Significance
Planar cell polarity (PCP) signaling coordinates cell polarization along an axis orthogonal to its apical–basal axis and is essential for both tissue development and function. In inner ear sensory epithelia, intercellular and cell-intrinsic PCP signaling controls the orientation and V shape of hair bundles atop sensory hair cells essential for hearing. Here we demonstrate that the conserved PDZ-domain scaffold protein Par3 plays an essential role in establishing PCP of sensory hair cells. Genetic mosaic analysis indicates that Par3 has both cell-autonomous and cell-nonautonomous functions in kinocilium positioning and hair bundle orientation. Mechanistically, Par3 interacts with and regulates localization of Rac guanine nucleotide exchange factors Tiam1 and Trio to activate Rac GTPases, thereby mediating both intercellular and cell-intrinsic PCP signaling.
Keywords: planar cell polarity, stereocilia, hair cell, Par3, microtubule
Abstract
In the inner ear sensory epithelia, stereociliary hair bundles atop sensory hair cells are mechanosensory apparatus with planar polarized structure and orientation. This is established during development by the concerted action of tissue-level, intercellular planar cell polarity (PCP) signaling and a hair cell-intrinsic, microtubule-mediated machinery. However, how various polarity signals are integrated during hair bundle morphogenesis is poorly understood. Here, we show that the conserved cell polarity protein Par3 is essential for planar polarization of hair cells. Par3 deletion in the inner ear disrupted cochlear outgrowth, hair bundle orientation, kinocilium positioning, and basal body planar polarity, accompanied by defects in the organization and cortical attachment of hair cell microtubules. Genetic mosaic analysis revealed that Par3 functions both cell-autonomously and cell-nonautonomously to regulate kinocilium positioning and hair bundle orientation. At the tissue level, intercellular PCP signaling regulates the asymmetric localization of Par3, which in turn maintains the asymmetric localization of the core PCP protein Vangl2. Mechanistically, Par3 interacts with and regulates the localization of Tiam1 and Trio, which are guanine nucleotide exchange factors (GEFs) for Rac, thereby stimulating Rac-Pak signaling. Finally, constitutively active Rac1 rescued the PCP defects in Par3-deficient cochleae. Thus, a Par3–GEF–Rac axis mediates both tissue-level and hair cell-intrinsic PCP signaling.
The sense of hearing and balance is critically dependent on the sensory hair cells of the inner ear. Hair cells are specialized epithelial cells that are polarized along both the apical–basal axis and within the plane of the epithelial cell sheet, referred to as planar cell polarity (PCP). Importantly, the hair bundle, the mechanotransduction organelle atop auditory hair cells, adopts a planar polarized structure with rows of stereocilia arranged in a staircase-like V shape, and thus is directionally sensitive to mechanical stimuli (1). Furthermore, planar polarity of the hair bundle is coordinated at the tissue level in all sensory epithelia. In the hearing organ, called the organ of Corti (OC), hair bundles are uniformly oriented with the vertices of the V pointing to the abneural or lateral edge. On the other hand, in the maculae, hair bundles show mirror-image polarity along the line of polarity reversal, which enables detection of movements in opposite directions (2).
These features of planar polarity are established during development, under genetic control by a multitiered signaling hierarchy. Upon hair cell differentiation, a cell-intrinsic machinery controls the migration and positioning of the kinocilium (and the associated basal body) at the lateral pole, where it is tethered to adjacent stereocilia at the vertex of the V-shaped hair bundle. Multiple signaling modules of the cell-intrinsic machinery have been discovered, including the small GTPase Rac1 and its effector p21-activated kinase (Pak) (3, 4), cdc42 and its downstream effector aPKC (5), as well as LGN–Gαi–dynein (6–9), a complex with an evolutionarily conserved role in mitotic spindle orientation. Recently, Dishevelled-associating protein with a high frequency of leucine residues (Daple) has been identified as another regulator of hair bundle shape and orientation through binding to Gαi (10). It has been suggested that these modules act in concert to provide subcellular landmarks and spatially control cortical capture of centriolar microtubules during kinocilium/basal body positioning, thereby influencing hair bundle shape and orientation.
At the organ level, an evolutionarily conserved noncanonical Wnt/PCP pathway mediates intercellular signaling between hair cells and supporting cells to coordinate hair bundle orientation (2, 11, 12). Components of this pathway include the “core” PCP genes, homologs of Drosophila Frizzled (Fzd3 and Fzd6), Van Gogh (Vangl1 and Vangl2), Flamingo (Celsr1 to 3), Dishevelled (Dvl1 to 3), Prickle, and Diego, as well as vertebrate-specific PCP genes, such as Ptk7 (13) and Scrib1 (14). Interestingly, in the maculae, the transcription factor Emx2 has been shown to reverse hair bundle polarity in its expression domain without affecting core PCP proteins (15, 16). These tissue-level regulators are not required for intrinsic bundle polarity, suggesting that the cell-intrinsic machinery can polarize individual hair cells independent of tissue-polarity cues. However, how this is achieved at the molecular level, and the precise mechanisms by which global PCP signals impinge on the cell-intrinsic machinery are incompletely understood.
To address these questions, here we investigated the role of Par3 (Pard3; Mouse Genome Informatics; www.informatics.jax.org) in hair cell PCP. Par3 encodes a PDZ-domain scaffold protein and is an evolutionarily conserved regulator of cell polarity (17). Central to its function in establishment of cell polarity, Par3 can self-associate to form oligomers and bind to membrane phospholipids and a diverse range of cell-polarity and cytoskeletal regulatory proteins. In mammalian epithelial cells, Par3 is localized to tight junctions, where it regulates the separation of apical and basolateral membrane domains (17). In Drosophila neuroblasts, the cortical Par3–Par6–aPKC complex recruits the LGN–Gαi–NuMA complex, thereby aligning the mitotic spindle to the cellular polarity axis (18). In this study, we found that Par3 is required for PCP but not apical–basal polarity in the OC. Par3 is asymmetrically localized during PCP establishment, which is regulated by the core PCP pathway. Deletion of Par3 disrupted microtubule organization and basal body positioning, leading to hair bundle shape and orientation defects. Surprisingly, Par3 has distinct localizations from its canonical partners Par6/aPKC and is not required for asymmetric localization of LGN/Gαi; instead, we present evidence that Par3 has both cell-autonomous and cell-nonautonomous functions in regulating hair bundle shape and orientation, and that Par3 mediates both tissue-level and hair cell-intrinsic PCP signaling through Rac GTPases.
Results
Par3 Is Asymmetrically Localized in the Developing OC.
To investigate the involvement of Par3 in hair cell PCP, we first analyzed Par3 protein localization in the OC at early stages of hair bundle morphogenesis. At embryonic day (E) 16.5, Par3 is localized to apical junctions of hair cells and supporting cells and significantly enriched along the lateral borders of hair cells (Fig. 1 A and B). By postnatal day (P) 0, when hair cell PCP has been established, Par3 could still be detected around cell junctions; however, its distribution no longer showed planar asymmetry (Fig. 1 E and F). Staining is absent from cochleae of inner ear-specific Par3 conditional knockout (Par3cKO) embryos mediated by Pax2-Cre (19), confirming antibody specificity (Fig. 1 C, D, G, and H). Therefore, transient asymmetric localization of Par3 at the onset of hair bundle morphogenesis suggests a role in hair cell PCP establishment.
Fig. 1.
Par3 is asymmetrically localized in the developing OC. Midbasal regions of the OC whole mount stained for Par3 (green). (A–D) At E16.5, Par3 is localized to hair cell-supporting cell contacts and enriched along the lateral borders of hair cells in the control (A and B) but absent in Par3cKO OC (C and D). (E–H) At P0, Par3 distribution at hair cell-supporting cell contacts is no longer planar polarized (E and F). Cell boundaries are labeled by phalloidin staining (F-actin, magenta). Arrowheads indicate the row of pillar cells. Brackets indicate OHC rows. (Scale bars: 6 µm.)
Par3 Regulates Cochlear Outgrowth and Hair Cell PCP.
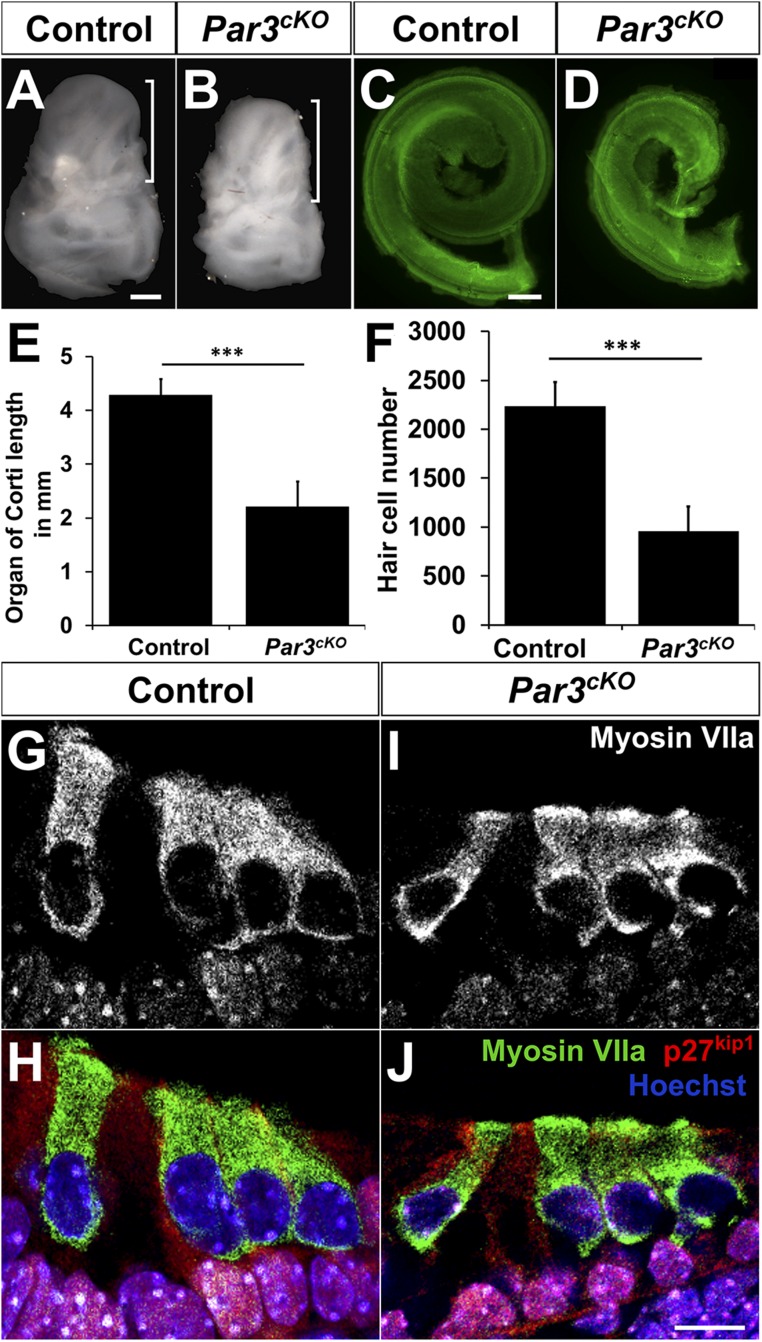
We next analyzed cochlear morphogenesis of Par3cKO mutants, which were alive at birth but died at P1. The mutant otic capsule was smaller in size compared with the control, with a shorter cochlear duct and decreased number of hair cells (Fig. 2 A–F). Cellular differentiation appeared normal, with the nuclei of inner hair cells (IHCs) and outer hair cells (OHCs) on top of the layer of the supporting cell nuclei (Fig. 2 G–J). Despite their decreased numbers, hair cells formed one row of IHCs and three rows of OHCs, interrupted occasionally by an extra OHC row (Figs. 1H and 2J). These results demonstrate a requirement of Par3 in cochlear outgrowth but not cell-fate determination in the OC.
Fig. 2.
Growth defects in Par3cKO cochleae. (A and B) P0 Par3cKO temporal bones (B) are smaller and misshapen compared with controls (A). Brackets indicate the cochlea. (C and D) The Par3cKO cochlear duct (D) is shorter in length compared with controls (C). Green, phalloidin staining. (E) Quantification of cochlear length. ***P < 0.001 (n = 6 each). (F) Quantification of hair cell numbers. ***P < 0.001 (n = 4 each). (E and F) Error bars represent SD. (G–J) Cross-sections from the midbasal region of P0 control (G and H) and Par3cKO cochleae (I and J) stained for the supporting cell marker p27kip1 (red), hair cell marker myosin VIIa (green), and nuclei (Hoechst, blue). (Scale bars: A and B, 500 µm; C and D, 100 µm; and G–J, 10 µm.)
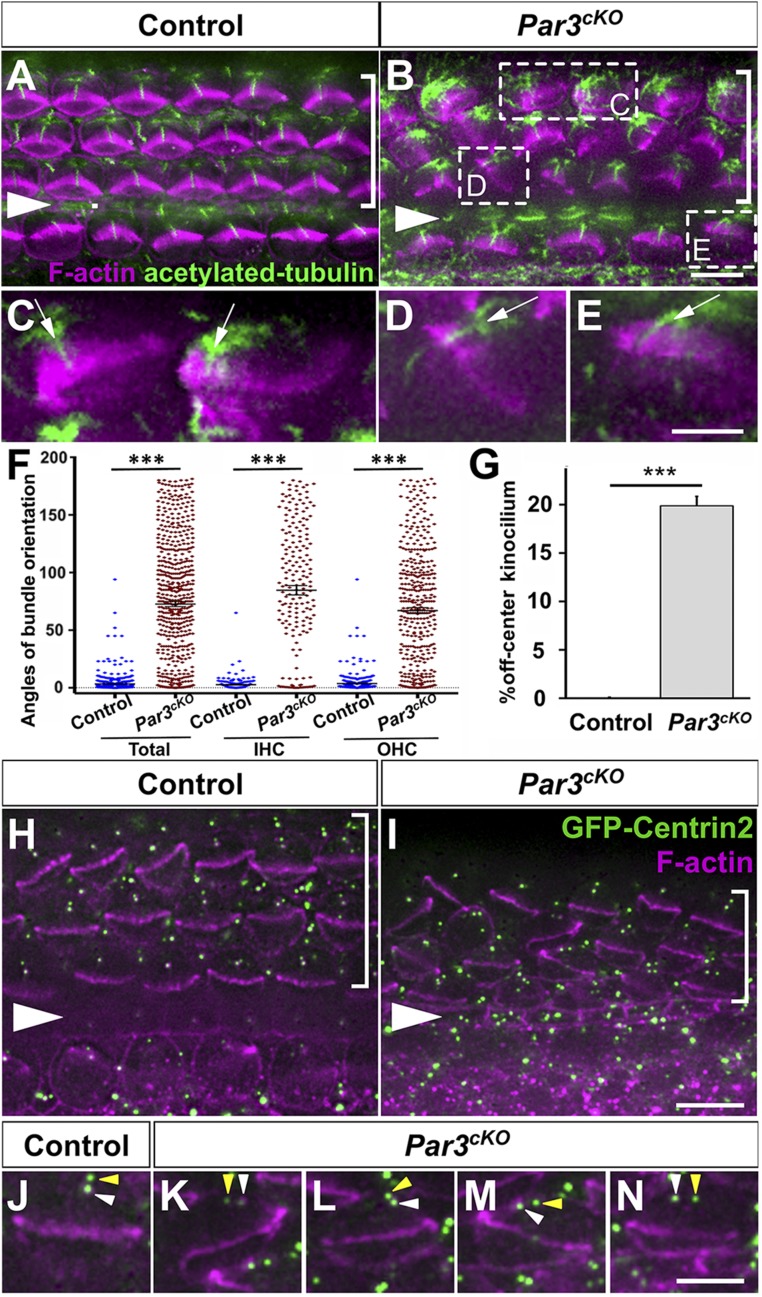
Next, we analyzed the establishment of hair cell PCP in the OC. In the control, hair bundles were uniformly oriented along the medial–lateral (or neural–abneural) axis of tissue polarity, with the kinocilium found at the vertex of the V-shaped hair bundle (Fig. 3A). By contrast, a significant number of IHCs and OHCs had misoriented hair bundles in Par3cKO OC (Fig. 3 B–F). Moreover, many hair bundles were misshapen, with “off-center” placement of the kinocilium away from the midpoint of the tallest row of stereocilia (Fig. 3 C–E, arrows; Fig. 3G). We also examined the positioning of hair cell basal bodies, which are marked by transgenic expression of GFP-Centrin2 (20). We have shown previously that centrioles in hair cells but not supporting cells are anchored at the lateral pole in a planar polarized manner (4). In control hair cells, the two centrioles are aligned along the medial–lateral axis at the lateral pole (Fig. 3 H and J). By contrast, many Par3cKO hair cells had a mispositioned basal body that correlated with hair bundle misorientation (Fig. 3I). Moreover, alignment of the two centrioles along the medial–lateral axis was also disrupted (Fig. 3 K–N, arrowheads). Together, defects in hair bundle shape and orientation, kinocilium positioning, and centriole planar polarity demonstrate a requirement of Par3 in both cell-intrinsic and tissue-level PCP establishment in the OC.
Fig. 3.
PCP defects in Par3cKO OC at P0. (A and B) Control (A) and Par3cKO (B) OC stained for acetylated tubulin (green) and F-actin (magenta). Examples of misshapen hair bundles with an off-center kinocilium are boxed in B. (C–E) Higher magnification of boxed hair cells in B. Arrows indicate off-center kinocilia. (F) Quantification of hair bundle orientation. n = 603 and 603 hair cells from three control and three mutant embryos, respectively. (G) Quantification of off-center kinocilium. n = 1,342 and 5,173 hair cells from six control and six mutant embryos, respectively. (F and G) Error bars represent SEM (F) and SD (G). ***P < 0.001. (H–N) Basal body positioning and centriole planar polarity are indicated by GFP-Centrin2 (green). White arrowheads indicate the basal body/mother centriole, and yellow arrowheads indicate the daughter centriole (J–N) Magenta, phalloidin staining. Arrowheads indicate the row of pillar cells. Brackets indicate OHC rows. Lateral side is up in all images. (Scale bars: A, B, H, and I, 6 μm; and C–E and J–N, 3 μm.)
The epithelial organization of the OC was further examined at E17.5 in whole-mount preparations. Localization of the adherens junction markers E-cadherin and β-catenin as well as the tight junction marker ZO-1 was largely unaffected in Par3cKO OC (SI Appendix, Fig. S1 A–F), indicating normal epithelial apical–basal polarity. Finally, the Nectin family of adhesion proteins interacts with Par3 in other epithelial cells (21) and regulates hair cell-supporting cell adhesion through heterotypic interactions (22, 23). We therefore examined the localization of Nectin family members. The overall staining patterns of Nectin1, 2, and 3 at hair cell-supporting cell contacts were largely unaffected (SI Appendix, Fig. S1 G–L). These results are consistent with Par3 acting downstream of or in parallel to the Nectins to mediate hair cell-supporting cell interactions in the OC.
Microtubule Defects in Par3cKO Cochleae.
We next sought to uncover the cellular events controlled by Par3 during PCP establishment in the OC. Accumulating evidence suggests that kinocilium/basal body positioning is achieved through interactions between the dynamic hair cell microtubule network and the hair cell cortex (4, 6). Microtubules are normally anchored at the basal body by their minor ends, while the free plus ends emanate out to form an aster-like network (Fig. 4A, Inset). In Par3cKO hair cells, the aster-like microtubule network became disorganized (Fig. 4B, Inset).
Fig. 4.
Microtubule defects in Par3cKO cochleae. (A and B) α-Tubulin staining in control (A) and Par3cKO OC (B). (A and B, Insets) Higher magnification of boxed hair cells. (C–F) EB1 immunostaining (magenta) in control (C and E) and Par3cKO OHCs (D and F). Cell boundaries are labeled by phalloidin staining (F-actin, green). (G–L) Cross-sections of P0 control and Par3cKO cochleae stained for acetylated tubulin (G and H; green in K and L), the hair cell marker myosin VI (I and J; magenta in K and L), and cell nuclei (K and L, blue). (M) Western blot analysis of acetylated tubulin and α-tubulin expression in E17.5 cochlear lysates. (N) Quantification showing increased levels of acetylated tubulin normalized to α-tubulin in Par3cKO cochleae (1.54 ± 0.05, SD; n = 3). Error bars represent SD. *P = 0.011. (Scale bars: A and B, 6 μm; C and F, 3 μm; and G–L, 10 μm.)
To determine how disorganized microtubules affected basal body positioning in Par3cKO hair cells, we used immunostaining of EB1, a plus end-tracking microtubule-binding protein, as a proxy for assaying interaction of microtubule plus ends with the hair cell cortex. In the control cochlea, there was significant enrichment of EB1 staining around the lateral hair cell cortex (Fig. 4 C and E, arrows), consistent with microtubule attachment at the lateral hair cell cortex. By contrast, EB1 staining was diffuse throughout the apical cytoplasm in Par3cKO hair cells, suggesting that microtubule cortical attachment is not preferentially stabilized at the lateral hair cell cortex in the absence of Par3 (Fig. 4 D and F).
In Par3cKO OC, microtubule disorganization was accompanied by increased staining intensity of acetylated tubulin (Fig. 3 A and B). This is confirmed in cochlear cross-sections, where increased staining intensity of acetylated tubulin was evident in hair cells (Fig. 4 H and L, arrowheads), pillar cells, and Deiters’ cells (Fig. 4 H and L, arrows). Furthermore, Western blot analysis of cochlear lysates demonstrated that the level of acetylated tubulin was increased by ∼50%, suggesting that microtubules are more stable in Par3cKO cochleae (Fig. 4 M and N).
Par3 Has Both Cell-Autonomous and Cell-Nonautonomous Functions in Regulating Hair Cell PCP.
Because we observed microtubule defects in both hair cells and supporting cells in the Par3cKO cochlea, we next performed genetic mosaic analysis to determine Par3 cell autonomy in hair cell PCP. Specifically, we used Fgf20Cre/+ to mediate Par3 deletion in the OC. Fgf20Cre/+ expresses Cre in the OC progenitor cells beginning at E10.5 (24, 25). Importantly, using the ROSA-mT/mG reporter for Cre activity (26), we found that Fgf20Cre/+-mediated recombination in the OC is mosaic, likely due to low expression level of Cre in those cells (Fig. 5A). Thus, we generated Fgf20Cre/+ Par3flox/flox; mTmG/+ mice, in which we identified both recombined hair cells (marked by membrane GFP; mG) and nonrecombined hair cells (marked by membrane Tomato; mT), and correlated the PCP phenotypes with the inferred Par3 genotype of individual hair cells. Of note, cochlear lengths of Fgf20Cre/+ Par3flox/flox; mTmG/+ mutants were similar to the control (SI Appendix, Fig. S2), suggesting that Fgf20Cre/+-mediated Par3 deletion did not grossly affect the number of OC progenitor cells or cochlear outgrowth. Both mG+ and mT+ hair cells showed hair bundle misorientation with similar severity, and had misshapen hair bundles with an off-center kinocilium (Fig. 5 B–J). Similar to Par3cKO, the planar polarity of hair cell centrioles was also disrupted in Fgf20Cre/+ Par3flox/flox OC (Fig. 5 K–P, arrowheads). We further correlated the hair bundle defects of mT+ hair cells with Cre expression of neighboring supporting cells, and found that every mT+ hair cell with a misoriented hair bundle and/or off-center kinocilium (n = 340) was surrounded by an average of 3.9 ± 0.50 (SD; range 2 to 5) mG+ supporting cells (Fig. 5 C′–E′). These results indicate that Par3 has a cell-nonautonomous function in supporting cells in regulating hair bundle orientation and kinocilium positioning.
Fig. 5.
Mosaic analysis of Par3 function in hair cell PCP. (A–B) Fgf20Cre/+-mediated mosaic OC at P0. (A) Both mT+ and mG+ hair cells in the control had normal PCP. (B) Both mT+ and mG+ hair cells in Par3-mosaic OC had misoriented and/or misshapen hair bundles. (C–H) Examples of mT+ (C–E) and mG+ (F–H) hair cells with a misoriented hair bundle and/or off-center kinocilium. Cyan, acetylated tubulin staining. Arrows indicate the kinocilium. (C′–H′) Single optical sections 4 μm below C–H showing supporting cells surrounding the hair cell. (I) Quantification of hair bundle orientation. n = 76 (mT+) and 81 (mG+) hair cells from three controls; n = 87 (mT+) and 91 (mG+) from three mutants. (J) Quantification of the off-center kinocilium. n = 544 (mT+) and 1,194 (mG+) hair cells from three embryos. Three types of defects were scored separately, namely misoriented hair bundle with normal shape (gray bars), misshapen hair bundle with normal orientation (magenta bars), and misoriented and misshapen hair bundle (blue bars). (I and J) Error bars represent SEM (I) and SD (J). ***P < 0.001. ns, not significant. (K–P) In contrast to the control (K), planar polarity of hair cell basal body/centrioles (marked by GFP-Centrin2) was disrupted in Fgf20Cre/+; Par3flox/flox OC (L–P). White arrowheads indicate the mother centriole/basal body, and yellow arrowheads indicate the daughter centriole. Lateral is up in all images. (Scale bars: A and B, 6 μm; and C–H′ and K–P, 3 μm.)
Furthermore, compared with mT+ hair cells, significantly more mG+ hair cells had an off-center kinocilium, either with or without hair bundle misorientation (Fig. 5J, blue and pink bars), suggesting that Par3 also plays a cell-autonomous role in regulating kinocilium positioning. Because most supporting cells had undergone recombination in these mosaic OCs, to more clearly establish the cell-autonomous function of Par3, we used another strategy for generating genetic mosaics. To this end, we generated Sox2CreER/+; Par3flox/flox; tdTomato/+ embryos and induced sparse recombination at E13.5 in OC progenitors (27) with a low-dose tamoxifen treatment. In the resulting mosaics, recombined hair cells and supporting cells (tdTomato+) were dispersed in the OC, and hair bundle orientation was largely normal, further supporting our conclusion that Par3 plays a cell-nonautonomous role in hair bundle orientation (SI Appendix, Fig. S3 A and B). Importantly, we found misshapen hair bundles with an off-center kinocilium in recombined hair cells that were surrounded entirely by nonrecombined supporting cells (SI Appendix, Fig. S3 D′′–F′′). Together, these results demonstrate that Par3 also has a cell-autonomous role in regulating kinocilium positioning and hair cell-intrinsic PCP.
Par3 Is an Effector and Regulator of Intercellular PCP Signaling.
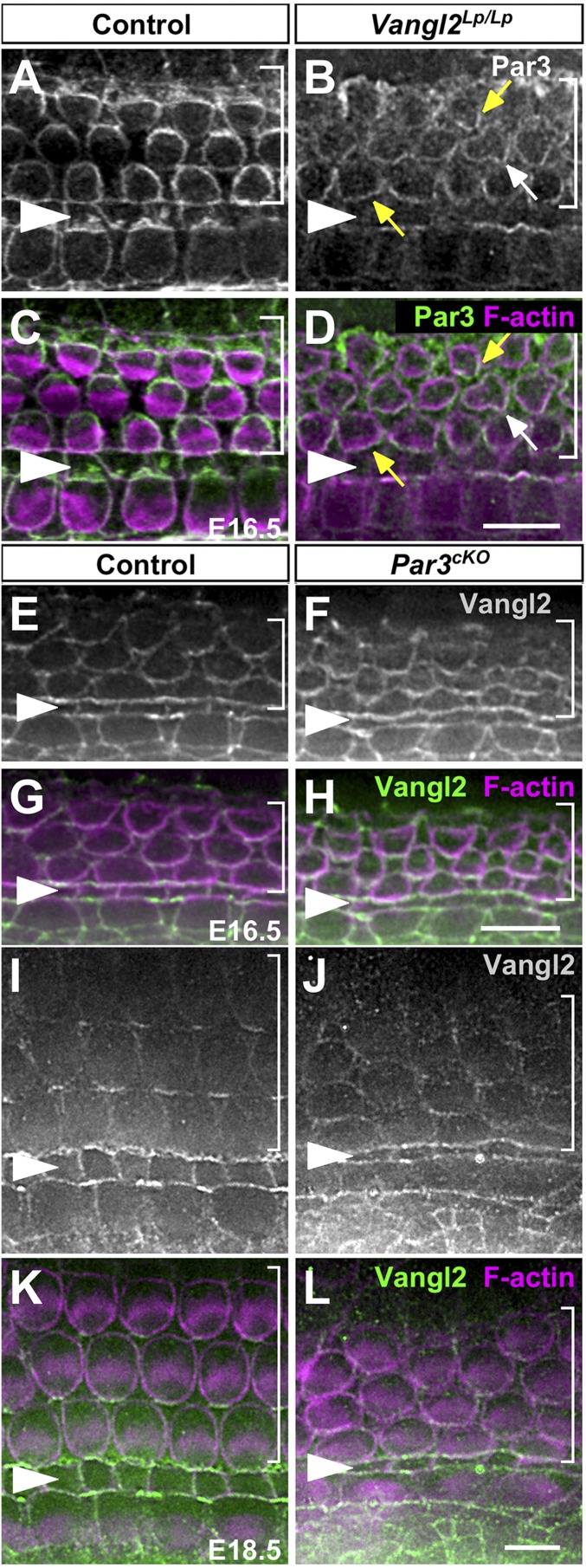
To investigate whether Par3 is a downstream effector of intercellular PCP signaling, we examined Par3 localization in Vangl2Lp/Lp and Ptk7−/− OCs at E16.5. In contrast to the control (Fig. 6 A and C), the enrichment of Par3 along the lateral border of hair cells was frequently misoriented or diminished in both Vangl2Lp/Lp (Fig. 6 B and D) and Ptk7−/− OCs (SI Appendix, Fig. S4 A–D). Thus, Par3 asymmetry is regulated by intercellular PCP signaling and is critical for hair bundle orientation.
Fig. 6.
Par3 is an effector and regulator of the core PCP pathway. (A–D) At E16.5, Par3 is enriched along the lateral borders of hair cells in the control (A and C). In Vangl2Lp/Lp OC (B and D), Par3 asymmetry is misoriented (yellow arrows) or diminished (white arrows). (E–H) At E16.5, Vangl2 was enriched along the medial borders of hair cells in both control (E and G) and Par3cKO OC (F and H). (I–L) At E18.5, asymmetric Vangl2 localization was reduced in Par3cKO OC (J and L) compared with the control (I and K). Cell boundaries are labeled by phalloidin staining (magenta). Arrowheads indicate the pillar cell row, and brackets indicate outer hair cell rows. (Scale bars: 6 μm.)
To further elucidate how Par3 regulates intercellular PCP signaling, we next asked whether Par3 regulates asymmetric localization of the core PCP protein Vangl2. At E16.5, Vangl2 was enriched on the supporting cell membranes abutting the medial borders of hair cells in both control and Par3cKO (Fig. 6 E–H). The localization of Ptk7 was also unaffected (SI Appendix, Fig. S4 E–H). However, at E18.5, asymmetric cortical localization of Vangl2 was significantly reduced in Par3cKO cochleae (Fig. 6 I–L). Thus, Par3 reinforces intercellular PCP signaling by maintaining asymmetric localization of the core PCP protein Vangl2.
Par3 Has Separate Functions from Par6 and aPKC During PCP Establishment.
The Par3–Par6–aPKC complex plays an evolutionarily conserved role in regulation of cell polarity in different contexts (17). To understand how Par3 regulates hair cell PCP, we next examined the localization of Par6 and aPKC in the developing OC. In contrast to Par3, neither Par6 nor aPKC was enriched along the lateral borders of hair cells at E16.5 (SI Appendix, Fig. S5 A–H). At P0, when Par3 is no longer asymmetrically localized in the OC, Par6 and aPKC were enriched on the medial side of hair cells. In Par3cKO OC, Par6 and aPKC were still asymmetrically localized, albeit disorganized and with decreased staining intensity (SI Appendix, Fig. S5 I–P). Thus, the asymmetric cortical localization of Par3 but not Par6 or aPKC at the onset of hair bundle development suggests that Par3 has separate functions from its canonical partners Par6 and aPKC during PCP establishment in the cochlea.
Par3 Is Not Required for Asymmetric Distribution of LGN or Gαi in Hair Cells.
A classic function of Par3 in mitotic spindle orientation is cortical recruitment of LGN through the adaptor protein Inscuteable (18). To determine whether Par3 plays a similar role in recruiting LGN to the lateral hair cell cortex during PCP establishment, we examined LGN and Gαi localization in Par3cKO cochleae at E17.5 (SI Appendix, Fig. S6 A–H) and P0 (SI Appendix, Fig. S6 I–P). Surprisingly, asymmetric LGN and Gαi localization at the bare zone and lateral cell cortex was robust and correlated with hair bundle orientation in both control and Par3cKO hair cells. These results indicate that Par3 is dispensable for the asymmetric localization of LGN and Gαi in hair cells during PCP establishment in the OC.
Par3 Stimulates Rac-PAK Signaling in the Cochlea Through Interactions with the Rac Guanine Nucleotide Exchange Factors Tiam1 and Trio.
Another regulator of hair cell PCP is the Rac1 GTPase, which induces autophosphorylation and activation of the downstream effector Pak. Localized Rac-Pak signaling has been shown to promote kinocilium positioning near the lateral hair cell cortex (3, 4). We therefore investigated whether Par3 regulates Rac-Pak signaling in the developing cochlea. Western blot analysis of E17.5 cochlear lysates using antibodies against phosphorylated, active Pak revealed a significant decrease in Pak activity levels in Par3cKO cochlear tissues compared with the control, indicating that Par3 positively regulates Rac-Pak signaling during PCP establishment (Fig. 7 A and B).
Fig. 7.
Par3 stimulates Rac-PAK activity by regulating Tiam1 and Trio in the cochlea. (A) Western blot analysis of pS141-Pak and Pak1 expression in E17.5 cochlear lysates. (B) Quantification showing decreased levels of pS141-Pak normalized to Pak1 in Par3cKO cochleae (0.53 ± 0.02, SD; n = 3), compared with the control. Error bars represent SD. **P = 0.002. (C and D) Par3 co-IP with Tiam1 (C) and Trio (D) in E16.5 mouse cochlear lysates. Normal rabbit IgG served as negative control. WB, Western blotting. (E–H) Compared with the control (E and G), Tiam1 staining intensity on hair cell microtubules was decreased in Par3cKO OC (F and H). (G and H) High-magnification images showing colocalization of Tiam1 (green) and acetylated tubulin (red). (I–L) Compared with the control (I and K), Trio staining was greatly reduced in Par3cKO OC (J and L). (K and L) High-magnification images showing Trio localization (green). Cell boundaries are labeled by phalloidin staining (blue). White arrowheads indicate the row of pillar cells. Brackets indicate OHC rows. (Scale bars: 6 μm.)
To determine the mechanisms by which Par3 promotes Rac-Pak signaling in the cochlea, we tested two guanine nucleotide exchange factors (GEFs) for Rac as candidates. Specifically, in other systems, Par3 has been shown to regulate Rac1 activity by directly interacting with Tiam1 and Trio (28–30). To determine whether Par3 interacts with Tiam1 and Trio in the developing cochlea, we performed coimmunoprecipitation (co-IP) experiments using pooled lysates of E16.5 wild-type cochleae. Indeed, Par3 was detected in the immunoprecipitates of anti-Tiam1 but not control antibodies, indicating that Par3 and Tiam1 form a complex in vivo (Fig. 7C). Likewise, co-IP assays demonstrated a Par3–Trio interaction in vivo (Fig. 7D).
Next, we examined Tiam1 and Trio localization in Par3cKO cochleae at E17.5. In the control, Tiam1 was localized to the cytoplasmic microtubule network and the kinocilium in hair cells (Fig. 7 E and G), as previously reported (6), while Trio was localized strongly to supporting cells and weakly to the microvillus zone on the medial side of hair cells (Fig. 7 I and K). In Par3cKO OC, both Tiam1 (Fig. 7 F and H) and Trio staining (Fig. 7 J and L) were significantly reduced. Taken together, these results suggest that Par3 promotes Rac signaling in the OC by regulating the localization and/or activity of Tiam1 and Trio.
Constitutively Active Rac1 Rescued the PCP Defects in Par3-Deficient Cochleae.
To further test whether Par3 mediates hair cell PCP through Rac signaling, we crossed a Cre-inducible, constitutively active Rac1 mutant transgene, ROSA-Rac1G12V (31), into Par3cKO to generate Par3cKO; Rac1G12V/+ embryos using Pax2-Cre. If Rac1 is a crucial downstream effector of Par3 in PCP signaling, then expression of Rac1G12V would be expected to restore Rac signaling and rescue the Par3cKO mutant phenotypes. Of note, Rac1G12V expression driven by Pax2-Cre by itself did not have significant deleterious effect on hair bundle morphogenesis or hair cell PCP, showing hair bundle defects only occasionally (Fig. 8 A and D). Remarkably, Rac1G12V expression in Par3cKO cochleae significantly rescued the defects in cochlear length, hair bundle shape and orientation, and kinocilium positioning (Fig. 8 B–G). Moreover, microtubule organization and centriole planar polarity were also largely restored (Fig. 8 H–K). Interestingly, the localization of the core PCP protein Vangl2 appeared increased and asymmetrically enriched on supporting cell membranes by Rac1G12V expression, regardless of the Par3 genotype (Fig. 8 L and M, arrows). Together, these results strongly suggest that Par3 mediates both intercellular and hair cell-intrinsic PCP signaling through Rac GTPases.
Fig. 8.
Constitutively active Rac1 rescued PCP defects of Par3-deficient cochleae. (A and B) P0 cochlea whole mount stained with phalloidin (magenta). (C) Quantification of cochlear lengths (n = 3 each). (D and E) P0 Rac1G12V/+ (D) and Par3cKO; Rac1G12V/+ OC (E) stained for acetylated tubulin (green) and F-actin (magenta). (F) Quantification of hair bundle orientation. Three embryos of each genotype were scored; n = 603 (Par3cKO), 293 (Rac1G12V/+), and 287 (Par3cKO; Rac1G12V/+) hair cells. (G) Quantification of kinocilium position index (Methods). Three embryos of each genotype were scored; n = 94 (control), 168 (Par3cKO), 241 (Rac1G12V/+), and 226 (Par3cKO; Rac1G12V/+) hair cells. (H–M) Hair cell microtubule organization (H and I), centriole (marked by GFP-Centrin2, green) planar polarity (J and K), and asymmetric localization of Vangl2 (L and M) in Rac1G12V/+ and Par3cKO; Rac1G12V/+ OC. (C, F, and G) Error bars represent SD (C) and SEM (F and G). ***P < 0.001. ns, not significant. Lateral side is up in all images. Arrowheads indicate the pillar cell row, and brackets indicate outer hair cell rows. (Scale bars: A and B, 300 µm; and D, E, and H–M, 6 μm.)
Discussion
Par3 Mediates Intercellular and Cell-Intrinsic PCP Signaling Through a Tiam1/Trio–Rac Axis.
Deletion of Par3 in the inner ear resulted in defects in both hair bundle orientation and kinocilium positioning within the hair bundle, indicating that it is a component of both intercellular and cell-intrinsic PCP signaling pathways. Several lines of evidence support a specific role of Par3 in PCP signaling in the OC, in addition to an early role in regulating OC progenitor cell numbers. First, asymmetric localization of Par3 is observed only after the onset of kinocilium migration in hair cells. Moreover, compared with Par3cKO, Fgf20Cre/+ Par3flox/flox mutants show a similar spectrum of PCP defects without affecting cochlear length, indicating that hair cell PCP defects are not a secondary effect of disrupted cochlear growth/extension.
Par3 has both cell-autonomous and cell-nonautonomous functions in PCP regulation. Interestingly, Par3 appears to function separate from its canonical partners Par6 and aPKC during the early phase of PCP establishment in the cochlea. One possible mechanism of regulating the dynamic interaction between Par3 and Par6/aPKC is Par3 protein phosphorylation (32, 33). Likewise, we found that Par3 is dispensable for asymmetric cortical localization of LGN and Gαi in hair cells, in contrast to its classic role in mitotic spindle orientation (18, 34).
Instead, Par3 deletion significantly decreased Rac-Pak activity levels in the developing cochlea, indicating a role in regulating Rac GTPase signaling. This is likely achieved through the Rac GEFs Tiam1 and Trio, both of which form a complex with Par3 in vivo and whose localization in the OC is regulated by Par3. Tiam1 and Trio are members of the Dbl family of Rac GEFs, and have been implicated in cytoskeletal dynamics, axon guidance, cell adhesion, and migration (35, 36). In the developing OC, Tiam1 expression is enriched in hair cells, while Trio is highly expressed in supporting cells. Thus, they are poised to mediate cell-autonomous and -nonautonomous functions of Par3 in PCP signaling. It has been reported that the GEF activity of Tiam1 and Trio is autoinhibited through intramolecular interactions (36, 37). As a scaffold protein, Par3 may mediate membrane translocation and release of autoinhibition of Tiam1 and Trio to activate Rac signaling in both hair cells and supporting cells. In support of a role of Tiam1 and Trio in PCP establishment, treatment of cochlear explants with NSC23766, which inhibits Rac binding to both Tiam1 and Trio (38), led to kinocilium/basal body positioning defects (4). Importantly, PCP defects in Par3cKO mutants were rescued by expression of constitutively active Rac1, indicating a key role of the Par3–Rac signaling axis in PCP establishment.
Par3 Regulates Microtubule Dynamics and Organization in the OC.
Loss of Par3 disrupted microtubule organization and cortical attachment in hair cells. A role of Par3 in stabilizing cortical attachment of astral microtubules has been shown in Caenorhabditis elegans zygotes (39), suggesting that this function of Par3 is evolutionarily conserved. The rescue results by Rac1G12V suggest that Par3 likely regulates microtubule dynamics through Rac signaling, which has been shown to locally regulate the activity of microtubule-associated proteins (MAPs) at the leading edge of migrating cells (40, 41). The MAPs targeted by Rac signaling in the OC during PCP establishment remain to be identified. Additional mechanisms by which Par3 regulates microtubule dynamics may involve Par3 association with microtubules or dynein (42, 43).
Par3 Integrates Multiple Pathways to Regulate Hair Bundle Shape and Orientation.
Intercellular PCP signaling coordinates hair bundle orientation through spatial regulation of the asymmetric distribution of PCP effectors such as Par3 (this study) and Rac-Pak signaling (3). A possible mechanism by which intercellular PCP signaling regulates Par3 localization is through Daple, which interacts with both Par3 (10) and the core PCP protein Dishevelled (44). Consistent with this, colocalization of Par3, Daple, and Dvl2 in the OC has been reported (10).
Interestingly, in contrast to Par3 and Rac1 mutants, core PCP mutants do not show an off-center kinocilium. This raises the possibilities that distinct subcellular pools of Par3 and Rac1 separately regulate hair bundle orientation and kinocilium positioning, and that, besides the core PCP pathway, an as-yet unidentified pathway converges on Par3 and Rac1 to position the kinocilium at the vertex of the V-shaped hair bundle.
Par3 reinforces intercellular PCP signaling by maintaining asymmetric localization of Vangl2. This regulation is specific for the core PCP proteins, as we show that Par3 deletion did not disrupt junctional localization of E-cadherin, ZO-1, Nectins, or Ptk7. Of note, constitutively active Rac1 largely restored Vangl2 localization in supporting cells of Par3cKO cochleae, suggesting Par3 regulates core PCP protein localization through Rac1 GTPase and its cytoskeletal targets (3, 45). These feedback regulations ensure the robust integration of cell-intrinsic and tissue-polarity signaling in the OC.
Methods
Mice.
To generate the Par3 conditional allele (designated Par3flox), we crossed Pard3<tm1a(KOMP)Wtsi>/H mice (46) with ROSA-FLPe mice (003946; Jackson Laboratory). Par3flox/flox animals were maintained on a C56Bl6 background and are viable and fertile. To generate various Par3 conditional mutants, Cre; Par3flox/+ males were mated with Par3flox/flox or Par3flox/+ females carrying the ROSA-mTmG, ROSA-Rac1*G12V, and/or GFP-Centrin2 transgene (007576, 012361, and 008234, respectively; Jackson Laboratory). The genotyping primers were 5′-AGAACCTGAAGATGTTCGCG-3′ and 5′-GGCTATACGTAACAGGGTGT-3′ (for Cre); and 5′-GCTGTTATCCTCCAAACCCTGAA-3′ and 5′-TACCGGAAATCTTTTTCTGACTTG-3′ (for Par3flox and Par3+).
For Sox2CreER-mediated mosaic analysis, Par3flox/flox females were mated with Sox2CreER/+; Par3flox/+; tdTomato/+ males and injected with 0.25 mg tamoxifen per 40 g body weight at E13.5, followed by embryo collection at P0. Sox2CreER (47) and the ROSA-tdTomato (48) reporter were obtained from the Jackson Laboratory (017593 and 007914, respectively).
Animal care and use were performed in compliance with NIH guidelines and the Animal Care and Use Committee at the University of Virginia. Mice were obtained from either the Jackson Laboratory or the referenced sources and maintained on a mixed genetic background. For timed pregnancies, the morning of the plug was designated as embryonic day 0.5, and the day of birth as postnatal day 0.
Quantification of Hair Cell Phenotypes and Statistical Analysis.
Cochlear length was determined from whole-mount phalloidin-stained images using ImageJ software (NIH). For quantification of hair bundle orientation, the angle formed by the intersection of a line connecting the center of the hair cell apex and the midpoint of the hair bundle and one parallel to the medial–lateral axis of the OC (assigned as 0°) was measured using ImageJ. To quantify kinocilium positioning within the hair bundle, we first measured the distances from the tip of the kinocilium to the two endpoints of the hair bundle, and then calculated their ratio (longer/shorter) as the “kinocilium position index.” An off-center kinocilium had an index value greater than 1.5. Hair cell differentiation follows a gradient from the base to the apex along the cochlear duct. Therefore, for analysis of protein localizations, care was taken to ensure that an equivalent midbasal region of the cochlea was compared between experimental groups. Experimental datasets were tested for significance using a Student’s t test or one-way ANOVA, and data are presented as mean ± SD, except for hair bundle angle and kinocilium index measurements, which are presented as mean ± SEM.
Antibodies, Immunohistochemistry, Western Blotting, and Co-IP.
Detailed protocols are described in SI Appendix, Methods.
Supplementary Material
Acknowledgments
We thank Wenxia Li, Michelle Lynskey, and Maxwell Madani for technical assistance; Drs. Kevin Pfister, Ann Sutherland (University of Virginia), and Andrew Ewald (Johns Hopkins University) for reagents; Dr. David M. Ornitz (Washington University School of Medicine) for Fgf20Cre mice; and Dr. Conor Sipe for critical reading of the manuscript. The Pard3<tm1a(KOMP)Wtsi>/H mice were obtained from MRC-Harwell, which is a participant of the International Mouse Phenotyping Consortium that funded the generation of the Pard3<tm1a(KOMP)Wtsi>/H mice. Funding and associated primary phenotypic information may be found at www.mousephenotype.org. This study was supported by NIH Grant R01DC013773 (to X.L.). Z.D. received a Harrison Undergraduate Research Award from the University of Virginia.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1816333116/-/DCSupplemental.
References
- 1.Schwander M, Kachar B, Müller U. Review series: The cell biology of hearing. J Cell Biol. 2010;190:9–20. doi: 10.1083/jcb.201001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deans MR. A balance of form and function: Planar polarity and development of the vestibular maculae. Semin Cell Dev Biol. 2013;24:490–498. doi: 10.1016/j.semcdb.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grimsley-Myers CM, Sipe CW, Géléoc GS, Lu X. The small GTPase Rac1 regulates auditory hair cell morphogenesis. J Neurosci. 2009;29:15859–15869. doi: 10.1523/JNEUROSCI.3998-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sipe CW, Lu X. Kif3a regulates planar polarization of auditory hair cells through both ciliary and non-ciliary mechanisms. Development. 2011;138:3441–3449. doi: 10.1242/dev.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirjavainen A, Laos M, Anttonen T, Pirvola U. The Rho GTPase Cdc42 regulates hair cell planar polarity and cellular patterning in the developing cochlea. Biol Open. 2015;4:516–526. doi: 10.1242/bio.20149753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sipe CW, Liu L, Lee J, Grimsley-Myers C, Lu X. Lis1 mediates planar polarity of auditory hair cells through regulation of microtubule organization. Development. 2013;140:1785–1795. doi: 10.1242/dev.089763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tarchini B, Jolicoeur C, Cayouette M. A molecular blueprint at the apical surface establishes planar asymmetry in cochlear hair cells. Dev Cell. 2013;27:88–102. doi: 10.1016/j.devcel.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Ezan J, et al. Primary cilium migration depends on G-protein signalling control of subapical cytoskeleton. Nat Cell Biol. 2013;15:1107–1115. doi: 10.1038/ncb2819. [DOI] [PubMed] [Google Scholar]
- 9.Bhonker Y, et al. The GPSM2/LGN GoLoco motifs are essential for hearing. Mamm Genome. 2016;27:29–46. doi: 10.1007/s00335-015-9614-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siletti K, Tarchini B, Hudspeth AJ. Daple coordinates organ-wide and cell-intrinsic polarity to pattern inner-ear hair bundles. Proc Natl Acad Sci USA. 2017;114:E11170–E11179. doi: 10.1073/pnas.1716522115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu X, Sipe CW. Developmental regulation of planar cell polarity and hair-bundle morphogenesis in auditory hair cells: Lessons from human and mouse genetics. Wiley Interdiscip Rev Dev Biol. 2016;5:85–101. doi: 10.1002/wdev.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodrich LV, Strutt D. Principles of planar polarity in animal development. Development. 2011;138:1877–1892. doi: 10.1242/dev.054080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu X, et al. PTK7/CCK-4 is a novel regulator of planar cell polarity in vertebrates. Nature. 2004;430:93–98. doi: 10.1038/nature02677. [DOI] [PubMed] [Google Scholar]
- 14.Montcouquiol M, et al. Identification of Vangl2 and Scrb1 as planar polarity genes in mammals. Nature. 2003;423:173–177. doi: 10.1038/nature01618. [DOI] [PubMed] [Google Scholar]
- 15.Holley M, et al. Emx2 and early hair cell development in the mouse inner ear. Dev Biol. 2010;340:547–556. doi: 10.1016/j.ydbio.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang T, Kindt K, Wu DK. Transcription factor Emx2 controls stereociliary bundle orientation of sensory hair cells. eLife. 2017;6:e23661. doi: 10.7554/eLife.23661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macara IG. Parsing the polarity code. Nat Rev Mol Cell Biol. 2004;5:220–231. doi: 10.1038/nrm1332. [DOI] [PubMed] [Google Scholar]
- 18.Yu F, Morin X, Cai Y, Yang X, Chia W. Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in Inscuteable apical localization. Cell. 2000;100:399–409. doi: 10.1016/s0092-8674(00)80676-5. [DOI] [PubMed] [Google Scholar]
- 19.Ohyama T, Groves AK. Generation of Pax2-Cre mice by modification of a Pax2 bacterial artificial chromosome. Genesis. 2004;38:195–199. doi: 10.1002/gene.20017. [DOI] [PubMed] [Google Scholar]
- 20.Higginbotham H, Bielas S, Tanaka T, Gleeson JG. Transgenic mouse line with green-fluorescent protein-labeled Centrin 2 allows visualization of the centrosome in living cells. Transgenic Res. 2004;13:155–164. doi: 10.1023/b:trag.0000026071.41735.8e. [DOI] [PubMed] [Google Scholar]
- 21.Takekuni K, et al. Direct binding of cell polarity protein PAR-3 to cell-cell adhesion molecule Nectin at neuroepithelial cells of developing mouse. J Biol Chem. 2003;278:5497–5500. doi: 10.1074/jbc.C200707200. [DOI] [PubMed] [Google Scholar]
- 22.Togashi H, et al. Nectins establish a checkerboard-like cellular pattern in the auditory epithelium. Science. 2011;333:1144–1147. doi: 10.1126/science.1208467. [DOI] [PubMed] [Google Scholar]
- 23.Fukuda T, et al. Aberrant cochlear hair cell attachments caused by Nectin-3 deficiency result in hair bundle abnormalities. Development. 2014;141:399–409. doi: 10.1242/dev.094995. [DOI] [PubMed] [Google Scholar]
- 24.Huh SH, Jones J, Warchol ME, Ornitz DM. Differentiation of the lateral compartment of the cochlea requires a temporally restricted FGF20 signal. PLoS Biol. 2012;10:e1001231. doi: 10.1371/journal.pbio.1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huh SH, Warchol ME, Ornitz DM. Cochlear progenitor number is controlled through mesenchymal FGF receptor signaling. eLife. 2015;4:e05921. doi: 10.7554/eLife.05921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 27.Kiernan AE, et al. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005;434:1031–1035. doi: 10.1038/nature03487. [DOI] [PubMed] [Google Scholar]
- 28.Chen X, Macara IG. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat Cell Biol. 2005;7:262–269. doi: 10.1038/ncb1226. [DOI] [PubMed] [Google Scholar]
- 29.Pegtel DM, et al. The Par-Tiam1 complex controls persistent migration by stabilizing microtubule-dependent front-rear polarity. Curr Biol. 2007;17:1623–1634. doi: 10.1016/j.cub.2007.08.035. [DOI] [PubMed] [Google Scholar]
- 30.Moore R, et al. Par3 controls neural crest migration by promoting microtubule catastrophe during contact inhibition of locomotion. Development. 2013;140:4763–4775. doi: 10.1242/dev.098509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srinivasan L, et al. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 2009;139:573–586. doi: 10.1016/j.cell.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakayama M, et al. Rho-kinase phosphorylates PAR-3 and disrupts PAR complex formation. Dev Cell. 2008;14:205–215. doi: 10.1016/j.devcel.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 33.Nagai-Tamai Y, Mizuno K, Hirose T, Suzuki A, Ohno S. Regulated protein-protein interaction between aPKC and PAR-3 plays an essential role in the polarization of epithelial cells. Genes Cells. 2002;7:1161–1171. doi: 10.1046/j.1365-2443.2002.00590.x. [DOI] [PubMed] [Google Scholar]
- 34.Williams SE, Ratliff LA, Postiglione MP, Knoblich JA, Fuchs E. Par3-mInsc and Gαi3 cooperate to promote oriented epidermal cell divisions through LGN. Nat Cell Biol. 2014;16:758–769. doi: 10.1038/ncb3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mertens AEE, Pegtel DM, Collard JG. Tiam1 takes PARt in cell polarity. Trends Cell Biol. 2006;16:308–316. doi: 10.1016/j.tcb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt S, Debant A. Function and regulation of the Rho guanine nucleotide exchange factor Trio. Small GTPases. 2014;5:e29769. doi: 10.4161/sgtp.29769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Z, Gakhar L, Bain FE, Spies M, Fuentes EJ. The Tiam1 guanine nucleotide exchange factor is auto-inhibited by its pleckstrin homology coiled-coil extension domain. J Biol Chem. 2017;292:17777–17793. doi: 10.1074/jbc.M117.799114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci USA. 2004;101:7618–7623. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Labbé J-C, Maddox PS, Salmon ED, Goldstein B. PAR proteins regulate microtubule dynamics at the cell cortex in C. elegans. Curr Biol. 2003;13:707–714. doi: 10.1016/s0960-9822(03)00251-3. [DOI] [PubMed] [Google Scholar]
- 40.Wittmann T, Waterman-Storer CM. Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3beta in migrating epithelial cells. J Cell Biol. 2005;169:929–939. doi: 10.1083/jcb.200412114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukata M, et al. Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell. 2002;109:873–885. doi: 10.1016/s0092-8674(02)00800-0. [DOI] [PubMed] [Google Scholar]
- 42.Chen S, et al. Regulation of microtubule stability and organization by mammalian Par3 in specifying neuronal polarity. Dev Cell. 2013;24:26–40. doi: 10.1016/j.devcel.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmoranzer J, et al. Par3 and dynein associate to regulate local microtubule dynamics and centrosome orientation during migration. Curr Biol. 2009;19:1065–1074. doi: 10.1016/j.cub.2009.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ishida-Takagishi M, et al. The Dishevelled-associating protein Daple controls the non-canonical Wnt/Rac pathway and cell motility. Nat Commun. 2012;3:859. doi: 10.1038/ncomms1861. [DOI] [PubMed] [Google Scholar]
- 45.Peng Y, Axelrod JD. Asymmetric protein localization in planar cell polarity: Mechanisms, puzzles, and challenges. Curr Top Dev Biol. 2012;101:33–53. doi: 10.1016/B978-0-12-394592-1.00002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arnold K, et al. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell. 2011;9:317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.