Abstract
Background
The recent description of the first plasmid-mediated colistin-resistant gene mcr-1, conferring transferable and low-level resistance to colistin, raised concern about the need to implement a rapid and reliable screening method to detect colistin-resistant clinical isolates. The only valid method to assess the MIC of colistin is the broth microdilution according to the joint CLSI-EUCAST Polymyxin Breakpoints Working Group. UMIC Colistine is a ready-to-use broth microdilution kit developed to easily assess colistin MIC by proposing unitary polystyrene strips containing 11 concentrations of dehydrated colistin. Here, we evaluated the UMIC Colistine kit on 235 Gram-negative rods (176 Enterobacterales, including 70 harboring a mcr gene, and 59 non-fermentative), through comparison to the reference broth microdilution method prepared in accordance with EN ISO 20776-1:2006 standard. Reproducibility of the UMIC Colistine was assayed with the three recommended quality control strains E. coli ATCC 25922, E. coli NCTC 13846 (mcr-1 positive), and P. aeruginosa ATCC 27853, as for stability testing.
Results
Categorical agreement was 100% with 63.4% (n = 149) of colistin-resistant strains, and 36.6% (n = 86) of colistin-susceptible strains with both methods (S ≤ 2 μg/mL and R > 2 μg/mL). No major error or very major error was reported. Essential agreement was 94.0% (n = 221), and 100% for detection of colistin-resistant strains as compared to the reference method. Pearson’s correlation between UMIC Colistine and the reference method was 0.98. Reproducibility of the UMIC Colistine system was 97.8% with MICs of the quality control strains within the target ranges. However, some isolates had lower MIC with UMIC Colistine, but that did not change their categorization as colistin-susceptible, and this phenomenon should be further explored.
Conclusions
The UMIC Colistine kit is an easy to perform unitary device that showed excellent results when compared to the reference method. The UMIC Colistine system is a rapid and reliable broth microdilution method that is suitable to assess the colistin MIC of clinical isolates in clinical microbiology laboratories.
Keywords: Colistin, Susceptibility testing, Polymyxin, UMIC, Enterobacteriaceae, Mcr-1, Broth microdilution, Detection method, Resistance, MIC
Background
The emergence of multi-drug resistant Gram-negative bacteria is a worldwide phenomenon and has led to the revival of old antibiotics as last resort treatments, including polymyxins [1]. Until 2015, all the described polymyxin-resistant genes were chromosomally encoded, including those coding for the two-component systems PmrA/PmrB, PhoP/PhoQ, and their negative regulator MgrB in the Klebsiella pneumoniae species [2]. In November 2015, the first plasmid-mediated colistin resistance gene was described and named mcr-1 [3]. The transferable mcr-1 gene has been detected in samples from all over the world and from various human and animal origins [4]. This discovery was followed by the description of other mcr genes: mcr-2 to mcr-8 [5–12]. This mobile colistin resistance that confers low levels of resistance with Minimal Inhibitory Concentrations (MIC) of colistin around 4 μg/mL [3] raised concerned about the capacity to detect colistin resistance in clinical microbiology laboratories [13].
Indeed, the clinical breakpoint of colistin, established by both the European Committee of Antibiotic Susceptibility Testing (EUCAST) and the Clinical and Laboratory Standards Institute (CLSI), is 2 μg/mL (resistant > 2 μg/mL and susceptible ≤2 μg/mL) for Enterobacteriaceae, P. aeruginosa and Acinetobacter spp. [14, 15]. More specifically, the CLSI recommends an Epidemiological Cut-off Value (ECV) and not a clinical breakpoint for the following Enterobacteriaceae species: Enterobacter aerogenes, Enterobacter cloacae, Escherichia coli, Klebsiella pneumoniae and Raoultella ornithinolytica.
Currently, the only available Antibiotic Susceptibility Testing (AST) method for colistin according to the joint CLSI-EUCAST Polymyxin Breakpoints Working Group is the EN ISO 20776-1:2006 standard Broth Microdilution method (BMD) [16] that has to be used with cation-adjusted Mueller-Hinton broth medium (MH2), untreated polystyrene trays without additives and colistine sulphate salt [17]. As BMD is a time-consuming technique, it is not suitable for clinical microbiology laboratories in public hospitals, and the establishment of a rapid and reliable method for colistin MIC determination is obviously warranted to control the spread of colistin resistance [13].
The UMIC Colistine kit (Biocentric, Bandol, France) was developed to easily determine colistin MIC with a ready-to-use device based on BMD method. The UMIC Colistine kit consists of unitary 12-wells polystyrene strips with 11 wells containing a range of dehydrated colistin concentrations from 0.06 to 64 μg/mL (with 2-fold dilutions between 2 consecutive wells), and one well for growth control.
Here, we evaluated the UMIC Colistine strips by comparison to the reference method prepared in accordance with the EN ISO 20776-1:2006 standard.
Methods
Bacterial strains
A total of 235 bacterial strains were used in this study, including 162 Enterobacteriaceae (77 Escherichia coli, 50 Klebsiella pneumoniae, 4 Klebsiella oxytoca, 18 Enterobacter cloacae, 4 Enterobacter aerogenes, 4 Enterobacter asburiae and 5 Salmonella enterica), 14 intrinsic colistin-resistant genera of Enterobacterales (9 Hafnia alvei, 1 Proteus mirabilis, 1 Morganella morganii, 1 Providencia alcalifaciens, 1 Providencia rettgeri and 1 Serratia marcescens), and 59 non-fermentative isolates (31 Pseudomonas sp., 18 Acinetobacter sp. and 10 Stenotrophomonas maltophilia) (Table 1) [18–23]. These microorganisms were isolated as part of standard care of patients or animals. 85 colistin-resistant isolates were well-characterized from previous studies, including 70 harboring a mcr gene (61 mcr-1, 1 mcr-2 and 8 mcr-3), with MICs ranging from 4 to 64 μg/mL, and their genotype are detailed in Table 1.
Table 1.
Colistin MICs obtained by broth microdilution comparing UMIC Colistine to the reference method (μg/mL). Discrepancies are indicated in bold
| Bacterial species | Isolates | Samples origins | Genotype | Colistin MIC (μg/mL) | |
|---|---|---|---|---|---|
| Reference | UMIC | ||||
| Escherichia coli | NCTC 13883 | Human, UK | mcr-1 | 4 | 4 |
| SE65 | Human, Algeria | mcr-1 | 4 | 4 | |
| 117R | Human, Saudi arabia | mcr-1 | 4 | 4 | |
| 1R2013 | Human, Saudi arabia | mcr-1 | 8 | 8 | |
| 1R 2104 | Human, Saudi arabia | mcr-1 | 8 | 4 | |
| 44A | Human, Saudi arabia | mcr-1 | 4 | 4 | |
| 6R | Human, Saudi arabia | mcr-1 | 4 | 4 | |
| 85R | Human, Saudi arabia | mcr-1 | 4 | 8 | |
| 95R | Human, Saudi arabia | mcr-1 | 4 | 8 | |
| 96R | Human, Saudi arabia | mcr-1 | 8 | 8 | |
| 134R | Human, Saudi arabia | mcr-1 | 4 | 4 | |
| 143R | Human, Saudi arabia | mcr-1 | 8 | 8 | |
| LH121 | Human, Laos | mcr-1 | 4 | 4 | |
| LH140 | Human, Laos | mcr-1, phoQ E375K | 8 | 8 | |
| LH257 | Human, Laos | mcr-1 | 16 | 8 | |
| LH57 | Human, Laos | mcr-1, phoQ E375K | 4 | 4 | |
| LH1 | Human, Laos | mcr-1 | 4 | 4 | |
| LH30 | Human, Laos | mcr-1 | 4 | 4 | |
| LH345.2 | Human, Laos | mcr-1 | 4 | 4 | |
| TH214 | Human, Thailand | mcr-1 | 4 | 8 | |
| TH99 | Human, Thailand | mcr-1 | 16 | 16 | |
| TH169.1 | Human, Thailand | mcr-1 | 4 | 4 | |
| TH259.1 | Human, Thailand | mcr-1 | 4 | 4 | |
| TH33.1 | Human, Thailand | mcr-1 | 4 | 4 | |
| TH44.1 | Human, Thailand | mcr-1 | 4 | 4 | |
| TH66.1 | Human, Thailand | mcr-1 | 4 | 8 | |
| TH134.1 | Human, Thailand | mcr-1 | 4 | 4 | |
| FHM128.1 | Human, France | mcr-1 | 4 | 4 | |
| FHM66.1 | Human, France | mcr-1 | 4 | 4 | |
| P4.5 t3 (4) | Pig, Lebanon | mcr-1 | 8 | 4 | |
| P1.2 (16) | Pig, Lebanon | mcr-1 | 4 | 4 | |
| P1.38 (18) | Pig, Lebanon | mcr-1 | 4 | 4 | |
| P1.5 t2 (8) | Pig, Lebanon | mcr-1 | 4 | 4 | |
| P2.12 (13) | Pig, Lebanon | mcr-1 | 4 | 4 | |
| P2.13 t1 (11) | Pig, Lebanon | mcr-1 | 4 | 4 | |
| P2.13 t2 (12) | Pig, Lebanon | mcr-1 | 4 | 4 | |
| P2.3 t2 (15) | Pig, Lebanon | mcr-1 | 4 | 4 | |
| P2.6 (14) | Pig, Lebanon | mcr-1 | 4 | 4 | |
| P4.21 t1 (7) | Pig, Lebanon | mcr-1 | 4 | 4 | |
| P4.5 t1 (1) | Pig, Lebanon | mcr-1 | 4 | 4 | |
| 12 | Environmental, Algeria | mcr-1 | 8 | 8 | |
| 14 | Environmental, Algeria | mcr-1 | 8 | 8 | |
| 28 | Environmental, Algeria | mcr-1 | 8 | 8 | |
| 31 | Environmental, Algeria | mcr-1 | 8 | 8 | |
| 3 | Environmental, Algeria | mcr-1 | 4 | 4 | |
| 5 | Environmental, Algeria | mcr-1 | 4 | 4 | |
| 10 | Environmental, Algeria | mcr-1 | 4 | 8 | |
| 15 | Environmental, Algeria | mcr-1 | 4 | 4 | |
| 39 | Environmental, Algeria | mcr-1 | 4 | 4 | |
| MCR-2 | Pig, Belgium | mcr-2 | 4 | 4 | |
| 16 | Environmental, Algeria | mcr-3 | 8 | 4 | |
| 8 | Environmental, Algeria | mcr-3 | 4 | 8 | |
| FHA102 | Human, France | pmrB A159V | 16 | 16 | |
| FHM19 | Human, France | pmrB P7-Q12 del (6 aa) | 16 | 8 | |
| FHA113 | Human, France | pmrB T156K | 8 | 8 | |
| NH94 | Human, Nigeria | pmrB I92 insertion | 16 | 16 | |
| LH345.1 | Human, Laos | 4 | 4 | ||
| LH53 | Human, Laos | 4 | 4 | ||
| TH176 | Human, Thailand | 8 | 8 | ||
| TH169.5 | Human, Thailand | 4 | 4 | ||
| LB4 | Human, France | 8 | 8 | ||
| 235 | Chicken, Algeria | mcr-1 | 4 | 4 | |
| P6 | Pig, Laos | mcr-1 | 4 | 4 | |
| P10 | Pig, Laos | mcr-1 | 4 | 8 | |
| P17 | Pig, Laos | mcr-1 | 8 | 4 | |
| P7 | Pig, Laos | 4 | 4 | ||
| ATCC 25922 | Human, Unknown | 0.5 | 0.25 | ||
| ATCC 35218 | Unknown | 1 | 0.5 | ||
| EC1 | Human, France | 0.5 | 0.25 | ||
| EC2 | Human, France | 0.5 | 0.5 | ||
| EC3 | Human, France | 0.5 | 0.25 | ||
| EC4 | Human, France | 1 | 0.5 | ||
| LH165S* | Human, Laos | 1 | 0.25 | ||
| TH77S | Human, Thailand | 0.5 | 0.25 | ||
| 282S | Chicken, Algeria | 1 | 1 | ||
| 161 | Chicken, Algeria | 1 | 0.5 | ||
| NDM-1 | Human, Israël | 0.5 | 0.25 | ||
| Klebsiella pneumoniae | FHA60 | Human, France | mcr-1 | 16 | 16 |
| FHM128 | Human, France | mcr-1 | 16 | 8 | |
| 119R | Human, Saudi arabia | mcr-1 | 8 | 8 | |
| LH131 | Human, Laos | mcr-1, mgrb stop | 32 | 32 | |
| LH61 | Human, Laos | mcr-1, mgrB sub A14S | 32 | 32 | |
| LH17 | Human, Laos | mcr-1, pmrB T157P | 32 | 32 | |
| LH92 | Human, Laos | mcr-1 | 16 | 16 | |
| LH94 | Human, Laos | mcr-3 | 32 | 32 | |
| TH68 | Human, Thailand | mcr-3 | 64 | 64 | |
| LH102 | Human, Lao | mcr-3 | 16 | 32 | |
| LH375 | Human, Lao | mcr-3 | 16 | 16 | |
| TH114 | Human, Thailand | mcr-3 | 16 | 16 | |
| TH164 | Human, Thailand | mcr-3 | 16 | 16 | |
| LB1 | Human, France | mgrB Stop | 64 | 64 | |
| FHM169 | Human, France | mgrB Stop | 16 | 16 | |
| LH12 | Human, Laos | mgrB Stop | 32 | 32 | |
| TH28 | Human, Thailand | mgrB IS2 | 32 | 32 | |
| TH54 | Human, Thailand | pmrB T157P | 16 | 16 | |
| TH224 | Human, Thailand | pmrB T157P | 8 | 16 | |
| TH205 | Human, Thailand | 8 | 8 | ||
| FHM120 | Human, France | 32 | 32 | ||
| FHA105 | Human, France | 64 | 64 | ||
| FHM77 | Human, France | 16 | 16 | ||
| LB3 | Human, France | > 64 | > 64 | ||
| SB11R | Human, France | > 64 | > 64 | ||
| SB12R | Human, France | > 64 | > 64 | ||
| LH140 | Human, Laos | 64 | 64 | ||
| KP1PC | Human, France | 16 | 16 | ||
| KP2PC | Human, France | 16 | 16 | ||
| 4321 | Human, UK | 32 | 32 | ||
| K39 | Human, Greece | > 64 | 64 | ||
| K76 | Human, Greece | > 64 | > 64 | ||
| 1172/0 | Human, Greece | 32 | 32 | ||
| 7E | Human, Greece | 32 | 32 | ||
| 18E | Human, Greece | 16 | 16 | ||
| 28E | Human, Greece | 16 | 16 | ||
| 9980 | Human, Greece | 16 | 16 | ||
| K77 | Human, Greece | 16 | 16 | ||
| 1E | Human, Greece | 8 | 8 | ||
| KAT3 | Human, Greece | 8 | 8 | ||
| 2017–10 | Human, Greece | 1 | 0.5 | ||
| 1678 | Human, Greece | 0.5 | 1 | ||
| 56 | Human, Greece | 0.5 | 0.25 | ||
| K72 | Human, Greece | 0.5 | 0.5 | ||
| KP1 | Human, France | 0.5 | 0.25 | ||
| KP6 | Human, France | 0.5 | 0.25 | ||
| TH28S | Human, Thailand | 0.5 | 0.25 | ||
| CIP 82.91 | Unknown | 0.25 | 0.25 | ||
| LB2* | Human, France | 1 | 0.25 | ||
| ATCC 700603 | Human, UK | 0.5 | 0.25 | ||
| Klebsiella oxytoca | FHA41 | Human, France | mgrB IS1 | 32 | 64 |
| FHA124 | Human, France | 16 | 32 | ||
| TH44* | Human, Thailand | 0.5 | 0.125 | ||
| KOX1 | Human, France | 0.5 | 0.25 | ||
| Enterobacter aerogenes | EA1509E | Human, France | pmrA G157A | > 64 | > 64 |
| SB7R | Human, France | 32 | 16 | ||
| EAE1 | Human, France | 0.5 | 0.25 | ||
| EAE2 | Human, France | 1 | 0.5 | ||
| Enterobacter asburiae | LH74 | Human, Laos | > 64 | > 64 | |
| TH66 | Human, Thailand | 32 | 64 | ||
| 1502 | Human, France | 0.5 | 0.25 | ||
| 1503 | Human, France | 0.5 | 0.25 | ||
| Enterobacter cloacae | SB1 | Human, France | mcr-1 | 4 | 4 |
| NH131 | Human, Nigeria | > 64 | > 64 | ||
| NH132 | Human, Nigeria | > 64 | > 64 | ||
| NH52 | Human, Nigeria | > 64 | > 64 | ||
| SB5R | Human, France | > 64 | > 64 | ||
| SB6R | Human, France | > 64 | > 64 | ||
| SB10R | Human, France | > 64 | 64 | ||
| SB4R | Human, France | 64 | 64 | ||
| TH66 | Human, Thailand | 32 | 32 | ||
| SB3R* | Human, France | 2 | 0.25 | ||
| SB2R | Human, France | 2 | 2 | ||
| SB5S | Human, France | 2 | 1 | ||
| SB1S* | Human, France | 1 | 0.25 | ||
| SB2S* | Human, France | 1 | 0.25 | ||
| SB3S* | Human, France | 1 | 0.25 | ||
| P7698* | Human, France | 1 | 0.25 | ||
| NH151 | Human, Nigeria | 0.5 | 0.25 | ||
| NH74 | Human, Nigeria | 0.5 | 0.25 | ||
| Salmonella enterica | 100RC3 | Human, Saudi Arabia | pmrB deletion (12aa) | 8 | 8 |
| 65R | Human, Saudi Arabia | pmrB deletion (12aa) | 8 | 8 | |
| 122R | Human, Saudi Arabia | 1 | 0.5 | ||
| 108R | Human, Saudi Arabia | 1 | 0.5 | ||
| 10A | Human, Saudi Arabia | 1 | 1 | ||
| Hafnia alvei | B42 | Bird, France | 8 | 8 | |
| P516 | Human, France | 8 | 8 | ||
| A63 | Bird, France | 4 | 4 | ||
| B11 | Bird, France | 4 | 8 | ||
| B21 | Bird, France | 4 | 8 | ||
| B47 | Bird, France | 4 | 4 | ||
| B59 | Bird, France | 4 | 4 | ||
| B02 | Bird, France | 4 | 4 | ||
| B04 | Bird, France | 4 | 4 | ||
| Morganella morgannii | FHA60 | Human, France | > 64 | > 64 | |
| Proteus mirabilis | NDM-1 | Human, Israel | > 64 | > 64 | |
| Providencia alcalifaciens | TH66 | Human, Thailand | > 64 | > 64 | |
| Providencia rettgeri | TH66 | Human, Thailand | > 64 | > 64 | |
| Serratia marcescens | P6 | Chicken, Algeria | > 64 | > 64 | |
| Stenotrophomonas maltophilia | SM10 | Human, France | > 64 | > 64 | |
| SM7 | Human, France | 32 | 16 | ||
| SM8 | Human, France | 32 | 32 | ||
| SM9 | Human, France | 32 | 32 | ||
| SM6 | Human, France | 16 | 16 | ||
| SM4 | Human, France | 8 | 8 | ||
| SM5 | Human, France | 8 | 8 | ||
| SM2 | Human, France | 4 | 4 | ||
| SM3 | Human, France | 4 | 4 | ||
| SM1 | Human, France | 1 | 1 | ||
| Pseudomonas aeruginosa | FHM-PACOLR1 | Human, France | > 64 | > 64 | |
| ATCC 27853 | Human, unknown | 1 | 1 | ||
| FHM_PA7 | Human, France | 1 | 1 | ||
| FHM-PA2 | Human, France | 2 | 2 | ||
| FHM-PA3 | Human, France | 2 | 2 | ||
| FHM-PA4 | Human, France | 1 | 1 | ||
| FHM-PA5 | Human, France | 1 | 1 | ||
| FHM-PA6 | Human, France | 1 | 1 | ||
| PA1 | Human, France | 0.5 | 0.5 | ||
| PA2 | Human, France | 1 | 1 | ||
| PA3 | Human, France | 1 | 2 | ||
| PA4 | Human, France | 0.5 | 0.25 | ||
| PA5 | Human, France | 1 | 1 | ||
| PA6 | Human, France | 0.5 | 0.25 | ||
| PA7 | Human, France | 1 | 1 | ||
| Pseudomonas putida | AEM06 | Environmental, France | 1 | 0.5 | |
| AEM10 | Environmental, France | 1 | 0.5 | ||
| AEM15 | Environmental, France | 1 | 0.5 | ||
| ETP11 | Environmental, France | 0.5 | 0.5 | ||
| AEM08 B | Environmental, France | 0.5 | 0.5 | ||
| AEM12 | Environmental, France | 0.5 | 0.5 | ||
| AEM13 | Environmental, France | 0.5 | 0.5 | ||
| AEM16 | Environmental, France | 0.5 | 0.5 | ||
| AEM17 A | Environmental, France | 0.5 | 0.5 | ||
| AEM17 B | Environmental, France | 0.5 | 0.25 | ||
| AEM19 | Environmental, France | 0.5 | 0.25 | ||
| PLC009 | Environmental, France | 0.5 | 0.25 | ||
| Pseudomonas stutzeri | AEM05 | Environmental, France | 0.25 | 0.5 | |
| Pseudomonas sp. | AEM08 A | Environmental, France | 1 | 0.5 | |
| AEM07 | Environmental, France | 0.5 | 0.5 | ||
| AEM20 | Environmental, France | 0.25 | 0.5 | ||
| Acinetobacter baumannii | ABIsac_ColiR | Human, France | pmrA E8D | 64 | 64 |
| AB3 | Human, France | 4 | 4 | ||
| AB9* | Human, France | 2 | 0.25 | ||
| 4322 | Human, UK | 1 | 1 | ||
| Big | Human, Iran | 1 | 0.5 | ||
| Small | Human, Iran | 1 | 0.5 | ||
| AB1 | Human, France | 1 | 0.5 | ||
| AB2 | Human, France | 1 | 0.5 | ||
| AB4 | Human, France | 1 | 0.5 | ||
| AB5 | Human, France | 1 | 0.5 | ||
| NDM-1* | Human, Lebanon | 1 | 0.25 | ||
| AB6* | Human, France | 1 | 0.25 | ||
| AB8* | Human, France | 1 | 0.25 | ||
| AB10* | Human, France | 1 | 0.25 | ||
| CR17 | Human, Spain | 0.5 | 0.5 | ||
| Acinetobacter nosocomialis | ABG13S | Human, Spain | 1 | 0.5 | |
| Acinetobacter pitti | G867* | Human, France | 1 | 0.25 | |
| Acinetobacter sp. | LH213 | Human, Laos | 1 | 1 | |
*Those strains have been tested in triplicate, details are explained in the text
The three Quality Control (QC) strains recommended by EUCAST for colistin susceptibility testing, Escherichia coli ATCC 25922, P. aeruginosa ATCC 27853 and E. coli NCTC 13846 (mcr-1 positive) were included in the study.
Broth microdilution plates preparation
The BMD reference method was prepared accordingly to the EN ISO 20776-1:2006 standard, with a stock solution of colistin prepared from colistin sulphate salt (MP Biomedicals, Illkirch, France) that was adjusted accordingly to the CLSI 2017 M100 guidelines [15]. The BBL™ Mueller-Hinton II Broth (Becton-Dickinson, Heidelberg, Germany) was used as MH2 for reference method and prepared following the manufacturer’s instructions. The stock solution of colistin was diluted in the MH2 medium in order to fill the 96-well polystyrene plates (ref. 3799, Corning, Hazebrouck, France) following the same scheme of UMIC Colistine strips (0.06 to 64 μg/mL of colistin), with a growth control well containing only MH2 medium. Stock solution and plates were freshly prepared every test day.
Colistin MIC testing
Each isolate was inoculated in parallel in both systems from the same 0.5 McFarland (McF) suspension in such a way as to obtain the same final inoculum of 5 × 105 CFU/mL (Colony Forming Unit / mL) or 5 × 104 CFU/well, by a 200-fold dilution. For UMIC Colistine, the 0.5 McF suspension was directly diluted in one of the MH2 tubes provided with the kits, of which 100 μL were added in each well of a unitary strip. For the reference method, an intermediate dilution was performed in the prepared MH2 medium then diluted in the 12 wells of a row of a freshly prepared plate. The QC strain E. coli NCTC 13846 was used as quality control each day of testing.
Results were read after incubation in aerobic atmosphere at 35 ± 1 °C for 18 ± 2 h, directly or after adding 50 μL of a prepared 5 mg/mL iodonitrotetrazolium chloride solution (Sigma-Aldricht, Illkirch, France), and could also be analyzed using ELX808 Ultra Microplate Readers (Biotek Instruments, Winooski, USA).
Reproducibility of UMIC Colistine
The reproducibility of the UMIC Colistine kit was assessed by testing the three QC strains in triplicate on 5 different days by 3 different laboratories, resulting in 45 values for each strain. Subcultures of each day’s samples were performed from the same primary culture as recommended.
Data analysis
Data were analyzed according to the EN ISO 20776-2:2007 standard [24] and EUCAST guidelines (resistant > 2 μg/mL or susceptible ≤2 μg/mL), using the MIC obtained with the prepared BMD method as reference MIC.
Categorical Agreement (CA, same clinical categorization), Essential Agreement (EA, MIC within ±1 doubling dilution from the reference MIC), Major Errors (ME, false resistant) and Very Major Errors (VME, false susceptible) were calculated by comparing the MICs obtained with UMIC Colistine to the reference MICs. Isolates with discrepant results were retested at least twice, and if not corrected, the values obtained from the first assay were kept. To be validated, the UMIC Colistine device should met the following criteria: CA ≥ 90%, EA ≥ 90%, ME ≤3% and VME ≤ 3%.
The correlation between the two systems was calculated using the Pearson method (value 128 was retained when the MIC was > 64 μg/mL).
The expected colistin MIC ranges for the QC strains are 0.25–1 μg/mL for E. coli ATCC 25922, 0.5–2 μg/mL for P. aeruginosa ATCC 27853 and 4 μg/mL with occasionally accepted values 2 and 8 μg/mL for E. coli NCTC 13846 [25]. The MIC values obtained for the three QC strains have to be in the acceptable ranges for ≥95% and reproducibility has to be comprised between ± one dilutions of the mode for ≥95% of the MIC results.
Stability of the UMIC Colistine
Stability assays on the UMIC Colistine strips were performed using the three QC strains. Stress test or shipping stability was assayed on strips that were previously incubated at 40 °C during 1 and 2 days, 1, 2, 3 and 4 weeks. Stability in use of UMIC Colistine was assayed by opening the package of the strips 1, 3, 6 and 24 h before use.
UMIC Colistine strips that were stored as recommended by the manufacturer were used as control. All tests were performed in triplicate and for each assay the same inoculum was used on all the strips tested for each strain.
Results
MIC results
The colistin MICs obtained for all isolates are summarized in Table 1: Categorical agreement was 100% with 63.4% (n = 149) of colistin-resistant strains, and 36.6% (n = 86) of colistin-susceptible strains with both methods, as highlighted in Fig. 1. No major error nor very major error was reported.
Fig. 1.
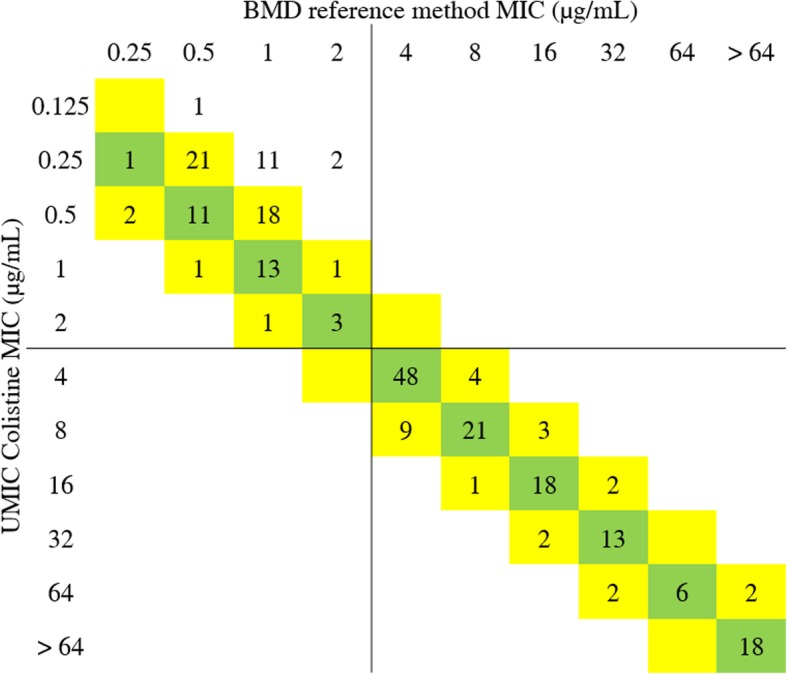
Categorical agreements for colistin MIC values between UMIC Colistine and reference method. Green shade is for identical values, and yellow shades for values within the essential agreement. Breakpoints are indicated with lines, according to CLSI recommendations (R > 2 μg/mL S ≤ 2 μg/mL)
Essential agreement was 94% (n = 221), with 64.7% (n = 152) identical values, and was 100% for colistin-resistant strains, including all the strains harboring the mcr genes. Indeed, fourteen strains classified as susceptible presented a lower MIC with UMIC Colistine (Fig. 1), and were distributed into different species: 1 E. coli, 1 K. pneumoniae, 1 K. oxytoca, 5 E. cloacae, 5 A. baumannii and 1 A. pitti (Table 1). Those strains were tested in triplicate, resulting in the reproducibility of the discrepancies, giving MICs of 1 or 2 μg/mL with reference method and 0.25 μg/mL with UMIC Colistine, except for K. oxytoca TH44 which gave a MIC of 0.5 or 1 μg/mL with reference method and 0.125 μg/mL with UMIC Colistine. However, their clinical categorization did not change as all the results obtained classified those isolates as colistin-susceptible and the correlation between UMIC Colistine and the reference method was 98.0% (Pearson’s r = 0.9801).
Reliability of UMIC Colistine
The reproducibility and quality performance of the UMIC Colistine system were both 97.8%, with only 3 values out of range for P. aeruginosa ATCC 27853 strain (MIC = 0.25 μg/mL). The QC strain E. coli NCTC 13846 always gave the recommended MIC of 4 μg/mL, except for 3 results at 2 μg/mL, which is occasionally acceptable according to EUCAST [26].
UMIC Colistine strips remained stable until 4 weeks of incubation at 40 °C and until 24 h after opening the package, obtaining the same MICs within the acceptable ranges at the different time points for the three QC strains as compared to control strips.
Usability
Manual preparation of BMD plates was time-consuming (about 1 h per day), needed a large amount of sterile material (sterilized MH medium, plates, colistin solution, etc.) and is a source of errors as it requires many steps: weighing, dissolving, diluting, distributing. The UMIC Colistine kit provided a complete assay that only requires traditional laboratory equipment. It is rapid and easy to use, and the skipped wells are mostly avoided as it is easy to check if the wells are empty or filled with a volume of 100 μL. The results reading was clear with flat-bottom wells.
Discussion
The need to implement a protocol to screen colistin-resistant isolates in clinical microbiology laboratories is urgent and requires a rapid and reliable method to replace BMD [27]. UMIC Colistine is easy to use and our study demonstrated its reliability to assess colistin susceptibility, as it could detect all the colistin-resistant isolates, notably all of the 70 mcr-positive strains tested. All the accuracy criteria were met, UMIC Colistine exhibited a high reproducibility, and quality performances where excellent even when testing strips that were stored at 40 °C to reflect the real conditions that could occur during storage and shipping of the device.
Discrepant results were obtained for some strains, mostly on Acinetobacter sp. and Enterobacter sp., but without impact on their categorization as colistin-susceptible. Those differences could be due to technical variations for some unknown reason, notably during the manual preparation of BMD with the possible loss of colistin, or to a particular phenotype of those isolates exhibited by the different features of the devices that could lead to the adhesion to the polystyrene surface of the wells. Indeed, microplates used for the reference method are tissue-culture treated, that should not impact on the colistin MIC, when UMIC Colistine strips are made of untreated polystyrene, corresponding to the recommendations on colistin susceptibility testing [17]. Additionally, the impact of the MH2 used was explored by testing the cation concentration of MH2 media used in this study. The results obtained were similar and acceptable, and the impact of the medium was eliminated: concentrations of Ca2+ were 22.1 mg/L for prepared MH2 and 22.06 mg/L for MH2 tubes provided with UMIC, and Mg2+ were 11.4 mg/L for both, when the required values are 20–25 mg/L for Ca2+ and 10–12.5 mg/L for Mg2+ according to EN ISO 20776-1:2006 standard.
Recently, the UMIC Colistine kit was evaluated together with other commercial colistine susceptibility testing devices in two studies that exhibited categorical agreements of 92 and 91.9% [26, 28]. The study performed by EUCAST obtained an essential agreement of 82% on 75 strains, with 3 VME and 3 ME, but the low number of isolates and species tested and the fact that the UMIC Colistine was not assayed with the same inoculum of the reference method can explain this lower agreement [26]. More recently, the evaluation of Jayol et al. [28] obtained 15 VME when testing 185 Gram-negative isolates: 2 H. alvei, 1 K. pneumoniae, 4 E. coli, 4 S. enterica and 4 S. maltophilia, including 5 mcr-positive isolates. There is no information on the essential agreement which seems to be over the 90% required. Concerning results were found for S. maltophilia isolates with high MIC but we did not find those discrepancies when testing 10 S. maltophilia strains. Moreover, all the H. alvei and mcr-positive isolates tested in our study were found to be colistin-resistant with UMIC Colistine. As the MH2 broth medium used for the reference method in these two studies was different from the MH2 medium used in our study, certainly explaining the differences observed in the results, it could be interesting to perform further studies evaluating different MH2 broth media for colistin susceptibility testing.
Finally, the UMIC Colistine kit has to be assayed on colistin-heteroresistant strains, that are also difficult to detect and often classified as susceptible [29].
Conclusion
The UMIC Colistine kit consists of an easy to perform technique that gave excellent results. UMIC Colistine is a reliable method to perform broth microdilution and assess the colistin MIC of clinical isolates in clinical microbiology laboratories.
Acknowledgements
We thank Biocentric Company for providing the kits. We also thank staff from Biolittoral and Synergie laboratories for technical experimentations in reproducibility testing. We thank Basil Britto Xavier for sending the mcr-2 positive strain. Finally, we thank A. Nicolay for testing the cation concentrations of the MH2 media.
Funding
This work was supported by the French Government under the « Investissements d’avenir » (Investments for the Future) program managed by the Agence Nationale de la Recherche (ANR, fr: National Agency for Research), (reference: Méditerranée Infection 10-IAHU-03). The funding body had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- AST
Antibiotic Susceptibility Testing
- ATCC
American Type Cultures Collection
- BMD
Broth Microdilution
- CA
Categorical Agreement
- CFU
Colony Forming Unit
- CLSI
Clinical and Laboratory Standards Institute
- EA
Essential Agreement
- ECV
Epidemiological Cut-off Values
- EN
European Norm
- EUCAST
European Committee of Antibiotic Susceptibility Testing
- ISO
International Organization for Standardization
- McF
McFarland
- ME
Major Error
- MH2
Mueller-Hinton
- MIC
Minimal Inhibitory Concentration
- NCTC
National Collection on Type Culture
- VME
Very Major Error
Authors’ contributions
LB conducted the experiments, analyzed the data and wrote the manuscript. LO participated in experiments and writing. SLP helped design the study. SB participated in the experiments and analysis of results. JMR designed the study and corrected the manuscript. All authors read and approved the manuscript.
Author’s informations
LB has been working at Biocentric Company since November 2, 2017, after the initial submission.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The Biocentric Company provided the UMIC Colistine kits to perform the study. LB has been working at Biocentric Company since November 2, 2017, after the initial submission.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Biswas S, Brunel J-M, Dubus J-C, Reynaud-Gaubert M, Rolain J-M. Colistin: an update on the antibiotic of the 21st century. Expert Rev Anti-Infect Ther. 2012;10:917–934. doi: 10.1586/eri.12.78. [DOI] [PubMed] [Google Scholar]
- 2.Baron S, Hadjadj L, Rolain J-M, Olaitan AO. Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int J Antimicrob Agents. 2016;48:583–591. doi: 10.1016/j.ijantimicag.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 4.Al-Tawfiq JA, Laxminarayan R, Mendelson M. How should we respond to the emergence of plasmid-mediated colistin resistance in humans and animals? Int J Infect Dis. 2017;54:77–84. doi: 10.1016/j.ijid.2016.11.415. [DOI] [PubMed] [Google Scholar]
- 5.Lu X, Hu Y, Luo M, Zhou H, Wang X, Du Y, et al. MCR-1.6, a new MCR variant carried by an IncP plasmid in a Colistin-resistant Salmonella enterica Serovar Typhimurium isolate from a healthy individual. Antimicrob Agents Chemother. 2017;61:e02632–e02616. doi: 10.1128/AAC.02632-16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 2016;21:6–11. doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 7.Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, et al. Novel plasmid-mediated Colistin resistance gene mcr-3 in Escherichia coli. MBio. 2017;8:e00543–e00517. doi: 10.1128/mBio.00543-17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, Luppi A, et al. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Eurosurveillance. 2017;22:30589. doi: 10.2807/1560-7917.ES.2017.22.31.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, Malorny B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother. 2017; September:1–8. doi:10.1093/jac/dkx327. [DOI] [PubMed]
- 10.AbuOun M, Stubberfield EJ, Duggett NA, Kirchner M, Dormer L, Nunez-Garcia J, et al. mcr-1 and mcr-2 (mcr-6.1) variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J Antimicrob Chemother. 2018. 10.1093/jac/dky272. [DOI] [PMC free article] [PubMed]
- 11.Yang Y-Q, Li Y-X, Lei C-W, Zhang A-Y, Wang H-N. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J Antimicrob Chemother. 2018;73:1791–1795. doi: 10.1093/jac/dky111. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Wang Y, Zhou Y, Li J, Yin W, Wang S, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect. 2018;7:122. doi: 10.1038/s41426-018-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters. Version. 2019:9 http://www.eucast.org.
- 15.Clinical and Laboratory Standards. M100-S27: Performance Standards for Antimicrobial Susceptibility Testing. 2017.
- 16.ISO 20776-1:2006 - Clinical laboratory testing and in vitro diagnostic test systems -- Susceptibility testing of infectious agents and evaluation of performance of antimicrobial susceptibility test devices -- Part 1: Reference method for testing the in vitro activity of antimicrobial agents against rapidly growing aerobic bacteria involved in infectious diseases. https://www.iso.org/standard/41630.html. Accessed 10 Jan 2018.
- 17.EUCAST. Recommendations for MIC determination of colistin (polymyxin E) As recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group. http://www.eucast.org. 2016; March, 22:2016. doi:22.03.16.
- 18.Dandachi I, Fayad E, El-Bazzal B, Daoud Z, Rolain J-M. Prevalence of extended-Spectrum Beta-lactamase-producing gram-negative bacilli and emergence of mcr-1 Colistin resistance gene in Lebanese swine farms. Microb Drug Resist 2018;:mdr.2018.0110. doi:10.1089/mdr.2018.0110. [DOI] [PubMed]
- 19.Rafei R, Dabboussi F, Hamze M, Eveillard M, Lemarié C, Mallat H, et al. First report of blaNDM-1-producing Acinetobacter baumannii isolated in Lebanon from civilians wounded during the Syrian war. Int J Infect Dis. 2014;21:21–23. doi: 10.1016/j.ijid.2014.01.004.. [DOI] [PubMed] [Google Scholar]
- 20.Lachish T, Elimelech M, Arieli N, Adler A, Rolain J-M, Assous MV. Emergence of New Delhi metallo-β-lactamase in Jerusalem, Israel. Int J Antimicrob Agents. 2012;40:566–567. doi: 10.1016/j.ijantimicag.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 21.Baron S, Bardet L, Dubourg G, Fichaux M, Rolain J-M. mcr-1 plasmid-mediated colistin resistance gene detection in an Enterobacter cloacae clinical isolate in France. J Glob Antimicrob Resist. 2017;10:35–36. doi: 10.1016/j.jgar.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 22.López-Rojas R, Jiménez-Mejías ME, Lepe JA, Pachón J. Acinetobacter baumannii resistant to colistin alters its antibiotic resistance profile: a case report from Spain. J Infect Dis. 2011;204:1147–1148. doi: 10.1093/infdis/jir476. [DOI] [PubMed] [Google Scholar]
- 23.Bardet L, Le Page S, Leangapichart T, Rolain J-M. LBJMR medium: a new polyvalent culture medium for isolating and selecting vancomycin and colistin-resistant bacteria. BMC Microbiol. 2017;17. [DOI] [PMC free article] [PubMed]
- 24.ISO 20776-2:2007 - Clinical laboratory testing and in vitro diagnostic test systems -- Susceptibility testing of infectious agents and evaluation of performance of antimicrobial susceptibility test devices -- Part 2: Evaluation of performance of antimicrobial susceptibility test devices. https://www.iso.org/standard/41631.html. Accessed 10 Jan 2018.
- 25.European Committee on Antimicrobial Susceptibility Testing (EUCAST). Routine and extended internal quality control for MIC determination and disk diffusion as recommended by EUCAST. Version. 2019:9 http://www.eucast.org.
- 26.Matuschek E, Åhman J, Webster C, Kahlmeter G. Antimicrobial susceptibility testing of colistin – evaluation of seven commercial MIC products against standard broth microdilution for Escherichia coli , Klebsiella pneumoniae , Pseudomonas aeruginosa , and Acinetobacter spp Clin Microbiol Infect 2017. doi:10.1016/j.cmi.2017.11.020. [DOI] [PubMed]
- 27.Osei Sekyere J. Mcr colistin resistance gene: a systematic review of current diagnostics and detection methods. Microbiologyopen. 2018:e00682. 10.1002/mbo3.682. [DOI] [PMC free article] [PubMed]
- 28.Jayol A, Nordmann P, André C, Poirel L, Dubois V. Evaluation of three broth microdilution systems to determine colistin susceptibility of gram-negative bacilli. J Antimicrob Chemother. 2018. 10.1093/jac/dky012. [DOI] [PubMed]
- 29.El-Halfawy OM, Valvano MA. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin Microbiol Rev. 2015;28:191–207. doi: 10.1128/CMR.00058-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


