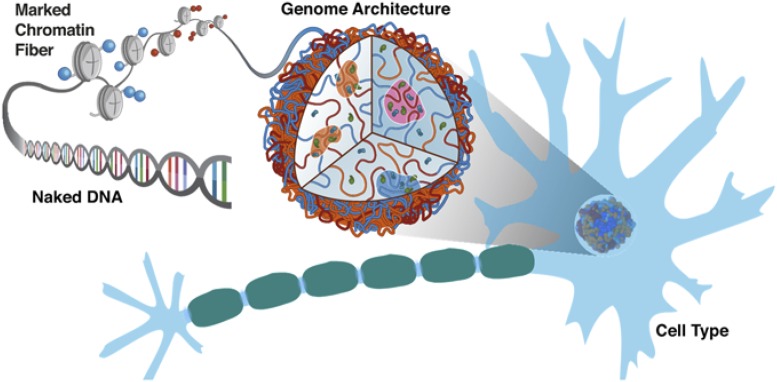
The eukaryotic genome is far more than a simple DNA polymer that encodes the sequences of proteins. The genome, or the ensemble of DNA together with all its associated proteins, is an information-processing machine that helps regulate the transcription of the very genes encoded within the DNA.
Chromosomes fold in three dimensions, bringing together segments of DNA separated by great genomic distances; this architecture plays an important role in controlling gene transcription. In different cell types within a single organism, the same chromosomes assume different spatial conformations. In PNAS, Bascom et al. (1) investigate how genes fold in three dimensions using data-driven physical simulations.
The elemental unit of chromosomal organization is the nucleosome: 147 bp of DNA tightly wrapped around eight histone proteins (two copies each of histones H2A, H2B, H3, and H4). Histone proteins undergo a vast repertoire of covalent posttranslational modifications on many of their residues, particularly along the flexible N-terminal domain typically referred to as the histone tail. These epigenetic modifications affect the structural dynamics of histones (2) and are known to be involved in gene regulation and to correlate with genome architecture (3). A DNA linker connects nucleosomes to one another; the length of the linker region is variable and also known to affect the structural organization of the genome. A third factor that affects the structural dynamics of the chromatin fiber is the binding of another protein of the histone family, the linker histone, to the DNA.
Bascom et al. (1) integrate the experimental data describing nucleosome positioning, histone tail acetylation, and linker histone binding to investigate the folding of a 55-kb chromosomal locus at nucleosome resolution. The locus chosen as a model system—the HOXC gene cluster—contains five genes encoding HOX proteins, master regulators of embryonic development. The choice of this locus is particularly interesting in light of the fact that for other genes involved in embryonic development, variations in 3D architecture have been observed to cause limb malformations in mammals (4). Bascom et al. (1) use experimental epigenetic data to build a detailed model of the gene hub in vivo in which nucleosomes are represented by a series of electrostatically charged beads, as are the histone tails and linker histone proteins. Linker DNA is instead represented by a wormlike chain whose length is also modeled based on experimental data. They then use a Monte Carlo scheme to simulate this model and study the structural ensemble of the HOXC gene hub and its biological functioning.
Although theoretical models have already shown that epigenetic profiles can be used to predict the global folding of chromosomes (5), Bascom et al. (1) advance the field by pushing the resolution of chromatin modeling to a level that allows us to study the inner workings of individual genes.
Using a mesoscale model, Bascom et al. (1) compute the structural ensemble of the HOXC gene hub from its basic physicochemical interactions, namely, polymer connectivity and steric and electrostatic interactions. Through physical simulations, they show how distinct epigenetic features cooperate to form an array of dynamic contacts bridging promoters in the HOXC gene hub. They find that histone tail acetylation leads to increased transient contacts between chromatin segments, supporting the hypothesis that phase separation of epigenetically modified chromatin may be one of the leading mechanisms of genome organization (6, 7). Bascom et al. also find that linker histone binding decreases long-range contacts. Interestingly, they observe that the combined presence of histone acetylation and linker histone binding instead promotes such long-range contacts. In the HOXC gene hub, these long-range transient DNA loops generate a contact probability map that exhibits “stripes” near promoter regions, a feature also observed in experimental contact probability maps (8) that is associated with transcriptional regulation.
In recent years, significant progress has been made in investigating nuclear architecture, leading to the generation of an unprecedented wealth of related experimental data. DNA–DNA proximity ligation assays on ensembles of cells report the frequency of contact between all pairs of genomic loci with kilobase resolution (3). At a lower resolution but within single cells, superresolution microscopy DNA tracing (9) as well as DNA cross-linking (10) report on the physical nature of contact domains and their shapes and the differences between homologous chromosomes. All of this knowledge is now being used to build realistic theoretical models of specific genes in specific cells, as opposed to modeling chromatin in its most ideal or generic form. The nucleosome resolution achieved in the study from Bascom et al. (1) finally allows the study of how individual genes fold and how gene folding affects gene regulation.
Proteins orchestrate the folding of genomes; as a consequence, protein positioning data obtained from chromatin immunoprecipitation is necessary to reconstitute the behavior of specific genomic loci. The patterns in which proteins bind along the DNA polymer determine chromosome conformation, much as the amino acid sequences of proteins determine their fold (Fig. 1). The molecular mechanism by which proteins act on DNA to fold it in three dimensions can be studied bottom-up with a physicochemical approach, as in the study from Bascom et al. (1). Alternatively, one could infer the energy landscape of gene folding from experimentally determined structural ensembles, using DNA–DNA ligation assays in the same way that X-ray crystallography has been used to crack the problem of protein folding (11). From the Bascom et al. study and other studies (6, 12–14), it is clear that chromosome structural ensembles are very different from those of proteins, with the latter dominated by one (or a few) native structures and the former characterized by transient contacts, more akin to a liquid with phase-separated compartments (6, 15) than to a crystal.
Fig. 1.
Naked DNA is decorated by proteins and epigenetic markings that differ from locus to locus and differ between cell types within the same organism. This ensemble of proteins that associates with the genome acts on DNA, shaping its 3D organization. In turn, the spatial architecture of the genome is involved in transcriptional regulation of the genes encoded in the DNA. Through data-driven physical simulations, Bascom et al. (1) show how epigenetic markers drive the formation of the HOXC gene hub. Image courtesy of Michele Di Pierro and Ryan Cheng (Rice University, Houston).
In PNAS, Bascom et al. investigate how genes fold in three dimensions using data-driven physical simulations.
The next few years reserve the challenge of combining the mesoscale approach of Bascom et al. (1) with data from immunoprecipitation, microscopy, and DNA ligation into a theoretical and computational framework to study the fluid structural ensembles of genes. This will also help us in developing a better understanding of the molecular origins of genome 3D organization.
In summary, the study from Bascom et al. (1) demonstrates that nucleosome-resolution modeling of specific genomic loci is now feasible. The mesoscale theoretical model of chromatin introduced in their study allows the investigation of the structural organization of genes in living cells and paves the way to studying the molecular mechanism by which 3D folding affects transcriptional regulation of genes. We look forward to the forthcoming development of a physical theory that integrates protein activity in the cellular nucleus, gene folding, and gene regulation.
Acknowledgments
M.D.P.’s research is supported by the Center for Theoretical Biological Physics sponsored by the National Science Foundation (Grants PHY-1427654 and NSF-CHE-1614101) and by the Welch Foundation (Grant C-1792).
Footnotes
The author declares no conflict of interest.
See companion article on page 4955.
References
- 1.Bascom GD, Myers CG, Schlick T. Mesoscale modeling reveals formation of an epigenetically driven HOXC gene hub. Proc Natl Acad Sci USA. 2019;116:4955–4962. doi: 10.1073/pnas.1816424116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Potoyan DA, Papoian GA. Energy landscape analyses of disordered histone tails reveal special organization of their conformational dynamics. J Am Chem Soc. 2011;133:7405–7415. doi: 10.1021/ja1111964. [DOI] [PubMed] [Google Scholar]
- 3.Rao SSP, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lupiáñez DG, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Pierro M, Cheng RR, Lieberman Aiden E, Wolynes PG, Onuchic JN. De novo prediction of human chromosome structures: Epigenetic marking patterns encode genome architecture. Proc Natl Acad Sci USA. 2017;114:12126–12131. doi: 10.1073/pnas.1714980114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Pierro M, Zhang B, Aiden EL, Wolynes PG, Onuchic JN. Transferable model for chromosome architecture. Proc Natl Acad Sci USA. 2016;113:12168–12173. doi: 10.1073/pnas.1613607113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jost D, Carrivain P, Cavalli G, Vaillant C. Modeling epigenome folding: Formation and dynamics of topologically associated chromatin domains. Nucleic Acids Res. 2014;42:9553–9561. doi: 10.1093/nar/gku698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vian L, et al. The energetics and physiological impact of cohesin extrusion. Cell. 2018;173:1165–1178.e20. doi: 10.1016/j.cell.2018.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nir G, et al. Walking along chromosomes with super-resolution imaging, contact maps, and integrative modeling. PLoS Genet. 2018;14:e1007872. doi: 10.1371/journal.pgen.1007872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan L, Xing D, Chang C-H, Li H, Xie XS. Three-dimensional genome structures of single diploid human cells. Science. 2018;361:924–928. doi: 10.1126/science.aat5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Pierro M, Cheng RR, Zhang B, Onuchic JN, Wolynes PG. Learning genomic energy landscapes from experiments. In: Tiana G, Giorgetti L, editors. Modeling the 3D Conformation of Genomes. CRC Press; Boca Raton, FL: 2019. [Google Scholar]
- 12.Fudenberg G, et al. Formation of chromosomal domains by loop extrusion. Cell Reports. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagano T, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanborn AL, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci USA. 2015;112:E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Pierro M, Potoyan DA, Wolynes PG, Onuchic JN. Anomalous diffusion, spatial coherence, and viscoelasticity from the energy landscape of human chromosomes. Proc Natl Acad Sci USA. 2018;115:7753–7758. doi: 10.1073/pnas.1806297115. [DOI] [PMC free article] [PubMed] [Google Scholar]