Significance
The alkaline pK for galactoside binding by the lactose permease of Escherichia coli correlates precisely with the pKa of Glu325, as determined by reaction-induced surface-enhanced infrared absorption spectroscopy (SEIRAS). Glu325 must be protonated for LacY to bind sugar effectively, but deprotonation is also essential for transport. SEIRAS is utilized to test the effect of mutating residues in the immediate neighborhood of Glu325 based on the rationale that interaction will alter the pKa. Neutral replacement of Arg302 with Ala has little or no effect, while replacement with positively charged Lys causes a two-pH unit acid shift. Since a number of other mutations in the vicinity have little effect, it is concluded that Arg302 is important for deprotonation of Glu325.
Keywords: transport, membrane proteins, permeases, protonation, surface-enhanced infrared spectroscopy
Abstract
Lactose permease is a paradigm for the major facilitator superfamily, the largest family of ion-coupled membrane transport proteins known at present. LacY carries out the coupled stoichiometric symport of a galactoside with an H+, using the free energy released from downhill translocation of H+ to drive accumulation of galactosides against a concentration gradient. In neutrophilic Escherichia coli, internal pH is kept at ∼7.6 over the physiological range, but the apparent pK (pKapp) for galactoside binding is 10.5. Surface-enhanced infrared absorption spectroscopy (SEIRAS) demonstrates that the high pKa is due to Glu325 (helix X), which must be protonated for LacY to bind galactoside effectively. Deprotonation is also obligatory for turnover, however. Here, we utilize SEIRAS to study the effect of mutating residues in the immediate vicinity of Glu325 on its pKa. The results are consistent with the idea that Arg302 (helix IX) is important for deprotonation of Glu325.
Glu325 plays a uniquely important role in the mechanism of coupling between galactoside and H+ translocation during symport by the lactose permease of Escherichia coli (LacY). Replacement with various neutral side chains leads to a symporter that is completely defective in all reactions involving coupled H+ translocation, with no effect on transmembrane galactoside exchange (1, 2) or binding (3, 4). Notably, the alternating access component of the mechanism, which is represented by transmembrane exchange of galactosides, is driven by sugar binding and dissociation, and not by the H+ electrochemical gradient (reviewed in ref. 5). Furthermore, galactoside binding exhibits an apparent pK (pKapp) of 10.5, which is abolished in mutant E325A, where high-affinity binding is observed up to pH 11, where LacY begins to destabilize (4). Thus, the galactoside-binding site is remarkably stable to alkaline pH, but Glu325 must be neutralized to elicit this property. Of paramount importance, it has also been demonstrated directly by surface-enhanced infrared absorption spectroscopy (SEIRAS) that the pKa of Glu325 itself is 10.5, the same as that obtained for galactoside binding, and that this perturbed pKa is due to a local hydrophobic environment (6). Thus, a main, unanticipated feature of coupling is that galactoside binding is dependent specifically upon protonation of Glu325, and protonation acts to neutralize an inhibitory negative charge on this side chain.
Of course, LacY must also deprotonate for turnover to occur. However, transmembrane exchange reactions, which represent alternating access, do not involve and exhibit pH profiles similar to that observed for galactoside binding (7, 8). Thus, during exchange, LacY remains protonated (5). In neutrophilic E. coli, internal pH is kept at ∼7.6 over the physiological range (9, 10) and the pKapp for galactoside binding is 10.5 (6, 11, 12). Therefore, to deprotonate only ∼50% of LacY, the pK would have to decrease by three orders of magnitude, which would be very inefficient. The alternative of decreasing cytosolic H+ concentration seems even more unlikely.
So how can deprotonation of LacY occur at physiological pH with a pKa of 10.5? One possibility is a structural change that exposes protonated Glu325 to a more aqueous local environment. Another is to bring Arg302 (helix IX) close to Glu325 (helix X). Although the two residues are 6–7 Å apart with the hydroxyl group of Tyr236 between them in the current structure, the double-Cys mutant R302C/E325C exhibits pyrene excimer fluorescence (13) and double-His R302H/E325H binds Mn(II) with micromolar affinity (14). Thus, Arg302 and Glu325 may be in closer proximity in another conformation of LacY. In this regard, like the Glu325 neutral mutants (2), neither R302S nor R302A LacY performs active transport, but both mutants bind galactoside and catalyze transmembrane exchange. For these reasons, it was suggested that positively charged Arg302 may be important with respect to deprotonation of Glu325 (15), and further evidence supporting this possibility has been presented (16).
Although reaction-induced SEIRAS may be useful for determining the pKas of acidic side chains (6, 17), other functional groups in proteins are often obscured by contributions from protein backbone reorganization caused by the induced reaction (18). In view of the critical importance of Glu325 and the possibility that Arg302 may be important for deprotonation, we have tested the effect of mutating Arg302 and other side chains in the immediate vicinity of Glu325 on its pKa based on the notion that side chains that interact chemically with Glu325 and/or change the local environment should alter the pKa (Figs. 1 and 2). The results provide further evidence that Arg302 may interact with Glu325 to drive deprotonation.
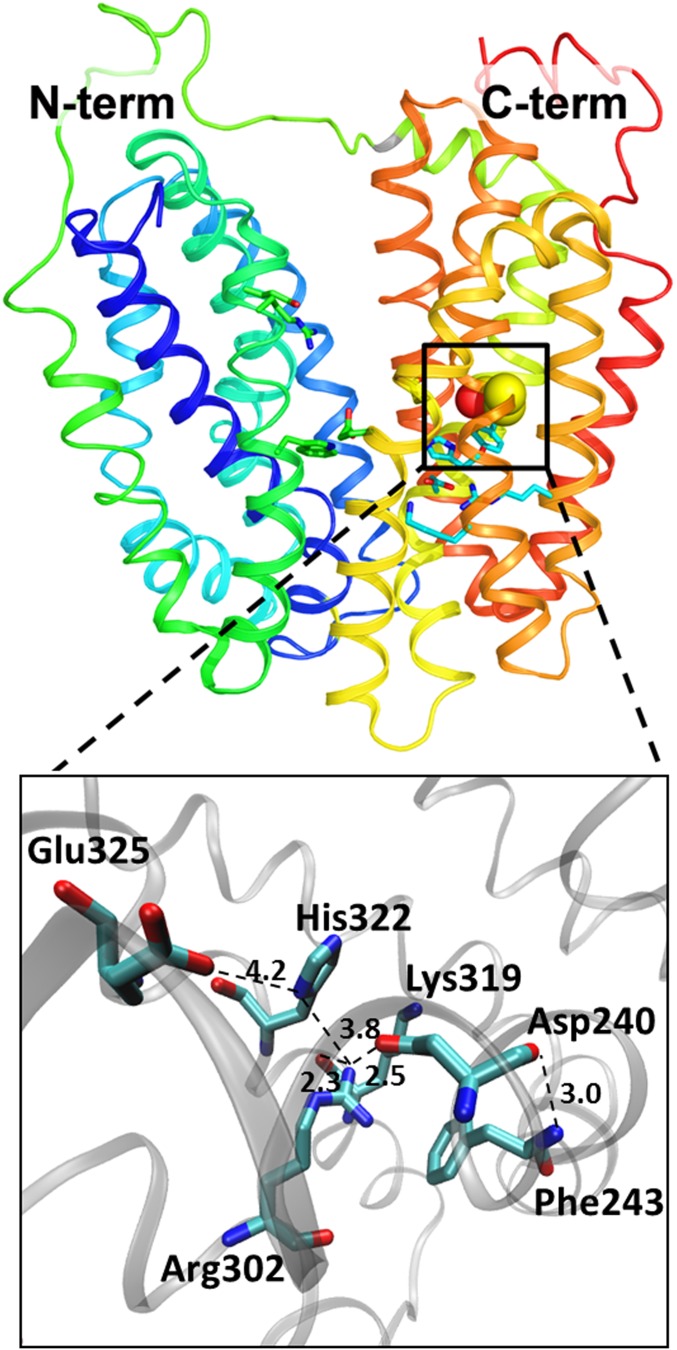
Fig. 1.
Cytoplasmic view of the E325 environment and of the positions of the mutations (Protein Data Bank ID code 2V8N).
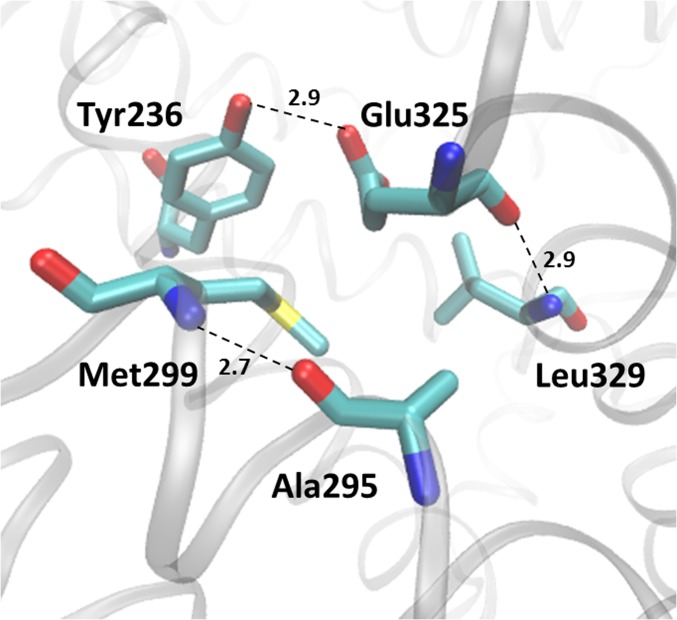
Fig. 2.
Hydrophobic pocket around Glu325 (Protein Data Bank ID code 2V8N).
Results
SEIRAS Spectra.
The spectra presented in Fig. 3A are difference spectra obtained by subtracting data obtained from samples equilibrated at pH 7.0 from samples obtained at a pH that completely or almost completely deprotonates Glu325 (pH 10.9). The difference spectra reflect all reorganization within LacY due to the shift in pH, including conformational changes in the backbone and changes in the protonation state of individual side chains, for example, the COOH vibrational mode of Glu325 assigned to 1,746 cm−1 (6). Positive and negative signals can be distinguished reflecting these conformational changes. The parts of the protein that do not change with pH are omitted from the difference spectra (17).
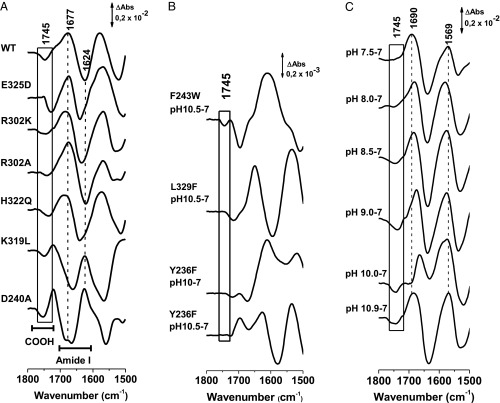
Fig. 3.
(A) pH-dependent difference SEIRAS spectra of LacY obtained by subtracting the samples equilibrated at pH 7 minus the samples equilibrated at high pH, inducing a deprotonation of Glu325 at 1,745 cm−1. Pseudo WT and mutants in G46W/G262W mutant background. (B) pH-dependent difference SEIRAS spectra of LacY by subtracting the samples equilibrated at pH 7 minus the samples equilibrated at high pH of the mutations in the LacY mutants in the G46W/G262W mutant background. (C) pH-dependent difference SEIRAS spectra of mutant LacYww/R302K equilibrated at pH 7.0 subtracted from the sample equilibrated at different pH values. ΔAbs, difference in absorption.
Data interpretation is possible because of the availability of a large number of infrared studies on proteins and model systems. The spectra are characterized generally by signals in the so-called “amide I” region between 1,690 and 1,620 cm−1 that involve backbone contributions, as well as contributions from individual side chains. The position of the amide I backbone signal is specific for the type of secondary structure, and signals at ∼1,650 cm−1 correspond to α-helices. These contributions are partially obscured by disordered structures observed at ∼1,642 cm−1 (19). The β-sheets are typically observed at ∼1,636 cm−1 and 1,670–1,690 cm−1. However, this type of difference spectrum may also shift due to a change in the environment of only part of the peptide backbone. So-called “doorway” shifts have also been described in KcsA K+ channels (20). In this case, the difference spectra are due to different ions, which reveal small structural changes within one type of secondary structure element. Thus, the difference mode observed between 1,700 and 1,610 cm−1 (Fig. 3A, amide I) is attributed to small changes in helices and small movements in the microenvironment of sites that may be involved in H+ transport, but are not due to changes in secondary structure.
In the amide II region near 1,570 cm−1, the contribution from the protein backbone includes in-plane N-H bending (40–60%) coupled to the ν(C-N) (20–40%) vibrational mode (19). Upon H/D exchange (SI Appendix, Fig. S1), the amide II band intensity at 1,571 cm−1 decreases, the in-plane N-H (N-D) bending mode uncouples and appears in the 940- to 1,040-cm−1 region, and the ν(C-N) moves to near 1,445 cm−1, mixing with other modes to form a new band called amide II*. The changes in the spectra upon H/D exchange confirm that most of the signals in the difference spectra at positions lower than 1,700 cm−1 originate from the protein backbone.
Effect of Mutations on Glu325 SEIRAS.
To identify side chains sufficiently close to Glu325 to perturb the pKa, a systematic study on the effect of potentially important mutations on the SEIRAS difference spectrum of Glu325 was undertaken (Fig. 2 and Table 1). Each mutant was introduced into the G46W/G262W background (LacYww) to ensure high stability at alkaline pH (21). Importantly, SEIRAS data for WT LacY and LacYww, as well as the corresponding E325A mutants, are indistinguishable (12). Difference spectra between pH 7.0 and an alkaline pH at which Glu325 is essentially completely deprotonated were carried out with each mutant (Fig. 3A): (i) pseudo-WT LacYww, (ii) LacYww/E325D, (iii) LacYww/R302K, (iv) LacYww/R302A, (v) LacYww/H322Q, (vi) LacYww/K319L, and (vii) LacYww/D240A. The alkaline pH for each mutant was selected to be as close to complete deprotonation as possible. As a result, the vibrational mode of the side chain of Glu325 is observed as a negative signal close to that observed for the pseudo-WT at 1,746 cm−1, a position that is boxed in Fig. 3A.
Table 1.
Overview of studied mutants
| Mutant | Peak position, cm−1 | pKa | Sugar/H+ symport | Discussed role of studied residue |
| LacY WT | 1,747 | 10.5 | Yes | |
| LacYww | 1,747 | 10.5 | No | |
| E325A | No signal | nd | No sugar/H+ symport, but transmembrane sugar exchange (1, 2, 15) | H+ translocation |
| Mutants | LacYww | |||
| E325A | No signal | nd | No sugar/proton symport, but transmembrane sugar exchange (1, 2) | H+ translocation |
| E325D | 1,730 | 8.3 | 15% of WT (8, 16) | H+ translocation |
| R302K | 1,758/1,743 | 8.4 | No | H+ translocation |
| R302A | 1,741 | 10.3 | No sugar/H+ symport, but transmembrane sugar exchange (15, 16) | H+ translocation |
| H322Q | 1,734 | 10.1 | No | Ligand to sugar |
| Y236F | — | — | Decreased binding affinity and little transport | Hydrogen bond donor to H322 stabilizing its conformation |
| D240A | 1,749 | 9.8 | No | Salt-bridged K319/D240 |
| F243W | 1,740 | 10.1 | 30% of WT | |
| K319L | 1,750 | 10 | Reduced binding | Salt-bridged K319/D240 |
| L329F | — | — | No | Part of hydrophobic pocket around E325 |
nd, not determined.
The difference spectrum of mutation E325D (Fig. 3A) exhibits a shift of the COOH vibrational mode to 1,728 cm−1, a position typical of a more hydrophilic environment. The COOH in Asp or Glu at position 325 does not have the same position, and a shift of 18 cm−1 is likely due to a change in microenvironment, hydrogen-bond strength, or a combination of both. This kind of shift was observed earlier for mutations from Glu → Asp and vice versa (22). The amide I/II region does not change significantly in difference spectra of the mutant, arguing against a change in the structural integrity of the protein. With mutations R302K and R302A, only small shifts are observed for the signal at 1,746 cm−1. However, with mutation R302K, the difference signal is significantly broader, an effect that may be due to a higher degree of freedom of Glu325 at this position. Mutation of residue H322Q (Fig. 3A) causes a shift in the Glu325 COOH vibration to 1,737 cm−1, which suggests stronger hydrogen bonding in the immediate environment of the residue without significant perturbation of the overall structure.
In the spectrum of the K319L mutant, the signal discussed is found at 1,750 cm−1, similar to the D240A mutation (Fig. 3A). The amide signature for these two mutants is clearly changed compared with the pseudo-WT. While there is a broad positive peak at 1,674 cm−1 with a shoulder at 1,722 cm−1 and a negative signal around 1,625 cm−1 for the pseudo-WT and the mutants discussed above, the signals are inverted here, suggesting a strongly modified change in conformational reorganization that takes place upon changing pH. Residues D240 and K319 are weakly salt-bridged both structurally (Fig. 1) and functionally (23). A strong perturbation of the salt bridge in the mutant can explain the conformational changes observed. Nevertheless, the influence on the Glu325 infrared signature is negligible.
Three additional mutants have been studied that show strongly perturbed spectra (Fig. 3B). In Y236F, a signal at 1,744 cm−1 was obtained for the step to pH 10.5, but the mutant is unstable and the amide I signature was different for each pH value studied. In L329F, no signal could be identified except for some conformational changes in the amide I/II range. Both residues are located in the hydrophobic patch surrounding Glu325 (Fig. 2). Finally, the effect of mutant Y236F was tested. Again, the signature of Glu325 was not identified, and for each pH step, the amide I signature was different, as can be seen when comparing the step to pH 10 and to pH 10.5. It is noted that Tyr236 is hydrogen-bonded to H322 (24). Mutations in the hydrophobic pocket of Glu325 perturb structure and the conformational flexibility of LacY.
Effect of Mutations on the pKa of Glu325.
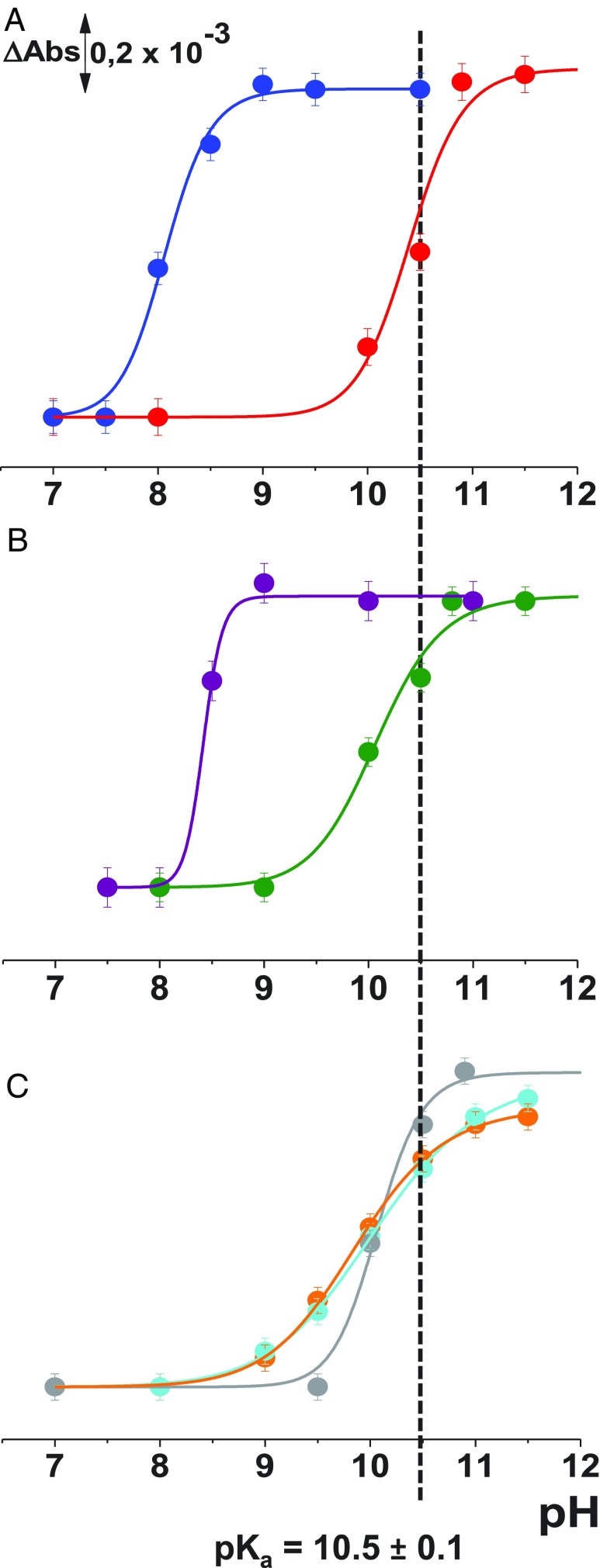
To test for an effect of each mutation on the pKa of Glu325, difference spectra were obtained for each mutant from pH 6.0–12.0. The pH-dependent increase in the negative signal at ∼1,745 cm−1 was plotted as a function of pH (Fig. 3A), except for mutation E325A, where no signal is observed (6). However, mutation E325D exhibits an acidic shift in the pKa from ∼10.5 to ∼8.3 (Fig. 4), consistent with a change in the microenvironment of the COOH. Mutation E325D retains 15–20% of WT transport activity with reduced affinity (4, 8, 16). Mutant R302A is also defective with respect to affinity for galactoside (4), but catalyzes transmembrane exchange (15), and the mutation has little or no effect on the pKa of Glu325 (Fig. 3A). However, mutation R302K is particularly interesting, as it causes the pKa of Glu325 to decrease to pH 8.3 (Figs. 3C and 4B), clearly an indication of interaction between the two side chains. The Glu325 signal at 1,745 cm−1 can be observed already at pH 8 and pH 8.5 in the difference spectra (Fig. 3C).
Fig. 4.
Titration curves for the ν(C = O) vibrational mode of Glu325 at 1,746 cm−1: pseudo- WT with (red) and LacYww/E325D (blue) (A); LacYww/R302K (purple) and LacYww/R302A (green) (B); and LacYww/D240A (orange), LacYww/K319L (cyan), and LacYww/H322Q (gray) (C).
Unexpectedly, mutation H322Q, which is only one helix turn from Glu325, has little or no effect on the pKa for Glu325 (Fig. 4). Since His322 is liganded to the galactopyranosyl moiety of LacY substrates, the binding affinity of the mutant and all transport reactions are strongly diminished (4). Mutations D240A and K319L disrupt a relatively weak salt bridge (23) and cause very small, but reproducible, acid shifts in the Glu325 titration. Finally, mutations Y236F (helix VII) and L329F (helix X) yield unstable Glu325 pH titrations as mentioned above, and the spectra differ at each pH tested. Both of these side chains comprise part of the hydrophobic pocket in which Glu325 is situated.
Discussion
LacY drives accumulation of lactose against a 50- to 100-fold concentration gradient (25), with a turnover number of ∼20 s−1. Both Δ (interior negative) and ΔpH (interior alkaline) have quantitatively the same effect on transport with a 50- to 100-fold decrease in Km and the same thermodynamic equilibrium. In opposition to time-honored conjecture, the Kd for galactosides on either side of the membrane is essentially the same in the absence or presence of (26). In the absence of , there is three- to fourfold inhibition of lactose transport in the presence of deuterium oxide (D2O), indicating that deprotonation is rate-limiting (7, 27–29). However, in the presence of , there is a driving force on the H+, deprotonation is no longer rate-limiting, and the transport rate is unaffected by D2O. It is particularly noteworthy that a number of experimental findings indicate strongly that functions kinetically as a driving force on the H+, while alternating access is driven by sugar binding and dissociation (30).
Clearly, a major problem for LacY turnover is deprotonation when the midpoint for both galactoside and H+ binding is pH 10.5, which translates into a Kd for H+ of ∼30 pM. As discussed, it is unlikely that either decreasing the cytosolic H+ concentration or the pKa of Glu325 is a realistic possibility. An increase in water accessibility to the hydrophobic site in which Glu325 is located is a possibility. However, as shown by three independent H/D exchange studies (31–33), the backbone amide protons in LacY are largely accessible to water. Another possibility is that Arg302 and Glu325 may be in closer proximity in another conformation of LacY (13, 14), and there is experimental evidence that distance between the two residues is important in this regard (16). The data obtained here indicate that Arg302 interacts with Glu325. While the R302A mutation does not significantly change the microenvironment of Glu325, introduction of a Lys induces a downshift of approximately two pH units and a broader COOH vibrational mode characteristic of a residue with significant rotational freedom. Although Arg and Lys are both positively charged, the distribution of the charge is different, and the delocalized charge on Arg is apparently critical for the pKa value measured here. However, neither the R302K mutation nor the R302A mutation causes gross alteration in structure, as is also the case for other mutations in the vicinity of Glu325 that disrupt a salt bridge or cause a change in conformation, but do not affect the pKa of Glu325.
Materials and Methods
Construction of Mutants, Purification of LacY, and Reconstitution.
Construction of mutants, expression in E. coli, and purification of LacY were performed as described before (4, 6). All constructs contained a C-terminal His6 tag that was used for affinity purification with Talon resin. Purified proteins (10–15 mg/mL) in 50 mM sodium phosphate [NaPi/0.02% DDM (pH 7.5)] were frozen in liquid nitrogen and stored at −80 °C until use.
Surface Modification of the Silicon Crystal and Protein Immobilization.
On the surface of a Si attenuated total reflection (ATR) crystal, a thin gold layer was formed by chemical deposition as described previously (6). The crystal was dried under an argon stream, and 40% (wt/vol) NH4F was added for 1 min to remove the Si oxide layer and to terminate with hydrogen; finally, the surface was rinsed and dried again. The crystal was heated at 65 °C for 10 min, together with the plating solution. The composition of the solution was a 1:1:1 mix (vol/vol/vol) of 15 mM NaAuCl4 + 150 mM Na2SO3 + 50 mM Na2S2O3 + 50 mM NH4Cl, and 2% HF (wt/vol; 1 mL). After reaching the temperature, the prism was covered with the solution for 40 s, and washed with water, followed by drying with a stream of argon.
After formation of a gold layer on the Si crystal, a nickel-nitrilotriacetic acid self-assembled monolayer was adapted from techniques reported by Grytsyk et al. (6), Ataka and Heberle (34), and Kriegel et al. (35). First, 1 mg/mL 3,3-dithiodipropionic acid di(N-hydroxysuccinimide ester) (DTSP) in dimethyl sulfoxide (DMSO) was allowed to self-assemble for 1 h. After monolayer formation, excess DTSP was washed away with DMSO and the crystal was dried under an argon stream. Afterward, the self-assembled monolayer was immersed in 100 mM Nα,Nα-bis(carboxymethyl)-l-lysine in 0.5 M K2CO3 at pH 9.8 for 3 h and rinsed with water. Finally, the surface was incubated in 50 mM Ni(ClO4)2 for 1 h.
Infrared Spectroscopy.
A configuration allowing the simultaneous acquisition of Fourier transform infrared (FTIR) spectra in the ATR mode with perfusion of solutions with well-defined composition was employed, including a silicon crystal with a 3-mm surface diameter as a multireflection ATR unit. All experiments were carried out with a Bruker Vertex 70 FTIR spectrometer (Globar source, KBr Beamsplitter, LN-MCT detector) at a scanner velocity of 40 kHz. The measurements were carried out at ∼7 °C. Solutions were kept in ice before use. For the data presented here, the pump speed was kept constant at a flow rate of 0.2 mL⋅min−1. Before each perfusion step, the input tube was carefully washed with water and buffer.
Difference Spectra.
To monitor pH-induced difference spectra, one perfusion buffer with a constant pH value of 7.0 was used as a reference (25 mM KPi/100 mM KCl/0.01% dodecyl-β,D-maltopyranoside) and a second perfusion solution with the same composition, but at different pH values ranging from 8.0 to 11.5, was employed. After equilibration for 30 min with the KPi buffer (pH 7.0) a reference spectrum was recorded as background and the perfusion solution was changed to the second solution (pH range: 8.0–11.5). After 20 min (pH 8.0–11.5) minus pH 7.0 difference spectra were recorded. The new state of the protein was recorded as background, and the solution was changed to pH 7.0. Finally, the pH 7.0 minus (pH 8.0–11.5) difference spectra were obtained. The same procedure was repeated at least five times, and the difference spectra were averaged. Baseline correction was done wherever necessary.
Difference Spectra of LacY After H/D Exchange.
The perfusion solution was prepared in D2O for the analogous pD values as described above. After perfusing with the pD 7 solution, the completed exchange was confirmed on the basis of the typical shift of the amide II band. The experiments were then performed like those carried out in H2O.
Supplementary Material
Acknowledgments
We thank Aurore Pereau (University of Strasbourg) for performing control experiments on mutant Y236F during her master internship. H.R.K. acknowledges the University of California, Los Angeles and a gift from Ruth and Bucky Stein. P.H. acknowledges the University of Strasbourg, Institute of Advanced Studies; the International Center Foundation Research in Chemistry (icFRC); and the CNRS and Initiative d’Excellence (IDEX) of the University of Strasbourg for financial support. The work was also supported by NIH Grant GM120043 and National Science Foundation Eager Grant MCB1747705 (to H.R.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820744116/-/DCSupplemental.
References
- 1.Carrasco N, Antes LM, Poonian MS, Kaback HR. Lac permease of Escherichia coli: Histidine-322 and glutamic acid-325 may be components of a charge-relay system. Biochemistry. 1986;25:4486–4488. doi: 10.1021/bi00364a004. [DOI] [PubMed] [Google Scholar]
- 2.Carrasco N, et al. Characterization of site-directed mutants in the lac permease of Escherichia coli. 2. Glutamate-325 replacements. Biochemistry. 1989;28:2533–2539. doi: 10.1021/bi00432a028. [DOI] [PubMed] [Google Scholar]
- 3.Smirnova IN, Kasho VN, Kaback HR. Direct sugar binding to LacY measured by resonance energy transfer. Biochemistry. 2006;45:15279–15287. doi: 10.1021/bi061632m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smirnova I, Kasho V, Sugihara J, Choe JY, Kaback HR. Residues in the H+ translocation site define the pKa for sugar binding to LacY. Biochemistry. 2009;48:8852–8860. doi: 10.1021/bi9011918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaback HR. A chemiosmotic mechanism of symport. Proc Natl Acad Sci USA. 2015;112:1259–1264. doi: 10.1073/pnas.1419325112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grytsyk N, Sugihara J, Kaback HR, Hellwig P. pKa of Glu325 in LacY. Proc Natl Acad Sci USA. 2017;114:1530–1535. doi: 10.1073/pnas.1621431114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Viitanen P, Garcia ML, Foster DL, Kaczorowski GJ, Kaback HR. Mechanism of lactose translocation in proteoliposomes reconstituted with lac carrier protein purified from Escherichia coli. 2. Deuterium solvent isotope effects. Biochemistry. 1983;22:2531–2536. doi: 10.1021/bi00279a034. [DOI] [PubMed] [Google Scholar]
- 8.Frillingos S, Kaback HR. Monoclonal antibody 4B1 alters the pKa of a carboxylic acid at position 325 (helix X) of the lactose permease of Escherichia coli. Biochemistry. 1996;35:10166–10171. doi: 10.1021/bi960995r. [DOI] [PubMed] [Google Scholar]
- 9.Ramos S, Schuldiner S, Kaback HR. The electrochemical gradient of protons and its relationship to active transport in Escherichia coli membrane vesicles. Proc Natl Acad Sci USA. 1976;73:1892–1896. doi: 10.1073/pnas.73.6.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zilberstein D, Schuldiner S, Padan E. Proton electrochemical gradient in Escherichia coli cells and its relation to active transport of lactose. Biochemistry. 1979;18:669–673. doi: 10.1021/bi00571a018. [DOI] [PubMed] [Google Scholar]
- 11.Smirnova I, et al. Sugar binding induces an outward facing conformation of LacY. Proc Natl Acad Sci USA. 2007;104:16504–16509. doi: 10.1073/pnas.0708258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smirnova IN, Kasho V, Kaback HR. Protonation and sugar binding to LacY. Proc Natl Acad Sci USA. 2008;105:8896–8901. doi: 10.1073/pnas.0803577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung K, Jung H, Wu J, Privé GG, Kaback HR. Use of site-directed fluorescence labeling to study proximity relationships in the lactose permease of Escherichia coli. Biochemistry. 1993;32:12273–12278. doi: 10.1021/bi00097a001. [DOI] [PubMed] [Google Scholar]
- 14.He MM, Voss J, Hubbell WL, Kaback HR. Use of designed metal-binding sites to study helix proximity in the lactose permease of Escherichia coli. 2. Proximity of helix IX (Arg302) with helix X (His322 and Glu325) Biochemistry. 1995;34:15667–15670. doi: 10.1021/bi00048a010. [DOI] [PubMed] [Google Scholar]
- 15.Sahin-Tóth M, Kaback HR. Arg-302 facilitates deprotonation of Glu-325 in the transport mechanism of the lactose permease from Escherichia coli. Proc Natl Acad Sci USA. 2001;98:6068–6073. doi: 10.1073/pnas.111139698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinglass AB, Smirnova IN, Kaback HR. Engineering conformational flexibility in the lactose permease of Escherichia coli: Use of glycine-scanning mutagenesis to rescue mutant Glu325→Asp. Biochemistry. 2001;40:769–776. doi: 10.1021/bi002171m. [DOI] [PubMed] [Google Scholar]
- 17.Zscherp C, Schlesinger R, Tittor J, Oesterhelt D, Heberle J. In situ determination of transient pKa changes of internal amino acids of bacteriorhodopsin by using time-resolved attenuated total reflection Fourier-transform infrared spectroscopy. Proc Natl Acad Sci USA. 1999;96:5498–5503. doi: 10.1073/pnas.96.10.5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barth A. Infrared spectroscopy of proteins. Biochim Biophys Acta. 2007;1767:1073–1101. doi: 10.1016/j.bbabio.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Arrondo JLR, Muga A, Castresana J, Goñi FM. Quantitative studies of the structure of proteins in solution by Fourier-transform infrared spectroscopy. Prog Biophys Mol Biol. 1993;59:23–56. doi: 10.1016/0079-6107(93)90006-6. [DOI] [PubMed] [Google Scholar]
- 20.Stevenson P, et al. Visualizing KcsA conformational changes upon ion binding by infrared spectroscopy and atomistic modeling. J Phys Chem B. 2015;119:5824–5831. doi: 10.1021/acs.jpcb.5b02223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smirnova I, Kasho V, Sugihara J, Kaback HR. Trp replacements for tightly interacting Gly-Gly pairs in LacY stabilize an outward-facing conformation. Proc Natl Acad Sci USA. 2013;110:8876–8881. doi: 10.1073/pnas.1306849110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hellwig P, Barquera B, Gennis RB. Direct evidence for the protonation of aspartate-75, proposed to be at a quinol binding site, upon reduction of cytochrome bo3 from Escherichia coli. Biochemistry. 2001;40:1077–1082. doi: 10.1021/bi002154x. [DOI] [PubMed] [Google Scholar]
- 23.Sahin-Tóth M, Dunten RL, Gonzalez A, Kaback HR. Functional interactions between putative intramembrane charged residues in the lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 1992;89:10547–10551. doi: 10.1073/pnas.89.21.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar H, et al. Structure of sugar-bound LacY. Proc Natl Acad Sci USA. 2014;111:1784–1788. doi: 10.1073/pnas.1324141111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robertson DE, Kaczorowski GJ, Garcia ML, Kaback HR. Active transport in membrane vesicles from Escherichia coli: The electrochemical proton gradient alters the distribution of the lac carrier between two different kinetic states. Biochemistry. 1980;19:5692–5702. doi: 10.1021/bi00566a005. [DOI] [PubMed] [Google Scholar]
- 26.Guan L, Kaback HR. Binding affinity of lactose permease is not altered by the H+ electrochemical gradient. Proc Natl Acad Sci USA. 2004;101:12148–12152. doi: 10.1073/pnas.0404936101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaiko O, Bazzone A, Fendler K, Kaback HR. Electrophysiological characterization of uncoupled mutants of LacY. Biochemistry. 2013;52:8261–8266. doi: 10.1021/bi4013269. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Celma JJ, Smirnova IN, Kaback HR, Fendler K. Electrophysiological characterization of LacY. Proc Natl Acad Sci USA. 2009;106:7373–7378. doi: 10.1073/pnas.0902471106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Celma JJ, Ploch J, Smirnova I, Kaback HR, Fendler K. Delineating electrogenic reactions during lactose/H+ symport. Biochemistry. 2010;49:6115–6121. doi: 10.1021/bi100492p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guan L, Kaback HR. Lessons from lactose permease. Annu Rev Biophys Biomol Struct. 2006;35:67–91. doi: 10.1146/annurev.biophys.35.040405.102005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.le Coutre J, Kaback HR, Patel CK, Heginbotham L, Miller C. Fourier transform infrared spectroscopy reveals a rigid alpha-helical assembly for the tetrameric Streptomyces lividans K+ channel. Proc Natl Acad Sci USA. 1998;95:6114–6117. doi: 10.1073/pnas.95.11.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patzlaff JS, Moeller JA, Barry BA, Brooker RJ. Fourier transform infrared analysis of purified lactose permease: A monodisperse lactose permease preparation is stably folded, alpha-helical, and highly accessible to deuterium exchange. Biochemistry. 1998;37:15363–15375. doi: 10.1021/bi981142x. [DOI] [PubMed] [Google Scholar]
- 33.Sayeed WM, Baenziger JE. Structural characterization of the osmosensor ProP. Biochim Biophys Acta. 2009;1788:1108–1115. doi: 10.1016/j.bbamem.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 34.Ataka K, Heberle J. Use of surface enhanced infrared absorption spectroscopy (SEIRA) to probe the functionality of a protein monolayer. Biopolymers. 2006;82:415–419. doi: 10.1002/bip.20501. [DOI] [PubMed] [Google Scholar]
- 35.Kriegel S, Uchida T, Osawa M, Friedrich T, Hellwig P. Biomimetic environment to study E. coli complex I through surface-enhanced IR absorption spectroscopy. Biochemistry. 2014;53:6340–6347. doi: 10.1021/bi500955a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.