Significance
Short peptides are encoded in genomes of all organisms and have important functions. Due to the small size of such open reading frames, they are frequently overlooked by automatic genome annotation. We investigated the gene that was misannotated as long noncoding RNA LINC00116 and demonstrated that this gene codes for a 56-amino-acid-long peptide, Mtln, which is localized in mitochondria. Inactivation of the Mtln coding gene leads to reduction of oxygen consumption attributed to respiratory complex I activity and perturbs lipid composition of the cell. This influence is mediated by Mtln interaction with NADH-dependent cytochrome b5 reductase. Disruption of the mitochondrial localization of the latter phenocopies Mtln inactivation.
Keywords: sORF, peptide, respiration, mitochondria, lipid metabolism
Abstract
Genes coding for small peptides have been frequently misannotated as long noncoding RNA (lncRNA) genes. Here we have demonstrated that one such transcript is translated into a 56-amino-acid-long peptide conserved in chordates, corroborating the work published while this manuscript was under review. The Mtln peptide could be detected in mitochondria of mouse cell lines and tissues. In line with its mitochondrial localization, lack of the Mtln decreases the activity of mitochondrial respiratory chain complex I. Unlike the integral components and assembly factors of NADH:ubiquinone oxidoreductase, Mtln does not alter its enzymatic activity directly. Interaction of Mtln with NADH-dependent cytochrome b5 reductase stimulates complex I functioning most likely by providing a favorable lipid composition of the membrane. Study of Mtln illuminates the importance of small peptides, whose genes might frequently be misannotated as lncRNAs, for the control of vitally important cellular processes.
Examination of eukaryotic cells’ transcriptome revealed a diversity of long transcripts exceeding an annotated set of protein coding genes by a factor of five (1). A number of those transcripts, designated long noncoding RNAs (lncRNAs), were extensively studied and ascribed important functions in chromatin remodeling, transcriptional control, miRNA sponging, transcription and splicing factor sequestration, and other functions described in recent reviews (2–5). However, with the development of a ribosome profiling method (6), a significant proportion of lncRNAs was found to be associated with ribosomes and presumably translated (7). Subsequently, it became apparent that short peptides whose genes might be overlooked by computational annotation pipelines due to their minimalistic size play a significant role in cells (8–10). For example, a peptide encoded by the tarsal-less (tal) lncRNA appeared to be important for epidermal differentiation in Drosophila (11). The Pri peptide, interacting with Ubr3 ubiquitin ligase, activates Svb processing by the proteasome (12). The pgc RNA encoded peptide (13) turned out to be crucial for RNAPII C-terminal domain Ser2 phosphorylation. The peptide interacts with P-TEFb, a key regulator of RNAPII-dependent transcription of most cellular genes (14), and prevents its recruitment to transcription sites.
Many small peptides are localized in mitochondria, an organelle whose main functions are respiration coupled to ATP production, Ca2+ homeostasis, cellular redox-potential maintenance, synthesis of steroids, heme, FeS clusters, induction of apoptosis, and many others, finally controlling cell fate and tissues’ functional integrity (15). The mammalian mitochondrial proteome is composed of over 1,100 proteins (16), 5% of that number being small proteins of less than 100 amino acids in length (17). Both mitochondrial (18, 19) and nuclear genomes code for the short mitochondrial peptides being integral components of oxidative phosphorylation (OXPHOS) complexes (17), or their assembly factors (20), and play significant roles in longevity (21), insulin resistance (19), and modulation of apoptosis (18). Mutations in genes coding for small peptides residing in mitochondria might have pathological outcomes, such as mitochondrial encephalomyopathies (22) and Leigh disease (23).
Here we describe a murine peptide encoded in a gene misannotated as 1500011k16Rik lncRNA, corresponding to human LINC00116. The peptide resides in mitochondria and is important for the activity of respiratory chain complex I. After our first report on the function of this peptide on the conference (24) and while this manuscript was under review two groups published similar conclusions on the functional role of this peptide (25, 26), named Mitoregulin, Mtln.
Results
Analysis of 1500011k16Rik Coding Potential.
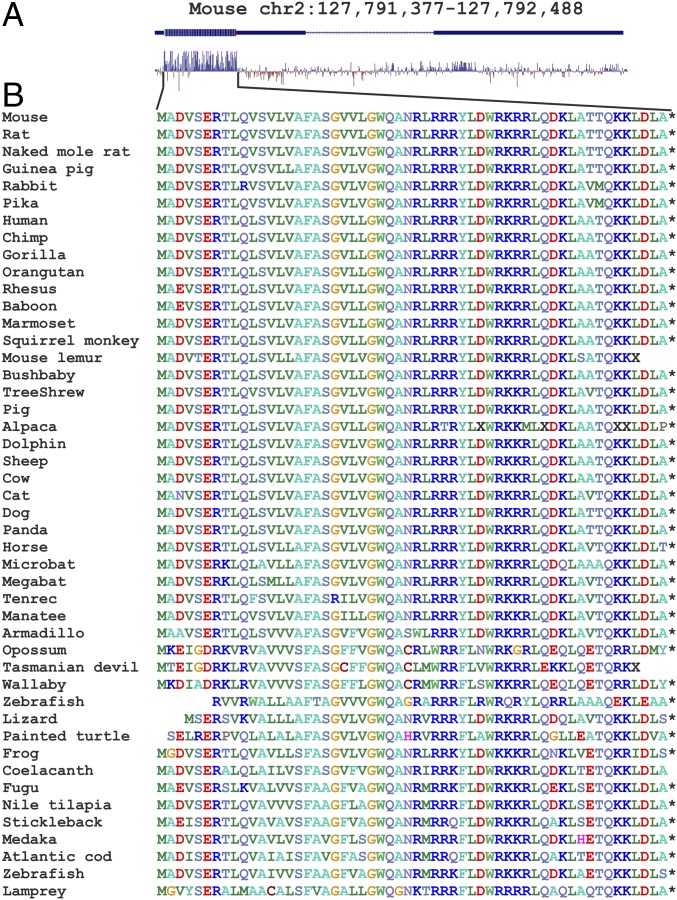
Many lncRNAs contain putative ORFs occurring by chance and not translated into a functional peptide entity. The murine 1500011k16Rik transcript contains a 56-amino acid ORF with a predicted single pass transmembrane segment (27), but not any detectable domain relative. Several sequence characteristics are indicative of coding potential. Analysis of nucleotide conservation of the genes homologous to 1500011k16Rik (Fig. 1A), among 60 vertebrate species (28), revealed that a region of putative ORF is the most, and almost only, conserved region of the gene. Analysis of the aggregated ribosome profiling data (29) revealed substantial ribosome coverage of the putative ORF (SI Appendix, Fig. S1A). Alignment of the putative products of ORF translation revealed a high degree of conservation at the amino acid level (Fig. 1B) with a high ratio of synonymous over nonsynonymous codon substitutions and absence of premature in-frame stop codons (SI Appendix, Fig. S1 B and C). As a result, we concluded that the transcript 1500011k16Rik is most likely to be translated into a 56-amino-acid-long peptide, which we will refer to as Mitoregulin (Mtln) (25) for consistency in the scientific literature.
Fig. 1.
Analysis of Mtln conservation. (A) Mouse genome area encompassing the 1500011k16Rik gene. Shown are exons and ORF location. Conservation diagram across vertebrates (28) is shown below the map. (B) Alignment of Mtln peptide sequences across the vertebrates.
The 1500011k16Rik Encoded Peptide Is Expressed and Localized in Mitochondria.
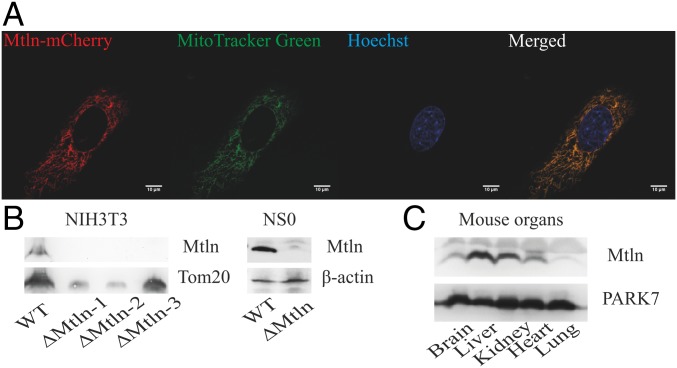
To confirm the presence of Mtln in the cultured murine cells, we applied CRISPR-Cas9 technology to inactivate 1500011k16Rik gene coding for Mtln in NIH 3T3 and NS0 cell lines (SI Appendix, Fig. S2) and to append the Mtln coding region by the C-terminal fluorescent protein mCherry tag in NIH 3T3 cell lines. The presence of Mtln-mCherry fusion protein, expressed at natural expression level from an endogenous Mtln promoter, could be detected in the cells by immunoblotting (SI Appendix, Fig. S3A) and immunocytochemistry (SI Appendix, Fig. S3B). To further determine exact peptide localization, we ectopically expressed Mtln-mCherry fusion and observed its colocalization with mitochondria using an organelle-specific dye, MitoTracker Green FM (Fig. 2A). Cell fractionation revealed that the Mtln peptide is always copurified with mitochondria similarly to the mitochondrial Tom20 protein (Fig. 2B, Left and SI Appendix, Fig. S3C). Our results perfectly match the conclusions of two independent groups, which showed that Mtln localizes in the inner membrane of mitochondria (25, 26). To exclude the possibility of any influence of the mCherry tag on the localization of Mtln, we raised polyclonal rabbit antibodies to Mtln and detected an endogenous peptide presence in mitochondria of the NIH 3T3 cell lines, but not in the generated cell lines with an inactivated Mtln gene (Fig. 2B, Left). To check that the Mtln peptide is present not only in the NIH 3T3 cell line but also other cell lines and living organisms, we performed immunoblotting analysis of Mtln’s presence in the NS0 murine myeloma cell line (Fig. 2B, Right) using the NS0 cell line with inactivated 1500011k16Rik gene coding for Mtln as a control and mouse tissues (Fig. 2C). Analysis revealed a high level of Mtln in the NS0 cell line and ubiquitous expression of Mtln in different mouse tissues.
Fig. 2.
1500011k16Rik encodes a novel polypeptide. (A) Confocal images of mCherry-tagged Mtln peptide; mitochondria were stained by MitoTracker Green FM and nuclei by Hoechst 33342. (Scale bar, 10 μm.) (B) Immunoblotting of isolated mitochondria from NIH 3T3 and cell lysates from NS0 cells for endogenous Mtln. Tom20 and β-actin were used as a loading control. ∆Mtln -1, -2, -3 are different knockouts for the NIH 3T3 cell line. (C) Immunoblotting of mouse organ lysates for endogenous Mtln. PARK7 was used as a loading control (67).
Respiratory Complex I Electron Transfer Activity Is Impaired in Knockout Cell Lines.
To test the influence of Mtln on the respiration of mitochondria, we compared NIH 3T3 and NS0 cells with their derivatives with inactivated 1500011k16Rik gene coding for Mtln (SI Appendix, Fig. S2A). Distribution of cells with high and low mitochondrial membrane potential was tested by the ratiometric fluorescent dye JC-1 and FACS analysis. The level of total reactive oxygen species (ROS) production in wild-type and mutant cells was estimated by FACS analysis with the help of 2′,7′ –dichlorofluorescin diacetate (DCFDA). In contrast to the results of Stein et al. (25) we do not see a significant difference between wild-type and knockout cells in ROS production along with membrane potential (SI Appendix, Fig. S4 A and B).
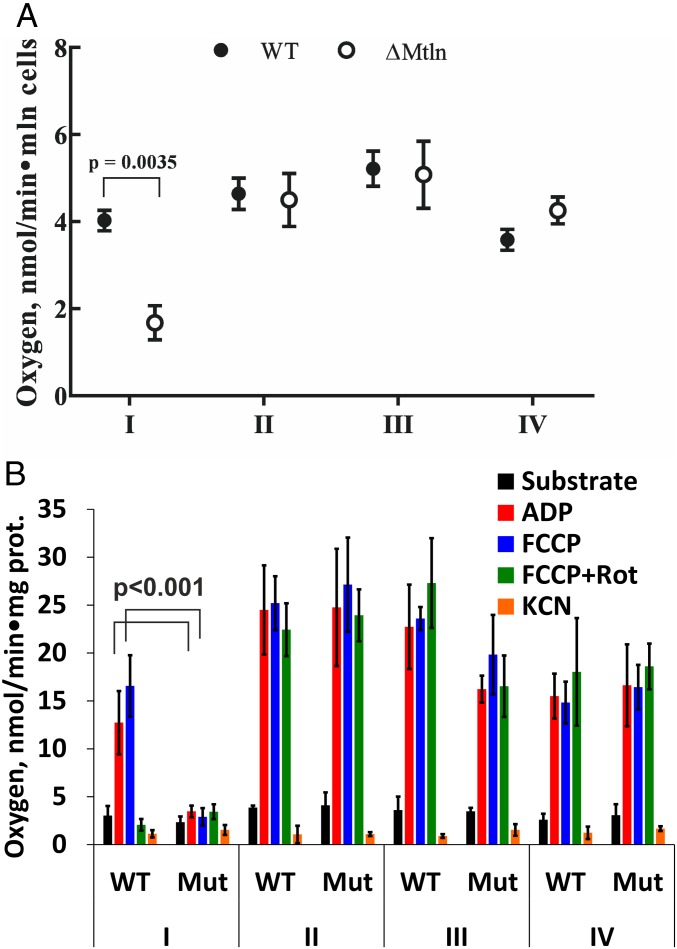
To check if mitochondrial Mtln peptide function is essential for the activity of any respiratory chain components, we compared respiratory efficiency of knockout cells with a parental cell line carrying functional Mtln. Oxygen consumption rates were measured to examine electron flow through respiratory complexes I–IV (Fig. 3 and SI Appendix, Fig. S5 A and B). Activity of respiratory complex I, NADH:ubiquinone oxidoreductase, measured on permeabilized cells as rotenone-sensitive oxygen reduction using glutamate and malate as substrates was found to depend on the presence of the Mtln peptide in both cell lines (Fig. 3A and SI Appendix, Fig. S5A). At the same time, measurements of oxygen consumption rates for isolated mitochondria revealed a significant decrease of complex I activity for both cell lines and a tendency to decrease for complex III (Fig. 3B and SI Appendix, Fig. S5B). The difference between the oxygen consumption rates by mitochondria isolated from the Mtln knockout and parental cell lines, attributed to complex I, is maximal upon the stimulation by ADP or uncoupler carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP). Nonstimulated respiration on NAD-linked substrates was not different for the mitochondria from mutant and parental cell lines likely due to the lack of energy-consuming processes. Complex I-associated respiration deficiency observed for mitochondria purified from Mtln knockout cell lines demonstrated that only perturbation of the mitochondria, but not of any other cellular compartment upon Mtln inactivation, is sufficient for the phenotypic manifestation.
Fig. 3.
Mtln inactivation reduces respiratory complex I activity. (A) Oxygen consumption rates for complexes I–IV for parental NIH 3T3 and ∆Mtln knockout cell lines. Values are average of three independent experiments done in triplicates. For the statistically significant changes, multiplicity adjusted P value (P < 0.05, Student’s t test, Holm–Sidak correction) is indicated. (B) Oxygen consumption rates for mitochondria isolated from parental NIH 3T3 and ∆Mtln cell lines. Values are average of three independent experiments done in triplicates. For the statistically significant changes, multiplicity adjusted P value (P < 0.05, Student’s t test) is indicated. For A and B data are presented as mean ± SD.
To check for a genetic complementation, we reintroduced either a copy of the Mtln gene or a luciferase to the knockout cell lines under a more potent synthetic promoter. Ectopic expression of Mtln, but not luciferase, restored complex I activity in both knockout cell lines (SI Appendix, Fig. S4C).
NADH:ubiquinone oxidoreductase is a large multisubunit complex which requires several additional assembly factors. To examine whether the Mtln peptide contributes to complex I activity directly or indirectly, we immunopurified complex I from the wild-type and Mtln knockout cells and measured its ability to oxidize NADH. Surprisingly, NADH dehydrogenase activity of the purified respiratory complex I appeared to be independent of Mtln (SI Appendix, Fig. S5 C and D). Additionally, complex I+III activity, measured as NADH-dependent cytochrome C reduction in the presence of potassium cyanide (KCN), turned out to be independent of Mtln presence (SI Appendix, Fig. S6). This result leads us to the hypothesis that Mtln manifests its influence on the function of respiratory complex I indirectly.
Our results are consistent with a recently published study (25), demonstrating that Mtln deletion does not affect assembly of Complex I, but impedes association of Complex I into respiratory chain supercomplexes.
Mtln Interacts with Cytochrome b5 Reductase 3.
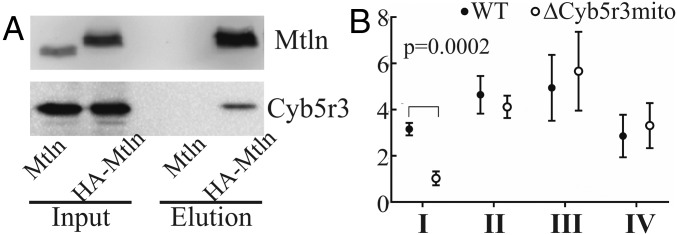
Since Mtln inactivation does not affect complex I activity directly, we suggested that the Mtln peptide may manifest its activity via interaction with some other enzyme whose function is needed for proper respiratory complex I functioning. To search for possible Mtln peptide partners, we immunoprecipitated the Mtln peptide from a NIH 3T3 cell line carrying ectopically expressed HA-Mtln. A single protein partner, that was confidently and reproducibly copurified with Mtln, was NADH cytochrome b5 reductase 3 (Cyb5r3) (Fig. 4A and SI Appendix, Fig. S7A). To further confirm Mtln interaction with Cyb5r3, we coprecipitated Cyb5r3 with Mtln using a different protein fusion, Mtln-mCherry, and anti-mCherry antibodies (SI Appendix, Fig. S7B). To check whether Mtln and Cyb5r3 are located in the same cellular compartment, we coexpressed Mtln-mCherry and Cyb5r3-eGFP protein fusions in the NIH 3T3 cells (SI Appendix, Fig. S7C) and confirmed their colocalization in mitochondria.
Fig. 4.
Mtln interacts with Cyb5r3. (A) Immunoprecipitation of HA-Mtln from NIH 3T3 cells followed by immunoblotting with anti-Mtln antibodies (Top) and anti-Cyb5r3 antibodies (Bottom). Cells expressing Mtln without tag were used as a control. (B) Activity of complexes I–IV in Cyb5r3 (Gly2 to Ala2) mutant cells (∆Cyb5r3mito). Values are average of four independent experiments done in triplicates. For the statistically significant change, multiplicity adjusted P value (P < 0.05, Student’s t test, Holm–Sidak correction) is indicated. Data are presented as mean ± SD.
Disruption of Cyb5r3 Mitochondrial Localization Phenocopies Mtln Inactivation.
Multiple localizations were reported for Cyb5r3, making its functional analysis somewhat complicated. Apart from the mitochondrial outer membrane, this enzyme was found in microsomal and cytoplasmic membranes. Luckily, it is known that Cyb5r3 must be N-myristoylated at Gly2 residue to get incorporated in the mitochondrial outer membrane (30). To check whether the Mtln peptide functions via its interaction with Cyb5r3, we decided to disrupt mitochondrial localization of the latter. We generated mutant NIH 3T3 cells with Gly2-to-Ala substitution (∆Cyb5r3mito), which appeared to be viable (SI Appendix, Fig. S7D). This mutation results in the decrease of Cyb5r3 protein levels in mitochondria (SI Appendix, Fig. S7E, Left). Oxygen consumption rate measurements in mutant cells (Fig. 4B) revealed that loss of Cyb5r3 localization in mitochondria altered the activity of respiratory complex I, similar to inactivation of its interacting peptide Mtln.
Further, to determine the functional relationship between Mtln and Cyb5r3, we generated Cyb5r3 Gly2-to-Ala mutant cells with Mtln deletion and overexpression (SI Appendix, Figs. S2B and S8A). To check whether Mtln absence or overexpression would affect the Cyb5r3 mutant cell’s phenotype, we compared oxygen consumption rates attributed to mitochondrial respiratory complexes (SI Appendix, Fig. S8B). We observed that inhibition of complex I activity caused by Cyb5r3 mutation could not be rescued or aggravated by the Mtln deletion or overexpression. Thereby, we concluded that it is likely that Mtln fulfills its function only via its interaction with Cyb5r3, the latter being the major functional component of the complex.
Mtln Deletion Does Not Change the Cytosolic NADH/NAD+ Balance.
Reduced contribution of NADH:ubiquinone oxidoreductase into mitochondrial respiration might be explained by a lower rate of NADH regeneration. To evaluate the influence of Mtln knockout on NADH/NAD+ balance, we applied SoNar fluorescent protein sensor (31). We found that in the steady-state condition, the wild-type and Mtln knockout cells do not demonstrate any significant difference in the cytoplasmic NADH/NAD+ redox balance (SI Appendix, Fig. S9A). In addition, we decided to assess the dynamics of redox balance changes in the cytosol affected by different metabolic substrates (SI Appendix, Fig. S9 B and C). The test revealed a lack of difference in the NADH/NAD+ dynamic between parental and Mtln-depleted cells. We can conclude that the cytosolic redox balance is not affected by Mtln ablation.
How Does Mtln Influence Cyb5r3?
Frequently, protein complex formation is necessary for the stabilization of components against proteolysis. However, it is not the case for Cyb5r3 interaction with Mtln, as revealed by immunoblotting of Cyb5r3 in Mtln knockout cells (SI Appendix, Fig. S10A). To evaluate any possible influence of Mtln on the localization of Cyb5r3, we used cell lines ectopically expressing the Cyb5r3-eGFP fusion protein. Localization of the Cyb5r3 fusion protein appeared to be mitochondrial in both the parental NIH 3T3 cell line and its derivative devoid of the Mtln peptide (SI Appendix, Fig. S10B). Moreover, we do not observe any difference in Mtln level or localization upon ∆Cyb5r3mito mutation (SI Appendix, Fig. S10 C and D).
Cyb5r3 catalyzes redox reactions with several different substrates, while it uses only NADH as a donor of an electron being one of the main consumers of the cytosolic NADH (32). To test an influence of Mtln on the ability of Cyb5r3 to oxidize NADH, we measured its NADH dehydrogenase activity in a reaction with K3[Fe(CN)6] as a spurious electron acceptor (SI Appendix, Fig. S10E). An ability of Cyb5r3 to extract an electron from NADH was found to be independent of the Mtln presence.
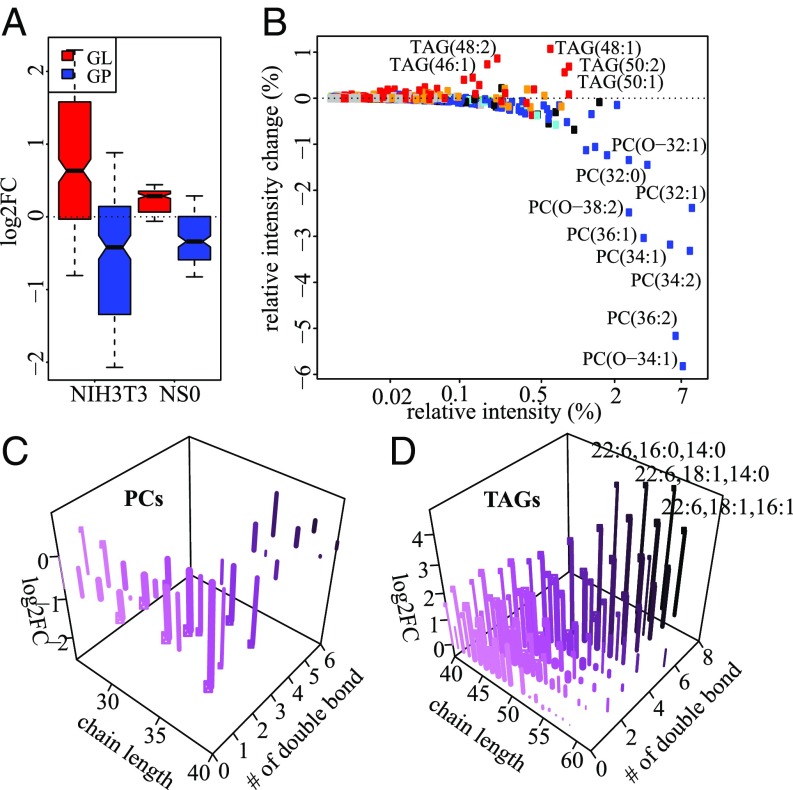
Two main processes that require the membrane-bound form of Cyb5r3 are Δ9 fatty acid desaturation (33) and cholesterol biosynthesis (34). For the evaluation of Mtln’s influence on Cyb5r3 activity in lipid metabolism, we performed LC-MS analysis of cell lines NIH 3T3 and NS0 deficient in Mtln. We quantified more than 1,000 lipids in each cell line (Materials and Methods). Quantities of hundreds of lipids were significantly and reproducibly (SI Appendix, Fig. S11A) changed upon Mtln depletion in two independently produced NIH 3T3-derived knockout cell lines. Most glycerolipids are overrepresented while most glycerophospholipids are underrepresented upon Mtln knockout in both NIH 3T3 and NS0 cell lines (Fig. 5A).
Fig. 5.
Mtln depletion alters level of phospholipids and triglycerides. (A) Distributions of log2 fold change (FC) for glycerolipids (GL, red), and glycerophospholipids (GP, blue) in ∆Mtln cell lines relative to the parental NIH 3T3 and NS0 cell lines. (B) Relationship between relative intensity in the wild type (x axis) and relative intensity change in ∆Mtln-2 (y axis) for TAGs (red) and PCs (blue). (C and D) Dependence of log2 FC of PCs (C) or TAGs (D) in ∆Mtln-2 and ∆Mtln-3 (z axis) on total chain length (x axis) and number of double bonds (y axis and color). Exact fatty chain composition is shown for three TAGs with the highest fold change. Concentration of TAGs and PCs in the wild type is shown by line width (in log scale). Lipids with statistically significant changes have a circle at the end of lines. Statistical significance was determined with Student’s t test (Benjamini–Hochberg-corrected P value <0.05).
While phosphatidylcholines (PCs) are the most abundant lipids in our dataset, their amounts are strongly reduced upon Mtln knockout, resulting in overall reduction of the PC fraction by about 20% (Fig. 5 B and C). The PCs whose quantity is most affected by Mtln knockout are one to three unsaturated with relatively long chains (Fig. 5C). Contrary to PCs, concentrations of most TAGs are elevated upon Mtln knockout and most affected species are either short or polyunsaturated with the highest effect observed for TAGs 52:6, 54:7, and 56:8 (Fig. 5 B and D and SI Appendix, Fig. S11B). MS-MS analysis of these TAGs revealed that all of them contain an acyl group of docosahexaenoic acid.
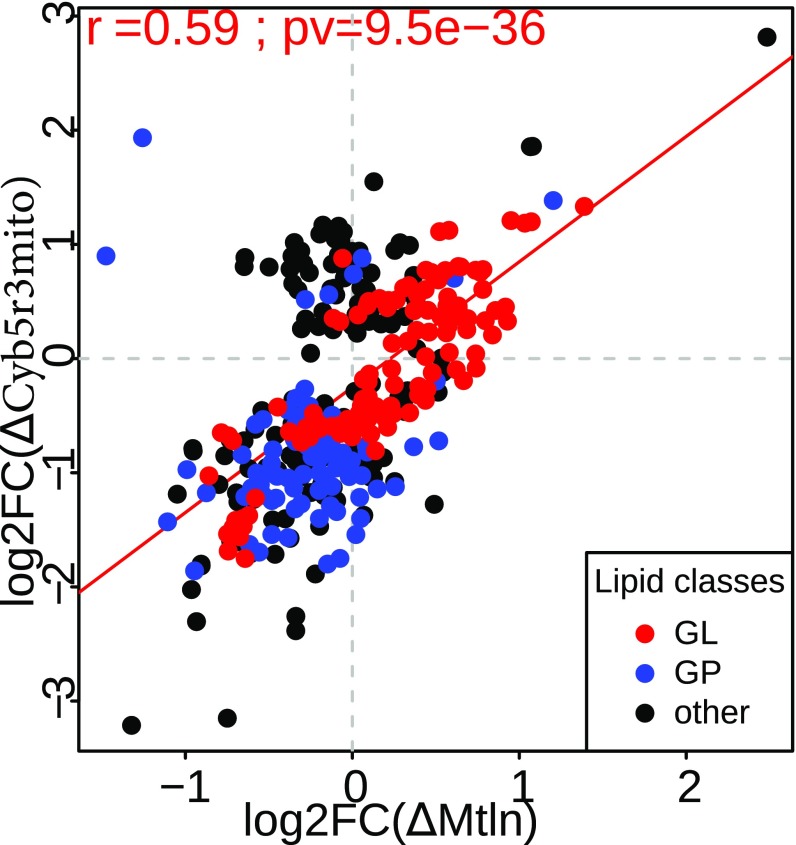
To check whether an influence of Mtln on cellular lipid composition is mediated by its interaction with Cyb5r3 we analyzed lipid composition of the cells possessing only the mutant Cyb5r3 form lacking mitochondrial localization. Lipidome analysis demonstrated a good correlation between lipid composition changes of Mtln knockout and ∆Cyb5r3mito mutant cells (Fig. 6). Moreover, we observe decrease in some PC fractions (SI Appendix, Fig. S12A) and increase in polyunsaturated TAGs (SI Appendix, Fig. S12B) for both mutant cell lines. These tendencies are reproduced but not aggravated in the Cyb5r3 mutant cells with additional Mtln deletion (SI Appendix, Figs. S13 A and B and S14). Analysis of lipid composition of ∆Cyb5r3mito mutant cells overexpressing the Mtln gene does not demonstrate a complete suppression of lipidome changes (SI Appendix, Figs. S13C and S14).
Fig. 6.
Lipid concentration changes caused by disruption of Cyb5r3 mitochondrial localization (∆Cyb5r3mito). Relationship between log2 fold changes induced by Mtln knockout (x axis) and ∆Cyb5r3mito mutant (y axis) compared with the wild type. Each point represents one lipid, glycerolipids (GL), glycerophospholipids (GP), and other lipid classes are shown in red, blue, and black, respectively. Least square regression line is shown in red. Pearson correlation coefficient, its 95% confidence interval and P value (t test) are shown in Upper Left corner.
Discussion
An assortment of short peptides encoded in genomes of higher organisms, such as mouse and human, could compose a nearly overlooked layer of regulatory molecules (11, 35, 36). In this work, we have appended the list of small functional peptide entities by a remarkable member, the Mtln mitochondrial peptide, in line with the results of other groups (25, 26) published while this work was under review. Mtln is highly conserved across vertebrates and, as it appears from our results, functions in the cell metabolism.
The majority of small peptides manifest their functions by binding to a partner protein or being a constituent of multiprotein complexes (10), as was shown for sarcolipin and phospholamban (37, 38), humanin (18), Toddler/ELABELA (39, 40), and DWORF (41). Only a few small peptides have separate enzymatic functions, such as 4-oxalocrotonate tautomerase, whose monomer contains only 62 amino acids (42). In line with this trend, we demonstrated that the Mtln peptide interacts with NADH:cytochrome b5 oxidoreductase 3 (Cyb5r3). Disruption of mitochondrial localization of the latter leads to the same phenotype as Mtln depletion in regard to the respiratory complex I attributed oxygen consumption and lipid composition change. Thus, Mtln may function by stimulation of Cyb5r3 activity in mitochondria necessary for maintenance of lipid homeostasis. Recent study demonstrated that Mtln may also interact with other mitochondrial proteins (26), such as HADHA and HADHB involved in fatty acid β-oxidation. Although particular Mtln interaction partners identified in our study and in the work of Makarewich et al. (26) are different, they both are attributed to related processes, which may indicate an involvement of Mtln in fatty acid metabolism with tissue or cell-type peculiarities.
This activity, as we demonstrated, is needed for respiratory complex I functioning in the context of the mitochondrial OXPHOS system, but not for NADH oxidase activity of isolated complex I or complexes I+III. This observation is in line with that of Stein et al. (25), demonstrating an influence of Mtln deficiency on the formation of respiratory chain supercomplexes, containing CI, but not onto the assembly of CI and association of CI and CIII complexes.
Multiple functions were ascribed to the Cyb5r3. Apart from the erythrocyte-specific methemoglobin reductase activity of the soluble Cyb5r3 isoform (43), which is unlikely to be relevant for our study, membrane-anchored Cyb5r3 is implicated in the desaturation of fatty acids (33), cholesterol biosynthesis (34), and the amidoxime system involved in lipogenesis (44–46) and drug metabolism.
Scenarios where Mtln may be involved in localization or stabilization of Cyb5r3 against partial or complete proteolysis were ruled out, since neither localization nor the protein level of Cyb5r3 are affected by Mtln, leaving the possibility that Mtln influences Cyb5r3 activity related to lipid metabolism. Lipid composition is known to be crucial for mitochondrial functioning and in particular to respiratory complex I activity (47–50). Relevance of Mtln for Cyb5r3 functioning in the lipid metabolism was deduced from the lipidome analysis of Mtln knockout cells. We found that most TAGs are overrepresented and most phospholipids are underrepresented upon knockout. The PCs are affected more than other types of lipids. The fact that disruption of mitochondrial Cyb5r3 localization leads to the lipidome change that correlate well with that caused by Mtln gene inactivation supports the idea that Mtln functions via its association with Cyb5r3. We speculate that the decrease in the phosphatidylcholine content observed in both the cell line devoid of Cyb5r3 mitochondrial localization and Mtln knockout cell line might be the cause of the observed decrease in respiratory complex I activity. Congruent results (50) demonstrated that removal of choline from the rat diet results in accumulation of triglycerides in liver and impaired respiratory function, particularly in regard to complex I-linked, NADH-dependent respiration. Also, the importance of PCs for complex I activity is corroborated by the facts that addition of l-alpha-glycerylphosphorylcholine (GPC) increased the efficacy of complex I-linked mitochondrial oxygen consumption (51) and catalytic activity of complex I is determined by the amounts of PC and phosphatidylethanolamine (PE) (52). According to published data, the decrease in mitochondrial PCs impairs oxygen consumption rate, affects formation of the mitochondrial supercomplexes, and leads to a disorganized cristae structure (53, 54). Recent studies revealed that deletion of Mtln results in disrupted cristae structure (26) and alters respiratory supercomplex formation (25). Supercomplexes reside on the cristae membrane (55), of which the positively curved leaflet consists of predominantly phosphatidylcholines (∼80%) (56). It is tempting to speculate that supercomplex formation can be affected by the shape of the cristae, which in turn depends on lipid composition. These observations imply that the possible mechanism of complex I inactivation in Mtln-deficient cells might be governed by the inactivation of mitochondria-related functioning of Cyb5r3, which, in turn, changes the lipid compositions of the mitochondrial membrane. The last event leads to a change in the shape of the cristae, which disrupts the distribution of supercomplexes and the interaction of their components, hence slowing down the inactive/active transition of complex I (57, 58) resulting in its inactivation.
Materials and Methods
Constructs for gene inactivation and genome editing were created on the basis of pX458 plasmid (59). Knockin constructs were created on the basis of Sleeping Beauty transposon vectors (60, 61). Monitoring of ROS production in adherent and suspension cells was performed according to the protocol proposed by Wojtala et al. (62). Oxygen consumption rates were measured as described (63). Mitochondria from cultured cells were isolated by differential centrifugation. Cyb5r3 enzymatic activity was determined in isolated mitochondria by monitoring NADH-dependent ferricyanide reduction (64). Metabolite extraction for lipid analysis was performed as described (65). Untargeted lipidome profiling was performed in positive ionization mode (66). The animal work was approved by the Lomonosov Moscow State University animal ethics committee.
More details on the study methodology are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Alex Lebedeff for his help in editing the manuscript; Ksenia Smirnova for her help in growing cells; Nikolai Anikanov and Elena Stekolschikova for lipid extraction and mass-spectrometry measurements for double knockout cell lines; and Dr. Yi Yang for providing the pcDNA3.1-SoNar and pcDNA3.1-iNapC vectors. A.C. thanks the Boehringer Ingelheim Fond for a travel grant to attend a European Molecular Biology Laboratory course. We also thank the Skolkovo Institute of Science and Technology for funding for the publication charges and open access fee. This work was supported by Russian Foundation for Basic Research (RFBR) Grants 17-04-01904 and 18-29-07005 (to P.S. and A.C.); Russian Science Foundation (RSF) Grant 17-75-30027 (to P.S.) for work related to genome editing; and the Moscow State University Scientific School (O.D.). Experiments on mice were supported by Russian Ministry of Science Grant 14.W03.31.0012. P.V. was supported by the President’s Scholarship (SP-4132.2018.4). The work in Mtln Deletion Does Not Change Cytosolic NADH/NAD+ Balance was supported by RSF Grant 17-15-01175 (to D.B.) and RFBR Grant 18-54-74003 (to V.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. I.U. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809105116/-/DCSupplemental.
References
- 1.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 2.Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16:167–179. doi: 10.1038/nrd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 5.St Laurent G, Wahlestedt C, Kapranov P. The landscape of long noncoding RNA classification. Trends Genet. 2015;31:239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingolia NT, et al. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pueyo JI, Magny EG, Couso JP. New peptides under the s(ORF)ace of the genome. Trends Biochem Sci. 2016;41:665–678. doi: 10.1016/j.tibs.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Chugunova A, Navalayeu T, Dontsova O, Sergiev P. Mining for small translated ORFs. J Proteome Res. 2018;17:1–11. doi: 10.1021/acs.jproteome.7b00707. [DOI] [PubMed] [Google Scholar]
- 10.Cabrera-Quio LE, Herberg S, Pauli A. Decoding sORF translation–From small proteins to gene regulation. RNA Biol. 2016;13:1051–1059. doi: 10.1080/15476286.2016.1218589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondo T, et al. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science. 2010;329:336–339. doi: 10.1126/science.1188158. [DOI] [PubMed] [Google Scholar]
- 12.Zanet J, et al. Pri sORF peptides induce selective proteasome-mediated protein processing. Science. 2015;349:1356–1358. doi: 10.1126/science.aac5677. [DOI] [PubMed] [Google Scholar]
- 13.Hanyu-Nakamura K, Sonobe-Nojima H, Tanigawa A, Lasko P, Nakamura A. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature. 2008;451:730–733. doi: 10.1038/nature06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 15.McBride HM, Neuspiel M, Wasiak S. Mitochondria: More than just a powerhouse. Curr Biol. 2006;16:R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 16.Palmfeldt J, Bross P. Proteomics of human mitochondria. Mitochondrion. 2017;33:2–14. doi: 10.1016/j.mito.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 17.Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: An updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44:D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo B, et al. Humanin peptide suppresses apoptosis by interfering with Bax activation. Nature. 2003;423:456–461. doi: 10.1038/nature01627. [DOI] [PubMed] [Google Scholar]
- 19.Zarse K, Ristow M. A mitochondrially encoded hormone ameliorates obesity and insulin resistance. Cell Metab. 2015;21:355–356. doi: 10.1016/j.cmet.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Floyd BJ, et al. Mitochondrial protein interaction mapping identifies regulators of respiratory chain function. Mol Cell. 2016;63:621–632. doi: 10.1016/j.molcel.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuku N, et al. The mitochondrial-derived peptide MOTS-c: A player in exceptional longevity? Aging Cell. 2015;14:921–923. doi: 10.1111/acel.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez-Moreira D, et al. X-linked NDUFA1 gene mutations associated with mitochondrial encephalomyopathy. Ann Neurol. 2007;61:73–83. doi: 10.1002/ana.21036. [DOI] [PubMed] [Google Scholar]
- 23.Hoefs SJ, et al. NDUFA2 complex I mutation leads to Leigh disease. Am J Hum Genet. 2008;82:1306–1315. doi: 10.1016/j.ajhg.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chugunova A, et al. LncRNAencoded short peptide is a functionally important component of mitochondria. FEBS J. 2017;284:74. [Google Scholar]
- 25.Stein CS, et al. Mitoregulin: A lncRNA-encoded microprotein that supports mitochondrial supercomplexes and respiratory efficiency. Cell Rep. 2018;23:3710–3720.e8. doi: 10.1016/j.celrep.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makarewich CA, et al. MOXI is a mitochondrial micropeptide that enhances fatty acid β-oxidation. Cell Rep. 2018;23:3701–3709. doi: 10.1016/j.celrep.2018.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.UniProt Consortium T. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018;46:2699. doi: 10.1093/nar/gky092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanchette M, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michel AM, et al. GWIPS-viz: Development of a ribo-seq genome browser. Nucleic Acids Res. 2014;42:D859–D864. doi: 10.1093/nar/gkt1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borgese N, Aggujaro D, Carrera P, Pietrini G, Bassetti M. A role for N-myristoylation in protein targeting: NADH-cytochrome b5 reductase requires myristic acid for association with outer mitochondrial but not ER membranes. J Cell Biol. 1996;135:1501–1513. doi: 10.1083/jcb.135.6.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Y, et al. SoNar, a highly responsive NAD+/NADH sensor, allows high-throughput metabolic screening of anti-tumor agents. Cell Metab. 2015;21:777–789. doi: 10.1016/j.cmet.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sottocasa GL, Kuylenstierna B, Ernster L, Bergstrand A. An electron-transport system associated with the outer membrane of liver mitochondria. A biochemical and morphological study. J Cell Biol. 1967;32:415–438. doi: 10.1083/jcb.32.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keyes SR, Cinti DL. Biochemical properties of cytochrome b5-dependent microsomal fatty acid elongation and identification of products. J Biol Chem. 1980;255:11357–11364. [PubMed] [Google Scholar]
- 34.Reddy VV, Kupfer D, Caspi E. Mechanism of C-5 double bond introduction in the biosynthesis of cholesterol by rat liver microsomes. J Biol Chem. 1977;252:2797–2801. [PubMed] [Google Scholar]
- 35.Makarewich CA, Olson EN. Mining for micropeptides. Trends Cell Biol. 2017;27:685–696. doi: 10.1016/j.tcb.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet. 2014;15:193–204. doi: 10.1038/nrg3520. [DOI] [PubMed] [Google Scholar]
- 37.Anderson DM, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magny EG, et al. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science. 2013;341:1116–1120. doi: 10.1126/science.1238802. [DOI] [PubMed] [Google Scholar]
- 39.Pauli A, et al. Toddler: An embryonic signal that promotes cell movement via apelin receptors. Science. 2014;343:1248636. doi: 10.1126/science.1248636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chng SC, Ho L, Tian J, Reversade B. ELABELA: A hormone essential for heart development signals via the apelin receptor. Dev Cell. 2013;27:672–680. doi: 10.1016/j.devcel.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 41.Nelson BR, et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351:271–275. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitman CP. The 4-oxalocrotonate tautomerase family of enzymes: How nature makes new enzymes using a beta-alpha-beta structural motif. Arch Biochem Biophys. 2002;402:1–13. doi: 10.1016/S0003-9861(02)00052-8. [DOI] [PubMed] [Google Scholar]
- 43.Passon PG, Hultquist DE. Soluble cytochrome b5 reductase from human erythrocytes. Biochim Biophys Acta. 1972;275:62–73. doi: 10.1016/0005-2728(72)90024-2. [DOI] [PubMed] [Google Scholar]
- 44.Neve EP, et al. Amidoxime reductase system containing cytochrome b5 type B (CYB5B) and MOSC2 is of importance for lipid synthesis in adipocyte mitochondria. J Biol Chem. 2012;287:6307–6317. doi: 10.1074/jbc.M111.328237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plitzko B, et al. Defining the role of the NADH-cytochrome-b5 reductase 3 in the mitochondrial amidoxime reducing component enzyme system. Drug Metab Dispos. 2016;44:1617–1621. doi: 10.1124/dmd.116.071845. [DOI] [PubMed] [Google Scholar]
- 46.Neve EP, et al. Expression and function of mARC: Roles in lipogenesis and metabolic activation of Ximelagatran. PLoS One. 2015;10:e0138487. doi: 10.1371/journal.pone.0138487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vázquez-Memije ME, Cárdenas-Méndez MJ, Tolosa A, Hafidi ME. Respiratory chain complexes and membrane fatty acids composition in rat testis mitochondria throughout development and ageing. Exp Gerontol. 2005;40:482–490. doi: 10.1016/j.exger.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 48.Ellis CE, et al. Mitochondrial lipid abnormality and electron transport chain impairment in mice lacking alpha-synuclein. Mol Cell Biol. 2005;25:10190–10201. doi: 10.1128/MCB.25.22.10190-10201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hagopian K, et al. Complex I-associated hydrogen peroxide production is decreased and electron transport chain enzyme activities are altered in n-3 enriched fat-1 mice. PLoS One. 2010;5:e12696. doi: 10.1371/journal.pone.0012696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hensley K, et al. Dietary choline restriction causes complex I dysfunction and increased H(2)O(2) generation in liver mitochondria. Carcinogenesis. 2000;21:983–989. doi: 10.1093/carcin/21.5.983. [DOI] [PubMed] [Google Scholar]
- 51.Strifler G, et al. Targeting mitochondrial dysfunction with L-alpha glycerylphosphorylcholine. PLoS One. 2016;11:e0166682. doi: 10.1371/journal.pone.0166682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharpley MS, Shannon RJ, Draghi F, Hirst J. Interactions between phospholipids and NADH:ubiquinone oxidoreductase (complex I) from bovine mitochondria. Biochemistry. 2006;45:241–248. doi: 10.1021/bi051809x. [DOI] [PubMed] [Google Scholar]
- 53.Horibata Y, et al. StarD7 protein deficiency adversely affects the phosphatidylcholine composition, respiratory activity, and cristae structure of mitochondria. J Biol Chem. 2016;291:24880–24891. doi: 10.1074/jbc.M116.736793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang L, et al. The phosphatidylcholine transfer protein Stard7 is required for mitochondrial and epithelial cell homeostasis. Sci Rep. 2017;7:46416. doi: 10.1038/srep46416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cogliati S, Enriquez JA, Scorrano L. Mitochondrial cristae: Where beauty meets functionality. Trends Biochem Sci. 2016;41:261–273. doi: 10.1016/j.tibs.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 56.Ikon N, Ryan RO. Cardiolipin and mitochondrial cristae organization. Biochim Biophys Acta Biomembr. 2017;1859:1156–1163. doi: 10.1016/j.bbamem.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Babot M, Galkin A. Molecular mechanism and physiological role of active-deactive transition of mitochondrial complex I. Biochem Soc Trans. 2013;41:1325–1330. doi: 10.1042/BST20130088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Babot M, et al. ND3, ND1 and 39kDa subunits are more exposed in the de-active form of bovine mitochondrial complex I. Biochim Biophys Acta. 2014;1837:929–939. doi: 10.1016/j.bbabio.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kowarz E, Löscher D, Marschalek R. Optimized sleeping beauty transposons rapidly generate stable transgenic cell lines. Biotechnol J. 2015;10:647–653. doi: 10.1002/biot.201400821. [DOI] [PubMed] [Google Scholar]
- 61.Mátés L, et al. Molecular evolution of a novel hyperactive sleeping beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 62.Wojtala A, et al. Methods to monitor ROS production by fluorescence microscopy and fluorometry. Methods Enzymol. 2014;542:243–262. doi: 10.1016/B978-0-12-416618-9.00013-3. [DOI] [PubMed] [Google Scholar]
- 63.Zhang J, et al. Measuring energy metabolism in cultured cells, including human pluripotent stem cells and differentiated cells. Nat Protoc. 2012;7:1068–1085. doi: 10.1038/nprot.2012.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin-Montalvo A, et al. Cytochrome b 5 reductase and the control of lipid metabolism and healthspan. NPJ Aging Mech Dis. 2016;2:16006. doi: 10.1038/npjamd.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sarafian MH, et al. Objective set of criteria for optimization of sample preparation procedures for ultra-high throughput untargeted blood plasma lipid profiling by ultra performance liquid chromatography-mass spectrometry. Anal Chem. 2014;86:5766–5774. doi: 10.1021/ac500317c. [DOI] [PubMed] [Google Scholar]
- 66.Bozek K, et al. Organization and evolution of brain lipidome revealed by large-scale analysis of human, chimpanzee, macaque, and mouse tissues. Neuron. 2015;85:695–702. doi: 10.1016/j.neuron.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 67.Wiśniewski JR, Mann M. A proteomics approach to the protein normalization problem: Selection of unvarying proteins for MS-based proteomics and western blotting. J Proteome Res. 2016;15:2321–2326. doi: 10.1021/acs.jproteome.6b00403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.