Significance
Drugs that cause epigenetic modification of DNA, such as 5-azacytidine (AZA), are used clinically to treat myelodysplastic syndromes and acute myeloid leukemia. In addition, AZA is being investigated for use against a range of different types of solid tumors, including lung and colorectal cancers. Treatment with AZA causes demethylation of DNA, thus increasing RNA synthesis, including the synthesis of double-stranded RNA, which is otherwise produced in virus-infected cells. We determined that cell death in response to AZA requires the antiviral enzyme RNase L. The results identify a drug target for enhancing the anticancer activity and reducing the toxicity of AZA and related drugs.
Keywords: DNA methyltransferase inhibitor, 5-azacytidine, OAS, RNase L, innate immunity
Abstract
Drugs that reverse epigenetic silencing, such as the DNA methyltransferase inhibitor (DNMTi) 5-azacytidine (AZA), have profound effects on transcription and tumor cell survival. AZA is an approved drug for myelodysplastic syndromes and acute myeloid leukemia, and is under investigation for different solid malignant tumors. AZA treatment generates self, double-stranded RNA (dsRNA), transcribed from hypomethylated repetitive elements. Self dsRNA accumulation in DNMTi-treated cells leads to type I IFN production and IFN-stimulated gene expression. Here we report that cell death in response to AZA treatment occurs through the 2′,5′-oligoadenylate synthetase (OAS)-RNase L pathway. OASs are IFN-induced enzymes that synthesize the RNase L activator 2-5A in response to dsRNA. Cells deficient in RNase L or OAS1 to 3 are highly resistant to AZA, as are wild-type cells treated with a small-molecule inhibitor of RNase L. A small-molecule inhibitor of c-Jun NH2-terminal kinases (JNKs) also antagonizes RNase L-dependent cell death in response to AZA, consistent with a role for JNK in RNase L-induced apoptosis. In contrast, the rates of AZA-induced and RNase L-dependent cell death were increased by transfection of 2-5A, by deficiencies in ADAR1 (which edits and destabilizes dsRNA), PDE12 or AKAP7 (which degrade 2-5A), or by ionizing radiation (which induces IFN-dependent signaling). Finally, OAS1 expression correlates with AZA sensitivity in the NCI-60 set of tumor cell lines, suggesting that the level of OAS1 can be a biomarker for predicting AZA sensitivity of tumor cells. These studies may eventually lead to pharmacologic strategies for regulating the antitumor activity and toxicity of AZA and related drugs.
DNA methyltransferase inhibitors (DNMTi’s) have been investigated as potential cancer therapeutic agents for at least 45 y (1). Methylation of cytosine residues in CpG islands of DNA results in epigenetic silencing of transcription, while removal of these methyl groups has the opposite effect of promoting transcription (2). Currently, the DNMTi’s 5-azacytidine (AZA) and 5-aza-2′-deoxycytidine (DAC) are approved for myelodysplastic syndromes, and also improve survival in acute myeloid leukemia (3–6). In addition, DNMTi’s are under investigation for the treatment of various solid tumors, including non–small-cell lung cancer (NSCLC) and breast and colorectal cancer (7, 8). DNMTi’s deplete cellular levels of DNA methyltransferases, leading to DNA hypomethylation and wide-ranging effects on cancer cells (9–12). However, there remains a critical need to better understand the molecular mechanisms for the antitumor and cytotoxic effects of DNMTi’s, to increase their clinical efficacy while minimizing toxicity.
Studies on the cancer therapeutic mechanisms of DNMTi’s, including AZA, have focused on the induction of tumor suppressor genes (8, 13) and, more recently, on the expression of self, double-stranded RNA (dsRNA). DNA demethylation caused by treatment with nanomolar levels of DNMTi’s induces the bidirectional transcription of human endogenous retrovirus (ERV)-like genes, leading to the formation of dsRNA, an IFN response, and inhibition of cell proliferation (14, 15). dsRNA, a common viral pathogen-associated molecular pattern, induces an innate immune response that includes the expression of type I IFN and IFN-stimulated genes (ISGs) (14, 15). Therefore, expression of self dsRNA accounts for the long-known observation that DNMTi’s induce ISGs (16). However, the connection between self dsRNA induction by AZA and tumor cell death induced by DNMTi’s is less clear.
The 2′,5′-oligoadenylate synthetase (OAS)-RNase L pathway is an IFN-stimulated antiviral response that requires dsRNA (17, 18). Type I and type III IFNs induce the transcription of a family of human OAS genes encoding enzymatically active OAS1 to 3 isozymes and enzymatically inactive OASL (19). dsRNA binds to and activates OAS1 to 3, which then use ATP to synthesize a series of 5′-triphosphorylated, 2′,5′-linked oligoadenylates (2-5As) (20). The only well-established function of 2-5A is activation of the single-stranded RNA-specific endoribonuclease RNase L (21). 2-5A binds to the inactive and latent monomeric form of RNase L, inducing dimerization and activation (also requiring ATP or ADP binding to the protein kinase-like domain of RNase L) (22–24). Activated RNase L cleaves viral and cellular RNAs, suppressing viral replication, in part by eliminating virus-infected cells by apoptosis (25, 26).
Self dsRNA accumulates during DNMTi treatment, and also when there is a mutation in the dsRNA-editing enzyme ADAR1, leading to the genetic disease Aicardi–Goutières syndrome (27). Previously, we reported that CRISPR-mediated knockout of RNase L prevents the lethal effect of ADAR1 deletion in A549 NSCLC cells (28). ADAR1 converts adenosine to inosine, destabilizing dsRNA and preventing or reducing the activation of OAS and, indirectly, also suppressing RNase L activation. Together, the presence of dsRNA, IFNs, and ISG expression (including OASs) in DNMTi-treated cells and the requirement for RNase L in cell death due to ADAR1 knockout led us to examine whether RNase L contributes to the elimination of cancer cells by DNMTi’s. Here we report that cells lacking RNase L or OAS1 to 3 are protected from the cytotoxic effects of AZA. Furthermore, small-molecule inhibitors of either RNase L or c-Jun NH2-terminal kinases (JNKs), which have been implicated in RNase L-mediated apoptosis, protected cells from AZA (29). Conversely, deletion of the 2′-phosphodiesterases (PDEs) that degrade 2-5A (PDE12 and AKAP7) (30, 31), deletion of ADAR1 that edits and destabilizes dsRNA (28), or ionizing radiation (IR) that activates IFN-dependent signaling (32) each increased the RNase L-dependent cytotoxic effects of AZA. Basal OAS1 expression in the NCI-60 set of human tumor cell lines correlated with AZA sensitivity. These findings suggest potential targets for enhancing the antitumor activity and mitigating the toxicity of AZA.
Results
Cells Lacking RNase L Are Resistant to AZA Treatment.
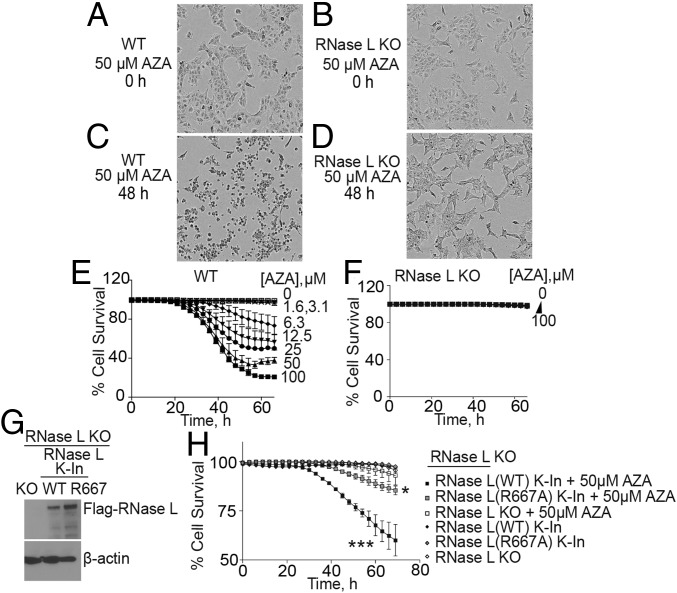
DNMTi’s induce the accumulation of self dsRNA in several different tumor cell types (7, 14, 15). We confirmed this observation in A549 NSCLC cells treated with 50 μM AZA for 48 h by visualizing cytoplasmic dsRNA accumulation with a monoclonal antibody against dsRNA (SI Appendix, Fig. S1). While there was no detectable dsRNA in untreated cells, accumulation of dsRNA in the cytoplasm was apparent after AZA treatment. To determine whether RNase L affects AZA sensitivity, we compared parental vector control A549 cells (wild type; WT) with RNase L knockout (KO) cells, previously generated with CRISPR-Cas9 technology (33) (SI Appendix, Fig. S2A). Cells treated with different amounts of AZA were monitored for survival as a function of time using an IncuCyte real-time cell imaging system (Essen Bioscience) and a dual-dye method. While the majority of WT cells appeared dead after 48 h of AZA treatment, the AZA-treated RNase L KO cells appeared normal (Fig. 1 A–D). Quantification of WT cell survival showed that 63 and 77% of the cells had died by 60 h in response to 50 or 100 μM AZA, respectively (Fig. 1E). Cell death in WT cells was observed at concentrations of 6 μM AZA or greater. In contrast, the RNase L KO cells were highly resistant to 100 μM AZA after 60 h, with little or no death (Fig. 1F).
Fig. 1.
RNase L mediates cell death in response to AZA treatment of A549 cells. (A–D) Phase-contrast images of (A and C) WT and (B and D) RNase L KO A549 cells at 0 and 48 h after adding AZA (50 μM) to media. (E and F) Kinetics of cell death of (E) WT and (F) RNase L KO cells after AZA treatment (1.6 to 100 μM). Percent cell survival was determined by real-time imaging with a dual-dye method (SI Appendix, Materials and Methods). Data are the averages ± SD from four identically treated replicates. Three biological replicates were performed, each with a minimum of three technical replicates. (G) RNase L KO cells were transduced with lentiviruses expressing either WT RNase L or a catalytically inactive mutant (R667A) for 72 h to obtain knockin cells. Western blots probed with anti-Flag M2 antibody (Upper), to detect Flag-RNase L, or anti–β-actin antibody (Lower) are shown. (H) Percent cell survival of RNase L KO cells in the absence or presence of lentivirus expressing either WT RNase L or a catalytically inactive mutant (R667A) and incubated without or with 50 μM AZA. Time 0 on the x axis is when AZA was added, 24 h after lentivirus infection. The data are the averages ± SD from three identically treated replicates. Two biological replicates were performed, each with a minimum of three technical replicates. *P < 0.05, ***P < 0.001.
To confirm that RNase L was responsible for AZA sensitivity, RNase L KO A549 cells were transiently transduced with lentiviral constructs encoding either WT or nuclease-dead mutant (R667A) RNase L (34) (Fig. 1G). Knockin (K-In) of WT RNase L sensitized the cells to AZA, leading to decreased cell survival, whereas introduction of mutant RNase L (R667A) had a diminished effect on cell survival (Fig. 1H). These results demonstrate that AZA-induced cell death requires RNase L with a functional catalytic domain.
Individual OAS1, 2, or 3 Isoforms Are Sufficient for Increasing AZA-Induced Cell Death.
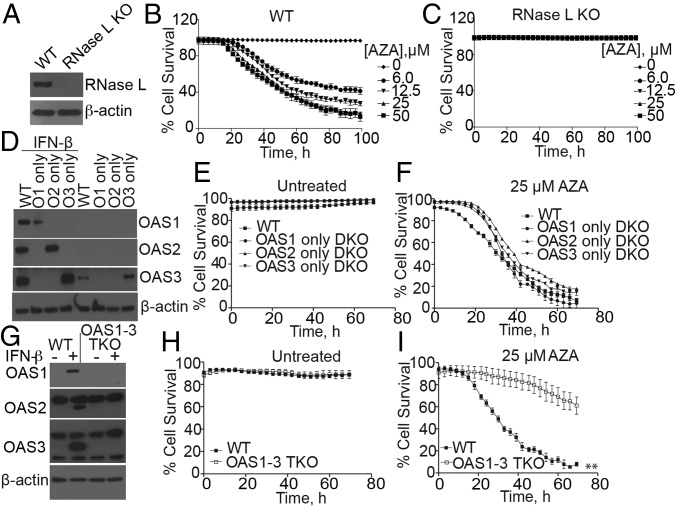
Immortalized, human mammary epithelial hTert-HME1 (HME) cells were also rendered highly resistant to AZA by knockout of RNase L (Fig. 2 A–C). The relative contributions of the different OAS isoforms to AZA-induced cell death were determined with double-knockout (DKO) HME cells, each expressing only one of the three enzymatically competent OAS isoforms. DKO cells lacking OAS2 and 3, OAS1 and 3, or OAS1 and 2 express only OAS1 (O1), 2 (O2), or 3 (O3), respectively (Fig. 2D). Survival of the different types of untreated cells was unaffected by OAS DKO or triple-knockout (TKO) genotypes (Fig. 2 E and H). Interestingly, expression of any of the three OAS isoforms rendered HME cells nearly as sensitive to AZA as the WT cells (Fig. 2F). However, compared with WT cells, the HME cells lacking all three OAS isoforms, namely TKO cells, were relatively resistant to AZA (Fig. 2 G and I), presumably because these cells are unable to synthesize 2-5A and are thus unable to activate RNase L. The observation that the OAS TKO cells are less resistant to AZA than RNase L KO cells suggests alternative functions of OAS isozymes that are not dependent on RNase L function (Fig. 2 C and I).
Fig. 2.
Involvement of OAS isozymes and RNase L in the death of HME cells in response to AZA treatment. (A) Western blots of WT and RNase L KO HME cells. (B and C) Percent survival of WT and RNase L KO HME cells, respectively, treated with different doses of AZA. The data are the averages ± SD of four identically treated replicates. Two biological replicates were performed, each with a minimum of three technical replicates. (D) Western blots of OASs from WT and OAS DKO HME cells incubated without or with IFN-β (200 IU/mL for 18 h), using OAS isozyme-specific antibodies. (E and F) Percent survival of WT and DKO cells that were either (E) untreated or (F) treated with 25 μM AZA. The data are the averages ± SD of three identical replicates. Three biological replicates were performed, each with a minimum of three technical replicates. (G) Western blots of WT and OAS1 to 3 TKO HME cells incubated without or with IFN-β (200 IU/mL for 18 h). (H and I) Percent cell survival of WT and TKO HME cells that were either (H) untreated or (I) treated with 25 μM AZA. The data are the averages ± SD of three identically treated technical replicates. Four biological replicates were performed, each with a minimum of three technical replicates. **P < 0.01.
Effect of MAVS on AZA Sensitivity.
dsRNA signaling to the type I IFN genes requires the MDA5-RIG-I/MAVS pathway (35). Therefore, to determine whether IFN production, with subsequent OAS induction, is required for AZA-induced cell death, A549 cells in which MAVS was knocked out individually or in combination with RNase L were used (SI Appendix, Fig. S2A). MAVS KO A549 cells were only slightly less sensitive to AZA treatment than WT cells, suggesting that, although IFN production may have contributed to AZA-mediated cell death, its effects were modest (SI Appendix, Fig. S2 B and C). However, the RNase L-MAVS DKO cells were resistant to AZA at concentrations as high as 50 μM (SI Appendix, Fig. S2D), similar to RNase L KO cells (Fig. 1F). These results suggest that basal levels of OAS1 to 3 in A549 cells, confirmed by gene expression analysis (SI Appendix, Table S1), are sufficient for the cell-lethal effect of AZA, through RNase L activation.
AZA Causes Apoptotic Death of A549 Cells.
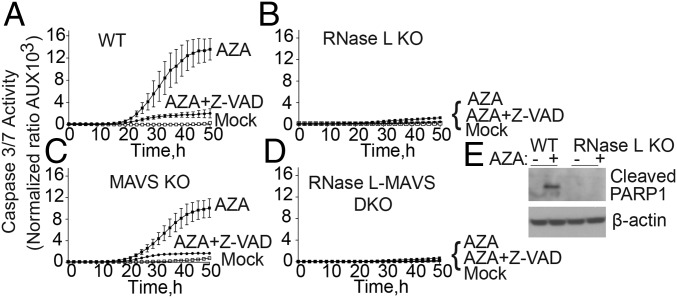
To determine the mode of cell death by AZA, caspase-3/7 activation was monitored as a marker of apoptosis by real-time imaging with the IncuCyte system and software (Fig. 3). While WT A549 cells activate caspase-3/7 strongly in response to AZA, the RNase L KO A549 cells were resistant to AZA (Fig. 3 A and B). MAVS KO cells showed a modest reduction in caspase-3/7 activation, whereas RNase L-MAVS DKO cells were highly resistant, similar to the RNase L single-KO cells (Fig. 3 B–D). As a positive control, the pan-caspase inhibitor ZVAD-FMK greatly suppressed caspase-3/7 activation, as expected.
Fig. 3.
Involvement of RNase L in AZA-induced apoptosis. (A–D) Caspase-3/7 activity in (A) WT, (B) RNase L KO, (C) MAVS KO, and (D) RNase L-MAVS DKO A549 cells mock-treated or treated with 50 µM AZA in the absence or presence of 50 μM ZVAD-FMK (a pan-caspase inhibitor). The data are the averages ± SD of three identically treated replicates. Two biological replicates were performed. (E) Western analyses of WT and RNase L KO A549 cells treated with 50 µM AZA or mock-treated for 48 h. The blots were probed with antibody specific for cleaved PARP1 and β-actin antibody. AU, arbitrary unit.
To confirm that AZA treatment induces apoptosis, PARP cleavage was monitored in Western assays. Treatment of WT A549 cells with 50 μM AZA for 48 h resulted in cleaved PARP (Fig. 3E). In contrast, identical treatment of RNase L KO A549 cells did not lead to PARP [poly-(ADP-ribose) polymerase] cleavage. Thus, AZA induction of both caspase-3/7 activation and PARP cleavage in WT but not in RNase L KO cells indicates cell death occurred through the proapoptotic activity of RNase L (25, 26, 36).
A Small-Molecule Inhibitor of RNase L Inhibits AZA-Induced Cell Death.
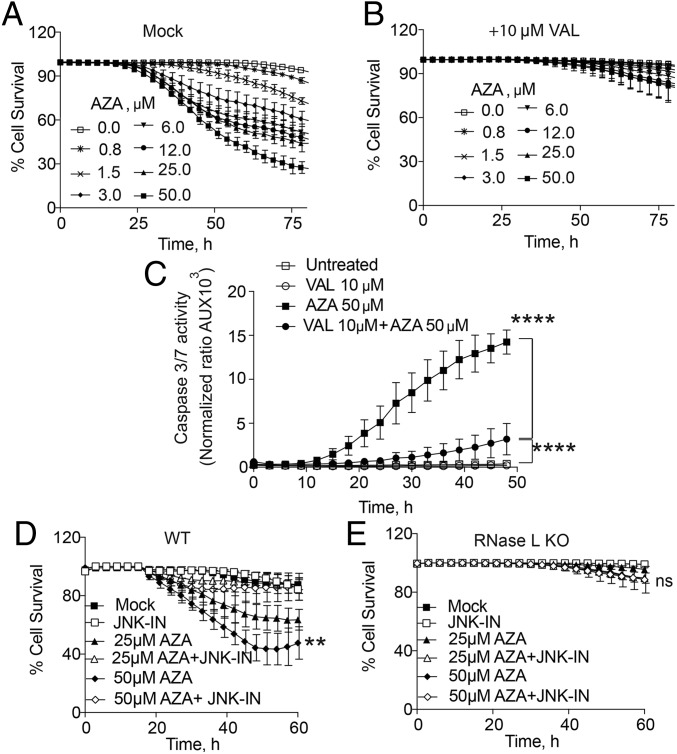
To further validate the involvement of RNase L in AZA-mediated cell death, a potent small-molecule inhibitor of RNase L, valoneic acid dilactone (VAL), was used. Treatment of A549 cells with 10 μM VAL protected them against the cytotoxic effect of AZA (Fig. 4 A and B). Similarly, VAL suppressed caspase-3/7 activation in response to AZA, showing that apoptosis was prevented by chemical inhibition of RNase L (Fig. 4C).
Fig. 4.
Small-molecule inhibitors of either RNase L or JNK inhibit AZA-induced cell death. (A and B) Percent survival of WT A549 cells after AZA treatment in the (A) absence or (B) presence of the RNase L inhibitor VAL. Data are the averages ± SD from five identically treated replicates. Three biological replicates were performed, each with a minimum of three technical replicates. (C) Caspase-3/7 activity in A549 cells in the presence or absence of AZA and VAL. Data are the averages ± SD from three identically treated replicates. Two biological replicates were performed. (D and E) Percent cell survival of WT and RNase L KO A549 cells, respectively, mock-treated or treated with 25 µM SP600125 (JNK inhibitor; JNK-IN) for 2 h and then incubated with AZA (25 or 50 μM). Data are the averages ± SD from five identically treated replicates. Four biological replicates were performed, each with a minimum of three technical replicates. **P < 0.01, ****P < 0.0001; ns, nonsignificant.
Previously, we reported that RNase L activity triggers the phosphorylation of JNKs, and also that JNK-deficient cells are resistant to RNase L-mediated apoptosis (29). Accordingly, AZA-induced cell death was inhibited by treating WT A549 cells with the JNK inhibitor SP600125 (Fig. 4D). The RNase L KO cells were resistant to AZA in the absence or presence of this inhibitor (Fig. 4E). The rescue from cell death by the JNK inhibitor implicates JNK in the RNase L-dependent mechanism of apoptosis in response to AZA treatment.
Knockouts of Host Phosphodiesterases That Degrade 2-5A Increase Cell Death in Response to AZA.
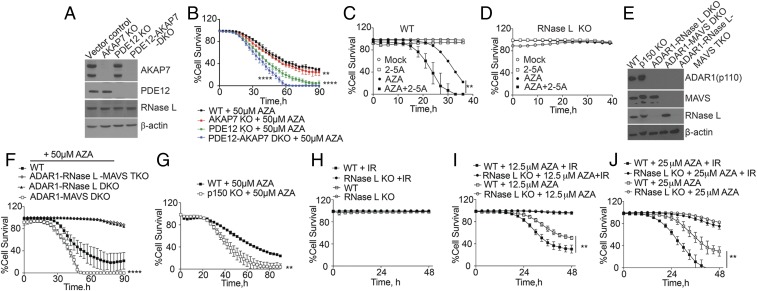
2-5A is degraded in cells through the action of the 2′,5′-phosphodiesterases AKAP7 and PDE12 (30, 31, 37). Therefore, to increase 2-5A accumulation and RNase L activity during AZA treatment, the genes encoding AKAP7 and PDE12 were knocked out in A549 cells, individually and in combination (Fig. 5A). Previously, ablation of either AKAP7 or PDE12 increased 2-5A levels by retarding its degradation (31, 37). Deletion of AKAP7 slightly increased cell-death rates in the presence of AZA, whereas PDE12 knockout had a much larger effect (Fig. 5B). The DKO for both AKAP7 and PDE12 greatly increased cell death in response to AZA (Fig. 5B). These results suggest that extending the half-life of 2-5A in cells by depleting or inhibiting 2-5A catabolic enzymes increases sensitivity to AZA.
Fig. 5.
Effect of RNase L on cell death during AZA treatment is increased by (A and B) KO of PDE12 and/or AKAP7, (C and D) 2-5A transfection, (E–G) KO of ADAR1 or its p150 isoform, or (H–J) IR. (A) Western blots of vector control WT, AKAP7 KO, PDE12 KO, and PDE12-AKAP7 DKO A549 cells, probed with the indicated antibodies. (B) Percent cell survival of WT, AKAP7 KO, PDE12 KO, and PDE12-AKAR7 DKO cells after AZA treatment. The data are the averages ± SD of three identical replicates. Three biological replicates were performed, each with a minimum of three technical replicates. (C and D) Percent survival of WT and RNase L KO cells with and without 1 μM 2-5A transfection. (E) Western blots of WT, ADAR1 p150 KO, ADAR1-RNase L DKO, ADAR1-MAVS DKO, and ADAR1-RNase L-MAVS TKO cells. (F and G) Percent cell survival after AZA treatment. The data are the averages ± SD of three identical replicates. Three biological replicates were performed, each with a minimum of three technical replicates. (H–J) Percent cell survival after IR (10 Gy) for 30 min, followed by AZA treatment in comparison with mock treatments. The data are the averages ± SD of three identical replicates. Two biological replicates were performed, each with a minimum of three technical replicates. **P < 0.01, ****P < 0.0001.
2-5A Increases the Sensitivity of A549 Cells to AZA.
To determine whether direct activation of RNase L would impact tumor cell killing by AZA, WT and RNase L KO A549 cells were treated with AZA alone, transfected with 2-5A, or treated with both agents (Fig. 5 C and D). Whereas transfection with a low dose of 2-5A (1 μM) by itself had no effect on WT cell survival and AZA (25 μM) by itself reduced cell survival, the combination of 2-5A (1 μM) and AZA (25 μM) eliminated nearly all cells (Fig. 5C). In contrast, RNase L KO cells were completely resistant to AZA, 2-5A, or the combination of both agents (Fig. 5D). These results demonstrate that exogenous 2-5A increases the cell-lethal effect of AZA and that the mechanism of action is entirely dependent on RNase L.
ADAR1 Knockout Increases the Susceptibility of Cells to AZA.
ADAR1 edits adenosine (A) to inosine (I) in dsRNA, which destabilizes dsRNA because A:U base pairs are more stable than I:U mismatches (38–40). Our prior study suggested that the accumulation of constitutively produced self dsRNA in ADAR1 KO cells leads to OAS-RNase L activation and cell death (28). Because the knockout of ADAR1 is lethal in A549 cells, it was necessary to use combination knockout cells for these experiments. To determine whether ADAR1 antagonizes the cell-lethal effect of AZA, A549 cells of different ADAR1, MAVS, and RNase L genotypes were treated with AZA. Western blots for these cell lines are shown in Fig. 5E and SI Appendix, Fig. S3 (in which IFN was used to induce levels of ADAR1 p150). ADAR1 p150, and to a lesser extent OAS1, was also induced by AZA (SI Appendix, Fig. S3B). Cells with ADAR1-RNase L-MAVS TKO or ADAR1-RNase L DKO were resistant to AZA (Fig. 5F). In contrast, ADAR1-MAVS DKO cells showed increased AZA sensitivity compared with WT cells, presumably because of increased accumulation of self dsRNA due to the absence of ADAR1 (Fig. 5F). To further determine the relative contribution of ADAR1 isoforms, A549 cells lacking the IFN-inducible ADAR1 p150 isoform were compared with WT A549 cells (Fig. 5G and SI Appendix, Fig. S3). ADAR1 p150 KO cells were more sensitive to AZA, suggesting that p150 is primarily involved in AZA-induced cytotoxicity. These results indicate that the AZA sensitivity of cells can be increased by ADAR1 deficiency.
IR Increases AZA-Induced Cell Death in an RNase L-Dependent Manner.
IR induces type I IFN signaling, and OAS genes are included in the IFN-related DNA damage resistance signature, whose expression correlates with radiation resistance in cancer cells (32, 41). To determine the effect of IR on AZA-mediated cell death through the OAS-RNase L pathway, WT and RNase L KO A549 cells were exposed to 10 Gy of IR and then either mock-treated or treated with 12.5 or 25 µM AZA (Fig. 5 H–J). The kinetics of cell death was determined by real-time cell imaging. There was no effect on cell viability of IR treatment alone in WT and RNase L KO cells, as A549 cells are relatively resistant to radiation (Fig. 5H). However, the combination of AZA and IR significantly increased the cell-death rates in WT cells compared with AZA treatment alone, whereas the RNase L KO cells were resistant to IR and AZA (Fig. 5 I and J). These results suggest that IR increases RNase L-dependent cell death triggered by AZA treatment.
OAS1 Expression in the NCI-60 Set of Human Tumor Cell Lines.
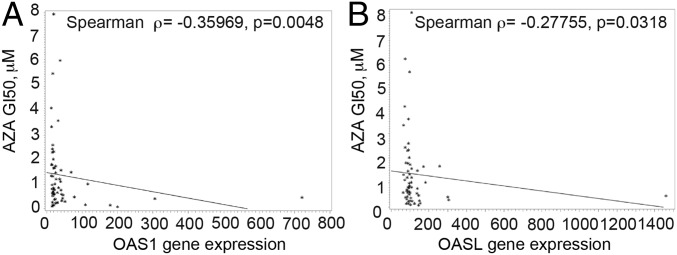
To determine whether AZA sensitivity is correlated with OAS-RNase L levels in different tumor cell types, we interrogated gene expression profiles of the NCI-60 database of 60 human tumor cell lines in the presence or absence of AZA (Fig. 6 and SI Appendix, Table S1). In these 60 cell lines, representative of the histologic and genetic diversity of cancer, the expression levels of OAS1 (Fig. 6A) and OASL (Fig. 6B) predict sensitivity to AZA; that is, the higher the expression levels of these enzymes, the greater the sensitivity of the cells to the lethal effect of AZA. These results suggest that OAS1 levels, in particular, can be a marker for sensitivity to AZA-induced cytotoxicity.
Fig. 6.
Basal OAS1 and OASL expression correlate with AZA sensitivity among NCI-60 tumor cell lines. Drug sensitivity to AZA is represented as GI50, the drug concentration resulting in a 50% growth reduction, quantified by measurement of total RNA at day 6 (raw data were downloaded from the National Cancer Institute Development Therapeutics Program; dtp.nci.nih.gov) (higher GI50 indicates less sensitivity to drug). GI50 was correlated with expression of OAS1 (A) and OASL (B) in the cell lines (gene expression values by microarray from the Gene Expression Omnibus database, accession no. GSE5846). Probe sets were 205552_s_at (for OAS1) and 210797_s_at (for OASL). The statistical method is Spearman’s ranked correlation coefficient test, calculated using SAS v9 software.
Discussion
The OAS-RNase L Pathway Mediates Tumor Cell Death in Response to AZA.
DNMTi’s have long been known to induce an IFN response that is characterized by ISG expression (16), although the molecular mechanism has only recently been elucidated. Hypomethylation of DNA resulting from DNMTi treatment leads to production of self dsRNA from ERVs, short interspersed nuclear elements (SINEs), and other repetitive DNA elements, triggering an innate immune response that resembles the response induced by viral infections, or by ADAR1 KO in the absence of viral infection (14, 15, 28, 42). dsRNA signals through the MDA5-RIG-I/MAVS/IRF3–IRF7 pathway to induce type I and III IFNs which, in turn, induce the expression of ISGs, including OAS1 to 3, that mediate most biological effects of these IFNs. For example, DAC was shown to induce an IFN response in colorectal cancer-initiating cells (CICs) through the MDA5/MAVS/IRF7 signaling pathway (14). Long-term growth of CICs was inhibited following transient treatment with a low dose of DAC. Similarly, the cellular response to DNMTi’s (AZA or DAC) was characterized by high expression of ERVs and IFN, which sensitized melanomas to immunotherapy with anti–CTLA-4 (15).
dsRNA also directly activates two types of IFN-induced enzymes, the protein kinase PKR, which blocks translational initiation, and OAS1 to 3, which synthesize 2-5A activators of RNase L (43). The only well-established function of 2-5A is activation of RNase L, an antiviral protein with proapoptotic activity (21, 25, 26, 36). Here we observe that deletion of RNase L or OAS1 to 3 isoforms renders cells highly resistant to AZA-induced death. These results are in contrast to the consequences of viral infections of cells, in which OAS3 is the dominant OAS isoform, at least in some cell types (33). In addition to demethylating DNA, AZA treatment leads to RNA demethylation (44), which increases RNA immune signaling through TLR3 (45). However, the dependence on the dsRNA-activated OAS-RNase L pathway for AZA-induced cell death suggests that it is DNA demethylation leading to dsRNA, and not RNA demethylation, that is responsible for the cytotoxic response. RNase L causes apoptosis through a ribotoxic stress pathway involving JNK-dependent signaling (29). Consistent with the involvement of RNase L in AZA-induced cell death is the observation that a small-molecule inhibitor of JNK suppresses apoptosis in response to AZA.
Previously, AZA-induced cytotoxicity was linked to reversible DNA damage stemming from the formation of covalent complexes between DNA and DNMTs (46, 47). Also, during DNA damage induced by IR or chemotherapeutic agents, U1 and U2 small noncoding RNAs accumulate in the cytoplasm, activating the pathogen recognition receptor RIG-I and IFN-dependent signaling (32). In support of this idea, we show here that IR increases the cytotoxicity of AZA through RNase L. Very interestingly, many different primary malignant tumors, including those of the head and neck, lung, prostate, breast, and high-grade gliomas, express high levels of OAS expression as part of the IFN-related DNA damage resistance signature (41). Therefore, AZA-induced cell death through OAS-RNase L is likely to preferentially target these cancer cells in vivo.
Chemically or Genetically Induced Self dsRNA Triggers Activation of OAS-RNase L and Cell Death.
Genetic deficiency in the dsRNA-editing enzyme ADAR1 can lead to RNase L-dependent death (28) or IFN-stimulated translational arrest through PKR (48). There are similarities between the cellular effects of AZA treatment and ADAR1 mutation, in that both lead to the formation of self dsRNA, transcribed from repetitive DNA elements, including Alu (SINE) and hERV elements (14, 15, 48). When ADAR1 is mutated, the deficiency in self dsRNA editing is believed to lead to increased activation of OAS-RNase L, whereas during AZA treatment it is likely that increased transcription of repetitive DNA elements causes OAS-RNase L activation. Since ADAR1 deletion is lethal in A549 cells, we used ADAR1-MAVS DKO A549 cells in our experiments (28). Because both ADAR1 knockout and AZA treatment result in the synthesis of self dsRNA, the combination of ADAR1 deficiency with AZA treatment was especially cytotoxic.
MAVS Is Not Required for AZA-Induced Cell Death.
Induction of type I IFNs by dsRNA occurs through the MDA5-RIG-I/MAVS/IRF3–IRF7-NF-κB signaling pathway (49). To determine whether the same pathway is required for AZA-induced cell death, MAVS KO cells were studied. MAVS deletion reduced but did not prevent cell death by OAS-RNase L in response to AZA treatment, a result similar to our findings in ADAR1 KO cells (28). Therefore, basal levels of OAS isozymes are sufficient to mediate cell death through RNase L activation (Fig. 1 and SI Appendix, Table S1). Prior studies on mouse hepatitis virus showed that, in mouse bone marrow macrophages, basal levels of OAS are sufficient for RNase L activation (33, 50). Taken together, these results reveal that IFN does not need to be induced for an antiviral or cell-death response through OAS-RNase L, provided that sufficient basal levels of OAS and RNase L are present. Furthermore, the correlation of basal OAS1 and OASL levels with AZA sensitivity in the NCI-60 tumor cell panel indicates that OAS1 (or OASL) can be a biomarker for AZA sensitivity in cancer.
DNMTi’s, RNase L, and Cancer.
Prior studies on the antitumor activities of AZA and DAC showed effects against melanoma through increased immune checkpoint therapy (15) and against colorectal cancer through targeting of colorectal cancer-initiating cells (14). Also, an earlier study showed that DAC could overcome resistance to IFN-induced apoptosis in renal carcinoma and melanoma (51). DNMTi treatment of mouse cells with mutant p53 led to massive dsRNA accumulation, originating at least in part from the transcription of repetitive DNA elements, culminating in a cell-suicidal type I IFN response (42). The transcribed repetitive DNA elements included SINEs, satellite repeats, endogenous IAP retroviral DNA, and noncoding RNA genes (42). However, A549 and HME cells are p53-wild type (52, 53), yet both are AZA-sensitive. Therefore, AZA sensitivity, and the involvement of RNase L in AZA-induced cell death, is not limited to cells with mutant p53, at least not in the human cell types examined in this study.
Our findings suggest strategies for increasing the anticancer effects of AZA. For instance, combining AZA with IR was highly effective in eliminating tumor cells in an RNase L-dependent manner. We also showed that knockout of PDE12 or AKAP7, which impair 2-5A degradation, increased tumor cell killing by AZA. Therefore, treatments that increase RNase L activity directly or indirectly, for instance by inhibiting PDE12 (30, 37) and/or AKAP7 (31), are predicted to increase the antitumor activities of AZA or DAC. Alternatively, inhibitors of RNase L, such VAL, are expected to mitigate the cytotoxicity of AZA. Therefore, our studies have therapeutic implications for either increasing the efficacy of DNMTi treatment of cancer or mitigating its toxicity.
Materials and Methods
Detailed materials and methods for construction of gene knockout cells, immunoblotting procedures, cell-death assays, caspase-3/7 assays, IR, immunofluorescence, and statistics are described in SI Appendix.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Awards R01AI104887 (to S.R.W. and R.H.S.) and R01AI135922 (to R.H.S.), a VeloSano Cancer Research Pilot Award (to R.H.S.), the Canadian Cancer Society (Impact Grant 704116 to F.S.), and Canadian Institute for Health Research (FDN143277 to F.S.).
Footnotes
Conflict of interest statement: S.B. and R.H.S. have a US patent application relating to this study. B.D. and R.H.S. receive royalties for the RNase L antibody used in this study. Y.S. has patents around tetrahydrouridine, decitabine, and 5-azacytidine; and consults for, and has equity and royalty rights with EpiDestiny.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815071116/-/DCSupplemental.
References
- 1.Weiss AJ, Stambaugh JE, Mastrangelo MJ, Laucius JF, Bellet RE. Phase I study of 5-azacytidine (NSC-102816) Cancer Chemother Rep. 1972;56:413–419. [PubMed] [Google Scholar]
- 2.Groudine M, Eisenman R, Weintraub H. Chromatin structure of endogenous retroviral genes and activation by an inhibitor of DNA methylation. Nature. 1981;292:311–317. doi: 10.1038/292311a0. [DOI] [PubMed] [Google Scholar]
- 3.Hamm CA, Costa FF. Epigenomes as therapeutic targets. Pharmacol Ther. 2015;151:72–86. doi: 10.1016/j.pharmthera.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Nazha A, et al. MDS Clinical Research Consortium Outcomes of patients with myelodysplastic syndromes who achieve stable disease after treatment with hypomethylating agents. Leuk Res. 2016;41:43–47. doi: 10.1016/j.leukres.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawson MA. The cancer epigenome: Concepts, challenges, and therapeutic opportunities. Science. 2017;355:1147–1152. doi: 10.1126/science.aam7304. [DOI] [PubMed] [Google Scholar]
- 6.Dombret H, et al. International phase 3 study of azacitidine vs conventional care regimens in older patients with newly diagnosed AML with >30% blasts. Blood. 2015;126:291–299. doi: 10.1182/blood-2015-01-621664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stone ML, et al. Epigenetic therapy activates type I interferon signaling in murine ovarian cancer to reduce immunosuppression and tumor burden. Proc Natl Acad Sci USA. 2017;114:E10981–E10990. doi: 10.1073/pnas.1712514114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiu X, Liang Y, Sellers RS, Perez-Soler R, Zou Y. Aerosol azacytidine inhibits orthotopic lung cancers in mice through its DNA demethylation and gene reactivation effects. PLoS One. 2014;9:e109874. doi: 10.1371/journal.pone.0109874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010;28:1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghoshal K, et al. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25:4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Li H, et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget. 2014;5:587–598. doi: 10.18632/oncotarget.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wrangle J, et al. Alterations of immune response of non-small cell lung cancer with azacytidine. Oncotarget. 2013;4:2067–2079. doi: 10.18632/oncotarget.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navada SC, Steinmann J, Lübbert M, Silverman LR. Clinical development of demethylating agents in hematology. J Clin Invest. 2014;124:40–46. doi: 10.1172/JCI69739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roulois D, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162:961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiappinelli KB, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162:974–986, and errata (2016) 164:1073 and (2017) 169:361. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karpf AR, et al. Inhibition of DNA methyltransferase stimulates the expression of signal transducer and activator of transcription 1, 2, and 3 genes in colon tumor cells. Proc Natl Acad Sci USA. 1999;96:14007–14012. doi: 10.1073/pnas.96.24.14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silverman RH, Weiss SR. Viral phosphodiesterases that antagonize double-stranded RNA signaling to RNase L by degrading 2-5A. J Interferon Cytokine Res. 2014;34:455–463. doi: 10.1089/jir.2014.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silverman RH. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol. 2007;81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kristiansen H, Gad HH, Eskildsen-Larsen S, Despres P, Hartmann R. The oligoadenylate synthetase family: An ancient protein family with multiple antiviral activities. J Interferon Cytokine Res. 2011;31:41–47. doi: 10.1089/jir.2010.0107. [DOI] [PubMed] [Google Scholar]
- 20.Kerr IM, Brown RE. pppA2′p5′A2 ′p5′A: An inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc Natl Acad Sci USA. 1978;75:256–260. doi: 10.1073/pnas.75.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou A, Hassel BA, Silverman RH. Expression cloning of 2-5A-dependent RNAase: A uniquely regulated mediator of interferon action. Cell. 1993;72:753–765. doi: 10.1016/0092-8674(93)90403-d. [DOI] [PubMed] [Google Scholar]
- 22.Dong B, Silverman RH. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J Biol Chem. 1995;270:4133–4137. doi: 10.1074/jbc.270.8.4133. [DOI] [PubMed] [Google Scholar]
- 23.Huang H, et al. Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity. Mol Cell. 2014;53:221–234. doi: 10.1016/j.molcel.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han Y, et al. Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response. Science. 2014;343:1244–1248. doi: 10.1126/science.1249845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou A, et al. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 1997;16:6355–6363. doi: 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castelli JC, et al. A study of the interferon antiviral mechanism: Apoptosis activation by the 2-5A system. J Exp Med. 1997;186:967–972. doi: 10.1084/jem.186.6.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice GI, et al. Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat Genet. 2012;44:1243–1248. doi: 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, et al. Ribonuclease L mediates the cell-lethal phenotype of double-stranded RNA editing enzyme ADAR1 deficiency in a human cell line. eLife. 2017;6:e25687. doi: 10.7554/eLife.25687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li G, Xiang Y, Sabapathy K, Silverman RH. An apoptotic signaling pathway in the interferon antiviral response mediated by RNase L and c-Jun NH2-terminal kinase. J Biol Chem. 2004;279:1123–1131. doi: 10.1074/jbc.M305893200. [DOI] [PubMed] [Google Scholar]
- 30.Kubota K, et al. Identification of 2′-phosphodiesterase, which plays a role in the 2-5A system regulated by interferon. J Biol Chem. 2004;279:37832–37841. doi: 10.1074/jbc.M400089200. [DOI] [PubMed] [Google Scholar]
- 31.Gusho E, et al. Murine AKAP7 has a 2′,5′-phosphodiesterase domain that can complement an inactive murine coronavirus ns2 gene. MBio. 2014;5:e01312-14. doi: 10.1128/mBio.01312-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ranoa DR, et al. Cancer therapies activate RIG-I-like receptor pathway through endogenous non-coding RNAs. Oncotarget. 2016;7:26496–26515. doi: 10.18632/oncotarget.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, et al. Activation of RNase L is dependent on OAS3 expression during infection with diverse human viruses. Proc Natl Acad Sci USA. 2016;113:2241–2246. doi: 10.1073/pnas.1519657113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong B, Niwa M, Walter P, Silverman RH. Basis for regulated RNA cleavage by functional analysis of RNase L and Ire1p. RNA. 2001;7:361–373. doi: 10.1017/s1355838201002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 36.Castelli JC, et al. The role of 2′-5′ oligoadenylate-activated ribonuclease L in apoptosis. Cell Death Differ. 1998;5:313–320. doi: 10.1038/sj.cdd.4400352. [DOI] [PubMed] [Google Scholar]
- 37.Wood ER, et al. The role of phosphodiesterase 12 (PDE12) as a negative regulator of the innate immune response and the discovery of antiviral inhibitors. J Biol Chem. 2015;290:19681–19696. doi: 10.1074/jbc.M115.653113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.George CX, John L, Samuel CE. An RNA editor, adenosine deaminase acting on double-stranded RNA (ADAR1) J Interferon Cytokine Res. 2014;34:437–446. doi: 10.1089/jir.2014.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomaselli S, Galeano F, Locatelli F, Gallo A. ADARs and the balance game between virus infection and innate immune cell response. Curr Issues Mol Biol. 2015;17:37–51. [PubMed] [Google Scholar]
- 40.Patterson JB, Samuel CE. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: Evidence for two forms of the deaminase. Mol Cell Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weichselbaum RR, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci USA. 2008;105:18490–18495. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leonova KI, et al. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc Natl Acad Sci USA. 2013;110:E89–E98. doi: 10.1073/pnas.1216922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaefer M, Hagemann S, Hanna K, Lyko F. Azacytidine inhibits RNA methylation at DNMT2 target sites in human cancer cell lines. Cancer Res. 2009;69:8127–8132. doi: 10.1158/0008-5472.CAN-09-0458. [DOI] [PubMed] [Google Scholar]
- 45.Karikó K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 46.Palii SS, Van Emburgh BO, Sankpal UT, Brown KD, Robertson KD. DNA methylation inhibitor 5-aza-2′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol Cell Biol. 2008;28:752–771. doi: 10.1128/MCB.01799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiziltepe T, et al. 5-Azacytidine, a DNA methyltransferase inhibitor, induces ATR-mediated DNA double-strand break responses, apoptosis, and synergistic cytotoxicity with doxorubicin and bortezomib against multiple myeloma cells. Mol Cancer Ther. 2007;6:1718–1727. doi: 10.1158/1535-7163.MCT-07-0010. [DOI] [PubMed] [Google Scholar]
- 48.Chung H, et al. Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell. 2018;172:811–824.e14. doi: 10.1016/j.cell.2017.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reikine S, Nguyen JB, Modis Y. Pattern recognition and signaling mechanisms of RIG-I and MDA5. Front Immunol. 2014;5:342. doi: 10.3389/fimmu.2014.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao L, et al. Cell-type-specific activation of the oligoadenylate synthetase-RNase L pathway by a murine coronavirus. J Virol. 2013;87:8408–8418. doi: 10.1128/JVI.00769-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reu FJ, et al. Overcoming resistance to interferon-induced apoptosis of renal carcinoma and melanoma cells by DNA demethylation. J Clin Oncol. 2006;24:3771–3779. doi: 10.1200/JCO.2005.03.4074. [DOI] [PubMed] [Google Scholar]
- 52.Lehman TA, et al. p53 mutations, ras mutations, and p53-heat shock 70 protein complexes in human lung carcinoma cell lines. Cancer Res. 1991;51:4090–4096. [PubMed] [Google Scholar]
- 53.Junk DJ, et al. Different mutant/wild-type p53 combinations cause a spectrum of increased invasive potential in nonmalignant immortalized human mammary epithelial cells. Neoplasia. 2008;10:450–461. doi: 10.1593/neo.08120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.