Significance
All eukaryotic cells initiate DNA replication at multiple genomic sites. For most cell types these sites lack a well-defined sequence signature, so it is not understood how they are selected. To analyze the factors that influence initiation site selection and determine the dynamics of replication throughout the genome, we developed an integrative computational model of DNA replication in the model organism Schizosaccharomyces pombe. This analysis showed that the locations of initiation sites are determined not only by the sequence preferences of the S. pombe origin recognition complex (ORC), but also by the interference of transcription with the formation of prereplication complexes. Our findings suggest that transcription has influenced evolution of the binding properties of ORC and replication dynamics in eukaryotes.
Keywords: DNA replication, replication origins, replication dynamics, transcriptional interference, replication timing
Abstract
Each genomic locus in a eukaryotic cell has a distinct average time of replication during S phase that depends on the spatial and temporal pattern of replication initiation events. Replication timing can affect genomic integrity because late replication is associated with an increased mutation rate. For most eukaryotes, the features of the genome that specify the location and timing of initiation events are unknown. To investigate these features for the fission yeast, Schizosaccharomyces pombe, we developed an integrative model to analyze large single-molecule and global genomic datasets. The model provides an accurate description of the complex dynamics of S. pombe DNA replication at high resolution. We present evidence that there are many more potential initiation sites in the S. pombe genome than previously identified and that the distribution of these sites is primarily determined by two factors: the sequence preferences of the origin recognition complex (ORC), and the interference of transcription with the assembly or stability of prereplication complexes (pre-RCs). We suggest that in addition to directly interfering with initiation, transcription has driven the evolution of the binding properties of ORC in S. pombe and other eukaryotic species to target pre-RC assembly to regions of the genome that are less likely to be transcribed.
Duplication of a eukaryotic genome is a complex process that must be completed in a timely and accurate manner once each cell division cycle. Although much has been learned about the biochemical pathways involved in DNA replication, less is known about the dynamics of the process at the genomic level. For a particular cell type, the average time of replication of each genomic locus has a characteristic and reproducible value, but the pattern of replication timing across the genome may change significantly during development and differentiation (1). There is strong evidence that the time of replication of different loci is correlated with the frequency of mutations of various kinds and that this is a significant factor in evolution and in the development of pathological conditions such as cancer (2–7). Thus, it is important to understand the features of the genome that determine the dynamics of replication.
Initiation of DNA replication in eukaryotic cells takes place in two temporally separated steps (8–10). From late M through G1 phase, prereplication complexes (pre-RCs) are assembled at multiple sites in the genome. Pre-RC assembly requires the origin recognition complex (ORC), Cdt1 and Cdc6, which function to load the replicative helicase Mcm2-7 onto DNA in an inactive form. During S phase a subset of the loaded Mcm2-7 helicases is activated in a series of steps leading to the formation of functional replisomes. We refer to this multistep process as pre-RC activation (or firing) in the remainder of this paper. The distribution of pre-RCs in the genome depends, at least in part, on the DNA binding properties of the ORC (11). However, the ORCs of most eukaryotic species do not bind to a highly specific recognition sequence, so the factors that determine the locations of pre-RCs are largely unknown. Because the activation of pre-RCs in S phase appears to be stochastic, the replication timing pattern may principally depend on the distribution of pre-RCs within the genome (12, 13).
A deeper understanding of the dynamics of eukaryotic DNA replication will require a quantitative model describing the spatial distribution of potential initiation sites and the time course of their activation during S phase. The fission yeast Schizosaccharomyces pombe represents a useful system for developing such a model because its genome has many characteristics in common with that of other eukaryotes. Early genetic studies identified segments of the S. pombe genome, called autonomously replicating sequence (ars) elements, that function as origins of DNA replication (14). These elements are large (>1 kb) and rich in A and T residues but do not contain a common sequence motif (15, 16). Subsequent characterization of S. pombe ORC (SpORC) revealed that the SpOrc4 subunit contains nine AT-hook motifs that are essential for binding of SpORC to ars elements in vitro and in vivo (17, 18). AT hooks bind to short stretches of AT-rich DNA via minor groove interactions and do not require a specific sequence for high-affinity binding (19).
The distribution of initiation sites in S. pombe has been probed by both single-molecule techniques (DNA combing) and genome-wide methods (mapping of nascent DNA and polymerase utilization). DNA combing experiments by Patel et al. (20) indicated that initiation events are independent of each other and that the distances between segments of nascent DNA have an exponential distribution, suggesting that initiation sites are distributed randomly over the genome. Subsequent experiments by Kaykov and Nurse (21) extended the DNA combing approach to very large DNA fragments that contain numerous replicated segments. Analysis of such fragments suggested that initiations may occur in clusters, implying that they may not be completely independent of each other.
The distribution of nascent DNA in the S. pombe genome during S phase has been probed by hybridization to microarrays or by deep sequencing (22–25). These studies provided considerable information about the distribution of potential initiation sites, but their resolution was limited by the size of the replicated segments. Although segments of nascent DNA must contain one or more initiation sites, the precise number and locations of such sites cannot be determined with certainty. An alternative approach to analyzing S. pombe DNA replication is profiling of DNA polymerase usage across the genome. Daigaku et al. (26) obtained estimates of the frequency of utilization of the leading and lagging strand DNA polymerases in 300-bp segments on both DNA strands. This rich dataset contains a great deal of information about the pattern of DNA synthesis in a population of S. pombe cells and has provided information about potential initiation sites at a higher resolution than data obtained by other approaches.
We have performed an integrated analysis of DNA combing and polymerase usage datasets for chromosome 2 of S. pombe (21, 26). We show that a simple model with only six parameters accurately describes the complex dynamics of DNA replication at high resolution. Our model suggests that there are many more potential sites of initiation in the genome than previously identified and that the probability of initiation at a given site primarily depends on two factors. The first factor is that SpORC binds preferentially to AT-rich sequences as previously described (17, 18, 27–30). The second factor is that initiations are largely excluded from active transcription units regardless of sequence, presumably because transcription interferes with assembly of pre-RCs or causes their dissociation. The model incorporates both of these factors in a probability distribution function describing the likelihood of a pre-RC at each position in chromosome 2. We suggest that in addition to its direct role in excluding pre-RCs, transcriptional activity has driven the evolution of the DNA binding properties of SpORC because AT-rich regions are mostly extragenic and less likely to be transcribed. Biasing pre-RC assembly to such regions would be expected to increase the efficiency of initiation by reducing futile assembly events. For similar reasons, we expect that other eukaryotic species have evolved mechanisms to reduce transcriptional interference by biasing ORC binding via interactions with sequence elements or other features of chromatin. Thus, in addition to providing an accurate picture of S. pombe DNA replication at high resolution, our model suggests general features of the initiation mechanism that are likely relevant to other eukaryotes including metazoans.
Results
Rationale and Approach.
Our goal was to create a quantitative model of the molecular events that give rise to the complex spatial and temporal patterns of DNA replication that have been observed in S. pombe cells. To define the critical parameters that control these patterns, we analyzed two large datasets from DNA combing and DNA polymerase usage experiments that offer different and largely complementary views of the replication process (21, 26). DNA combing of large molecules provides information about the spacing of initiation sites and the number of active forks at different times during S phase but little information about the features of the genome that determine the probability of initiation at different sites. Conversely, DNA polymerase usage data provide highly detailed information relevant to discovering the mechanisms that determine the local probability of initiation but little direct information about the time course of replication or the absolute number of initiation events. As we show below, by using a simple probabilistic model of DNA replication to analyze both datasets, it is possible to obtain an accurate description of the dynamics of S. pombe DNA replication.
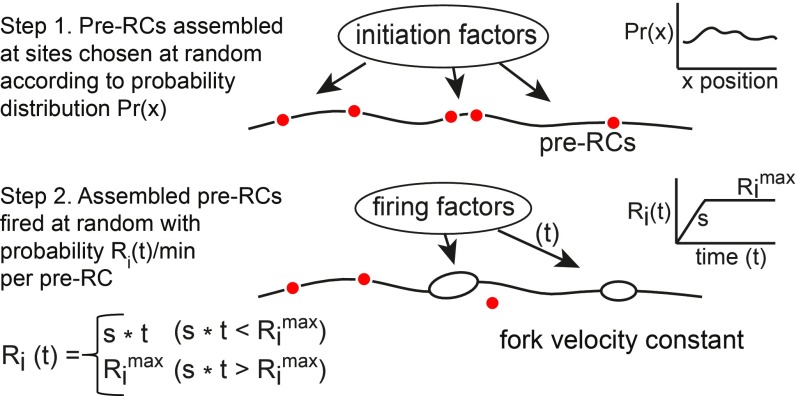
The main features of the model are as follows (Fig. 1): A fixed number of pre-RCs are assembled at sites chosen at random according to a probability function Pr(x) that describes the likelihood of a pre-RC at each position x in a chromosome during S phase. Following assembly, the pre-RCs are activated (fired) at random at a rate of Ri(t) initiations per min per pre-RC at time t during S phase. Based on the observation that the rate of initiation increases during S phase (21), we described Ri(t) by two parameters: the rate of increase in the firing rate (S initiations per min2 per pre-RC) and the maximum firing rate (Rimax initiations per min per pre-RC). The activation of a pre-RC generates two replication forks that move in opposite directions at a constant average velocity. Forks terminate synthesis when they meet forks moving in the opposite direction (or reach the end of the chromosome). Any unfired pre-RCs encountered by forks are disassembled.
Fig. 1.
Model for eukaryotic DNA replication. In step 1, ORC and associated initiation factors bind to sites in the genome in G1 phase and catalyze pre-RC assembly. The distribution of pre-RCs is described by a probability distribution giving the likelihood of stable assembly at each genomic position. A fixed number of pre-RCs per cell is assembled at sites chosen at random from this distribution. In step 2, assembled pre-RCs are fired at random at an increasing rate during S phase. Replication forks move bidirectionally from each initiation site at a constant rate. Any unfired pre-RCs encountered by forks are disassembled.
The model was implemented in the Java programming language with a DNA synthesis engine that updates initiations and fork movements every 0.01 min. In most experiments, 1,000–2,000 molecules were replicated in silico, and the average values of various replication variables were calculated as a function of time. Our implementation shares some features with previous models (31–33) but defines a unique, mechanistically based function to describe the probability of pre-RC assembly and firing, rather than employing empirically derived origin efficiencies or average firing times.
Simulation of DNA Combing Experiments.
We first tested the model using data from DNA combing experiments (21). In these experiments, two-color fluorescence imaging was used to visualize newly replicated DNA segments within large chromosomal DNA fragments. Because of their large size (2 Mb on average), each combed molecule contained a number of replicated segments, making it possible to estimate the average density of replication forks as a function of the percentage of the molecule that had completed replication.
Patel et al. (20) observed that the distances between replicated segments in combed DNA molecules exhibit an exponential distribution. In keeping with this observation, we modeled the combing data by randomly placing pre-RC sites in the genome, a procedure that generates an exponential distribution of the intervals between pre-RCs. The resulting model has only four parameters: the number of pre-RCs that are assembled in each cell, the rate of increase in the rate of firing of assembled pre-RCs during S phase, the maximum rate of firing, and the rate of fork movement. The available estimates of the average rate of fork movement in eukaryotic cells range between 1 and 3 kb/min (34–36). In our initial simulations, we assumed a rate of 2 kb/min in the middle of this range, which is close to the average of 2.8 kb/min estimated by DNA combing (21). We optimized the remaining three parameters by minimizing the mean square difference between the number of forks per Mb predicted by the simulation and the number of forks per Mb observed in 160 combed molecules in the Kaykov and Nurse (21) dataset.
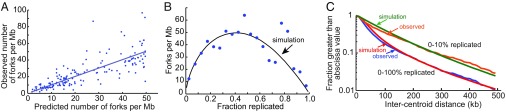
Fig. 2A shows a plot of the observed fork density (forks per Mb) vs. the fork density predicted by the model after optimization of the parameters. A linear regression gives a slope of 0.97 with an R2 value of 0.63. Using these optimized parameters, we predicted the average number of forks per Mb as a function of the fraction of the genome replicated (Fig. 2B). The simulation agrees well with the observed fork density of the combed molecules. We also predicted the distribution of the distances between centroids of the replicated segments in the genome (Fig. 2C). The lower curves in Fig. 2C show the complementary cumulative frequency distributions of the intercentroid distances for all 160 molecules in the dataset (0–100% replicated). There is excellent agreement between prediction and experiment with the two curves exhibiting very similar slopes over a range encompassing 99% of the 3,200 intercentroid distances observed by Kaykov and Nurse (21). The dataset of combed molecules is heterogeneous containing molecules with widely different extents of replication and therefore widely different average intercentroid distances. We plotted the complementary cumulative frequency distribution of a subset of molecules whose percent replication was less than 10% (Fig. 2C; 0–10% replicated). The distribution of the latter molecules has a shallower initial slope because the average intercentroid distance is larger in molecules that have not yet fired many pre-RCs. Again, there is good agreement between prediction and experiment. We conclude that the model provides an accurate and quantitative description of DNA replication in S. pombe.
Fig. 2.
Simulation of DNA combing data. Simulations of S. pombe DNA replication based on the model described in Fig. 1 were performed with optimized firing parameters assuming an exponential distribution of the intervals between pre-RCs. The values of the parameters, optimized as described in Methods, were as follows: pre-RC density, 80 per Mb; rate of increase of firing probability per min, 0.022 events per min2 per pre-RC; maximum firing rate, 0.3 events per min per pre-RC; fork velocity, 2 kb/min. (A) Comparison of the observed density of replication forks with that predicted by the simulation. Each point represents a DNA molecule in the Kaykov and Nurse (21) dataset. The value on the ordinate is the observed density of forks in each molecule, and the value on the abscissa is the predicted density of forks for a molecule with the same percent replication. Slope of the regression line = 0.97; R2 = 0.63. (B) Comparison of the observed and predicted density of replication forks as a function of the fraction of the genome replicated. The observed fork densities (blue points) are the average observed values in windows of 5% replicated. The black line gives the fork densities predicted by the simulation. (C) Distributions of predicted and observed intercentroid distances [the distances between centers of replicated segments, referred to as interorigin distances in Kaykov and Nurse (21)]. The distributions are shown as semilog plots of the fraction of intercentroid distances greater than the value on the abscissa. Lower curves include 160 combed DNA molecules in the dataset of Kaykov and Nurse (21) with extents of replication ranging from 0 to 100%. Upper curves include a subset of molecules replicated from 0 to 10%.
Independence of Initiation Events.
It was previously suggested that the curvature of the cumulative frequency distribution of intercentroids in a semilog plot is caused by clustering of initiation events, implying that firing of nearby pre-RCs is coordinated in some way (21). However, curvature is also evident in the simulation based on a random probability distribution for pre-RC sites and stochastic firing, so clustering is not required to explain this phenomenon (Fig. 2C). The curvature is a natural consequence of the fact that the dataset contains molecules with widely different average intercentroid distances (Fig. 2C), so the cumulative distribution function is not a pure exponential but a sum of exponentials with different decay constants. Thus, we find no evidence for coordination of initiation events, in agreement with the results of Patel et al. (20).
The Number of Pre-RCs.
During the optimization of the replication parameters, we noted that the number of assembled pre-RCs per cell is not constrained at the upper bound. Values of pre-RC density of 80 per Mb and higher are equally consistent with the combing data because the increases in pre-RC density can be compensated by a decrease in the optimized firing rate per pre-RC (SI Appendix, Fig. S1). The supernumerary pre-RCs are largely eliminated by disassembly of pre-RCs encountered by forks, so the number of initiations remains relatively constant (SI Appendix, Fig. S1B). A pre-RC density of 80 per Mb corresponds to an average distance between pre-RCs of 12.5 kb or 1,100 pre-RCs per genome. We used this density for our simulations, but we cannot rule out the possibility that the actual pre-RC density may be somewhat greater. Direct measurement of the average number of assembled pre-RCs is needed to resolve this issue.
Dynamics of DNA Replication.
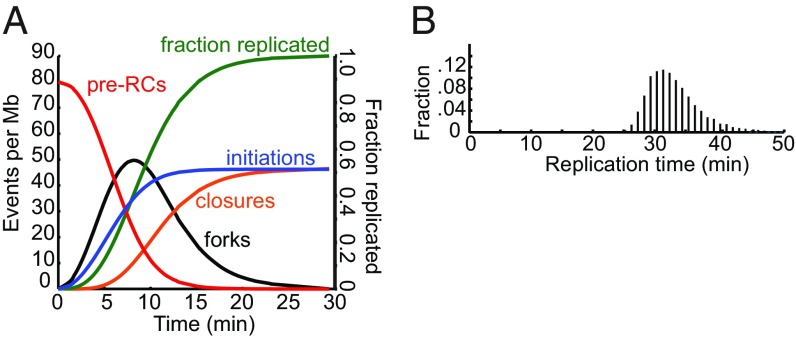
The estimates of the replication parameters from the DNA combing data allowed us to examine the global dynamics of DNA replication in detail (Fig. 3A). In this analysis, the initial number of pre-RCs assembled in each cell is 1,090, corresponding to an average density of 80 pre-RCs per Mb. The number of replication forks reaches a maximum of about 50 per Mb, in good agreement with the maximum observed experimentally. The number of initiations increases throughout S phase, although at a decreasing rate due to the exhaustion of unfired pre-RCs. The average number of initiation events reaches 47 per Mb, so ∼40% of the assembled pre-RCs are disassembled by forks before they can fire.
Fig. 3.
Dynamics of S. pombe DNA replication. Simulation of S. pombe DNA replication with the optimized parameters given in the legend of Fig. 2. (A) Values of replication variables as a function of time during S phase. (B) Frequency distribution of the time required to complete DNA replication.
The simulated time course of DNA replication shows a number of interesting features. Approximately 90% of initiations occur during the first third of S phase. By this time most assembled pre-RCs have either fired or been disassembled by forks. Thus, the remaining two-thirds of S phase is mostly devoted to DNA chain elongation by forks established earlier in S phase. Because the distances between initiation sites follow an exponential distribution, the number of forks, and hence the average rate of DNA synthesis, also undergo a slow exponential decline over time (20, 31). It takes approximately twice as long for the cell to complete the replication of the second half of the genome as the first. Because the rate of DNA synthesis is very low for a long period of time, it seems likely that measurements of the average duration of S phase based on flow cytometry or other relatively insensitive methods have significantly underestimated the length of S phase. Consistent with this suggestion, SI Appendix, Fig. S2 shows that DNA synthesis can be detected by pulse labeling with EdU well into period of the cell cycle generally considered to be the postreplicative G2 phase.
One additional predicted feature of the replication dynamics of S. pombe is that the duration of S phase in a population of cells has a relatively broad distribution (Fig. 3B). Although the average duration of S phase at a fork rate of 2 kb per min is 32 min, it is more than 42 min in about 3% of the cells. This is a direct consequence of the stochastic nature of DNA initiation. By chance, some cells will have a few very large genomic intervals lacking initiations, requiring forks to move great distances. The predicted heterogeneity in the duration of S phase was confirmed by the pulse labeling experiments (SI Appendix, Fig. S2).
As noted above, the average fork rate in S. pombe and other eukaryotic cells is not known with high precision, so we considered the effects of changing the fork rate from the assumed value of 2 kb/min. In our model the fork velocity is effectively a scale factor that relates the fraction of the genome replicated per unit time to the instantaneous number of forks, so its value only affects the average duration of S phase, not the shape of the temporal profile of DNA replication shown in Fig. 3. At the assumed average fork rate of 2 kb/min the simulation predicts that the average duration of S phase is 32 min. If the average fork rate were as low as 1 kb/min or as high as 3 kb/min, the predicted average durations of S phase are 64 and 21 min, respectively.
Polymerase Usage Experiments.
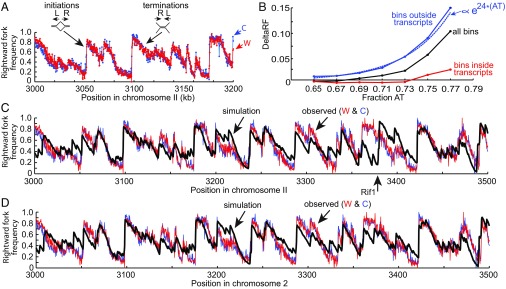
Although application of the model to the DNA combing data provides an accurate description of the global dynamics of DNA replication, a complete picture requires incorporation of information about the distribution of initiation sites as a function of position in the genome. As noted above, the most detailed high-resolution view of S. pombe DNA replication is a genome-wide study of DNA polymerase usage (26). This so-called Pu-seq study employed mutant forms of the leading (epsilon) and lagging (delta) strand polymerases that incorporate ribonucleotides at high frequency. By localizing the positions of ribonucleotide incorporation via a high-throughput sequencing strategy, Daigaku et al. (26) quantified the average utilization of DNA polymerases epsilon and delta on both the Watson and Crick strands in 300-bp segments across the S. pombe genome. This information is sufficient to determine the fraction of rightward-moving forks (or, reciprocally, leftward-moving forks) in each segment of the genome and thus encodes the fine details of initiations and terminations of DNA replication averaged over a population of cells (Fig. 4A).
Fig. 4.
Simulation of polymerase usage. (A) Polymerase usage data for a segment of chromosome 2 from Daigaku et al. (26). Rightward fork frequency increases to the right at sites of initiation. Rightward fork frequency decreases to the right at sites of termination. (B) Initiations are excluded from transcribed regions. DeltaRF is defined as the change in rightward fork frequency in a 300-bp segment relative to that of the segment immediately to its left. Black line indicates deltaRF for all 300-bp segments (bins) in chromosome 2 as a function of AT content, blue line indicates deltaRF for 300-bp segments outside transcription units, red line indicates deltaRF for 300-bp segments inside transcription units, and blue dotted line indicates exponential fit. (C) Comparison of observed rightward fork frequencies in a segment of chromosome 2 (red, Watson strand; blue, Crick strand) with the rightward fork frequencies predicted by simulation of replication (black). The firing parameters for the simulation were the same as in Fig. 2. The relative probability of pre-RC assembly at each site in the genome, obtained as described in the text, was , where AT(x) is the fraction AT in a 25-bp window centered at position x and T(x) = 0 if the window overlaps a transcription unit, T(x) = 1 otherwise. (D) Comparison of observed rightward fork frequencies with rightward fork frequencies predicted by a simulation that disallows firing of pre-RCs at the Rif1 binding site (position 3380 kb). See text for details.
Changes in the rightward fork frequency reflect the probability of initiation or termination events. In a segment of the genome that has a high probability of initiation, the average fraction of rightward forks will increase toward the right, whereas in a segment that has no probability of initiation, the fraction of rightward forks will decrease due to terminations (Fig. 4A). The observed pattern of rightward forks as a function of position is extremely complex showing significant changes over distances of even a few kb, indicating that the probability of initiation and termination can change dramatically over short distances (26). We reasoned that if we could discover a probability distribution for sites of pre-RC assembly that allowed the model to reproduce these data, it would reveal the underlying features of the genome that promote (or suppress) pre-RC assembly.
Initiation Events Occur at AT-Rich Sequences but Are Excluded from Transcribed Regions of the Genome.
As an initial approach to studying the influence of genomic features on the probability of initiation, we defined a heuristic measure of initiation frequency, deltaRF, as the change in the rightward fork frequency in each 300-bp segment relative to that of the segment immediately preceding it. Roughly speaking, deltaRF is a measure of the fraction of initiations minus the fraction of terminations in each 300-bp segment of the genome. We first examined the effect of AT content on deltaRF in 300-bp segments of chromosome 2 of S. pombe (Fig. 4B). As expected, the average value of deltaRF increases dramatically with the AT content of a segment, indicating that the Pu-seq data are consistent with previous work showing that AT content is a strong predictor of SpORC localization (17, 18, 29, 30).
In addition to examining the role of the sequence preferences of SpORC, we explored the possibility that transcriptional activity might affect the initiation frequency (37–41). For this purpose, we divided the population of 300-bp segments in chromosome 2 into segments that are in annotated transcription units and segments that are in nontranscribed regions. The striking result of this analysis is shown in Fig. 4B. The average deltaRF for 300-bp segments in nontranscribed regions showed an even stronger dependence on AT content than segments as a whole, increasing by a factor of ∼10 for each 0.1 increase in the fraction AT. By contrast, the average deltaRF for segments in transcribed segments was negative or close to zero at all AT contents. This finding indicates that initiations are largely excluded from transcribed regions of the S. pombe genome. The fact that the exclusion is independent of AT content suggests that it is an intrinsic feature of transcribed regions and is not simply explained by the possibility that such regions have no affinity for SpORC. We suggest that transcription antagonizes pre-RC assembly, perhaps by causing the dissociation of assembly intermediates or assembled pre-RCs. This hypothesis is consistent with experiments in Saccharomyces cerevisiae suggesting that transcription through an origin reduces ORC and minichromosome maintenance (MCM) binding and inhibits origin activity (refs. 39 and 41; Discussion).
Simulation of Pu-Seq Experiments.
Based on the analysis above we postulated a simple probability distribution for pre-RC sites with only two parameters to describe the effects of transcription and base composition. The relative probability Pr(x) of a pre-RC at position x in the genome is given by
| [1] |
where AT(x) is the fraction AT in a 25-bp window centered at position x, C is a constant, and T(x) = 0 if the window overlaps a transcription unit, T(x) = 1 otherwise. To optimize the parameter C, we minimized the mean square deviation between the rightward fork frequency observed in the Pu-seq experiment and that predicted by simulations based on this probability distribution and the firing parameters deduced from the DNA combing experiments. A value of C = 21 was obtained, corresponding to an eightfold increase in the probability of pre-RC assembly for each 0.1 increase in fraction AT.
We used the complete set of optimized parameters to simulate DNA replication of chromosome 2 of S. pombe. A comparison of the predicted and observed rightward fork frequencies for a 500-kb region of chromosome 2 is shown in Fig. 4C. The Pu-seq data provides two complementary estimates of the rightward fork frequency based on DNA polymerase usage on the Watson and Crick strands. Both are plotted in Fig. 4C. The agreement between the simulation and the experimental data is remarkably good (mean square deviation from the average of the experimental data for W and C is 0.0268). The simulation predicts initiation at more than 90% of the positions where the observed rightward fork frequency increases significantly (ref. 26 and Fig. 4 C and D), indicating that the optimized probability distribution in expression 1 provides an accurate description of the likelihood of pre-RC assembly as a function of genome position. Consistent with the results shown in Fig. 4B, a simulation that included the effects of both AT content and transcription had a smaller average deviation from the experimental data than simulation based on AT content alone (mean square deviation 0.0317).
Effect of Rif1 on Firing Rate.
Although the agreement between simulation and experiment in Fig. 4C is generally good, the simulation predicts a strong initiation site near position 3,380 kb in chromosome 2 that was not observed in the Pu-seq data (arrow below the plot in Fig. 4C). This site was shown to be a binding site for the Rif1 protein by ChIP assay (25). Rif1 is a regulatory factor that inhibits activation of pre-RCs at least in part by promoting dephosphorylation of firing factors (42). Although Rif1 localizes preferentially to subtelomeric regions where it contributes to late replication, Hayano et al. (25) identified a number of putative Rif1 sites scattered over the remainder of chromosome 2. The site at position 3,380 kb of chromosome 2 does not function efficiently as an initiation site in wild-type S. pombe but does so in mutant cells that lack Rif1. However, consistent with the optimized probability distribution in expression 1, pre-RCs are assembled at position 3,380 kb in wild-type cells, so the inhibitory effect of Rif1 is mediated at the firing step (25, 43). When we modified the simulation to prevent firing of any pre-RCs assembled in the Rif1-binding region containing position 3,380 kb, the agreement between predicted and observed right fork frequencies improved dramatically in the vicinity of this site (Fig. 4D).
We detected two other positions in chromosome 2 where the simulation predicted efficient initiation that was not observed in the Pu-seq experiment (positions 888 kb and 2,340 kb). Both of these positions correspond to sites where deletion of Rif1 increases the efficiency of initiation in vivo (25). Like the site at 3,380 kb, modification of the simulation to suppress pre-RC firing in these sites significantly improved the local agreement between predicted and observed rightward fork frequency (Fig. 5). Of the remaining Rif1 sites outside of the subtelomeres in chromosome 2, only three were associated with detectable discrepancies between the simulation and the observed Pu-seq data, and the discrepancies were quite minor. Thus, our results suggest that Rif1 has significant effects on the dynamics of DNA replication at relatively few sites outside of the subtelomeric regions and that these effects are highly localized. The assumption of our model that all pre-RCs have the same global firing rate at any given time during S phase appears to be consistent with the dynamics of replication at most sites.
Fig. 5.
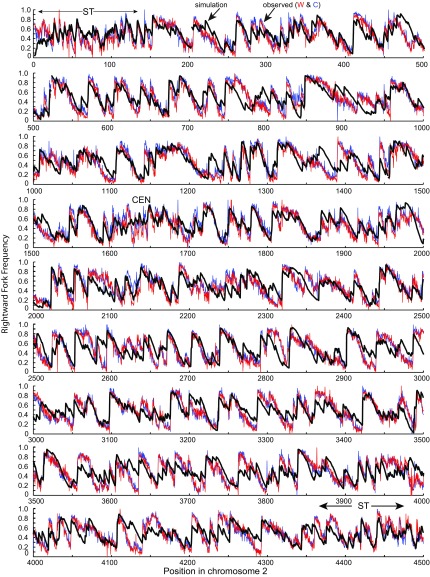
Replication fork directionality across S. pombe chromosome 2. The observed rightward fork frequencies in 300-bp segments of the 4.54-Mb chromosome 2 (red, Watson strand; blue, Crick strand) are from the polymerase usage data of Daigaku et al. (26). The predicted rightward fork frequencies (black line) are from a simulation with the parameters as described in the legend of Fig. 4. The simulation disallows firing of pre-RCs at six Rif1 sites in chromosome 2 (see text).
A complete comparison of the predicted and observed rightward fork frequencies over the entire chromosome is shown in Fig. 5. The simulation predicts much of the detailed structure of the observed Pu-seq data, indicating that the underlying model captures the principal factors that shape the pattern of initiation and termination events.
Prediction of Replication Timing.
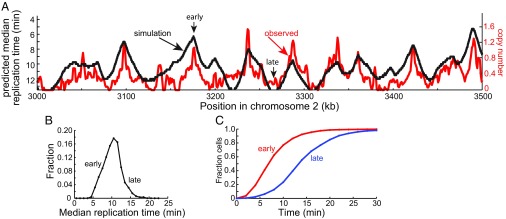
A number of experimental approaches have been used to estimate the relative timing of the replication of different positions in the genome. Many of these are dependent on cell synchronization methods or treatment with replication inhibitors. Daigaku et al. (26) probed replication timing by three different approaches: deep sequencing of genomic DNA from cell populations synchronized by centrifugal elutriation, deep sequencing of S phase cells enriched by cell sorting (Sort-seq), and mathematical analysis of Pu-seq data. The estimates of relative timing of various segments in the S. pombe genome derived from the three independent methods were highly correlated. We compared the median replication times predicted by our model with the normalized copy numbers observed in the Sort-seq experiment because it required minimal perturbation of the cell population (Fig. 6A). An excellent correlation was observed indicating that the model accurately predicts the relative timing of replication of different regions of the genome.
Fig. 6.
Prediction of replication timing. (A) Comparison of the observed replication timing profile of a segment of chromosome 2 with that predicted by simulation of DNA replication. The red line shows the copy number of each 1-kb segment of the genome obtained by deep sequencing of DNA from S phase cells [Sort-seq data of Daigaku et al. (26)]. This profile is taken as a measure, not necessarily linear, of the relative timing of replication of each segment. The predicted median times of replication of each 300-bp segment are from a simulation with the parameters described in Fig. 2. (B) Frequency distribution of predicted median replication times for all 300-bp segments of chromosome 2. (C) Time courses of replication of typical early and late replicating segments (from the positions indicated by arrows in A). The ordinate shows the fraction of cells that have replicated the segment at a given time.
The simulations also showed that the frequency distribution of median replication times of 300-bp segments across chromosome 2 is quite heterogeneous (Fig. 6B). The mean of the distribution was 10.2 min, and the SD was 2.4 min. We examined the detailed kinetics of replication of an early replicating segment (median replication time 6.2 min) and a late replicating segment (median replication time 13.6). As shown in Fig. 6C, ∼75% of the cells in the population replicated the early segment by 10 min, which is about one-third of the duration of S phase. This was expected because as shown in Fig. 3 above, the vast majority of initiation events in the cell population have occurred by this time. Thus, any region of the genome that has a relatively high probability of pre-RC assembly will be close to one or more initiation sites in nearly every cell and will complete replication early in S phase. By contrast, the time of replication of the late replicating region in the cell population was spread over most of S phase. This behavior is also easy to understand. Regions of the genome that have a relatively low density of potential pre-RC assembly sites will likely be distant from sites of initiation. Because of the stochastic nature of pre-RC assembly and firing, the distance of such regions from the nearest initiation event will vary over a large range in individual cells, and their times of replication will show similar variance. Although the average time of replication of late regions is greater than early regions, it is important to recognize that late regions can be just as likely to replicate in the first half of S phase as the second half of S phase (Fig. 6C). For completeness, we also predicted the fraction of cells that had replicated each 300-bp segment in the region of chromosome 2 shown in Fig. 6A as a function of time in S phase (SI Appendix, Fig. S3).
Distribution of Initiations and Terminations.
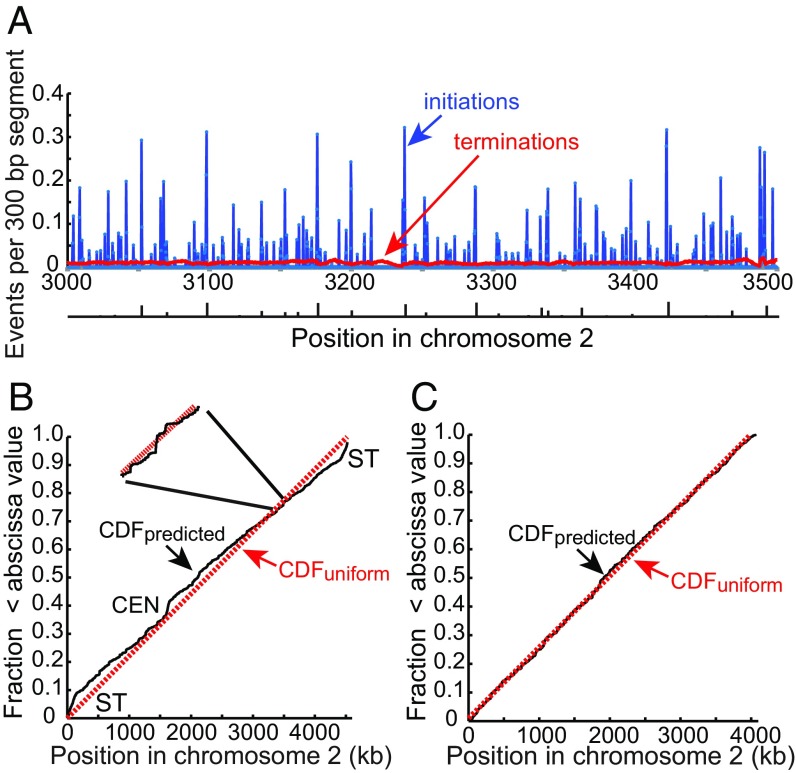
We have shown that a relatively simple distribution function based on two genome features, base composition and transcriptional activity, is sufficient to describe the probability of initiation in the S. pombe genome. To quantify the average number of initiation and termination events per cell cycle in each 300-bp segment of the genome, we simulated the replication of 10,000 molecules and accumulated all initiation and termination sites. Fig. 7A shows the results for the 500-kb region of chromosome 2 shown in the preceding figures. Initiations occur in many 300-bp segments, but the average number of initiations per segment per cell is generally less than 0.35. For comparison, the positions identified as potential origins by Daigaku et al. (26) are shown at the bottom of Fig. 7A. These correspond to major transitions in DNA polymerase usage and are identified by the model as well, but the model predicts many more potential initiation sites. Termination events, defined as positions where replication forks converge, are spread over the entire chromosomal segment with an average of about 0.01 terminations per 300-bp segment per cell. The frequency distribution of predicted initiations and terminations per cell per 300-bp segment of chromosome 2 is shown in SI Appendix, Fig. S4. A higher-resolution view of terminations is shown in SI Appendix, Fig. S5. The distribution of terminations is not completely uniform, because segments close to regions with high frequency of initiation have fewer than the average number of terminations, and segments between regions with high frequencies of initiation have more than the average number of terminations.
Fig. 7.
Initiations and terminations. (A) Average number of initiations and terminations per cell cycle for each 300-bp segment in the same 500-kb region of chromosome 2 as in Figs. 4 and 6. The frequency of initiations and terminations in each segment of the genome was determined by simulations with a total of about 2M initiation events. The origins identified from polymerase usage data by Daigaku et al. (26) are shown below the figure with the height of each bar indicating the relative efficiency of use. (B) Probability of pre-RCs in 300-bp bins across chromosome 2 plotted as a cumulative distribution function (CDF). The probabilities were calculated from the function given in the legend of Fig. 4. The red dotted line is the CDF of a uniform distribution. Inset shows the local fluctuation in the probability of pre-RC assembly in a short segment of the chromosome. ST, subtelomere; CEN, centromere. (C) CDF of pre-RC site probability after elimination of centromere and subtelomeric regions, amounting to about 8% of the genome.
The Nature of the Probability Distribution for Pre-RC Assembly Sites.
The predicted cumulative probability distribution (CDF) for pre-RC assembly sites on chromosome 2, based on expression 1, is plotted in Fig. 7B together with the CDF for a uniform probability distribution. As shown in Fig. 7B, Inset, the predicted CDF shows a great deal of local fluctuation over short distances due to inhomogeneities in the local distribution of blocks of transcribed and nontranscribed DNA and the distribution of AT content in nontranscribed DNA. Over longer distances, the predicted CDF is close to the CDF of a uniform distribution, indicating that the density of potential initiation sites is similar over most of the genome. The exceptions to this pattern are the subtelomeres and the centromere where the slope of the CDF is steeper, indicating that the predicted density of pre-RCs is somewhat higher in these regions due to their relative paucity of transcripts and high AT contents. This prediction is consistent with relatively high density of MCM binding observed in these regions (e.g., refs. 24 and 25). However, as mentioned above, the density of pre-RCs is not the sole determinant of replication timing in centromeres and subtelomeres. Subtelomeric regions are late-replicating in part because firing of the assembled pre-RCs is inhibited by Rif1 (25), and centromeres are early-replicating, in part because firing factors are recruited by centromeric Swi6 protein (HP1) (44). Thus, the dynamics of DNA replication in these regions differ from those of the bulk of the genome. If the centromere and subtelomere regions, which account for less than 8% of chromosome 2, are removed from the analysis, the resulting CDF is very close to uniform over the remainder of the genome (Fig. 7C).
Discussion
In this study we constructed a quantitative model of the molecular events that govern the genome-wide dynamics of DNA replication in S. pombe. The model makes a small number of assumptions, requires only a few parameters, and incorporates growing evidence that the selection of initiation sites is probabilistic. It accurately captures the details of two large and high-quality experimental views of the replication process and has a number of features that may generalize to other eukaryotes.
The model posits that the number of potential initiation sites in the fission yeast genome is much larger than the number predicted by previous studies (Fig. 7). In this view the peaks of replicated DNA observed in origin-mapping experiments to date do not represent single discrete origins but are the cumulative product of multiple initiations at different sites in different cells. The probabilities of initiation at different genomic sites fall on a continuum, but the probability at any particular site is very low. For example, our simulations suggest that only about 3% of the 300-bp segments have a total probability of initiation greater than 0.2 per cell (SI Appendix, Fig. S4). Similarly, estimates of origin efficiency, usually defined as the fraction of cells that initiate DNA replication at a particular site, do not reflect some intrinsic property of a single origin but are simply a summation of the probabilities of initiations over different local sites. This interpretation is consistent with several observations. Genetic analysis of S. pombe ars sequences indicated that they contain multiple partially redundant elements that contribute to their activity (16). In subsequent biochemical studies, we showed that SpORC binds with similar affinity to several different nonoverlapping fragments of the 1.2-kb ars1 element and suggested that the high affinity of SpORC for ars elements is due to the cumulative effect of multiple potential binding sites, each of which could serve as an initiation site (18, 30). We also estimated that at least half of intergenes in the S. pombe genome contain potential initiation sites (30). In addition, a close examination of the polymerase utilization data indicates that large increases in rightward fork frequency associated with initiation events do not generally occur in a single 300-bp segment but in incremental steps over several segments, indicative of multiple initiations spanning a short interval (Fig. 4A). For example, the large increase in rightward fork frequency near position 3,100 kb in Fig. 4A occurs over three adjacent 300-bp segments. Finally, simulations of DNA replication based on our model predict that by the time 10% of the genome has been replicated, the pattern of newly synthesized DNA has consolidated into peaks much fewer in number than the underlying initiation sites. The locations of these peaks correspond to major peaks of nascent DNA observed in origin-mapping experiments carried out in the presence of hydroxyurea (SI Appendix, Fig. S3, and ref. 22).
A second important feature of our model is that the locations of initiation sites are determined not only by the strong preference of SpORC for AT tracts (17, 18, 27–30) but also by the inhibitory effect of transcription (Fig. 4B). This fact emerged from our analysis of the polymerase usage data, which revealed that initiations do not occur in transcription units even if they contain AT-rich sequences that could be recognized by SpORC. Although we cannot rule out other possibilities, the simplest explanation for this exclusion is that the transcription machinery destabilizes pre-RCs and/or intermediates in pre-RC assembly. Such a mechanism may be analogous to the disassembly of unfired pre-RCs induced by the replication machinery itself. The exclusion of initiations from transcription units confines initiation events to circumscribed regions and, thus, is a significant contributor to the observed fluctuations in the probability of initiation along the genome.
Several studies in S. cerevisiae have provided evidence that transcription can interfere with initiation at origins of DNA replication (37–41, 45). In particular, it was shown that transcription through a yeast origin driven by an inducible GAL promoter blocked origin function (41). Subsequent analysis of the mechanism of this effect demonstrated that transcription reduces ORC binding and pre-RC assembly (39, 40). It is likely that transcriptional interference with initiation also occurs in metazoan cells. For example, initiation events at the DHFR locus of CHO cells were mapped to multiple sites in a 55-kb intergenic region adjacent to the DHFR gene but were never observed in the body of the gene itself. However, when transcription of the DHFR gene was abrogated by deletion of the promoter, initiation of DNA replication was observed in the body of the gene (46).
It is expected that dissociation of ORC, pre-RCs, or assembly intermediates displaced from the DNA by transcription would reduce the overall efficiency of initiation. Displacement of pre-RCs may not be irrevocable if it occurs during the active period of pre-RC assembly in G1 (39), but pre-RCs displaced during S phase cannot be replaced because new pre-RC assembly is precluded. Because intergenic DNA in S. pombe has a significantly higher average AT content than genes, the AT hooks in SpORC have the effect of biasing binding toward nontranscribed DNA where stable assembly of pre-RCs can take place unimpeded (30). Thus, we suggest that the bias of SpORC binding to AT-rich DNA serves to increase the overall efficiency of utilization of cellular initiation factors by reducing futile assembly events.
Transcriptional interference with initiation may contribute significantly to the changes in the pattern of DNA replication that have been observed to occur during cell differentiation and development. One early and informative example in yeast is a change in origin utilization that accompanies entry into the meiotic program from the mitotic cell cycle (40). It was observed that the activity of ARS605, a highly efficient origin of DNA replication in the mitotic cell cycle, is completely suppressed in premeiotic S phase. ARS605 is located in the body of the MSH4 gene, which is specifically transcribed in early meiosis. Detailed analysis revealed that transcription of MSH4 blocked origin function by reducing ORC binding. Although there are many examples of changes in the pattern of initiation of DNA replication timing during metazoan development (see ref. 47 for review), a classic case is the change that occurs at the midblastula transition of Xenopus embryos. In early embryos, transcription of the zygotic genome is repressed, and potential origins of DNA replication are distributed uniformly (48). At the midblastula transition, when transcription of the genome ensues, initiation becomes more localized to specific regions. Analysis of the rRNA loci showed that the localization was a result of reduced initiation in transcription units (49). It was suggested that this effect was most likely a result of changes in the local chromatin landscape associated with transcription, but direct interference of transcription with initiation could not be ruled out.
Although it appears likely that transcriptional interference is a major factor determining the distribution of pre-RCs, our assumption of complete exclusion from annotated transcription units is probably an oversimplification, albeit a necessary one, given the limitations in our current knowledge of the rate and timing of transcription of various genes. We might expect that the extent of transcriptional interference with initiation would be a function of the frequency of transcription of a gene during G1 and S phases of the cell cycle. Thus, the accuracy of the model may improve as more quantitative information accumulates about the activity of transcription units.
Our model is consistent with the view that the timing of replication of the vast majority of positions in the genome is determined largely by the local probability density of potential pre-RC sites. This hypothesis was suggested previously based in part on the correlation of early replication with the average number of ORC binding sites (50–54). Our simulations show that the probabilistic nature of initiation has two important consequences for replication timing. Positions that replicate on average late in S phase have a very broad distribution of replication times, so they replicate early in a significant fraction of the cells in the population. Conversely, any position in the genome has some probability of replicating late in S phase because, by chance, nearby initiations may not occur even in regions with a high probability of pre-RC assembly.
The timing of replication of genomic sequences is biologically important because late replication is correlated with mutation frequency (2–5). The correlation appears to shape mutational processes during evolution and is also a consistent feature of human cancer cells. The rate of base substitution mutation may be higher in late replicating DNA because the expression and/or activity of mutagenic translesion DNA polymerases is confined to a period in late S/G2 phase (4, 55, 56). In the case of S. pombe, we have reported that when cells commit to mitosis, they lose the capacity for homology-dependent postreplication repair, which is largely error-free, and switch to a mutagenic mechanism requiring DNA polymerase zeta and rev1 (56). Late replication may also be associated with a higher rate of deletion mutations and rearrangements. For example, common fragile sites (CFSs), which have been associated with copy number variants in human cancers, occur in very large, actively transcribed genes that are late replicating because of a paucity of initiation events (57, 58). It has been suggested that the reduced frequency of initiation at CFSs might be a result of eviction of pre-RCs by transcription (59). Our model is consistent with this possibility, and simulation of S. pombe replication predicts a positive correlation between the average replication time of the midpoints of transcription units and the size of such units (SI Appendix, Fig. S6).
Although the variation in AT content and exclusion of pre-RCs from actively transcribed DNA result in large short-range fluctuations in the probability of stable pre-RC assembly, our model predicts that the average density of potential initiation sites in longer intervals is similar over most of the genome (Fig. 7). This is a consequence of the fact that nontranscribed regions and associated AT-rich tracts are distributed relatively evenly. The heterochromatic centromere and subtelomeric DNA are exceptions to this rule, presumably because their genetic organization and chromatin structure are different from the bulk of the genome. The replication of heterochromatin in metazoans also appears to differ from the rest of the genome (reviewed in ref. 60). Heterochromatic regions replicate very late in S phase after most euchromatic sequences. It is possible that like the subtelomeric regions of the S. pombe, the rate of firing of pre-RCs in metazoan heterochromatin is reduced by some inhibitory mechanism.
It seems likely that transcriptional interference is a significant factor affecting the distribution of initiation sites in all eukaryotic cells. Given the potential effect of this phenomenon on the efficiency of initiation, we speculate that eukaryotic cells evolved mechanisms to bias ORC binding to avoid transcriptional interference. Although ORC binding to AT-rich DNA provides this function in S. pombe, the interactions with DNA or chromatin that drive biased ORC binding may be different in other species. In budding yeast, ORC is targeted to nontranscribed or weakly transcribed genomic loci by highly specific interactions with the ars consensus sequence (39). In metazoans ORC may be targeted by interactions with chromatin marks or some feature of so-called open-chromatin sites (52, 53, 61, 62). Future modeling studies of DNA replication of metazoans at high resolution may help to illuminate these issues.
Methods
DNA replication in an S. pombe cell was modeled in silico as a two-step process. In the first step, pre-RCs were assembled at a fixed number of sites in the genome. The positions of the pre-RCs were chosen at random according to a probability distribution that incorporated the sequence preferences of SpORC and the influence of transcriptional interference. In the second step, assembled pre-RCs were fired, and new DNA strands were synthesized. The assumptions of the model, as well as the details of its in silico implementation, are described in SI Appendix. Source code for the replication simulation software has been posted at https://github.com/tomkelly-mskcc/Spombe-Replication-Dynamics.
Supplementary Material
Acknowledgments
We thank Kenneth Marians, John Petrini, Brendan Cormack, and Iestyn Whitehouse for comments on the manuscript.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 4776.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818680116/-/DCSupplemental.
References
- 1.Rhind N, Gilbert DM. DNA replication timing. Cold Spring Harb Perspect Biol. 2013;5:a010132. doi: 10.1101/cshperspect.a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stamatoyannopoulos JA, et al. Human mutation rate associated with DNA replication timing. Nat Genet. 2009;41:393–395. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen CL, et al. Impact of replication timing on non-CpG and CpG substitution rates in mammalian genomes. Genome Res. 2010;20:447–457. doi: 10.1101/gr.098947.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lang GI, Murray AW. Mutation rates across budding yeast chromosome VI are correlated with replication timing. Genome Biol Evol. 2011;3:799–811. doi: 10.1093/gbe/evr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woo YH, Li WH. DNA replication timing and selection shape the landscape of nucleotide variation in cancer genomes. Nat Commun. 2012;3:1004. doi: 10.1038/ncomms1982. [DOI] [PubMed] [Google Scholar]
- 6.Liu L, De S, Michor F. DNA replication timing and higher-order nuclear organization determine single-nucleotide substitution patterns in cancer genomes. Nat Commun. 2013;4:1502. doi: 10.1038/ncomms2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bleichert F, Botchan MR, Berger JM. Mechanisms for initiating cellular DNA replication. Science. 2017;355:eaah6317. doi: 10.1126/science.aah6317. [DOI] [PubMed] [Google Scholar]
- 9.Bell SP, Labib K. Chromosome duplication in Saccharomyces cerevisiae. Genetics. 2016;203:1027–1067. doi: 10.1534/genetics.115.186452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelly T. Historical perspective of eukaryotic DNA replication. Adv Exp Med Biol. 2017;1042:1–41. doi: 10.1007/978-981-10-6955-0_1. [DOI] [PubMed] [Google Scholar]
- 11.Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 12.Bechhoefer J, Rhind N. Replication timing and its emergence from stochastic processes. Trends Genet. 2012;28:374–381. doi: 10.1016/j.tig.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Czajkowsky DM, Liu J, Hamlin JL, Shao Z. DNA combing reveals intrinsic temporal disorder in the replication of yeast chromosome VI. J Mol Biol. 2008;375:12–19. doi: 10.1016/j.jmb.2007.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maundrell K, Hutchison A, Shall S. Sequence analysis of ARS elements in fission yeast. EMBO J. 1988;7:2203–2209. doi: 10.1002/j.1460-2075.1988.tb03059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubey DD, Kim SM, Todorov IT, Huberman JA. Large, complex modular structure of a fission yeast DNA replication origin. Curr Biol. 1996;6:467–473. doi: 10.1016/s0960-9822(02)00514-6. [DOI] [PubMed] [Google Scholar]
- 16.Clyne RK, Kelly TJ. Genetic analysis of an ARS element from the fission yeast Schizosaccharomyces pombe. EMBO J. 1995;14:6348–6357. doi: 10.1002/j.1460-2075.1995.tb00326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chuang RY, Kelly TJ. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc Natl Acad Sci USA. 1999;96:2656–2661. doi: 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chuang RY, Chretien L, Dai J, Kelly TJ. Purification and characterization of the Schizosaccharomyces pombe origin recognition complex: Interaction with origin DNA and Cdc18 protein. J Biol Chem. 2002;277:16920–16927. doi: 10.1074/jbc.M107710200. [DOI] [PubMed] [Google Scholar]
- 19.Reeves R, Nissen MS. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J Biol Chem. 1990;265:8573–8582. [PubMed] [Google Scholar]
- 20.Patel PK, Arcangioli B, Baker SP, Bensimon A, Rhind N. DNA replication origins fire stochastically in fission yeast. Mol Biol Cell. 2006;17:308–316. doi: 10.1091/mbc.E05-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaykov A, Nurse P. The spatial and temporal organization of origin firing during the S-phase of fission yeast. Genome Res. 2015;25:391–401. doi: 10.1101/gr.180372.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heichinger C, Penkett CJ, Bähler J, Nurse P. Genome-wide characterization of fission yeast DNA replication origins. EMBO J. 2006;25:5171–5179. doi: 10.1038/sj.emboj.7601390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu J, et al. Genome-wide identification and characterization of replication origins by deep sequencing. Genome Biol. 2012;13:R27. doi: 10.1186/gb-2012-13-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi M, et al. Genome-wide localization of pre-RC sites and identification of replication origins in fission yeast. EMBO J. 2007;26:1327–1339, and erratum (2007) 26:2821. doi: 10.1038/sj.emboj.7601585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayano M, et al. Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev. 2012;26:137–150. doi: 10.1101/gad.178491.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daigaku Y, et al. A global profile of replicative polymerase usage. Nat Struct Mol Biol. 2015;22:192–198. doi: 10.1038/nsmb.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong D, DePamphilis ML. Site-specific DNA binding of the Schizosaccharomyces pombe origin recognition complex is determined by the Orc4 subunit. Mol Cell Biol. 2001;21:8095–8103. doi: 10.1128/MCB.21.23.8095-8103.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JK, Moon KY, Jiang Y, Hurwitz J. The Schizosaccharomyces pombe origin recognition complex interacts with multiple AT-rich regions of the replication origin DNA by means of the AT-hook domains of the spOrc4 protein. Proc Natl Acad Sci USA. 2001;98:13589–13594. doi: 10.1073/pnas.251530398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segurado M, de Luis A, Antequera F. Genome-wide distribution of DNA replication origins at A+T-rich islands in Schizosaccharomyces pombe. EMBO Rep. 2003;4:1048–1053. doi: 10.1038/sj.embor.7400008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai J, Chuang RY, Kelly TJ. DNA replication origins in the Schizosaccharomyces pombe genome. Proc Natl Acad Sci USA. 2005;102:337–342. doi: 10.1073/pnas.0408811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lygeros J, et al. Stochastic hybrid modeling of DNA replication across a complete genome. Proc Natl Acad Sci USA. 2008;105:12295–12300. doi: 10.1073/pnas.0805549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrick J, Jun S, Bechhoefer J, Bensimon A. Kinetic model of DNA replication in eukaryotic organisms. J Mol Biol. 2002;320:741–750. doi: 10.1016/s0022-2836(02)00522-3. [DOI] [PubMed] [Google Scholar]
- 33.Yang SC, Rhind N, Bechhoefer J. Modeling genome-wide replication kinetics reveals a mechanism for regulation of replication timing. Mol Syst Biol. 2010;6:404. doi: 10.1038/msb.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Méchali M. Eukaryotic DNA replication origins: Many choices for appropriate answers. Nat Rev Mol Cell Biol. 2010;11:728–738. doi: 10.1038/nrm2976. [DOI] [PubMed] [Google Scholar]
- 35.Raghuraman MK, et al. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- 36.Técher H, et al. Replication dynamics: Biases and robustness of DNA fiber analysis. J Mol Biol. 2013;425:4845–4855. doi: 10.1016/j.jmb.2013.03.040. [DOI] [PubMed] [Google Scholar]
- 37.Donato JJ, Chung SC, Tye BK. Genome-wide hierarchy of replication origin usage in Saccharomyces cerevisiae. PLoS Genet. 2006;2:e141. doi: 10.1371/journal.pgen.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gros J, et al. Post-licensing specification of eukaryotic replication origins by facilitated Mcm2-7 sliding along DNA. Mol Cell. 2015;60:797–807. doi: 10.1016/j.molcel.2015.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lõoke M, et al. Relicensing of transcriptionally inactivated replication origins in budding yeast. J Biol Chem. 2010;285:40004–40011. doi: 10.1074/jbc.M110.148924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mori S, Shirahige K. Perturbation of the activity of replication origin by meiosis-specific transcription. J Biol Chem. 2007;282:4447–4452. doi: 10.1074/jbc.M609671200. [DOI] [PubMed] [Google Scholar]
- 41.Snyder M, Sapolsky RJ, Davis RW. Transcription interferes with elements important for chromosome maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2184–2194. doi: 10.1128/mcb.8.5.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davé A, Cooley C, Garg M, Bianchi A. Protein phosphatase 1 recruitment by Rif1 regulates DNA replication origin firing by counteracting DDK activity. Cell Rep. 2014;7:53–61. doi: 10.1016/j.celrep.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu PY, Nurse P. Establishing the program of origin firing during S phase in fission yeast. Cell. 2009;136:852–864. doi: 10.1016/j.cell.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayashi MT, Takahashi TS, Nakagawa T, Nakayama J, Masukata H. The heterochromatin protein Swi6/HP1 activates replication origins at the pericentromeric region and silent mating-type locus. Nat Cell Biol. 2009;11:357–362. doi: 10.1038/ncb1845. [DOI] [PubMed] [Google Scholar]
- 45.Nieduszynski CA, Blow JJ, Donaldson AD. The requirement of yeast replication origins for pre-replication complex proteins is modulated by transcription. Nucleic Acids Res. 2005;33:2410–2420. doi: 10.1093/nar/gki539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saha S, Shan Y, Mesner LD, Hamlin JL. The promoter of the Chinese hamster ovary dihydrofolate reductase gene regulates the activity of the local origin and helps define its boundaries. Genes Dev. 2004;18:397–410. doi: 10.1101/gad.1171404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nordman J, Orr-Weaver TL. Regulation of DNA replication during development. Development. 2012;139:455–464. doi: 10.1242/dev.061838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahbubani HM, Paull T, Elder JK, Blow JJ. DNA replication initiates at multiple sites on plasmid DNA in Xenopus egg extracts. Nucleic Acids Res. 1992;20:1457–1462. doi: 10.1093/nar/20.7.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hyrien O, Maric C, Méchali M. Transition in specification of embryonic metazoan DNA replication origins. Science. 1995;270:994–997. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- 50.Rhind N. DNA replication timing: Random thoughts about origin firing. Nat Cell Biol. 2006;8:1313–1316. doi: 10.1038/ncb1206-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rhind N, Yang SC, Bechhoefer J. Reconciling stochastic origin firing with defined replication timing. Chromosome Res. 2010;18:35–43. doi: 10.1007/s10577-009-9093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacAlpine HK, Gordân R, Powell SK, Hartemink AJ, MacAlpine DM. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 2010;20:201–211. doi: 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miotto B, Ji Z, Struhl K. Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers. Proc Natl Acad Sci USA. 2016;113:E4810–E4819. doi: 10.1073/pnas.1609060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Das SP, et al. Replication timing is regulated by the number of MCMs loaded at origins. Genome Res. 2015;25:1886–1892. doi: 10.1101/gr.195305.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waters LS, Walker GC. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G(2)/M phase rather than S phase. Proc Natl Acad Sci USA. 2006;103:8971–8976. doi: 10.1073/pnas.0510167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Callegari AJ, Kelly TJ. Coordination of DNA damage tolerance mechanisms with cell cycle progression in fission yeast. Cell Cycle. 2016;15:261–273. doi: 10.1080/15384101.2015.1121353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Letessier A, et al. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470:120–123. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- 58.Wilson TE, et al. Large transcription units unify copy number variants and common fragile sites arising under replication stress. Genome Res. 2015;25:189–200. doi: 10.1101/gr.177121.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pentzold C, et al. FANCD2 binding identifies conserved fragile sites at large transcribed genes in avian cells. Nucleic Acids Res. 2018;46:1280–1294. doi: 10.1093/nar/gkx1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gilbert DM, et al. Space and time in the nucleus: Developmental control of replication timing and chromosome architecture. Cold Spring Harb Symp Quant Biol. 2010;75:143–153. doi: 10.1101/sqb.2010.75.011. [DOI] [PubMed] [Google Scholar]
- 61.Pourkarimi E, Bellush JM, Whitehouse I. Spatiotemporal coupling and decoupling of gene transcription with DNA replication origins during embryogenesis in C. elegans. eLife. 2016;5:e21728. doi: 10.7554/eLife.21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gindin Y, Valenzuela MS, Aladjem MI, Meltzer PS, Bilke S. A chromatin structure-based model accurately predicts DNA replication timing in human cells. Mol Syst Biol. 2014;10:722. doi: 10.1002/msb.134859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.