Summary
Truncating mutations in the sarcomere protein titin cause dilated cardiomyopathy due to sarcomere insufficiency. However, it remains mechanistically unclear how these mutations decrease sarcomere content in cardiomyocytes. Utilizing human induced pluripotent stem cell derived cardiomyocytes, CRISPR/Cas9, and live microscopy, we characterize the fundamental mechanisms of human cardiac sarcomere formation. We observe that sarcomerogenesis initiates at protocostameres, sites of cell-extracellular matrix adhesion, where nucleation and centripetal assembly of α-actinin-2-containing fibers provide a template for the fusion of Z-disk precursors, Z-bodies, and subsequent striation. We identify that β-cardiac myosin-titin-protocostamere form an essential mechanical connection that transmits forces required to direct α-actinin-2 centripetal fiber assembly and sarcomere formation. Titin propagates diastolic traction stresses from β-cardiac myosin, but not α-cardiac myosin or non-muscle myosin II, to protocostameres during sarcomerogenesis. Ablating protocostameres or decoupling titin from protocostameres abolishes sarcomere assembly. Together these results identify the mechanical and molecular components critical for human cardiac sarcomerogenesis.
Keywords: sarcomerogenesis, induced pluripotent stem cells IPSCs, cardiomyocyte, mechanotransduction, focal adhesion, cardiomyopathy, titin, β-cardiac myosin
Introduction
Mutations in a number of proteins associated with sarcomeres, the fundamental contractile unit of cardiac myocytes, lead to various forms of human cardiomyopathy (Fatkin et al., 2014; Harvey and Leinwand, 2011). These cardiomyopathies are accompanied by changes in cardiomyocyte morphology and sarcomere content (Harvey and Leinwand, 2011; Kresh and Chopra, 2011). Therefore, it seems that the building of sarcomeres is critical to cardiomyocyte structural adaptation during both development and disease.
Our current understanding of sarcomere assembly is incomplete, based mostly on observations from immunohistochemistry analysis in vertebrate models of either cardiomyocytes or skeletal muscle cells with limited perturbations to critical sarcomeric protein components (Rhee et al., 1994; Sanger et al., 2009; Schultheiss et al., 1990; White et al., 2014). Cardiomyocytes are thought to initiate myofibrillar assembly at the cell periphery from pre-myofibrils consisting of non-muscle myosin II (NMM II) and α-actinin-2 fibers, providing a template for subsequent recruitment of titin, α-and/or β-cardiac myosins to form mature myofibrils (Dabiri et al., 1997; Rhee et al., 1994). The observation that cardiac myofibrillar assembly begins at the cell periphery also has led to the hypothesis that protocostameres, sites of cell-extracellular matrix (ECM) adhesion that resemble focal adhesions, may be involved (Sparrow and Schock, 2009). Protocostameres share a similar molecular composition to classical focal adhesions, and include proteins such as integrins, paxillin, vinculin, and focal adhesion kinase (FAK), and overtime mature into specialized cell-ECM junctions, costameres, that overlap with the Z-disks at the plasma membrane (Gehmlich et al., 2007). Additionally, the role of myosin generated force is increasingly being recognized as essential for myofibrillar assembly in both skeletal and cardiac muscles in vivo and in vitro (Geach et al., 2015; Majkut et al., 2013; Weitkunat et al., 2014). However, it remains controversial what role specific myosins, or other sarcomeric components such as titin, play in initiating the sarcomere assembly process (Dabiri et al., 1997; Myhre et al., 2014; Tullio et al., 1997).
To date, studies describing the process of sarcomerogenesis have been limited to non-human cardiomyocytes. Cardiomyocytes sourced from such vertebrates, like human cardiomyocytes, have limited or no proliferative potential and therefore are not readily amenable to the genetic manipulation and long term culture needed to interrogate individual molecular contributions to the process of sarcomerogenesis. Additionally, in the context of understanding human cardiomyopathies, it is attractive to have a human cell model to study the mechanisms of sarcomerogenesis. Advances in genome engineering technologies and induced pluripotent stem cell (iPSC) derived cardiomyocyte differentiation protocols have provided a new source of human cardiomyocytes that have recently been used to model normal cells and various cardiomyopathies in vitro (Birket et al., 2015; Chong et al., 2014; Hinson et al., 2015; Kodo et al., 2016; Lundy et al., 2013; Sun et al., 2012; Wang et al., 2014). These iPSC-derived cardiomyocytes (iPSC-CMs) express a similar RNA profile to that of embryonic day 14.5–18.5 mice, a developmental time when cardiac myofibrillar assembly is most active (DeLaughter et al., 2016; Kuppusamy et al., 2015). Thus, iPSC-CMs offer the potential to model cardiac sarcomere assembly and reveal its underlying mechanisms.
We have recently shown that human iPSC-CMs reprogrammed from dilated cardiomyopathy patients harboring heterozygous truncating mutations in the giant sarcomere protein titin, considered to be the molecular ruler for defining the length of mature sarcomeres (Anderson and Granzier, 2012). These exhibit compromised contractile activity due to sarcomere insufficiency, which could be a direct result of the mutant cells’ inability to assemble sarcomeres or the instability of the existing sarcomeres (Herman et al., 2012; Hinson et al., 2015; Lundy et al., 2013). Others have similarly reported loss of sarcomeres in vertebrate models harboring titin truncations (Gotthardt et al., 2003) and in titin-depleted muscle cells (Miller et al., 2003; Person et al., 2000). However, despite such evidence, it remains unclear how titin truncations disrupt sarcomere assembly or stability to cause decreased sarcomere content.
In this work, we combine human iPSC-CMs, CRISPR/Cas9, and live microscopy technologies to develop a model of de novo human cardiac sarcomerogenesis and uncover that titin truncating mutations lead to impairment of sarcomere formation and subsequent myofibrillar assembly in human cardiomyocytes. Furthermore, we delve into the fundamental mechanics of the sarcomere assembly process, elucidating key mechanical and molecular players critical for generating sarcomeres in human cardiac cells. We report that (i) the presence of full length titin is necessary to generate the basal cardiomyocyte tension that is required to initiate sarcomere assembly from the cell periphery, (ii) in contrast to previous findings, we show that β-cardiac myosin, but not NMM II, generates the required basal tension to direct sarcomere assembly, and (iii) protocostameres are the sites of initiation for sarcomere assembly and the coupling of titin to protocostameres is critical to drive assembly.
Results
De novo sarcomere assembly is impaired in cardiomyocytes with titin truncations (TTNtv)
Sarcomeres are rapidly disassembled when cardiomyocytes are detached from the ECM and held in suspension (Figure S1A). Therefore, to examine the influence of mutations that truncate titin (TTNtv) on sarcomere assembly over time, we suspended and then replated heterozygous A-band TTNtv cardiomyocytes (Figure S1B) to trigger new sarcomere assembly and immunostained at different time points for cardiac α-actinin-2, subsequently referred to as actinin, a primary Z-disk component of sarcomeres. Quantification of sarcomere content revealed not only decreased end-point sarcomere content (Figure 1A), but also consistently reduced sarcomere content over a 96 hour time course of assembly (Figure 1B), suggesting that TTNtv indeed leads to defects in sarcomere assembly or stability.
Figure 1: TTNtvs lead to sarcomere insufficiency due to impaired sarcomerogenesis.
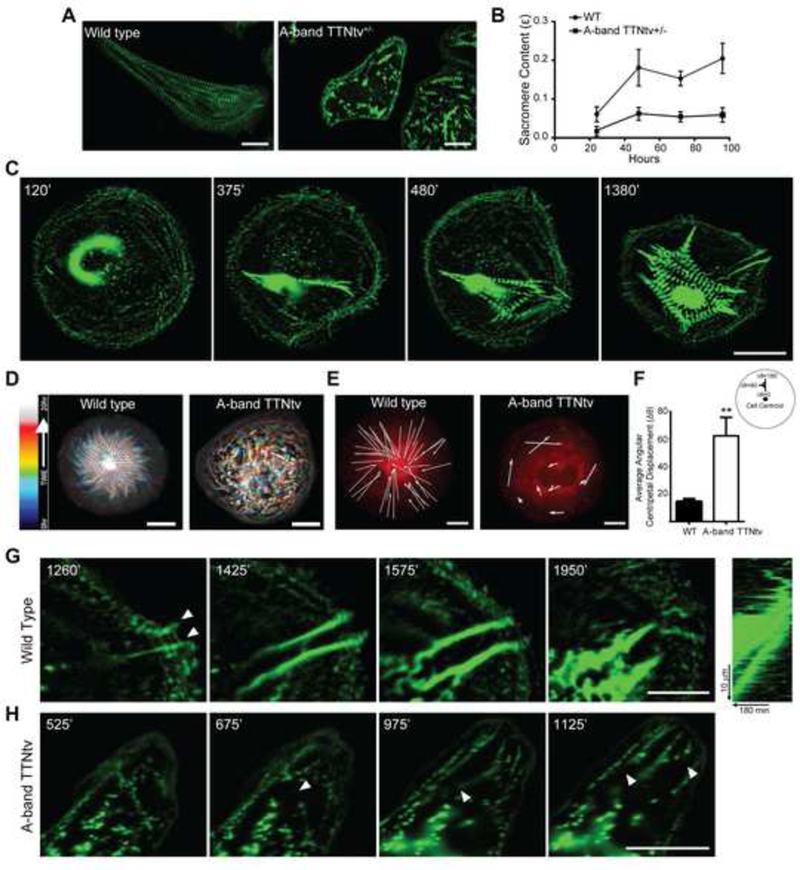
(A) Immunostaining for Z-disc protein α-actinin-2 (green) shows decreased striation in heterozygous A-band TTNtv mutant cells compared to wild type, representative images of cells fixed and stained for actinin after 96 hours of culture, scale bars 20 μm. (B) Quantification of average sarcomere content in A-band TTNTv+/− and wild type cells over time; n≥8 WT cells, n≥9 A-band TTNtv+/− cells for each time point; across three independent experiments; mean ± s.e.m. (C) Time lapse images of GFP-actinin stably expressed in wild type cells shows de-novo sarcomerogenesis. Left to right, initial appearance of centripetal actinin fibers at the cell periphery leads to formation of sarcomeres and progresses with expansion of striation throughout the cell, scale bar 20 μm. (D) Temporal-color coded hyperstack of representative sarcomere assembly time lapse images showing the emergence of patterned myofibrils in wild type cells and an absence in A-band TTNtv cells. Scale bars 20 μm. (E) Representative displacement vectors of actinin puncta over time in wild type and A-band TTNtv cells. Scale bars 10 μm. (F) Quantification of angular centripetal displacement of actinin puncta towards the cell centroid in wild type and TTNtv cells. n=2256 tracked actinin particles across five WT and n=1683 actinin particles across five TTNtv cells; across three independent experiments; mean ± s.e.m.; t-test, **P<0.01. (G) Representative time lapse images of actinin puncta in wild type cells forming centripetal actinin fibers. Arrows indicate the emanation of actinin puncta from cell adhesion sites. Scale bar 20 μm. (H) Representative time lapse images of actinin puncta in TTNtv cells, arrows indicate the rupturing events of existing actinin fibers at the cell edge. Scale bar 20 μm. See also Figure S1.
To address whether TTNtv affected sarcomere stability or assembly, we generated cardiomyocytes that stably express fluorescently labelled actinin to monitor assembly dynamics in real time. Because cell geometry can affect the organization of sarcomeres (Bray et al., 2008), we plated cells onto isotropic circular ECM patterns to consistently quantify sarcomere assembly without confounding cell shape effects. Time lapse imaging of sarcomerogenesis visualized via actinin in wild type cardiomyocytes revealed the dynamics of how sarcomeres form (Figure 1C; Movie S1). We observed that sarcomerogenesis was driven by two distinct flow patterns of actinin, which emanated from the cell periphery and processed towards the cell interior where sarcomere compaction occurred. Centripetal actinin fibers nucleated at sites resembling protocostameres and registered the initial template fibers for Z-body fusion (Figure 1C, G, arrows; Movie S1). Transverse actinin flow, resembling classical actin retrograde flow (Gardel et al., 2008; Tee et al., 2015), led to expansion and fusion of actinin into Z-bodies and their incorporation into existing centripetal actinin fibers (Figure S1C; Movie S1 ). Over time sarcomere units formed myofibril bundles which compacted and stabilized in the cell interior, permitting further Z-body formation and fusion at the periphery (Figure 1C, D, Movie S1). Though more difficult to quantify, the observed coordination of actinin fiber formation and transverse flow leading to sarcomere assembly proceeded similarly on anisotropic rectangular substrates (Movies S2, S3) and on substrates of physiologically relevant stiffness (4kPa) (Movie S3) as well as in 3D cardiac tissues (Movie S4, Figure S1E), un-patterned substrates and on other geometrical shapes (Figure S1D).
Having established the capacity to monitor sarcomere assembly dynamics in living cells, we investigated the effects of the predominant pathogenic TTNtv mutation in the myosin binding A-band region (Herman et al., 2012; Hinson et al., 2015), except where indicated homozygous TTNtv cells are reported. Visualizing actinin in cardiomyocytes containing A-band TTNtv revealed a breakdown of sarcomerogenesis, with a complete loss of coordinated movements of actinin puncta, centripetal fiber nucleation, Z-body fusion, and myofibril compaction (Figure 1D-H, Movie S5). Similarly sarcomere assembly was significantly impaired, though to a slightly lesser extent, in heterozygous A-band TTNtv cardiomyocytes (Movie S6). Particle tracking of actinin puncta showed coordinated procession towards the cell center in wild type cardiomyocytes (Figure 1E-G) whereas in A-band TTNtv cells actinin movement had decreased centripetal persistence (Figure 1E,F,H). Although disoriented formations of centripetal actinin fibers were occasionally observed in A-band TTNtv mutant cardiomyocytes, their existence was transient with frequent rupturing events (Figure 1H, arrows; Movie S3). Thus, truncations of the A-band result in a loss of directional actinin flow, centripetal fiber nucleation, Z-body fusion, and myofibril compaction, suggesting that titin is critical for the initial stages of sarcomere assembly.
Titin and myosin-dependent basal force generation is required for sarcomerogenesis
Sarcomere remodeling during heart development and disease are accompanied by changes in cardiomyocyte force generation (Kresh and Chopra, 2011). Cardiomyocytes exert two contractile tensions, active/systolic and resting/diastolic tension. Previously we demonstrated a deficit in systolic tension in I or A-band TTNtvs (Hinson et al., 2015), but the impact of titin truncation on diastolic tension generation and its role during sarcomerogenesis remain unexplored. Surprisingly, computation of diastolic stresses using traction force microscopy (TFM) revealed a dramatic decrease in diastolic traction stresses in A-band TTNtv cardiomyocytes (Figure 2A, B). Since titin normally links myosin to actinin, and A-band TTNtvs eliminate this coupling, we asked whether this myosingenerated contractility was important for sarcomere assembly. Treatment of wild type cardiomyocytes with the myosin ATPase inhibitor blebbistatin decreased diastolic traction stresses and decreased end-point sarcomere content (Figure 2A-E). Comparing the levels of diastolic force generation to sarcomere content across wild type, TTNtv, and blebbistatin conditions suggested that a basal level of tension is required to efficiently initiate and assemble sarcomeres (Figure 2C).
Figure 2: Titin and myosin-dependent basal force generation is essential for sarcomerogenesis.
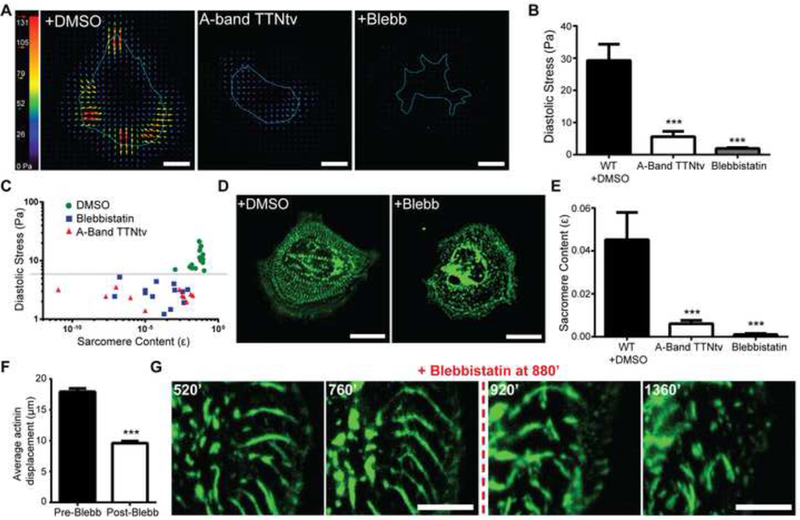
(A) Representative traction force microscopy stress maps of wild type, A-band TTNtv, and the myosin ATPase inhibitor blebbistatin treated cells. Scale bars 20 μm. (B) Average root mean square (RMS) diastolic traction stress (Pa) in wild type with DMSO, A-band TTNtv and 50 μM blebbistatin± treated cells. n=20 WT cells, n=15 A-band TTNtv cells, n=20 blebbistatin treated cells; across three independent experiments; mean ± s.e.m.; one-way ANOVA with Bonferroni multiple-comparison, ***P<0.0005. (C) Logarithmic plot of RMS diastolic traction stress (Pa) versus sarcomere content for wild type, A-band TTNtv, and blebbistatin treated cells. Each data point represents the sarcomere content and RMS diastolic traction stress exerted by the same cell. n=17 WT cells, n= 11 A-band TTNtv cells and n=16 blebbistatin treated cells; across three independent experiments. (D) Representative image of wild type cells stably expressing mApple-actinin treated with DMSO or 50μM blebbistatin±. Scale bars 20 μM (E) Quantification of sarcomere content comparing WT with DMSO to A-band TTNtv, and blebbistatin treated cells. n=20 WT cells, n=15 A-band TTNtv cells and n=20 blebbistatin treated cells; across three independent experiments; mean ± s.e.m.; one-way ANOVA with Bonferroni multiple-comparison, ***P<0.0005 (F) Quantitative comparison of average total actinin puncta displacement before and after blebbistatin treatment of the same cells. n=1034 pre-blebbistatin and n=934 post-blebbistatin actinin particles from five cells; across three independent experiments; mean ± s.e.m.; t-test, ***P<0.0005. (G) Representative time lapse images of mApple-actinin expressing wild type cell before and after blebbistatin treatment, indicating the disruption of existing centripetal actinin fibers and inhibition of sarcomerogenesis. Scale bars 20 μm.
Interestingly, quantification of mApple-tagged actinin dynamics revealed that blocking myosin activity resulted in abrogation of directed actinin flow and myofibrillar assembly (Figure 2D-F; Movie S2). Time lapse imaging of cardiomyocytes before and after addition of blebbistatin revealed that existing centripetal actinin fibers were acutely disrupted and formation of nascent centripetal fibers was prevented (Figure 2G; Movie S2). Together, these data indicate that transmission of myosin-generated contractility is required to initiate and maintain the process of actinin centripetal flow and sarcomere assembly.
β-cardiac myosin generated forces drive sarcomere assembly independent of nonmuscle myosin II
While blebbistatin abrogated the earliest stages of sarcomerogenesis, it remained unclear which of the specific myosin(s) are involved. Human cardiomyocytes express non-muscle myosin (NMM) IIA and IIB and α-and β-cardiac myosin (MHC) isoforms (Figure S2A-C), all of which are inhibited by blebbistatin (Limouze et al., 2004; Straight et al., 2003). Immunofluorescent localization during sarcomere assembly revealed that NMM IIA and IIB were restricted to the cell periphery (Figure 3A, Figure S2B, C), whereas the localization of MHC myosin was distributed, with a concentration in the cell center and extensions to the cell periphery (Figure S2B). Using CRISPR/Cas9-mediated genetic ablation, we individually knocked out NMM IIA or IIB and observed no defects in sarcomere assembly or content (Figure S2A, C). To account for any functional redundancy between the two isoforms, we generated cardiomyocytes with a dual knock out of NMM IIA and IIB. Surprisingly, loss of both NMM IIA and IIB also did not significantly impair sarcomere assembly or content (Figure 3A, B) compared to control, but did drastically alter cardiomyocyte morphology consistent with their known role in cortical stability (Vicente-Manzanares et al., 2007). These results indicate that non-muscle myosins IIA and IIB are not required for cardiomyocyte sarcomerogenesis, but do contribute to the maintenance of overall cardiomyocyte morphology, as was previously identified in NMM IIB-null mice (Tullio et al., 1997).
Figure 3: β-cardiac myosin generated forces drive sarcomere assembly.
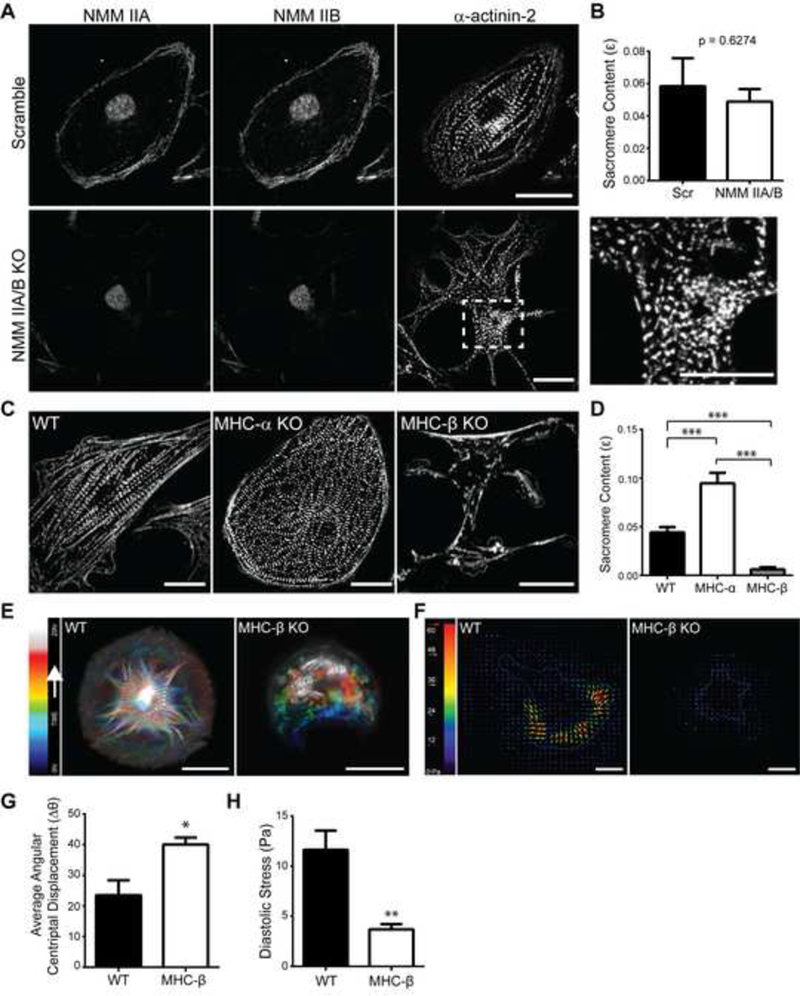
(A) Top and bottom panel (left to right) representative images of scramble (Scr) control cells and non-muscle myosin (NMM) IIA/B knock out cells stained for NMM IIA, NMM IIB, and actinin. (Inset) The presence of striated myofibrils in NMM IIA/B knock out cells. Scale bars 20 μm. (B) Quantification of sarcomere content comparing scramble and NMM IIA/B dual knock out cells. n=8 Scr cells, n=6 NMM IIA/B KO cells; across three independent experiments; mean ± s.e.m.; t-test. (C) Representative images of wild type, myosin heavy chain MHC-α and MHC-β knock out cells stained for actinin. Scale bars 20 μm. (D) Quantification of sarcomere content comparing wild type, MHC-a KO and MHC-β KO cells. n=18 WT cells, n=25 MHC-β KO cells and n=19 MHC-α KO cells; across three independent experiments; mean ± s.e.m.; one-way ANOVA with Bonferroni multiple-comparison, ***P<0.0005. (E) Temporal-color coded hyperstack of representative sarcomere assembly time lapse images of wild type and MHC-β KO cells stably expressing mApple-actinin shows the formation of patterned myofibrils in wild type and an absence in MHC-β KO cells. Scale bars 20 μm. (F) Representative stress maps of wild type and MHC-β KO cells. Scale bars 20 μm. (G) Quantification of angular centripetal displacement of actinin puncta towards the cell centroid in wild type and MHC-β KO cells. n=1184 tracked actinin particles across four wild type cells and n=962 particles across five MHC-β KO cells; across three independent experiments; mean ± s.e.m.; t-test, *P<0.05. (H) Average root mean square (RMS) diastolic traction stress (Pa) in wild type and MHC-β KO cells. n=15 WT cells and n=11 MHC-β KO cells; across three independent experiments; mean ± s.e.m.; t-test, **P<0.01. See also Figure S2.
We next investigated the roles of MHC-α and MHC-β, which are regulated during cardiac development and disease (Lyons et al., 1990), but their contributions to sarcomerogenesis remain unestablished. Using CRISPR/Cas9-mediated genetic ablation, we individually knocked out MHC-α or MHC-β in cardiomyocytes. Deletion of MHC-α did not impair sarcomere assembly or content in comparison to control cardiomyocytes (Figure 3C, D). However, ablation of MHC-β significantly inhibited both sarcomere assembly and content (Figure 3C, D, Figure S2D). Quantification of actinin dynamics and sarcomere assembly in MHC-β knock outs revealed decreased persistence of actinin flow and nearly a complete loss of centripetal actinin fiber nucleation (Figure 3E, G, Movie S7), similar to what was observed for blebbistatin treated and A- band TTNtv mutant cardiomyocytes. We also noted by TFM analysis that MHC-β was responsible for generating the basal diastolic tension we previously identified as critical for initiating and propagating sarcomere assembly (Figure 3F, H). Taken together these results indicate that MHC-β is the critical myosin motor that generates the tension propagated through titin to drive sarcomerogenesis.
Titin is a critical mechanical link between protocostameres and β-cardiac myosin generated forces to drive sarcomere assembly
Sarcomere assembly, while dependent on myosin-generated forces, appeared to initiate at centripetal actinin fibers emanating from peripheral sites that resembled sites of cell-matrix adhesion, protocostameres. Given the similarities in structure and function of protocostameres and focal adhesions, and the shared dependency on force for sarcomere assembly observed here and for focal adhesion assembly (Burridge and Guilluy, 2016), we investigated whether protocostameres were integral to sarcomerogenesis. Cardiomyocytes expressing tagged actinin were fixed during assembly and stained for paxillin. We observed a distinct co-localization of paxillin-containing protocostameres at distal origins of centripetal actinin fibers (Figure 4A), suggesting that protocostameres are the anchoring points for centripetal fiber nucleation. Indeed, time lapse imaging of cells expressing tagged forms of both paxillin and actinin revealed dynamic coordination and co-localization of centripetal fiber formation and protocostameres (Movie S8). A similar localization of paxillin containing protocostameres were observed in cardiomyocytes in 3D extracellular matrices (Figure S3D). We observed co-localization of MHC-β, but not NMM IIA/B, with actinin and paxillin at sites of centripetal fiber formation (Figure S3B). Additionally, immunostaining for paxillin in A-band TTNtv and MHC-β knockout cardiomyocytes showed a reduction in total protocostamere area (Figure 4A-C, Figure S2D), commensurate with the lower diastolic forces observed in these cells (Figure 2A, B, 3F,H). Taken together, these results suggest that the formation of protocostameres requires the coupling of MHC-β force generation through titin, and that these protocostameres may contribute to sarcomerogenesis.
Figure 4: Titin is a critical mechanical link between protocostameres and myosin generated forces to drive the sarcomere assembly.
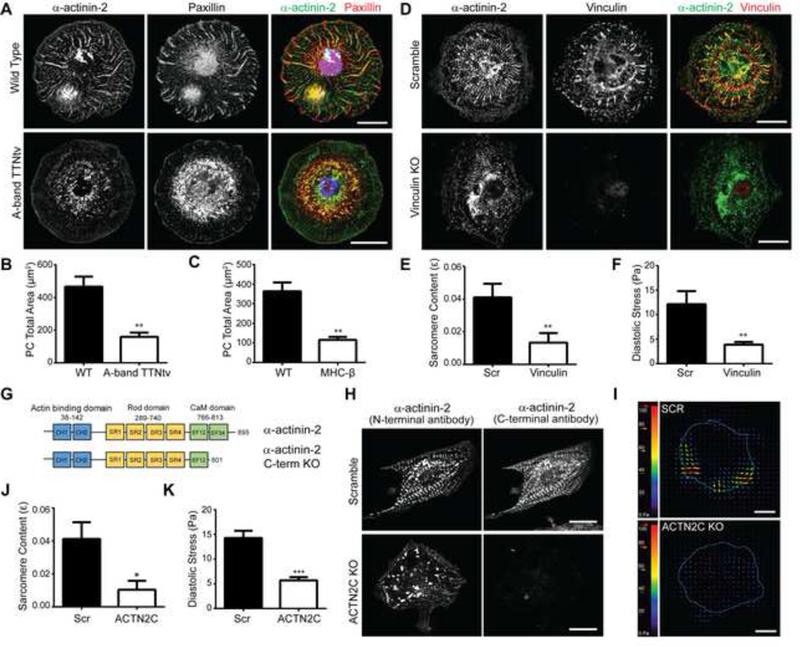
(A) Top and bottom panel (left to right) representative images of the basal surface of cells during nascent centripetal fiber formation stained for actinin (green) and paxillin (red) in wild type and A-band TTNtv cells, merge shows nuclear stain with DAPI in blue. Scale bars 20 μm. (B) Quantitative bar graph comparisons of average total protocostamere (PC) area present in WT vs A-band TTNtv cells and (C) WT vs MHC-P KO cells. n=21 wild type cells, n=16 A-band TTNtv cells and n=34 wild type cells, n=34 cells MHC-β KO cells; across three independent experiments; mean ± s.e.m.; t-tests, **P<0.01. (D) Top and bottom panel (left to right) representative images of cells stained for actinin (green) and vinculin (red) in scramble control and vinculin KO cells. Scale bars 20 μm. (E) Quantitative bar graph comparing the sarcomere content in scramble and vinculin KO cells. n=11 scramble cells, n=8 vinculin KO cells; across three independent experiments; mean ± s.e.m.; t-test, **P<0.01.(F) Average root mean square (RMS) diastolic traction stress (Pa) in scramble and vinculin KO cells. n=12 scramble cells, n=13 vinculin KO cells; across three independent experiments; mean ± s.e.m.; t-test, **P<0.01. (G) Diagram showing the CRISPR- mediated c-terminal actinin truncation of the titin binding domain. (H) Top and bottom panel (left to right) representative images of cells stained for N-terminal actinin (green) and C-terminal actinin (red) in scramble and actinin c-terminal knock out cells. Scale bars 20 μm. (I) Representative stress maps of scramble and ACTN2-C KO cells. Scale bars 20 μm. (J) Quantification of sarcomere content comparing wild type and ACTN2-C KO cells. n=10 scramble cells, n=16 ACTN2-C KO cells; across three independent experiments; mean ± s.e.m.; t-test, *P<0.05. (k) Average root mean square (RMS) diastolic traction stress (Pa) in scramble and ACTN2-C KO cells. n=15 Scr cells and n=12 ACTN2-C KO cells; across three independent experiments; mean ± s.e.m.; t-test, ***P<0.0005. See also Figure S3.
To test whether global disruption of protocostameres impaired sarcomerogenesis, we used CRISPR/Cas9 to deplete vinculin, a mechanosensitive scaffolding protein that is critical for focal adhesion maturation (Grashoff et al., 2010; Humphries et al., 2007), links actinin to protocostameres in cardiomyocytes (Lu et al., 1992; Zemljic-Harpf et al., 2007), and is a genetic cause of cardiomyopathy (Harvey and Leinwand, 2011) (Figure 4D, Figure S3A). Indeed, knock out of vinculin resulted in a loss of sarcomere content (Figure 4E). Imaging of actinin dynamics in vinculin knockout cells revealed a loss of centripetal fiber nucleation, similar to A-band TTNtv and MHC-β knock out cardiomyocytes (Movie S9). A loss in diastolic tractions stress was also observed with vinculin depletion (Figure 4F, Figure S3C). These results suggest that protocostameres are critical nucleation sites for sarcomerogenesis that provide mechanical resistance against MHC-β generated forces required to drive centripetal actinin fiber formation.
We next tested whether titin is the molecular link that couples MHC-β force generation to protocostamere-anchored actinin polymerization. To decouple titin binding from protocostameres, we used CRISPR/Cas9 genomic editing to truncate actinin at the C-terminal titin binding EF3/4 domain (Ribeiro Ede et al., 2014; Young and Gautel, 2000) in cardiomyocytes (denoted ACTN2-C) (Figure 4G, H, Figure S3A). While actinin localized to protocostameres, we observed a significant loss in sarcomere content in ACTN2-C cardiomyocytes similar to that observed for A-band TTNtv and MHC-β knock out cells (Figure 4H,J). Similarly, assembly dynamics were impaired due to the inability to form stable centripetal actinin fibers (Movie S10). Diastolic traction stresses exerted by ACTN2-C cells were significantly lower than controls, comparable to A-band TTNtv and MHC-β knockout cells (Figure 4I, K). Taken together, these results suggest that titin forms an essential mechanical connection between the force generating MHC-β and the protocostamere-actinin (Z-body) complex during sarcomerogenesis. This mechanical connection provides the force transmission needed to promote and direct centripetal actinin fiber nucleation and assembly from protocostameres, ultimately leading to Z-body fusion and striation.
Discussion
Earlier studies of cardiac myofibrillar assembly used non-human (Dabiri et al., 1997; Ehler et al., 1999; Ferrari et al., 1998; Gerull et al., 2002; Schultheiss et al., 1990; Tokuyasu and Maher, 1987; Tullio et al., 1997; van der Ven et al., 2000) or non-cardiac cell (Hill et al., 1986; Miller et al., 2003; Sanger et al., 2009; White et al., 2014) models wherein de novo sarcomere assembly dynamics have not been easily observed. Here we use live imaging of genetically modified human stem cell derived cardiomyocytes to demonstrate that sarcomere assembly is a highly dynamic and coordinated process. While recognizing that iPSC-derived cardiomyocytes have limitations and cannot fully recapitulate in vivo processes, the immaturity of these cells is particularly suited to studying the embryonic process of sarcomere formation (DeLaughter et al., 2016; Kuppusamy et al., 2015). We identify that nucleation and centripetal assembly of actinin fibers at protocostameres is a primary, critical step in sarcomerogenesis. Interestingly, similar tension bearing centripetal fiber-like structures containing skeletal muscle myosin have been observed in an in vivo Drosophila flight muscle model before the appearance of mature striated myofibrils (Weitkunat et al., 2014). Previous studies, including our publications have speculated that A-band truncations of titin result in sarcomere insufficiency in part through changes in signal transduction (Gerull et al., 2002; Hinson et al., 2015; Musa et al., 2006; van der Ven et al., 2000). Here, our findings suggest that A-band TTNtvs impair sarcomerogenesis specifically by disrupting mechanical force transmission from myosin, through titin and actinin, to protocostameres. This mechanical model of centripetal assembly provides a new starting point to better understand the sarcomere assembly process.
The role of specific myosins during sarcomere assembly has until now remained unclear (Dabiri et al., 1997; Tullio et al., 1997). Previous studies altering substrate stiffness and utilizing blebbistatin have indirectly alluded to NMM II as being the primary motor responsible for early stage sarcomerogenesis, but this attribution has not been genetically demonstrated (Chopra et al., 2011; Majkut et al., 2013; Ribeiro et al., 2015). In contrast to these studies, using CRISPR-mediated genomic deletion in a model of de novo sarcomere assembly, we show for the first time that MHC-β, but not NMM II or MHC-α, is the key myosin that generates force critical for sarcomere assembly. Importantly, this finding sheds mechanistic insight onto the fact that nearly all pathogenic myosin mutations are in MHC-β, and that these mutations encode missense residues (Fatkin et al., 2014). Based on these studies, we suggest that there is substantial evolutionary constraint against MHC-β truncations, unlike other sarcomere proteins. Interestingly, while we identify that NMM II is dispensable for sarcomerogenesis, we identify a clear function in regulating cardiomyocyte morphology and cortical stability.
By disrupting the connection to MHC-β force generation (TTNtv) or to protocostameres (ACTN2-C), we identify that titin provides a critical, bi-directional mechanical link between MHC-β and protocostameres to drive sarcomere assembly. Consistent with our results, knocking out vinculin in mice embryos was also shown to form a non-beating cardiac region without any apparent sarcomeric structures (Xu et al., 1998). Importantly, the identified mechanism offers new possibilities into how physical changes in the cardiac microenvironment such as hemodynamic load or matrix stiffness are transduced through costameres into changes in cardiac structure and sarcomere organization. We suggest that these mechanisms can enable improved maturation of cardiomyocytes derived from iPSCs. The combination of genome editing, human iPSC technology and real time visualization of sarcomere assembly dynamics utilized here is a powerful new approach to assess the mechanistic implications of pathogenic mutations on cardiac sarcomere structure and motivates new therapeutic approaches to their intervention.
STAR METHODS
Contact For Reagent and Resource Sharing
Further information and request for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Christopher S. Chen (chencs@bu.edu).
Experimental Model and Subject Details
All patient samples were obtained, cryopreserved and reprogrammed after informed written consent using protocols that were approved by the Institutional Review Board of Partners HealthCare. Heterozygous A-band TTNtv t-cells were taken from patient I.D. MEK-111, male 15 years of age. Endogenously tagged GFP-paxillin expressing iPSCs were provided by the Allen Institute for Cell Science Human iPSC Collection, parental iPSC’s isolated from fibroblasts, male 30 years of age. Wild type cells I.D. Personal Genome Project (PGP1) were taken from isolated from fibroblasts, male 53 years of age.
Method Details
iPSC production and cardiomyocyte differentiation
Patient-specific iPSCs harboring heterozygous A-band TTNtv mutations as well as the wild types were produced from cryopreserved T-cells using STEMCCA lentivirus as described previously (Hinson et al., 2015). iPSCs were screened for copy number variants and virtual karyotyping using Illumina HumanOmniExpress-12v1 arrays. iPSCs were maintained in complete mTeSR1 medium (Stem Cell) and differentiated to the CM lineage in RPMI 1640 medium (Gibco) supplemented with B27 minus insulin (ThermoFisher) by sequential targeting of the WNT pathway - activating WNT pathway using 12μM of CHIR99021 (Tocris) in Day1 and inhibiting WNT pathway using 5μM of IWP4 (Tocris) in Day3 and Day4. CMs were isolated after showing spontaneous beating (usually between Day9 to Day14) using metabolic selection by adding 4mM of DL-lactate (Sigma) in glucose free DMEM medium (ThermoFisher) for two days and CM purity was determined by FACS analysis (>90% Troponin T+). Following selection, CMs were maintained and assayed in RPMI 1640 medium supplemented with B27 (ThermoFisher) and studied on Day30 post initiation of differentiation. Endogenously tagged GFP-paxillin expressing iPSCs were provided by the Allen Institute for Cell Science Human iPSC Collection and cultured as described above.
CRISPR/Cas9 iPSC gene editing, recombinant cloning
CRISPR/Cas9-mediated genetic modifications were generated using both scarless iPSC clonal selection and lentiviral batch infection approaches. For homozygous A-band TTNtv, MYH6, and MYH7 knockouts, the scarless, iPSC gene-editing protocol was used as described previously (Hinson et al., 2015). iPSCs were sorted by FACS for GFP+, clonally expanded, and Sanger sequenced for genotyping. For scramble control, MYH9, MYH10, VCL, and ACTN2 C-terminal knockouts, stable CRISPR knockout cell lines were generated using the lentiCRISPRv2 system (gift from Feng Zhang, Addgene plasmid #52961). Specific gRNAs were cloned into the BsmBI site of plentiCRISPRv2. Individual gRNA-containing plentiCRISPRv2 plasmids were co-transfected with pVSVG, pRSV-REV, and pMDLg/pRRE packaging plasmids into HEK-293T cells using calcium phosphate transfection. After 48 hours, viral supernatants were collected, concentrated using PEG-IT viral precipitator (SBI), and re-suspended in PBS. Cardiomyocytes were transduced in growth media overnight and media was replaced the following morning. 48 hours post infection; cells were selected with 2.5 μg/ml puromycin for 3–4 days. All CRISPR modifications were verified by western blot.
The following guide RNAs were used: Scramble: 5’-GCACTACCAGAGCTAACTCA-3’, MYH9: 5’-TCAAGGAGCGTTACTACTCA-3’, MYH10: 5’-TGGATTCCATCAGAACGCCA-3’, VCL: 5’-GCGCACGATCGAGAGCATCC-3’, ACTN2 C KO: 5’-ATTTGCCCGCATTATGACCC-3’, MYH6: 5’-CTGCCGGTGTACAATGCCG-3’, MYH7: 5’-ACTCAGCCGACCTGCTCAA-3’.
Human actinin-2 (Addgene #52669) was PCR amplified and used to make N-terminal GFP and mApple-tagged constructs in a modified pRRL lentiviral expression vector. For live GFP-paxillin movies, iPSCs with endogenously tagged GFP-paxillin were utilized. All constructs were expressed via lentiviral transduction as described for CRISPR knockouts.
Microcontact printing
Micropatterened substrates were prepared as described previously(Hinson et al., 2015). Briefly, PDMS stamps were created from patterned silicon masters that were generated using photolithography. Glass substrates were spin coated with PDMS and polymerized in a 60°C oven overnight. PDMS stamps were coated with 50μg/ml fibronectin for 1 hr in room temperature and stamped onto PDMS coated glass substrates that were pre-activated in a UV-Ozone cleaner (Jelight) for 7 minutes. Patterned substrates were treated with 0.2% pluronic F127 (Sigma) for 1 hour before cell seeding to prevent non-specific adhesion of cells to the un-patterned areas of the substrate.
Antibodies and inhibitors
Anti-paxillin (349, 1:100) was from BD Biosciences. Anti-vinculin (hVin1, 1:5000) was from Sigma. Anti-actinin2 (ab9465, 1:500) was from Abcam. Anti-actinin2 c-terminal (70R-1068, 1:200) was from Fitzgerald. Anti-NMM IIA (PRB-440P, 1:1000) and NMM IIB (PRB-445P, 1:1000) were from Biolegend. Anti-NMM IIB was directly conjugated with Alexa Fluor 568 (ThermoFisher), allowing the simultaneous immunofluorescent staining of NMM IIA and NMM IIB (Figure S2b). Anti-MHC (MF-20, 1:1000) was from DSHB. Anti-MHC-α (22281–1-AP, 1:5000) and Anti-MHC-β (22280–1-AP, 1:5000) were from Protein Tech Group (Chicago, IL). Blebbistatin± (racemic) was from CalBioChem.
Immunofluorescence and western blotting
Cardiomyocytes cultured in growth medium were perm/fixed with 1% paraformaldehyde/0.03% Triton X-100 in PBS containing calcium and magnesium (PBS+) at 37C for 90 seconds. Cells were immediately post-fixed in 4% paraformaldehyde in PBS+ at 37C for 15 minutes. Cells were rinsed three times with PBS+ and permeabilized with 0.5% Triton X-100 in PBS+ for 10 minutes. Cells were blocked with 2% BSA in PBS+. Primary and secondary antibodies were applied in 2% BSA in PBS+ and rinsed three times over 30 min with PBS+ between each treatment. For western blot analyses, CMs were lysed 2X NuPAGE® LDS Sample Buffer (Life Technologies) containing 100 mM DTT and analyzed by SDS-PAGE and immunoblotting with chemiluminescent HRP detection. Immunofluorescent images and western blots were adjusted for brightness and contrast using ImageJ software (NIH, Bethesda, MD).
Microscopy
For live cell imaging, cardiomyocytes were plated onto fibronectin micropatterned substrates in growth media containing OxyFluor (Oxyrase). 24 to 48 hours post plating, CMs were transferred to a temperature and CO2 equilibrated environmental microscope chamber and imaged every 20 minutes for 24 hours on either a Nikon Ti Eclipse epifluorescence microscope with a Yokogawa CSU-21/Zeiss Axiovert 200M inverted spinning disk microscope with a Zeiss LD C-Apochromat 40x or 63x, 1.1 N.A. water-immersion objective and an Evolve EMCCD camera (Photometrics) or a Nixon Plan Fluor 40x, 1.3 N.A. and ORCA-100 camera (Hamamatsu).
Polyacrylamide gel for traction force microscopy
Polyacrylamide gels of desired stiffness were made by adjusting acrylamide and bisacrylamide stock solution (Bio-Rad Laboratories, Hercules, CA) concentrations (Chopra et al., 2011). A solution of 40% acrylamide, 2% bisacrylamide and 1xPBS was polymerized by adding tetramethylethylene diamine (Fisher BioReagents) and 1% ammonium persulfate. A droplet of the gel solution supplemented with 0.2 μm fluorescent beads solution (Molecular Probe, Fisher Scientific) was deposited on a quartz slide (Fisher Scientific) and covered with a 25-mm glass (Fisher) coverslip pretreated with 3-aminopropyltrimethoxysilane (Sigma-Aldrich) and glutaraldehyde (Sigma-Aldrich). After polymerization, the gel surface attached to the quartz slide was functionalized with fibronectin via EDC-NHS chemistry. Briefly, the gel surface was activated in a UV-Ozone cleaner (Jelight) for 2 minutes, detached from the quartz slide, soaked in a solution with EDC and NHS for 15 minutes and incubated with 50 μg/mL fibronectin solution at 37°C for 2 hours. The gel was sterilized and stored in 1X PBS before cell seeding.
Micropatterened polyacrylamide hydrogels
Micropatterned polyacrylamide hydrogels were prepared by adopting a previously described method(Polio et al., 2012). Briefly, PDMS stamps were coated with Matrigel (VWR) overnight at 4°C and stamped onto plasma activated 25-mm glass (Fisher) substrate. Polyacrylamide gel solution of desired stiffness (4kPa) was made as described in above section. A droplet of the pre-polymer polyacrylamide solution was sandwiched in between the patterned glass substrate and a 25-mm glass (Fisher) coverslip pretreated with 3-aminopropyltrimethoxysilane (Sigma-Aldrich) and glutaraldehyde (Sigma-Aldrich). After polymerization, the gel was soaked in IxPBS for 1 hour before being detached from the pre-patterned glass substrate. The gel was sterilized and stored in 1X PBS before cell seeding.
Engineered heart microtissues (utissues)
Cardiac μtissues were prepared as previously described(Boudou et al., 2012). Briefly, PDMS microfabricated tissue gauges (μTUG) substrates were molded from the SU-8 masters, with embedding fluorescent microbeads (Fluoresbrite 17147; Polysciences, Inc.) onto the cantilevers. μTUG substrates were treated with 0.2% pluronic F127 for 30 minutes to prevent cell-extracellular matrix iPSC-CMs were dissociated after trypsin digestion and mixed with stromal cells (human mesenchymal stem cells, hMSCs) to enable tissue compaction. The amount of stromal cells added was 30% of the total myocyte population. A suspension of ~20,000 cells within reconstitution mixture, consisting of 2.25mg/mL liquid neutralized collagen I (BD Biosciences) and 0.5mg/mL human fibrinogen (Sigma-Aldrich), was added to the substrate. The device was centrifuged to drive the cells into the micropatterned wells. The cell density was kept low to prevent high tissue displacement due to cardiomyocyte contraction. Within 24 hours hMSCs compacted tissues. Only in compacted tissues did we observe sarcomere assembly in cardiomyocytes. For live imaging cardiomyocytes were imaged as mentioned previously (See microscopy and image analysis section). Image slices at 1μm were taken with a total thickness of 15μm.
Quantification and Statistical Analysis
Image analysis and quantification
For quantifying protocostamere area, cardiomyocytes were immunostained for paxillin and acquired at 63x. Paxillin images for all conditions were equally intensity thresholded and adhesions areas were quantified using ImageJ.
For tracking of actinin puncti, time lapse recordings of GFP/mApple labelled actinin expressing cardiomyocytes were obtained as described. Centroid coordinates for individual cardiomyocytes were calculated over time using custom MATLAB scripts (Mathworks, Natick, MA). Time lapse image stacks were imported into IMARIS software (Bitplane AG, Zurich, Switzerland). Using IMARIS particle tracking algorithm, actinin puncta were first detected, equally size and intensity thresholded, and the displacement vectors of each puncta within a cardiomyocyte were calculated overtime. Tracking was kept consistent between each tested condition and image sets were unbiasedly batch processed in IMARIS. The Cartesian coordinates for the displacement of tracked actinin puncti over time for each cardiomyocytes were processed using a custom routine in MATLAB. To calculate the angular displacement, the Cartesian coordinates of each puncta were converted to polar coordinates. The centroid computed for each cardiomyocyte was set as the origin of the polar coordinate system in order to compute the angular displacement (Δθ) of each particle towards the cell center. Particle angular displacement Δθ >180° was normalized by subtracting from 360°.
Sarcomere content analysis (FFT)
Images of single cardiomyocytes on fibronectin coated micropatterned substrate and polyacrylamide gels expressing tagged α-actinin-2 or stained for α-actinin-2 were processed by a custom routine in ImageJ (NIH, Bethesda, MD) and MATLAB (Mathworks, Natick, MA), as described previously (Hinson et al., 2015). Briefly, the spectra of 2D Fourier transforms of α-actinin-2 images were converted to a 1D representation by summing the intensity as a function of distance from the center of the 2D image. The 1D spectra were then normalized and fit to a function composed with an aperiodic component to account for irregular α-actinin-2 cellular structures and a periodic component that represents the regularly spaced sarcomeres. The normalized area under the first nonzero peak of the periodic component was taken as a measure of the regularity of the sarcomere structure.
Traction stress measurements
The traction forces exerted iPS-CMs on the gel substrates were computed by measuring the displacement of fluorescent beads embedded within the gel as described previously(Tseng et al., 2012) (Software available at https://sites.google.com/site/qingzongtseng/tfm). Briefly, images of bead motion near the substrate surface, distributed in and around the contact region of a single cell (before and after cell detachment with 10% sodium dodecyl sulfate, were acquired with Yokogawa CSU-21/Zeiss Axiovert 200M inverted spinning disk microscope with a Zeiss LD C-Apochromat 40×, 1.1 N.A water-immersion objective and an Evolve EMCCD camera (Photometrics). The traction stress vector fields were generated using an open source package of Fiji plugins (developed by Tseng, Q.(Tseng et al., 2012)). Root mean squares of the magnitudes of single cell stress vectors were computed using the formula: , in which Sn is a single stress vector.
Statistics
Sample sizes and P values are reported in each figure legend and statistical analyses were performed using GraphPad Prism 5.0. In the figures, unless otherwise noted, graphs show mean ± standard error of the mean (s.e.m.). When experiments involved only a single pair of conditions, statistical differences between the two sets of data were analyzed with a two-tailed, unpaired Student t-test assuming unequal variances. For data sets containing more than two samples, one-way ANOVA with a classical Bonferroni multiple-comparison post-test was used to determine adjusted P values. Sample sizes of sufficient power were chosen on the basis of similar published research and were confirmed statistically by appropriate tests. When representative images are shown, these represent at least three independent samples.
Supplementary Material
Movie S1. Related to Figure 1. Dynamic sarcomerogenesis in human cardiomyocytes.
Representative wild type cardiomyocyte expressing GFP-actinin was plated onto an isotropic fibronectin-coated circular pattern overnight. Sarcomere assembly time lapse imaging was started the following morning for 24 hours. 20 minute frame rate, accelerated 12,000x for display. Scale bar indicates 20 μm, timescale hours:minutes.
Movie S9. Related to Figure 4. Protocostameres are required for sarcomerogenesis.
Representative vinculin knock out cardiomyocyte expressing GFP-actinin was plated onto an isotropic fibronectin-coated circular pattern overnight. Sarcomere assembly time lapse imaging was started the following morning. Ablating protocostameres leads to an absence of centripetal fiber formation. 20 minute frame rate, accelerated 12,000x for display. Scale bar indicates 20 μm, timescale hours:minutes.
Movie S10. Related to Figure 4. Mechanical linking of titin to protocostameres drives sarcomerogenesis.
Representative ACTN2-C knock out cardiomyocyte expressing GFP-actinin was plated onto an isotropic fibronectin-coated circular pattern overnight. Sarcomere assembly time lapse imaging was started the following morning. 20 minute frame rate, accelerated 12,000x for display. Scale bar indicates 20 μm, timescale hours:minutes.
Movie S2. Related to Figures 1 and 2. Inhibition of myosin contractility acutely impairs sarcomerogenesis.
Wild type cardiomyocytes expressing mApple-actinin were plated onto fibronectin-coated isotropic circular pattern (left panel) or anisotropic rectangular pattern (right panel) overnight. Note the presence of transverse and centripetal actinin fibers on the rectangular substrate. Sarcomere assembly time lapse imaging was started the following morning. Blebbstatin± (50 μM) was added during the time lapse (+blebbistatin) leading to acute breakdown of existing centripetal fibers and inhibition of sarcomerogenesis. 20 minute frame rate, accelerated 12,000x for display. Scale bars indicate 20 μm, timescale hours:minutes.
Movie S3. Related to Figure 1. Dynamic sarcomerogenesis in human cardiomyocytes on substrates of physiologically relevant stiffness
Representative wild type cardiomyocyte expressing GFP-actinin was plated onto a fibronectin-coated rectangular pattern on compliant polyacrylamide gel (4kPa) overnight. Centripetal and transverse fibers contribute to myofibril assembly from points of adhesion, though Z-bodies along the sides of these cells tended to fuse and align along the sides of the long, parallel myofibrils, while Z-bodies at the ends tended to extended existing myofibrils. Sarcomere assembly time lapse imaging was started the following morning for 24 hours. 20 minute frame rate, accelerated 12,000x for display. Scale bar indicates 20 μm, timescale hours:minutes.
Movie S4. Related to Figure 1. Dynamic sarcomerogenesis in human cardiomyocytes within 3D collagen in engineered heart microtissues.
Representative wild type cardiomyocytes expressing GFP-actinin embedded within 3D collagen matrix in an engineered heart microtissue. Sarcomere assembly time lapse imaging was started 24 hours after making the microtissue. Large black arrows indicate transverse actinin flow, and small gray arrows indicate centripetal actinin fibers. 20 minutes frame rate, accelerated 12,000x for display. Scale bar indicates 20 μm, timescale hours:minutes.
Movie S5. Related to Figure 1. Titin truncation mutations abrogate sarcomere assembly.
Representative A-band TTNtv cardiomyocyte expressing GFP-actinin was plated onto an isotropic fibronectin-coated circular pattern overnight. Transient centripetal fibers were observed occasionally with frequent rupturing events (left panel) and in many cases the presence of centripetal fibers was completely absent (right panel), all cases had no initiation of sarcomere assembly. Few organized actinin puncta or Z-bodies were observed and transverse actinin flow was absent. Sarcomere assembly time lapse imaging was started the following morning. 20 minute frame rate, accelerated 12,000x for display. Scale bar indicates 20 μm, timescale hours:minutes.
Movie S6. Related to Figure 1. Patient-derived cardiomyocytes with heterozygous titin truncations have impaired sarcomere assembly.
Representative patient derived cardiomyocyte harboring heterozygous A-band TTNtv expressing GFP-actinin was plated onto an isotropic fibronectin-coated circular pattern overnight. Transverse actinin flow and relatively stable centripetal fibers were observed compared to homozygous A-band TTNtv mutations, however given the slow rate of sarcomere assembly relatively few or no myofibirils could be observed. Sarcomere assembly time lapse imaging was started the following morning. 20 minute frame rate, accelerated 12,000x for display. Scale bar indicates 20 μm, timescale hours:minutes.
Movie S7. Related to Figure 3. β-cardiac myosin is critical for sarcomerogenesis.
Representative MHC-β knock out cardiomyocyte expressing GFP-actinin was plated onto an isotropic fibronectin-coated circular pattern overnight. Centripetal and transverse actinin fiber assembly was disrupted leading to no sarcomere assembly. Sarcomere assembly time lapse imaging was started the following morning. 20 minute frame rate, accelerated 12,000x for display. Scale bar indicates 20 μm, timescale hours:minutes.
Movie S8. Related to Figure 4. Dynamic role of protocostomeres during sarcomerogenesis.
Representative wild type cardiomyocyte endogenously expressing GFP-paxillin (left panel, shown in red in merge right panel) and transduced to co-express mApple-actinin (middle panel, shown in green in merge right panel) was plated onto an isotropic fibronectin-coated circular pattern overnight. Sarcomere assembly time lapse imaging was started the following morning. Dynamic co-localization of paxillin and actinin is observed, with centripetal fibers emanating from paxillin-containing protocostameres. 20 minute frame rate, accelerated 12,000x for display. Scale bar indicates 20 μm, timescale hours:minutes.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-paxillin | BD Biosciences | 610051, RRID:AB_397463 |
| Mouse anti-vinculin | Sigma-Aldrich | V9131, RRID:AB_477629 |
| Rabbit anti-non-muscle myosin IIA | Biolegend | 909801, RRID:AB_2565100 |
| Rabbit anti-non-muscle myosin IIB | Biolegend | 909901, RRID:AB_2565101 |
| Mouse anti-cardiac myosin heavy chain | Developmental Studies hybridoma bank | MF-20, RRID:AB_2147781 |
| Rabbit anti-cardiac myosin heavy chain α | Proteintech | 22281–1-AP |
| Rabbit anti-cardiac myosin heavy chain β | Proteintech | 22280–1-AP |
| Rabbit anti-beta Tubulin | Abcam | ab6046, RRID:AB_2210370 |
| Rabbit anti-actinin2 (c-terminal) | Fitzgerald | 70R-1068, RRID:AB_10809287 |
| Mouse anti-actinin2 | Abcam | ab9465, RRID:AB_307264 |
| Alexa 488-goat anti Mouse IgG | Thermo Fisher | R37120, RRID:AB_2556548 |
| Alexa 647-goat anti Rabbit IgG | Thermo Fisher | A27040, RRID:AB_2536101 |
| HRP-donkey anti Mouse IgG | Fitzgerald | 43-GM30, RRID:AB_1287764 |
| HRP-donkey anti Rabbit IgG | Fitzgerald | 43C-CB1134, RRID:AB_1287213 |
| Bacterial and Virus Strains | ||
| Stbl3 Competent Cells | Thermo Fisher | C737303 |
| Biological Samples | ||
| Cryopreserved T-cells | Hinson et.al Science, 2015 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Blebbistatin | Calbiochem | 203390, CAS# 856925–71–8 |
| CHIR99021 | Tocris | 4423, CAS# 25291706–9 |
| IWP4 | Tocris | 5214, CAS#686772–17–8 |
| DL-lactate | Sigma | L7900, CAS# 72–173 |
| DAPI | Sigma | 10236276001, CAS#28718–90–3 |
| Fibronectin | BD Biosciences | 356009 |
| 40% acrylamide solution | BIO-RAD | 1610140 |
| 2% Bis Solution | BIO-RAD | 1610143 |
| N,N,N′,N′-Tetramethylethylenediamine | Sigma-Aldrich | T9281, CAS#110–18–9 |
| Ammonium persulfate | Sigma-Aldrich | A3678, CAS#7727–54–0 |
| NHS (N-hydroxysuccinimide) | Thermo Fisher | 24500 |
| EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride) | Thermo Fisher | 22980, CAS#25952–53–8 |
| Fluospheres Carboxylate-Modified Microspheres, 0.2 μm | Thermo Fisher | F8807 |
| Alexa 488-phalloidin | Thermo Fisher | A12379 |
| Alexa 568-phalloidin | Thermo Fisher | A12380 |
| Alexa 568-NHS Ester | Thermo Fisher | A2003 |
| Pierce™ ECL Western Blotting Substrate | Thermo Fisher | 32106 |
| Pluronic F127 | Sigma | P2443, CAS# 9003–11–6 |
| SU8 photoresist | Thermo Fisher | NC0060520 |
| Experimental Models: Cell Lines | ||
| Human Induced pluripotent stem cells (hIPSC) | Hinson et.al Science, 2015 | N/A |
| hIPSC MYH7−/− | This paper | N/A |
| hIPSC MYH6−/− | This paper | N/A |
| HEK 293T | Clonetech | 632180 |
| EGFP tagged Paxillin hIPSC | Allen Institute | N/A |
| Oligonucleotides | ||
| Scramble gRNA (5’-GCACTACCAGAGCTAACTCA-3’) | This paper | N/A |
| MYH9 gRNA (5’-TCAAGGAGCGTTACTACTCA-3’) | This paper | N/A |
| MYH10 gRNA (5’-TGGATTCCATCAGAACGCCA-3’) | This paper | N/A |
| VCL gRNA (5’-GCGCACGATCGAGAGCATCC-3’) | This paper | N/A |
| MYH6 gRNA (5’-CTGCCGGTGTACAATGCCG-3’) | This paper | N/A |
| MYH7 gRNA (5’-ACTCAGCCGACCTGCTCAA-3’) | This paper | N/A |
| ACTN2C gRNA (5’-ATTTGCCCGCATTATGACCC-3’) | This paper | N/A |
| Recombinant DNA | ||
| plentiCRISPR v2 | Addgene | 52961 |
| pLenti CMV GFP Puro | Addgene | 17448 |
| pEGFP-ACTN2 | Addgene | 52669 |
| pLenti CMV GFP-ACTN2 Puro | This paper | N/A |
| pLenti CMV mApple-ACTN2 Puro | This paper | N/A |
| plentiCRISPR v2 MYH9 gRNA | This paper | N/A |
| plentiCRISPR v2 MYH10 gRNA | This paper | N/A |
| plentiCRISPR v2 ACTN2C gRNA | This paper | N/A |
| plentiCRISPR v2 VCL gRNA | This paper | N/A |
| Software and Algorithms | ||
| Matlab R2015B | MathWorks (https://www.mathworks.com/) | RRID:SCR_001622 |
| Graphpad Prism 6 | http://www.graphpad.com | RRID:SCR_002798 |
| Imaris | BITPLANE (http://www.bitplane.com/imaris/imaris) | RRID:SCR_007370 |
| FIJI (ImageJ) | https://fiji.sc/# | RRID:SCR_002285 |
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (HL084553 and HL080494 to C.E.S. and J.G.S and HL125807 to J.T.H) and the Engineering Research Centers Program of the National Science Foundation (No. EEC-1647837). A.C. acknowledges fellowship support from the American Heart Association, M.L.K acknowledges fellowship support from The Hartwell Foundation and from the NIH through the Translational Research in Regenerative Medicine Training program (T32 EB005583), K.Z. acknowledges fellowship support from the American Heart Association, and W.J.P. acknowledges financial support from a Ruth L. Kirchstein National Research Service Award (HL129733). C.E.S. and J.G.S. acknowledge support from the Leducq Fondation and C.E.S. from the Howard Hughes Medical Institute. C.C.S. acknowledges fellowship support from the Sarnoff Foundation. Endogenously tagged GFP-paxillin iPSCs were generously provided by the Allen Institute for Cell Science Human iPSC Collection.
Footnotes
Conflict of Interest Disclosures: JGS and CES are founder and owns shares in Myokardia Inc., a startup company that is developing therapeutics that target the sarcomere. CSC is a founder and share owner of Innolign Biomedical, a company that is developing engineered organ models for pharmaceutical research and development. TH receives grant funding from Myokardia to study dilated cardiomyopathy.
REFERENCES
- Anderson BR, and Granzier HL (2012). Titin-based tension in the cardiac sarcomere: molecular origin and physiological adaptations. Prog Biophys Mol Biol 110, 204–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birket MJ, Ribeiro MC, Kosmidis G, Ward D, Leitoguinho AR, van de Pol V, Dambrot C, Devalla HD, Davis RP, Mastroberardino PG, et al. (2015). Contractile Defect Caused by Mutation in MYBPC3 Revealed under Conditions Optimized for Human PSC-Cardiomyocyte Function. Cell Rep 13, 733–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudou T, Legant WR, Mu A, Borochin MA, Thavandiran N, Radisic M, Zandstra PW, Epstein JA, Margulies KB, and Chen CS (2012). A microfabricated platform to measure and manipulate the mechanics of engineered cardiac microtissues. Tissue Eng Part A 18, 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray MA, Sheehy SP, and Parker KK (2008). Sarcomere alignment is regulated by myocyte shape. Cell motility and the cytoskeleton 65, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K, and Guilluy C (2016). Focal adhesions, stress fibers and mechanical tension. Exp Cell Res 343, 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JJ, Yang X, Don CW, Minami E, Liu YW, Weyers JJ, Mahoney WM, Van Biber B, Cook SM, Palpant NJ, et al. (2014). Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature 510, 273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra A, Tabdanov E, Patel H, Janmey PA, and Kresh JY (2011). Cardiac myocyte remodeling mediated by N-cadherin-dependent mechanosensing. American journal of physiology Heart and circulatory physiology 300, H1252–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabiri GA, Turnacioglu KK, Sanger JM, and Sanger JW (1997). Myofibrillogenesis visualized in living embryonic cardiomyocytes. Proceedings of the National Academy of Sciences of the United States of America 94, 9493–9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaughter DM, Bick AG, Wakimoto H, McKean D, Gorham JM, Kathiriya IS, Hinson JT, Homsy J, Gray J, Pu W, et al. (2016). Single-Cell Resolution of Temporal Gene Expression during Heart Development. Dev Cell 39, 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehler E, Rothen BM, Hammerle SP, Komiyama M, and Perriard JC (1999). Myofibrillogenesis in the developing chicken heart: assembly of Z-disk, M-line and the thick filaments. J Cell Sci 112 (Pt 10), 1529–1539. [DOI] [PubMed] [Google Scholar]
- Fatkin D, Seidman CE, and Seidman JG (2014). Genetics and disease of ventricular muscle. Cold Spring Harbor perspectives in medicine 4, a021063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari MB, Ribbeck K, Hagler DJ, and Spitzer NC (1998). A calcium signaling cascade essential for myosin thick filament assembly in Xenopus myocytes. J Cell Biol 141, 1349–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardel ML, Sabass B, Ji L, Danuser G, Schwarz US, and Waterman CM (2008). Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J Cell Biol 183, 999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geach TJ, Hirst EM, and Zimmerman LB (2015). Contractile activity is required for Z-disc sarcomere maturation in vivo. Genesis 53, 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehmlich K, Pinotsis N, Hayess K, van der Ven PF, Milting H, El Banayosy A, Korfer R, Wilmanns M, Ehler E, and Furst DO (2007). Paxillin and ponsin interact in nascent costameres of muscle cells. Journal of molecular biology 369, 665–682. [DOI] [PubMed] [Google Scholar]
- Gerull B, Gramlich M, Atherton J, McNabb M, Trombitas K, Sasse-Klaassen S, Seidman JG, Seidman C, Granzier H, Labeit S, et al. (2002). Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat Genet 30, 201–204. [DOI] [PubMed] [Google Scholar]
- Gotthardt M, Hammer RE, Hubner N, Monti J, Witt CC, McNabb M, Richardson JA, Granzier H, Labeit S, and Herz J (2003). Conditional expression of mutant M-line titins results in cardiomyopathy with altered sarcomere structure. J Biol Chem 278, 6059–6065. [DOI] [PubMed] [Google Scholar]
- Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, et al. (2010). Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 466, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey PA, and Leinwand LA (2011). The cell biology of disease: cellular mechanisms of cardiomyopathy. J Cell Biol 194, 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman DS, Lam L, Taylor MR, Wang L, Teekakirikul P, Christodoulou D, Conner L, DePalma SR, McDonough B, Sparks E, et al. (2012). Truncations of titin causing dilated cardiomyopathy. N Engl J Med 366, 619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CS, Duran S, Lin ZX, Weber K, and Holtzer H (1986). Titin and myosin, but not desmin, are linked during myofibrillogenesis in postmitotic mononucleated myoblasts. J Cell Biol 103, 2185–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson JT, Chopra A, Nafissi N, Polacheck WJ, Benson CC, Swist S, Gorham J, Yang L, Schafer S, Sheng CC, et al. (2015). HEART DISEASE. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science 349, 982–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries JD, Wang P, Streuli C, Geiger B, Humphries MJ, and Ballestrem C (2007). Vinculin controls focal adhesion formation by direct interactions with talin and actin. J Cell Biol 179, 1043–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodo K, Ong SG, Jahanbani F, Termglinchan V, Hirono K, InanlooRahatloo K, Ebert AD, Shukla P, Abilez OJ, Churko JM, et al. (2016). iPSC-derived cardiomyocytes reveal abnormal TGF-beta signalling in left ventricular non-compaction cardiomyopathy. Nat Cell Biol 18, 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresh JY, and Chopra A (2011). Intercellular and extracellular mechanotransduction in cardiac myocytes. Pflugers Archiv : European journal of physiology 462, 75–87. [DOI] [PubMed] [Google Scholar]
- Kuppusamy KT, Jones DC, Sperber H, Madan A, Fischer KA, Rodriguez ML, Pabon L, Zhu WZ, Tulloch NL, Yang X, et al. (2015). Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes. Proceedings of the National Academy of Sciences of the United States of America 112, E2785–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limouze J, Straight AF, Mitchison T, and Sellers JR (2004). Specificity of blebbistatin, an inhibitor of myosin II. J Muscle Res Cell Motil 25, 337–341. [DOI] [PubMed] [Google Scholar]
- Lu MH, DiLullo C, Schultheiss T, Holtzer S, Murray JM, Choi J, Fischman DA, and Holtzer H (1992). The vinculin/sarcomeric-alpha-actinin/alpha-actin nexus in cultured cardiac myocytes. J Cell Biol 117, 1007–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundy SD, Zhu WZ, Regnier M, and Laflamme MA (2013). Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cells Dev 22, 1991–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons GE, Schiaffino S, Sassoon D, Barton P, and Buckingham M (1990). Developmental regulation of myosin gene expression in mouse cardiac muscle. J Cell Biol 111, 2427–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majkut S, Idema T, Swift J, Krieger C, Liu A, and Discher DE (2013). Heart-specific stiffening in early embryos parallels matrix and myosin expression to optimize beating. Curr Biol 23, 2434–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Musa H, Gautel M, and Peckham M (2003). A targeted deletion of the C-terminal end of titin, including the titin kinase domain, impairs myofibrillogenesis. J Cell Sci 116, 4811–4819. [DOI] [PubMed] [Google Scholar]
- Musa H, Meek S, Gautel M, Peddie D, Smith AJ, and Peckham M (2006). Targeted homozygous deletion of M-band titin in cardiomyocytes prevents sarcomere formation. J Cell Sci 119, 4322–4331. [DOI] [PubMed] [Google Scholar]
- Myhre JL, Hills JA, Prill K, Wohlgemuth SL, and Pilgrim DB (2014). The titin A-band rod domain is dispensable for initial thick filament assembly in zebrafish. Dev Biol 387, 93–108. [DOI] [PubMed] [Google Scholar]
- Person V, Kostin S, Suzuki K, Labeit S, and Schaper J (2000). Antisense oligonucleotide experiments elucidate the essential role of titin in sarcomerogenesis in adult rat cardiomyocytes in long-term culture. J Cell Sci 113, 3851–3859. [DOI] [PubMed] [Google Scholar]
- Polio SR, Rothenberg KE, Stamenovic D, and Smith ML (2012). A micropatterning and image processing approach to simplify measurement of cellular traction forces. Acta Biomater 8, 82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee D, Sanger JM, and Sanger JW (1994). The premyofibril: evidence for its role in myofibrillogenesis. Cell motility and the cytoskeleton 28, 1–24. [DOI] [PubMed] [Google Scholar]
- Ribeiro AJ, Ang YS, Fu JD, Rivas RN, Mohamed TM, Higgs GC, Srivastava D, and Pruitt BL (2015). Contractility of single cardiomyocytes differentiated from pluripotent stem cells depends on physiological shape and substrate stiffness. Proceedings of the National Academy of Sciences of the United States of America 112, 12705–12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro Ede A Jr., Pinotsis N, Ghisleni A, Salmazo A, Konarev PV, Kostan J, Sjoblom B, Schreiner C, Polyansky AA, Gkougkoulia EA, et al. (2014). The structure and regulation of human muscle alpha-actinin. Cell 159, 1447–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger JW, Wang J, Holloway B, Du A, and Sanger JM (2009). Myofibrillogenesis in skeletal muscle cells in zebrafish. Cell motility and the cytoskeleton 66, 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss T, Lin ZX, Lu MH, Murray J, Fischman DA, Weber K, Masaki T, Imamura M, and Holtzer H (1990). Differential distribution of subsets of myofibrillar proteins in cardiac nonstriated and striated myofibrils. J Cell Biol 110, 1159–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow JC, and Schock F (2009). The initial steps of myofibril assembly: integrins pave the way. Nature reviews Molecular cell biology 10, 293–298. [DOI] [PubMed] [Google Scholar]
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, and Mitchison TJ (2003). Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science 299, 1743–1747. [DOI] [PubMed] [Google Scholar]
- Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, Navarrete EG, Hu S, Wang L, Lee A, et al. (2012). Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med 4, 130–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee YH, Shemesh T, Thiagarajan V, Hariadi RF, Anderson KL, Page C, Volkmann N, Hanein D, Sivaramakrishnan S, Kozlov MM, et al. (2015). Cellular chirality arising from the self-organization of the actin cytoskeleton. Nat Cell Biol 17, 445–457. [DOI] [PubMed] [Google Scholar]
- Tokuyasu KT, and Maher PA (1987). Immunocytochemical studies of cardiac myofibrillogenesis in early chick embryos. I. Presence of immunofluorescent titin spots in premyofibril stages. J Cell Biol 105, 2781–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Q, Duchemin-Pelletier E, Deshiere A, Balland M, Guillou H, Filhol O, and Thery M (2012). Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proceedings of the National Academy of Sciences of the United States of America 109, 1506–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullio AN, Accili D, Ferrans VJ, Yu ZX, Takeda K, Grinberg A, Westphal H, Preston YA, and Adelstein RS (1997). Nonmuscle myosin II-B is required for normal development of the mouse heart. Proceedings of the National Academy of Sciences of the United States of America 94, 12407–12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ven PF, Bartsch JW, Gautel M, Jockusch H, and Furst DO (2000). A functional knock-out of titin results in defective myofibril assembly. J Cell Sci 113 (Pt 8), 1405–1414. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Zareno J, Whitmore L, Choi CK, and Horwitz AF (2007). Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J Cell Biol 176, 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, Yuan H, Jiang D, Zhang D, Zangi L, et al. (2014). Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med 20, 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitkunat M, Kaya-Copur A, Grill SW, and Schnorrer F (2014). Tension and force-resistant attachment are essential for myofibrillogenesis in Drosophila flight muscle. Curr Biol 24, 705–716. [DOI] [PubMed] [Google Scholar]
- White J, Barro MV, Makarenkova HP, Sanger JW, and Sanger JM (2014). Localization of sarcomeric proteins during myofibril assembly in cultured mouse primary skeletal myotubes. Anatomical record 297, 1571–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Baribault H, and Adamson ED (1998). Vinculin knockout results in heart and brain defects during embryonic development. Development 125, 327–337. [DOI] [PubMed] [Google Scholar]
- Young P, and Gautel M (2000). The interaction of titin and alpha-actinin is controlled by a phospholipid-regulated intramolecular pseudoligand mechanism. EMBO J 19, 6331–6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemljic-Harpf AE, Miller JC, Henderson SA, Wright AT, Manso AM, Elsherif L, Dalton ND, Thor AK, Perkins GA, McCulloch AD, et al. (2007). Cardiac-myocyte-specific excision of the vinculin gene disrupts cellular junctions, causing sudden death or dilated cardiomyopathy. Mol Cell Biol 27, 7522–7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Related to Figure 1. Dynamic sarcomerogenesis in human cardiomyocytes.
Representative wild type cardiomyocyte expressing GFP-actinin was plated onto an isotropic fibronectin-coated circular pattern overnight. Sarcomere assembly time lapse imaging was started the following morning for 24 hours. 20 minute frame rate, accelerated 12,000x for display. Scale bar indicates 20 μm, timescale hours:minutes.
Movie S9. Related to Figure 4. Protocostameres are required for sarcomerogenesis.
Representative vinculin knock out cardiomyocyte expressing GFP-actinin was plated onto an isotropic fibronectin-coated circular pattern overnight. Sarcomere assembly time lapse imaging was started the following morning. Ablating protocostameres leads to an absence of centripetal fiber formation. 20 minute frame rate, accelerated 12,000x for display. Scale bar indicates 20 μm, timescale hours:minutes.
Movie S10. Related to Figure 4. Mechanical linking of titin to protocostameres drives sarcomerogenesis.
Representative ACTN2-C knock out cardiomyocyte expressing GFP-actinin was plated onto an isotropic fibronectin-coated circular pattern overnight. Sarcomere assembly time lapse imaging was started the following morning. 20 minute frame rate, accelerated 12,000x for display. Scale bar indicates 20 μm, timescale hours:minutes.
Movie S2. Related to Figures 1 and 2. Inhibition of myosin contractility acutely impairs sarcomerogenesis.
Wild type cardiomyocytes expressing mApple-actinin were plated onto fibronectin-coated isotropic circular pattern (left panel) or anisotropic rectangular pattern (right panel) overnight. Note the presence of transverse and centripetal actinin fibers on the rectangular substrate. Sarcomere assembly time lapse imaging was started the following morning. Blebbstatin± (50 μM) was added during the time lapse (+blebbistatin) leading to acute breakdown of existing centripetal fibers and inhibition of sarcomerogenesis. 20 minute frame rate, accelerated 12,000x for display. Scale bars indicate 20 μm, timescale hours:minutes.
Movie S3. Related to Figure 1. Dynamic sarcomerogenesis in human cardiomyocytes on substrates of physiologically relevant stiffness
Representative wild type cardiomyocyte expressing GFP-actinin was plated onto a fibronectin-coated rectangular pattern on compliant polyacrylamide gel (4kPa) overnight. Centripetal and transverse fibers contribute to myofibril assembly from points of adhesion, though Z-bodies along the sides of these cells tended to fuse and align along the sides of the long, parallel myofibrils, while Z-bodies at the ends tended to extended existing myofibrils. Sarcomere assembly time lapse imaging was started the following morning for 24 hours. 20 minute frame rate, accelerated 12,000x for display. Scale bar indicates 20 μm, timescale hours:minutes.
Movie S4. Related to Figure 1. Dynamic sarcomerogenesis in human cardiomyocytes within 3D collagen in engineered heart microtissues.
Representative wild type cardiomyocytes expressing GFP-actinin embedded within 3D collagen matrix in an engineered heart microtissue. Sarcomere assembly time lapse imaging was started 24 hours after making the microtissue. Large black arrows indicate transverse actinin flow, and small gray arrows indicate centripetal actinin fibers. 20 minutes frame rate, accelerated 12,000x for display. Scale bar indicates 20 μm, timescale hours:minutes.
Movie S5. Related to Figure 1. Titin truncation mutations abrogate sarcomere assembly.
Representative A-band TTNtv cardiomyocyte expressing GFP-actinin was plated onto an isotropic fibronectin-coated circular pattern overnight. Transient centripetal fibers were observed occasionally with frequent rupturing events (left panel) and in many cases the presence of centripetal fibers was completely absent (right panel), all cases had no initiation of sarcomere assembly. Few organized actinin puncta or Z-bodies were observed and transverse actinin flow was absent. Sarcomere assembly time lapse imaging was started the following morning. 20 minute frame rate, accelerated 12,000x for display. Scale bar indicates 20 μm, timescale hours:minutes.
Movie S6. Related to Figure 1. Patient-derived cardiomyocytes with heterozygous titin truncations have impaired sarcomere assembly.
Representative patient derived cardiomyocyte harboring heterozygous A-band TTNtv expressing GFP-actinin was plated onto an isotropic fibronectin-coated circular pattern overnight. Transverse actinin flow and relatively stable centripetal fibers were observed compared to homozygous A-band TTNtv mutations, however given the slow rate of sarcomere assembly relatively few or no myofibirils could be observed. Sarcomere assembly time lapse imaging was started the following morning. 20 minute frame rate, accelerated 12,000x for display. Scale bar indicates 20 μm, timescale hours:minutes.
Movie S7. Related to Figure 3. β-cardiac myosin is critical for sarcomerogenesis.
Representative MHC-β knock out cardiomyocyte expressing GFP-actinin was plated onto an isotropic fibronectin-coated circular pattern overnight. Centripetal and transverse actinin fiber assembly was disrupted leading to no sarcomere assembly. Sarcomere assembly time lapse imaging was started the following morning. 20 minute frame rate, accelerated 12,000x for display. Scale bar indicates 20 μm, timescale hours:minutes.
Movie S8. Related to Figure 4. Dynamic role of protocostomeres during sarcomerogenesis.
Representative wild type cardiomyocyte endogenously expressing GFP-paxillin (left panel, shown in red in merge right panel) and transduced to co-express mApple-actinin (middle panel, shown in green in merge right panel) was plated onto an isotropic fibronectin-coated circular pattern overnight. Sarcomere assembly time lapse imaging was started the following morning. Dynamic co-localization of paxillin and actinin is observed, with centripetal fibers emanating from paxillin-containing protocostameres. 20 minute frame rate, accelerated 12,000x for display. Scale bar indicates 20 μm, timescale hours:minutes.


