Significance
The precise role of epigenetic factors in controlling chromatin structure is not well understood. Here, we use publicly available data to build and “fold” a nucleosome-resolution mesoscale model of the developmentally regulated HOXC gene locus that incorporates histone tail acetylation, linker histone binding, and nucleosome occupancy. We discover a spontaneous contact hub that bridges promoters and exon/intron junctions. Our work emphasizes how epigenetic factors are coordinated to influence chromatin architecture and opens the way for nucleosome resolution modeling of epigenetically regulated genes.
Keywords: chromatin modeling, chromatin folding, gene structure, chromatin loop domains, contact hub
Abstract
Gene expression is orchestrated at the structural level by nucleosome positioning, histone tail acetylation, and linker histone (LH) binding. Here, we integrate available data on nucleosome positioning, nucleosome-free regions (NFRs), acetylation islands, and LH binding sites to “fold” in silico the 55-kb HOXC gene cluster and investigate the role of each feature on the gene’s folding. The gene cluster spontaneously forms a dynamic connection hub, characterized by hierarchical loops which accommodate multiple contacts simultaneously and decrease the average distance between promoters by 100 nm. Contact probability matrices exhibit “stripes” near promoter regions, a feature associated with transcriptional regulation. Interestingly, while LH proteins alone decrease long-range contacts and acetylation alone increases transient contacts, combined LH and acetylation produce long-range contacts. Thus, our work emphasizes how chromatin architecture is coordinated strongly by epigenetic factors and opens the way for nucleosome resolution models incorporating epigenetic modifications to understand and predict gene activity.
Developmental regulation in eukaryotic organisms depends on chromatin fibers looping across large genomic distances spanning kilobases to megabases (1). Such looping depends on epigenetic factors that are tightly regulated during development and differentiation stages of the cell (2, 3). Chromatin fibers are made up of nucleosomes, where each nucleosome consists of 147 bp of DNA wrapped tightly around eight core histone proteins (two copies each of H2A, H2B, H3, and H4) (4–6). Each histone protein has flexible N-terminal tail domains that extend away from the nucleosome into the surrounding solvent (7). Among many epigenetic chromatin modifications, acetylation of histone tails, nucleosome positioning, and linker histone (LH) binding are known to strongly affect chromatin structure.
Histone tail acetylation—the addition of acetyl groups to the lysine residues within N-terminal tails—is one of many reversible, posttranslational modifications (PTMs) that modulate chromatin architecture and play key roles in regulating gene expression. The modified residues can participate in hydrogen bonding, promoting more stably folded secondary structures of the respective tail (9–11). Acetylation of lysine 16 on the H4 tail (H4K16ac) decreases kilobase-range contacts and is associated with linear fiber decondensation (i.e., fiber lengthening along its principal axis) (10, 12). Acetylation in living chromatin is often found in small islands that range from 1 to 5 kb (13).
Nucleosome positions, often measured as nucleosome repeat length (NRL), or the amount of DNA wrapped around the parent nucleosome (147 bp) plus the linker length, are nonuniform in eukaryotic fibers; in mouse embryonic stem (mES) cells, NRL follows a distribution that can be approximated by positions (where ), with 4 or 5 most common (14). Regions near the 5′ end of each gene-encoding region, commonly referred to as transcription start sites (TSSs), are generally depleted of nucleosomes (also known as nucleosome-free regions, or NFRs) (15). Studies of chromatin fibers reconstituted in vitro and mesoscale modeling show that DNA linker lengths can strongly affect the structure of chromatin fibers, where short linkers (i.e., NRL 162–182 bp) are associated with stiff, straight fibers, and longer linkers are associated with more disordered globules (16, 17).
Besides tail acetylation and nonuniform nucleosome positions, the LH protein strongly affects fiber secondary structure through binding DNA dynamically at various concentrations, on or off the dyad axis formed by entering and exiting DNA (18–24), leading to significant linear compaction (i.e., a shortening of the fiber along its principal axis) (25). Each LH contains a small, uncharged N-terminal domain, a globular head (GH) that binds nucleosomal DNA (19), and a long, positively charged and intrinsically disordered C-terminal domain (CTD) that binds to the entering or exiting linker DNA (26). Chromatin immunoprecipitation (ChIP) combined with sequencing assays (ChIP-Seq) shows that LH localizes to specific regions of chromatin fibers, often anticorrelated with specific PTMs such as acetylation and gene-encoding regions (27). LH knockout studies in mice have shown that while LH silences many genes, it is also necessary to maintain active transcription of some genes (28). LH also has roles in development regulation and diseases (29, 30).
Changes in epigenetic features are part of the regulation of development (31), differentiation (32), and cancer progression (33). Information regarding epigenetic features as a function of genome position is publicly available for many organisms and cell lines [e.g., ENCODE (34)], but combining these features into a unified structural model is not straightforward. Here, we integrate all available data to build and “fold” a mesoscale chromatin model of the 55-kb HOXC gene cluster and systematically investigate the effect of LH, acetylation, NFRs, and DNA linker length distributions on chromatin folding through reference systems that contain different subsets of the gene cluster’s epigenetic features. Our model of the HOXC gene locus includes five genes (containing 11 exons), 284 nucleosomes, five acetylation islands, and 55 kb of DNA.
We find that epigenetically modified chromatin (with LH, acetylation, and NFRs) in this context cooperates to compact a gene hub, forming a series of densely packaged hierarchical loops. This increased density is associated with a dynamic kilobase-range contact hub that bridges promoters within the locus, specifically bringing into contact an LH-rich and acetylation-rich region, as well as “stripe” regions in the nucleosome contact maps, indicating sampling around promoter regions. Our results indicate that nucleosome resolution models that explicitly incorporate epigenetic features derived from experimental data can explain and help predict the genetic regulation of various gene loci as a function of epigenetic modifications.
Results
In addition to the HOXC gene model shown in Fig. 1 B and C, which contains life-like linker length distribution with LH, acetylation islands, and NFRs, we define five other reference systems that contain a subset of the HOXC gene cluster’s factors: Uniform, UniformNFR, Life-Like, Life-LikeAc, and Life-LikeLH (Materials and Methods and SI Appendix). The Uniform fiber has uniform linker lengths without NFRs. The UniformNFR fiber has uniform linker lengths and NFRs. Life-Like fibers have a distribution of linker lengths modeled after living systems. We use this term as in prior work (35) to distinguish these from fibers that have uniform or heterogeneous nucleosome positions. Life-LikeAc fibers have the same life-like linker length distribution plus acetylation islands with acetylated histone tails. Finally, Life-LikeLH fibers contain a life-like linker length distribution with LH placed in intergenic regions.
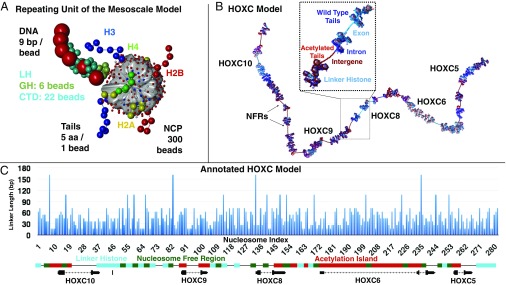
Fig. 1.
Mesoscale model and HOXC system setup. (A) The repeating unit of the mesoscale model consists of a rigid nucleosome core with wrapped DNA represented by 300 point charges and beads for linker DNA, LH, and the flexible histone tails. See details in ref. 8. NCP, nucleosome core particle. (B) A rendering of the HOXC gene cluster model demonstrates gene positions and epigenetic elements in a 3D model. Acetylated tails are red, and WT tails are blue. Intron DNA is drawn in dark blue, exon DNA is colored light blue, and intergenic DNA is dark red. (C) DNA linker lengths used in the HOXC model are plotted against the nucleosome index, along with annotated gene locations and epigenetic features, with acetylation islands in red, LH in teal, and NFRs in green.
Contact Probability Matrices Show Increased Contact Densities in HOXC Fibers.
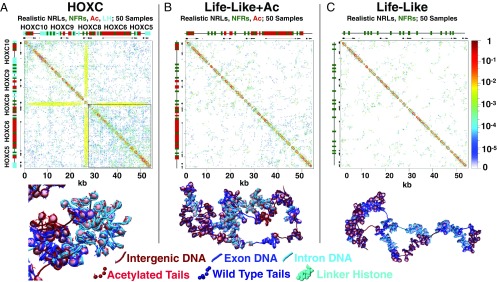
In Fig. 2, we show the 2D contact probability matrix for the HOXC system compared with Life-LikeAc and Life-Like reference systems. HOXC contact maps show a significant increase in contact density and coverage, with two discernible contact domains: LH-rich (top left of contact map) and acetylation-rich region (bottom right of contact map). The latter region is more dense than the former. In fact, stripes or lines running horizontally or vertically, originating near NFRs, demarcate these two subdomains (also see SI Appendix, Fig. S1). Two stripes originating at the HOXC8 promoter indicate contact between this promoter and all other portions of the fiber. This is also evident in a fiber image shown in Fig. 2. The Life-LikeAc (Fig. 2B) and Life-Like (Fig. 2C) reference fibers show homogeneously distributed contacts where acetylation leads to increased contacts of low intensity, but do not form stripes, nor do they show the separation of two domains. Life-Like fibers show the least amount of long-range contacts, with a uniformly distributed contact map.
Fig. 2.
Fiber renderings and contact probability matrices for HOXC and reference fibers. Black bars (top and sides) show the positions of HOXC genes. (A) HOXC system. (B) Reference system with life-like nucleosome positions, NFRs, and acetylation marks. (C) Reference system with life-like nucleosome positions and NFRs.
The epigenetic features also emerge from a close-up of the 30- to 55-kb region (acetylation-rich) (SI Appendix, Fig. S2). This subregion shows increased contacts bridging HOXC5 and HOXC6 genes within the acetylated region. Contact matrices for additional reference systems are shown in SI Appendix, Figs. S4 and S5.
HOXC Gene System Forms Dense Globules with Extensive Hierarchical Looping.
Previously, we have shown that compact chromatin fibers at metaphase tend to fold into hierarchical loops, leading to a compact 3D structure where loops stack in space like rope flaking (8, 36, 37). While we showed that these loops can reproduce contacts observed in chromatin conformation capture (3C) experiments (8), we had not included the effects of life-like nucleosome placement, acetylation, NFRs, or LH.
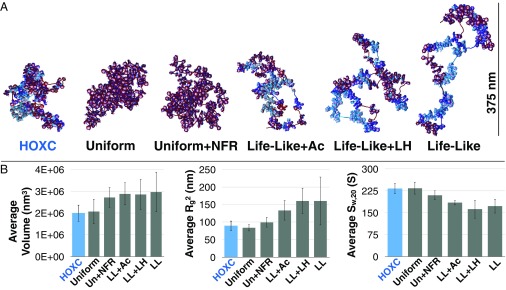
Fig. 3 shows fiber renderings of all systems studied, with average volumes, radii of gyration (), and sedimentation coefficients (). HOXC fibers form the most dense globules with extensive hierarchical looping (8). Uniform and UniformNFR fibers display similarly condensed fibers with extensive hierarchical looping, where NFRs increase the amount of DNA exposed to solvent. Life-LikeLH fibers behave similarly to Life-Like fibers, with some increased linear compaction in LH-rich regions and LH/NFR looping, but LH does not significantly affect the overall volume when compared with Life-Like fibers. Life-LikeAc fibers form slightly more compact globules that Life-Like fibers, but occupy a significantly larger volume than HOXC fibers. Life-Like fibers, which contain 20% short linkers (where NRL = 173 bp), produce stiff fiber segments near NFRs, forming nucleosome “clutches” and intraclutch short-range interactions similar to those observed in mES cells (38). These fibers produce the largest volume of those studied (Fig. 3).
Fig. 3.
Epigenetic factors control fiber morphology and density. (A) Fiber renderings of HOXC, Uniform, UniformNFR, Life-LikeAc, Life-LikeLH, and Life-Like systems. (B) The volume, , and measurements as averaged across all 50 trajectories.
Acetylation and LH Cooperate to Form a Dynamic Hub that Bridges Gene Promoters.
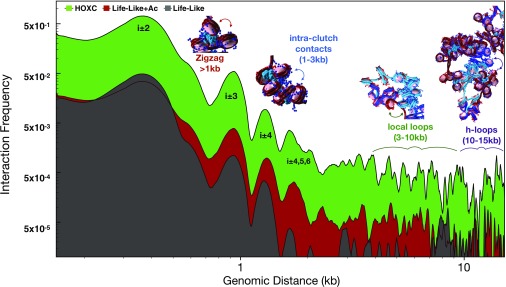
To demonstrate the cooperative nature of LH and acetylation in forming kilobase-range contacts and illustrate the various folding motifs, we analyzed the total interaction probability as a function of genome position in Fig. 4. The largest peak corresponds to next-neighboring nucleosomes at 250 bp, indicative of zigzag fiber folding. Experimental evidence for zigzag morphology has been observed in situ in human cells via electron microscopy-assisted nucleosome interaction capture (EMANIC) (36) and genome-wide probing of short-range internucleosomal interactions with ionizing radiation (39), as well as in situ yeast chromatin via Micro-C data (40). Several near-neighbor nucleosome peaks in the 1- to 5-kb range arise from intraclutch interactions, observed in living cells by EMANIC (36). Next, contacts in the 5- to 10-kb range arise from local looping and near-neighbor clutch interactions, and interactions in the 10- to 20-kb range arise from large loops or hierarchical loops. All contacts are clearly elevated for HOXC fibers (green curve) compared with the reference fibers. Life-Like fibers (with no LH or acetylation; gray) show the least amount of contacts in the 1- to 20-kb range.
Fig. 4.
Analysis of internucleosome interaction contacts for HOXC and reference systems. We annotate structural peaks as follows. Short-range contacts (1 kb) measure next-neighbor interactions common in zigzag fibers. Contacts in the 1- to 2-kb range arise from intraclutch interactions. Chromatin loops between neighboring clutches account for 5- to 10-kb contacts, and hierarchical loops account for 10- to 20-kb contacts.
Each Epigenetic Feature Imparts Distinct Aggregation Tendencies to the Fiber.
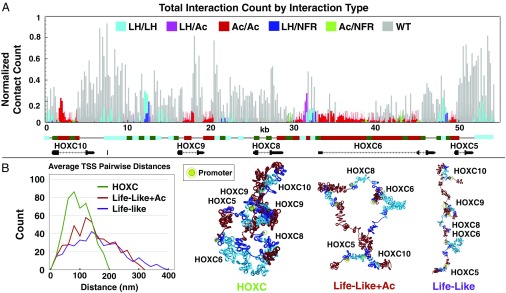
To further distinguish the nature of contacts observed in contact maps, we plotted the contact count of six distinct contact types as a function of genomic position. Fig. 5A shows the normalized contact count observed in HOXC fibers between NFRs and LH regions (NFR/LH), NFRs and acetylation regions (NFR/Ac), LH regions (LH/LH), acetylation and LH regions (Ac/LH), and acetylation/acetylation regions (Ac/Ac). Interactions involving unmodified elements (WT) make up the majority of contacts within the hub. Among epigenetically modified elements, however, the strongest interactions are between LH and LH-dense regions, forming loops between intergenic regions. The second-strongest loop type formed is between acetylation and LH regions, particularly in the intergenic region between HOXC8 and HOXC6 genes, where acetylation islands are found immediately on either side of the LH. There are also moderate interactions between acetylation regions and one NFR in the region between HOXC9 and HOXC10, where an LH-dense region is immediately surrounded by NFRs. Finally, acetylation islands form weakly interacting regions near most of the TSSs. Regions with high amounts of acetylation and nucleosome depletion show some weak Ac/NFR interactions, particularly along the HOXC6 exon region. NFRs interacting with NFRs define the weakest form of epigenetic interaction.
Fig. 5.
Formation of dynamic contact hub depends on epigenetic features. (A) Number of contacts observed in HOXC fiber system between LH/LH (teal), LH/Ac (pink), Ac/Ac (red), LH/NFR (blue), Ac/NFR (green), and any interaction that involves unmodified regions (WT; gray) as a function of genomic position. (B, Left) Plot of average pairwise distance count between gene promoters in the HOXC system vs. two reference systems. (B, Right) Renderings of chromatin configurations with promoter contacts (green/yellow highlight).
The Contact Hub Decreases Average Distance Between Promoters.
To characterize the nature of connections between gene pairs, we calculated the Cartesian distance between pairs of 1 nucleosome cores flanking TSSs between genes in the system. Fig. 5B shows the average pairwise distance counts between all promoter pairs for HOXC fibers, Life-Like fibers, and Life-LikeAc fibers, with a rendering of the DNA and promoter regions for each system. The average distance between gene promoters decreased from 150 to 50 nm, indicating the increased proximity between these sites and the formation of a contact hub.
Hierarchical Looping Allows for Many Dynamic Connections Within the Hub.
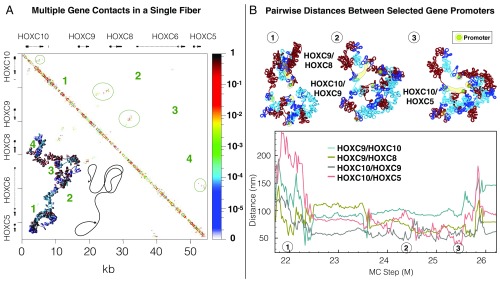
Finally, we show in Fig. 6 that the strongly interacting regions of the connection hub within HOXC fibers are dynamic, where hierarchical looping leads to the formation of multiple contacts simultaneously. Fig. 6A shows an example where an individual folded HOXC gene fiber forms four simultaneous contacts, involving intragene, contiguous-gene, and noncontiguous gene contacts. The corresponding contact matrix is annotated by contact locations. Note how this folding motif allows for nonexclusive contacts between neighboring pairs, next-neighbor pairs, or nonadjacent pairs of genes without tangling the overall fiber.
Fig. 6.
Multiple dynamic contacts form within the hub. (A) Hierarchical looping accommodates four simultaneous loops in an individual fiber. One folded HOXC gene cluster fiber is shown with a sketch demonstrating the fiber folding and the genes and fiber-folding pattern and the corresponding contact probability matrix. (B) Cartesian distances between promoter regions of four gene pairs are plotted as a function of simulation length in a single trajectory. Fiber snapshots, with the promoters highlighted in green and yellow, demonstrate the formation of transient connections between promoters. MC, Monte Carlo.
These dynamic connections can also be seen from Fig. 6B, which shows the distance between gene-promoter pairs for HOXC10/HOXC5, HOXC10/HOXC9, HOXC9/HOXC8, and HOXC9/HOXC5 genes as a function of simulation step, with three accompanying fiber snapshots. All four gene pairs, which represent both neighboring gene segments and gene pairs on the opposite side of the gene neighborhood, transiently form contacts where their distance is close to 50 nm, but no single pair forms a contact that persists throughout the simulation.
Discussion
It is well known that epigenetic features lead to distinct chromatin morphologies both in vitro (41) and in vivo (42), suggesting that active, euchromatin states may spontaneously segregate from inactive, heterochromatin states when modified. Our mesoscale model allows for nucleosome resolution study of the folding and formation of one such activation hub, providing unique insight into the mechanics of gene regulation.
Nucleosome Resolution Epigenetic Models Are Now Possible.
Previously, we have shown that chromatin fibers spanning 10 kb can form hierarchical loops while maintaining zigzag topology, in good agreement with EMANIC cross-linking data of interphase and metaphase fibers in HeLa cells (36). We also showed that this effect can accommodate looping at the 80-kb scale found by 3C experiments at developmentally regulated genes such as the GATA4 gene locus (8). Although sophisticated models have been developed that use epigenetic profiles to predict global chromatin-folding features (43), computational models that represent epigenetic factors such as acetylation and LH explicitly are not available, to our knowledge. Our results indicate that epigenetic factors affect the kilobase-scale chromatin folding through modulating the formation and dynamics of hierarchical loops, providing a structural interpretation of the role of epigenetic features on fiber contact morphology. Additionally, ChIP with paired-end tags data (44), circular chromosome conformation capture (4C), and a high-resolution Hi-C variant, Bridge Linker-Hi-C (BL-Hi-C), developed by Liang et al. (45) show looping among HOXC10, HOXC9, and HOXC6 genes. Such contacts are evident in our HOXC fibers.
Epigenetic Features Cooperate to Form a Dynamic Connection Hub Bridging Local Promoters.
Our results suggest that each epigenetic feature imparts complex folding properties that cannot easily be generalized at all genomic scales. For example, nucleosome depletion at specific regions separates fiber clutches along the genomic sequence. Life-like linker length distributions (or nucleosome positions) further serve to separate neighboring clutches by stiffening individual clutches, but this stiffening comes at a cost, decreasing kilobase-range contacts and leading to a less dynamic fiber. Histone tail acetylation near such short linkers reduces this stiffness, leading to a more stochastic structure with transient contacts at long distances. LH cooperates with acetylation, acting to compact the total polymer by attracting NFRs neighboring acetylated regions, where the linker DNA polyelectrolyte is less effectively screened by strongly folded acetylated tails. Together, these features serve to condense the fiber into dynamic globules with volumes and densities similar to those observed experimentally, significantly increasing coverage of contacts in the 10- to 20-kb range.
Attraction between the LH- and acetylation-rich regions brings the HOXC8 promoter into contact with all other genes. We also see an NFR-rich region just downstream of HOXC10 forming strong contacts with HOXC9, HOXC8, and HOXC6 genes (Fig. 2). However, these features only become apparent in aggregate ensembles or long time scales. While we observed some simultaneous multiloop contacts in single structures (Fig. 6A), the increased propensity to bridge multiple promoters arises on the ensemble level, reflecting the cumulative effects of transient contacts forming between promoters and other regions (Fig. 5A). Thus, stripe patterns in the contact maps may help explain how the cell facilitates loading and efficient diffusion of proteins, by forming strong connections between hubs while maintaining local sampling within the hub. This additionally helps explain the stochastic nature of gene expression observed in eukaryotes (46).
The hypothesis that epigenetic marks can act as boundary insulators is thus supported by our results. CHiP-Seq assays show that acetylation is sparse in the region downstream of the HOXC genes, which does not interact with the connection hub. Thus, it is plausible that heavily modified chromatin spontaneously separates from nearby unmodified chromatin. Our data indicate, however, that the boundaries resulting from such a mechanism are dynamic and only arise as a function of aggregate probability across dynamic samples, as opposed to precisely positioned boundary elements along the genome. Of course, other factors not modeled here, such as CTCF (and loop extrusion), long-noncoding RNAs, and transcription factors, likely play similarly complex roles.
Conclusion
A current attractive model for chromatin structure involves phase transitions (i.e., liquid to gas-like states) as a function of elevated transcription, explaining the spontaneous separation of active from inactive compartments, as seen in Hi-C experiments (47–49). However, as the majority of these models incorporate this assumption implicitly, it is important to develop nucleosome-resolution models that produce these patterns from basic physical variables in the living system. Here, we show that epigenetic features modeled based in a physical–chemical model can fold a gene, helping to interpret biological function of the HOXC gene locus. Specifically, LH, histone tail acetylation, and nucleosome placement cooperate to control the dynamic folding landscape of chromatin. In particular, that LH and acetylation reverse the trend that each factor imparts alone to promote and target kilobase-range contacts is both surprising and indicative of the complex orchestration of many factors on the structural and biophysical levels of the genome.
While our HOXC fibers provide a model system for studying the structure of genes in mammals, many more structural epigenetic features need to be investigated. In particular, the DNA CpG methylation, transcription factor binding, long noncoding RNAs, and CTCF factors missing from our current model likely play important roles in modulating structure. In addition, because epigenetic genomic data based on DNA sequencing generally represent ensembles which are averaged across heterogenous cell populations, and some nucleosome positions can have poor resolution depending on the methodology and ensemble averaging involved (50), more work on proper interpretation of such data for incorporation into structural models is needed. Both improvements in modeling and experiments and in their joint interpretation will be important for moving toward higher-resolution views of genes and genome architecture. Nonetheless, our current mesoscale approach represents a link between structural experiments and the functions of genes in living tissues, while retaining predictive power of the underlying models. As we have shown here, incorporating publicly available epigenetic profiles into in silico structural models makes it feasible now to fold into 3D and investigate the structure and function of various genes in model organisms or cell lines and, ultimately, to obtain a precise description of the dynamic 3D topology of the genomic code.
Materials and Methods
Mesoscale Model.
Our mesoscale chromatin model (16, 51, 52) is composed of four distinct components: a modified worm-like chain polymer to represent linker DNA as beads, partially charged beads representing the electrostatically charged surface of the nucleosome determined by our Discrete Surface Charge Optimization algorithm (53), coarse-grained beads for histone tails, and coarse-grained beads for LH residues (see ref. 8 and SI Appendix for full details of the model and parameters).
Building the HOXC Gene Locus.
To obtain realistic nucleosome positions, acetylation island positions, and LH distributions, we used various experimental data to model each parameter and produce the model of nucleosome positions, acetylation islands, and LH binding regions shown in Fig. 1 B and C. The University of California, Santa Cruz (UCSC) genome browser was used to annotate gene locations and acetylation peak locations (by H3K27ac Chip-Seq) (54). NFRs were modeled based on micrococcal nuclease sequencing (MNase-Seq) data of H9 embryonic stem cells from Yazdi et al. (55), downloaded from the Gene Expression Omnibus (accession code GSM1194221) (56) and visualized by using the Integrated Genome Browser (57). Using these data, we identified the locations of 17 NFRs, which we classified as being either “long” or “short.” The former group has 4 NFRs with 162-bp linkers, and the latter has 13 NFRs with 108-bp linkers. To model realistic nucleosome positions, we used the distribution of linker lengths from chemical mapping in mES cells (14), where we placed nucleosomes such that the overall distribution observed by chemical mapping matched that of our model (Fig. 1C). LHs were placed to emulate trends seen in mES cells (27), where they are found in intergenic and nonacetylated regions only.
Simulation Parameters and Data Analysis.
In addition to the HOXC gene model shown in Fig. 1 B and C, we defined five other reference systems of 55 kb in length that we called Uniform, UniformNFR, Life-Like, Life-LikeLH, and Life-LikeAc (SI Appendix). These systems were designed to test for the effect of each parameter in comparison with the real system. The Uniform fiber system has a uniform linker length of 53 bp without NFRs, whereas the UniformNFR fiber system has a uniform linker length of 53 bp interrupted by NFRs, modeled either as a 108- or 612-bp linker length. The positions of these NFRs were placed as above, and the percentages of 108-bp vs. 162-bp NFRs were placed as above. Life-Like fibers have a distribution of linker lengths (SI Appendix) modeled after living mammalian genome nucleosome positions by chemical mapping with NFRs (14). Life-LikeAc fibers have a life-like linker length distribution with acetylation peaks placed according to data taken from the UCSC genome browser (58). Life-LikeLH fibers contain a life-like linker length distribution with LH placed in intergenic regions. The positions of NFRs, acetylation, and LH are given for all reference systems in SI Appendix. An idealized zigzag starting structure was used for all simulations. See details in SI Appendix. Our LH model was based on the rat H1d LH structure predicted by Bharath et al. through fold recognition and molecular modeling (59). LH was rigidly fixed and placed on the dyad axis separated by a distance of 2.6 nm. We showed previously that, despite LH positioning in the dyad axis, the asymmetric structure of the GH and nucleosome interaction leads to an asymmetric chromatosome organization (60).
All systems were simulated for 60 million Monte Carlo steps or more. The Uniform, UniformNFR, and Life-LikeLH systems were simulated with 10 independent trajectories, and the other systems were simulated with 50 independent trajectories. Contact maps were normalized across each trajectory for each system and summed for the final contact map. Volume measurements were calculated from a convex bounding surface enclosing all elements of the fiber. A sample surface is shown in SI Appendix, Fig. S3. For a detailed description of calculating fiber volume, radius of gyration (), and sedimentation coefficient (), see SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health, National Institute of General Medical Sciences Awards R01-GM055264 and R35-GM122562; and Phillip-Morris USA and Phillip-Morris International (T.S.). Computing was performed on the New York University high-performance computing cluster Prince and the private cluster Schulten.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 4774.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1816424116/-/DCSupplemental.
References
- 1.Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 2007;26:2041–2051. doi: 10.1038/sj.emboj.7601654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Gen. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 3.Ong C-T, Corces VG. Enhancer function: New insights into the regulation of tissue-specific gene expression. Nat Rev Gen. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finch JT, et al. Structure of nucleosome core particles of chromatin. Nature. 1977;269:29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- 5.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 6.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 7.Luger K, Richmond TJ. The histone tails of the nucleosome. Curr Opin Gen Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 8.Bascom GD, Sanbonmatsu KY, Schlick T. Mesoscale modeling reveals hierarchical looping of chromatin fibers near gene regulatory elements. J Phys Chem B. 2016;120:8642–8653. doi: 10.1021/acs.jpcb.6b03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Potoyan DA, Papoian GA. Energy landscape analyses of disordered histone tails reveal special organization of their conformational dynamics. J Am Chem Soc. 2011;133:7405–7415. doi: 10.1021/ja1111964. [DOI] [PubMed] [Google Scholar]
- 10.Collepardo-Guevara R, et al. Chromatin unfolding by epigenetic modifications explained by dramatic impairment of internucleosome interactions: A multiscale computational study. J Am Chem Soc. 2015;137:10205–10215. doi: 10.1021/jacs.5b04086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Q, Yang R, Korolev N, Liu CF, Nordenskiöld L. Regulation of nucleosome stacking and chromatin compaction by the histone H4 N-terminal tail–H2A acidic patch interaction. J Mol Biol. 2017;14:2075–2096. doi: 10.1016/j.jmb.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 12.Zhang R, Erler J, Langowski J. Histone acetylation regulates chromatin accessibility: Role of H4K16 in inter-nucleosome interaction. Biophys J. 2017;112:450–459. doi: 10.1016/j.bpj.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roh T, Wei G, Farrell CM, Zhao K. Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res. 2007;17:74–81. doi: 10.1101/gr.5767907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voong LN, et al. Insights into nucleosome organization in mouse embryonic stem cells through chemical mapping. Cell. 2016;67:1555–1570.e15. doi: 10.1016/j.cell.2016.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu B, Charvin G, Siggia ED, Cross FR. Nucleosome-depleted regions in cell-cycle-regulated promoters ensure reliable gene expression in every cell cycle. Dev Cell. 2010;18:544–555. doi: 10.1016/j.devcel.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perišić O, Collepardo-Guevara R, Schlick T. Modeling studies of chromatin fiber structure as a function of DNA linker length. J Mol Biol. 2010;403:777–802. doi: 10.1016/j.jmb.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Correll SJ, Schubert MH, Grigoryev SA. Short nucleosome repeats impose rotational modulations on chromatin fibre folding. EMBO J. 2012;31:2416–2426. doi: 10.1038/emboj.2012.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Y-B, Gerchman SE, Ramakrishnan V, Travers A, Muyldermans S. Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature. 1998;395:402–405. doi: 10.1038/26521. [DOI] [PubMed] [Google Scholar]
- 20.Thomas JO. Histone H1: Location and role. Curr Opin Cell Biol. 1999;11:312–317. doi: 10.1016/S0955-0674(99)80042-8. [DOI] [PubMed] [Google Scholar]
- 21.Fan Y, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 22.Woodcock CL, Skoultchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 23.Fyodorov DV, Zhou B-R, Skoultchi AI, Bai Y. Emerging roles of linker histones in regulating chromatin structure and function. Nat Rev Mol Cell Biol. 2018;19:192–206. doi: 10.1038/nrm.2017.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ali Öztürk M, Cojocaru V, Wade RC. Toward an ensemble view of chromatosome structure: A paradigm shift from one to many. Structure. 2018;26:1050–1057. doi: 10.1016/j.str.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Bednar J, et al. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci USA. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caterino TL, Hayes JJ. Structure of the H1 C-terminal domain and function in chromatin condensation. Biochem Cell Biol. 2010;89:35–44. doi: 10.1139/O10-024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao K, et al. High-resolution mapping of H1 linker histone variants in embryonic stem cells. PLoS Genet. 2013;9:e1003417. doi: 10.1371/journal.pgen.1003417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geeven G, et al. Local compartment changes and regulatory landscape alterations in histone H1-depleted cells. Genome Biol. 2015;16:289. doi: 10.1186/s13059-015-0857-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu J, et al. Dynamic placement of the linker histone H1 associated with nucleosome arrangement and gene transcription in early Drosophila embryonic development. Cell Death Dis. 2018;9:765. doi: 10.1038/s41419-018-0819-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popova EY, et al. Developmentally regulated linker histone H1c promotes heterochromatin condensation and mediates structural integrity of rod photoreceptors in mouse retina. J Biol Chem. 2013;288:17895–17907. doi: 10.1074/jbc.M113.452144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noordermeer D, et al. The dynamic architecture of Hox gene clusters. Science. 2011;334:222–225. doi: 10.1126/science.1207194. [DOI] [PubMed] [Google Scholar]
- 32.Bonev B, et al. Multiscale 3D genome rewiring during mouse neural development. Cell. 2017;171:557–572.e24. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salzberg AC, et al. Genome-wide mapping of histone H3K9me2 in acute myeloid leukemia reveals large chromosomal domains associated with massive gene silencing and sites of genome instability. PLoS One. 2017;12:e0173723. doi: 10.1371/journal.pone.0173723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.ENCODE Project Consortium The ENCODE (ENCyclopedia of DNA elements) project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 35.Bascom GD, Kim T, Schlick T. Kilobase pair chromatin fiber contacts promoted by living-system-like DNA linker length distributions and nucleosome depletion. J Phys Chem B. 2017;121:3882–3894. doi: 10.1021/acs.jpcb.7b00998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grigoryev SA, et al. Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes. Proc Natl Acad Sci USA. 2016;113:1238–1243. doi: 10.1073/pnas.1518280113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gavin B, Schlick T. Linking chromatin fibers to gene folding by hierarchical looping. Biophys J. 2017;112:434–445. doi: 10.1016/j.bpj.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ricci MA, Manzo C, García-Parajo MF, Lakadamyali M, Cosma MP. Chromatin fibers are formed by heterogeneous groups of nucleosomes . Cell. 2015;160:1145–1158. doi: 10.1016/j.cell.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 39.Risca VI, Denny SK, Straight AF, Greenleaf WJ. Variable chromatin structure revealed by in situ spatially correlated DNA cleavage mapping. Nature. 2017;541:237–241. doi: 10.1038/nature20781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsieh TH, et al. Mapping nucleosome resolution chromosome folding in yeast by Micro-C. Cell. 2015;162:108–119. doi: 10.1016/j.cell.2015.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev. 2011;21:175–186. doi: 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boettiger AN, et al. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature. 2016;529:418–422. doi: 10.1038/nature16496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Pierro M, Cheng RR, Lieberman Aiden E, Wolynes PG, Onuchic JN. De novo prediction of human chromosome structures: Epigenetic marking patterns encode genome architecture. Proc Natl Acad Sci USA. 2017;114:12126–12131. doi: 10.1073/pnas.1714980114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ernst J, Kellis M. ChromHMM: Automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang Z, et al. BL-Hi-C is an efficient and sensitive approach for capturing structural and regulatory chromatin interactions. Nat Commun. 2017;8:1622. doi: 10.1038/s41467-017-01754-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blake WJ, Kærn M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- 47.Barbieri M, et al. Complexity of chromatin folding is captured by the strings and binders switch model. Proc Natl Acad Sci USA. 2012;109:16173–16178. doi: 10.1073/pnas.1204799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jost D, Pascal C, Cavalli G, Vaillant C. Modeling epigenome folding: Formation and dynamics of topologically associated chromatin domains. Nucleic Acids Res. 2014;42:9553–9561. doi: 10.1093/nar/gku698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Pierro M, Zhang B, Aiden EL, Wolynes PG, Onuchic JN. Transferable model for chromosome architecture. Proc Natl Acad Sci USA. 2016;113:12126–12131. doi: 10.1073/pnas.1613607113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schöpflin R, et al. Modeling nucleosome position distributions from experimental nucleosome positioning maps. Bioinformatics. 2013;29:2380–2386. doi: 10.1093/bioinformatics/btt404. [DOI] [PubMed] [Google Scholar]
- 51.Collepardo-Guevara R, Schlick T. Chromatin fiber polymorphism triggered by variations of DNA linker lengths. Proc Natl Acad Sci USA. 2014;111:8061–8066. doi: 10.1073/pnas.1315872111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arya G, Schlick T. Role of histone tails in chromatin folding revealed by a mesoscopic oligonucleosome model. Proc Natl Acad Sci USA. 2006;103:16236–16241. doi: 10.1073/pnas.0604817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beard DA, Schlick T. Modeling salt-mediated electrostatics of macromolecules: The discrete surface charge optimization algorithm and its application to the nucleosome. Biopolymers. 2001;58:106–115. doi: 10.1002/1097-0282(200101)58:1<106::AID-BIP100>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 54.Karolchik D, et al. The UCSC genome browser database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yazdi PG, et al. Increasing nucleosome occupancy is correlated with an increasing mutation rate so long as DNA repair machinery is intact. PLoS One. 2015;10:e0136574. doi: 10.1371/journal.pone.0136574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nicol JW, Helt GA, Blanchard SG, Jr, Raja A, Loraine AE. The Integrated Genome Browser: Free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bharath MMS, Chandra NR, Rao MRS. Molecular modeling of the chromatosome particle. Nucleic Acids Res. 2003;31:4264–4274. doi: 10.1093/nar/gkg481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luque A, Collepardo-Guevara R, Grigoryev S, Schlick T. Dynamic condensation of linker histone C-terminal domain regulates chromatin structure. Nucleic Acids Res. 2014;42:7553–7560. doi: 10.1093/nar/gku491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.