The DNA genome must be completely duplicated with exquisite accuracy before a cell divides. The origin of replication (the place replication starts) is a single unique DNA sequence in a bacterial genome (1). By contrast, eukaryotic chromosomes have numerous initiation start sites, but these sites are not defined by a particular sequence and they change location in each cell cycle (2, 3). How such a vital process as DNA replication is orchestrated in seemingly random fashion is a mystery. In PNAS, Kelly and Callegari (4) devise a simple mathematical model that largely describes global chromosome replication dynamics in the fission yeast Saccharomyces pombe, using extensive global datasets from Kaykov and Nurse (5) and from Daigaku et al. (6) of the Carr laboratory. Kelly and Callegari’s model requires few parameters and assumes that selection of initiation sites is stochastic. Their mathematical modeling depends on two main features: (i) AT-rich DNA to which the S. pombe origin recognition complex (ORC) binds, and (ii) DNA that is outside transcription units. The ability to describe the global landscape of replication over the S. pombe genome gives hope that the approach may apply to higher eukaryotes such as ourselves.
The quest to identify replication origin sequences in eukaryotes has a long and torturous history (3). Over three decades, many laboratories have tried to locate the elusive origin sequence in mammals, but this has often led to conflicting conclusions and hotly debated results. A main region of study was the 55-kb intergenic region between the dihydrofolate reductase (DHFR) and 2BE2121 loci of Chinese hamster ovary cells, a region that amplifies in response to methotrexate treatment (7). Dissection of this initiation region mostly resulted in less and less frequent initiations, although a few regions appeared to hold promise, including a region as small as 500 bp (8). While the search for defined origin sequences in eukaryotes finally came up empty-handed, it reinforced an emerging view that eukaryotic initiation-site selection and timing of firing is a stochastic process.
Defined origins do exist in one of the smallest eukaryotes, the budding yeast Saccharomyces cerevisiae (1, 2), but exactly which origins are used in a given cell cycle and the mechanisms that determine when they fire is not yet understood. The discovery of defined autonomously replicating origin sequences in budding yeast has led to a detailed understanding of the biochemistry of origin activation. The six-subunit ORC was first isolated by Bell and Stillman (9). Since then, contributions of many laboratories over two dozen years has led to the detailed mechanistic picture in which ORC, along with Cdc6 and Cdt1, loads two head-to-head stacked Mcm2-7 rings onto DNA in G1 phase, referred to as a pre-replicative complex (pre-RC) (10, 11). At the G1-to-S transition, the pre-RC is activated by several proteins and two kinases to assemble Cdc45 and GINS onto the minichromosome maintenance (MCM) protein complexes to form the active Cdc45/Mcm2-7/GINS (CMG) helicase identified by Ilves et al. (12) and Moyer et al. (13), both of the Botchan laboratory. The two CMG helicases generate bidirectional replication forks.
Dividing replication into two phases explained the “licensing” phenomenon identified by Blow and Laskey (14). Thus, PreRCs can only be assembled, or licensed, in G1 phase (i.e., the PreRC), and can only be fired in S phase, explaining how replication of a chromosome with numerous start sites is limited to only once per cell cycle. For the job of DNA synthesis per se, eukaryotes use two different DNA polymerases (Pols). Studies by Burgers and Kunkel (15) (and their collaborators) using mutant Pols revealed that Pol ε replicates the leading strand and Pol δ replicates the lagging strand. Importantly, while other eukaryotes lack well-defined origins, they contain the same replication machinery as budding yeast. However, the global dynamics that underlie replication of chromosomes was still not understood for any eukaryote.
The fission yeast S. pombe is an attractive model to study replication because it has a small (13.6-Mb) genome dispersed on three chromosomes, yet its replication pattern is like that in Xenopus, mice, and humans. Thus, S. pombe lacks defined origins and initiates in AT-rich sites that are used inefficiently and, when selected, fire stochastically. Chuang and Kelly (16) previously identified that S. pombe ORC subunit 4 (Orc4) has nine AT-hook domains, strongly biasing it to AT-rich sequences. To gain insight into the global replication dynamics in S. pombe (i.e., the problem of origin selection and timing), Kelly and Callegari (4) utilize two extensive global datasets from refs. 5 and 6.
In the study by Kaykov and Nurse (5), single-molecule DNA combing was used to map the locations of initiation sites and their timing over the entire genome. Importantly, the study greatly advanced DNA combing techniques to examine individual DNA molecules of great lengths, up to 5 Mb, thus observing numerous origins in one DNA molecule. Consistent with stochastic events, examination of the same sequence from different cells showed distinct patterns of initiation. In fact, descendants from the same cell gave different patterns of origin selection/timing, indicating that inherited epigenetic marks do not dictate the S. pombe replication program. Interestingly, the frequency of initiation sites gradually increased fourfold over the first half of S phase, revealing that S phase is not a sharp transition from G1, but instead is a more gradual process. Initiation sites were mapped to 2-kb resolution, predicting about 1,200 initiation sites per cell, with an average distance of about 11 kb between them. While exciting, the new data did not explain the underlying mechanism of initiation-site selection and their firing time.
Daigaku et al. (6) were interested in the usage of Pol ε and Pol δ over the genome and confirmed that Pol ε and Pol δ are largely confined to leading and lagging strands, respectively, over the entire genome (with minor exceptions). To perform global mapping of Pol usage, they developed a Pol usage sequencing (Pu-seq) method to map global use of Pols δ and ε during replication. The Pu-seq technique uses certain mutants of each Pol that incorporate ribo-NTPs at elevated frequency, followed by alkaline cleavage at ribo-NMP incorporation sites that were mapped at nucleotide resolution by high-throughput sequencing. The data were binned to 300 bp, giving a very high-resolution map of global genome replication.
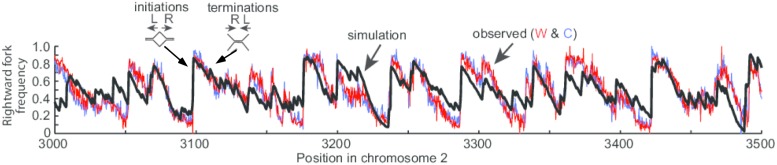
Kelly and Callegari (4) combine these global genome datasets along with key insights from other studies. A main lead was data indicating that initiation sites occur outside transcription units. For example, transcription had been inferred to prevent sites of replication initiation, and one of the earlier such studies placed a Gal promotor near an efficient origin in S. cerevisiae, which showed that transcription interferes with origin activity (17). Moreover, it had been noted that mammalian initiation sites in the classic DHFR locus did not occur in the body of the transcription unit, but upon inactivating the DHFR promotor, initiation sites were now observed to occur in the body of the gene (7). Moreover, the Pol usage data by Daigaku et al. (6) revealed that in S. pombe, many AT-rich sites that should bind ORC do not initiate if they are in transcription units. Thus, Kelly and Callegari (4) incorporate into the mathematical model that transcription interferes with pre-RCs, probably by either displacing pre-RCs or preventing their assembly. The model further incorporates AT-rich enhancement of S. pombe ORC (SpORC) binding in a probability distribution function that was optimized by the global experimental datasets. The model accurately describes the Pu-seq data in chromosome 2 at high resolution (Fig. 1).
Fig. 1.
Example of Pu-seq data and mathematical model fit. The black line is the mathematical model fit to experimental Pu-seq data [red, Watson (W) strand (Pol ε); blue Crick (C) strand (Pol δ)]. Rightward fork frequency increases to the right (R) of an initiation site, and decreases to the right of a termination site. L, left. Adapted from figure 4 of ref. 4.
The report in PNAS by Kelly and Callegari demonstrates that the complex task of selecting replication initiation sites and timing of their firing in S. pombe can be described by a simple stochastic mathematical model with surprisingly few variables.
There are interesting predictions of the mathematical model, some expected and others more surprising. It has long been known that cells contain many more MCM proteins than origins, often referred to as the “MCM paradox.” Excess MCM proteins have been assumed to form pre-RCs that do not fire but are held in reserve to initiate synthesis for replication forks that stop prematurely in S phase. Consistent with this, the model suggests that excess pre-RCs may be loaded but are knocked off DNA by replication forks or transcription before they get a chance to fire. The model also predicts that very late replicating sequences extend beyond the normal S phase and may explain DNA synthesis in G2 phase as a longer time that is needed to complete replication of regions with a scarcity of pre-RCs (4).
The model further suggests that SpORC evolved to bind extragenic regions, which are typically rich in AT sequence. This suggests that similar evolutionary pressures may also apply to ORCs of higher eukaryotes. Although higher eukaryotes lack the AT hook in Orc4, some metazoan ORC subunits contain domains that bind histone modifications; thus, particular chromatin states may factor into higher eukaryotic replication dynamics (3, 18). One may expect that several other variables will be needed to explain global replication dynamics of higher eukaryotes, considering their greater complexity compared with yeast. For example, a particular metazoan cell line has a reproducible initiation-site profile, but different developmentally derived cells show quite different profiles (18), suggesting that 3D physical structure of the chromatin, transcription profile, or other developmental changes may be important determinants of initiation-site selection and timing. A classic case is Xenopus, in which global transcription is suppressed in embryos and potential initiation sites are uniformly distributed (19), but changes at the midblastula stage result in nonuniform and stochastic initiation sites.
Of possible medical importance, mutations are associated with late-replicating regions (20). Thus, mathematical prediction of late-replicating regions may promote understanding of some types of genomic instability that lead to pathological states, including cancer. Interestingly, earlier studies showed that in S. pombe, repair of DNA lesions can occur by homologous recombination, a relatively error-free process, but this process is shut down in G2 phase, and repair shifts to error-prone processes like translesion DNA Pols (21) which, if general, may help explain mutagenesis during late replication.
In overview, the report in PNAS by Kelly and Callegari (4) demonstrates that the complex task of selecting replication initiation sites and timing of their firing in S. pombe can be described by a simple stochastic mathematical model with surprisingly few variables and, thus, provides a view that the stochastic replication program of higher eukaryotic cells may also be understood and described by mathematical modeling in the future.
Footnotes
The authors declare no conflict of interest.
See companion article on page 4973.
References
- 1.O’Donnell M, Langston L, Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol. 2013;5:a010108. doi: 10.1101/cshperspect.a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bleichert F, Botchan MR, Berger JM. Mechanisms for initiating cellular DNA replication. Science. 2017;355:eaah6317. doi: 10.1126/science.aah6317. [DOI] [PubMed] [Google Scholar]
- 3.Prioleau MN, MacAlpine DM. DNA replication origins—Where do we begin? Genes Dev. 2016;30:1683–1697. doi: 10.1101/gad.285114.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly T, Callegari AJ. Dynamics of DNA replication in a eukaryotic cell. Proc Natl Acad Sci USA. 2019;116:4973–4982. doi: 10.1073/pnas.1818680116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaykov A, Nurse P. The spatial and temporal organization of origin firing during the S-phase of fission yeast. Genome Res. 2015;25:391–401. doi: 10.1101/gr.180372.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daigaku Y, et al. A global profile of replicative polymerase usage. Nat Struct Mol Biol. 2015;22:192–198. doi: 10.1038/nsmb.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamlin JL, Mesner LD, Dijkwel PA. A winding road to origin discovery. Chromosome Res. 2010;18:45–61. doi: 10.1007/s10577-009-9089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burhans WC, Vassilev LT, Caddle MS, Heintz NH, DePamphilis ML. Identification of an origin of bidirectional DNA replication in mammalian chromosomes. Cell. 1990;62:955–965. doi: 10.1016/0092-8674(90)90270-o. [DOI] [PubMed] [Google Scholar]
- 9.Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 10.Evrin C, et al. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci USA. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Remus D, et al. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blow JJ, Laskey RA. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature. 1988;332:546–548. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- 15.Burgers PMJ, Kunkel TA. Eukaryotic DNA replication fork. Annu Rev Biochem. 2017;86:417–438. doi: 10.1146/annurev-biochem-061516-044709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chuang RY, Kelly TJ. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc Natl Acad Sci USA. 1999;96:2656–2661. doi: 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snyder M, Sapolsky RJ, Davis RW. Transcription interferes with elements important for chromosome maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2184–2194. doi: 10.1128/mcb.8.5.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nordman J, Orr-Weaver TL. Regulation of DNA replication during development. Development. 2012;139:455–464. doi: 10.1242/dev.061838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahbubani HM, Paull T, Elder JK, Blow JJ. DNA replication initiates at multiple sites on plasmid DNA in Xenopus egg extracts. Nucleic Acids Res. 1992;20:1457–1462. doi: 10.1093/nar/20.7.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu L, De S, Michor F. DNA replication timing and higher-order nuclear organization determine single-nucleotide substitution patterns in cancer genomes. Nat Commun. 2013;4:1502. doi: 10.1038/ncomms2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Callegari AJ, Kelly TJ. Coordination of DNA damage tolerance mechanisms with cell cycle progression in fission yeast. Cell Cycle. 2016;15:261–273. doi: 10.1080/15384101.2015.1121353. [DOI] [PMC free article] [PubMed] [Google Scholar]