Significance
Glia and neurons, the two principal cell types of the nervous system, interact intimately and are critical for normal development and function of the nervous system. While much is known about glia interaction with axons, the architecture and importance of glia interaction with the somatodendritic neuronal compartment is practically unknown. Using Drosophila as a model system, we have identified important and previously unknown roles for glia interaction with the somatodendritic compartment in dendrite structure, activity, and regeneration. Furthermore, recent clinical studies have shown patients with ATRX mutations exhibit white matter and glial abnormalities in their central nervous system, underscoring the significance of our study that uncovers an essential role for glial-expressed ATRX in mediating proper glial development and function.
Keywords: glia–neuron interaction, sensory neurons, ATRX
Abstract
Sensory neurons perceive environmental cues and are important of organismal survival. Peripheral sensory neurons interact intimately with glial cells. While the function of axonal ensheathment by glia is well studied, less is known about the functional significance of glial interaction with the somatodendritic compartment of neurons. Herein, we show that three distinct glia cell types differentially wrap around the axonal and somatodendritic surface of the polymodal dendritic arborization (da) neuron of the Drosophila peripheral nervous system for detection of thermal, mechanical, and light stimuli. We find that glial cell-specific loss of the chromatin modifier gene dATRX in the subperineurial glial layer leads to selective elimination of somatodendritic glial ensheathment, thus allowing us to investigate the function of such ensheathment. We find that somatodendritic glial ensheathment regulates the morphology of the dendritic arbor, as well as the activity of the sensory neuron, in response to sensory stimuli. Additionally, glial ensheathment of the neuronal soma influences dendritic regeneration after injury.
Neurons of the peripheral nervous system (PNS) have diverse structural and functional specializations to detect and process sensory modalities, such as temperature, pain, and touch. In addition to the intrinsic properties of sensory neurons, their intimate interaction with glia is essential for normal development and function of sensory neurons (1). A unique feature of sensory neurons of the PNS in both vertebrates as well as invertebrates, such as Drosophila, is the complete ensheathment of neuronal soma by glial cell membranes (2, 3). This close apposition between neuronal soma and glia raises the possibility that somatodendritic glial interaction might influence neuronal development, activity, and remodeling during development or under injurious conditions.
Drosophila dendritic arborization (da) neurons of the class IV type are sensory neurons with extensive dendritic arbors that cover the body wall of the animal (4) and are required for thermal (5), light (6), and mechanical sensation (7–9). Peripheral glial cells are critical for embryonic development of sensory neurons, as their genetic ablation results in neuronal death as well as axonal pathfinding defects (1, 10, 11). During embryonic development in the fly PNS, glial cell membranes migrate upwards from the axon toward the neuronal cell body of sensory neurons, eventually completely ensheathing the soma as well as the proximal dendrites (3, 12). Once wrapped, the somatodendritic glial ensheathment of sensory neurons persists throughout larval development until metamorphosis. A dramatic pruning and glial remodeling process then ensues during metamorphosis (13). The class IV da neurons undergo pruning to sever all their distal dendrites (14), leaving the glia-covered soma, axon, and proximal dendrites intact (3, 15). Postpruning, the glial covering over neuronal soma is briefly unraveled to allow regrowth of dendrites followed by glial rewrapping, which remains intact in adulthood (3, 14). This intimate and dynamic interaction of the glia with the somatodendritic neuronal surface of da neurons is similar to the glial ensheathment of vertebrate sensory neuronal soma by satellite glial cells; hence, the Drosophila da neurons are well suited for the investigation of neuron–glia interactions (16, 17). Ultrastructural studies of cross-sections of peripheral nerve bundles have revealed that the axons are systematically wrapped in three glial layers composed of structurally and molecularly distinct glial cell types (18, 19), namely the wrapping glia, subperineurial glia, and perineurial glia. Wrapping glia form the innermost layer and interact directly with the neuronal surface, wrapping each axon in the nerve bundle individually, thereby insulating it from other axons (20, 21). The subperineurial glial layer lies in between the perineurial and wrapping glial layers, forms septate junctions, and contributes to the formation of the blood–brain barrier (19). These two layers are further enveloped by the perineurial glia layer (22). In contrast to the axon–glia interaction, the architecture of glial membranes surrounding the somatodendritic surface of sensory neurons is not known. It is also unknown as to whether somatodendritic glial ensheathment is essential for neuronal structure or regenerative potential after injury. In addition, whether glial somatodendritic ensheathment affects basal or evoked neuronal activity is largely unknown, given the challenging requirement of investigating the functional involvement of somatodendritic glial ensheathment without disrupting axonal–glial interaction.
In this study, we focus on how glia interact with the class IV da neuron, a multimodal sensory neuron of the Drosophila PNS with an elaborate dendritic arbor that detects harmful stimuli, such as high temperature (5), UV light (6), and harsh mechanical stimulus (7–9). We first define the identity of the glial cells that ensheath the somatodendritic surface of class IV da sensory neurons. We find that, among the three glial cell types in the PNS, only the perineurial and the subperineurial glial layer ensheath the soma and proximal dendrites of the da neurons, while the wrapping glia exclusively wrap the axon. A screen for glial-expressed factors important for somatodendritic glial wrapping led to the identification of the chromatin remodeler dATRX/XNP, a homolog of the intellectual disability gene ATRX, as a prerequisite for somatodendritic wrapping of the sensory neuron. We show that conditional glial-specific RNA interference (RNAi) mediated depletion of dATRX, or glial-specific expression of a dominant-negative dATRX, leads to an almost complete elimination of somatodendritic ensheathment of da neurons while the axons remain wrapped by glia. This finding provided a unique genetic handle to investigate the function of somatodendritic glial ensheathment of sensory neurons without removing axonal ensheathment. We show that glial somatodendritic ensheathment is important for proper dendritic morphology and activity of the class IV da sensory neuron in response to light stimuli. Additionally, dendritic regeneration after induced injury is stunted in absence of glial somatodendritic interaction. We show that glial-specific loss of the chromatin remodeler dATRX leads to caspase activation in glial cells. Finally, we find that dATRX knockdown in the subperineurial glial layer is sufficient for the loss of somatodendritic ensheathment. These studies thus uncover an important role for somatodendritic glial ensheathment in the development and activity of sensory neurons as well as in its regenerative response to injury.
Results
Distinct Modes of Glial Ensheathment of Somatodendritic and Axonal Compartments.
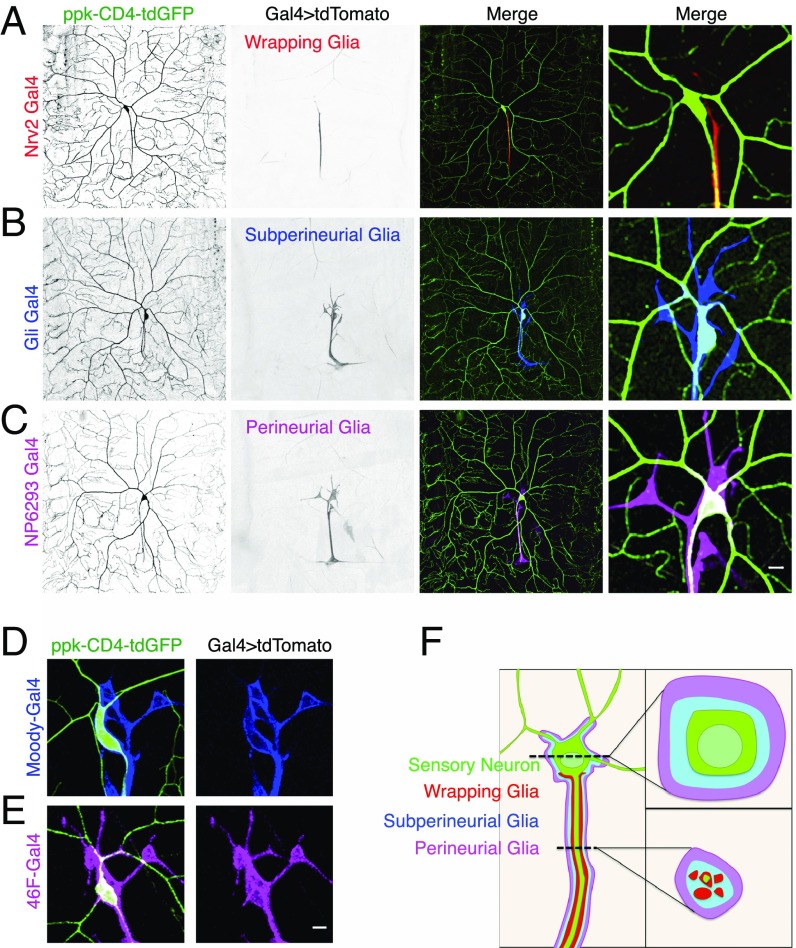
To understand the cellular basis of neuronal–glia interaction, it is essential to examine the spatiotemporal distribution of glial cells and how their membrane processes interact with neurons. To determine the pattern of glial interaction with the somatodendritic and axonal surface of the sensory da neurons, we visualized how each of the three principal peripheral glia cell types interacts with different compartments of the class IV da sensory neuron. Using a marker for the class IV sensory neuron, ppk-CD4-tdTomato, and membrane-tagged GFP expression in glia driven by previously reported cell type-specific glia Gal4 drivers, allowed us to simultaneously visualize sensory neurons and glia and their interaction. We found that all three glia cell types wrap the axon of the class IV da neurons (Fig. 1 A–C). Surprisingly, we found that the somatodendritic surface of the class IV da neuron was wrapped by only the perineurial and subperineurial glial layers as visualized by membrane-tagged GFP driven by 6293-Gal4 and Gli-Gal4 (Fig. 1 B–D). The wrapping glia, visualized by Nrv2-Gal4, exclusively interacts with the axonal surface of the sensory neurons but does not interact with the somatodendritic surface of sensory neurons (Fig. 1 A and F). We tested other independent Gal4 drivers that mark the perineurial and subperineurial glial layers, namely previously characterized 46F-Gal4 for the perineurial layer (23) and Moody-Gal4 for the subperineurial layer (24, 25), and confirmed that both the perineurial and subperineurial layers cover the somatodendritic surface of sensory da neurons (Fig. 1 D–F). Taken together, these results suggest that distinct molecular mechanisms for neuronal–glia interaction determine the glial ensheathment of axonal and somatodendritic neuronal surfaces.
Fig. 1.
Differential architecture of glia–neuron interaction in the somatodendritic and axonal compartments of sensory neurons. Class IV da neurons expressing the neuronal marker ppk-CD4-GFP along with UAS-CD4-tdTomato driven by glial layer-specific Gal4. (A) The Nrv2–Gal4 drives expression in the wrapping glia (red) and the neuron is marked by ppk-CD4-GFP (green). (B) The Gli-Gal4 drives expression in the subperineurial glia (blue) that is in between the perineurial and wrapping glia layers. (C) The perineurial glia layer is visualized using the NP6293-Gal4 (magenta). The extent of neuronal surface ensheathed by each layer of glia is observable in the merge panel. (Scale bar, 10 μm.) (D and E) Independent Gal4 lines Moody-Gal4 and 46F-Gal4 driving UAS-CD4-tdTomato in the subperineurial (blue) and perineurial glial cells (magenta), respectively, can be visualized interacting with the class IV da neurons marked by ppk-CD4-GFP. (Scale bar, 10 μm.) (F) Schematic represents the overall architecture of the three glial layers as well as depicts the cross-sectional view of neuron–glia interaction at the somatodendritic and axonal compartments of da sensory neurons.
Glial-Specific Expression of dATRX Is Essential for Somatodendritic Wrapping of Sensory Neurons.
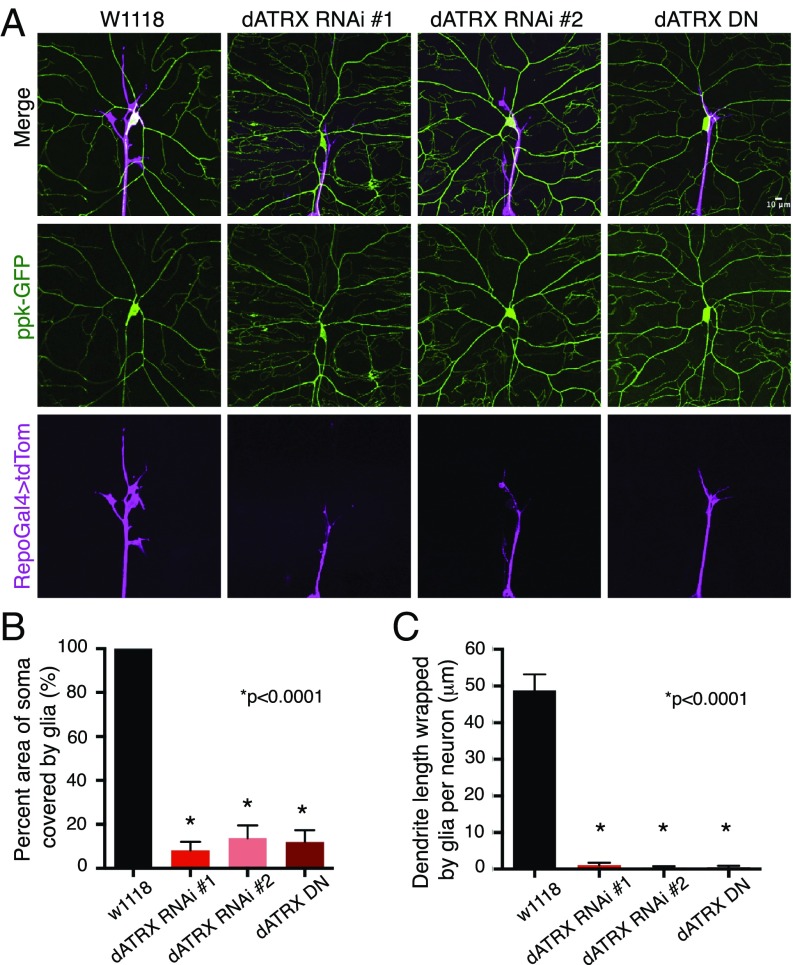
While glia ensheathment is important for insulation of axons, the importance of glia–somatodendritic interaction is an open question. In a screen designed to identify factors required for glial ensheathment of the somatodendritic but not axonal compartment, we discovered a glial-specific role of the chromatin remodeler and ATRX syndrome gene dATRX. Initially known as XNP, dATRX is expressed in both neuron and glia during embryonic and larval development (26). We disrupted expression of dATRX in all three layers of glia through RNAi, and visualized glia by using reversed polarity (Repo) for a pan-glial specific Gal4 driver that was recombined with a UAS-CD4-tdTomato construct. We found that glia-specific knockdown of dATRX using two independent lines led to a failure in glial ensheathment of the somatodendritic surface of class IV da neurons in third-instar larvae, while the glia wrapping of axons in these larvae remained (Fig. 2A). Furthermore, glial-specific expression of a dominant-negative mutant of dATRX (K495R) resulted in a similar phenotype where the somatodendritic surface of da neurons was not covered by glial membranes while the axonal ensheathment remained (Fig. 2A). The surface area of the neuronal soma wrapped by glia decreased from 100% in control larvae (average area = 58.93 μm2, n = 15) to about 8% in absence of dATRX (average area = 4.15 μm2, n = 15) expression in glia (Fig. 2B). The proximal part of primary dendrites were wrapped by glia in control larvae measuring (48.84 ± 4.33 μm, n = 15); however, when dATRX expression in glia was perturbed by either RNAi (n = 15) or by dominant-negative dATRX expression (n = 11), the proximal dendrites were completely devoid of glial ensheathment (Fig. 2C). Importantly, knockdown of dATRX in neurons had no effect on the glial–neuronal interaction, indicating that dATRX functions cell-autonomously in glial cells to regulate neuronal wrapping. These data suggest a glial-specific role of dATRX in normal ensheathment of the somatodendritic surface of sensory neurons.
Fig. 2.
Glial expression of ATRX syndrome gene dATRX/XNP is required for somatodendritic wrapping of sensory neurons. (A) Third-instar larvae expressing ppk-CD4-tdGFP (green) and Repo-Gal4 > CD4-tdTomato (magenta) to visualize neuron and glia, respectively, either alone (W1118), with UAS-dATRX RNAi, or UAS-dATRX dominant-negative (DN) (K495R) are shown. Extent of glial coverage of the somatodendritic compartment of class IV neurons is evident in the merged image. (Scale bar, 10 μm.) (B) Percent area of somatodendritic neuronal surface wrapped by glia in control, two independent dATRX RNAi lines, and dATRX DN third-instar larvae is plotted (n = 15, 15, 11, and 11, respectively, t test, P < 0.0001). (C) Total length of dendrite per neuron wrapped by glia in control, two independent dATRX RNAi lines and dATRX DN third-instar larvae (n = 15, 15, 11, and 11, respectively, t test, P < 0.0001) is plotted.
Somatodendritic Glial Compartment Influences the Shape of the Dendritic Arbor.
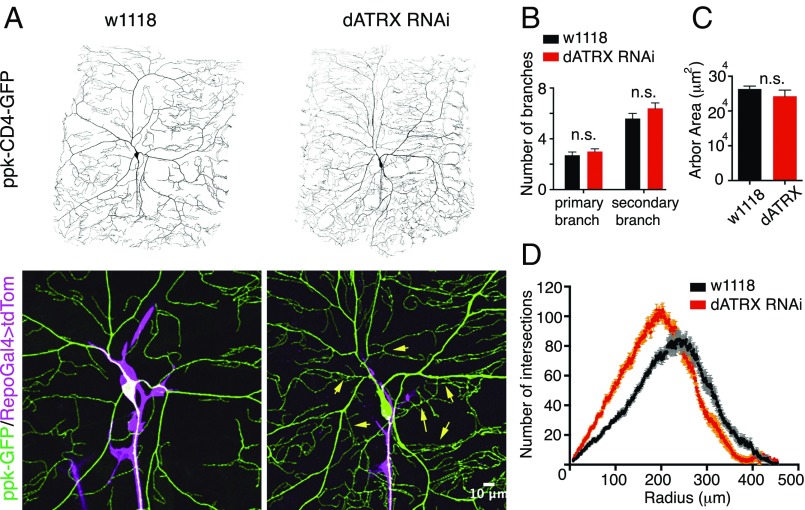
Given the intimate ensheathment of glia with the somatodendritic neuronal surface, we next investigated whether the glial compartment of the somatodendritic region contributes to the development of the dendritic arbor and morphology of the da sensory neurons. To test whether the development of dendritic arbors was perturbed in neurons without somatodendritic glial ensheathment, we imaged third-instar larvae expressing the class IV da neuronal marker ppk-CD4-tdGFP along with either RepoGal4 > CD4-tdTomato or RepoGal4 > CD4-tdTomato along with dATRX RNAi. The dendritic arbor in the absence of somatodendritic glial ensheathment grew to complete coverage of its territory and did not manifest any defects in the number of primary and secondary branches and the area of arbor covered; however, the morphology and the density of the arbor, especially of the terminal branches, was significantly affected (Fig. 3 A–C). Sholl analysis of neuronal dendrites with or without somatodendritic ensheathment revealed that the dendritic density and number of intersections proximal to the soma increased in absence of glial ensheathment (Fig. 3D). We found that the number of primary and secondary branches was unaltered, but the tertiary dendritic branches increased in number and density close to the soma. These data indicate that in the absence of glial wrapping of its somatodendritic surface, the neuron maintains a higher density of dendritic branches closer to the soma.
Fig. 3.
Somatodendritic glial wrapping modulates neuronal morphology. (A) Dendritic arbor of third-instar larvae expressing ppk-CD4-tdGFP and RepoGal4 > CD4-tdTomato either alone or with UAS-dATRX-RNAi is shown. Merged magnified image depicts the defect in the somatodendritic glial ensheathment in dATRX knockdown animals, as well as the change in the dendritic arbor morphology. Numerous ectopic terminal branches close to the soma can be visualized in the Lower Right image (yellow arrows). (Scale bar, 10 μm.) (B) Bar graph shows the number of primary (n = 15, 12, respectively, t test, P = 0.176) and secondary branches (n = 15, 12, respectively, t test, P = 0.087) in control and RepoGal4 > dATRX-RNAi larvae, which are not significantly altered. (C) Total dendritic arbor area covered by neurons in presence and absence of somatodendritic glial wrapping is not significantly (n.s.) affected and is plotted as a bar graph (n = 13, 10, respectively, t test, P > 0.05). (D) Sholl analysis reveals a leftward curve shift due to the large number of terminal branches closer to the soma in RepoGal4 > dATRX-RNAi larvae (peak = 203.6 μm from soma at 104.5 intersections) compared with control larvae (peak = 239.9 μm from soma at 84.75 intersections).
Neuronal Activity in Response to Light Stimuli Is Regulated by Somatodendritic Neuron–Glia Interaction.
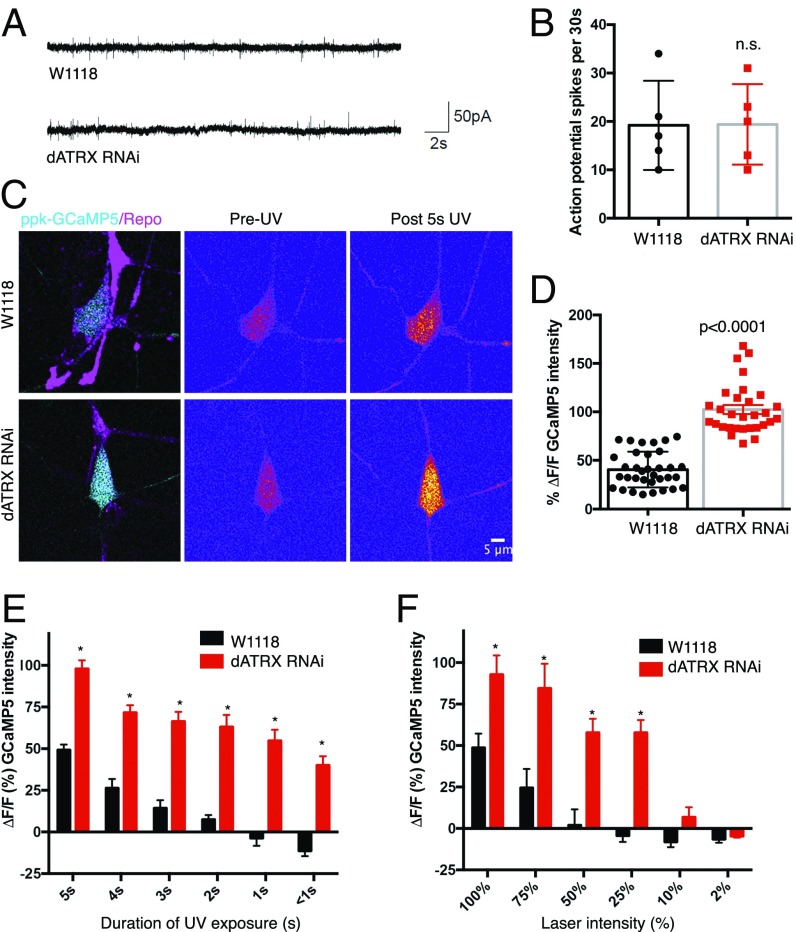
We next tested whether somatodendritic neuron–glial interaction influences the electrophysiological property of sensory neurons. First, by performing extracellular recordings in filleted larval preparations, we tested whether the spontaneous activity of class IV da neurons was affected by removing somatodendritic glial wrapping. We found that the frequency of action potentials fired by the class IV da neurons in animals expressing either RepoGal4 > tdTomato alone or RepoGal4 > tdTomato along with dATRX RNAi were indistinguishable (Fig. 4 A and B). Second, we tested whether the evoked neuronal activity in response to sensory stimuli was affected by somatodendritic neuron–glial interaction. Class IV da neurons are activated in response to UV light and the influx of calcium in response to light stimuli can be visualized using a genetically encoded calcium reporter (6). To visualize the evoked response of class IV da neurons to UV light in intact live animals, we used third-instar larvae expressing GCaMP5 in class IV da neurons (ppk-GCaMP5) along with either RepoGal4 > CD4-tdTomato or with RepoGal4 > Cd4-tdTomato and dATRX RNAi. The in vivo change in intracellular calcium levels on exposure to a UV stimulus lasting 5 s was imaged using a confocal microscope. We found that upon light stimulation, the class IV da neurons in control animals showed an increase of intracellular calcium by 40.72 ± 3.12% (n = 34), while those in RepoGal4 > dATRX RNAi animals showed an increase of 102.4 ± 4.71% (n = 30) (Fig. 4 C and D). We found that the decay of calcium transients in the class IV da neurons after the 5-s light stimulus was much faster in the control animals than in larvae with the RepoGal4 > dATRX RNAi. The calcium influx was abnormally high and persisted for a longer duration in neurons lacking somatodendritic glial wrapping. Given the increased response to light stimuli, we next investigated whether neurons in the absence of glial wrapping were more sensitive to UV by measuring the strength of evoked response to light stimuli of varying intensity and time duration. We varied the time of UV exposure from <1 s to 5 s while keeping the UV intensity constant (at 100%), and found that control neurons did not respond to stimuli of less than 2 s at this maximum laser strength, while the dATRX RNAi animals lacking somatodendritic–glia wrapping responded to even UV stimulus lasting <1 s (at 100% laser strength of 50 mW) (Fig. 4E). When we varied the intensity of the laser while keeping the time of stimulus constant at 5 s, we observed that neurons in control animals did not respond to less than 75% laser power, while neurons in RepoGal4 > dATRX RNAi animals were responsive even at 25% of laser strength (Fig. 4F). These findings suggest that while the spontaneous neuronal activity was unaffected in neurons lacking glial somatodendritic wrapping, both the amplitude and sensitivity of evoked responses to sensory stimuli are affected by glia–neuron interaction at the somatodendritic surface.
Fig. 4.
Somatodendritic glial wrapping affects neuronal activity in response to evoked stimuli. (A) Representative tracings of extracellular recordings obtained from third-instar larvae expressing ppk-CD4-tdGFP;RepoGal4 > CD4-tdTomato (control) or ppk-GFP;RepoGal4 > tdTomato along with UAS-dATRX RNAi. (B) Number of action potential spikes in 30-s period is plotted for control and RepoGal4 > dATRX RNAi larvae (n = 5 each, t test, *P = 0.972). (C) Third-instar larvae expressing ppk-GCaMP5;RepoGal4 > CD4-tdTomato (control) or ppk-GCaMP5;RepoGal4 > CD4-tdTomato along with UAS-dATRX RNAi were exposed to 5-s UV stimulus at 100% laser intensity. Representative images depict baseline GCaMP5 fluorescence (pre) and increase in fluorescence on UV exposure (post). (D) Bar graph plot shows the change in fluorescence intensity of the calcium reported GCaMP5 in class IV da neurons of control (black) and RepoGal4 > dATRX RNAi (red) third-instar larvae (n = 34, 30, respectively, t test P < 0.0001). (E) Changes in GCaMP5 fluorescence intensity on varying duration of UV exposure from <1 s to 5 s while keeping the laser-intensity constant at 100% laser power is depicted for control and RepoGal4 > dATRX RNAi larvae (n = 7 each, paired t test, *P < 0.001). (F) Changes in GCaMP5 fluorescence intensity on varying the laser intensity from 10 to 100% laser power while keeping the duration of UV exposure constant at 5 s is depicted (n = 6 each, paired t test, *P < 0.0001).
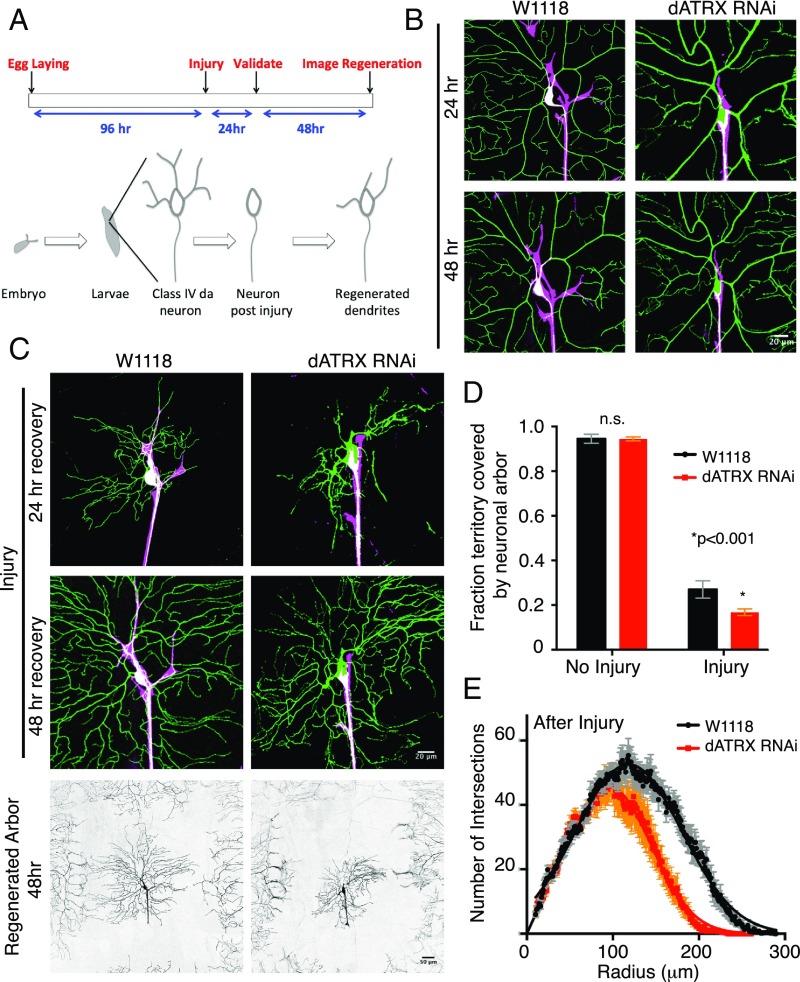
Dendritic Regeneration Is Stunted in Absence of Somatodendritic Glial Ensheathment.
Glia play an essential role in regeneration of axons after injury (27), but whether glial somatodendritic ensheathment contributes to dendritic regeneration is unclear. We tested whether somatodendritic glial wrapping of sensory neurons is required for the ability of class IV da neurons to regenerate their dendrites after injury. To assess the regeneration capacity after dendrite injury, we utilized the “balding injury” paradigm, which involves precise laser-mediated removal of all primary dendrites of the class IV da neuron (28, 29). This procedure leads to degeneration of the dendritic arbor while sparing the axon and soma, which then sprouts new dendrites. Larvae were collected 96 h after synchronized egg-laying, and subjected to either the balding injury or “no injury,” which served as a control. At 24 h after injury, larvae were imaged to ensure that dendrites had indeed been severed, and then they were imaged again 48 h after injury to analyze the extent of regeneration of the dendritic branches (Fig. 5A). We found that the extent of dendritic growth without injury was unchanged in the absence of somatodendritic–glia wrapping. Dendritic growth was quantified as the fraction of total hemisegment area covered by the dendritic arbor 48 h after the no-injury control manipulation was indistinguishable between control (0.94 ± 0.020, n = 9) and RepoGal4 > dATRX RNAi (0.94 ± 0.009, n = 13) -expressing animals (Fig. 5 B and D). In contrast, in the injury condition when all dendrites were severed, the extent of dendritic regeneration after injury was substantially reduced in absence of somatodendritic glial wrapping (Fig. 5 C and D). The territory covered by the regenerated dendritic arbor in RepoGal4 > dATRX RNAi-expressing animals was reduced by 38% compared with control. The fraction of total area that was occupied by the regenerated dendritic arbor 48 h after injury decreased from 0.27 ± 0.03 (n = 7) in control to 0.16 ± 0.14 (n = 7) in RepoGal4 > dATRX RNAi-expressing animals. To identify the global changes in the dendritic arbor morphology after regeneration, we performed Sholl analysis on the regenerated dendrites of control and RepoGal4 > dATRX RNAi-expressing animals. While we found no significant difference in the number of intersections close to the soma, the overall area covered by the dendrites was reduced and the extent of growth was stunted in neurons that lacked somatodendritic glial ensheathment (Fig. 5 C and E). These data together suggest that glia wrapping the somatodendritic region of sensory neurons contributes to the regenerative capacity of dendrites after injury by impacting dendritic growth.
Fig. 5.
Glial wrapping of somatodendritic surface promotes dendritic regeneration. (A) Schematic representation of the timeline for the balding injury paradigm. (B) Class IV da neuron (green) of RepoGal4 > CD4-tdTomato control larvae and of larvae expressing RepoGal4 > CD4-tdTomato, dATRX RNAi at 24 h and 48 h after no injury. Glia membranes are shown in magenta. (Scale bar, 20 μm.) (C) Dendrites (green) of control and RepoGal4 > CD4-tdTomato, dATRX RNAi expressing larvae at 24 h and 48 h after injury. Glia membranes are shown in magenta. (Scale bar, 20 μm.) The Bottom row shows the entire regenerated arbor of control and RepoGal4 > CD4-tdTomato, dATRX RNAi expressing larvae in grayscale. (Scale bar, 50 μm.) (D) Total coverage of regenerated dendritic arbor in control (black) and RepoGal4 > dATRX RNAi (red) expressing larvae normalized to hemisegment area 48 h after no injury (n = 9, 13 respectively, t test, P > 0.05) and injury paradigm (n = 7 each, t test, P < 0.001). (E) Sholl analysis depicting the number of intersections made by regenerated dendritic branches as a function of radius is shown for the regenerated dendritic arbor 48 h after injury of control RepoGal4 > w1118 (black) and RepoGal4 > dATRX RNAi (red) expressing larvae (n = 7 per condition). The total area under the curve decreased in RepoGal4 > tdTomato, dATRX RNAi larvae compared with control (n = 10 each, 7704 μm2 to 5288 μm2, P < 0.001).
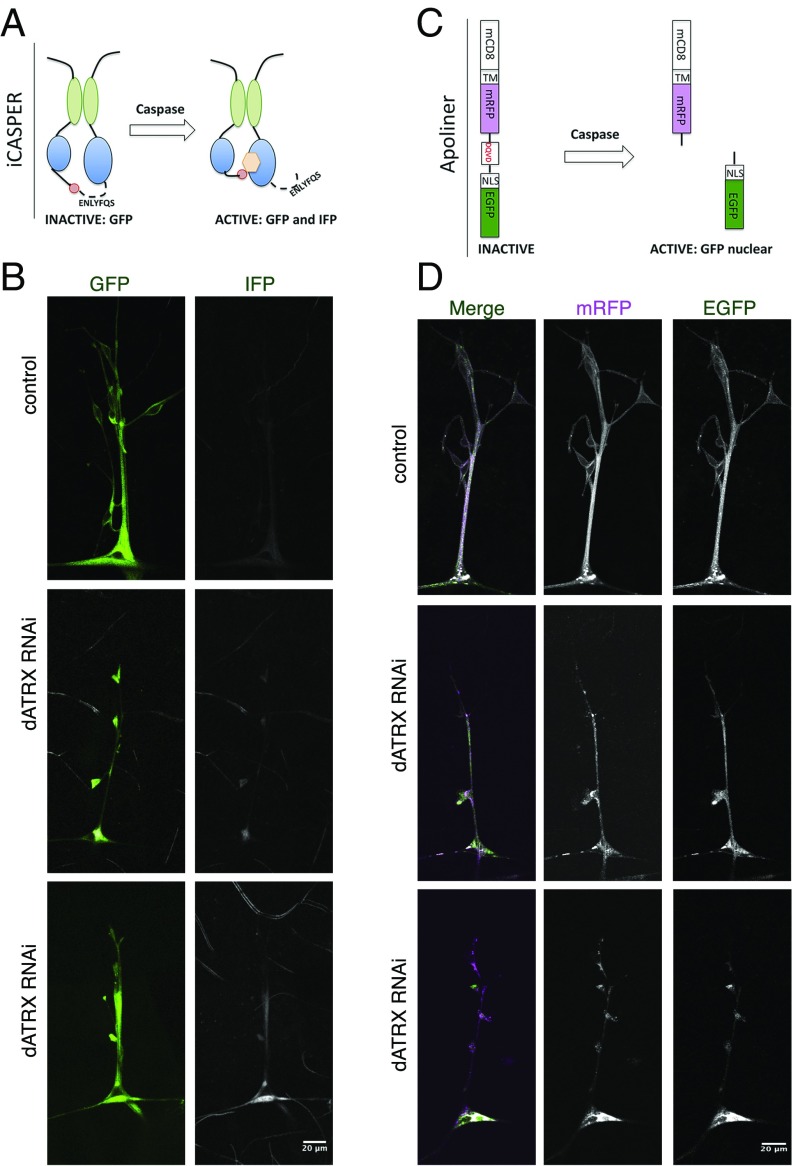
Disrupted Somatodendritic Neuron–Glia Interaction in Absence of dATRX Is Due to Caspase Activation in Glial Cells.
We tested the mechanism through which dATRX contributes to somatodendritic–glia interaction. Given the essential role of ATRX in cell survival (30–32), we hypothesized that absence of dATRX expression in glial cells could either lead to glial membrane retraction or glial cell death, thereby exposing the somatodendritic surface of the neuron. To test whether glial cells undergo caspase activation in the absence of dATRX, we used two independent sensors of caspase activity in vivo in the fly nervous system. The first sensor, iCasper, is an infrared fluorescent executioner-caspase reporter (Fig. 6A) (33). We found that third-instar larvae with UAS-iCasper expression driven by Repo-Gal4 exhibited characteristic GFP fluorescence in the glial membranes surrounding the sensory neurons but showed no fluorescence in the infrared wavelength, with the absence of infrared fluorescent protein (IFP) activation indicative of little or no caspase activity in these glia. In contrast, larvae with both UAS-iCasper and UAS-ATRX RNAi expression driven by RepoGal4 showed a strong IFP signal, indicating that glial cells were undergoing apoptosis (Fig. 6B). The second sensor is Apoliner, a reporter construct based on differential localization of two fluorescent proteins linked by a caspase-sensitive site (34) (Fig. 6C). With activation of caspases, this site is cleaved, leading to nuclear localization of GFP while the mRFP reporter stays membrane-bound. Apoliner is specific and sensitive, detectable before other markers of apoptosis, such as immunostaining for cleaved caspase3, and it does not perturb normal development in Drosophila (34). Fluorescence imaging of third-instar larvae with Repo-Gal4–driven UAS-Apoliner expression revealed that both GFP and mRFP were present throughout the glial membranes. This complete colocalization indicative of lack of cleavage suggests that caspase activity in glia was undetectable. In contrast, in animals with RepoGal4-driven expression of both dATRX RNAi and UAS-Apoliner, there was considerable cleavage so that GFP and mRFP showed distinct distributions, indicating the presence of caspase activation in the absence of dATRX (Fig. 6D). These studies, using the in vivo fluorogenic apoptosis sensors iCasper and Apoliner, show that in the absence of dATRX, a subpopulation of glial cells undergo caspase activation, thereby contributing to the disruption of somatodendritic ensheathment of sensory neurons by glia while sparing the glial wrapping of axons.
Fig. 6.
Loss of dATRX expression in glia leads to caspase activation. (A) Schematic depicts the iCASPER construct, composed of split-GFP domains as well as a caspase cleavage site that controls the positioning of chromophore for an infrared fluorescent protein. Caspase activity leads to conformation change that permits chromophore incorporation and IFP fluorescence. GFP fluorescence is seen in both the presence and absence of caspase activity. (B) Third-instar larvae expressing RepoGal4 > iCasper or RepoGal4 > iCasper, dATRX RNAi were imaged in the GFP and far red channel to visualize glia membranes (green) and the caspase activity detected by the iCasper sensor (gray). (Scale bar, 20 μm.) (C) Schematic representation of the Apoliner construct. In absence of caspase the GFP and mRFP are linked together through a short peptide and retained at the plasma membrane due to the transmembrane domain attached to mRFP. In the presence of active caspase, the linker is cleaved leading to GFP translocation from the plasma membrane while the mRFP is still attached to the membrane. The lack of colocalization of mRFP and GFP indicates caspase activity. (D) Third-instar larvae expressing RepoGal4 > Apoliner or RepoGal4 > Apoliner, dATRX RNAi were imaged in the GFP and RFP channel to visualize activity of caspase in glia membranes. GFP and mRFP and completely colocalized in control animals; however, in dATRX knockdown larvae, GFP and mRFP are no longer localized indicating caspase activation. (Scale bar, 20 μm.)
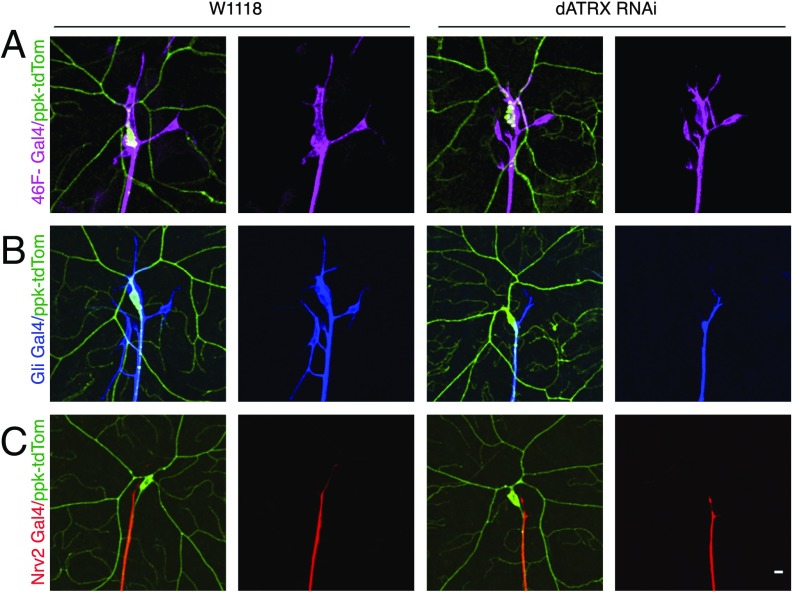
Loss of dATRX in Subperineurial Layer Is Sufficient to Disrupt Somatodendritic Neuronal Ensheathment.
Using the marker for the class IV da sensory neurons, ppk-CD4-tdTomato, and dATRX RNAi expression in glia driven by previously reported glial cell-type–specific Gal4 drivers, allowed us to test if ATRX was required in a particular glia layer for its function in maintaining somatodendritic neuron–glia interaction, or whether it was functionally required in all layers. We found that the somatodendritic–glia covering in sensory neurons was unaffected when dATRX was knocked down in the perineurial layer specifically by expression of 46F-Gal4 > CD4-tdGFP along with UAS-dATRX RNAi, compared with control 46F-Gal4 > CD4-tdGFP animals (Fig. 7A). However, dATRX knockdown in the subperineurial layer using Gli-Gal4 > CD4-tdGFP, UAS-dATRX lead to the uncovering of the somatodendritic glial ensheathment, while the soma was completely wrapped in control animal expressing Gli-Gal4 > CD4-tdGFP (Fig. 7B). Therefore, depletion of dATRX in the subperineurial layer is sufficient to disrupt the somatodendritic neuron–glia interaction, implying that the subperineurial covering of the neuron is a prerequisite for the association of the perineurial glia cells with the somatodendritic surface of the neuron. As expected from earlier results, knockdown of dATRX in the wrapping glia using the Nrv2-Gal4 that exclusively covers the axon did not affect the axonal wrapping (Fig. 7C).
Fig. 7.
Loss of dATRX in subperineurial layer is sufficient to disrupt somatodendritic neuronal ensheathment. Class IV da neurons expressing the neuronal marker ppk-CD4-tdTomato (green) along with UAS-CD4-tdGFP with or without UAS-dATRX RNAi driven by glial layer specific Gal4. (A) The 46F-Gal4 drives expression in the perineurial glia (magenta) and the neuron is marked by ppk-CD4-tdTomato (green). The extent of somatodendritic glial coverage is unaffected by dATRX knockdown in perineurial glia. (B) The effect of dATRX knockdown in subperineurial glia layer is visualized using the Gli-Gal4 driving expression of membrane tagged GFP and UAS-dATRX RNAi in the subperineurial glia (blue). (C) The wrapping glia layer visualized using the Nrv2-Gal4 (red) is unaffected by dATRX knockdown. The extent of neuronal surface ensheathed by each layer of glia is observable in the merge panels. (Scale bar, 10 μm.)
Discussion
This study defines the architecture and function of glia interaction with the somatodendritic compartment of the da sensory neurons in the Drosophila peripheral nervous system. We show that the somatodendritic compartment of each da neuron is ensheathed by two distinct glial membrane layers, in contrast to the axonal wrapping, where an additional third layer is provided by a glial cell type that exclusively wraps axons but not the somatodendritic surface. Glia-specific knockdown of the chromatin remodeler dATRX causes a dramatic loss of glial ensheathment of somatodendritic neuronal surface while maintaining axonal glial wrapping. This genetic manipulation allowed us to address whether glial ensheathment of the somatodendritic neuronal surface is important for neuronal development and function. We show that neuronal development, neuronal activity in response to stimuli, and regenerative capacity of dendrites on injury are affected by the glial interaction with the somatodendritic compartment of the neuron.
Glial Ensheathment of Distinct Neuronal Compartments.
Glial cells ensheath a variety of different neuronal compartments, raising the question of whether distinct molecular and cellular mechanisms exist for compartment-specific interactions. For example, Schwann cells in the peripheral nervous system ensheath the processes of the dorsal root ganglion neurons exclusively, but not the soma (35). Satellite glia cells, on the other hand, envelop the somatic compartment of the dorsal root ganglion neurons but do not wrap the neuronal processes. Oligodendrocytes myelinate the axonal compartment but do not interact with the somatodendritic region of motor neurons in the spinal cord, and interestingly, this property has recently been attributed to an inhibitory signal in the somatodendritic region that limits oligodendrocyte–neuron interaction to the axonal compartment (35). Cortex glia in the fly CNS do not associate with the synaptic neuropil but associate selectively with neuronal cell bodies (36). Astrocyte membrane processes wrap synapses composed of presynaptic and postsynaptic membranes in the mammalian CNS, where astrocytes can regulate synapse formation, maturation, function, and elimination (37). In Drosophila, several distinct neuron–glia interaction types have been defined, including some that have important parallels to the mammalian architecture (16). The fly peripheral nerve bundles are encased in three layers of glial cell membranes. The inner most, wrapping glia, individually wraps axons and is encased by another glial layer, the subperineurial layer. Ultrastructural analyses have revealed that septate junctions formed between cells of the subperineurial layer by neurexin IV, contactin, and neuroglian are the basis of the blood–nerve barrier in flies (19, 38). These three proteins are remarkably conserved and serve similar functions in murine paranodal junctions in myelinated axons (19, 39). Our study shows that glial membranes interact differentially with the axonal and somatodendritic compartments of sensory neurons, such that the wrapping glia does not ensheath the somatodendritic compartment of sensory neurons. The sequential timing and development of glial ensheathment is quite interesting, where the subperineurial sheath envelops the axons before they are individually wrapped from the inside by wrapping glia (38). Based on these observations, we speculate three possibilities: (i) the presence of an inhibitory cue limited to the somatodendritic compartment that prevents wrapping glia from interacting with the soma; (ii) presence of a cell adhesion molecule exclusively expressed by axons that maintains the wrapping glia interaction; or (iii) a cell adhesion molecule exclusively present in the somatodendritic compartment of sensory neurons that interacts directly with the subperineurial layer, possibly leading to the formation of glia–neuronal septate junctions. Testing these hypotheses in the future will provide important insight into the cytoarchitecture of glia–neuron communication.
Glia–Neuron Interaction in Neuronal Structure, Activity, and Regeneration.
Dynamic and reciprocal interactions between glia and neurons are important for neuronal survival, development, and function (22, 40–42). There is growing appreciation for the role of glial cells in sculpting neuronal shape and function. For example, glia play an important role in maintaining the shape of the receptive endings of sensory neurons in Caenorhabditis elegans (43). In the fly sensory neurons of the PNS, neuron–glia interaction mediated by neuroglian, an Ig-like molecule closely related to the vertebrate neural adhesion molecule, together with Ankyrin contributes to axonal and dendritic morphogenesis (44, 45). Our results show that glial ensheathment of the somatodendritic compartment is important for the shape of the dendritic arbor. We find that while the total territory covered by the neurons and the number of primary and secondary dendrites is unchanged, the number of tertiary branches around the soma increased in neurons without glial wrapping of their somatodendritic compartments. The dendrites of da neurons form intimate interactions with the extracellular matrix of the epidermal cells of the body wall via integrins; in some cases parts of dendrites are completely encased in the extracellular matrix (46–49). It raises an intriguing question as to whether dendrites extend additional terminal branches to compensate for the absence of glial interaction by increasing their interaction with the epidermis. Therefore, we speculate that glial wrapping of the neuron, unlike neuronal–epidermal interactions, is inhibitory to dendritic branching and sprouting, and that signaling mechanisms must exist that determine the boundary on the primary dendrites where the dendritic glial wrapping ends and neuronal–epidermal interactions take over. Interplay of interactions between neuron, glia, and epidermal cells likely shapes and refines the dendritic arbor during neuronal development.
Glia modulates neuronal activity through neurotransmitter uptake as well as bidirectional calcium signaling (50); and conversely, neuronal activity can influence glial development and function (51–53). Calcium signaling in glia can either enhance or suppress neuronal signaling in a context-dependent manner. For example, increase of intracellular calcium in cortical glia leads to enhanced seizure susceptibility, as does acute induction of calcium influx through overexpression of transient receptor potential channels in cortical glia (54), whereas acute calcium influx in astrocyte-like glia cells in flies causes paralysis and a rapid loss of neuronal activity (55). Dysfunction in glia can drive hyper/hypoexcitability of neurons and can be neuropathological (56). Our results suggest that the glial membrane interaction with the da sensory neurons does not affect spontaneous activity, but profoundly impacts evoked neuronal activity in response to external sensory stimuli. Without glial ensheathment of the soma and dendrites of sensory neurons, these neurons are hyperactive and hypersensitive to UV stimulation. This inhibitory role of glia might be neuroprotective and could be a beneficial in constraining the neuronal response to noxious and damaging stimuli, thereby preventing excitatory neurotoxicity. This finding warrants further exploration. The genetic tractability and the tools available to manipulate the ensheathing glia and nociceptive neurons make the fly PNS an excellent system to study the glial contribution to nociception as well as development of neuropathies and chronic pain.
Dendritic regeneration is genetically and intrinsically controlled, and is dependent on both neuronal cell type and the developmental timing of injury (27). Extrinsic factors important for regeneration could include clearance of debris formed due to degeneration, injury, or developmental pruning, and the involvement of epidermal cells and glia in promoting regeneration (3, 46). We found that elimination of somatodendritic ensheathment of neurons by glia led to stunted dendritic regeneration. In pupae after dendrite pruning, glial unwrapping of the somatodendritic neuronal surface precedes formation of new primary dendrite branches (3); hence, it is likely that glial unwrapping facilitates initial phases of dendrite growth. Our data suggest a supportive role of glia wrapping in neuronal regeneration after dendrite injury. Absence of somatodendritic–glia wrapping diminishes the extent of overall arbor regeneration, suggesting that prolonged unwrapping after the primary dendrites have extended minimizes the dendritic arbor regenerative capacity. It is possible that glia provide growth-supporting neurotrophic factors directly promoting regeneration, as in the case of Schwann cells in the mammalian PNS (57). It is also conceivable that glia contribute to dendritic regeneration by providing neurons protection from the surge of neuronal calcium influx in response to injury. Our recordings indicate that glia moderates neuronal activity in response to UV stimuli. One intriguing possibility for future investigation concerns the involvement of calcium signaling through the somatodendritic neuron–glia interface in the neuronal response to injury and the regenerative potential of dendrites. Our results suggest that glia play a primary and unique role in supporting neuronal physiology pertaining to both evoked neuronal activity and regeneration after injury, and that this glial function cannot be compensated or substituted by epithelial support cells.
Role of ATRX in Glial Development and Mental Illness.
Our study identifies an important role of glia-specific expression of dATRX in affecting glia–neuron interaction. Mutations in the human homolog ATRX manifest clinically as severe intellectual disability, motor deficits, seizures, microcephaly, and α-thalassemia (58–61). Remarkably, human mutations associated with mental retardation alter residues lying in the SNF2 and helicase chromatin-modifying domains that are conserved in the fly dATRX (26). Most of the studies have focused on the role of ATRX in neurons. For example, loss of ATRX in neurons is associated with neuronal death, while in other cell types loss of ATRX predisposes cells to DNA damage-induced apoptosis via the p53 pathway (32). Neuron-specific ATRX conditional knockout mice show defect in lamination of the neocortex due to apoptosis of the neuroprogenitor cells, leading to extensive hypocellularity (62, 63). Neonatal lethality and defective neurogenesis manifested by increased cell death in ATRX forebrain conditional knockout mice is rescued by inhibiting the tumor-suppressor protein p53 (63). The fly homolog dATRX is required for both CNS glial survival and longitudinal axon formation (26). Interestingly, it appears that overexpression of the fly dATRX can also induce neuronal apoptosis via the JNK pathway, indicating that correct gene dosage of ATRX is required for normal neuronal development (31, 64). While several studies have delineated the function of ATRX function in neuronal differentiation and proliferation, its function in glia remains unclear. Our studies show that glial-specific knockdown of dATRX leads to caspase activation in glia that exposes the somatodendritic neuronal surface without removing the axonal wrapping. We further found that dATRX is specifically required in the subperineurial glial cells that form septate junctions with each other, as well as directly interacts with the neuronal somatodendritic surface. In contrast, dATRX knockdown in the wrapping glia and perineurial glia layers, or in neurons via ppk-Gal4, did not affect somatodendritic–glia interaction. Why some cell types are more sensitive to the absence of ATRX is an open question. It has been reported that diverse cell types respond differently to the loss of ATRX in mice with conditional ablation of ATRX (32). Notably, recent studies employing magnetic resonance imaging to investigate structural changes in human ATRX patients have revealed severe glial defects, such as abnormal myelination and disruption of white matter (59, 65), highlighting the need to further investigate the role of ATRX in glia to fully understand the contribution of glial defects in neurological disorders.
Methods
Fly Stocks.
Lines obtained from the Bloomington Drosophila Stock Center include Nrv2-Gal4 (B#6797), UAS-dATRX/XNP RNAi lines (B#29444, B#32894), UAS-WT-dATRX (B#26645), UAS-DN-dATXR (B#26646), and UAS-Apoliner (B#32122). The NP6293-Gal4 line is from the Kyoto Stock Center (105188). 46F-Gal4 and Moody-Gal4 lines are gifts from Graeme Davis, University of California, San Francisco, CA, and Vanessa Auld, University of British Columbia, Vancouver, Canada, respectively. Fly line ppk-CD4-tdGFP;RepoGal4 > CD4-tdtomato has been previously described (3). Calcium imaging was performed by recombining a ppk-GCaMP5 fusion fly line with RepoGal4 > CD4-tdTomato. The UAS-Casper (33) fly line was a generous gift from Xiaokun Shu, University of California, San Francisco, CA.
Imaging and Image Analysis.
An upright Leica SP5 laser-scanning confocal microscope equipped with laser lines 405, 488, 568, and 630 nm, was used for all imaging experiments. Larvae were mounted on glass slides using glycerol with silicon gel as a spacer and a glass coverslip was placed on top to cover before imaging. All image analysis was performed using ImageJ/Fiji.
Sholl Analysis.
An outline encompassing the entire dendritic arbor of the neuron was drawn and then pasted on a blank image to enable analysis without interference from neighboring arbors. Sholl analysis function of Fiji was used for analysis of dendritic arbor on images after applying the intensity threshold, such that all fine terminal braches were recognized. The soma center was used as the center and concentric circles were drawn at 10 μm away from the center with a distance of 1 μm between each concentric circle. The number of intersections was plotted as a function of radius.
Electrophysiology.
Larval fillet preparations were made by dissecting third instar larvae in hemolymph-like saline containing: 103 mM NaCl, 3 mM KCl, 5 mM TES, 10 mM trehalose, 10 mM glucose, 7 mM sucrose, 26 mM NaHCO3, 1 mM NaH2PO4, and 4 mM MgCl2, adjusted to pH 7.25 and 310 mOsm. Next, 2 mM Ca2+ was added before use. Removing muscles with a fine forceps exposed neurons for recording. Glass electrodes for electrophysiological recording were pulled with a P-97 puller (Sutter Instruments) from thick-wall borosilicate glass, and filled with external saline solution. Extracellular recording was achieved on class IV da neurons identified by GFP fluorescence. Spontaneous action potentials were recorded with a sampling rate of 10 kHz and low-pass–filtered at 1 kHz. Multiclamp 700B amplifier, DIGIDITA 1440A, and Clampex 10.3 software (Molecular Devices) were used to acquire and analyze the data.
Measuring UV Response of Class IV da Neurons.
The Leica SP5 microscope equipped with a 405 laser line (50 mW) was used at 100% laser power to deliver a 5-s exposure to da neurons. Images were acquired in the GFP channel using the 20× objective and 2.5× optical magnification at one frame per second. In experiments where the laser intensity or exposure time was varied, laser intensity was set through the Leica software between 10% and 100% and exposure time was manually controlled between 1–5 s. Exposures that were shorter than the scanning rate of 1 fps were categorized as <1-s UV exposures. Data analysis was performed using Fiji measure function to quantify the GCaMP5 intensity before and after the UV exposure.
Regeneration Assay.
A Chameleon two-photon 930-nm laser mounted on a custom-built Zeiss fluorescence microscope was used to severe dendrites from da neurons by focusing at every primary branch point proximal to the cell body, as previously described (29). Neurons were imaged ∼24 h after injury to confirm that dendrite branches had been severed and then 48 h after injury to assess regeneration. Uninjured neurons from larvae of the same genotype were used as the no injury control.
Data Analysis and Statistics.
All data were statistically analyzed using Microsoft Excel and GraphPad Prism software, as described in the detailed figure legends.
Acknowledgments
Funding was provided by Grant K99/R00 NIH Pathway to Independence Award 5K99MH108648 (to S.Y.) and NIH Grant 1R35NS97227 (to Y.N.J.). Y.N.J. and L.Y.J. are investigators at the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
References
- 1.Sepp KJ, Auld VJ. Reciprocal interactions between neurons and glia are required for Drosophila peripheral nervous system development. J Neurosci. 2003;23:8221–8230. doi: 10.1523/JNEUROSCI.23-23-08221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanani M. Satellite glial cells in sensory ganglia: From form to function. Brain Res Brain Res Rev. 2005;48:457–476. doi: 10.1016/j.brainresrev.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Han C, Jan LY, Jan Y-N. Enhancer-driven membrane markers for analysis of nonautonomous mechanisms reveal neuron-glia interactions in Drosophila. Proc Natl Acad Sci USA. 2011;108:9673–9678. doi: 10.1073/pnas.1106386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jan Y-N, Jan LY. Branching out: Mechanisms of dendritic arborization. Nat Rev Neurosci. 2010;11:316–328. doi: 10.1038/nrn2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tracey WD, Jr, Wilson RI, Laurent G, Benzer S. painless, a Drosophila gene essential for nociception. Cell. 2003;113:261–273. doi: 10.1016/s0092-8674(03)00272-1. [DOI] [PubMed] [Google Scholar]
- 6.Xiang Y, et al. Light-avoidance-mediating photoreceptors tile the Drosophila larval body wall. Nature. 2010;468:921–926. doi: 10.1038/nature09576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang RY, et al. Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr Biol. 2007;17:2105–2116. doi: 10.1016/j.cub.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong L, Hwang RY, Tracey WD. Pickpocket is a DEG/ENaC protein required for mechanical nociception in Drosophila larvae. Curr Biol. 2010;20:429–434. doi: 10.1016/j.cub.2009.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SE, Coste B, Chadha A, Cook B, Patapoutian A. The role of Drosophila Piezo in mechanical nociception. Nature. 2012;483:209–212. doi: 10.1038/nature10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sepp KJ, Schulte J, Auld VJ. Peripheral glia direct axon guidance across the CNS/PNS transition zone. Dev Biol. 2001;238:47–63. doi: 10.1006/dbio.2001.0411. [DOI] [PubMed] [Google Scholar]
- 11.Edenfeld G, Stork T, Klämbt C. Neuron-glia interaction in the insect nervous system. Curr Opin Neurobiol. 2005;15:34–39. doi: 10.1016/j.conb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Sepp KJ, Schulte J, Auld VJ. Developmental dynamics of peripheral glia in Drosophila melanogaster. Glia. 2000;30:122–133. doi: 10.1002/(sici)1098-1136(200004)30:2<122::aid-glia2>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 13.Subramanian A, et al. Remodeling of peripheral nerve ensheathment during the larval-to-adult transition in Drosophila. Dev Neurobiol. 2017;77:1144–1160. doi: 10.1002/dneu.22502. [DOI] [PubMed] [Google Scholar]
- 14.Kuo CT, Jan LY, Jan Y-N. Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin-proteasome, and ecdysone signaling. Proc Natl Acad Sci USA. 2005;102:15230–15235. doi: 10.1073/pnas.0507393102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams DW, Truman JW. Cellular mechanisms of dendrite pruning in Drosophila: Insights from in vivo time-lapse of remodeling dendritic arborizing sensory neurons. Development. 2005;132:3631–3642. doi: 10.1242/dev.01928. [DOI] [PubMed] [Google Scholar]
- 16.Rodrigues F, Schmidt I, Klämbt C. Comparing peripheral glial cell differentiation in Drosophila and vertebrates. Cell Mol Life Sci. 2011;68:55–69. doi: 10.1007/s00018-010-0512-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu Y, et al. Neuronal soma-satellite glial cell interactions in sensory ganglia and the participation of purinergic receptors. Neuron Glia Biol. 2010;6:53–62. doi: 10.1017/S1740925X10000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banerjee S, Bhat MA. Glial ensheathment of peripheral axons in Drosophila. J Neurosci Res. 2008;86:1189–1198. doi: 10.1002/jnr.21574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banerjee S, Pillai AM, Paik R, Li J, Bhat MA. Axonal ensheathment and septate junction formation in the peripheral nervous system of Drosophila. J Neurosci. 2006;26:3319–3329. doi: 10.1523/JNEUROSCI.5383-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Hilchen CM, Bustos AE, Giangrande A, Technau GM, Altenhein B. Predetermined embryonic glial cells form the distinct glial sheaths of the Drosophila peripheral nervous system. Development. 2013;140:3657–3668. doi: 10.1242/dev.093245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matzat T, et al. Axonal wrapping in the Drosophila PNS is controlled by glia-derived neuregulin homolog Vein. Development. 2015;142:1336–1345. doi: 10.1242/dev.116616. [DOI] [PubMed] [Google Scholar]
- 22.Limmer S, Weiler A, Volkenhoff A, Babatz F, Klämbt C. The Drosophila blood-brain barrier: Development and function of a glial endothelium. Front Neurosci. 2014;8:365. doi: 10.3389/fnins.2014.00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie X, Auld VJ. Integrins are necessary for the development and maintenance of the glial layers in the Drosophila peripheral nerve. Development. 2011;138:3813–3822. doi: 10.1242/dev.064816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwabe T, Bainton RJ, Fetter RD, Heberlein U, Gaul U. GPCR signaling is required for blood-brain barrier formation in drosophila. Cell. 2005;123:133–144. doi: 10.1016/j.cell.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 25.Stork T, Bernardos R, Freeman MR. Analysis of glial cell development and function in Drosophila. Cold Spring Harb Protoc. 2012;2012:1–17. doi: 10.1101/pdb.top067587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun X, Morozova T, Sonnenfeld M. Glial and neuronal functions of the Drosophila homolog of the human SWI/SNF gene ATR-X (DATR-X) and the jing zinc-finger gene specify the lateral positioning of longitudinal glia and axons. Genetics. 2006;173:1397–1415. doi: 10.1534/genetics.106.057893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song Y, et al. Regeneration of Drosophila sensory neuron axons and dendrites is regulated by the Akt pathway involving Pten and microRNA bantam. Genes Dev. 2012;26:1612–1625. doi: 10.1101/gad.193243.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stone MC, Albertson RM, Chen L, Rolls MM. Dendrite injury triggers DLK-independent regeneration. Cell Rep. 2014;6:247–253. doi: 10.1016/j.celrep.2013.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson-Peer KL, DeVault L, Li T, Jan LY, Jan Y-N. In vivo dendrite regeneration after injury is different from dendrite development. Genes Dev. 2016;30:1776–1789. doi: 10.1101/gad.282848.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ritchie K, Watson LA, Davidson B, Jiang Y, Bérubé NG. ATRX is required for maintenance of the neuroprogenitor cell pool in the embryonic mouse brain. Biol Open. 2014;3:1158–1163. doi: 10.1242/bio.20148730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee NG, et al. dXNP, a Drosophila homolog of XNP/ATRX, induces apoptosis via Jun-N-terminal kinase activation. FEBS Lett. 2007;581:2625–2632. doi: 10.1016/j.febslet.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Conte D, et al. Loss of Atrx sensitizes cells to DNA damaging agents through p53-mediated death pathways. PLoS One. 2012;7:e52167. doi: 10.1371/journal.pone.0052167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.To T-L, et al. Rationally designed fluorogenic protease reporter visualizes spatiotemporal dynamics of apoptosis in vivo. Proc Natl Acad Sci USA. 2015;112:3338–3343. doi: 10.1073/pnas.1502857112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bardet P-L, et al. A fluorescent reporter of caspase activity for live imaging. Proc Natl Acad Sci USA. 2008;105:13901–13905. doi: 10.1073/pnas.0806983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Redmond SA, et al. Somatodendritic expression of JAM2 inhibits oligodendrocyte myelination. Neuron. 2016;91:824–836. doi: 10.1016/j.neuron.2016.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coutinho-Budd JC, Sheehan AE, Freeman MR. The secreted neurotrophin Spätzle 3 promotes glial morphogenesis and supports neuronal survival and function. Genes Dev. 2017;31:2023–2038. doi: 10.1101/gad.305888.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen NJ, Eroglu C. Cell biology of astrocyte-synapse interactions. Neuron. 2017;96:697–708. doi: 10.1016/j.neuron.2017.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stork T, et al. Organization and function of the blood-brain barrier in Drosophila. J Neurosci. 2008;28:587–597. doi: 10.1523/JNEUROSCI.4367-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Einheber S, et al. The axonal membrane protein Caspr, a homologue of neurexin IV, is a component of the septate-like paranodal junctions that assemble during myelination. J Cell Biol. 1997;139:1495–1506. doi: 10.1083/jcb.139.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scholes J. Glial-neuronal interaction during development. Ann N Y Acad Sci. 1991;633:169–173. doi: 10.1111/j.1749-6632.1991.tb15606.x. [DOI] [PubMed] [Google Scholar]
- 41.Volkenhoff A, et al. Glial glycolysis is essential for neuronal survival in Drosophila. Cell Metab. 2015;22:437–447. doi: 10.1016/j.cmet.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 42.Corty MM, Freeman MR. Cell biology in neuroscience: Architects in neural circuit design: Glia control neuron numbers and connectivity. J Cell Biol. 2013;203:395–405. doi: 10.1083/jcb.201306099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singhvi A, et al. A glial K/Cl transporter controls neuronal receptive ending shape by chloride inhibition of an rGC. Cell. 2016;165:936–948. doi: 10.1016/j.cell.2016.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamamoto M, Ueda R, Takahashi K, Saigo K, Uemura T. Control of axonal sprouting and dendrite branching by the Nrg-Ank complex at the neuron-glia interface. Curr Biol. 2006;16:1678–1683. doi: 10.1016/j.cub.2006.06.061. [DOI] [PubMed] [Google Scholar]
- 45.Bieber AJ, et al. Drosophila neuroglian: A member of the immunoglobulin superfamily with extensive homology to the vertebrate neural adhesion molecule L1. Cell. 1989;59:447–460. doi: 10.1016/0092-8674(89)90029-9. [DOI] [PubMed] [Google Scholar]
- 46.Han C, et al. Epidermal cells are the primary phagocytes in the fragmentation and clearance of degenerating dendrites in Drosophila. Neuron. 2014;81:544–560. doi: 10.1016/j.neuron.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meltzer S, et al. Epidermis-derived semaphorin promotes dendrite self-avoidance by regulating dendrite-substrate adhesion in Drosophila sensory neurons. Neuron. 2016;89:741–755. doi: 10.1016/j.neuron.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han C, et al. Integrins regulate repulsion-mediated dendritic patterning of drosophila sensory neurons by restricting dendrites in a 2D space. Neuron. 2012;73:64–78. doi: 10.1016/j.neuron.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim ME, Shrestha BR, Blazeski R, Mason CA, Grueber WB. Integrins establish dendrite-substrate relationships that promote dendritic self-avoidance and patterning in Drosophila sensory neurons. Neuron. 2012;73:79–91. doi: 10.1016/j.neuron.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma Z, Stork T, Bergles DE, Freeman MR. Neuromodulators signal through astrocytes to alter neural circuit activity and behaviour. Nature. 2016;539:428–432. doi: 10.1038/nature20145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hasel P, et al. Neurons and neuronal activity control gene expression in astrocytes to regulate their development and metabolism. Nat Commun. 2017;8:15132. doi: 10.1038/ncomms15132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishibashi T, et al. Astrocytes promote myelination in response to electrical impulses. Neuron. 2006;49:823–832. doi: 10.1016/j.neuron.2006.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stevens B, Porta S, Haak LL, Gallo V, Fields RD. Adenosine: A neuron-glial transmitter promoting myelination in the CNS in response to action potentials. Neuron. 2002;36:855–868. doi: 10.1016/s0896-6273(02)01067-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Melom JE, Littleton JT. Mutation of a NCKX eliminates glial microdomain calcium oscillations and enhances seizure susceptibility. J Neurosci. 2013;33:1169–1178. doi: 10.1523/JNEUROSCI.3920-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang YV, Ormerod KG, Littleton JT. Astrocyte Ca(2+) influx negatively regulates neuronal activity. eNeuro. 2017;4:ENEURO.0340–16.2017. doi: 10.1523/ENEURO.0340-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robel S, Sontheimer H. Glia as drivers of abnormal neuronal activity. Nat Neurosci. 2016;19:28–33. doi: 10.1038/nn.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barton MJ, John JS, Clarke M, Wright A, Ekberg J. The glia response after peripheral nerve injury: A comparison between Schwann cells and olfactory ensheathing cells and their uses for neural regenerative therapies. Int J Mol Sci. 2017;18:287. doi: 10.3390/ijms18020287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Villard L, et al. Splicing mutation in the ATR-X gene can lead to a dysmorphic mental retardation phenotype without alpha-thalassemia. Am J Hum Genet. 1996;58:499–505. [PMC free article] [PubMed] [Google Scholar]
- 59.Wada T, et al. Neuroradiologic features in X-linked α-thalassemia/mental retardation syndrome. AJNR Am J Neuroradiol. 2013;34:2034–2038. doi: 10.3174/ajnr.A3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shioda N, et al. Aberrant calcium/calmodulin-dependent protein kinase II (CaMKII) activity is associated with abnormal dendritic spine morphology in the ATRX mutant mouse brain. J Neurosci. 2011;31:346–358. doi: 10.1523/JNEUROSCI.4816-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hamzeh AR, et al. A novel missense mutation in ATRX uncovered in a Yemeni family leads to alpha-thalassemia/mental retardation syndrome without alpha-thalassemia. Ir J Med Sci. 2017;186:333–337. doi: 10.1007/s11845-016-1418-6. [DOI] [PubMed] [Google Scholar]
- 62.Bérubé NG, et al. The chromatin-remodeling protein ATRX is critical for neuronal survival during corticogenesis. J Clin Invest. 2005;115:258–267. doi: 10.1172/JCI22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seah C, et al. Neuronal death resulting from targeted disruption of the Snf2 protein ATRX is mediated by p53. J Neurosci. 2008;28:12570–12580. doi: 10.1523/JNEUROSCI.4048-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hong YK, et al. dXNP/DATRX increases apoptosis via the JNK and dFOXO pathway in Drosophila neurons. Biochem Biophys Res Commun. 2009;384:160–166. doi: 10.1016/j.bbrc.2009.04.112. [DOI] [PubMed] [Google Scholar]
- 65.Lee JS, et al. Alpha-thalassemia X-linked intellectual disability syndrome identified by whole exome sequencing in two boys with white matter changes and developmental retardation. Gene. 2015;569:318–322. doi: 10.1016/j.gene.2015.04.075. [DOI] [PubMed] [Google Scholar]