Abstract
Understanding protein kinase family members that lack key catalytic residues – or pseudokinases – is a major challenge in cell signaling. In this issue of Cell, Sreelatha et al. (2018) describe how one pseudokinase transfers adenosine monophosphate (AMP) rather than phosphate to protein substrates, revealing unexpected catalytic diversity for the kinase fold.
The protein kinase superfamily, or ‘kinome’ numbers over 500 proteins in humans that catalyze protein phosphorylation events involved in all facets of cell signaling. Many are now targets of important drugs, mostly in cancer. As with other enzyme families, however, function does not always follow form and approximately 10% of the human protein kinome lacks one or more key conserved catalytic residues – leading to their classification as ‘pseudokinases’ (Murphy et al., 2017). Several studies in the 2000s showed how some pseudokinases nonetheless retain phosphotransfer activity – adopting unique structures to compensate for the noted deficits. Perhaps the most famous is WNK kinase (Min et al., 2004). Named ‘with no lysine (K)’ because it lacks this key residue, WNK activity is rescued by a lysine elsewhere in the sequence. Many others have been reported not even to bind ATP (Murphy et al., 2014), however, arguing that they may truly be catalytically ‘dead’. Pseudokinases of this type are thought to function allosterically, often as scaffolds for regulated complex formation (Murphy et al., 2017). New structural studies are advancing our understanding of allosteric pseudoenzyme function, which can take many forms. But, we should not ignore the possibility that some pseudoenzymes may have been repurposed to catalyze other reactions. Indeed, in this issue of Cell, work by Sreelatha et al. (2018) reminds us not to let our thinking about possible enzyme activities be constrained by the predicted protein fold.
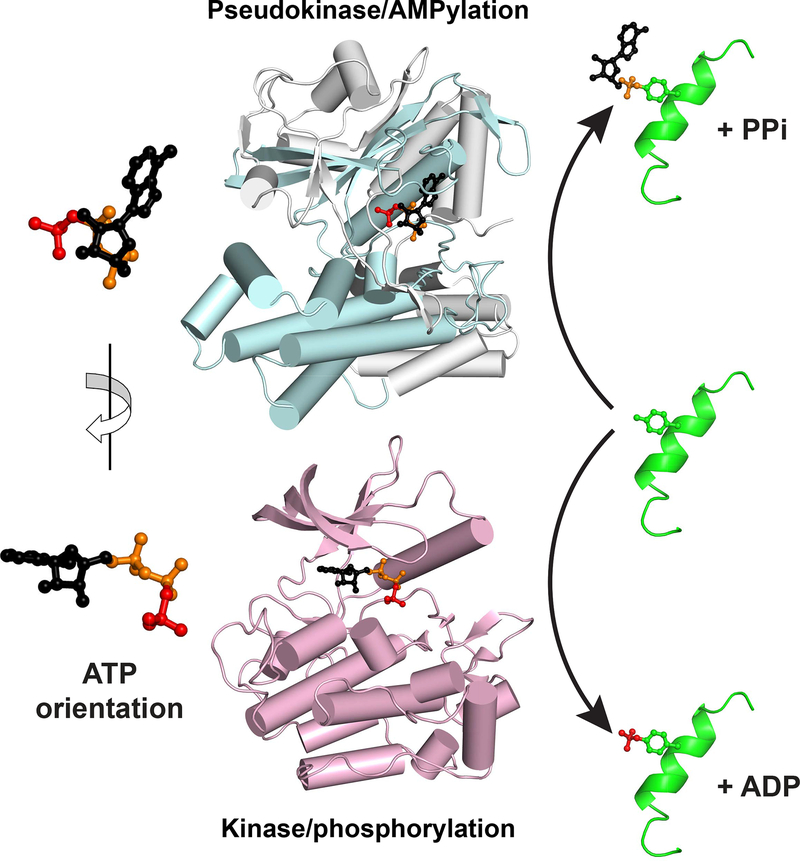
The work of Sreelatha et al. reveals that, rather that transferring the terminal γ-phosphate of ATP to its protein substrates, the conserved pseudokinase selenoprotein-O (SelO) instead transfers AMP – as part of a cellular response to oxidative stress. SelO is one of 25 human proteins that incorporates the amino acid selenocysteine (Sec) into its polypeptide chain, and was predicted to adopt a kinase-like fold (Dudkiewicz et al., 2012). The crucial catalytic base aspartate in the kinase ‘His-Arg-Asp’ motif is replaced with a valine in SelO proteins, classifying SelO as a pseudokinase. Sreelatha et al. determined a 2.3 Å crystal structure of a bacterial SelO ortholog bound to the non-hydrolyzable ATP analog AMP-PNP. The structure reveals a clear kinase fold at the protein’s core – as predicted – set within a larger structure through extensions at the N- and C-termini (Figure 1). The bound ATP analog is located in the predicted active site but – remarkably – is flipped almost 180° relative to its orientation in all other known protein kinase structures, and is held in place by unique insertions in loops between secondary structure elements. The flipped ATP orientation led Sreelatha et al. to hypothesize that – instead of transferring the terminal phosphate – SelO might transfer adenosine monophosphate (AMP) onto substrate proteins. Consistent with this, they found that SelO from humans, yeast, and E. coli could incorporate radioactive label from the α- but not γ-phosphate of ATP. Parallel mass spectrometry studies further demonstrated AMPylation of threonines in SelO proteins and other substrates. They also noticed that SelO has a potential catalytic base aspartate (D252) adjacent to the α-phosphate of the bound AMP-PNP that could ‘replace’ the traditional catalytic base aspartate in the kinase ‘His-Arg-Asp’ motif (which lies close to the γ-phosphate). Indeed, mutation of D252 to alanine in the bacterial SelO protein abolishes its AMPylation activity.
Figure 1. ATP Binds to Kinase Domain Folds in Flipped Orientations for AMPylation and Phosphorylation.
The structure of SelO (upper), from PDB entry 6EAC, is show in pale cyan and grey, with the kinase fold colored cyan and the SelO-specific components grey. In the lower panel, the structure of the insulin receptor kinase in its active conformation (PDB entry 1IR3) is shown in pale magenta. Bound ATP is shown for each protein, with the γ-phosphate colored red, the α- and β-phosphates orange, and the rest of the molecule black. As depicted with the magnified molecules at left, the ATP orientation is flipped by ~180° about a vertical axis between the two proteins. As a result, whereas the γ-phosphate (red) faces ‘out’ of the kinase active site to be appended to substrate (right), AMP instead faces out of the SelO active site for substrate AMPylation (top right).
AMPylation is not a new activity. It is also called adenylylation, occurs primarily on serine, threonine, and tyrosine side-chains, and is already known also to be catalyzed by the largely helical adenylyl transferase (ATase) and ‘filamentation induced by cAMP’, or Fic domains (Casey and Orth, 2018). Important roles for AMPylation in bacterial homeostasis have been described, particularly in regulation of metabolism and cell stress. Bacterial pathogens also employ AMPylation to modify their host cell activities. For example, Legionella pneumophila enzymes AMPylate Rab-family small G-proteins to promote formation of replication vacuoles that protect the bacteria within the infected host (Müller et al., 2010). In eukaryotes, AMPylation is employed to regulate activity of the endoplasmic reticulum (ER) BiP chaperone with changing ER stress (Preissler et al., 2017). Sreelatha et al. suggest that the kinase-fold AMPylating enzyme SelO is also involved in cellular responses to stress – in this case oxidative stress. They showed that the activity of purified bacterial SelO can be modulated by redox regulation of an intramolecular disulfide bond. Moreover, in a search for E. coli proteins that are AMPylated by SelO in vitro and in vivo, they found the glutathione-dependent redox enzyme glutaredoxin (grx). They hypothesize that, by AMPylating a tyrosine close to the redox active site in grx, SelO reduces its ability to reverse S-glutathionylation of proteins – protecting against oxidative stress. Indeed, loss of SelO reduces levels of global protein S-glutathionylation in yeast and E. coli. Sreelatha et al. also show that SelO levels are elevated in S. cerevisiae grown on non-fermentable carbon sources, and that the protein AMPylation activity of SelO protects yeast from oxidative stress – apparently at least in part through grx-mediated effects on protein S-glutathionylation.
Just as SelO represents a kinase fold that can catalyze AMPylation, so are some Fic domain-containing AMPylating enzymes capable of catalyzing phosphorylation, UMPylation, or phosphocholination – depending on the particular protein and substrate (Garcia-Pino et al., 2014). Intriguingly, and paralleling the comparison for the kinase fold shown in Figure 1, a Fic domain that phosphorylates targets binds ATP in an orientation flipped compared with that seen for Fic domains that catalyze AMPylation (Garcia-Pino et al., 2014). Adding further to this intrigue, a Legionella de-AMPylating enzyme shows strong structural similarity to serine/threonine specific protein phosphatases (Chen et al., 2013). The lesson from all of these studies, underlined in bold by the paper by Sreelatha et al., is that we cannot simply judge an enzyme by its fold. Non-canonical activities have grown as a theme in kinase – and enzyme – biology in recent years (Murphy et al., 2017), but catalysis of alternative reactions has been less of a focus than non-catalytic functions. The story of SelO – and indeed of Fic domains – suggests that this might be short-sighted and that we should anticipate several pseudokinases, pseudophosphatases, and other pseudoenzymes rising from the dead with fascinating lessons about the evolution of biological chemistry.
ACKNOWLEDGMENTS
This material is based upon work supported in part by the National Science Foundation Graduate Research Fellowship under Grant No. DGE1122492 to J.B.S. and the National Institutes of Health (R35-GM122485) to M.A.L.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Casey AK, and Orth K (2018). Enzymes Involved in AMPylation and deAMPylation. Chem. Rev 118, 1199–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Tascón I, Neunuebel MR, Pallara C, Brady J, Kinch LN, Fernández-Recio J, Rojas AL, Machner MP, and Hierro A (2013). Structural basis for Rab1 de-AMPylation by the Legionella pneumophila effector SidD. PLoS Pathog. 9, e1003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudkiewicz M, Szczepińska T, Grynberg M, and Pawłowski K (2012). A novel protein kinase-like domain in a selenoprotein, widespread in the tree of life. PLoS One 7, e32138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Pino A, Zenkin N, and Loris R (2014). The many faces of Fic: structural and functional aspects of Fic enzymes. Trends Biochem. Sci 39, 121–129. [DOI] [PubMed] [Google Scholar]
- Min X, Lee BH, Cobb MH, and Goldsmith EJ (2004). Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension. Structure 12, 1303–1311. [DOI] [PubMed] [Google Scholar]
- Müller MP, Peters H, Blümer J, Blankenfeldt W, Goody RS, and Itzen A (2010). The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329, 946–949. [DOI] [PubMed] [Google Scholar]
- Murphy JM, Mace PD, and Eyers PA (2017). Live and let die: insights into pseudoenzyme mechanisms from structure. Curr. Opin. Struct. Biol 47, 95–104. [DOI] [PubMed] [Google Scholar]
- Murphy JM, Zhang Q, Young SN, Reese ML, Bailey FP, Eyers PA, Ungureanu D, Hammaren H, Silvennoinen O, Varghese LN, et al. (2014). A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem. J 457, 323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissler S, Rohland L, Yan Y, Chen R, Read RJ, and Ron D (2017). AMPylation targets the rate-limiting step of BiP’s ATPase cycle for its functional inactivation. Elife 6, e29428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreelatha A, Yee SS, Lopez VA, Park BC, Kinch L, Pilch S, Servage KA, Zhang J, Jiou J, Karasiewicz M, Łobocka M, Grishin N, Orth K, Kucharczyk R, Pawłowski K, Tomchick DR, and Tagliabracci VS (2018) Protein AMPylation by an evolutionarily conserved pseudokinase. Cell 175, this issue, XXX–YYY [DOI] [PMC free article] [PubMed] [Google Scholar]