Significance
Grain number is a key determinant of cereal grain yield, but its underlying genetic basis in wheat remains undefined. This study demonstrates a direct association between increased floret fertility, higher grain number per spike, and higher plot yields of field-grown wheat. The GNI1 gene, encoding an HD-Zip I transcription factor, was identified as responsible for increased floret fertility. The WT allele acts specifically during rachilla development, with its product serving to lower grain yield potential; in contrast, the reduced-function variant increased both floret and grain number. GNI1 evolved through gene duplication in the Triticeae, and its mutations have been under parallel selection in both wheat and barley over the course of domestication.
Keywords: floret fertility, grain number, duplication, HD-Zip I transcription factor, wheat
Abstract
Floret fertility is a key determinant of the number of grains per inflorescence in cereals. During the evolution of wheat (Triticum sp.), floret fertility has increased, such that current bread wheat (Triticum aestivum) cultivars set three to five grains per spikelet. However, little is known regarding the genetic basis of floret fertility. The locus Grain Number Increase 1 (GNI1) is shown here to be an important contributor to floret fertility. GNI1 evolved in the Triticeae through gene duplication. The gene, which encodes a homeodomain leucine zipper class I (HD-Zip I) transcription factor, was expressed most abundantly in the most apical floret primordia and in parts of the rachilla, suggesting that it acts to inhibit rachilla growth and development. The level of GNI1 expression has decreased over the course of wheat evolution under domestication, leading to the production of spikes bearing more fertile florets and setting more grains per spikelet. Genetic analysis has revealed that the reduced-function allele GNI-A1 contributes to the increased number of fertile florets per spikelet. The RNAi-based knockdown of GNI1 led to an increase in the number of both fertile florets and grains in hexaploid wheat. Mutants carrying an impaired GNI-A1 allele out-yielded WT allele carriers under field conditions. The data show that gene duplication generated evolutionary novelty affecting floret fertility while mutations favoring increased grain production have been under selection during wheat evolution under domestication.
The tribe Triticeae (subfamily Pooideae, family Poaceae) encompasses ∼30 genera and 360 species, including the economically important cereal crops bread wheat (Triticum aestivum), durum wheat (Triticum turgidum ssp. durum), barley (Hordeum vulgare), and rye (Secale cereale) (1). Triticeae plants produce an unbranched inflorescence, referred to as a spike. Whereas the majority of species, including wheat, develop a single spikelet on each rachis node, some species produce two or more (2). The wheat spike is made up of a number of spikelets, with a terminal spikelet at its apex; each spikelet generates an indeterminate number of florets attached to a secondary axis, the rachilla (3, 4). The number of grains set per spikelet is determined by the fertility of each floret (5, 6). At the white anther stage, a wheat spikelet normally produces up to 12 floret primordia (Fig. 1A); however, during development, more than 70% of the florets abort (6, 7). Despite its importance for grain number determination and the potential for grain yield improvement, the genetic basis of floret fertility in wheat is largely unknown.
Fig. 1.
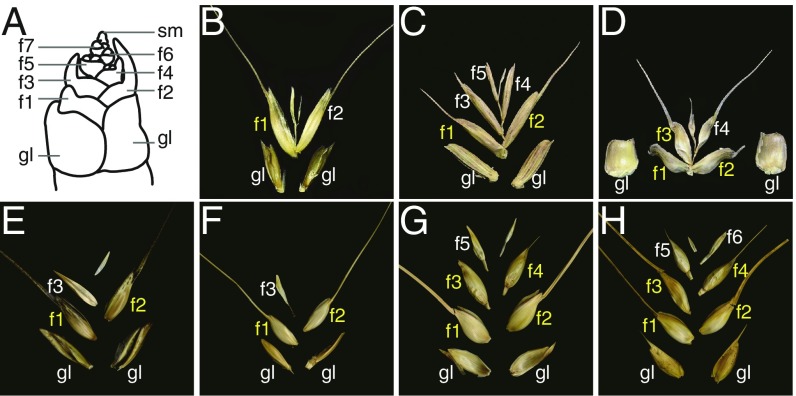
Structure of the wheat spikelet. (A) A schematic model illustrating the spike at the white anther stage. (B–D) The diploid progenitors of bread wheat: (B) Triticum urartu, (C) Aegilops speltoides, (D) Ae. tauschii. (E–G) Tetraploid wheats: (E) wild emmer (T. turgidum ssp. dicoccoides), (F) domesticated emmer (T. turgidum ssp. dicoccum), (G) durum (T. turgidum ssp. durum). (H) Hexaploid bread wheat (T. aestivum). Fertile florets are marked in yellow. f, floret; gl, glume; sm, spikelet meristem.
Two polyploidization events have been responsible for the appearance of bread wheat, an allohexaploid which harbors the three subgenomes B, A, and D (8, 9). The first event led to the formation of the allotetraploid wild emmer (Triticum turgidum ssp. dicoccoides, genome formula BBAA) 0.36 to 0.5 Mya. Its A subgenome was inherited from the diploid species Triticum urartu and its B subgenome from Aegilops speltoides or an extinct closely related species. Domesticated emmer (Triticum turgidum ssp. dicoccum) was selected by early farmers from stands of wild emmer and is the progenitor of modern durum wheat, currently the most widely cultivated tetraploid wheats. The second polyploidization event, which occurred ∼7,000 y ago, involved domesticated tetraploid wheat and the D subgenome donor Aegilops tauschii. As the ploidy level increased, the spikes evolved to produce a larger number of florets per spikelet: Diploid wheats (T. urartu and Triticum monococcum) set one or two grains per spikelet, tetraploid wheats two or three, and hexaploid wheats more than three (Fig. 1 B–H) (10).
The genetic diversity of grass inflorescences determines its reproduction and therefore, the resulting number of branches, flowers, and grains (11). Grass inflorescences take the form of either racemes (a single central monopodial axis), panicles (primary and secondary branches), or spikes (lacking a pedicel). Following the domestication of the cereals, inflorescence architecture has been improved by encouraging the formation of a higher number of reproductive branches (spikelets) (12). Since inflorescence architecture is a target for selection, a better understanding of the genetic mechanisms underlying spikelet development may help increase cereal grain yield. Floret number per spikelet is a major determinant of spikelet architecture. The spikelets of rice (Oryza sativa), barley, sorghum (Sorghum bicolor), and maize (Zea mays) are classified as determinate. They produce one floret each in rice and barley, but two in sorghum and maize. On the other hand, an indeterminate number of florets per spikelet are produced by both wheat and oat (Avena sativa). Sterile florets are a common feature, independent of spikelet determinacy; thus, there are two lateral florets formed in two-rowed barleys, a lower floret in maize and sorghum, and several apical florets in wheat and oat.
Recent studies have suggested that wheat grain yield is affected more by variation in grain number per spike than by variation in grain size (13, 14). A number of quantitative trait loci (QTL) affecting grain number per spike have been mapped in wheat; however, the gene(s) underlying these loci have yet to be identified (15–18). Genome-wide association analyses of European winter bread wheats have revealed a QTL responsible for an enhanced grain number per spikelet on chromosome arm 2AL (19); however, the underlying gene is unknown. The present study investigated natural variation for grain number per spikelet in polyploid wheats and their wild relatives and identified a gene underlying floret fertility and grain number. Additionally, the evolutionary trajectory of floret fertility in wheats was explored.
Results
Cloning of a QTL for Grain Number per Spikelet.
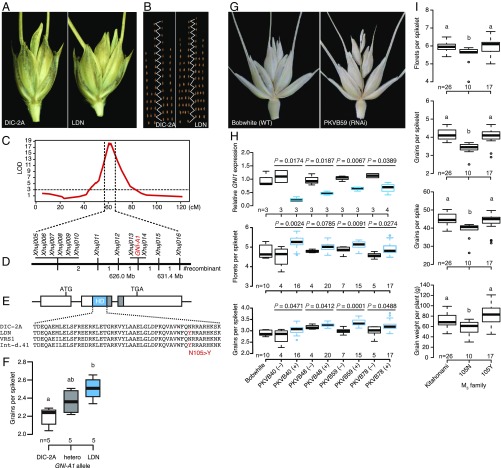
To reveal the genetic basis of the number of fertile florets formed per spikelet, a population of recombinant inbred substitution lines (RISLs), derived from a cross between durum wheat cultivar (cv.) Langdon (LDN) and the line DIC-2A, were characterized. DIC-2A harbors a copy of chromosome 2A inherited from the wild emmer wheat accession ISR-A in the genetic background of LDN (20); it produced an average of two grains per spikelet whereas LDN produced 2.4 (Fig. 2A). The increased grain number per spikelet in LDN was largely driven by the higher number of grains set in the basal and central parts of the spike (Fig. 2B). A single major QTL, associated with a log10 odds (LOD) score of 18.71 was mapped to chromosome 2AL; it accounted for 61% of the phenotypic variance (Fig. 2C). To further narrow the target genomic region, a backcross recombinant line population was developed, which allowed the locus to be mendelized as the gene Grain Number Increase 1-A (GNI-A1) (Fig. 2D). Fine mapping located GNI-A1 within a 5.4-Mbp region which harbors 26 putative genes, including one encoding an HD-Zip I transcription factor, the closest wheat homolog to the barley Six-rowed spike 1 gene (vrs1) (SI Appendix, Table S1) (21). A sequence comparison of the two parental GNI-A1 alleles revealed a polymorphism responsible for a single amino acid substitution (N105Y: 105 asparagine to tyrosine) within the highly conserved homeodomain (Fig. 2E). The recombinant plants carrying the LDN allele (generating the 105Y variant) displayed a significantly higher grain number per spikelet than those carrying the DIC-2A allele (105N variant, ancestral) (Fig. 2F). Notably, the mutation in LDN was identical to that found in the barley six-rowed spike mutant Int-d.41 allele at vrs1 (21), suggesting that the function of the resulting HD-Zip I protein was lost or attenuated in LDN.
Fig. 2.
The genes responsible for increasing grain number per spikelet. (A) Representative spikelets from DIC-2A and LDN. (B) The number of grains along the spike. (C) A locus controlling grain number per spikelet maps to chromosome arm 2AL. (D) Fine mapping of the GNI-A1 locus. (E) Gene structure of GNI-A1. The LDN and Int-d.41 allele encode a single amino acid substitution in the homeodomain (HD). (F) The additive effect of the GNI-A1 alleles. (G) Spikelet morphology of cv. Bobwhite and of a transgenic derivative harboring a GNI-A1 RNAi construct. (H) The abundance of the GNI1 transcript (Upper), the number of florets (Middle), and the number of grains (Bottom) per spikelet of the T1 generation transgenic (+) and nontransgenic (−) plants. P values were determined using the Student’s t test. (I) The phenotype of TILLING mutants in the M3 generation. “105N” indicates the mutant (functional) allele, and “105Y” the WT (reduced-function) variant in cv. Kitahonami. Box edges represent the 25% and 75% quantiles, with the median values indicated by bold lines. Whiskers indicate 1.5 times the interquartile range, and the remaining data are indicated by circles. Mean values marked with different letters in F and I differ significantly (P ≤ 0.05) from one another, as determined by Tukey’s honest significant difference test.
To verify the inhibitory role, GNI-A1 was silenced using RNA interference (RNAi). The relevant RNAi construct was transformed into the hexaploid wheat variety Bobwhite, which carries the 105N allele. Four independent transgenic events were obtained: All of the plants produced a higher number of florets and of grains per spikelet on average than did sibling construct-negative plants (Fig. 2 G and H). A decreased abundance of GNI1 transcript was associated with the presence of each of the four transgenes (Fig. 2H). These results supported the hypothesis that a functional copy of GNI-A1 inhibits floret development in wheat. No significant effect of the transgene was observed for plant height, spike number, spike length, spikelet number, or grain size, indicating that the gene’s function is likely spatially specific (SI Appendix, Fig. S1).
The Reduced-Function Allele of GNI-A1 Enhances Yield.
The Japanese high yielding bread wheat cv. Kitahonami carries the GNI-A1 allele which encodes the 105Y variant; it sets on average 4.26 grains per spikelet (Fig. 2I). Pedigree analysis revealed that cultivars carrying the 105Y variant produced a significantly higher number of grains per spikelet than did those encoding either the 105N or the 105K variant (4.03 vs. 3.21 and 3.07 grains per spikelet, respectively) and that the 105Y allele in cv. Kitahonami probably arose from the United Kingdom bread wheat cv. Norman (SI Appendix, Fig. S2). TILLING of cv. Kitahonami generated a number of heterozygous M2 plants harboring a Y105N mutation. As predicted, 105N homozygous progeny of these plants produced significantly fewer florets per spikelet and grain per spike than did 105Y homozygous progeny, as well as a lower grain weight per plant (Fig. 2I). The change in grain number per spikelet was mainly confined to the basal and central parts of the spike, as was also the case in tetraploid wheat. No significant variation was associated with the GNI-A1 allele for other traits (SI Appendix, Fig. S3). Thus, the N105Y mutation appears to contribute to a high number of grains per spikelet due to its ensuring a lower rate of apical floret abortion.
Yield tests were conducted to investigate the effect of the GNI-A1 allele on grain yield in the field (SI Appendix, Fig. S4). The performance of M4-derived cv. Kitahonami TILLING (encoding either the 105N or the 105Y variants) was compared at a site in both Kitami and Naganuma (Hokkaido, Japan), with four replications at the former site and three at the latter. Plants carrying the 105Y allele enjoyed a yield advantage of 10 to 30% at both sites. Their grain number per spike was slightly increased, but there was no change with respect to either grain size (1,000 grain weight) or the number of spikes per plant (SI Appendix, Fig. S4). The biomass of plants carrying the 105N variant was significantly lowered at Kitami. The indication was that the GNI-A1 105Y allele made a positive contribution to grain yield.
GNI1 Transcript Accumulates in the Distal End of the Spikelet and the Rachilla.
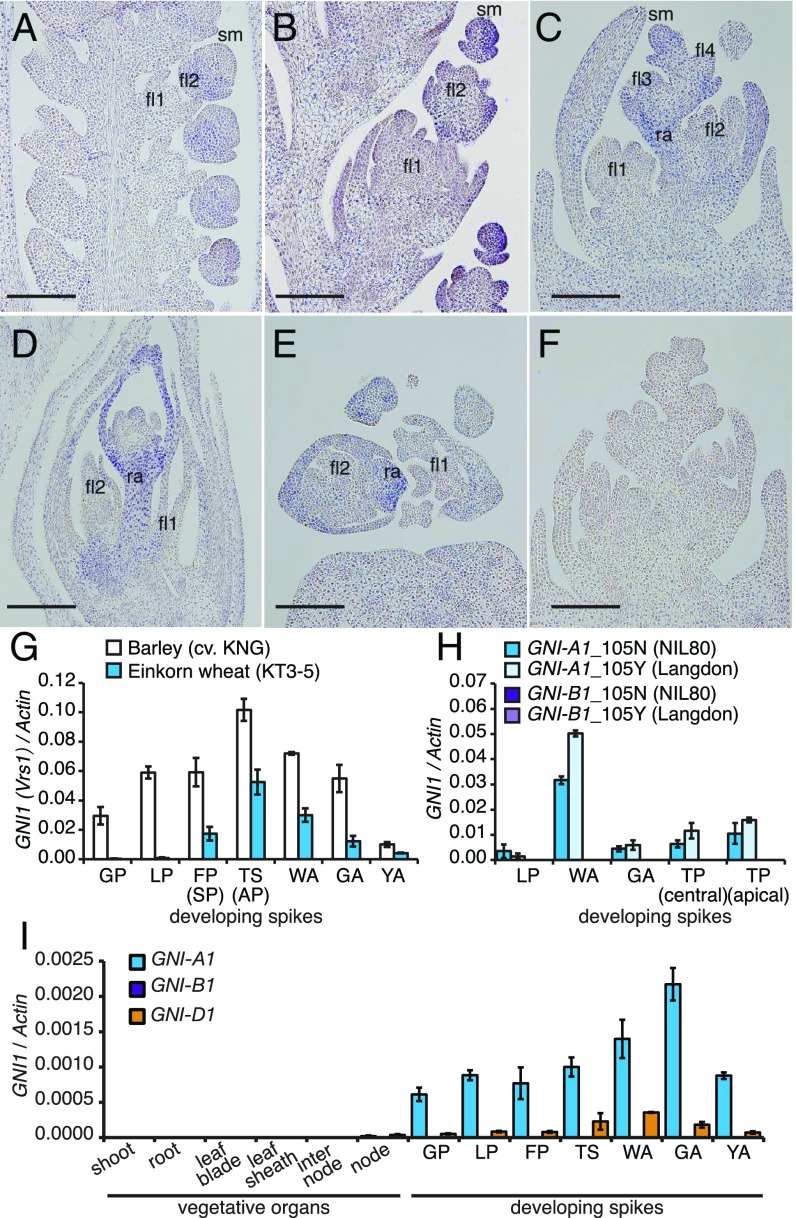
GNI1 mRNA was localized using in situ hybridization in the spikelet meristem of einkorn wheat (T. monococcum), which generates floret meristems on its lower flank (Fig. 3 A–F). Einkorn wheat was selected for this experiment because it exhibits a particularly high abundance of the GNI1 transcript (Fig. 3G). Following the differentiation of the second and third floret primordia during the terminal spikelet stage, GNI1 transcripts were detected in the spikelet meristem and apical floret (Fig. 3B). At the white anther stage, GNI1 mRNA was present in the spikelet meristem and in the rachilla bearing the florets and floret primordia, except for the first floret, which is usually fertile (Fig. 3 C–E). Thus, as development progressed, GNI1 transcription was diminished in the more basal florets within a spikelet. These observations implied that GNI1 expression inhibited apical floret development at the distal end of the spikelets and in part rachilla growth and development.
Fig. 3.
Expression profiling of GNI1. (A–F) In situ hybridization used to determine the sites of expression of GNI1 during the development of the einkorn wheat spike. (A–D) Longitudinal sections prepared at (A) the floret primordium stage, (B) the terminal spikelet stage, (C) the white anther stage, and (D) the green anther stage. (E) A cross-section made at the white anther stage. (F) Control hybridization using the sense probe. fl, floret; ra, rachilla; sm, spikelet meristem. (Scale bars: 200 µm.) (G–I) Transcription profiling of GNI1 in einkorn wheat and barley (Vrs1) (G), in tetraploid wheat (H), and in bread wheat cv. Bobwhite (I). Values shown as mean ± SE (n = 3). AP, awn primordium stage; FP, floret primordium stage; GA, green anther stage; GP, glume primordium stage; LP, lemma primordium stage; SP, stamen primordium stage; TP, tipping stage; TS, terminal spikelet stage; WA, white anther stage; YA, yellow anther stage.
Quantitative real-time PCR (qRT-PCR) analysis revealed that GNI-A1 was predominantly transcribed in immature spikes (Fig. 3 G–I). Transcript abundance peaked between the white and the green anther stages in tetraploid and hexaploid wheats, corresponding to the presence of the maximum number of floret primordia (6). A minor difference in GNI-A1 transcript level was observed between tetraploid wheats carrying the 105N and those carrying the 105Y allele. The result suggests that the N105Y change is a causal mutation and not linked with regulatory change (Fig. 3H). A slightly higher expression in the 105Y allele could be the result of a mild negative autoregulation. The abundance of the GNI-B1 transcript was negligible in the floral organs of both tetraploid and hexaploid wheat, despite its nucleotide sequence being very similar to that of GNI-A1 (Fig. 3 H and I and SI Appendix, Fig. S5). The abundance of the GNI-D1 transcript was lower than that of GNI-A1 (Fig. 3I).
RNA-seq profiles were generated from contrasting allelic forms selected in the cv. Kitahonami TILLING population to gain a better understanding of the molecular basis of the effect of the GNI-A1 mutation. The abundance of the GNI-A1 transcript was slightly higher in plants carrying the 105Y than in those carrying the 105N allele (SI Appendix, Fig. S6B), supporting the notion that suppression of grain number by the 105N allele is not caused by the expression abundance of GNI-A1. The results revealed that genes involved in nitrogen and sucrose metabolism, as well as in G protein beta/gamma-subunit complex binding, were more strongly transcribed in plants carrying the 105Y variant, consistent with the increased grain number associated with this allele (SI Appendix, Fig. S6A and Datasets S1 and S2). Consistent also with the improved floret fertility shown by the 105Y variant, there was a greater abundance of transcript generated from the Flowering locus T homolog FT-D1 in the 105Y spike although there was no evidence for the differential transcription of either of its homoeoalleles FT-A1 and FT-B1, either at white or green anther stages (SI Appendix, Fig. S6C). Inspection of a public RNA-seq database showed that similar profiles have been reported elsewhere (SI Appendix, Fig. S7). Given that the product of FT1 likely acts as a floral promoting factor during early floret development, the differential transcription of FT-D1 may represent the consequence of a greater level of floral activity occurring in plants carrying the GNI-A1 105Y allele. These plants reached heading on average 3 to 5 d earlier than those carrying the GNI-A1 105N allele, but there was no difference evident in the number of days required to reach maturity.
Allelic Variation of GNI-A1 in Wheat.
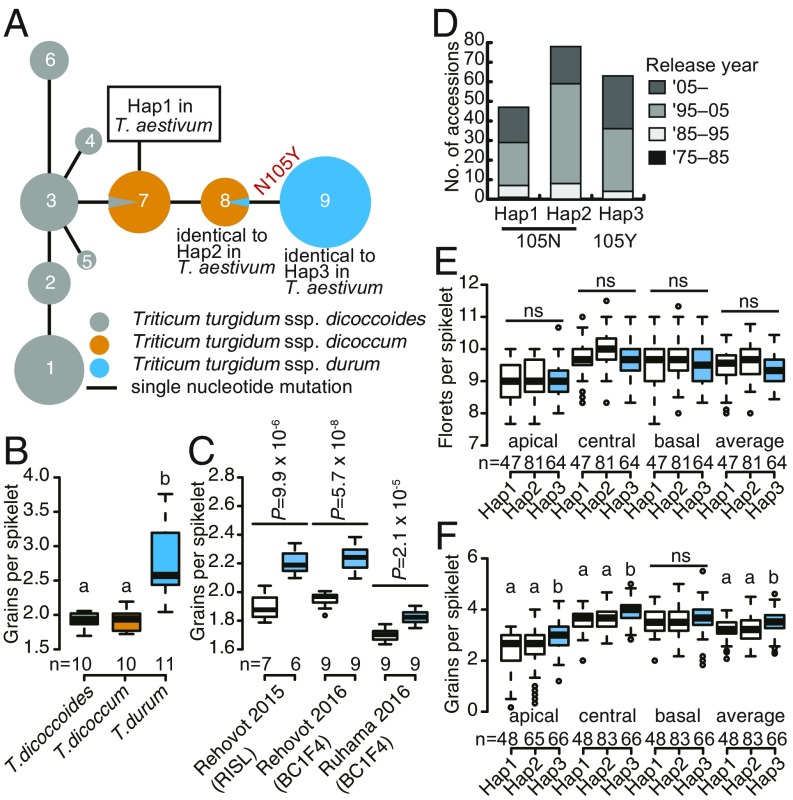
Natural variation within the GNI-A1 locus was investigated by resequencing the allele present in a set of 72 tetraploid wheats, including both wild and domesticated emmer and durum wheat entries. The analysis revealed nine haplotypes (Fig. 4A and SI Appendix, Table S2). The 105Y variant was restricted to the durum wheat entries, which featured a significantly higher number of grains per spikelet than the emmer wheat entries (Fig. 4B). When the number of grains per spikelet was measured in plants grown in three different environments, it was clear that lines carrying the 105Y allele consistently produced a higher number: The broad-sense heritability of the trait was 0.8, and the trait was stably expressed, even in the relatively low-yielding environment of Ruhama (Fig. 4C). Resequencing among a panel of 210 European winter bread wheat cultivars (19) revealed three haplotypes: Hap1 and -2 included the 105N variant while Hap3 included the 105Y allele (Fig. 4D and SI Appendix, Table S3). The number of florets present at the green anther stage, indicative of the potential maximum grain number, did not differ significantly among the three haplotypes (Fig. 4E). Cultivars carrying Hap3 produced more grains per spikelet at apical and central positions of the spike, as well as on average (2.97, 3.95, and 3.52, respectively), compared with cultivars carrying Hap1 (2.45, 3.58, and 3.18, respectively) and Hap2 (2.53, 3.64, and 3.22, respectively) (Fig. 4F). Hap3 cultivars produced more grains per spike and exhibited a higher spike fertility index. Their ratio of spike dry weight to stem dry weight was also higher, due to their investment of less biomass in the production of stem and leaf tissue (SI Appendix, Fig. S8).
Fig. 4.
Allelic variation at GNI-A1. (A) Haplotype network analysis in tetraploid wheat. The median-joining network was based on the resequencing of 35 entries of wild emmer, 14 of domesticated emmer, and 23 of durum wheat. Numbers 1 to 9 indicate haplotypes. Circle sizes correspond to the frequency of individual haplotypes. The lengths of the lines represent the genetic distance between pairs of haplotypes. (B) Grain number per spikelet. (C) The effect of allelic status at GNI-A1 across environments. P values were determined using the Student’s t test. (D) The frequency of three haplotypes in a panel of 210 European winter bread wheat cultivars. (E) The number of florets per spikelet at green anther stage. (F) Hap3 (105Y) cultivars form a higher number of grains per spikelet. The letters appearing in B, E, and F are used to indicate where means differed from one another significantly (P ≤ 0.05) as determined by Tukey’s honest significant difference test. ns, nonsignificant.
Evolution of GNI1 in Triticeae.
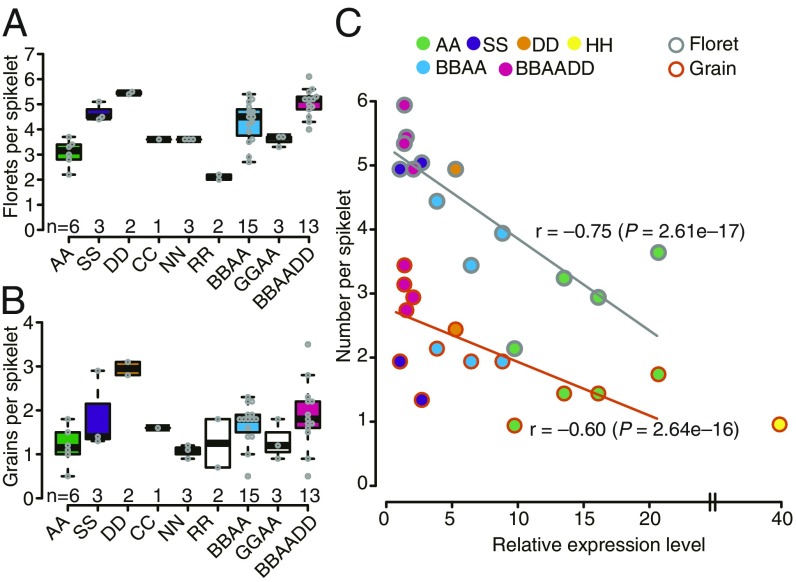
To clarify the evolution of GNI1 (i.e., HOX1) in the Triticeae, the gene was resequenced in a diverse collection of wild species. All 14 genera examined harbored a HOX2 homolog, in line with the known conservation of this gene among the grasses (22). Only Hordeum, Dasypyrum, Secale, Taeniatherum, Aegilops, Amblyopyrum, and Triticum species harbored copies of both HOX2 and HOX1 (SI Appendix, Fig. S9). The association between the number of fertile florets and ploidy level was examined by testing a set of diploid, tetraploid, and hexaploid wheat relatives. As anticipated, ploidy level was in general positively correlated with floret fertility although this was not the case among Aegilops species. While the diploid species T. urartu produced three florets and a single grain per spikelet, the Aegilops diploids (both S and D genome carriers) produced three to six florets and one to four grains per spikelet (Fig. 5 A and B). Tetraploid wheats produced four to five florets and two to three grains per spikelet while the hexaploids produced five to seven florets and three to five grains per spikelet (Fig. 5 A and B). A qRT-PCR experiment targeting GNI1 revealed a negative association between the abundance of GNI1 transcript and either the number of florets or grains per spikelet (Fig. 5C), implying that a lower level of GNI1 transcription enhances floret fertility in both diploid and polyploid wheat.
Fig. 5.
The function of GNI1 in diploid, tetraploid, and hexaploid Triticeae species. (A) Floret number per spikelet. (B) Grain number per spikelet. (C) The correlation between either trait and the abundance of the GNI1 transcript relative to barley (HH). AA, T. urartu/monococcum; BBAA, T. turgidum; GGAA, T. timopheevii; BBAADD, T. aestivum; CC, Ae. markgrafii; DD, Ae. tauschii; NN, Ae. uniaristata; RR, S. cereale; SS, Ae. speltoides/longissima.
Discussion
Here, it has been demonstrated that the GNI1 allele 105Y is a key determinant of grain number in wheat, achieved by its control over floret fertility. The gene encodes an HD-Zip I transcription factor which is most strongly active during the growth and development of the apical florets and the rachilla. The reduced-function mutation (105Y) has the effect of increasing the grain number, without any adverse effect on either the number of spikes or spikelets per spike, or on grain size. The suggestion is that improving floret fertility by reducing floret abortion could represent a promising breeding strategy for enhancing grain yield in plants which form nonbranched spikes, such as in wheat. An increase in floret number has been associated with mutations of certain members of the APETALA2 (AP2) transcription factor family (such as ids1 in maize and Q in wheat), the products of which control the fate of the spikelet meristem; however, the floret fertility of these mutants is relatively low (23, 24). This indicates that developing an appropriate number of floret primordia would be helpful for improving grain number. Optimizing the functionality of GNI1 and AP2 genes may help improve floret fertility, but their interaction remains unclear. Identifying a network of genes controlled by GNI1 and investigating other genes responsible for increased floret fertility may generate further opportunities for enhancing grain yield in wheat.
HD-Zip class I genes, including GNI1 in wheat, have evolved through a series of gene duplications, functionalization, and mutation (25–27). The progenitor of GNI1 experienced duplication following the separation of the Triticeae tribe from Bromeae. All species had a HOX2 homolog while only seven genera, including Hordeum, Secale, and Triticum, retained its paralog HOX1, which therefore appeared polyphyletic across the Triticeae phylogeny (28). This suggests that HOX1, for which the sequence is otherwise well conserved, was either independently lost or pseudogenized in the few lineages that diverged between Hordeum and the common ancestor of Secale and Triticum. It might be possible that our method, which can handle up to 25% of mismatches, did not capture the sequence because of pseudogenization, but then orthology would not be guaranteed. Notably, only plants which maintained the duplication have been successfully domesticated to become cereal crop species.
GNI1 is an ortholog of the barley Vrs1 gene, which controls lateral floret fertility. The product of the Vrs1 gene inhibits the development of lateral florets, particularly the pistil (21, 27), whereas loss-of-function mutants can form up to three times as many grains per rachis node. While Vrs1 mRNA accumulates in the rachilla and pistil (27), the GNI1 transcript is highly abundant in the rachilla and apical florets, suggesting that expression in the rachilla is its ancestral feature. In addition, barley Vrs1 appears to have acquired its specific expression pattern, particularly in the pistil of lateral spikelets, over the course of the evolution of the Hordeum inflorescence. The present analysis suggests that, in S genome Aegilops species, these genes are likely to have been converted to pseudogenes whereas those in the diploid Triticum (A genome) and Hordeum (H genome) species have remained functional (27). The GNI1/Vrs1-mediated floral changes are consistent with the idea of the genetic hotspot hypothesis, in which evolutionary relevant mutations tend to accumulate in specific genes and at specific positions within genes (29). In the case of GNI1/Vrs1, the coding sequence changes and loss-of-function alleles were selected in parallel. This observation can have important contributions to morphological evolution in opposition to the prevailing regulatory evolution view that regulatory changes, which minimize pleiotropic effects while simultaneously promoting adaptation, are important (30).
The single amino acid substitution (N105Y) present in a conserved domain of GNI-A1, which has led to a reduction in functionality, was selected post the divergence of durum wheat. A previous study has demonstrated that six-rowed barley originated from the domesticated two-rowed type via mutations in Vrs1 (21). Together, these observations suggest that mutations for increased grain number in wheat and barley have undergone parallel selection post domestication (31). The very high allele frequency (96%) of the 105Y allele among durum wheats reflects strong selection pressure toward increased grain number; meanwhile, its absence in both wild and domesticated emmer wheat germplasm implies that the mutation has some negative effect on fitness. During the evolution and domestication of wheat, the mutant allele probably became increasingly favored in the farming environment since it delivers increased grain yield. In European winter bread wheat germplasm, the 105N and 105Y alleles were represented in a 2:1 ratio, suggesting that the latter allele probably entered breeding populations only relatively recently. The high heritability and stability of the GNI1 allele (Fig. 4C and SI Appendix, Fig. S4) underline the potential utility of the 105Y allele to increase grain yield in wheat breeding programs around the world.
Materials and Methods
Details about plant materials, QTL mapping, fine mapping, transformation, TILLING, yield trials, quantitative RT-PCR, in situ hybridization, RNA-seq, haplotype analysis, and phylogenetic analysis are in SI Appendix and Materials and Methods. Gene sequences generated in this study are available from the DNA Data Bank of Japan (DDBJ) under accession numbers AB711370–AB711394 (http://getentry.ddbj.nig.ac.jp/getentry/na/AB711370 and http://getentry.ddbj.nig.ac.jp/getentry/na/AB711394) and AB711888–AB711913 (http://getentry.ddbj.nig.ac.jp/getentry/na/AB711888 and http://getentry.ddbj.nig.ac.jp/getentry/na/AB711913) and from the NCBI GenBank under accession numbers MH134165–MH134483. The RNA-seq data have been submitted to the European Nucleotide Archive under accession number PRJEB25119.
Supplementary Material
Acknowledgments
Grains of the RISL population were kindly provided by S. Xu and J. Faris [US Department of Agriculture (USDA) Agricultural Research Service (ARS)]. Y. Ono, F. Kobayashi, and H. Handa (National Agriculture and Food Research Organization) made available the DNA samples and grains of the cv. Kitahonami TILLING population. The winter bread wheat panel materials were a gift of M. Röder [Leibniz Institute of Plant Genetics and Crop Plant Research (IPK)]. We thank H. Tsuji (Yokohama City University) for providing the pANDA-b vector and the following institutions for providing germplasm: National BioResource Project-Wheat (Kyoto, Japan), the Nordic Genetic Resources Center (Alnarp, Sweden), IPK (Gatersleben, Germany), USDA (Aberdeen, ID), and the International Maize and Wheat Improvement Center (El Batán, Mexico). We acknowledge the excellent technical support given by S. Kikuchi (Chiba University), H. Koyama, M. Sakuma (National Institute of Agrobiological Sciences), and C. Trautewig and A. Fiebig (IPK). This research was financially supported by Genomics for Agricultural Innovation Program of the Ministry of Agriculture, Forestry, and Fisheries of Japan Grant TRS1002 (to S.S. and T.K.); a grant-in-aid from the Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellow for Research Abroad (to S.S.); Grant-in-Aid for Young Scientists (B) 16K18635 (to S.S.); Chief Scientist of the Israel Ministry of Agriculture and Rural Development Grant 20-10-0066 (to Z.P.); US Agency for International Development Middle East Research and Cooperation Grant M34-037 (to Z.P.); and German Research Foundation Grant BL462/10. During parts of this study, T.S. received financial support from HEISENBERG Program of the German Research Foundation (DFG) Grant SCHN 768/8-1 and the IPK core budget.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Gene sequences generated in this study have been deposited in the DNA Data Bank of Japan (DDBJ) (accession nos. AB711370–AB711394 and AB711888–AB711913) and in the NCBI GenBank database (accession nos. MH134165–MH134483). The RNA-seq data have been deposited in the European Nucleotide Archive (accession no. PRJEB25119).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815465116/-/DCSupplemental.
References
- 1.Barkworth EM, Bothmer R. Scientific names in the Triticeae. In: Muehlbauer GJ, Feuillet C, editors. Genetics and Genomics of the Triticeae. Vol 7. Springer; New York: 2009. pp. 3–30. [Google Scholar]
- 2.Sakuma S, Salomon B, Komatsuda T. The domestication syndrome genes responsible for the major changes in plant form in the Triticeae crops. Plant Cell Physiol. 2011;52:738–749. doi: 10.1093/pcp/pcr025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonnett OT. Inflorescences of Maize, Wheat, Rye, Barley and Oats: Their Initiation and Development. University of Illinois College of Agriculture, Agricultural Experimental Station; Urbana, IL: 1966. [Google Scholar]
- 4.Hanif M, Langer RHM. The vascular system of the spikelet in wheat (Triticum aestivum) Ann Bot. 1972;36:721–727. [Google Scholar]
- 5.Sreenivasulu N, Schnurbusch T. A genetic playground for enhancing grain number in cereals. Trends Plant Sci. 2012;17:91–101. doi: 10.1016/j.tplants.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Guo Z, Schnurbusch T. Variation of floret fertility in hexaploid wheat revealed by tiller removal. J Exp Bot. 2015;66:5945–5958. doi: 10.1093/jxb/erv303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo Z, Slafer GA, Schnurbusch T. Genotypic variation in spike fertility traits and ovary size as determinants of floret and grain survival rate in wheat. J Exp Bot. 2016;67:4221–4230. doi: 10.1093/jxb/erw200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.International Wheat Genome Sequencing Consortium (IWGSC) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345:1251788. doi: 10.1126/science.1251788. [DOI] [PubMed] [Google Scholar]
- 9.Marcussen T, et al. International Wheat Genome Sequencing Consortium Ancient hybridizations among the ancestral genomes of bread wheat. Science. 2014;345:1250092. doi: 10.1126/science.1250092. [DOI] [PubMed] [Google Scholar]
- 10.Shitsukawa N, Kinjo H, Takumi S, Murai K. Heterochronic development of the floret meristem determines grain number per spikelet in diploid, tetraploid and hexaploid wheats. Ann Bot. 2009;104:243–251. doi: 10.1093/aob/mcp129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bommert P, Satoh-Nagasawa N, Jackson D, Hirano HY. Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol. 2005;46:69–78. doi: 10.1093/pcp/pci504. [DOI] [PubMed] [Google Scholar]
- 12.Gross BL, Olsen KM. Genetic perspectives on crop domestication. Trends Plant Sci. 2010;15:529–537. doi: 10.1016/j.tplants.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynch JP, et al. The impact of variation in grain number and individual grain weight on winter wheat yield in the high yield potential environment of Ireland. Eur J Agron. 2017;87:40–49. [Google Scholar]
- 14.Feng F, et al. The effect of grain position on genetic improvement of grain number and thousand grain weight in winter wheat in North China. Front Plant Sci. 2018;9:129. doi: 10.3389/fpls.2018.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sukumaran S, Lopes M, Dreisigacker S, Reynolds M. Genetic analysis of multi-environmental spring wheat trials identifies genomic regions for locus-specific trade-offs for grain weight and grain number. Theor Appl Genet. 2018;131:985–998, and correction (2018) 131:999. doi: 10.1007/s00122-017-3037-7. [DOI] [PubMed] [Google Scholar]
- 16.Bhusal N, Sarial AK, Sharma P, Sareen S. Mapping QTLs for grain yield components in wheat under heat stress. PLoS One. 2017;12:e0189594. doi: 10.1371/journal.pone.0189594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu G, et al. Mapping QTLs of yield-related traits using RIL population derived from common wheat and Tibetan semi-wild wheat. Theor Appl Genet. 2014;127:2415–2432. doi: 10.1007/s00122-014-2387-7. [DOI] [PubMed] [Google Scholar]
- 18.Wu J, et al. The introgression of chromosome 6P specifying for increased numbers of florets and kernels from Agropyron cristatum into wheat. Theor Appl Genet. 2006;114:13–20. doi: 10.1007/s00122-006-0405-0. [DOI] [PubMed] [Google Scholar]
- 19.Guo Z, et al. Genome-wide association analyses of 54 traits identified multiple loci for the determination of floret fertility in wheat. New Phytol. 2017;214:257–270. doi: 10.1111/nph.14342. [DOI] [PubMed] [Google Scholar]
- 20.Joppa LR. Chromosome engineering in tetraploid wheat. Crop Sci. 1993;33:908–913. [Google Scholar]
- 21.Komatsuda T, et al. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci USA. 2007;104:1424–1429. doi: 10.1073/pnas.0608580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakuma S, Pourkheirandish M, Matsumoto T, Koba T, Komatsuda T. Duplication of a well-conserved homeodomain-leucine zipper transcription factor gene in barley generates a copy with more specific functions. Funct Integr Genomics. 2010;10:123–133. doi: 10.1007/s10142-009-0134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Debernardi JM, Lin H, Chuck G, Faris JD, Dubcovsky J. microRNA172 plays a crucial role in wheat spike morphogenesis and grain threshability. Development. 2017;144:1966–1975. doi: 10.1242/dev.146399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chuck G, Meeley RB, Hake S. The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Genes Dev. 1998;12:1145–1154. doi: 10.1101/gad.12.8.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vlad D, et al. Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science. 2014;343:780–783. doi: 10.1126/science.1248384. [DOI] [PubMed] [Google Scholar]
- 26.González-Grandío E, et al. Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proc Natl Acad Sci USA. 2017;114:E245–E254. doi: 10.1073/pnas.1613199114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakuma S, et al. Divergence of expression pattern contributed to neofunctionalization of duplicated HD-Zip I transcription factor in barley. New Phytol. 2013;197:939–948. doi: 10.1111/nph.12068. [DOI] [PubMed] [Google Scholar]
- 28.Escobar JS, et al. Multigenic phylogeny and analysis of tree incongruences in Triticeae (Poaceae) BMC Evol Biol. 2011;11:181. doi: 10.1186/1471-2148-11-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stern DL, Orgogozo V. Is genetic evolution predictable? Science. 2009;323:746–751. doi: 10.1126/science.1158997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stern DL. The genetic causes of convergent evolution. Nat Rev Genet. 2013;14:751–764. doi: 10.1038/nrg3483. [DOI] [PubMed] [Google Scholar]
- 31.Abbo S, et al. Plant domestication versus crop evolution: A conceptual framework for cereals and grain legumes. Trends Plant Sci. 2014;19:351–360. doi: 10.1016/j.tplants.2013.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.