Significance
All living things must sense and react to their environment. Many single-celled organisms do so by using two-component systems, most simply consisting of a sensor histidine kinase and a response regulator. These systems are involved in pathogenicity pathways and can be targeted by new antibiotics. However, the molecular mechanisms used by histidine kinases to translate sensing into responses are not well understood. To probe this general question, we apply a combination of biophysical techniques to a monomeric histidine kinase that senses blue light to determine the structural changes occurring upon activation. We find these changes to be similar to those predicted for the common dimeric histidine kinases, illustrating that the mechanism of activation is conserved regardless of oligomeric state.
Keywords: sensor histidine kinase, HDX-MS, ESR spectroscopy, two-component system, photoreceptor
Abstract
Translation of environmental cues into cellular behavior is a necessary process in all forms of life. In bacteria, this process frequently involves two-component systems in which a sensor histidine kinase (HK) autophosphorylates in response to a stimulus before subsequently transferring the phosphoryl group to a response regulator that controls downstream effectors. Many details of the molecular mechanisms of HK activation are still unclear due to complications associated with the multiple signaling states of these large, multidomain proteins. To address these challenges, we combined complementary solution biophysical approaches to examine the conformational changes upon activation of a minimal, blue-light–sensing histidine kinase from Erythrobacter litoralis HTCC2594, EL346. Our data show that multiple conformations coexist in the dark state of EL346 in solution, which may explain the enzyme’s residual dark-state activity. We also observe that activation involves destabilization of the helices in the dimerization and histidine phosphotransfer-like domain, where the phosphoacceptor histidine resides, and their interactions with the catalytic domain. Similar light-induced changes occur to some extent even in constitutively active or inactive mutants, showing that light sensing can be decoupled from activation of kinase activity. These structural changes mirror those inferred by comparing X-ray crystal structures of inactive and active HK fragments, suggesting that they are at the core of conformational changes leading to HK activation. More broadly, our findings uncover surprising complexity in this simple system and allow us to outline a mechanism of the multiple steps of HK activation.
While all living things sense and respond to their environment, this process is especially important to single-celled organisms. To do so, these organisms often use two-component systems, consisting of a sensor histidine kinase (HK), which autophosphorylates in response to specific stimuli, and a response regulator (RR), which is then phosphorylated by the HK and activates downstream responses (1, 2). Despite recent structural studies of HKs motivated by general interest in signal transduction mechanisms and more specific goals of designing new antibiotic/antifungal agents (3, 4), the detailed mechanisms of HK activation have been inferred chiefly from static snapshots of active and inactive states (5–9).
Aside from notable exceptions (10, 11), HKs are canonically viewed as dimeric proteins with modular architecture that includes an extracytoplasmic sensor region, transmembrane (TM) helices, various signal transduction domains, and a kinase domain consisting of dimerization and histidine phosphotransfer (DHp) and catalytic ATP-binding (CA) subdomains (12, 13). After the sensor detects its stimulus, the signal propagates through the TM and other intervening domains via a variety of motions alternatively described as piston-like motions, scissoring, or helical rotations (12), to the DHp helices. The CA domain then catalyzes the transfer of a phosphoryl group from the bound ATP substrate to the phosphoacceptor histidine located on the first DHp helix. We term this process, consisting of the two steps of (i) stimulus detection and (ii) increase of autophosphorylation enzymatic activity, HK “activation.” The molecular details of this process are somewhat unclear due to the disparate truncated protein constructs used in different studies, although it is apparent that activation is associated with breaking the dimeric symmetry observed in inactive states (14, 15).
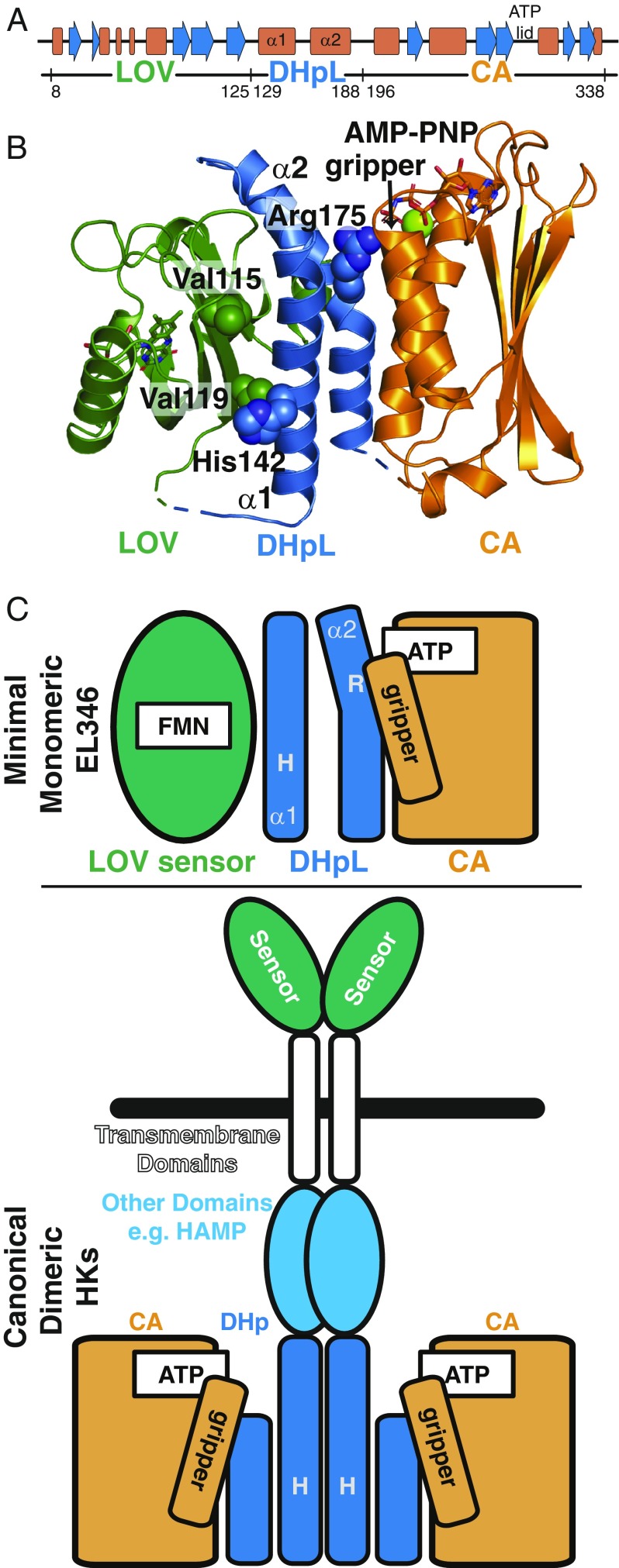
In contrast, the soluble, minimal HK EL346 consists simply of a sensor domain [a Light-Oxygen-Voltage (LOV) domain that responds to blue light] and kinase domain (10, 16). Since EL346 contains only the minimum modules needed to sense and respond to its stimulus, and light is easy to apply in a laboratory setting, this protein is well-suited for mechanistic studies of signal transduction. In addition, from the sensor point of view, one well-established mechanism of LOV domain signaling involves release of a C-terminal helix (17–19). Interestingly, the EL346 kinase functions as a monomer, with the standard DHp dimerization interface blocked by a direct interaction with the sensor LOV domain in the dark, inactive state (10) (Fig. 1). Another useful feature of this kinase is a collection of point mutations that alter dark and light state activity, including constitutively inactive (V119A) and active (V115A) mutants, which tune the basal dark-state activity of EL346 by modulating of the LOV-DHpL interface (10).
Fig. 1.
Dark-state structure and domains of EL346. (A) Secondary structure and domain boundaries of EL346, derived from the dark-state crystal structure in B. (B) Dark-state structure of EL346 (PDB ID code 4R3A) displayed as a cartoon, showing LOV (green), DHpL (blue), and CA (orange) domains. Important residues V115, V119, H142, and R175 are shown as spheres. Ligands are shown as sticks (RBF and AMP-PNP) or spheres (Mg2+). (C) Domain-level schematic diagram of EL346 in the dark state (Top), with colors as in B, and canonical dimeric transmembrane HKs (Bottom).
These practical advantages of EL346 make it an ideal system to probe the conformational changes that accompany HK activation in a natural, full-length context. Thus, we studied the conformations of this enzyme in solution in the dark state, upon light sensing, and as a result of ATP hydrolysis. To do so, we applied biophysical methods that are especially useful for highly dynamic systems, such as hydrogen-deuterium exchange by mass spectrometry (HDX-MS) and pulsed electron spin resonance (ESR) spectroscopy. Intriguingly, we found that, in the dark state, the enzyme exists in both inactive and active conformations, a balance that is shifted during activation. In agreement with our proposed model (10), we found that light-induced changes in the sensor domain are propagated to the kinase domain via a disruption of direct interactions between the domains. Finally, we directly detect the destabilization of the central DHpL helices upon illumination and loss of their interaction with the gripper helix (15) in the catalytic domain upon ATP hydrolysis, confirming and more fully characterizing previously inferred HK activation-induced changes.
Results
EL346 Is Monomeric Regardless of Illumination State.
We subjected EL346, without and with prior illumination, to size-exclusion LC-MS in nondenaturing conditions (20, 21). Both unilluminated and illuminated proteins eluted at times corresponding to a 39-kDa globular protein (consistent with monomeric EL346) without sign of dimers or higher-order oligomers in any of the chromatograms. However, illumination resulted in a small but reproducible shift in elution time and broadening of the elution peak (SI Appendix, Fig. S1A). This shift may suggest a change in conformation or a decrease in number of conformations sampled. The mass spectra of unilluminated and illuminated protein samples were almost identical, with charge states and m/z values corresponding to monomeric protein, as well as low-intensity dimers and trimers (SI Appendix, Fig. S1B). Since these oligomers coelute with monomeric protein, they likely result from nonspecific protein association in the electrospray droplets (20, 22). The masses of the major peaks match the predicted molecular weight of EL346 plus that of the flavin chromophores riboflavin (RBF), FMN, and FAD (SI Appendix, Fig. S1C).
DHpL α1 and α2 Helices Are Exposed upon Activation by Light.
To more precisely characterize the structural rearrangements upon light sensing, we used HDX-MS, which reports on changes in amide protection and, by extension, changes in protein structure (23). Deuterium uptake can mark transiently populated states for detection on the peptide level (“local HDX-MS”), making it a powerful tool that has been applied to studying dynamic signaling states of photoreceptors (24, 25) and histidine kinases (26–28). Our protocol (Fig. 2A and SI Appendix, SI Methods) reproducibly resulted in coverage of about 67% of the protein sequence, with extensive representation of sites in the DHpL and CA domains. Most of the missing peptides reside in the LOV domain (and notably in the ATP lid of the catalytic CA domain); however, as experiments are performed on full-length protein, we can still detect the effects exerted by the LOV on the kinase domain. By studying mutations in the LOV domain (see below), we were also able to examine communication between the LOV domain and the rest of the protein.
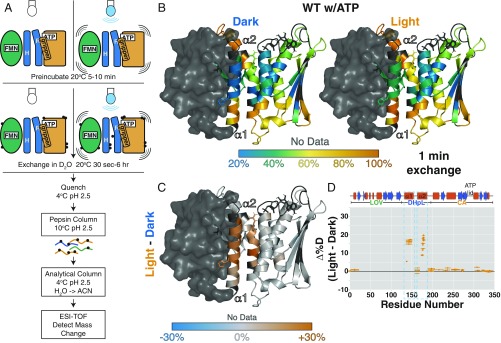
Fig. 2.
Illumination increases exposure of DHpL helices in HDX-MS. (A) Schematic of local HDX-MS experiment. See SI Appendix, SI Methods, for details. Black circles represent protons exchanged for deuterons. (B) Percentage of deuteration at each amide site after 1 min of exchange (20 °C, pH 7.5) in the dark (Left) and light (Right) with ATP mapped onto the EL346 structure. The structure is based on the dark-state crystal structure with loops modeled in by MMM software. The LOV domain, with little peptide coverage, is shown as a surface. Residues with no data are dark gray. (C) Difference between percentage of deuteration at each amide site after 1 min of exchange in the light and in the dark for WT EL346 with ATP mapped onto the EL346 structure. Residues with no data are dark gray. (D) Difference between percentage of deuteration at each peptide after 1 min of exchange in the light and in the dark for WT EL346 with ATP plotted against the EL346 primary sequence. Helices (red boxes) and strands (blue arrows) are shown above.
Overall, we see that ATP-bound EL346 has a central core that is well protected from HDX in both dark and light states (Fig. 2B). However, many helices in the DHpL and CA domains display fraying at the ends. For example, the N-terminal end of the α2 helix in the DHpL domain, which interacts with the LOV domain β-sheet surface in the dark-state crystal structure (10) (SI Appendix, Fig. S2), is not protected from exchange even after 1 min, suggesting that regions of the protein visit other conformations in solution.
Upon illumination, the overall pattern of exchange remains similar, but there are differences in several regions anticipated to be involved in HK regulation. Subtracting the extent of deuteration in the dark from that in the light illuminates three specific regions of increased exchange: the central regions of both DHpL helices, namely (i) α1, which harbors the phosphoacceptor H142; (ii) α2, near the pronounced kink and nucleotide-contacting R175; and (iii) the gripper helix of the CA domain (Fig. 2C), implicating these three helices in EL346 activation by light, as elaborated below. It is important to point out that the conformational changes elicited by illumination include two molecular mechanisms: (i) conformational response to light and (ii) conformational changes resulting in increased autophosphorylation rate. From the comparison of the extent of deuteration in the light and dark discussed above, we detected a combination of these changes localized to the α1, α2, and gripper helices.
The EL346 Structure Is More Dynamic in the Absence of the Nucleotide γ-Phosphate.
Concerned that we might be observing a heterogeneous mix of ATP- and ADP-bound conformations in these experiments, we repeated the local HDX-MS experiments with ADP. Strikingly, exchange throughout the DHpL and CA domains increases greatly with ADP compared with ATP (Fig. 3A), even in the dark, suggesting that the protein structure is stabilized when bound to ATP. These data provide important solution-state validation of our dark-state crystal structure, where the side chain of R175 (DHpL) directly contacts the γ-phosphate of AMP-PNP in a functionally critical way (10) (Fig. 1B). However, it is not only the vicinity of R175 that is destabilized upon ADP binding, but also both DHpL helices and the gripper helix (Fig. 3 B and C), suggesting that the γ-phosphate stabilizes this interaction and implicating these regions specifically in the ATP hydrolysis reaction catalyzed by the kinase.
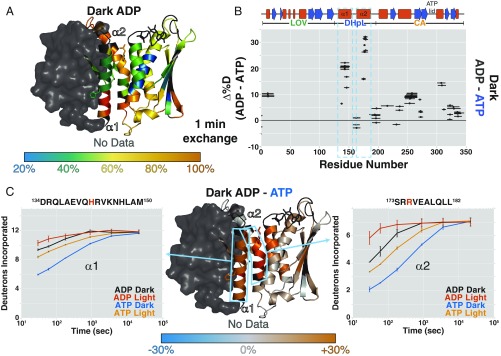
Fig. 3.
ADP decreases EL346 stability overall. (A) Percentage of deuteration at each amide site after 1 min of exchange in the dark with ADP mapped onto the EL346 structure. AMP-PNP is converted to ADP in PyMOL (51) for visualization. Residues with no data are dark gray. (B) Differences between the percentage of deuteration at each peptide after 1 min of exchange in the dark with ADP and ATP plotted against the EL346 primary sequence. Helices (red boxes) and strands (blue arrows) are shown above. (C) Difference between percentage of deuteration at each amide site after 1 min of exchange in the dark with ADP and ATP mapped onto the EL346 structure (Center). Residues with no data are dark gray. Sky blue boxes highlight representative peptides from DHpL helices α1 (Left) and α2 (Right) shown with uptake plots in the dark and light with ATP or ADP. ADP, dark (black); ADP, light (vermillion); ADP, dark (blue); and ATP, light (orange). Phosphoacceptor H142 and ATP-coordinating R175 are highlighted in vermillion in the peptide sequence.
We obtained a similar result using solution NMR spectroscopy with a C-terminally truncated EL346(1-338) construct that gave rise to higher-quality spectra (SI Appendix, SI Methods) but did not affect the overall structure of EL346 (SI Appendix, Fig. S3 A and B). A comparison of fingerprint NMR spectra [transverse relaxation-optimized spectroscopy heteronuclear single quantum coherence (15N/1H-TROSY-HSQC)] of EL346(1-338) in the dark with AMP-PNP and ADP indicated decreased stability with ADP, with a loss of cross-peaks likely arising from well-folded β-strand residues (SI Appendix, Fig. S3C).
Comparing the 2H uptake plots of DHpL helix peptides reveals that, with ADP, these helices are less protected in the dark than the corresponding peptides with ATP even in the light (Fig. 3C). Both helices are destabilized upon illumination, indicating that their stability is coupled. The overall rank order of exchange (ATP dark < ATP light < ADP dark < ADP light) argues that the ADP-bound conformation is a postchemistry state with a destabilized kinase domain ready to facilitate nucleotide exchange for another round of phosphorylation.
Comparison of Inactive and Active Mutants Confirms That Enzymatic Activation Involves DHpL and Gripper Helix Destabilization.
Experiments on both ATP- and ADP-bound EL346 implicate destabilization of the DHpL helices and the gripper helix in response to light. To separate out the conformational changes correlated to enzymatic activation (increased autophosphorylation rate) from those involved in light sensing, we turned to two previously characterized mutants of EL346: the constitutively inactive V119A and the constitutively active V115A (10). These two hydrophobic residues on the LOV domain directly contact ends of the stable core of the DHpL domain, which includes the phosphoacceptor H142, as seen in Figs. 2B and 3A.
We measured the extent of HDX in the inactive V119A and active V115A mutants of EL346 in the dark. Compared with wild type (WT), the inactive V119A mutant displayed protection throughout the DHpL and CA domains in the dark (Fig. 4A). The regions with the biggest differences between the mutant and WT were the α1 and α2 helices of the DHpL domain. The helices and C-terminal region of the CA domain also showed some protection. In active V115A, the DHpL helices showed increased exchange, as did the gripper helix (Fig. 4B). Other regions of the CA domain displayed some protection, similar to the inactive V119A mutant. By comparing patterns of protection and exposure in the WT and mutant proteins, we identify three regions where the extent of exchange correlates with the increased autophosphorylation rate: the two DHpL helices and the gripper helix in CA (arrows in Fig. 4). These data are entirely consistent with autophosphorylation activities measured for the V119A (reduced activity in the dark and light correlates with reduced exchange in these three regions) and V115A (increased activity in the dark and light correlates with increased exchange in these three regions) mutants. These regions also show increased exchange in the WT protein upon illumination (Fig. 4C).
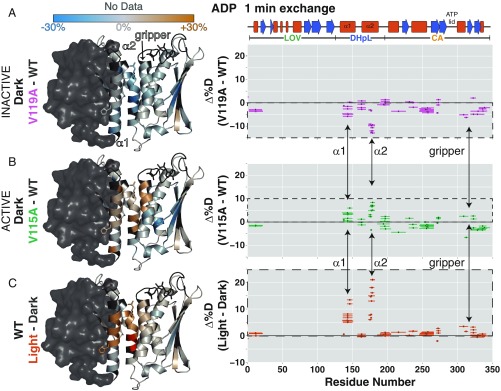
Fig. 4.
Comparison of inactive (V119A) and active (V115A) mutants also implicates DHpL helix exposure in activation. Difference between the extent of deuteration after 1 min of exchange between inactive V119A and WT EL346 (A) in the dark, active V115A and WT (B) in the dark, and in the light and dark for WT (C), with ADP, depicted as the difference between the percentage of deuteration at each amide site mapped onto the EL346 structure (Left) and the difference between the percentage of deuteration at each peptide plotted against the EL346 primary sequence (Right). Residues with no data are dark gray. Helices (red boxes) and strands (blue arrows) are shown above. Opposing changes in exchange localize to the DHpL helices and gripper helix and are shown with arrows.
The lack of a drastic increase in exchange in the mutants suggests that each mutant protein largely adopts the same structure as the WT. This is corroborated by size-exclusion chromatography (SEC) data showing very similar elution volumes for the WT protein and two point mutants (SI Appendix, Fig. S4). A slight shift of elution volume toward earlier elution supports the existence of a marginally more expanded conformation in the cases of both valine-to-alanine mutants, as we originally proposed (10). This more expanded conformation may also be reflected in the decreased exchange in peptides originating from the A′α and Aβ regions of the LOV domain in both mutants (Fig. 4 A and B). Additionally, each valine-to-alanine mutation affects both α1 and α2 helices, confirming that the stability of one DHpL helix is coupled to the other.
Inactive and Active EL346 Mutants Still Exhibit Structural Response to Light.
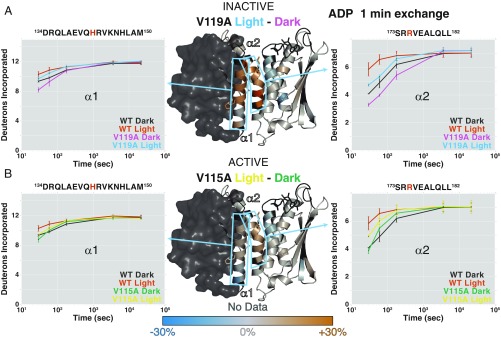
Individual peptide uptake plots confirm that the DHpL helices exchange to a lesser extent in V119A and a greater extent in V115A, when dark states are compared (Fig. 5). However, light does have some effect on the structural level, despite having minimal effect on the autophosphorylation rate in these constitutively (in)active mutants, more so in the inactive V119A than active V115A (Fig. 5). Interestingly, there is no light-induced destabilization of the gripper helix, in contrast to the light-induced changes in the WT protein (Fig. 5 A and B vs. Fig. 4C). This comparison allows us to more directly link changes in the gripper helix to increased autophosphorylation rather than light sensing and suggests that these two steps in the activation mechanism can be decoupled by mutations (10).
Fig. 5.
Light still increases exposure of DHpL helices in mutants with altered light-responsive behavior. (Center) Difference between the percentage of deuteration at each amide site after 1 min of exchange in the light and in the dark for inactive V119A (A) and active V115A (B) mutants with ADP mapped onto the EL346 structure. Residues with no data are dark gray. Sky blue boxes highlight representative peptides from DHpL helices α1 (Left) and α2 (Right) shown with uptake plots for inactive V119A (A) and active V115A (B) compared with WT EL346 in the dark and light with ADP. WT, dark (black); WT, light (vermillion); V119A, dark (purple); V119A, light (sky blue); V115A, dark (green); and V115A, light (yellow). Phosphoacceptor H142 and ATP-coordinating R175 are highlighted in vermillion in the peptide sequence.
Multiple Conformations Exist in the Dark State of EL346.
To get a better idea of the structural changes in the dynamic conformations of EL346, we used site-directed spin-labeling (SDSL) and double electron-electron resonance (DEER) ESR spectroscopy to measure distances between EL346 domains in the dark and light states (29, 30). One key advantage of DEER is that samples are frozen from solution, allowing the distance distribution to report on the multiple populations present before freezing.
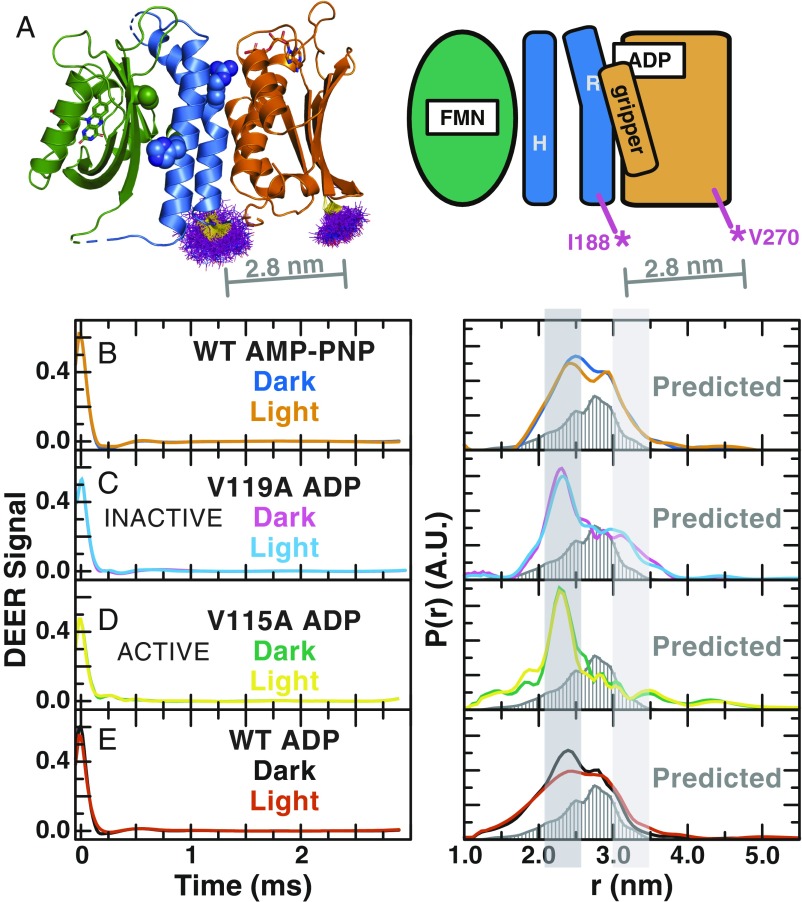
We introduced cysteine residues at positions 188 and 270, allowing measurement of a key distance between the DHpL and CA domains (Fig. 6A). After labeling these two cysteine residues with paramagnetic 1-oxyl-2,2,5,5-tetramethyl-pyrroline-3-methyl)-methanethiosulfonate (MTSL), we confirmed that the MTSL-labeled mutant protein remained active. The initial rate of the autophosphorylation reaction for the labeled mutant protein was determined by a 32P autokinase assay (10) to be 2.4× faster in the dark and 3.4× faster in the light than the WT protein, with a net 1.7× increase upon illumination (SI Appendix, Fig. S5). We then measured the DEER signal in the dark with AMP-PNP and determined the distance distribution between these sites, avoiding a mix of hydrolyzed and nonhydrolyzed ATP. Despite a good signal-to-noise ratio, we observed broad and overlapped peaks in the distance distribution calculated via Tikhonov regularization and maximum entropy method (SI Appendix, Fig. S6), suggesting that EL346 adopts multiple conformations in the dark state. To improve the sensitivity and resolution of these spectra, we subjected the DEER signal to wavelet denoising and singular value decomposition reconstruction (Fig. 6 B–E) (31–34).
Fig. 6.
DEER distance reveals multiple conformations in dark and light states of EL346. (A) MTSL label rotamer distributions (purple lines) at positions 188 and 270 in the EL346 dark-state crystal structure (PDB ID code 4R3A) calculated by mtsslWizard (52) and schematic diagram of label sites I188C and V270C in EL346 showing LOV (green), DHpL (blue), and CA (orange) domains. The average predicted distance of 2.8 nm is shown as a gray line. (B–E) Baseline-subtracted wavelet-denoised time domain DEER signal (Left) and distance distributions of wavelet-denoised DEER data (Right) for EL346 WT with AMP-PNP (B) in the dark (blue) and light (orange), inactive V119A with ADP (C) in the dark (purple) and light (sky blue), active V115A with ADP (D) in the dark (green) and light (yellow), and WT with ADP (E) in the dark (black) and light (vermillion) labeled at positions 188 (DHpL) and 270 (CA). All Right panels also show the predicted distance distribution (dark gray, bars) generated by MMM software (35) from the crystal structure of dark-state EL346 as well as highlighted peaks assigned to different states at ∼2.3 nm (dark gray, broken) and >3.0 nm (light gray, inactive).
We observed two major peaks in the distance distribution of AMP-PNP bound EL346 in the dark, centered at about 2.4 and 2.8 nm (Fig. 6B). The 2.8-nm distance agrees very well with the maximum of the distance distribution predicted by the MMM software (35) from the EL346 dark-state crystal structure (10). After illumination, the distance distribution undergoes a minor perturbation (Fig. 6B), evidenced by a small change in relative intensities of the two major peaks. However, the constitutively inactive V119A mutant shows a strikingly different distance distribution, with the 2.4-nm peak shifting slightly to 2.3 nm and becoming the dominant peak, and the 2.8-nm peak greatly receding in intensity (Fig. 6C). Nevertheless, notable signal from distances above 3.0 nm is still present in the inactive mutant. In the active V115A mutant, the shorter 2.3-nm peak remains the major peak, but all distances at 2.8 nm and above are greatly reduced in intensity. The presence of a large peak at 2.3/2.4 nm in all distance distributions measured from all of our MTSL-labeled proteins, coupled with the increased autophosphorylation activity that we measured in the MTSL-labeled WT EL346, suggests that this distance reports on a “broken” state induced by either the removal of native cysteine residues or the introduction of new cysteine residues and their labeling with MTSL, leading to slightly increased autophosphorylation activity. By contrast, the 2.8 nm and longer distances likely arise from inactive conformations, such as that seen in the dark-state crystal structure. In the presence of ADP, the change upon illumination manifests in broader distance peaks (Fig. 6E), indicative of a highly dynamic state, in agreement with our local HDX-MS data.
With our assignments in hand, we can conclude that multiple conformations are present in the dark state, likely indicating both catalytically active and inactive states in the presence of either ADP or AMP-PNP. The presence of a notable proportion of active conformation in the dark state is supported by our autophosphorylation assays indicating higher activity of the MTSL-labeled mutant protein in the dark. Local HDX-MS data support the presence of two different conformations in the dark state through the appearance of bimodal exchange patterns for several peptides (exemplified in SI Appendix, Fig. S7). In the dark state of the WT protein, these peptides exist in two populations that interconvert in the EX1 regime more slowly than the intrinsic rate of HDX of ∼10 s−1 under these conditions (36). This behavior, indicating exchange between a more protected and a more exposed conformation, virtually disappears in the light and active V115A mutant, with only the more exposed conformation remaining. In the inactive V119A mutant, we again see two interconverting states in solution. Thus, this interconversion with a more protected state in HDX-MS peptides seems to correlate with the presence of DEER distances between 2.8 and 3.2 nm. Since the ratios of the two states in this measurement depend on rates of exchange and not equilibrium populations, we underscore that they need not match the relative populations estimated at equilibrium from the DEER distance distributions.
Discussion
We have shown that activation of the blue-light–sensing HK EL346, whether by the native stimulus of light or by artificial “decoupling” mutations in the LOV domain, proceeds via the destabilization of the LOV-DHpL interface before propagating to the catalytic domain. We can rationalize these observations in the context of the dark-state crystal structure, in which the DHpL domain serves as a “bridge” with large interfaces to both the sensory LOV and catalytic CA domains. We note that the ∼2,000 Å2 DHpL/CA interface is important in this control, particularly via the 600-Å2 interactions between the DHpL helices and CA domain gripper helix. In crystal structures of truncated dimeric HKs, repositioning of this interaction and some unfolding of the gripper helix accompanies autophosphorylation (6, 8, 15, 37). Thus, our data confirm these activation-coupled structural rearrangements more directly and further allow us to separate the effects of light sensing and increase in autophosphorylation rate (see below).
The destabilization of DHpL helices likely indicates interconversion between two conformations, as illustrated by our HDX-MS data showing EX1 behavior in specific DHpL peptides, and not complete unfolding of the helices. This behavior is reminiscent of similar conformational exchange between active and inactive states in other LOV proteins, including an isolated LOV-Jα construct (38) and an engineered LOV-Jα-GTPase (25). While calculation of interconversion rates awaits further kinetic modeling studies, a rough estimate obtained from the expected rates of HDX under these conditions suggests slower interconversion rates for EL346 than the simpler LOV-Jα protein (∼10 s−1 for EL346; ∼1,000 s−1 for LOV-Jα) (36, 38). Interestingly, the “decoupled” mutants, both active V115A and inactive V119A, have a small structural response to light. The α2 helix of the DHpL, which normally contacts the nucleotide, is especially sensitive. This finding argues that the mechanisms of light sensing (in the LOV domain but communicated to the DHpL helices) can be decoupled from the mechanism of autophosphorylation (mainly in the kinase domain) with the DHpL domain serving as a central point of communication for these two mechanisms.
We also find that multiple conformations are present in both the dark and light states of the protein. These states are apparent in three major distances between spin labels in the DHpL CA domains. The longest of these distances, ∼3.2 nm, is assigned to an inactive conformation, while the shortest, 2.3/2.4 nm, is assigned to a broken state arising from the mutations or spin labels introduced into the DEER sample, manifesting in increased enzymatic activity of the labeled mutant protein (SI Appendix, Fig. S5). The middle distance, ∼2.8 nm, corresponds to the maximum of the predicted distance from the dark-state crystal structure and likely represents an inactive conformation as well. The presence of the broken state distance in all datasets likely masks other conformations of the protein, but we can conclude that ADP-loaded samples of both dark and light states of the protein have large proportions of broken and inactive states, as well as presumably an active state. The simultaneous presence of active and inactive conformations in both dark and light states agrees with, and explains, the findings that EL346 displays residual dark-state autophosphorylation (10) and our observation of increased activity in MTSL-labeled protein (SI Appendix, Fig. S5). An equilibrium between active and inactive states has also been proposed for the oxygen-sensing HK AfGcHK based on HDX-MS data (28). Additionally, we show that our previous crystal structure does not fully describe the conformation of EL346 in the dark in solution with the coexistence of active and inactive states and detectable helical fraying in all conformations.
The interactions of LOV domains with C-terminal helices are a major signaling mechanism for both natural and engineered LOV-containing proteins (18, 39). In Avena sativa phototropin LOV2 (AsLOV2), the N-terminal A′α and C-terminal Jα helices are bound to the LOV β-sheet surface in the dark and released in the light (17, 18, 40). The EL346 DHpL helices occupy the same binding sites on the LOV β-sheet surface (SI Appendix, Fig. S2), suggesting a similar signaling mechanism for EL346 light response. Previous removal of the entire LOV domain released these putative inhibitory LOV-DHpL contacts and resulted in a massive rearrangement of DHpL helices [Protein Data Bank (PDB) ID code 4R39], while more subtle approaches to disrupt these interactions via the V115A and V119A mutations led to decoupling of light sensing and activity (10). In addition, our previous limited proteolysis data indicated that the α2 helix near R175 became accessible to trypsinolysis upon light activation. The HDX-MS data presented here are completely consistent with this model, with DHpL helices α1 and α2 both showing increased exchange and presumably detachment from the LOV surface in enzymatically active states generated by illumination or point mutation. These data complement previous findings of changes in stability of helical domains N-terminal to signaling kinases—from destabilization of helices leading into DHp domains correlated with higher kinase activity in the dimeric transmembrane HKs DesK and BvgS (41–43) to increased stability in the helical domains of cytosolic fragments of EnvZ (44) and the Asp receptor (45). The implication of this structural rearrangement in activation across HKs with different sensor domains and oligomeric states strongly argues for a conserved universal role of changes in DHp helix stability in activation. However, technical limitations with respect to activation and study of full-length signaling proteins in vitro currently do not allow for a comprehensive comparative study across systems.
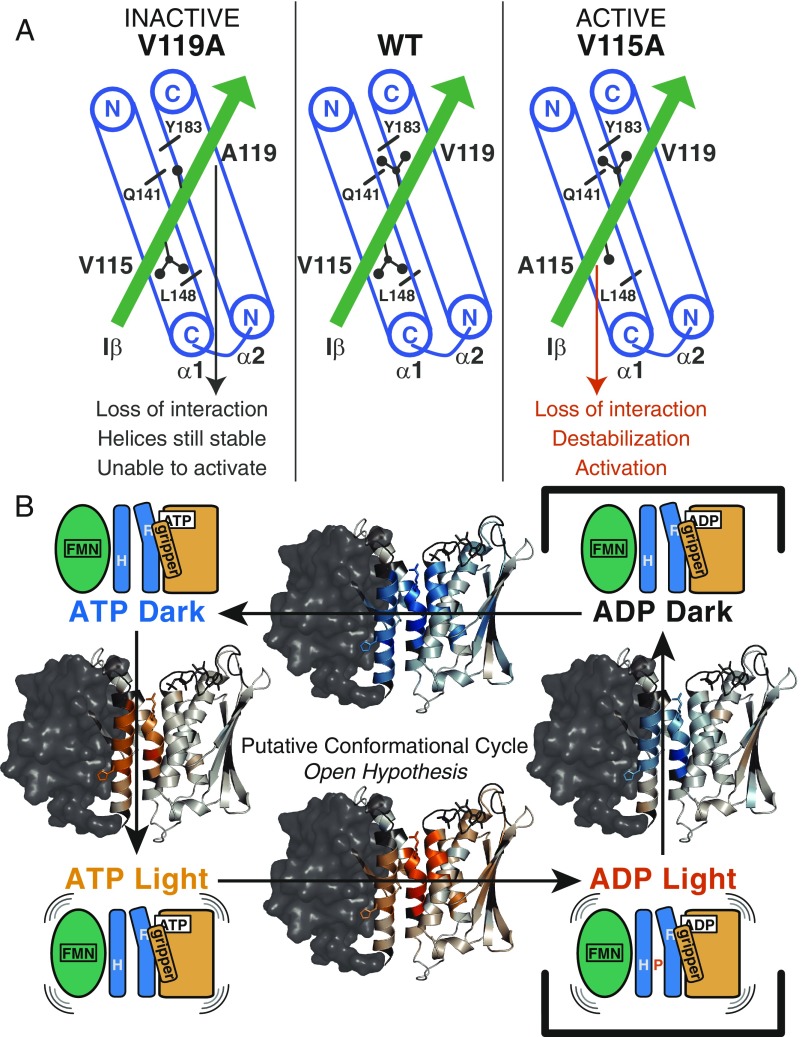
Although both valine-to-alanine mutations are expected to perturb the hydrophobic interactions between LOV and DHpL domains, they have opposing effects on activity (Fig. 7A). Altering the interaction of V119, which contacts both DHpL helices, retains DHpL helix stability and prevents enzymatic activation, while the same change at V115, which contacts the α1 helix, destabilizes both DHpL helices and promotes autophosphorylation. The greater destabilizing effect of V115A could be due to its interaction with the α1 helix C terminus, loss of which may be propagated to the α2 helix immediately C-terminal to it, or its position on the same end of the molecule as the nucleotide-binding site. Alternatively, the V119A mutation could strengthen the DHpL-CA interaction due to “knobs-into-holes” packing. Interestingly, both mutations affect both DHpL helices, indicating that they are coupled in their signaling mechanism. Other effects that both mutations have in common are increased protection of the A′α helix in the LOV domain and some regions in the CA domain seen in our HDX-MS data (Fig. 4 A and B) and the slight shift in elution time to a larger size/shape for the mutants (SI Appendix, Fig. S4). These data hint at a loss of LOV-DHpL contacts that may be responsible for the abrogated light response of the mutants.
Fig. 7.
Disruption of LOV-DHpL interface leads to activation transitions. (A) In the WT EL346 protein (Center), V115 interacts with the C-terminal part of the α1 helix and V119 interacts with both α1 (N-terminal) and α2 (C-terminal) helices. The central regions of the DHpL helices between these hydrophobic residues make up the stable core of the DHpL domain, including H142, as seen in HDX-MS data. In the inactive V119A mutant (Left), the interaction of V119 with Y183 (α2) and Q141 (α1) is likely lost, but the DHpL helices remain stable and autophosphorylation cannot be activated. In the active V115A mutant (Right), the interaction of V115 with L148 (α1) is likely lost, leading to destabilization of the central core of the DHpL helices and activation of autophosphorylation. (B) A proposed reaction cycle with overlaid HDX-MS difference maps shows that (counterclockwise from Top Left) photoactivation of the ATP-bound state results in DHpL helix destabilization, followed by further destabilization of the DHpL and large changes in the CA domain, especially the gripper helix, upon autophosphorylation (ATP to ADP). The sequence of steps to return to the dark state and bind ATP is unknown.
This increased conformational flexibility of the DHpL helices is likely transmitted to the CA domain by a loss of their interaction with the gripper helix, which is illustrated by comparing light- and nucleotide-induced changes in HDX profiles in a putative conformational cycle (Fig. 7B). The change upon illuminating the ATP-bound dark state is mostly limited to the DHpL helices, while the CA domain is mostly untouched. However, concomitant with the loss of the γ-phosphate from ATP, there is increased exchange in both the DHpL helices and the gripper helix and, to a lesser extent, other regions of the CA domain. This role of the gripper helix is supported by changes in protection of that region in the inactive and active mutants, but not in response to light. This different response allows us to separate the structural changes accompanying light sensing and enzymatic activation leading to autophosphorylation. The light response consists of DHpL helix destabilization, likely due to release of inhibitory contacts with the LOV domain, while the changes leading to increased autophosphorylation are in the DHpL helices and gripper helix, likely involving breaking of that interface. These two components of HK activation can be separated out using our nucleotide and mutant data.
While we underscore that this proposed cycle of conformational changes (Fig. 7) remains an open hypothesis to guide future work, our proposal is supported by the involvement of the exact protein regions that were identified from comparing available crystal structures of active and inactive variants of dimeric HK fragments. Our direct interrogation of these structural rearrangements during the two steps of HK activation (stimulus detection and autophosphorylation) in solution and a full-length native HK confirms that they are central to the general mechanism of HK activation. Interestingly, in all these structures, only one protomer in each dimeric assembly is in the active state, resulting in an asymmetric dimer. Such breaking of symmetry is thought to be essential for activation of HisKA-type HKs (8, 9, 14, 15). Much of this view is rooted in the fact that most HKs are dimeric; however, it is becoming clear that a wider variety of oligomeric assemblies are used in HK signaling—from the monomeric EL346 to the trimeric SasA component of cyanobacterial signaling systems (11) to the monomeric/dimeric acid-sensing EvgS in Escherichia coli (46). We assert that this variety of quaternary structure used for HK regulation suggests that the root element—destabilization of the DHp (or DHpL) interactions with the CA domain—is conserved, even if the precise mechanism of triggering that destabilization is different (i.e., symmetry-breaking vs. sensor/DHpL interface breaking).
Conclusions.
Using a minimal, light-sensing histidine kinase, EL346, we have uncovered aspects of both the inactive and the active states of HKs. Our HDX-MS and DEER data show that the crystal structure of EL346 does not fully represent the dark-state structure in solution in that multiple conformations coexist with certain regulatory helices undergoing substantial dynamics. These findings may explain the detectable dark-state activity of EL346 and raise questions about the conformations of other HKs in inactive states. We also show that, in response to light, the DHpL domain helices are freed from LOV inhibitory contacts and visit less-protected conformations, and, upon ATP hydrolysis, the DHpL interaction with the gripper helix in the CA domain is destabilized. This mechanism holistically integrates findings from crystal structures of fragments of canonical dimeric HKs, suggesting that it is a general conformational change leading to HK activation.
Materials and Methods
We expressed WT and mutant EL346 from the pHis-GB1-Parallel1 vector in E. coli BL21 (DE3) cells (Novagen/Millipore Sigma) and purified the proteins by affinity and size exclusion chromatography in the dark under dim red light. We determined protein concentration from an approximate absorption coefficient ε446 = 11,800 M−1·cm−1 for flavin-containing proteins, assuming all flavin was protein-bound. All structural analyses were performed at pH 7.5 with 10 mM MgCl2 and 5 mM AMP-PNP, ADP, or ATP. For SEC coupled to native MS, we injected 30 μM EL346 with ADP into a SEC column equilibrated with 50 mM ammonium acetate solution (47). The HDX-MS experimental workflow is summarized in Fig. 2A. We diluted 50 μM EL346 1/100 in 100% D2O buffer, incubated for a set amount of time, and quenched the exchange with ice-cold low-pH quench buffer before subjecting the protein to peptic digestion and peptide separation on a reversed-phase column, followed by mass detection. Time between quench and elution was 5–10 min, at 0–4 °C and pH 2.5, to minimize back-exchange. We collected 15N/1H TROSY HSQC spectra at 25 °C on dark-adapted (4 h) EL346 1–338 with 5 mM nucleotide using a Bruker AVANCE III HD 600 MHz spectrometer with a cryogenically cooled TCI probe. For SDSL and DEER experiments, we first removed all cysteine residues aside from the flavin-binding cysteine (Cys55) in WT and V115A EL346 and then introduced the I188C and V270C mutations. We expressed and purified the mutant proteins as above, except that we retained the His6-Gβ1 tag on V115A EL346 I188C V270C due to low expression and labeled the proteins with MTSL. The MTSL-labeled I188C V270C protein was confirmed to have similar activity to the WT protein with a 32P autophosphorylation assay as in ref. 10. We performed DEER experiments on labeled proteins at 50–100 μM in buffer containing 80% D2O and 20% d8-glycerol (both Cambridge Isotope Labs) at cryogenic temperature (60 K) on a home-built 17.3 GHz Ku-band pulsed ESR spectrometer as previously described (48, 49) using a standard four-pulse DEER sequence (50). To remove noise from DEER signals, we applied a recently developed wavelet denoising method (31, 32) to the original data before subtracting background in the log domain and reconstructing distance distributions by singular value decomposition (33, 34). Detailed procedures are found in SI Appendix, SI Methods.
Supplementary Material
Acknowledgments
We thank Katie Bunde for performing initial V115A HDX-MS experiments and other K.H.G. laboratory members for fruitful discussions. This work was supported by NIH Grants R01 GM106239 (to K.H.G.), F32 GM119311 (to I.D.), and P41 GM103521 (to J.H.F.), as well as a Burroughs Wellcome Fund 2016 Collaborative Research Travel grant (to I.D.). HDX-MS and NMR data were collected at the CUNY Advanced Science Research Center Biomolecular Mass Spectrometry and Biomolecular NMR Spectroscopy Facilities, respectively, and DEER data were collected at the National Biomedical Center for Advanced ESR Technologies (ACERT, Cornell University).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813586116/-/DCSupplemental.
References
- 1.Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu Rev Microbiol. 2012;66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu Rev Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tiwari S, et al. Two-component signal transduction systems of pathogenic bacteria as targets for antimicrobial therapy: An overview. Front Microbiol. 2017;8:1878. doi: 10.3389/fmicb.2017.01878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov. 2010;9:117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 5.Albanesi D, et al. Structural plasticity and catalysis regulation of a thermosensor histidine kinase. Proc Natl Acad Sci USA. 2009;106:16185–16190. doi: 10.1073/pnas.0906699106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casino P, Miguel-Romero L, Marina A. Visualizing autophosphorylation in histidine kinases. Nat Commun. 2014;5:3258. doi: 10.1038/ncomms4258. [DOI] [PubMed] [Google Scholar]
- 7.Ferris HU, et al. Mechanism of regulation of receptor histidine kinases. Structure. 2012;20:56–66. doi: 10.1016/j.str.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 8.Mechaly AE, Sassoon N, Betton JM, Alzari PM. Segmental helical motions and dynamical asymmetry modulate histidine kinase autophosphorylation. PLoS Biol. 2014;12:e1001776. doi: 10.1371/journal.pbio.1001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trajtenberg F, et al. Regulation of signaling directionality revealed by 3D snapshots of a kinase: Regulator complex in action. eLife. 2016;5:e21422. doi: 10.7554/eLife.21422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rivera-Cancel G, Ko WH, Tomchick DR, Correa F, Gardner KH. Full-length structure of a monomeric histidine kinase reveals basis for sensory regulation. Proc Natl Acad Sci USA. 2014;111:17839–17844. doi: 10.1073/pnas.1413983111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valencia S J, et al. Phase-dependent generation and transmission of time information by the KaiABC circadian clock oscillator through SasA-KaiC interaction in cyanobacteria. Genes Cells. 2012;17:398–419. doi: 10.1111/j.1365-2443.2012.01597.x. [DOI] [PubMed] [Google Scholar]
- 12.Gushchin I, Gordeliy V. Transmembrane signal transduction in two-component systems: Piston, scissoring, or helical rotation? Bioessays. 2018;40:1700197. doi: 10.1002/bies.201700197. [DOI] [PubMed] [Google Scholar]
- 13.Zschiedrich CP, Keidel V, Szurmant H. Molecular mechanisms of two-component signal transduction. J Mol Biol. 2016;428:3752–3775. doi: 10.1016/j.jmb.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neiditch MB, et al. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–1108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhate MP, Molnar KS, Goulian M, DeGrado WF. Signal transduction in histidine kinases: Insights from new structures. Structure. 2015;23:981–994. doi: 10.1016/j.str.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Correa F, Ko WH, Ocasio V, Bogomolni RA, Gardner KH. Blue light regulated two-component systems: Enzymatic and functional analyses of light-oxygen-voltage (LOV)-histidine kinases and downstream response regulators. Biochemistry. 2013;52:4656–4666. doi: 10.1021/bi400617y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harper SM, Christie JM, Gardner KH. Disruption of the LOV-Jalpha helix interaction activates phototropin kinase activity. Biochemistry. 2004;43:16184–16192. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- 18.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 19.Losi A, Gardner KH, Möglich A. Blue-light receptors for optogenetics. Chem Rev. 2018;118:10659–10709. doi: 10.1021/acs.chemrev.8b00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muneeruddin K, Thomas JJ, Salinas PA, Kaltashov IA. Characterization of small protein aggregates and oligomers using size exclusion chromatography with online detection by native electrospray ionization mass spectrometry. Anal Chem. 2014;86:10692–10699. doi: 10.1021/ac502590h. [DOI] [PubMed] [Google Scholar]
- 21.Abzalimov RR, et al. Studies of pH-dependent self-association of a recombinant form of arylsulfatase A with electrospray ionization mass spectrometry and size-exclusion chromatography. Anal Chem. 2013;85:1591–1596. doi: 10.1021/ac302829k. [DOI] [PubMed] [Google Scholar]
- 22.Sun N, Soya N, Kitova EN, Klassen JS. Nonspecific interactions between proteins and charged biomolecules in electrospray ionization mass spectrometry. J Am Soc Mass Spectrom. 2010;21:472–481. doi: 10.1016/j.jasms.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Oganesyan I, Lento C, Wilson DJ. Contemporary hydrogen deuterium exchange mass spectrometry. Methods. 2018;144:27–42. doi: 10.1016/j.ymeth.2018.04.023. [DOI] [PubMed] [Google Scholar]
- 24.Lindner R, Heintz U, Winkler A. Applications of hydrogen deuterium exchange (HDX) for the characterization of conformational dynamics in light-activated photoreceptors. Front Mol Biosci. 2015;2:33. doi: 10.3389/fmolb.2015.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winkler A, et al. Structural details of light activation of the LOV2-based photoswitch PA-Rac1. ACS Chem Biol. 2015;10:502–509. doi: 10.1021/cb500744m. [DOI] [PubMed] [Google Scholar]
- 26.Guo Y, Iavarone AT, Cooper MM, Marletta MA. Mapping the H-NOX/HK binding interface in Vibrio cholerae by hydrogen/deuterium exchange mass spectrometry. Biochemistry. 2018;57:1779–1789. doi: 10.1021/acs.biochem.8b00027. [DOI] [PubMed] [Google Scholar]
- 27.Koshy SS, Eyles SJ, Weis RM, Thompson LK. Hydrogen exchange mass spectrometry of functional membrane-bound chemotaxis receptor complexes. Biochemistry. 2013;52:8833–8842. doi: 10.1021/bi401261b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stranava M, et al. Coordination and redox state-dependent structural changes of the heme-based oxygen sensor AfGcHK associated with intraprotein signal transduction. J Biol Chem. 2017;292:20921–20935. doi: 10.1074/jbc.M117.817023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiang YW, Borbat PP, Freed JH. The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J Magn Reson. 2005;172:279–295. doi: 10.1016/j.jmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Nohr D, Rodriguez R, Weber S, Schleicher E. How can EPR spectroscopy help to unravel molecular mechanisms of flavin-dependent photoreceptors? Front Mol Biosci. 2015;2:49. doi: 10.3389/fmolb.2015.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srivastava M, Anderson CL, Freed JH. A new wavelet denoising method for selecting decomposition levels and noise thresholds. IEEE Access. 2016;4:3862–3877. doi: 10.1109/ACCESS.2016.2587581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srivastava M, Georgieva ER, Freed JH. A new wavelet denoising method for experimental time-domain signals: Pulsed dipolar electron spin resonance. J Phys Chem A. 2017;121:2452–2465. doi: 10.1021/acs.jpca.7b00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Srivastava M, Freed JH. Singular value decomposition method to determine distance distributions in pulsed dipolar electron spin resonance. J Phys Chem Lett. 2017;8:5648–5655. doi: 10.1021/acs.jpclett.7b02379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srivastava M, Freed JH. Singular value decomposition method to determine distance distributions in pulse dipolar electron spin resonance: II. Estimating uncertainty. J Phys Chem A. 2019;123:359–370. doi: 10.1021/acs.jpca.8b07673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeschke G. MMM: A toolbox for integrative structure modeling. Protein Sci. 2018;27:76–85. doi: 10.1002/pro.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C, et al. Mechanistic insights revealed by the crystal structure of a histidine kinase with signal transducer and sensor domains. PLoS Biol. 2013;11:e1001493. doi: 10.1371/journal.pbio.1001493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao X, Rosen MK, Gardner KH. Estimation of the available free energy in a LOV2-J alpha photoswitch. Nat Chem Biol. 2008;4:491–497. doi: 10.1038/nchembio.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu YI, et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halavaty AS, Moffat K. N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry. 2007;46:14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- 41.Lesne E, et al. Balance between coiled-coil stability and dynamics regulates activity of BvgS sensor kinase in Bordetella. MBio. 2016;7:e02089. doi: 10.1128/mBio.02089-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lesne E, et al. Coiled-coil antagonism regulates activity of Venus flytrap-Domain-containing sensor kinases of the BvgS family. MBio. 2018;9:e02052-17. doi: 10.1128/mBio.02052-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saita E, et al. A coiled coil switch mediates cold sensing by the thermosensory protein DesK. Mol Microbiol. 2015;98:258–271. doi: 10.1111/mmi.13118. [DOI] [PubMed] [Google Scholar]
- 44.Wang LC, Morgan LK, Godakumbura P, Kenney LJ, Anand GS. The inner membrane histidine kinase EnvZ senses osmolality via helix-coil transitions in the cytoplasm. EMBO J. 2012;31:2648–2659. doi: 10.1038/emboj.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koshy SS, Li X, Eyles SJ, Weis RM, Thompson LK. Hydrogen exchange differences between chemoreceptor signaling complexes localize to functionally important subdomains. Biochemistry. 2014;53:7755–7764. doi: 10.1021/bi500657v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sen H, et al. Structural and functional analysis of the Escherichia coli acid-sensing histidine kinase EvgS. J Bacteriol. 2017;199:e00310-17. doi: 10.1128/JB.00310-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Konermann L. Addressing a common misconception: Ammonium acetate as neutral pH “buffer” for native electrospray mass spectrometry. J Am Soc Mass Spectrom. 2017;28:1827–1835. doi: 10.1007/s13361-017-1739-3. [DOI] [PubMed] [Google Scholar]
- 48.Borbat PP, Crepeau RH, Freed JH. Multifrequency two-dimensional fourier transform ESR: An X/Ku-band spectrometer. J Magn Reson. 1997;127:155–167. doi: 10.1006/jmre.1997.1201. [DOI] [PubMed] [Google Scholar]
- 49.Georgieva ER, Borbat PP, Norman HD, Freed JH. Mechanism of influenza A M2 transmembrane domain assembly in lipid membranes. Sci Rep. 2015;5:11757. doi: 10.1038/srep11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-time free measurement of dipole-dipole interactions between electron spins. J Magn Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 51.Schrödinger LLC. 2018. The PyMOL Molecular Graphics System (Schrödinger, LLC, New York), Version 2.2.
- 52.Hagelueken G, Abdullin D, Schiemann O. mtsslSuite: Probing biomolecular conformation by spin-labeling studies. Methods Enzymol. 2015;563:595–622. doi: 10.1016/bs.mie.2015.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.