Significance
Gaucher disease is a rare genetic disorder that has crippling health consequences. Mutations in the GBA1 gene are known to disrupt the enzyme glucocerebrosidase-1, but it is not known, at atom-level detail, as to how enzyme function is lost. This study uses multiscale simulations and deep learning to define precisely the mechanism underlying the disruption of glucocerebrosidase-1 and, in particular, its interaction with the facilitator protein, saposin C.
Keywords: multiscale simulations, gene mutations, lysosomal storage disease, rare disease
Abstract
The lysosomal enzyme glucocerebrosidase-1 (GCase) catalyzes the cleavage of a major glycolipid glucosylceramide into glucose and ceramide. The absence of fully functional GCase leads to the accumulation of its lipid substrates in lysosomes, causing Gaucher disease, an autosomal recessive disorder that displays profound genotype–phenotype nonconcordance. More than 250 disease-causing mutations in GBA1, the gene encoding GCase, have been discovered, although only one of these, N370S, causes 70% of disease. Here, we have used a knowledge-based docking protocol that considers experimental data of protein–protein binding to generate a complex between GCase and its known facilitator protein saposin C (SAPC). Multiscale molecular-dynamics simulations were used to study lipid self-assembly, membrane insertion, and the dynamics of the interactions between different components of the complex. Deep learning was applied to propose a model that explains the mechanism of GCase activation, which requires SAPC. Notably, we find that conformational changes in the loops at the entrance of the substrate-binding site are stabilized by direct interactions with SAPC and that the loss of such interactions induced by N370S and another common mutation, L444P, result in destabilization of the complex and reduced GCase activation. Our findings provide an atomistic-level explanation for GCase activation and the precise mechanism through which N370S and L444P cause Gaucher disease.
The enzyme glucocerebrosidase-1 (GCase) catalyzes the cleavage of the major glycolipid glucosylceramide (GL-1) into glucose and ceramide and the minor lipid glucosylsphingosine (Lyso-GL-1) into sphingosine and water (1–4). The lipid tails of both glycolipids are embedded within the intralysosomal membrane, such that both substrates are inaccessible and require the assistance of an 84-residue facilitator protein saposin C (SAPC), a member of the sphingolipid activator protein family (5–8). There is experimental evidence that both GCase and SAPC associate in the intralysosomal membrane, but the mechanism through which SAPC destabilizes intralysosomal vesicles to make lipids accessible to GCase is not well understood (2, 9).
Loss-of-function mutations of the GBA1 gene encoding GCase result in a crippling human disorder, Gaucher disease (10). Despite being a monogenic disorder, Gaucher disease presents with extreme phenotypic variability, ranging from an asymptomatic form to disease characterized by severe organ damage (11). Hepatosplenomegaly, anemia, thrombocytopenia, osteoporosis, and bone-marrow infiltration are hallmarks, with neurodegeneration noted with certain mutations. Although ∼250 GBA1 mutations have been reported thus far (12), just one, N370S, is responsible for >70% of the cases of Gaucher disease type 1 in the Ashkenazi Jewish population (13, 14). Another mutation, L444P, accounts for ∼40% of Gaucher disease types 2 and 3 worldwide (15–19). Our earlier studies have recapitulated the entire human phenotype in mice through the selective deletion of GBA1 in cells of the hematopoietic and mesenchymal cell lineages using the Cre-lox technology (20–22). However, there is no clear explanation yet as to how N370S and L444P mutations cause human disease and whether there is a role for SAPC in this process (13, 14, 23, 24).
SAPC not only mediates the contact of the GCase with its natural substrates, but is also known to induce a conformational change to stimulate enzyme activity directly (6, 9, 25). As a consequence, mutations in GCase that affect its association with SAPC would result not only in diminished GCase activity, but also in vulnerability of GCase to early degradation. Likewise, mutations in the PSAP gene that causes malfunction or absence of SAPC in the lysosomal compartment cause a juvenile form of Gaucher disease (7, 8).
The interaction between GCase and SAPC has been modeled earlier, and although this serves as a good starting point, the study has considerable limitations (26). First, the available model is unable to account for experimental data (27, 28), a limitation in itself. Second, there is no structural information on how the GCase–SAPC complex interacts with the membrane. In separate studies, selected mutants have been modeled through molecular-dynamics (MD) simulations (29, 30), but these studies lack information on the GCase–SAPC interface, specifically, membrane anchoring and the influence of membrane lipid and substrate on the complex.
Here, we report a model of GCase in complex with SAPC, which has been constructed by employing structural bioinformatics, including knowledge-based protein–protein docking (PPD). Multiscale MD simulations were conducted to understand the structural mechanism underlying the association of GCase with SAPC in its membrane environment. The results from our deep-learning approach explain the activation mechanism of GCase by SAPC and provide a structural explanation at the atomistic level on how the two most commonly occurring mutations N370S and L444P cause Gaucher disease.
Results
The GCase–SAPC Complex.
GCase is a globular protein composed of three domains (SI Appendix, Fig. S1) (31). Domain I (residues 1–27 and 383–414) is a small three-stranded antiparallel β-sheet; domain II (residues 30–75 and 431–497) is an independent eight-stranded β-barrel; and domain III (residues 76–381 and 416–430) is an (α/β) and triose-phosphate isomerase (TIM) barrel, containing the active site. Domains I and III interact tightly and are linked by one of the loops at the entrance of the binding site. Domains II and III are separated by a long loop that acts as a hinge, with structural folds that are similar to other hydrolases, such as α-galactosidase (31, 32). The active site, containing two catalytic residues—namely, E340 (catalytic nucleophile) and E235 (acid-base residue)—lies in a cavity formed at the center of the TIM barrel motif surrounded by residues R120, D127, F128, W179, N234, Y244, F246, Y313, C342, S345, W381, N396, F397, and V398. Loops 1–5, containing residues 311–319, 345–349, 394–399, 237–248, and 283–288, respectively, presenting at the entrance of the active site, can rearrange in different conformations to regulate substrate accessibility (4, 31, 32). Two different conformations of loop 1 have been reported—namely, extended and helical. Notably, in the active state, loop 1 is in a helical conformation with the side chain of residue D315 pointing toward residue N370, while in the inactive state, it adopts an extended conformation with residue D315 pointing toward loop 2. Furthermore, the bulky side chain of W348 (loop 2) is oriented toward outside the binding site in the active state, whereas in the inactive enzyme, it points toward the entrance of the active site and thus blocks it. Likewise, in the inactive state, the side chain of R395 (loop 3) orients toward the catalytic residue E340, but points outside the binding site when the enzyme is active (4, 31, 32).
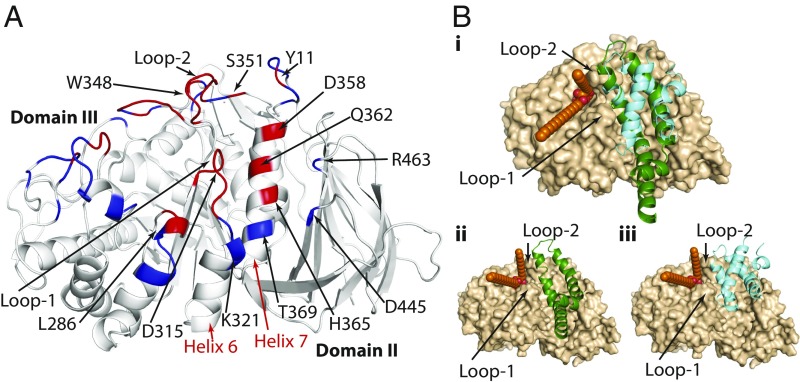
To understand the structural mechanism of GCase activation and the role of SAPC binding, a GCase–SAPC complex was generated by employing a knowledge-based protocol. By using the CPORT algorithm (33), the SAPC binding site was predicted to be located on helix 7, flanked by helix 6, loop 1, loop 2 at the entrance of the active site, and domain II (Fig. 1A). This predicted location is consistent with experimental evidence, which identified the N370S binding site to be positioned on helix 7 (34). The PPD program Hex (35) was used to generate the model, which was corroborated with an alternative program, Haddock (36). The complexes were constructed by using the crystal structures of the active and inactive conformations of the enzyme GCase (4, 32) and that of SAPC in its closed and open conformations (Fig. 1B) (5, 37). Molecular-docking studies demonstrated that the extended loop 1 clashes with the binding site. Moreover, the lipid substrate glucosylceramide (GluCer) could not be properly positioned within the binding site when the loop 1 was in the extended conformation. Three residues from loop 3 (R395, N396, and F397) play important roles in substrate accessibility to the active site (31).
Fig. 1.
The predicted GCase–SAPC interface. (A) Residues in red are those considered by CPORT to take part in protein–protein binding, and those marked in blue can potentially intervene in the binding. The protein–protein binding site was identified over helix 7 of domain III, flanked by helix 6 and domain II. (B, i) Superimposition of the poses of GCase in complex with SAPC in open (green) and closed (cyan) conformations. (B, ii and iii) GCase in complex with open SAPC (green; ii) or with closed SAPC (cyan; iii) and GluCer (orange spheres). GCase has been illustrated in surface representation and is colored in light brown; GluCer is colored in orange.
After applying screening criteria to the top 20 solutions of each docking run, only 1 docking solution was identified that was common to both active and inactive complexes using both docking programs. This docked pose was among the top results after selection using different correlation methods. We noted that the SAPC-binding site on GCase lay between domain III loops 1 (H311–P319) and 2 (S345–S351) at the entrance of the active site, helix 6 (K321–L330), helix 7 (W357–L372), and domain II (T43–S45, Q440–D445, L461–S465, and Y487). SI Appendix, Table S1 shows the residues involved in the protein–protein binding and electrostatic interactions.
Coarse-Grained MD.
To understand the structural basis of the catalytic function of GCase, we studied its dynamics and the conformational changes arising from interactions with other components, such as the lipid bilayer, SAPC, and GluCer. We thus used coarse-grained MD (CG-MD) to study lipid self-assembly, specifically, to determine how the complex anchors to the bilayer. GCase was simulated to position the complex on the lipid-bilayer interface in the absence or presence of GluCer and SAPC. A total of five 1.2-µs-long CG simulations were conducted employing the Martini coarse-grained force field (Table 1). Membrane assembly occurred between 40 and 120 ns in all simulations (SI Appendix, Fig. S2). The proteins/complexes became inserted into the lipid membrane immediately after their formation and remained anchored throughout the course of the simulations. Importantly, the entrance of the GCase active site oriented toward the bilayer, consistent with allowing GluCer access into the binding site anchored from within the membrane. The orientation of SAPC anchoring to the membrane was also consistent with that observed in experimental studies (37). Membrane anchoring became stronger during the initial equilibration phase, in all of the instances. The distance between the centers of mass between GCase and the lipid bilayer decreased as the equilibration progressed and thereafter remained stable. Furthermore, in atomistic simulations of the complex (3a and 3b in Table 2), the equilibrium distance to the membrane increased as SAPC became positioned between GCase and the membrane (SI Appendix, Fig. S3).
Table 1.
Summary of the coarse-grained simulations
| Simulation | System | PDB ID | Length (µs) | DPPC | Waters |
| 1: GG | GCase | 1OGS | 1.20 | 300 | 5,000 |
| 2: CG | GCase + GluCer | 1OGS | 1.20 | 338 | 6,431 |
| 3: CG | CPX | 2NSX + 2GTG (pose 5) | 1.20 | 414 | 8,500 |
| 4A: CG | SapC (closed) | 2GTG | 1.20 | 250 | 4,000 |
| 4B: CG | SapC (open) | 2QYP | 1.20 | 250 | 4,000 |
Five different systems were inserted into the membrane via self-assembly simulations. They include GCase (1), (ii) GCase bound to GluCer (2), GCase bound to Sap-C and GluCer (3), and Sap-C in closed (4A) and open (4B) conformations.
Table 2.
List of AT-MD simulations
| Simulation | System | a | b | No. of atoms | AT, ns |
| 1 | GCase | — | Inactive | 20,070 | 500 |
| 2a | GCase + GluCer | Active | 22,084 | 1,000 | |
| 2b | GCase + GluCer | Inactive | 22,082 | 1,000 | |
| 3a | CPX | Active | 26,540 | 1,000 | |
| 3b | CPX | Inactive | 26,537 | 1,000 | |
| 4 | Sap-C | — | — | 13,242 | 500 |
| 5a | CPX (N370S) | Active | — | 26,536 | 1,000 |
| 5b | CPX (N370S) | Inactive | 26,536 | 1,000 | |
| 6a | CPX (L444P) | Active | — | 26,535 | 1,000 |
| 6b | CPX (L444P) | — | Inactive | 26,535 | 1,000 |
Atomistic MD of Wild-Type GCase.
Atomistic MD (AT-MD) allowed us to study, at atom-level detail, the dynamics of the interactions between different components in the system. Once the membrane–protein complexes were assembled in CG-MD (above), they were converted to atomistic models and simulated by using classical AT-MD. A total of 10 simulations were performed (Table 2). To evaluate conformational stability of the complex over time, root-mean-square deviation (rmsd) of Cα atoms from the initial structure (SI Appendix, Fig. S4) and rms fluctuation (rmsf) per residue (SI Appendix, Fig. S5) were calculated for simulations 2a (active, no SAPC), 2b (inactive, no SAPC), 3a (active complex), and 3b (inactive complex). All four configurations were found to be stable over the simulated period, with each reaching equilibration at ∼250 ns.
Conformational changes at the protein–protein interface and at the surface were monitored by following changes in surface electrostatics in both active and inactive GCase conformations in the complex (SI Appendix, Figs. S6–S8). In simulation 3a, the electrostatic surface of GCase did not alter significantly throughout the simulation. During simulation 3b, the change in the electrostatic surface was found to be prominent. Notably, at its start, the SAPC binding region on GCase was positively charged. The area of SAPC was flat when compared in simulation 3a, in which GCase showed more cavities and was noted as slightly more negative. Toward the end of simulation 3b, the binding area of SAPC appeared more negative and irregular, with some cavities appearing that were equivalent to those observed in simulation 3a. There was also considerable difference in the electrostatic surface in the catalytic pocket. In simulation 3a, the catalytic pocket was deeper and wider than in the first part of 3b. Toward the end of 3b, however, the electrostatics of the catalytic site had changed, appearing wider and similar to that of simulation 3a. We postulate that changes in electrostatic surface pattern in GCase are a result of conformational changes and could possibly be under the influence of SAPC binding.
Active-Site Loop Dynamics of Wild-Type GCase.
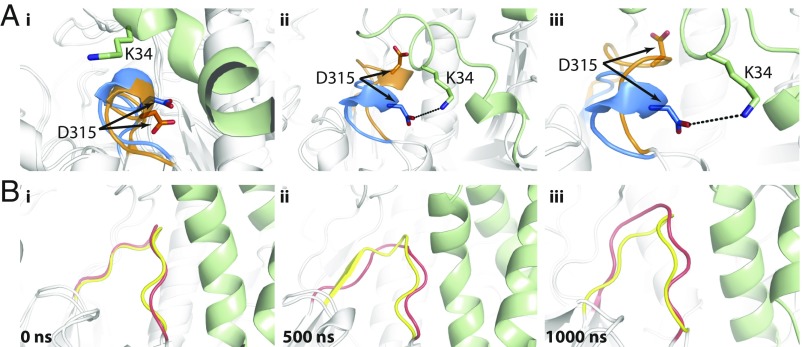
Analysis of the loop dynamics at the entrance of the active site shed light on the activation mechanism of the enzyme. In simulation 2a (no SAPC), loop 1 lost its helical structure as the simulation progressed. However, in simulation 3a (active complex), when SAPC was present, the interaction between D315 of GCase and K34 of SAPC stabilized the helical conformation of loop 1, and this configuration was maintained over the simulation (Fig. 2A). In inactive-state complex (simulation 3b), residue K34 of SAPC interacted with the backbone atoms of L314 and Y373, which are maintained throughout the simulation and force loop 1 to adopt a near-helical conformation. It is important to note that the latter two residues surround D315, a key residue that forms interactions with SAPC (31). Of note also is that the impetus for loop 1 to adopt a helical state is absent without SAPC in simulation 2b (Fig. 2B).
Fig. 2.
A hydrogen bond between GCaseD315 and SAPCK33 maintains helical conformation of loop 1. (A) Shown are snapshots of conformations extracted from simulation 3a (active complex) at 0 (i), 500 (ii), and 1,000 (iii) ns. GCase, blue; SAPC, green. Comparison of conformations adopted by loop 1 in simulation 2a (GCase, orange) has been made at equivalent time and superimposed on that of 3a. (B) Comparison of conformations adopted by loop 1 in simulations 2b (no SAPC; red) and 3b (inactive complex; yellow) at 0 (i), 500 (ii), and 1,000 (iii) ns. Loop 1 in simulation 2b extends toward helix 7. The interaction of residue SAPCK34 with the neighboring GCaseD315 in simulation 3b influences loop 1 to adopt a helical conformation.
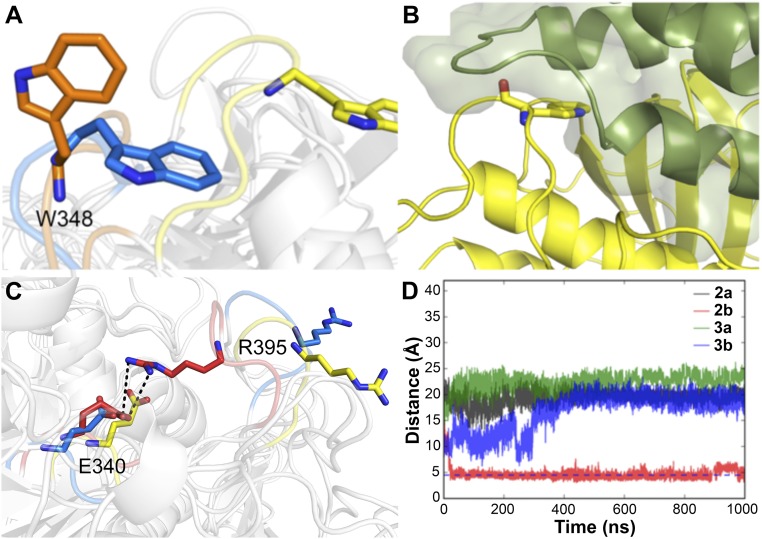
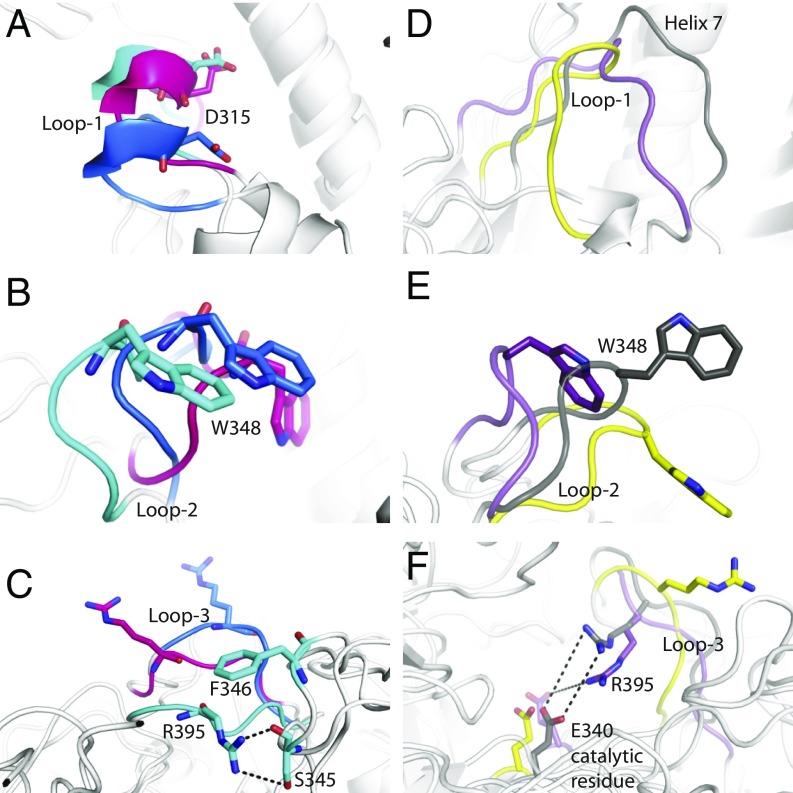
We also observed differences in the conformation of loops 2 and 3 in the presence or absence of SAPC (Fig. 3A). In both simulations 3a and 3b, the side chain of W348, located in loop 2 of GCase, was oriented toward the outside of the binding site. In the active complex (simulation 3a), the side chain of W348 was tucked in a hydrophobic pocket formed by SAPC (Fig. 3B). However, in simulation 2b (inactive GCase, no SAPC), the bulky indole side chain of W348 was found to partially obstruct the entrance to the binding site, while in simulation 2a, W348 became embedded in the membrane. In the inactive state (simulation 2b), residue R395 (loop 3) and catalytic residue E340 formed a stable hydrogen bond, which occluded the entrance to the active site, thus preventing substrate access (Fig. 3C). This interaction was not observed in simulation 3b (inactive complex), in which R395 oriented toward the outside of the binding site, with a final orientation as observed in the active state (Fig. 3C).
Fig. 3.
Conformation of the loops at the entrance of the binding site. (A) Conformation of loop 2 in simulations 2a (active GCase, no SAPC; orange), 3a (active complex; yellow), and 3b (inactive complex; blue) at 1,000 ns. (B) Snapshot of GCase–SAPC (green) complex at 1,000 ns in simulation 3b. SAPC stabilizes the active form of loop 2, where residue GCaseW348 is tucked in a hydrophobic pocket formed in SAPC. (C) Conformation of loop 3, highlighting the orientation of side chains of R395–E340 in different simulations at 1,000 ns. (D) Distance between specific atoms in the side chains of residues R395 and E340 of GCase in simulations 2a, 2b, 3a, and 3b (as shown).
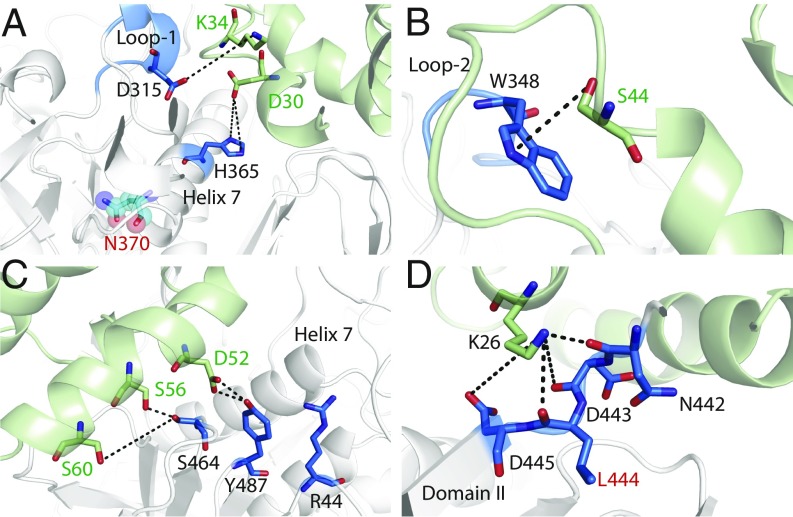
We found that a number of protein–protein interactions (PPIs) stabilized the GCase–SAPC complex. In simulation 3a (active complex; Fig. 4 and SI Appendix, Fig. S9), residue K34 of SAPC formed a stable hydrogen bond with residue D315, which is essential in maintaining the helicity of loop 1. There was a stable interaction in loop 2 between residues S44 (SAPC) and W348 (GCase). In helix 7, there were PPIs between residues D30 (SAPC) and H365 (GCase). Finally, interactions in domain II of GCase included those between D52 (SAPC) and R44 and Y487 (GCase); S60 (SAPC) and S464 (GCase); S60 (SAPC) and S464 (GCase); and K26 (SAPC) and N442, D445, D443 (backbone), and L444 (backbone) of GCase.
Fig. 4.
PPIs in simulation 3a (active complex) at 1,000 ns. Loop 1 and helix 7 (A), loop 2 (B), and domain II (C and D) are shown. SAPC is colored in green, and interacting residues in GCase are colored blue. The position of residue N370 has been represented with spheres and is colored in cyan. Distances of the interactions, over the course of the simulation, have been illustrated in SI Appendix, Fig. S9.
In simulation 3b (inactive complex), residues T24 and K34 of SAPC formed stable interactions with loop 1, as well as with the side chain and backbone of residues K321 and L314, respectively. In loop 2, two stable PPIs formed between residues S44 (SAPC) and E349 (GCase) and between S37 (SAPC) and K346 (GCase). In helix 7, there were PPIs between residues D33 (SAPC) and H365 (GCase), D30 (SAPC) and Y373 (GCase), and K34 (SAPC) and Y373 (GCase). Finally, interactions within domain II of GCase included Q48 (SAPC) with S45 (GCase); D52 (SAPC) with R44 and S465 (GCase); S56 (SAPC) with S465 and S464 (GCase); and K26 (SAPC) and D443, L444, with D445 (GCase) (SI Appendix, Figs. S10 and S11).
AT-MD of Mutant GCases.
AT-MD simulations were also performed for two of the most clinically prevalent Gaucher mutations in GCase—namely, N370S and L444P. Mutant GCases were simulated in complex with SAPC in an intraluminal membrane environment, using both active and inactive conformations (Table 2). Cα-rmsd of GCase was calculated in all four simulations and compared with the wild type. Notably, there was an overall conformational stability of GCase in the three active-state simulations, where loop 1 adopted a helical conformation (simulations 3a, 5a, and 6a; SI Appendix, Fig. S12). The equilibration time in the wild-type simulation (3a) was shorter (∼100 ns) than in GCaseN370S and GCaseL444P mutants (∼250 ns) (5a and 6a). In simulation 3a, the average Cα-rmsd from the end of the equilibration was lower in simulation 3a (2.4 ± 0.1 Å) than in simulation 5a (3.1 ± 0.2 Å) or 6a (3.4 ± 0.1 Å), indicating a more stable wild-type GCase in the complex. Analysis of rmsds of simulations containing the extended, inactive-state conformation of loop 1—namely, simulations 3b, 5b, and 6b—showed a similar trend after equilibration (SI Appendix, Fig. S13). In simulation 3b, unlike its active counterpart, the average rmsd value from the end of the equilibration was slightly higher (3.8 ± 0.1 Å) than in simulation 5b (3.6 ± 0.2 Å) and similar to the average in simulation 6b (3.8 ± 0.1 Å). In simulation 6b, GCase exhibited the greatest conformational drift compared with all other simulations.
However, the mutant GCaseN370S–SAPC and GCaseL444P–SAPC complexes were unstable, thus affecting the conformation of GCase over the course of the simulations. These simulations also showed that point mutations affected loop dynamics. In the first 300 ns of simulation 5a (GCaseN370S–SAPC), the helical conformation of loop 1 was lost. This helicity, however, partly recovered when interactions between K34 (SAPC) and the side chain of D315 (GCase) occurred during the second half of simulation 5a (Fig. 5).
Fig. 5.
GCase loop dynamics. (A–C) Comparison of loop conformations at the entrance of the binding site in simulations 3a (active complex; blue), 5a (GCaseN370S active state; pink), and 6a (GCaseL444P active state; cyan). Loops 1, 2, and 3 are illustrated. (A) Loop 1 maintains the helical conformation due to the influence of SAPC. (B) Due to the instability of the protein–protein binding, W348 (loop 2) does not remain inserted in the hydrophobic pocket in the mutants. (C) Loop 3 closes toward the binding site in simulation 6a. (D–F) Dynamic evolution of the loops at the entrance of the active site in simulations 3b (inactive complex; yellow), 5b (GCaseN370S inactive state; gray), and 6b (GCaseL444P inactive state; purple). (D) Loop 1 extends toward helix 7 in the mutants. (E) Poor binding between the two proteins prevents residue W348 from occupying the hydrophobic pocket in SAPC. (F) Loop 3 adopts a closed conformation in the mutants. Snapshots were taken at 1,000 ns.
In simulation 6a (GCaseN370S–SAPC), loop 1 retained the helical conformation, although the helix was deformed and moved toward loop 2. In both mutants, the poor coupling between GCase and SAPC rendered loop 2 free, unlike in the wild-type simulations, where loop 2 remained tucked in a hydrophobic pocket formed in SAPC. The evolution of loop 3 was also different in the two mutants. While in the wild type, residue R395 was oriented toward the outside of the active site, in simulation 5b (GCaseN370S–SAPC, inactive state), it was oriented toward the inside, creating interactions with residue S350 of loop 2. This interaction lies adjacent to a bulky phenylalanine side chain that impeded the return of loop 3 to an open conformation. In simulation 6b (GCaseL444P–SAPC, inactive state), the guanidinium side chain of residue R395 pointed toward the exterior of the binding pocket, although loop 3 was more closed than in the wild type.
A comparison of active-site loop dynamics in simulations of the inactive state (loop 1, extended form) of wild-type and mutant GCase also highlights some important differences (Fig. 5 D–F). In simulations 5b (GCaseN370S–SAPC, inactive state) and 6b (GCaseL444P–SAPC, inactive state), loop 1 extended toward helix 7. Residue W348 did not remain consistently tucked in the hydrophobic pocket on SAPC, as was noted in the wild-type simulation 3b. In simulations 5b and 6b, loop 3 adopted a closed conformation, whereby residue R395 interacted with the catalytic residue E340. This interaction completely obstructed the binding site and was similar to that observed in simulation 2b, where inactive-state GCase (loop 1 extended conformation) was simulated without SAPC.
PPIs were also affected in mutant complex simulations, with the disruption of many interactions identified in the wild-type GCase–SAPC complex. These differences were most pronounced in simulations 5a and 5b, where residue N370 was mutated to serine (SI Appendix, Fig. S14). In active wild-type GCase–SAPC complex (simulation 3a), the interaction between residue H365 in helix 7 and D30 of SAPC was stable throughout the simulation. This interaction was completely lost in the GCaseN370S–SAPC mutant complex. In active-state simulation 5a, a PPI between D315 of GCase and K34 of SAPC was formed at ∼400 ns and partially recovered loop-1 helicity. PPIs between residue K26 of SAPC and residue N442 and D443 in the proximities of L444 were disrupted, while those between K26 (SAPC) and L444 and D445 (GCase) were maintained (SI Appendix, Fig. S15). Other disrupted PPIs included those between residue W348 (GCase) and S44 (SAPC) and Q440 (GCase) and E64 (SAPC). In inactive GCaseN370S–SAPC (simulation 5b), SAPC became loosely attached to the GCaseN370S after ∼400 ns. At this point, SAPC was positioned near a completely deformed loop 1, and the PPIs formed were between residue K321 near loop 1 and residues D30 and E27 of SAPC. Toward the end of the simulation, new PPIs between the GCaseN370S and SAPC formed; these, however, did not involve helix 7 (which contains residue 370). Finally, interactions between K26 and L444 and surrounding residues were completely abrogated in this GCaseN370S–SAPC mutant simulation. The interaction between D30 of SAPC and Y373 in the proximities of N370 was also disrupted; this is otherwise stable in the corresponding wild-type protein complex (SI Appendix, Fig. S16).
Equally prominent differences were noted when L444 was mutated to proline in simulations 6a and 6b (SI Appendix, Fig. S17). In the active-state GCaseL444P–SAPC (simulation 6a), interactions between residues K26 (SAPC) and P444 and D445 (GCase) were disrupted from ∼600 ns onward, although the interaction with residue D443 was maintained beginning at ∼500 ns. Interactions of residues in SAPC with loop 1 of GCase were almost nonexistent toward the end of the simulation, but some interactions between SAPC and domains I and II of GCase remained stable from 500 ns onward (SI Appendix, Fig. S18). In the inactive-state GCaseL444P–SAPC simulation 6b, interactions between residue K26 (SAPC) and residues P444 and other surrounding residues, including D445 and D443, were completely lost. The disruption of these interactions makes SAPC partially detached and translates toward the end of helix 7 near domain I. Interactions with loop 1 were almost nonexistent. Stable interactions that remained included those between S44 (SAPC) and Q350 (GCase) and between D52 (SAPC) and R353 or W357 (backbone) of GCase (SI Appendix, Fig. S19).
Deep Clustering of AT-MD Simulations.
Using the AT-MD simulations, we next probed how the wild-type and mutant simulations differ with respect to their dominant motions. We posited that the conformational motions of GCase, especially when subjected to interactions with SAPC, would be nonlinear. Hence, linear models such as principal component analysis (PCA) may not sufficiently capture the conformational diversity in these simulations (38, 39). To account for the nonlinearity in protein conformational fluctuations, we recently developed a deep-clustering approach to identify intermediate states from folding trajectories (Methods) (40). We examined whether our deep-clustering approach based on a convolutional variational autoencoder (CVAE; Methods) could elucidate (i) the differences in the conformational motions between the active and inactive states of the wild-type and mutant GCase, and (ii) the different conformational states that are influenced by the motions within the wild-type and mutant GCase AT-MD. We used contact matrices from GCase as a starting point for our analysis.
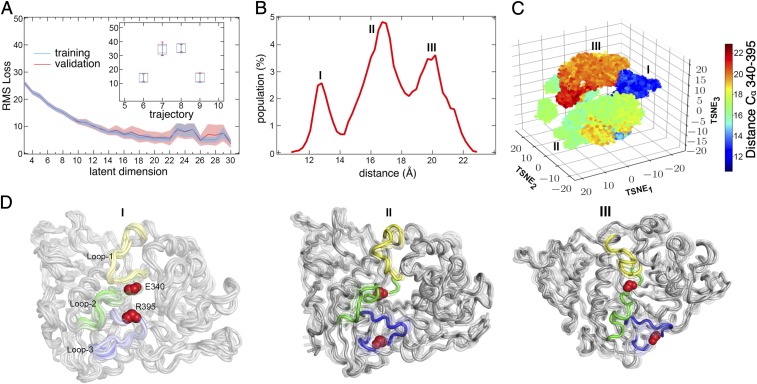
We first examined how many intrinsic latent dimensions are necessary to describe the conformational diversity observed from the wild-type AT-MD simulations. To estimate this, we plotted the overall loss (L) as a function of the number of dimensions in the latent space (Fig. 6A). This is similar to the cumulative variance plots used to estimate the total number of principal modes needed to describe the observed variance in the simulations for techniques such as PCA (38). Our results for the CVAE show that, as the number of intrinsic dimensions increase, the rms loss also decreases. As shown in Fig. 6A, however, the loss in the validation dataset increased beyond 14 dimensions, indicating that the CVAE is overfitting. Hence, for the GCase system, we can use a 14-dimensional latent space to describe the conformational motions sampled in the simulations.
Fig. 6.
Differences in GCase conformational states identified by deep learning. (A) The rms loss between the training and validation datasets from the trajectories (2a, 2b, 3a, and 3b) modeled using the CVAE are shown. The optimum number of dimensions is determined as 14, based on the rms-loss metric, beyond which the rms loss of the validation set is larger than the training data. A, Inset represents the rms loss with respect to the mutant simulations (5a, 5b, 6a, and 6b)—notably, the mutant simulations have a higher rms-loss value, indicating distinct differences in the conformational motions sampled by the MD simulations. (B) Histogram of the distance between residues E340 and R395 occupy three distinct peaks, indicative of at least three conformational states sampled in the MD simulations. (C) CVAE-learned representation of the conformational motions embedded in a 3D space using t-SNE (Methods) depicting three distinct conformational states. (D) Cartoon representations of the three states shown as an ensemble. Ensemble members were picked with respect to the peaks in B, illustrating E340 and R395 residues as red spheres for easy identification. Notably, the three ensembles highlight the separation between E340 and R395, indicating the distinct conformations of loops 1 (yellow), 2 (green), and 3 (purple) in the open state of GCase.
We evaluated the performance of CVAE on the mutant AT-MD simulations. As shown in Fig. 6 A, Inset, the rms loss for the mutant simulations was higher on average compared with the wild-type AT-MD simulations (average rms loss of 7.93 in wild-type vs. 24.77 in mutant AT-MD simulations). This difference in rms loss allowed us to posit that the conformational motions in the wild-type and mutant AT-MD simulations are different. Note that this is significant, given that we used contact matrices from GCase (without the substrate or SAPC included) to build the latent space representations, and the rms loss captures how well the model trained on the wild-type AT-MD simulations captures the conformational motions in the mutant simulations.
To understand how the conformational motions in the GCase simulations are different, we next examined the CVAE latent space. Given that a 14-dimensional latent space is difficult to visualize, we used the t-distributed stochastic neighborhood embedding (t-SNE) to examine the latent space in three dimensions. As shown in Fig. 6C, the t-SNE–based visualization allowed us to distinguish the conformational states visited by the simulations—especially in the context of the distance between the Cα atoms of residues 340 and 395. The distance between these two residues is critical, considering the formation of the ion–pair interaction between E340–R395 obstructs the binding pocket and locks it down in a closed conformation. A histogram of the distances between these two residues (Fig. 6B) represents the presence of three distinct states that are labeled I–III—we examined if the CVAE-based clustering can recapitulate these states. The CVAE clustering of the AT-MD simulations showed a clear distinction between the loop conformations as depicted in the 3D representations (Fig. 6D). Note that the CVAE representation does not use the distance between these residues as input, but discovers these as a consequence of the differences in the conformational motions. Corresponding representations of these sample conformations are shown in cartoon form in Fig. 6D.
Discussion
GCase–SAPC protein–protein binding sites have been defined both experimentally and in silico. Based upon the competition of synthetic lipids, two binding sites located at positions 6–27 and 45–60 and two activation sites at positions 27–34 and 41–48 on SAPC have been defined in an earlier study (28). In a separate study, chimeric saposins were used to identify a single activation site between residues 47 and 62 (27). Together, these studies led to two sets of possibilities: One was that the two activation sites lay adjacent to the loops at the entrance of the active site in GCase and exerted actions on the surrounding environment, and the second was that the SAPC activation site lay adjacent to the loops at the entrance of the active site in GCase. A pose consistent with the first premise was identified via Hex docking when active and inactive conformations of GCases were used. This pose was also identified in results from Haddock docking, albeit being the only plausible pose. In this study, we have followed a knowledge-based docking protocol to characterize the complete GCase–SAPC protein–protein interface in depth, which satisfies all of the requirements to be an optimal pose. The predicted binding site is in agreement with experimental data (27, 28) and lies adjacent to the loops at the entrance of the active site of GCase (31). CG-MD was further used to characterize the association of the GCase–SAPC complex in the membrane, providing an opportunity to observe the lipid self-assembly process. Quality controls demonstrated that the membrane was formed correctly and that wild-type and mutant GCase–SAPC complexes anchored to the membrane as peripheral membrane proteins in all of the simulations.
To understand the conformational changes within the GCase–SAPC complexes in depth, the CG coordinates of all simulations were transformed to atomistic, with further bursts of 1,000-ns simulations, with a total sampling time of 9 μs. Notably, the values of rmsd, which reflected conformational drifts in the systems, were more stable when GCase was simulated along with SAPC than when simulated alone (simulations 3a and 3b, respectively), suggesting that SAPC normally stabilizes GCase. In contrast, the mutated GCaseN370S– and GCaseL444P–SAPC complexes were unstable in both inactive and active states. Furthermore, when GCase was simulated in an inactive state (loop 1 extended), it exhibited higher rmsd values than in an active state (loop 1 helical). A comparison of interactions made by N370 and L444 in the wild type (SI Appendix, Figs. S20–S23) has been made with N370S and L444P mutants (SI Appendix, Figs. S24–S31).
We also analyzed rmsf values to study loop dynamics, interactions within the binding site, and PPIs. The highest rmsf peaks corresponded to surface loops, whereas the core structure was stable, with some differences in the loops at the active-site entrance (SI Appendix, Figs. S5 and S32–S33). Loop 1 partially lost its helical form in the active state (simulation 2a), whereas it extended toward helix 7 in the inactive state (simulation 2b). In the active-state complex (simulation 3a), loop 1 conserved its helicity during the course of the entire simulation due to the restraint placed by interaction with residue K34 of SAPC. In the inactive-state complex (simulation 3b), however, loop 1 did not change its extended conformation, and loop 2 displayed rmsf values >2 Å in two simulations, 2a and 3b. In simulation 2a (active state, no SAPC), the loop moved from a helical conformation to become embedded inside the lipid membrane, and in 3b (inactive complex), the loop was tucked in a hydrophobic pocket present in SAPC. Loop 3 displayed high mobility only during simulation 3b, moving from a closed conformation, where the side chain of residue R395 pointed toward the interior of the binding pocket, to an open conformation.
Loop dynamics at the entrance of the active site in GCase shed further light on the GCase–SAPC interactions and their aberration with the two disease-causing mutations GCaseN370S and GCaseL444P. Loop 1 (residues 311–319) normally adopts a helical conformation in the active state when simulated as a complex (simulation 3a), specifically because of interactions between D315 (GCase) and K34 (SAPC). The helicity of loop 1 is lost partly when GCase is simulated without SAPC in simulation 2a (no SAPC), wherein loop 1 establishes interactions with residues of loop 2—namely, W348 and K346. The simulation of mutant GCaseN370S– and GCaseL444P–SAPC complexes (simulations 5a and 6a; SI Appendix, Fig. S34) mimicked the simulation of GCase without SAPC. The mutants showed a complete loss of helical conformation, with partial recovery once an interaction between D315 and K34 was established. Subsequent stability resulted from interactions between H365 and S366 in helix 7.
Furthermore, in simulations with the wild-type GCase–SAPC complex (3a and 3b), residue W348 is tucked in a hydrophobic pocket on SAPC at the protein–protein interface (SI Appendix, Fig. S35). In simulations with both mutants, except in active-state GCaseN370S–SAPC (simulation 5a), W348 is not tucked inside this hydrophobic pocket. Among all of the simulations of inactive states, the open conformation of loop 3 is identified only in simulation 3b. Notably, in the rest of the simulations of inactive states of both mutant complexes and wild-type GCase (without SAPC)—namely 5b, 6b, and 2b, respectively—residue R395 in loop 3 forms an H-bond with E340, which completely obstructs the binding pocket and locks it in a closed conformation. Together, the data provide the structural basis for poor accessibility of lipid substrates to the GCase catalytic site in both mutations, N370S and L444P, even in the presence of unaltered SAPC.
AT-MD, although necessary, is not sufficient to derive a complete picture of the free-energy surface of a protein (41). Traditional MD analyses measure conformational drifts, such as rmsd or radius of gyration, and therefore cannot be used to infer dominant motions accountable for protein function (42). Furthermore, given the nature of complex interactions of GCase with SAPC and its substrates, we expected the conformational motions to be nonlinear. Hence, we used a deep-clustering approach that indicates that loop 1 was functionally relevant, with secondary roles of other loops. However, we noted that we used only the contact maps of GCase (generated from the trajectories) as inputs for our analysis. Our analysis was able to identify three substates in our simulations, corresponding to (i) the inactive enzyme, (ii) an intermediate, and (iii) an active conformation. The inactive conformation passed through an intermediate state to result in the active conformation. Of note is that the intermediate state was the same conformation as observed toward the end of simulation 3b (inactive complex), whereby W348 (loop 2) was inserted in a hydrophobic pocket on SAPC. This substate enabled the stable binding of K34 (SAPC) with D315 (loop 1) and, in turn, influenced loop 3 to orient away from the binding site. The conformation was observed only in the GCase–SAPC complex.
The structural differences identified at the GCase–SAPC interface and the stability of interactions in different simulations together reflect the dynamics of the protein–protein recognition. Proteins do not fit in a static manner as building blocks, but do so via a flexible and evolving process. The disruption of some of these interactions can alter this evolution, thereby making recognition at the protein–protein interface unfavorable, best exemplified by GCaseL444P. Notably, the mutation of L444 to proline, a cyclic and more rigid residue, prevents its interaction with residue K26 of SAPC, essentially abrogating GCase–SAPC interactions. In contrast, in the N370S mutation, loop 1 extended toward helix 7, resulting in the loss of its interaction with SAPC, which then destabilizes the complex.
Methods
PPD.
Prior information about interactions at the protein–protein interface can limit docking sampling and increase the chance of obtaining accurate results. Here, we have used the CPORT (33) interface predictor to propose potential sites of PPIs on GCase and SAPC surface. A model of GCase in complex with SAPC was generated by using a knowledge-based protocol. The complex was constructed by docking the X-ray crystal structures of active [Protein Data Bank (PDB) ID code 2NSX] (4) or inactive (PDB ID code 1OGS) (32) conformation of GCase with the X-ray crystal structures of SAPC in its closed (PDB ID code 2GTG) (5) and open (PDB ID code 2QYP) conformations (37). The PPD program Hex (35, 43, 44) was used to generate the model, which was corroborated with a second docking program, Haddock (36).
Docking calibration was performed in two sets of docking experiments. In the first set, geometrical parameters of the program were adjusted. The combination of correlation type and postprocessing procedure was optimized in a second set of calibration experiments. Once the most suitable parameters were identified, PPD experiments were conducted in combinations of different conformations of both partner proteins. For each combination, two series of seven docking runs were carried out, using the parameters obtained from the calibration experiments. In the first series, the center of mass of each protein was used as a centroid or origin of the geometrical operations. In the second series, residue H365 of GCase was used as a centroid or origin. The docking poses obtained were screened per different criteria, including the relative position of the proteins and number of electrostatic interactions, taking into account results of protein–protein interface predictors and known experimental data—namely, binding and activation regions of SAPC. A total of six docking poses were selected for energy minimization using the MD engine AMBER 12 (45). A second docking program, Haddock, was also used to validate docking datasets. A total of five runs were conducted, making different selections for passive and active residues of both proteins.
CG-MD.
A total of five CG-MD simulations were carried out to study the insertion of the proteins and their complexes in a lipid bilayer representative of a lysosomal membrane. These included (i) inactive GCase, (ii) inactive GCase with GluCer in its binding site, and (iii) active GCase with GluCer in complex with SAPC and SAPC alone in open (4A) and closed (4B) conformations. The systems were parameterized by using the Martini Forcefield in the Gromacs MD engine to group atoms in clusters of four (“beans”) for evaluation of their physicochemical properties (46).
For parameterization, the atomistic models were converted by using the maritinize.py script, with side-chain beads generated employing the elastic network option (elastic bond force = 500 kJ⋅mol−1⋅nm−2, with lower and upper elastic cutoffs at 0.5 and 0.9 nm, respectively). In simulations 2 and 3, where GluCer was simulated, the parameters for the substrate were obtained from the Martini website. The substrate was manually positioned in the active site, using atomistic coordinates extracted from the crystal structure. Lipid tails of the substrate were extended, as the parameters identified accounted for a molecule with shorter acyl tails.
The majority of phospholipids in lysosomal membranes are phosphatidylcholine, typified by dipalmitoyl phosphatidylcholine (DPPC), the most widely used phosphatidylcholine for simulations. A box of DPPC was generated using CG parameters from a single DPPC molecule. The optimum number of lipids for each system was identified by using trial and error.
Systems containing proteins or their corresponding complexes and the correct number of DPPC molecules were energy-minimized and solvated in alternate runs until the desired portion of water/DPPC was obtained. Energy minimization was performed in two consecutive steps by employing the steepest-descent and conjugate-gradient (1,000 cycles) algorithms. MD-CG simulations were run for 1.2 μs with a time step for integration of 0.003 ns. Standard cutoff schemes for nonbonded interactions were used to conduct the simulations. Lennard–Jones interactions were shifted to zero between 0.9 and 1.2 nm, and electrostatic interactions were shifted to zero between 0 and 1.2 nm. The nonbonded neighbor list cutoff was set to 0.14 nm to improve energy conservation, and the list was updated every 10 steps. Temperature was coupled separately for protein/complexes, lipids, and solvent by using the Berendsen algorithm at 325 K, using a time constant for coupling of 1.5 ps. Pressure of the system was coupled semiisotropically by using Berendsen algorithm at 1 bar, a compressibility of 3 × 10−5, and a time constant for coupling of 3.0 ps.
To convert CG to atomistic coordinates, a snapshot, representative of the protein/complex, inserted in the self-assembled membrane was selected. The Sugarpie script was chosen to carry out the CG to atomistic conversion after water and GluCer were removed from the CG coordinates. The atomistic structures used as templates for the conversion were same as those used for atomistic to CG conversion. After the conversion, GluCer was repositioned in the binding site by performing an alignment with the docked structure.
AT-MD.
Ten simulations (total sampling time 9 μs) were run with different systems that were converted from CG-MD (Table 2). Active and inactive states were identified by surveying different crystalline forms of GCase in the PDB. The inactive state was defined when loop 1 adopted an extended conformation and the R395–E340 ion pair occluded the entrance of the binding site, while the active state was defined when loop 1 was in a helical conformation and the ion-pair interaction was lost. The systems were built by using the Gromos53A6 force field (47), and Gromacs was used as the MD engine.
For AT parameterization, force-field-compliant topologies were generated by using proteins from the converted models. The substrate GluCer was added in some simulations, by aligning the converted models to the initial docked structure. CG models were converted by using the same AT coordinates that were used to generate them. The converted models were, in some cases, used to obtain different GCase conformations or mutants by aligning to the desired structures. Simulation 2-CG was used to obtain the atomistic coordinates for simulations 2a and 2b, and simulation 3-CG was used to obtain the coordinates for simulations 3a, 3b, 5a, 5b, 6a, and 6b. The mutant GCases, GCaseN370S and GCaseL444P, were generated by using the mutagenesis program in the ICM-Pro molecular modeling package, using the same PDB structures as that of the wild type. The atomistic mutant simulations were set up by converting CG structures of the complex extracted from 3-CG simulation, using the mutated proteins instead of the wild type.
The models were solvated by using single point charge water and energy-minimized by using 5,000 steps of the steepest-descent method. Counterions were added to neutralize the systems. A second round of energy minimization was conducted by employing an additional 5,000 steps of the steepest-descent method. Two rounds of equilibration were carried out: (i) 0.1 ns of constant number, volume, and temperature equilibration with time steps of 0.002 ns, using the V-rescale algorithm for temperature coupling (separately for protein/complexes, lipids ,and solvent) at 323 K, using a time constant for coupling of 0.5 ps; and (ii) 1 ns of constant number, pressure, and volume equilibration with a time step of 0.002 ns, using Nose–Hoover temperature coupling and Parrinello–Rahman for pressure coupling. The pressure of the system was coupled semiisotropically by using the Berendsen algorithm at 1 bar, a compressibility of 4.5 × 10−5 and a time constant for coupling of 5.0 ps. The production run was carried out for 1,000 ns without any restraints with a time step of 0.002. A cutoff of 1.2 nm was chosen for neighbor-list generation and coulomb and Lennard–Jones interactions. Particle–Mesh–Ewald summation was chosen for electrostatic interactions. For those systems with two proteins or ones that included the substrate, strong position restraints were applied for energy-minimization runs and soft restraints for equilibration-phase simulations. There were no restraints on the system during the production run.
Deep Learning.
Each trajectory was processed by using the MDAnalysis library (48) to extract the contact matrices using the Cα atoms; a distance cutoff of 8 Å or less was used to define two residues to be in contact. Contact matrices offer a natural advantage: They are independent of rotation/translation issues, which can be problematic while analyzing MD trajectories. We then used a CVAE to capture the large-scale conformational motions within a low-dimensional latent space in an unsupervised fashion (40). Autoencoders typically have an hourglass-like architecture, where the data (from MD trajectories) are compressed into a lowdimensional latent space and then reconstructed by using successive “output” layers. Variational autoencoders (VAEs) force the latent space to be normally distributed (49). This constraint provides a way to overcome the issues with sparsity in latent dimensions and forces the latent representation to utilize all of the information within the MD trajectories.
We used convolutional layers as inputs to the VAE (and, thus, the term CVAE) since sliding filter maps can better describe secondary and tertiary structure interactions, as quantified from the contact-map dynamics from the MD trajectories. We trained the CVAE on the wild-type AT-MD simulations; we then used the mutant AT-MD simulations as testing data, while simultaneously inferring how these simulations may be different from the wild-type simulations. Although in unsupervised learning applications, we do not require a cross-validation step, we used 60/40 split in the training/validation data (wild-type AT-MD simulations) to assess the quality of the CVAE build. This also allowed us to estimate the number of intrinsic latent dimensions required to sufficiently describe the conformational motions observed in the wild-type AT-MD simulations. The objective of the CVAE is to reduce the loss (L), which is composed of two terms: (i) the reconstruction loss, Er, which measures the ability of the CVAE in reconstructing the input contact matrix, calculated as the cross-entropy loss between f(z), which indicates the reconstructed probability of contact between two Cα atoms and the original X conformations from the simulation, which indicate the existence of contact between two Cα atoms; and (ii) the latent loss, El, which measures the loss as a consequence of constraining the latent space to be normal distribution. The latent loss is defined as a regularizing constraint that forces the latent embeddings z to conform to a Gaussian distribution; this is calculated as the Kullback–Leibler (KL) divergence between the latent embeddings z and a normal distribution with mean 0 and SD 1 (40).
Details of the reconstruction loss and the latent loss are described in our previous work (40). We used the RMSProp algorithm to train the CVAE and trained the models for 50 epochs (SI Appendix, Figs. S36 and S37). Similar to PCA (50), the projections of the simulations onto the CVAE latent space representation provide information on the dominant motions (represented as VAEi, where i represents the particular index to the latent space) sampled in the simulations. However, these modes are not ordered—for convenience, we just order the modes based on the variance accounted for in the simulations.
Supplementary Material
Acknowledgments
We thank Dr. Pramod Mistry (Yale) for his invaluable advice. A. Ramanathan and D.B. were supported in part by the Laboratory Directed Research and Development Program of Oak Ridge National Laboratory, managed by UT-Battelle, LLC, for US Department of Energy Grant DE-AC05-00OR22725. M.Z. was supported by National Institutes of Health Grants R01 AG23176, R01 AR65932, and R01 AR67066 (to M.Z.), and DK113627 (to M.Z. and L.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818411116/-/DCSupplemental.
References
- 1.Kolter T, Sandhoff K. Lysosomal degradation of membrane lipids. FEBS Lett. 2010;584:1700–1712. doi: 10.1016/j.febslet.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 2.Schulze H, Kolter T, Sandhoff K. Principles of lysosomal membrane degradation: Cellular topology and biochemistry of lysosomal lipid degradation. Biochim Biophys Acta. 2009;1793:674–683. doi: 10.1016/j.bbamcr.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 3.Vasella A, Davies GJ, Böhm M. Glycosidase mechanisms. Curr Opin Chem Biol. 2002;6:619–629. doi: 10.1016/s1367-5931(02)00380-0. [DOI] [PubMed] [Google Scholar]
- 4.Lieberman RL, et al. Structure of acid beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease. Nat Chem Biol. 2007;3:101–107. doi: 10.1038/nchembio850. [DOI] [PubMed] [Google Scholar]
- 5.Ahn VE, Leyko P, Alattia JR, Chen L, Privé GG. Crystal structures of saposins A and C. Protein Sci. 2006;15:1849–1857. doi: 10.1110/ps.062256606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fabbro D, Grabowski GA. Human acid beta-glucosidase. Use of inhibitory and activating monoclonal antibodies to investigate the enzyme’s catalytic mechanism and saposin A and C binding sites. J Biol Chem. 1991;266:15021–15027. [PubMed] [Google Scholar]
- 7.Tamargo RJ, Velayati A, Goldin E, Sidransky E. The role of saposin C in Gaucher disease. Mol Genet Metab. 2012;106:257–263. doi: 10.1016/j.ymgme.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tylki-Szymańska A, et al. Gaucher disease due to saposin C deficiency, previously described as non-neuronopathic form–No positive effects after 2-years of miglustat therapy. Mol Genet Metab. 2011;104:627–630. doi: 10.1016/j.ymgme.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 9.Sun Y, Qi X, Grabowski GA. Saposin C is required for normal resistance of acid beta-glucosidase to proteolytic degradation. J Biol Chem. 2003;278:31918–31923. doi: 10.1074/jbc.M302752200. [DOI] [PubMed] [Google Scholar]
- 10.Sidransky E. Gaucher disease: Insights from a rare mendelian disorder. Discov Med. 2012;14:273–281. [PMC free article] [PubMed] [Google Scholar]
- 11.Grabowski GA. Phenotype, diagnosis, and treatment of Gaucher’s disease. Lancet. 2008;372:1263–1271. doi: 10.1016/S0140-6736(08)61522-6. [DOI] [PubMed] [Google Scholar]
- 12.Hruska KS, LaMarca ME, Scott CR, Sidransky E. Gaucher disease: Mutation and polymorphism spectrum in the glucocerebrosidase gene (GBA) Hum Mutat. 2008;29:567–583. doi: 10.1002/humu.20676. [DOI] [PubMed] [Google Scholar]
- 13.Charrow J, et al. The Gaucher registry: Demographics and disease characteristics of 1698 patients with Gaucher disease. Arch Intern Med. 2000;160:2835–2843. doi: 10.1001/archinte.160.18.2835. [DOI] [PubMed] [Google Scholar]
- 14.Taddei TH, et al. The underrecognized progressive nature of N370S Gaucher disease and assessment of cancer risk in 403 patients. Am J Hematol. 2009;84:208–214. doi: 10.1002/ajh.21362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horowitz M, et al. Prevalence of nine mutations among Jewish and non-Jewish Gaucher disease patients. Am J Hum Genet. 1993;53:921–930. [PMC free article] [PubMed] [Google Scholar]
- 16.Stone DL, et al. Glucocerebrosidase gene mutations in patients with type 2 Gaucher disease. Hum Mutat. 2000;15:181–188. doi: 10.1002/(SICI)1098-1004(200002)15:2<181::AID-HUMU7>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 17.Koprivica V, et al. Analysis and classification of 304 mutant alleles in patients with type 1 and type 3 Gaucher disease. Am J Hum Genet. 2000;66:1777–1786. doi: 10.1086/302925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eto Y, Ida H. Clinical and molecular characteristics of Japanese Gaucher disease. Neurochem Res. 1999;24:207–211. doi: 10.1023/a:1022553819241. [DOI] [PubMed] [Google Scholar]
- 19.Jeong SY, Park SJ, Kim HJ. Clinical and genetic characteristics of Korean patients with Gaucher disease. Blood Cells Mol Dis. 2011;46:11–14. doi: 10.1016/j.bcmd.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 20.Mistry PK, et al. Glucocerebrosidase gene-deficient mouse recapitulates Gaucher disease displaying cellular and molecular dysregulation beyond the macrophage. Proc Natl Acad Sci USA. 2010;107:19473–19478, and erratum (2012) 109:9220. doi: 10.1073/pnas.1003308107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Offman MN, Krol M, Silman I, Sussman JL, Futerman AH. Molecular basis of reduced glucosylceramidase activity in the most common Gaucher disease mutant, N370S. J Biol Chem. 2010;285:42105–42114. doi: 10.1074/jbc.M110.172098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, et al. Gaucher disease gene GBA functions in immune regulation. Proc Natl Acad Sci USA. 2012;109:10018–10023. doi: 10.1073/pnas.1200941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bendikov-Bar I, Ron I, Filocamo M, Horowitz M. Characterization of the ERAD process of the L444P mutant glucocerebrosidase variant. Blood Cells Mol Dis. 2011;46:4–10. doi: 10.1016/j.bcmd.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 24.Sun QY, et al. Glucocerebrosidase gene L444P mutation is a risk factor for Parkinson’s disease in Chinese population. Mov Disord. 2010;25:1005–1011. doi: 10.1002/mds.23009. [DOI] [PubMed] [Google Scholar]
- 25.Berent SL, Radin NS. Mechanism of activation of glucocerebrosidase by co-beta-glucosidase (glucosidase activator protein) Biochim Biophys Acta. 1981;664:572–582. doi: 10.1016/0005-2760(81)90134-x. [DOI] [PubMed] [Google Scholar]
- 26.Atrian S, et al. An evolutionary and structure-based docking model for glucocerebrosidase-saposin C and glucocerebrosidase-substrate interactions—Relevance for Gaucher disease. Proteins. 2008;70:882–891. doi: 10.1002/prot.21554. [DOI] [PubMed] [Google Scholar]
- 27.Qi X, Qin W, Sun Y, Kondoh K, Grabowski GA. Functional organization of saposin C. Definition of the neurotrophic and acid beta-glucosidase activation regions. J Biol Chem. 1996;271:6874–6880. doi: 10.1074/jbc.271.12.6874. [DOI] [PubMed] [Google Scholar]
- 28.Weiler S, Kishimoto Y, O’Brien JS, Barranger JA, Tomich JM. Identification of the binding and activating sites of the sphingolipid activator protein, saposin C, with glucocerebrosidase. Protein Sci. 1995;4:756–764. doi: 10.1002/pro.5560040415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Offman MN, et al. Comparison of a molecular dynamics model with the X-ray structure of the N370S acid-beta-glucosidase mutant that causes Gaucher disease. Protein Eng Des Sel. 2011;24:773–775. doi: 10.1093/protein/gzr032. [DOI] [PubMed] [Google Scholar]
- 30.Zubrzycki IZ, Borcz A, Wiacek M, Hagner W. The studies on substrate, product and inhibitor binding to a wild-type and neuronopathic form of human acid-beta-glucosidase. J Mol Model. 2007;13:1133–1139. doi: 10.1007/s00894-007-0232-5. [DOI] [PubMed] [Google Scholar]
- 31.Lieberman RL. A guided tour of the structural biology of Gaucher disease: Acid-β-glucosidase and saposin C. Enzyme Res. 2011;2011:973231. doi: 10.4061/2011/973231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dvir H, et al. X-ray structure of human acid-beta-glucosidase, the defective enzyme in Gaucher disease. EMBO Rep. 2003;4:704–709. doi: 10.1038/sj.embor.embor873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Vries SJ, Bonvin AM. CPORT: A consensus interface predictor and its performance in prediction-driven docking with HADDOCK. PLoS One. 2011;6:e17695. doi: 10.1371/journal.pone.0017695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salvioli R, et al. The N370S (Asn370–>Ser) mutation affects the capacity of glucosylceramidase to interact with anionic phospholipid-containing membranes and saposin C. Biochem J. 2005;390:95–103. doi: 10.1042/BJ20050325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ritchie DW, Kemp GJL. Protein docking using spherical polar Fourier correlations. Proteins. 2000;39:178–194. [PubMed] [Google Scholar]
- 36.de Vries SJ, van Dijk M, Bonvin AM. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 37.Rossmann M, et al. Crystal structures of human saposins C and D: Implications for lipid recognition and membrane interactions. Structure. 2008;16:809–817. doi: 10.1016/j.str.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 38.Stein SAM, Loccisano AE, Firestine SM, Evanseck JD. Principal components analysis: A review of its application on molecular dynamics data. Annu Rep Comput Chem. 2006;2:233–261. [Google Scholar]
- 39.Doerr S, Ariz-Extreme I, Harvey MJ, De Fabritiis G. 2017. Dimensionality reduction methods for molecular simulations. arXiv:1710.10629v2. Preprint, posted November 2, 2017.
- 40.Bhowmik D, Gao S, Young MT, Ramanathan A. Deep clustering of protein folding simulations. BMC Bioinformatics. 2018;19:484. doi: 10.1186/s12859-018-2507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 42.Noé F, Schütte C, Vanden-Eijnden E, Reich L, Weikl TR. Constructing the equilibrium ensemble of folding pathways from short off-equilibrium simulations. Proc Natl Acad Sci USA. 2009;106:19011–19016. doi: 10.1073/pnas.0905466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ritchie DW. Evaluation of protein docking predictions using Hex 3.1 in CAPRI rounds 1 and 2. Proteins. 2003;52:98–106. doi: 10.1002/prot.10379. [DOI] [PubMed] [Google Scholar]
- 44.Ritchie DW. Recent progress and future directions in protein-protein docking. Curr Protein Pept Sci. 2008;9:1–15. doi: 10.2174/138920308783565741. [DOI] [PubMed] [Google Scholar]
- 45.Case DA, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI force field: Coarse grained model for biomolecular simulations. J Phys Chem B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 47.Oostenbrink C, Soares TA, van der Vegt NF, van Gunsteren WF. Validation of the 53A6 GROMOS force field. Eur Biophys J. 2005;34:273–284. doi: 10.1007/s00249-004-0448-6. [DOI] [PubMed] [Google Scholar]
- 48.Michaud-Agrawal N, Denning EJ, Woolf TB, Beckstein O. MDAnalysis: A toolkit for the analysis of molecular dynamics simulations. J Comput Chem. 2011;32:2319–2327. doi: 10.1002/jcc.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doersch C. 2016. Tutorial on variational autoencoders. arXiv:1606.05908. Preprint, posted June 19, 2016.
- 50.Duan M, Fan J, Li M, Han L, Huo S. Evaluation of dimensionality-reduction methods from peptide folding-unfolding simulations. J Chem Theory Comput. 2013;9:2490–2497. doi: 10.1021/ct400052y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.