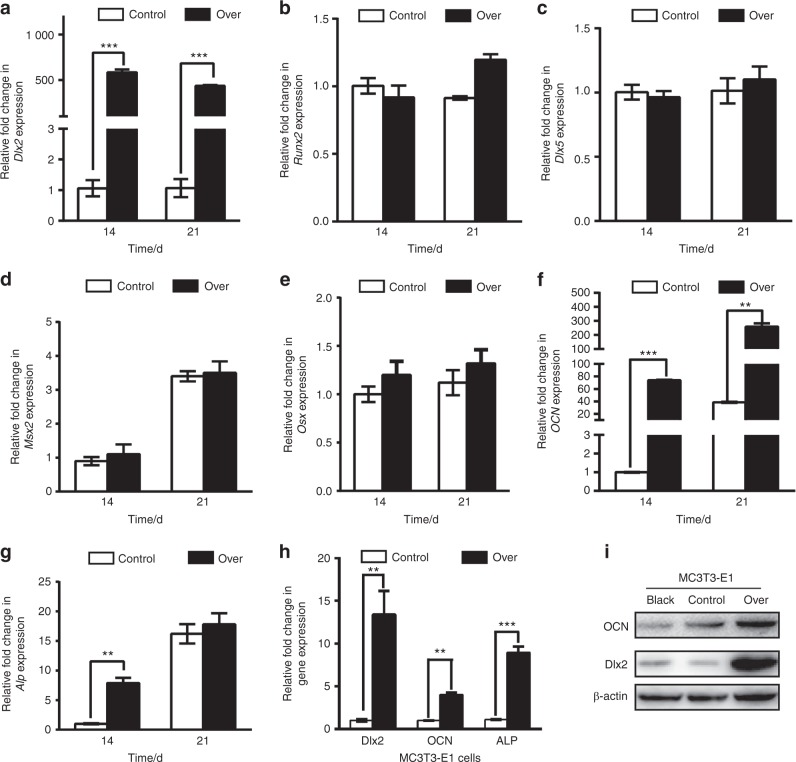
To gain an insight into the mechanism of Dlx2-induced osteogenesis, we examined the expression of a panel of osteogenesis-associated genes by RT-qPCR in Dlx2-overexpressing BMSCs and control BMSCs cultured in OIM. As shown in Fig. 4a, RT-qPCR analysis revealed that transduction of Lenti-DLX2 OE into BMSCs resulted in a 526.2-fold and 481.3-fold increase in Dlx2 expression at days 14 and 21 of osteogenic differentiation, respectively. Unexpectedly, there was no difference in Runx2, Dlx5, Msx2, and Osx expression between Dlx2-overexpressed BMSCs and mock-transduced BMSCs at 14 and 21 days after osteogenic induction (Fig. 4b–e) nor at 1, 2, and 7 days after osteogenic induction (data not shown), indicating that these genes might not be involved in the Dlx2-induced promotion of osteogenesis. Importantly, forced overexpression of Dlx2 led to upregulation of OCN at days 14 and 21 after osteogenic induction and upregulation of Alp at day 14 (Fig. 4f, g), consistent with the ALP staining and Alizarin red staining results (Fig. 2a–c). In summary, we found, for the first time, that forced overexpression of Dlx2 in BMSCs leads to upregulation of Alp and OCN, instead of Runx2, Dlx5, Msx2, and Osx.
Fig. 4.
Dlx2 overexpression in BMSCs has no impact on Runx2, Dlx5, Msx2 and Osxexpression. RT-qPCR analysis was performed to evaluate the expression levels of Dlx2 (a), Runx2 (b), Dlx5 (c), Msx2 (d), Osx (e), OCN (F), and Alp (g) in BMSCs transduced with Lenti-DLX2 OE (over) or Lenti-CTRL (control) at 14 and 21 days after osteogenic induction. Gapdh was used as an internal control. Statistical significance was determined as described in Fig. 1. h RT-qPCR analysis was used to evaluate the expression level of OCN and Alp upon forced overexpression of Dlx2 in MC3T3-E1 cells. i Western blot analysis was performed to measure the protein levels of OCN upon forced overexpression of Dlx2 in MC3T3-E1. β-Actin was used as an internal control. Over, BMSCs transduced with Lenti-DLX2 OE; control, BMSCs transduced with Lenti-CTRL
Dlx2 overexpression leads to increased expression of OCN and Alp in MC3T3-E1 cells
We next tested whether Dlx2 overexpression could also lead to upregulation of OCN and Alp in other osteoblast precursor cell lines. Compared with mock-transduced cells, we observed enhanced mRNA and protein levels of OCN in Dlx2-overexpressing MC3T3-E1 cells after osteogenic induction (Fig. 4h, i). Dlx2-overexpressing MC3T3-E1 cells also exhibited higher Alp transcription (Fig. 4h) and enhanced ALP activity (Fig. S1A). Moreover, as in BMSCs, we observed no significant difference in Msx2, Dlx5, Runx2, and Osx expression after Lenti-DLX2 OE transduction in MC3T3-E1 cells after osteogenic induction, as shown in the Figure S2.
Taken together, these data demonstrate that forced overexpression of Dlx2 induces upregulation of OCN and Alp upon osteogenic induction both in BMSCs and MC3T3-E1 cell lines. Given that the expression levels of Runx2, Dlx5, Msx2, and Osx remained unchanged upon Dlx2 overexpression, we speculate that OCN and Alp are the direct target genes of Dlx2.
Characterization of the mouse OCN promoter and identification of its Dlx2-response element
Previous genetic studies have shown that through direct binding to the promoter of OCN, homeodomain (HD) proteins Msx2, Dlx3, and Dlx5 regulate the expression of OCN in osteogenic cells. Dlx3 binds the OCN promoter to stimulate its expression, while the binding of Msx2 and the recruitment of Dlx5 represses OCN expression. Moreover, Dlx5 and Msx2 regulate Alp expression by directly binding to its promoter.15,16 Considering that Dlx2, an HD protein, shares strong sequence similarity with Dlx5 and Dlx3, we speculated that Dlx2 might also be involved in the regulation of OCN and Alp transcription in osteogenic cells by binding to their promoters.
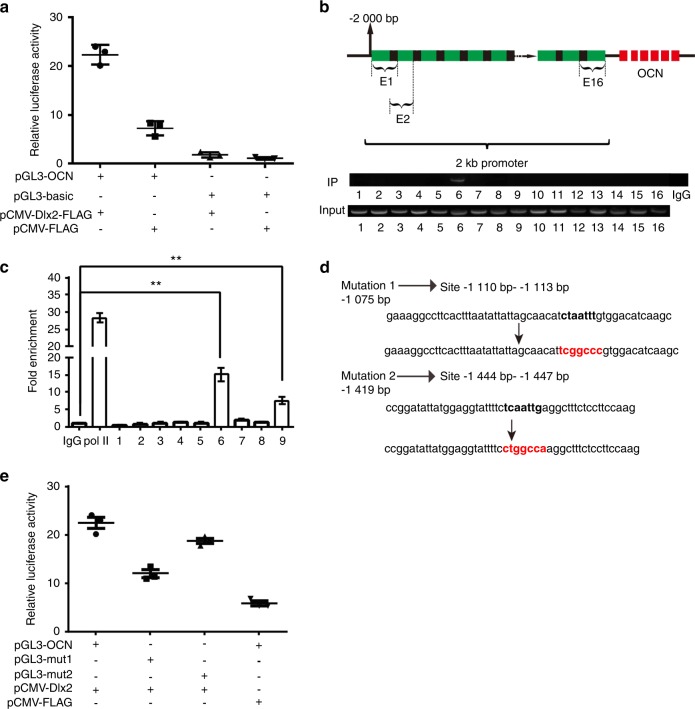
We first determined whether OCN is directly regulated by Dlx2, we analyzed its promoter in MC3T3-E1 cells. We inserted the whole promoter region of OCN into the pGL3-basic plasmid, generating pGL3-OCN, and transferred this vector into MC3T3-E1 cells along with pCMV-Dlx2-FLAG to allow the overexpression of Dlx2. As shown in Fig. 5a, we observed an ~3.2-fold increase in the transcriptional activity of pGL3-OCN after introduction of pCMV-Dlx2-FLAG into MC3T3-E1 cells, suggesting that the OCN promoter contains at least one Dlx2-response element (RE).
Fig. 5.
Characterization of the mouse OCN promoter and identification of its Dlx2-response elements. a The basal luciferase activity of the whole OCN promoter construct (pGL3-OCN) and that of the empty (pGL3-basic) construct in MC3T3-E1 cells was determined by transfecting the cells with each promoter reporter construct along with the Dlx2 overexpression vector (pCMV-Dlx2-FLAG) or the empty vector (pCMV-FLAG). Cells were then harvested 24 h after the transfection, and luciferase activity was measured and normalized to the protein concentration in the cell lysate. b ChIP analysis was performed to determine the Dlx2-response elements in the OCN promoter. MC3T3-E1 cells were transfected with pCMV-Dlx2-FLAG. Semi-quantitative PCR was performed using overlapping and closely spaced primer pairs to dissect the whole OCN promoter region into 16 short (~ 175 bp) overlapping parts for identification of the bound protein. Normal IgG (2 μg) was used as control. The PCR products were then separated by electrophoresis through a 2% agarose gel. c ChIP analysis followed by RT-qPCR was performed using the same primers described in b. Statistical significance was determined as described in Fig. 1. d The sequences of the nucleotides whose sequences contain the AATT element in the OCN promoter and the sequences of two mutants. The mutant binding sites are marked in red, and the putative Dlx2-binding sites are indicated in parentheses. e The luciferase activity of wild-type OCN promoter constructs (pGL3-OCN) and the mutated ones (pGL3-mut1 and pGL3-mut2) were determined by transfecting these vectors into MC3T3-E1 cells along with pCMV-Dlx2-FLAG or pCMV-FLAG. The OCN promoter construct bearing approximately −2000 to 0 bp region was subjected to site-directed mutagenesis to substitute the AATT sequence (pGL3-OCN) with either mutation1 (pGL3-mut1) or mutation2 (pGL3-mut2). The luciferase activity was measured 24 h later
To further identify the Dlx2-RE(s) in the OCN promoter, we performed ChIP analysis in MC3T3-E1 cells transfected with pCMV-Dlx2-FLAG. Semi-quantitative PCR analysis showed one strong signal in the E6 region (−1 311 bp to −1 175 bp) matching the predicted ~175 bp size of the E6 PCR product (Fig. 5b). Considering the low sensitivity of semi-quantitative PCR, we next carried out RT-qPCR. Consistent with the above findings, qPCR also revealed that Dlx2 was highly enriched at region E6 (−1 311 to −1 175 bp) and at region E9 (−1 073 bp to – 932 bp) (Fig. 5c). The qPCR results of region E10 to region E16 are not shown since the signals at these regions were as weak as the negative control in semi-quantitative PCR results. In addition, the occupancy of Dlx2 in the OCN promoter was correlated with increased transcription represented by elevated occupancy of RNA polymerase II (Pol II) (Fig. 5c), while the negative control (cells introduced with pCMV-FLAG) showed very weak signal (data not shown). These results indicate that there is a Dlx2-RE in the OCN promoter in region E6 (in primer set 6) and E9 (in primer set 9). Consistent with this, bioinformatics analysis (JASPAR database) indicated that the promoter region of OCN contains potential Dlx2-REs (−1 447 bp to −1 444 bp and −1 113 bp to −1 110 bp), both of which have the ATTA sequence.
We next checked whether Dlx2 can directly bind to the two predicted Dlx2-REs in the OCN promoter. To do this, we inserted mutated OCN promoter into the pGL3-basic plasmid in which the ATTA sequence was partly mutated, generating pGL3-mut1 (upstream RE mutant) and pGL3-mut2 (downstream RE mutant) (Fig. 5d). As shown in Fig. 5e, we found that mutation of either the upstream RE or downstream RE abrogated the ability of Dlx2 to regulate its activity. These observations demonstrate that in MC3T3-E1 cells, Dlx2 directly binds to the upstream RE and downstream RE in the OCN promoter to positively regulate its transcription.
Dlx2 upregulates Alp expression by directly binding to its promoter
Next, we tried to investigate whether Alp was regulated by Dlx2 in a similar way by binding to its promoter. To verify this transcriptional regulation, we first cloned and inserted the whole promoter region of Alp into the pGL3-basic plasmid, generating pGL3-ALP. This vector was introduced into MC3T3-E1 cells along with pCMV-Dlx2-FLAG to allow overexpression of Dlx2. As shown in Fig. 6a, we observed an ~6.7-fold increase in the transcriptional activity of pGL3-ALP after introduction of pCMV-Dlx2-FLAG into MC3T3-E1 cells, indicating that there is at least one Dlx2-RE in the Alp promoter. Supporting this notion is that bioinformatics analysis (JASPAR database) indicated that the promoter region of Alp contains potential Dlx2-RE (−1 194 bp to −1 187 bp, region A1).
Fig. 6.
Characterization of the mouse Alp promoter and identification of its Dlx2-response element. a The basal luciferase activity of the whole Alp promoter construct (pGL3-ALP) and that of the empty (pGL3-basic) construct in MC3T3-E1 cells was determined by transfecting the cells with each promoter reporter construct along with the Dlx2 overexpression vector (pCMV-Dlx2-FLAG) or the empty vector (pCMV-FLAG). Cells were then harvested 24 h after the transfection and luciferase activity was measured and normalized to the protein concentration in the cell lysate. b ChIP-qPCR analysis was performed to determine the Dlx2-response elements in the Alp promoter. MC3T3-E1 cells were transfected with pCMV-Dlx2-FLAG. Normal IgG (2 μg) was used as control. Pol II was used as a positive control. c ChIP analysis followed by RT-PCR was performed using the same primers described in b. d The sequence of the nucleotides whose sequences contain attaatt sequence in the Alp promoter and the sequence of the mutant. The mutant binding sites are marked in red and the putative Dlx2-binding sites are indicated in parentheses. e The luciferase activity of wild-type Alp promoter constructs (pGL3-ALP) and the mutated ones (pGL3-mutationA1) were determined by transfecting these vectors into MC3T3-E1 cells along with pCMV-Dlx2-FLAG or pCMV-FLAG. The Alp promoter construct bearing ~−2000 to 0 bp region was subjected to site-directed mutagenesis to substitute the original sequence (pGL3-ALP) with mutation (pGL3-mutationA1). The luciferase activity was measured 24 h later. f Schematic illustration of the regulation of OCN and Alp by Dlx2
Next, we checked whether Dlx2 can bind directly to the predicted Dlx2-REs (A1) in the Alp promoter. ChIP analysis was performed in MC3T3-E1 cells transfected with pCMV-Dlx2-FLAG or pCMV-FLAG, and primer set was designed to amplify the region A1. As shown in Fig. 6b, c, both qPCR and semi-quantitative PCR revealed that Dlx2 was highly enriched in region A1. In addition, elevated occupancy of Pol II in the Alp promoter was associated with increased occupancy of Dlx2 in this region, while the negative control (MC3T3-E1 cells introduced with pCMV-FLAG) showed very weak signal (data not shown).
To determine whether A1 is the Dlx2-binding site in the Alp promoter, we carried out site-directed mutagenesis in A1 region. We inserted mutated Alp promoter into the pGL3-basic plasmid, generating pGL3-mutationA1 (Fig. 6d). As shown in Fig. 6e, we found that mutation of A1 significantly attenuated the ability of Dlx2 to regulate its activity. These findings demonstrated that Dlx2 directly binds to region A1 in the Alp promoter to positively regulate its expression (Fig. 6e). Taken together, all these data proved that Dlx2 promotes OCN and Alp expression by directly binding to their promoters, and then regulates osteogenic differentiation in vitro and accelerates bone formation in vivo.
Discussion
Dlx gene family plays a critical role in osteogenesis. Previous study showed that Dlx2, Dlx5, and Dlx6 are upregulated in immature osteoblasts, while the expression of Dlx3 is elevated in differentiated osteoblasts and osteocytes. Dlx3 and Dlx5 potently transactivates osteoblastic marker genes15; Dlx6 also has stimulatory effects on osteogenic differentiation.17 These findings indicate that as a member of Dlx gene family, Dlx2 may be also involved in the osteogenic differentiation.
Dlx2, a member of vertebrate Dlx gene family, is expressed in the epithelium and mesenchyme of the mandible and maxilla.18 Previous studies have shown that newborn Dlx2−/− mice die immediately after birth and have abnormal craniofacial bones originating from the first branchial arch maxillary process,9 while Dlx2 overexpression in cranial neural crest cell (CNCC) leads to premaxillary hypoplasia and spinal deformities in mice.9 Although Dlx2 knockout and overexpression models demonstrate a crucial role of Dlx2 in promoting skeleton formation, the molecular and cellular mechanisms underlying the regulation of osteogenic differentiation by Dlx2 still remain unclear.
Here, we present evidence that forced overexpression of Dlx2 enhances the osteogenic differentiation potential of mouse BMSCs and MC3T3-E1 in vitro and accelerates bone formation in vivo by directly regulating OCN and Alp. Our finding is consistent with the previous study in SCAPs that overexpression of Dlx2 also enhanced osteogenic differentiation.19 SCAPs are mesenchymal-like stem cells that are able to differentiate into multiple linages, including odontoblastic and osteoblastic lineage, and do not undergo adipogenic differentiation, while BMSCs are able to undergo osteogenic, chondrogenic, and adipogenic differentiation.20 Relative to BMSCs, SCAPs display elevated secretion of proteins involved in metabolic processes, chemokines, and neutrophins, whereas BMSCs secret much more proangiogenic factors and ECM proteins. Therefore, overexpression of Dlx2 in SCAPs and BMSCs could be used in dentin regeneration and bone formation, respectively.21
The rodent Dlx5 and Dlx2 HD transcription factors are critical for bone development. During osteogenesis, Dlx5 upregulates Alp expression while suppresses OCN transcription.22 On the contrary, we found that Dlx2 transactivates OCN and ALP by directly binding to their promoters. Moreover, we identified two Dlx2-REs in OCN promoter, and one Dlx2-RE in Alp promoter involved in Dlx2-induced Alp expression. These findings demonstrate that Dlx2 is a crucial regulator regulating the osteogenic differentiation potential of both mesenchymal stem cells and osteogenic cells.
BMP-2 is one of the most important cytokines promoting differentiation of mesenchymal cells into osteoblasts.23 Stimulated by BMP-2, the transcription factors Dlx5, Msx2, and Runx2 work coordinately to regulate osteogenic differentiation.24 Both Runx2 and Dlx3 positively regulate OCN expression while Dlx5 represses its expression; Dlx5 stimulates ALP expression, whereas Msx2 depresses its expression.25 At the onset of osteogenic differentiation, Msx2 is released from the promoter of OCN, while Dlx3, Dlx5, and Runx2 are recruited. The released Msx2 then binds to the Alp promoter to upregulate its expression. At a later stage of osteogenic differentiation, during matrix mineralization, Dlx5 replaces Dlx3 to regulate OCN expression.26 Runx2 and Osx are another key transcription factors that are necessary for osteogenesis.27 After differentiating into pre-osteoblasts, Runx2 and Osx promote the cells to produce bone matrix. Dlx gene family is involved in the regulation of Runx2 and OSX transcription. Dlx5 induces expression of Runx2 and Osx, which work sequentially to stimulate the expression of OCN and Alp. Dlx3 also contributes to the activation of Runx2 during osteogenic differentiation.28 These findings indicate that Dlx gene family plays a crucial role in expression of osteogenic-associated genes. The results of the present study showed that overexpression of Dlx2 showed no significant effects on Runx2, Msx2, and Dlx5 expression upon osteogenic induction, but stimulated OCN and Alp expression, indicating that Runx2, Msx2, and Dlx5 may not participate in Dlx2-induced osteogenesis; Dlx2 may directly upregulate OCN and Alp to promote osteogenic differentiation.
Alp and OCN are two key marker genes of osteoblastic cells. ALP plays a critical role in early osteogenesis and hydrolyzes various types of phosphates to promote cell maturation and calcification, while OCN promotes the later stage of osteogenesis through combining with minerals.29,30 Both Dlx3 and Dlx5 directly upregulate Alp expression, while OCN is activated by Dlx3 but suppressed by Dlx5.16,31 Moreover, forced overexpression of Dlx5 in BMSCs led to a reduction in the mineralized matrix deposition, and impaired the ability of these cells to develop to the final stages of osteogenesis, and severely affected in vivo bone formation in immunodeficient mice.13 Although Dlx2 shares a strong sequence similarity with Dlx5, we found that Dlx2 positively regulates both Alp and OCN expression in BMSCs and MC3T3-E1 cells.
A previous study showed that loss of Dlx1/2−/− leads to abnormal bone formation of the upper jaw, while Dlx5/6−/− deficient mice exhibit profound abnormalities of the lower jaw tissue.22 However, piles of studies revealed that Dlx5 is the master regulator of osteogenic differentiation, since it directly controls the transcription of multiple osteogenic-associated genes, including Alp, OCN, Runx2, OSX, and Smads family, affecting the whole process of bone formation.32,33 Therefore, Dlx5 is also involved in the development of the upper jaw, but may be not as important as Dlx2. On the other hand, the maxilla is only formed by intramembranous ossification of the craniofacial mesenchyme, while the mandible can be formed by both intramembranous and endochondral ossification.34 Our previous study has shown that Dlx2 is involved in endochondral ossification.9,35 Therefore, Dlx2 is also involved in the bone formation of lower jaw, but is not as important as Dlx5. Together, both Dlx2 and Dlx5 are involved in the development of mandible and maxilla. Considering the fact that a variety of homeobox genes work coordinately during the bone formation,36 further investigations are required to find out how Dlx2 is involved during the bone formation in mandible and maxilla.
In conclusion, our data demonstrated for the first time that forced overexpression of Dlx2 enhances the osteogenic differentiation potential of BMSCs and MC3T3-E1 cells by directly upregulating OCN and Alp (Fig. 6f). We also presented evidence that there are Dlx2-REs in mouse OCN and Alp promoter that mediate the regulation of Dlx2 on OCN or Alp expression. This study may present a promising future strategy for the treatment of bone defects with Dlx2-overexpressing BMSCs.
Material and methods
Isolation and culture of mouse BMSCs
All animal experiments were performed according to guidelines of the Institutional Animal Care and Use Committees of the Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine. All experimental protocols were reviewed and approved by the Institutional Animal Care and Use Committees of the Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China. BMSCs were isolated from the tibias of 6-week-old male C57/BL6 mice ((10 ± 0.5) g) and cultured according to previous study.37
An MC3T3-E1 cell line was obtained from the Cell Bank of the Chinese Academy of Science (Shanghai, China), C3H10 T1/2 cell line from American Type Culture Collection (Rockville, MD, USA), and a Human embryonic kidney 293T (HEK 293T) cell line from American Type Culture Collection (Rockville, MD, USA). The MC3T3-E1, C3H10 T1/2 cell line, and HEK 293T cells were cultured as described previously.38,39 OIM contained 50 mg· L−1 ascorbic acid, 1 × 10−7 mol· L−1 dexamethasone, and 50 mg· L−1 β-glycerophosphate plus α-MEM (Sigma-Aldrich Corp. (St. Louis, MO, USA)). HEK 293T cell line was utilized for packaging viral constructs.
Lentiviral construction and transduction
The lentiviral expression system overexpressing Dlx2 was termed as Lenti-Dlx2 OE. The open reading frame of mice Dlx2 (NM_010054) was synthesized and cloned into pL/IRES/GFP plasmid (Novobio, Shanghai, China) for generating pL/IRES/GFP-DLX2. The empty lentiviral expression system without insertion was termed as Lenti-CTRL and used as the control. 293T cells were then transfected with plasmids listed above. The transfection and lentiviral transduction was done as described previously.40,41
ALP, Alizarin red staining, and semi-quantitative analysis
Transduced BMSCs or MC3T3-E1 cells were first cultured in OIM for 14 or 21 days. ALP and Alizarin red staining were carried out as described previously.40 ALP staining was carried out with BCIP/NBT Alkaline Phosphatase Color Development Kit (Beyotime Institute of Biotechnology, China), and semi-quantitative analysis of ALP activity was performed using p-nitrophenyl phosphate (p-NPP) (Sigma-Aldrich) as the substrate. For Alizarin red staining, cells were first fixed with 70% ethanol. Afterward, the fixed cells were stained with 2% Alizarin Red (Sigma-Aldrich), according to the previous study.19
Semi-quantitative RT-PCR, RT-qPCR, and Western blot analysis
RT-PCR, RT-qPCR, and western blot analysis were performed as described previously.40 Total RNA was extracted from cultured cells using TRIzol RNA Isolation reagent (Takara, Tokyo, Japan), according to the manufacturer’s instruction. Three independent cultures were used for RNA preparations. First-strand cDNA was generated with High-Capacity cDNA Reverse Transcription Kit, (Applied Biosystems, San Diego, CA), and one microliter of each RT reaction mixture was amplified with Ex Taq DNA polymerase (Takara, Tokyo, Japan). As for RT-qPCR, cDNA was amplified using premix SYBR Green Ex Taq reagent kit (DRR820A, Takara) with a STEP ONE PLUS real-time PCR system (Applied Biosystems, Forster City, CA), according to the manufacturer’s instruction. All the primers used in this study are listed in Table 1. As for western blotting, anti-Dlx2 (1:800; ab85995, Abcam, Cambridge, UK), anti-OCN (1:1 000; ab93876, Abcam), and anti-β-actin (1:3 000; EPR16769, Abcam, Cambridge, UK) were used for the detection of Dlx2, OCN, and β-actin, respectively. The secondary antibodies used this study were bought from Sigma-Aldrich and conjugated to horseradish peroxidase (anti-rabbit (1:5 000, A0545) or anti-mouse (1:5 000, SAB3701214)).
Table 1.
Primers used in this study
| Primer | Sequence | Details | Reference |
|---|---|---|---|
| Primers for RT-qPCR | |||
| ep Dlx2 FW | CATGGGCTCCTACCAGTACCAC | qPCR primer for Dlx2 | 8 |
| ep Dlx2 RV | TCGGATTTCAGGCTCAAGGTC | qPCR primer for Dlx2 | 8 |
| ep Gapdh FW | GGTGAAGGTCGGTGTGAACG | qPCR primer for Gapdh | 8 |
| ep Gapdh RV | CTCGCTCCTGGAAGATGGTG | qPCR primer for Gapdh | 8 |
| ep ALP FW | TGGGCATTGTGACTACCACTCGG | qPCR primer for Alp | 8 |
| ep ALP RV | CCTCTGGTGGCATCTCGTTATCC | qPCR primer for Alp | 8 |
| ep OCN FW | GGACCATCTTTCTGCTCACTCTG | qPCR primer for Ocn | 8 |
| ep OCN RV | GTTCACTACCTTATTGCCCTCCTG | qPCR primer for Ocn | 8 |
| ep Runx2 FW | AACTTCCTGTGCTCCGTGCTG | qPCR primer for Runx2 | 8 |
| ep Runx2 Rv | TCGTTGAACCTGGCTACTTGG | qPCR primer for Runx2 | 8 |
| ep Msx2 FW | GGAGCACCGTGGATACAGGA | qPCR primer for Msx2 | 8 |
| ep Msx2 RV | AGGCTAGAAGCTGGGATGTGG | qPCR primer for Msx2 | 8 |
| ep OSX FW | CCCTTCTCAAGCACCAATGG | qPCR primer for Osx | 41 |
| ep OSX RV | AAGGGTGGGTAGTCATTTGCATA | qPCR primer for Osx | 41 |
| Primers for ChIP analysis | |||
| Primer-F16 | gaggctgagagagagagcaca | For ChIP-qPCR | This study |
| Primer-R16 | tgctgtggtaggtgattgc | For ChIP-qPCR | This study |
| Primer-F15 | gcagacactgaaaatcacagg | For ChIP-qPCR | This study |
| Primer-R15 | tgtgctctctctctcagcctc | For ChIP-qPCR | This study |
| Primer-F14 | cataaaactaaccaggacactcc | For ChIP-qPCR | This study |
| Primer-R14 | gcctgtgattttcagtgtctgc | For ChIP-qPCR | This study |
| Primer-F13 | ccacaatgggctaggctct | For ChIP-qPCR | This study |
| Primer-R13 | catccagtgggggtgtgt | For ChIP-qPCR | This study |
| Primer-F12 | acacaagcagggctagaacc | For ChIP-qPCR | This study |
| Primer-R12 | agcccattgtgggagctac | For ChIP-qPCR | This study |
| Primer-F11 | ttgtggacatcaagcggg | For ChIP-qPCR | This study |
| Primer-R11 | aggttctagccctgcttgtg | For ChIP-qPCR | This study |
| Primer-F10 | tttcatttccacctagagcaag | For ChIP-qPCR | This study |
| Primer-R10 | tcgggggtatctggtttcag | For ChIP-qPCR | This study |
| Primer-F9 | gtttggcatggagcctttg | For ChIP-qPCR | This study |
| Primer-R9 | tgtgttacagtcacttgctctagg | For ChIP-qPCR | This study |
| Primer-F8 | gccctcttctagtgtgtctgaa | For ChIP-qPCR | This study |
| Primer-R8 | cccaagttcaaaggctccat | For ChIP-qPCR | This study |
| Primer-F7 | cacagttggactggggaggt | For ChIP-qPCR | This study |
| Primer-R7 | cttcagacacactagaagagggc | For ChIP-qPCR | This study |
| Primer-F6 | tcaccagcgcaaatcacac | For ChIP-qPCR | This study |
| Primer-R6 | ctgtgtgctttcttattcacctacc | For ChIP-qPCR | This study |
| Primer-F5 | aagggaaacaccacccactc | For ChIP-qPCR | This study |
| Primer-R5 | atttgcgctggtgaggac | For ChIP-qPCR | This study |
| Primer-F4 | cctccactgcctcaagaact | For ChIP-qPCR | This study |
| Primer-R4 | aaggctggaaaggagtgggt | For ChIP-qPCR | This study |
| Primer-F3 | acagaaggcaggtcagctaca | For ChIP-qPCR | This study |
| Primer-R3 | gggaagagtaaggtggaggtt | For ChIP-qPCR | This study |
| Primer-F2 | ttccccctagccgaacaag | For ChIP-qPCR | This study |
| Primer-R2 | acctgccttctgtgatcctct | For ChIP-qPCR | This study |
| Primer-F1 | agaggatcacagaaggcaggt | For ChIP-qPCR | This study |
| Primer-R1 | aaaggatgctgtggttggtgattg | For ChIP-qPCR | This study |
| Primer-A1F | tgcctgggtttgttttcatttgt | For ChIP-qPCR | This study |
| Primer-A1R | caagggaaatttcctagcac | For ChIP-qPCR | This study |
In vivo osteogenic differentiation
The osteogenic differentiation potential of transduced BMSCs was evaluated with in vivo ectopic bone formation analysis, as described previously.13 Briefly, osteogenic-induced cells of passage 3 were injected into the β-TCP (Shanghai Rebone Biomaterials Co., Ltd., Shanghai, China) with a syringe, and appropriate volume of OIM was added to cover the BMSCs/ β-TCP constructs. After 7 days of culturing in vitro, the constructs were implanted subcutaneously in the back of immunocompromised female nude mice (CD-1 Nu/Nu, 10-week-old, Charles River). At each time point (6 and 8 weeks after implantation), six mice for each group were sacrificed with an overdose of pentobarbital (210 mg/kg intraperitoneally) to retrieve the BMSCs/β-TCP constructs. Micro-CT images of each cellular construct were taken with a micro-CT system (SMX-100CT-SV3; Shimadzu, Japan). Radiological density of each cellular construct was measured in Hounsfield density units.
Luciferase assay
The whole mouse OCN promoter (2000 bp) was subcloned into the pGL3 basic vector (Promega), generating pGL3-OCN. To produce constructs that contain mutation in E6 region, we carried out mutagenic PCR with Mt1 primer and Mt2 primer, generating pGL3-mut1. Likewise, Mt3 and Mt4 primers were used for generating pGL3-mut2 (Table 1). The open reading frame of mice Dlx2 (NM_010054) was synthesized and cloned into pCMV-FLAG, generating pCMV-Dlx2-FLAG for Dlx2 overexpressing. Transfection assay was performed as described previously.31 As for each transfection assay, 0.5 μg of the Dlx2 overexpression vector (pCMV-Dlx2-FLAG) or pCMV-FLAG and 0.5 μg of the luciferase reporter vector were transfected into MC3T3-E1 cells. Similarly, we inserted the whole Alp promoter into the pGL3-basic vectors, generating pGL3-ALP, and produced a vector that contains mutated Alp promoter, generating pGL3-mutationA1. Afterward, pCMV-Dlx2-FLAG or pCMV-FLAG was introduced into MC3T3-E1 cells along with pGL3-ALP or pGL3-mutationA1.
ChIP analysis
ChIP analysis was carried out according to the standard protocol.15 MC3T3-E1 cells transfected with pCMV-Dlx2-FLAG or pCMV-FLAG were fixed with 1% formaldehyde and this cross-linking was quenched with glycine (0.125 mol· L−1 for final concentration). After the cells were lysed and homogenized with a Dounce homogenizer, the nuclei were collected by centrifuging. The nuclei pellet was then resuspended in sonication buffer followed by sonication. Chromatin were incubated with anti-FLAG antibodies (F3165, Sigma-Aldrich), mouse IgG and anti-RNA-polymerase II (PLA0292, Sigma-Aldrich), and this immune complex were incubated with protein G sepharose (17-0618-01, GE Amersham). The DNA fragments in this immune complex were purified with phenol–chloroform extraction. ChIP samples were then analyzed with RT-PCR and RT-qPCR. Pol II was used as a positive control, as described in a previous study.15 The primers used in ChIP analysis are listed in Table 1.
Supplementary information
Acknowledgements
The authors thank Dr. Wenbin Zhang, Dr. Jianjun Zhang, Dr. Ming Yan and Dr. Xu Wang from the Shanghai Ninth People’s Hospital for helping us with the technique. We would like to thank the members of the Shen- and Chen-labs for helpful advice and discussion. We also thank Native EE for its linguistic assistance during the preparation of this manuscript. This work is supported by grant (81771036) from National Natural Science Foundation of China (to S.G.S.), grant (81741028) from National Natural Science Foundation of China (to J.D.) and grant (17410710500) Shanghai International Scientific and Technological Cooperation Projects Laser Micro-machine and Vascularization of TCP/PCL Scaffolds (to W.Z.).
Competing interests
The authors declare no competing interests.
Contributor Information
Xudong Wang, Email: xudongwang70@hotmail.com.
Steve Guofang Shen, Phone: +86 21 23271699, Email: maxillofacsurg@163.com.
Supplementary information
The online version of this article (10.1038/s41368-019-0046-1) contains supplementary material, which is available to authorized users.
References
- 1.Ulsamer A, et al. BMP-2 induces Osterix expression through up-regulation of Dlx5 and its phosphorylation by p38. J. Biol. Chem. 2008;283:3816–3826. doi: 10.1074/jbc.M704724200. [DOI] [PubMed] [Google Scholar]
- 2.Stock DW, et al. The evolution of the vertebrate Dlx gene family. Proc. Natl Acad. Sci. USA. 1996;93:10858–10863. doi: 10.1073/pnas.93.20.10858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee MH, Kwon TG, Park HS, Wozney JM, Ryoo HM. BMP-2-induced Osterix expression is mediated by Dlx5 but is independent of Runx2. Biochem. Biophys. Res. Commun. 2003;309:689–694. doi: 10.1016/j.bbrc.2003.08.058. [DOI] [PubMed] [Google Scholar]
- 4.Shirakabe K, Terasawa K, Miyama K, Shibuya H, Nishida E. Regulation of the activity of the transcription factor Runx2 by two homeobox proteins, Msx2 and Dlx5. Genes Cells. 2001;6:851–856. doi: 10.1046/j.1365-2443.2001.00466.x. [DOI] [PubMed] [Google Scholar]
- 5.Depew MJ, Lufkin T, Rubenstein JL. Specification of jaw subdivisions by Dlx genes. Science. 2002;298:381–385. doi: 10.1126/science.1075703. [DOI] [PubMed] [Google Scholar]
- 6.Ferguson CA, Tucker AS, Sharpe PT. Temporospatial cell interactions regulating mandibular and maxillary arch patterning. Development. 2000;127:403–412. doi: 10.1242/dev.127.2.403. [DOI] [PubMed] [Google Scholar]
- 7.Merlo GR, et al. Multiple functions of Dlx genes. Int. J. Dev. Biol. 2004;44:619–626. [PubMed] [Google Scholar]
- 8.Sun H, Liu Z, Li B, Dai J, Wang X. Effects of DLX2 overexpression on the osteogenic differentiation of MC3T3‑E1 cells. Exp. Ther. Med. 2015;9:2173–2179. doi: 10.3892/etm.2015.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai J, et al. The effect of overexpression of Dlx2 on the migration, proliferation and osteogenic differentiation of cranial neural crest stem cells. Biomaterials. 2013;34:1898–1910. doi: 10.1016/j.biomaterials.2012.11.051. [DOI] [PubMed] [Google Scholar]
- 10.Pittenger MF, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 11.Kuznetsov SA, et al. Single‐colony derived strains of human marrow stromal fibroblasts form bone after transplantation in vivo. J. Bone Mineral. Res. 1997;12:1335–1347. doi: 10.1359/jbmr.1997.12.9.1335. [DOI] [PubMed] [Google Scholar]
- 12.Krebsbach PH, et al. Bone formation in vivo: comparison of osteogenesis by transplanted mouse and human marrow stromal fibroblasts. Transplantation. 1997;63:1059–1069. doi: 10.1097/00007890-199704270-00003. [DOI] [PubMed] [Google Scholar]
- 13.Muraglia A, et al. DLX5 overexpression impairs osteogenic differentiation of human bone marrow stromal cells. Eur. J. Cell Biol. 2008;87:751–761. doi: 10.1016/j.ejcb.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Dalby MJ, et al. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat. Mater. 2007;6:997. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 15.Hassan MQ, et al. Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol. Cell. Biol. 2004;24:9248–9261. doi: 10.1128/MCB.24.20.9248-9261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryoo HM, et al. Stage-specific expression of Dlx-5 during osteoblast differentiation: involvement in regulation of osteocalcin gene expression. Mol. Endocrinol. 1997;11:1681–1694. doi: 10.1210/mend.11.11.0011. [DOI] [PubMed] [Google Scholar]
- 17.Pfeffer U, et al. The coding region of the human DLX6 gene contains a polymorphic CAG/CCG repeat. Int. J. Oncol. 2001;18:1293–1297. doi: 10.3892/ijo.18.6.1293. [DOI] [PubMed] [Google Scholar]
- 18.Depew MJ, Simpson CA, Morasso M, Rubenstein JLR. Reassessing the Dlx code: the genetic regulation of branchial arch skeletal pattern and development. J. Anat. 2005;207:501–561. doi: 10.1111/j.1469-7580.2005.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qu B, et al. Distal-less homeobox 2 promotes the osteogenic differentiation potential of stem cells from apical papilla. Cell Tissue Res. 2014;357:133–143. doi: 10.1007/s00441-014-1833-9. [DOI] [PubMed] [Google Scholar]
- 20.Huang GTJ, et al. Stem/progenitor cell–mediated de novo regeneration of dental pulp with newly deposited continuous layer of dentin in an in vivo model. Tissue Eng. Part A. 2009;16:605–615. doi: 10.1089/ten.tea.2009.0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu S, Zhao Y, Ma Y, Ge L. Profiling the secretome of human stem cells from dental apical papilla. Stem Cells Dev. 2016;25:499–508. doi: 10.1089/scd.2015.0298. [DOI] [PubMed] [Google Scholar]
- 22.Samee, N., De Vernejoul, M.-C. & Levi, G. Role of DLX regulatory proteins in osteogenesis and chondrogenesis. Crit. Rev. Eukaryot. Gene Expr.17, 173–186 (2007). [DOI] [PubMed]
- 23.Hanai Ji, et al. Interaction and functional cooperation of PEBP2/CBF with smads synergistic induction of the immunoglobulin germline Cα promoter. J. Biol. Chem. 1999;274:31577–31582. doi: 10.1074/jbc.274.44.31577. [DOI] [PubMed] [Google Scholar]
- 24.Nishimura R, Hata K, Harris S, Ikeda F, Yoneda T. Core-binding factor α1 (Cbfa1) induces osteoblastic differentiation of C2C12 cells without interactions with Smad1 and Smad5. Bone. 2002;31:303–312. doi: 10.1016/S8756-3282(02)00826-8. [DOI] [PubMed] [Google Scholar]
- 25.Javed A, et al. runt homology domain transcription factors (Runx, Cbfa, and AML) mediate repression of the bone sialoprotein promoter: evidence for promoter context-dependent activity of Cbfa proteins. Mol. Cell. Biol. 2001;21:2891–2905. doi: 10.1128/MCB.21.8.2891-2905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langenbach F, Handschel J. Effects of dexamethasone, ascorbic acid and β-glycerophosphate on the osteogenic differentiation of stem cells in vitro. Stem Cell Res. Ther. 2013;4:117. doi: 10.1186/scrt328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang C. Transcriptional regulation of bone formation by the osteoblast-specific transcription factor Osx. J. Orthop. Surg. Res. 2010;5:37. doi: 10.1186/1749-799X-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hassan, M. Q. et al. BMP2 commitment to the osteogenic lineage involves activation of Runx2 by DLX3 and a homeodomain transcriptional network. J. Biol. Chem.281, 40515–40526 (2006). [DOI] [PubMed]
- 29.Glowacki J, Rey C, Glimcher M, Cox K, Lian J. A role for osteocalcin in osteoclast differentiation. J. Cell. Biochem. 1991;45:292–302. doi: 10.1002/jcb.240450312. [DOI] [PubMed] [Google Scholar]
- 30.Beck GR, Zerler B, Moran E. Phosphate is a specific signal for induction of osteopontin gene expression. Proc. Natl Acad. Sci. USA. 2000;97:8352–8357. doi: 10.1073/pnas.140021997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim YJ, Lee MH, Wozney JM, Cho JY, Ryoo HM. Bone morphogenetic protein-2-induced alkaline phosphatase expression is stimulated by Dlx5 and repressed by Msx2. J. Biol. Chem. 2004;279:50773–50780. doi: 10.1074/jbc.M404145200. [DOI] [PubMed] [Google Scholar]
- 32.Lee MH, et al. BMP-2-induced Runx2 expression is mediated by Dlx5, and TGF-β1 opposes the BMP-2-induced osteoblast differentiation by suppression of Dlx5 expression. J. Biol. Chem. 2003;278:34387–34394. doi: 10.1074/jbc.M211386200. [DOI] [PubMed] [Google Scholar]
- 33.Robledo RF, Rajan L, Li X, Lufkin T. The Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial, and appendicular skeletal development. Genes Dev. 2002;16:1089–1101. doi: 10.1101/gad.988402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tortelli F, Tasso R, Loiacono F, Cancedda R. The development of tissue-engineered bone of different origin through endochondral and intramembranous ossification following the implantation of mesenchymal stem cells and osteoblasts in a murine model. Biomaterials. 2010;31:242–249. doi: 10.1016/j.biomaterials.2009.09.038. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, Zhang W, Shi J, Dai J, Shen SG. Dlx2 overexpression enhanced accumulation of type II collagen and aggrecan by inhibiting MMP13 expression in mice chondrocytes. Biochem. Biophys. Res. Commun. 2018;503:528–535. doi: 10.1016/j.bbrc.2018.05.066. [DOI] [PubMed] [Google Scholar]
- 36.Thesleff I. Homeobox genes and growth factors in regulation of craniofacial and tooth morphogenesis. Acta Odontol. Scand. 1995;53:129–134. doi: 10.3109/00016359509005962. [DOI] [PubMed] [Google Scholar]
- 37.Russo FP, et al. The bone marrow functionally contributes to liver fibrosis. Gastroenterology. 2006;130:1807–1821. doi: 10.1053/j.gastro.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto K, Arakawa T, Ueda N, Yamamoto S. Transcriptional roles of nuclear factor B and nuclear factor-interleukin-6 in the tumor necrosis factor-dependent induction of cyclooxygenase-2 in MC3T3-E1 cells. J. Biol. Chem. 1995;270:31315–31320. doi: 10.1074/jbc.270.52.31315. [DOI] [PubMed] [Google Scholar]
- 39.Heim M, et al. Phytanic acid, a natural peroxisome proliferator-activated receptor (PPAR) agonist, regulates glucose metabolism in rat primary hepatocytes. FASEB J. 2002;16:718–720. doi: 10.1096/fj.01-0816fje. [DOI] [PubMed] [Google Scholar]
- 40.Xie Q, et al. The role of miR-135-modified adipose-derived mesenchymal stem cells in bone regeneration. Biomaterials. 2016;75:279–294. doi: 10.1016/j.biomaterials.2015.10.042. [DOI] [PubMed] [Google Scholar]
- 41.Cheng SL, et al. MSX2 promotes osteogenesis and suppresses adipogenic differentiation of multipotent mesenchymal progenitors[J] J. Biol. Chem. 2003;278:45969–45977. doi: 10.1074/jbc.M306972200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.