Abstract
While licensed vaccines elicit protective antibody responses against a variety of viral infections, an effective vaccine for hepatitis C virus (HCV) has remained elusive. The extraordinary genetic diversity of HCV and the ability of the virus to evade the immune response have hindered vaccine development efforts. However, recent studies have greatly expanded the number of well-characterized broadly neutralizing human monoclonal antibodies (bNAbs) against HCV. These bNAbs target relatively conserved HCV epitopes, prevent HCV infection in animal models, and are associated with spontaneous clearance of human HCV infection. In this review, recent high resolution bNAb epitope mapping and structural analysis of bNAb-epitope complexes that may serve as a guide for vaccine development are discussed along with major obstacles.
Keywords: Hepatitis C virus, broadly neutralizing antibodies, HCV vaccine
Why is an HCV vaccine necessary in an era of effective antiviral therapy?
Hepatitis C virus (HCV) infection is a global health crisis, with approximately 71 million people infected worldwide [1]. HCV infection is largely asymptomatic, but over time can lead to cirrhosis, end-stage liver disease, and hepatocellular carcinoma in chronically infected individuals [2, 3]. While direct-acting antiviral (DAA) therapy has revolutionized care for patients with HCV, infection incidence is on the rise in the United States [4]. Less than 5% of the world’s HCV-infected population and only 50% of the United States’ HCV-infected population are aware that they are infected [5, 6]. Only a small minority of those people who are aware of their infection have access to treatment, and those untreated remain at risk for transmitting the infection to others and developing HCV-related complications [7]. Those who receive DAA therapy are still at risk for reinfection [8]. For these reasons, development of a prophylactic vaccine remains a critical component of HCV eradication efforts.
The challenge of HCV genetic diversity
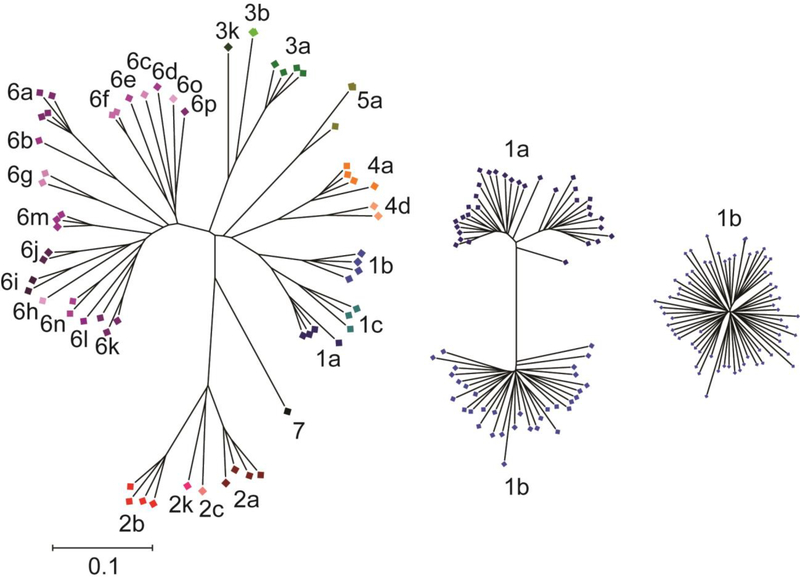
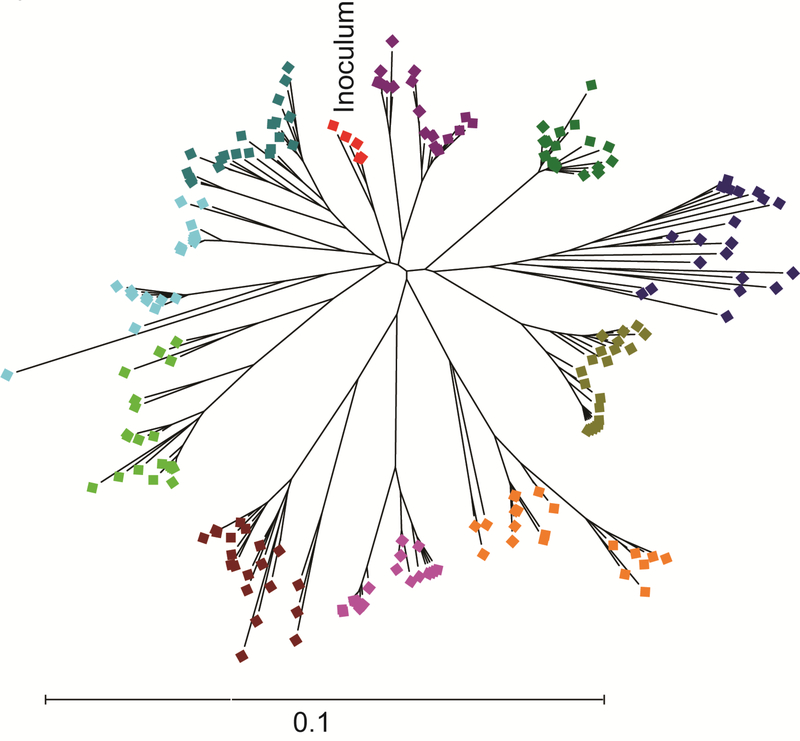
The immense genetic diversity of HCV is a major challenge to vaccine development efforts. HCV can be divided into seven genotypes that exhibit approximately 30% inter-genotypic amino acid variation in their envelope genes (E1 and E2). There are more than 60 known subtypes, and subtypes within each genotype differ at approximately 15% of their E1E2 amino acids [9] (Fig. 1). Importantly, even viral strains within the same subtype differ at up to 10% of their E1E2 amino acids. In addition to inter and intra-genotypic diversity, the error-prone polymerase of the virus coupled with immune selection leads to generation of a diverse viral swarm within each infected individual [10] (Fig. 2). Due to this viral diversity, neutralizing antibodies that are protective against one HCV strain may not be protective against strains from other genotypes, or even other strains from the same subtype. Therefore, vaccine strategies will need to generate a broad immune response capable of inhibiting infection by viruses that are extremely genetically diverse.
Figure 1. Genetic diversity of HCV E1E2 across genotypes, subtypes, and within a single subtype.
Phylogenetic trees of reference E1E2 amino acid sequences downloaded from the LANL HCV sequence database. Trees were inferred using the Neighbor-Joining method, with branch lengths drawn to scale. Subtypes are labelled and indicated with different colors. All three trees are on the same scale. Distances were computed using the JTT method. Analyses were performed in MEGA7 [90]. Not all known subtypes are shown, and the number of sequences shown for each subtype does not reflect worldwide prevalence.
Figure 2. Genetic diversity of HCV E1E2 sequences from a subtype 1b single-source outbreak.
Phylogenetic analysis of E1E2 amino acid sequences of 10 individuals from the Irish anti-D cohort, who were all infected from the same inoculum (red symbols) between 1977–1978 [14]. Sequences of virus from each individual are a different color. Phylogenetic trees were inferred using the Neighbor-Joining method, with branch lengths drawn to scale. Distances were computed using the JTT method. Analyses were performed in MEGA7 [90].
Evidence for antibody-mediated control of HCV infection.
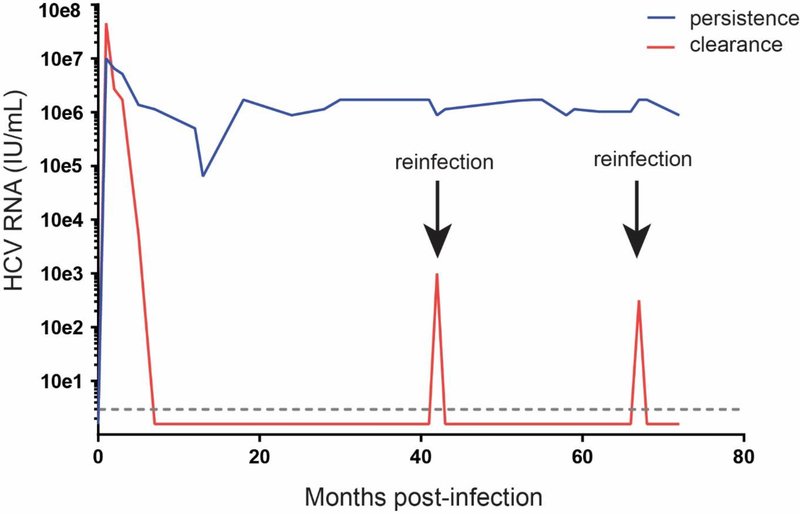
Despite HCV genetic diversity, there is evidence from studies of HCV infection of humans and animal models that an antibody-based vaccine could be effective. While most people who are infected with HCV remain chronically infected, approximately 25% of individuals clear the infection without treatment [11]. Individuals who clear HCV infection are sometimes re-infected with different HCV strains, suggesting that clearance is not always indicative of sterilizing immunity. However, individuals who clear initial infections clear subsequent infections more than 80% of the time [11]. Relative to first infections, these reinfections are characterized by shorter duration and lower peak viremia, indicating that adaptive immunity is conferring protection (Fig. 3). Similarly, chimpanzees who spontaneously clear primary HCV infection are also protected from both homologous and heterologous viral challenge, with lower magnitude and shorter duration of viremia with re-challenge [12].
Figure 3. Typical HCV RNA levels in humans with clearance of multiple infections or persistence of infection.
Representative graph demonstrating the history of viremia in chronically infected individuals (shown in blue) and in individuals who clear repeated HCV infections with heterologous strains of HCV (shown in red). The dotted line designates the limit of detection of standard HCV RNA detection methods, and all timepoints plotted below that line are HCV RNA negative.
Notably, clearance of primary human HCV infection is also associated with the early development of a broadly-neutralizing plasma response against HCV [13–16]. In addition, we and others have isolated broadly neutralizing monoclonal antibodies (bNAbs) from individuals who cleared HCV infection without treatment [17, 18]. Unexpectedly, the bNAbs that we isolated from two individuals after HCV clearance displayed minimal somatic hypermutation, suggesting that the barrier to vaccine-induction of similar bNAbs may be lower than previously thought [17]. In addition to a role in clearance of infection, neutralizing antibodies also likely play a role in suppressing chronic infection. Development of broadly neutralizing plasma in chronic infection is associated with lower viral loads, and slower disease progression [19]. In contrast, hypogammaglobulinemia is associated with increased rates of disease progression [20].
Antibodies are also protective against HCV infection in animal models. Infusion of bNAbs can prevent infection in humanized mice [21, 22] and chimpanzees [23], and combinations of bNAbs also abrogated established HCV infection in a humanized mouse model [24]. In vaccinated chimpanzees, a sustained antibody response to the HCV envelope correlated with a reduction in viral load [25]. Given this strong evidence for the role of neutralizing antibodies in control of HCV, more studies are needed to identify and characterize the neutralizing breadth, neutralizing titer, and target epitopes of antibodies in broadly neutralizing human plasma. Defining these correlates of natural protection will help to define the antibody response that needs to be elicited by a vaccine in order for it to protect against HCV infection.
Anti-HCV bNAbs target conserved envelope protein domains
The targets of the neutralizing antibody (NAb) response against HCV are the viral envelope glycoproteins, E1 and E2. While E2 is the main target of NAbs against the virus, it is also the most diverse region of the HCV genome [26]. Hypervariable Region 1 (HVR1), a 27-amino acid region at the amino-terminus of E2, is an immunodominant epitope that is capable of rapid evolution [27, 28]. Thus, while most HCV-infected individuals develop strain-specific NAbs against HVR1, viral escape mutations confer rapid resistance to these antibodies [29–32]. Notably, an HVR1-binding monoclonal antibody (mAb) isolated recently from an individual who cleared HCV infection displayed significant neutralizing breadth against multiple genotype 1 isolates, suggesting that not all HVR1-binding NAbs are strain-specific [17].
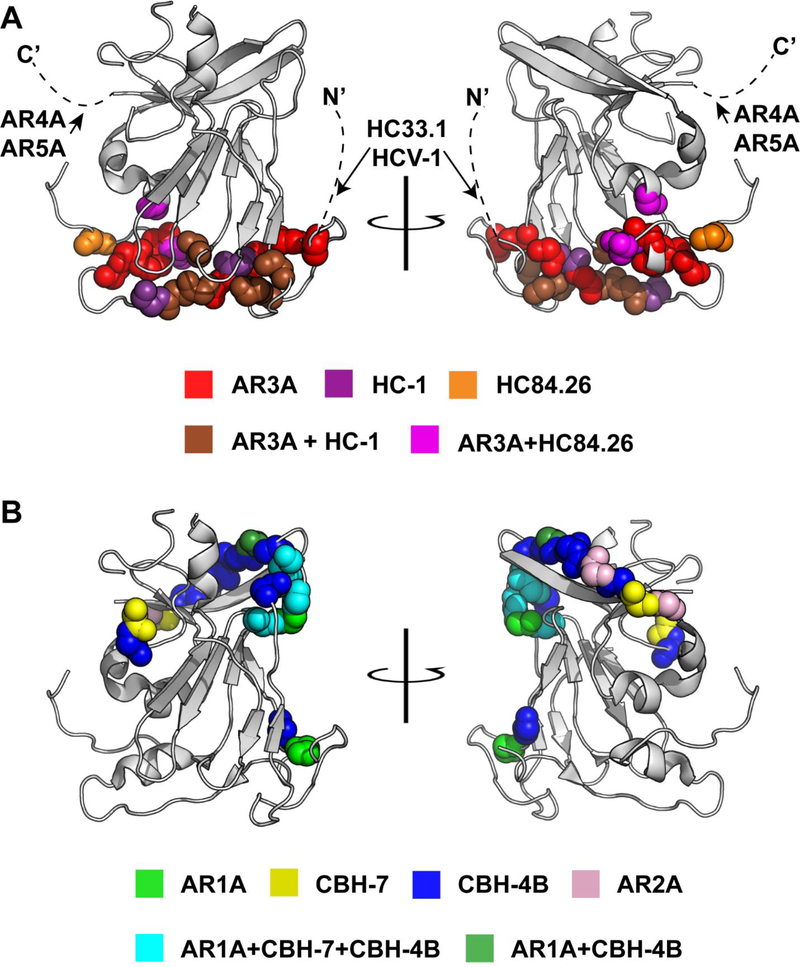
To date, dozens of bNAbs have been isolated which target conserved epitopes outside of HVR1, providing insight into the antibody response that should be elicited by a vaccine [17, 21, 33–41]. Relationships between binding epitopes of many of these bNAbs have historically been somewhat unclear due to incomplete epitope mapping and diverse nomenclature used to identify antigenic sites. However, in two recent studies, Pierce, et al. [42] and Gopal, et. al. [43] performed alanine scanning mutagenesis across all amino acids of E2 or E1E2, respectively, coupled with E1E2 binding assays using panels of HCV-specific mAbs. These studies, along with prior work, have elucidated the amino acids critical for native envelope protein folding, as well as the relationships between the binding epitopes of diverse bNAbs. These studies reveal that most cross-reactive mAbs against HCV target one of six overlapping antigenic sites on E2 (Fig. 4 and Table 1).
Figure 4. Critical binding residues of representative cross-reactive anti-HCV human mAbs.
The crystallized structure of the HCV E2 protein, strain H77, from Kong, et al. [50] PDB accession 4MWF, modified in Pymol, v1.8.6.2. The E2 structure is in gray, with critical binding residues of single or multiple mAbs identified by alanine scanning mutagenesis (See Table 1) marked with colored spheres. Dashed lines indicate regions missing from the crystal structure, which contain putative binding epitopes of HC33.1, HCV-1, AR4A, and AR5A. (A) bNAbs targeting the CD81 binding site of E2 (AR3A, HC-1, HC84.26, HC33.1, and HCV-1) or the E1E2 interface (AR4A and AR5A). (B) Weakly neutralizing mAbs targeting the beta sheet and back layer of E2.
Table 1.
Representative human mAbs with cross-reactivity against multiple HCV strains
| Antigenic Site | Antigenic Domain1 | mAb | Binding residues2 | comments | refs |
|---|---|---|---|---|---|
| CD81 binding site-front layer | AR3 | AR3A | 425, 427, 428, 429, 436, 437, 438, 440, 441, 442, 485, 503, 518, 520, 529, 530, 535, 616 |
Broadly neutralizing | [21, 43] |
| HEPC33 | 425, 427, 428, 436, 437, 438, 441, 442, 452, 503, 517, 520, 529, 535, 558 |
Broadly neutralizing | [17] | ||
| HEPC743 | 424, 425, 427, 428, 429, 436, 437, 452, 499, 503, 518, 519, 523, 530, 535 |
Broadly neutralizing | [17] | ||
| Domain B | HC-1 | 426, 428, 429, 430, 503, 529, 530, 535 |
Broadly neutralizing | [42] | |
| Domain D | HC84.26 | 441, 442, 446, 616 |
Broadly neutralizing | [42] | |
| CD81 binding site-Amino Acids 412–423 | Domain E | HC33.1 | 413, 418, 420 | Broadly neutralizing | [42] |
| AS412 | HCV-1 | 413, 420 | Broadly neutralizing | [46] | |
| E1E2-interface4 | AR4 | AR4A5 | 201, 205, 459, 486, 487, 543, 545, 569, 585, 594, 597, 652, 677, 679, 698 |
Broadly neutralizing | [37, 43] |
| AR5 | AR5A5 | 201, 205, 459, 486, 513, 543, 569, 585, 594, 597, 639, 652, 657, 677, 679 |
Broadly neutralizing | [37, 43] | |
| Amino Acids 544–549 | AR1 | AR1A | 495, 519, 544, 545, 547, 548, 549, 632 |
Weakly neutralizing | [21, 43] |
| Domain C | CBH-7 | 544, 545, 547, 549, 597, 626 |
Weakly neutralizing, broadly reactive | [42] | |
| N/A | HEPC46 | 452, 541, 543, 544, 545, 546, 547, 548, 549, 569, 594, 597, 598, 631, 633 |
Weakly neutralizing | [17] | |
| Amino Acids 629–633 | Domain A | CBH-4B3 | 503, 517, 543, 544, 545, 547, 549, 594, 598, 627, 629, 630, 631, 632, 633 |
Non-neutralizing | [42] |
| Back layer | AR2 | AR2A | 625, 628 | Narrow breadth | [21, 43] |
based on E1E2 protein binding competition analysis
Binding residues from Gopal, et al. defined as alanine mutations that reduced binding to <25% of wild type, but control antibodies maintained binding >75%. Binding residues from Pierce, et al. and Bailey, et al. defined as alanine mutations that reduced binding to <50% of wild type, but control antibodies maintained binding >75%. Binding residues from Broering, et al. defined as alanine mutations that reduced binding to <25% of wild type.
Mutations at >15 residues reduced binding to <50%, so only the 15 sites with greatest effect on binding are listed.
It is not known which of these mutations are directly involved in envelope-mAb contact, and which are required for E1E2 interaction.
Mutations at >15 residues reduced binding to <25%, so only the 15 sites with greatest effect on binding are listed.
Some of the most potent anti-HCV bNAbs isolated to date target a conserved region at the CD81 binding site of E2 (CD81bs) (Fig. 4A). These include antibodies targeting overlapping epitopes in what are commonly called Domain B, Domain D, Antigenic Region 3 (AR3), or “Epitope II” [17, 21, 33, 35, 44]. BNAbs from this group have been isolated from chronically infected individuals and from individuals who spontaneously cleared HCV, generally neutralizing multiple viral strains from multiple genotypes [17, 21, 38]. Notably, despite overlapping binding epitopes, these mAbs vary in their neutralizing breadth and in the HCV strains that they neutralize most and least potently [45].
A second group of broadly neutralizing CD81bs antibodies targets a continuous epitope spanning amino acids 412–423, commonly referred to as Antigenic Domain E, AS412, or “Epitope I”. This group of antibodies includes rodent-derived antibodies AP33, 3a/11, and H77.39 as well as human antibodies HCV-1 and HC33.1 [36, 46–48]. A third binding site of bNAbs is the E1E2 interface, which is targeted by bNAbs designated AR4A and AR5A. These bNAbs bind to epitopes that are still ill-defined. AR4A and AR5A do not compete for E1E2 binding, but both appear to bind near the C-terminus of E2, and require complexed E1 and E2 for binding [24, 37].
Other mAbs targeting what are commonly referred to as AR1, Domain C, Domain A, and AR2 are generally weakly-neutralizing or non-neutralizing (Fig 4B). AR1-specific antibodies, designated AR1A and AR1B, bind E2 at epitopes including amino acids 544–549, but fail to neutralize most strains [21]. AR1A and AR1B epitopes overlap significantly with the epitope of another broadly-reactive but weakly neutralizing antibody, the Domain C antibody designated CBH-7 [34]. Notably, Domain A antibodies, such as the mAb designated CBH-4B, also appear to have critical binding residues at 544–549, but unlike AR1 and Domain C mAbs, they appear to also rely on amino acids 629–633 in the E2 back layer for binding, and they are non-neutralizing [34]. AR2 mAbs, which bind primarily to the back layer of E2, also display relatively narrow neutralizing breadth [21].
To date, few bNAbs against E1 have been isolated. However, human monoclonal antibodies that target E1, including a mAb designated IGH526, were isolated from an HCV-infected individual [49]. While these mAbs were able to neutralize genotype 1a, 1b, 4, 5, and 6 strains, they were unable to potently neutralize genotype 2 or 3 strains. In summary, recent high resolution epitope mapping of anti-HCV mAbs has demonstrated that most cross-reactive mAbs against HCV target one of six overlapping antigenic sites, with the majority of bNAbs targeting the CD81bs of E2.
Structural analysis of bNAb epitopes
X-ray crystal structures of bNAb-epitope complexes have provided the first glimpse of the three-dimensional structures that would be necessary to induce bNAbs with a vaccine. These studies have also identified challenges that may limit induction of bNAbs, including complex conformational epitopes, structural flexibility of envelope proteins, and functional differences between mAbs binding to closely related binding epitopes.
The crystal structure of a truncated strain H77 E2 “core” complexed with an AR3-specific bNAb, designated AR3C, demonstrated that the AR3C conformational epitope is structurally complex, with extensive AR3C contact residues in multiple E2 domains, including the front layer and the CD81 binding loop. This structure also suggested that some regions of E2 are flexible, since deletion of some regions was necessary to facilitate crystallization, and other regions were disordered in the structure [50]. Several other mAbs binding to epitopes overlapping with that of AR3C have also been crystallized with peptides spanning their epitopes. HC84–1 and HC84–27 are both broadly neutralizing, and they bind to the same side of the E2 front layer alpha helix as AR3C [38]. In contrast, a weakly neutralizing mAb, designated mAb#8, also binds to a similar epitope, but with a very different angle of approach, which may explain its poor neutralizing activity [51].
Crystal structures of the bNAb epitope spanning amino acids 412 to 423 (Domain E/Epitope I/AS412), in complex with mAbs AP33, HCV-1, 3/11, and HC33.1, demonstrate that mAbs can also bind to this epitope with different angles of approach [40, 52–55]. In addition, the epitope assumes a beta hairpin conformation in some complexes and a more extended conformation in others, indicating that it is structurally flexible. Other studies have also demonstrated that this region is flexible even when E2 is properly folded. [54, 56]. Overall, these structural studies help to define the bNAb epitopes that should be incorporated into a vaccine, but they also identify the challenge of complex, conformational epitopes that might be difficult to reproduce for a vaccine, and structural flexibility of these epitopes that could limit bNab induction. To facilitate rational vaccine design, many more structures of bNAbs in complex with genetically diverse E2 or E1E2 proteins are needed.
The challenge of bNAb resistance.
While many bNAbs neutralize multiple diverse strains of HCV, mutations conferring resistance to many individual bNAbs and groups of bNAbs have been identified. Some bNAb resistance mutations fall within identified bNAb epitopes. These include N415D and N417S mutations that confer resistance to HCV1 as well as F442L/I mutations that confer resistance to HC84.26 and other Domain D antibodies [45, 57].
Notably, several studies have demonstrated no association between the level of bNAb resistance of natural HCV strains and mutations within the epitopes of those bNAbs [13, 45]. Other studies have identified few mutations in bNAb epitopes in HCV-infected individuals [58], even though bNAbs commonly develop during chronic infection. This observation may be explained in part by the discovery that mutations outside of bNAb epitopes are capable of conferring bNAb resistance.[45, 59, 60]. Mutations in the central beta sheet of E2, distant from known binding epitopes, confer resistance to Domain B/AR3 bNAbs [45, 61], and a mutation in HVR1 that modulates E2-scavenger receptor-B1 interaction confers resistance to bNAbs AR4A and HC33.4, even if their binding epitopes are fully conserved [59].
In addition to these extra-epitopic resistance mutations, HCV E1E2 also features a glycan shield [62], and is associated with apolipoproteins [63, 64] that can partially obscure conserved neutralization epitopes. Also, some non-neutralizing or poorly-neutralizing antibodies bind to epitopes adjacent to bNAb epitopes, antagonizing bNAb neutralizing activity [65, 66].
The role of CD4+ T cell responses.
The role of CD4+ T cells in priming the anti-HCV antibody response is poorly understood. The presence of a vigorous and multispecific proliferative CD4+ T cell response against HCV proteins is a strong immunological correlate of spontaneous control of acute HCV infection [67–70]. In vaccinated chimpanzees, control of heterologous HCV infection was associated with the quality of the peripheral T-helper immune response, suggesting the importance of CD4+ T cell responses in clearance of HCV infection [71]. However, the chimpanzees who cleared infection did not have any measurable difference in antibody response. Given the critical nature of CD4+ T cells in antibody maturation, CD4+ T cells would need to be primed in an antibody-based vaccine. Anti-HCV CD4+ T cell responses, which are primed in human acute infection, disappear in chronic infection [72]. Avoiding this CD4+ T cell exhaustion phenotype in vaccinated subjects would be critical to eliciting a potent antibody-based vaccine response. Therefore, inclusion of an effective antigen to elicit CD4+ T cell responses should also be a vaccination consideration.
Selection of effective vaccine antigens
Purified recombinant full-length E1E2 protein from a single HCV strain has been most extensively studied as an antibody-inducing vaccine antigen. This vaccine induced fairly strong heterologous neutralizing activity in guinea pigs [73], and was protective against homologous HCV challenge in chimpanzees [74]. It also led to reduced rates of persistence in chimpanzees after challenge with a heterologous, albeit neutralization sensitive, HCV strain [75]. However, immunization of healthy human volunteers induced detectable bNAb titers in only three of sixteen vaccinees [76], suggesting that further optimization of the vaccine antigen or of the vaccination strategy may be needed.
Some studies have demonstrated that HVR1 may occlude conserved bNAb epitopes, suggesting that deletion of HVR1 might enhance vaccine induction of antibodies against these epitopes [77]. In support of this approach, fairly high titers of bNAbs were induced in guinea pigs by high molecular weight complexes of E2 protein with three variable regions deleted [78]. Alternatively, molecular scaffolds have been developed to present single conserved epitopes [79]. However, combinations of human NAbs targeting multiple epitopes, including an epitope in HVR1, were recently found to display complementary neutralizing breadth and in some cases neutralizing synergy, suggesting that full-length E1E2 may also have advantages as a vaccine antigen relative to truncated proteins [80]. Overall, more work is needed to identify ideal vaccine antigens. In particular, more vaccine trials using human subjects will be critical, as it is unclear that anti-HCV antibody responses induced in animals are predictive of responses that would be induced by the same vaccine in humans.
Lessons from Dengue, Zika, and HIV vaccination efforts
Recent success with vaccine development for other viruses from the flaviviridae family should provide encouragement for HCV vaccine development efforts. Recently, a live-attenuated dengue vaccine elicted complete protection against dengue virus in a human challenge model [81, 82]. This vaccine induced neutralizing antibodies against all four dengue virus serotypes. Recent Zika virus vaccine testing has also shown protection in mice and rhesus monkeys [83, 84]. Notably, all flavivirus vaccines to date use neutralizing antibodies as their surrogate of protection [85, 86].
While HIV-1 vaccine development is still ongoing, many of the strategies in development for anti-HIV bNAb induction may also be applicable to HCV vaccine efforts, since both viruses have highly variable envelope proteins. The unmutated common ancestors (UCA) of some anti-HIV bNAbs do not bind to native HIV envelope trimers, suggesting that somatic mutations are critical for antigen recognition [87]. However, the UCA of a bNAb isolated from one individual avidly bound the envelope protein of the transmitted/founder HIV-1 strain from the same subject. Similarly, we found that the UCA of an anti-HCV bNAb that we isolated from a subject who cleared HCV infection bound a transmitted/founder HCV strain from the same subject [17], suggesting that careful selection of natural transmitted/founder viral strains may be a promising vaccine strategy for both HIV-1 and HCV [88]. Another strategy that has shown some promise for HIV-1 vaccine development is the use of sequential vaccinations with diverse envelope proteins to drive maturation from UCA antibodies to mature bNAbs [89]. Sequential vaccination could also be considered as a strategy to induce higher titer, more potent bNAbs against HCV.
Concluding Remarks and Future Perspectives
HCV has immense genetic diversity, and the ability of the virus to rapidly evade the immune response presents a challenge to vaccine development efforts. However, recent studies have greatly expanded the number of well-characterized bNAbs against HCV. These bNAbs target relatively conserved HCV epitopes, are associated with spontaneous clearance of human HCV infection, and prevent HCV infection in animal models. Recent high resolution bNAb epitope mapping, and structural analysis of bNAb-epitope complexes, can guide vaccine development by defining potential vaccine antigens. However, obstacles remain. Viral envelope mutations both within and outside of bNAb binding epitopes can confer resistance, and vaccine studies to date have failed to elicit high titers of bNAbs. Ongoing characterization of the antibody responses in individuals who naturally clear HCV infection will better define correlates of immune protection. In addition, further study of bNAb-envelope structural interactions is needed to define the molecular interactions necessary for broad neutralization. These studies, coupled with vaccine trials in animals and humans, could lead to a much-needed prophylactic vaccine stimulating protective antibody responses against HCV.
Highlights
HCV is a global health crisis, and vaccine development is critical for HCV eradication efforts.
Despite extensive HCV genetic diversity, broadly neutralizing antibodies (bNAbs) are associated with control of human HCV infection, and protection in animal models of HCV infection.
Most cross-reactive anti-HCV monoclonal antibodies (mAbs) target one of six domains on E1E2, with the most broadly-neutralizing mAbs targeting the CD81 binding site of E2 or the E1E2 interface.
Structural features of HCV E2, including complex conformational epitopes and structural flexibility, pose challenges for vaccine design.
Outstanding Questions
What are the neutralizing breadth, neutralizing titer, and target epitopes of antibodies associated with natural control of human HCV infection?
What are the structural similarities and differences between E2-bNAb complexes comprising diverse E2 strains?
What are the critical genetic and structural features of an HCV antigen that would elicit bNAbs in human vaccinees?
References
- 1.WHO, Global Hepatitis Report, 2017. 2017, World Health Organization. [Google Scholar]
- 2.Lagging LM, et al. , Progression of fibrosis in untreated patients with hepatitis C virus infection. Liver, 2002. 22: p. 136–144. [DOI] [PubMed] [Google Scholar]
- 3.Hoofnagle JH, Course and outcome of hepatitis C. Hepatology, 2002. 36: p. S21–S29. [DOI] [PubMed] [Google Scholar]
- 4.Suryaprasad AG, et al. , Emerging epidemic of hepatitis C virus infections among young nonurban persons who inject drugs in the United States, 2006–2012. Clin Infect Dis, 2014. 59(10): p. 1411–9. [DOI] [PubMed] [Google Scholar]
- 5.Holmberg SD, et al. , Hepatitis C in the United States. N Engl J Med, 2013. 368(20): p. 1859–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gravitz L, Introduction: a smouldering public-health crisis. Nature, 2011. 474(7350): p. S2–4. [DOI] [PubMed] [Google Scholar]
- 7.Sulkowski MS, et al. , Daclatasvir plus sofosbuvir for previously treated or untreated chronic HCV infection. N Engl J Med, 2014. 370(3): p. 211–21. [DOI] [PubMed] [Google Scholar]
- 8.Franco S, et al. , Detection of a sexually transmitted hepatitis C virus protease inhibitor-resistance variant in a human immunodeficiency virus-infected homosexual man. Gastroenterology, 2014. 147(3): p. 599–601.e1. [DOI] [PubMed] [Google Scholar]
- 9.Smith DB, et al. , Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology, 2014. 59(1): p. 318–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martell M, et al. , Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. Journal of Virology, 1992. 66: p. 3225–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osburn WO, et al. , Spontaneous Control of Primary Hepatitis C Virus Infection and Immunity Against Persistent Reinfection. Gastroenterology, 2010. 138: p. 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanford RE, et al. , Cross-genotype immunity to hepatitis C virus. J Virol., 2004. 78: p. 1575–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osburn WO, et al. , Clearance of hepatitis C infection is associated with the early appearance of broad neutralizing antibody responses. Hepatology, 2014. 59(6): p. 2140–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bailey JR, et al. , Constraints on viral evolution during chronic hepatitis C virus infection arising from a common-source exposure. J.Virol, 2012. 86: p. 12582–12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pestka JM, et al. , Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc.Natl.Acad.Sci.U.S.A, 2007. 104: p. 6025–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Logvinoff C, et al. , Neutralizing antibody response during acute and chronic hepatitis C virus infection. Proc.Natl.Acad.Sci.U.S.A, 2004. 101: p. 10149–10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailey JR, et al. , Broadly neutralizing antibodies with few somatic mutations and hepatitis C virus clearance. JCI Insight, 2017. 2(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merat SJ, et al. , Hepatitis C virus Broadly Neutralizing Monoclonal Antibodies Isolated 25 Years after Spontaneous Clearance. PLoS One, 2016. 11(10): p. e0165047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swann RE, et al. , Broad Anti-Hepatitis C Virus (HCV) Antibody Responses Are Associated with Improved Clinical Disease Parameters in Chronic HCV Infection. J Virol, 2016. 90(9): p. 4530–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bjoro K, et al. , Long-term outcome of chronic hepatitis C virus infection in primary hypogammaglobulinaemia. QJM, 1999. 92: p. 433–441. [DOI] [PubMed] [Google Scholar]
- 21.Law M, et al. , Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat.Med, 2008. 14: p. 25–27. [DOI] [PubMed] [Google Scholar]
- 22.Keck ZY, et al. , Affinity maturation of a broadly neutralizing human monoclonal antibody that prevents acute HCV infection. Hepatology, 2016: p. 1922–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morin TJ, et al. , Human Monoclonal Antibody HCV1 Effectively Prevents and Treats HCV Infection in Chimpanzees. PLoS.Pathog, 2012. 8: p. e1002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Jong YP, et al. , Broadly neutralizing antibodies abrogate established hepatitis C virus infection. Sci Transl Med, 2014. 6(254): p. 254ra129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Youn JW, et al. , Sustained E2 antibody response correlates with reduced peak viremia after hepatitis C virus infection in the chimpanzee. Hepatology, 2005. 42: p. 1429–1436. [DOI] [PubMed] [Google Scholar]
- 26.Kao JH, et al. , Quasispecies of hepatitis C virus and genetic drift of the hypervariable region in chronic type C hepatitis. J.Infect.Dis, 1995. 172: p. 261–264. [DOI] [PubMed] [Google Scholar]
- 27.Ray SC, et al. , Acute hepatitis C virus structural gene sequences as predictors of persistent viremia: hypervariable region 1 as a decoy. J.Virol, 1999. 73: p. 2938–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimizu YK, et al. , A hyperimmune serum against a synthetic peptide corresponding to the hypervariable region 1 of hepatitis C virus can prevent viral infection in cell cultures. Virology, 1996. 223: p. 409–412. [DOI] [PubMed] [Google Scholar]
- 29.Dowd KA, et al. , Selection Pressure from Neutralizing Antibodies Drives Sequence Evolution during Acute Infection with Hepatitis C Virus. Gastroenterology, 2009. 136: p. 2377–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farci P, et al. , Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc.Natl.Acad.Sci.USA, 1994. 91: p. 7792–7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimizu YK, et al. , Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. Journal of Virology, 1994. 68: p. 1494–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Hahn T, et al. , Hepatitis C virus continuously escapes from neutralizing antibody and T-cell responses during chronic infection in vivo. Gastroenterology, 2007. 132: p. 667–678. [DOI] [PubMed] [Google Scholar]
- 33.Keck ZY, et al. , Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV e2 with resistance to neutralization escape in a genotype 2a isolate. PLoS.Pathog, 2012. 8: p. e1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hadlock KG, et al. , Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J.Virol, 2000. 74: p. 10407–10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keck ZY, et al. , A point mutation leading to hepatitis C virus escape from neutralization by a monoclonal antibody to a conserved conformational epitope. J.Virol, 2008. 82: p. 6067–6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keck Z, et al. , Cooperativity in virus neutralization by human monoclonal antibodies to two adjacent regions located at the amino terminus of hepatitis C virus E2 glycoprotein. J.Virol, 2013. 87: p. 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giang E, et al. , Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc.Natl.Acad.Sci.U.S.A, 2012. 109: p. 6205–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krey T, et al. , Structural basis of HCV neutralization by human monoclonal antibodies resistant to viral neutralization escape. PLoS Pathog, 2013. 9(5): p. e1003364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johansson DX, et al. , Human combinatorial libraries yield rare antibodies that broadly neutralize hepatitis C virus. Proc.Natl.Acad.Sci.U.S.A, 2007. 104: p. 16269–16274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kong L, et al. , Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1. Proc.Natl.Acad.Sci.U.S.A, 2012. 109: p. 9499–9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Owsianka A, et al. , Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. J.Virol, 2005. 79: p. 11095–11104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pierce BG, et al. , Global mapping of antibody recognition of the hepatitis C virus E2 glycoprotein: Implications for vaccine design. Proc Natl Acad Sci U S A, 2016: p. E6946–E6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gopal R, et al. , Probing the antigenicity of hepatitis C virus envelope glycoprotein complex by high-throughput mutagenesis. PLoS Pathog, 2017. 13(12): p. e1006735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giang E, et al. , Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc Natl Acad Sci U S A, 2012. 109(16): p. 6205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bailey JR, et al. , Naturally selected hepatitis C virus polymorphisms confer broad neutralizing antibody resistance. J Clin Invest, 2015. 125(1): p. 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Broering TJ, et al. , Identification and characterization of broadly neutralizing human monoclonal antibodies directed against the E2 envelope glycoprotein of hepatitis C virus. J.Virol, 2009. 83: p. 12473–12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sabo MC, et al. , Neutralizing monoclonal antibodies against hepatitis C virus E2 protein bind discontinuous epitopes and inhibit infection at a postattachment step. J.Virol, 2011. 85: p. 7005–7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarr AW, et al. , Characterization of the hepatitis C virus E2 epitope defined by the broadly neutralizing monoclonal antibody AP33. Hepatology, 2006. 43: p. 592–601. [DOI] [PubMed] [Google Scholar]
- 49.Meunier JC, et al. , Isolation and characterization of broadly neutralizing human monoclonal antibodies to the e1 glycoprotein of hepatitis C virus. J Virol, 2008. 82(2): p. 966–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kong L, et al. , Hepatitis C virus E2 envelope glycoprotein core structure. Science, 2013. 342(6162): p. 1090–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deng L, et al. , Structural evidence for a bifurcated mode of action in the antibody-mediated neutralization of hepatitis C virus. Proc Natl Acad Sci U S A, 2013. 110(18): p. 7418–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Potter JA, et al. , Toward a hepatitis C virus vaccine: the structural basis of hepatitis C virus neutralization by AP33, a broadly neutralizing antibody. J Virol, 2012. 86(23): p. 12923–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kong L, et al. , Structure of hepatitis C virus envelope glycoprotein E2 antigenic site 412 to 423 in complex with antibody AP33. J Virol, 2012. 86(23): p. 13085–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meola A, et al. , Structural flexibility of a conserved antigenic region in hepatitis C virus glycoprotein E2 recognized by broadly neutralizing antibodies. J Virol, 2015. 89(4): p. 2170–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y, et al. , Structural basis for penetration of the glycan shield of hepatitis C virus E2 glycoprotein by a broadly neutralizing human antibody. J Biol Chem, 2015. 290(16): p. 10117–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kong L, et al. , Structural flexibility at a major conserved antibody target on hepatitis C virus E2 antigen. Proc Natl Acad Sci U S A, 2016: p. 12768–12773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chung RT, et al. , Human monoclonal antibody MBL-HCV1 delays HCV viral rebound following liver transplantation: a randomized controlled study. Am J Transplant, 2013. 13(4): p. 1047–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodrigo C, et al. , Limited naturally occurring escape in broadly neutralizing antibody epitopes in hepatitis C glycoprotein E2 and constrained sequence usage in acute infection. Infect Genet Evol, 2017. 49: p. 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.El-Diwany R, et al. , Extra-epitopic hepatitis C virus polymorphisms confer resistance to broadly neutralizing antibodies by modulating binding to scavenger receptor B1. PLoS Pathog, 2017. 13(2): p. e1006235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carlsen TH, et al. , Breadth of neutralization and synergy of clinically relevant human monoclonal antibodies against HCV genotypes 1a, 1b, 2a, 2b, 2c, and 3a. Hepatology, 2014: p. 1551–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keck ZY, et al. , Mutations in hepatitis C virus E2 located outside the CD81 binding sites lead to escape from broadly neutralizing antibodies but compromise virus infectivity. J.Virol, 2009. 83: p. 6149–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Falkowska E, et al. , Hepatitis C virus envelope glycoprotein E2 glycans modulate entry, CD81 binding, and neutralization. J.Virol, 2007. 81: p. 8072–8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang H, et al. , Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc.Natl.Acad.Sci.U.S.A, 2007. 104: p. 5848–5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fauvelle C, et al. , Apolipoprotein E Mediates Evasion From Hepatitis C Virus Neutralizing Antibodies. Gastroenterology, 2016. 150(1): p. 206–217 e4. [DOI] [PubMed] [Google Scholar]
- 65.Zhang P, et al. , Depletion of interfering antibodies in chronic hepatitis C patients and vaccinated chimpanzees reveals broad cross-genotype neutralizing activity. Proc.Natl.Acad.Sci.U.S.A, 2009. 106: p. 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Keck ZY, et al. , Antibody Response to Hypervariable Region 1 Interferes with Broadly Neutralizing Antibodies to Hepatitis C Virus. J Virol, 2016. 90(6): p. 3112–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schulze zur Wiesch J, et al. , Broadly directed virus-specific CD4+ T cell responses are primed during acute hepatitis C infection, but rapidly disappear from human blood with viral persistence. J.Exp.Med, 2012. 209: p. 61–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Diepolder HM, et al. , The role of hepatitis C virus specific CD4+ T lymphocytes in acute and chronic hepatitis C. J Mol Med (Berl), 1996. 74(10): p. 583–8. [DOI] [PubMed] [Google Scholar]
- 69.Chang KM, et al. , Differential CD4(+) and CD8(+) T-cell responsiveness in hepatitis C virus infection. Hepatology, 2001. 33(1): p. 267–76. [DOI] [PubMed] [Google Scholar]
- 70.Rehermann B, Hepatitis C virus versus innate and adaptive immune responses: a tale of coevolution and coexistence. J Clin Invest, 2009. 119(7): p. 1745–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rollier C, et al. , Control of heterologous hepatitis C virus infection in chimpanzees is associated with the quality of vaccine-induced peripheral T-helper immune response. J.Virol, 2004. 78: p. 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schulze Zur Wiesch J, et al. , Broadly directed virus-specific CD4+ T cell responses are primed during acute hepatitis C infection, but rapidly disappear from human blood with viral persistence. J Exp Med, 2012. 209(1): p. 61–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stamataki Z, et al. , Hepatitis C virus envelope glycoprotein immunization of rodents elicits cross-reactive neutralizing antibodies. Vaccine, 2007. 25(45): p. 7773–84. [DOI] [PubMed] [Google Scholar]
- 74.Choo QL, et al. , Vaccination of chimpanzees against infection by the hepatitis C virus. Proceedings of the National Academy of Sciences of the United States of America, 1994. 91: p. 1294–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Houghton M, Prospects for prophylactic and therapeutic vaccines against the hepatitis C viruses. Immunol Rev, 2011. 239(1): p. 99–108. [DOI] [PubMed] [Google Scholar]
- 76.Law JL, et al. , A hepatitis C virus (HCV) vaccine comprising envelope glycoproteins gpE1/gpE2 derived from a single isolate elicits broad cross-genotype neutralizing antibodies in humans. PLoS One, 2013. 8(3): p. e59776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Prentoe J, et al. , Hypervariable region 1 differentially impacts viability of hepatitis C virus strains of genotypes 1 to 6 and impairs virus neutralization. J Virol, 2011. 85(5): p. 2224–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vietheer PT, et al. , The core domain of hepatitis C virus glycoprotein E2 generates potent cross-neutralizing antibodies in guinea pigs. Hepatology, 2017. 65(4): p. 1117–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pierce BG, et al. , Structure-Based Design of Hepatitis C Virus Vaccines That Elicit Neutralizing Antibody Responses to a Conserved Epitope. J Virol, 2017. 91(20). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mankowski MC, et al. , Synergistic anti-HCV broadly neutralizing human monoclonal antibodies with independent mechanisms. Proc Natl Acad Sci U S A, 2018. 115(1): p. E82–E91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kirkpatrick BD, et al. , The live attenuated dengue vaccine TV003 elicits complete protection against dengue in a human challenge model. Sci Transl Med, 2016. 8(330): p. 330ra36. [DOI] [PubMed] [Google Scholar]
- 82.Durbin AP, et al. , A 12-Month-Interval Dosing Study in Adults Indicates That a Single Dose of the National Institute of Allergy and Infectious Diseases Tetravalent Dengue Vaccine Induces a Robust Neutralizing Antibody Response. J Infect Dis, 2016. 214(6): p. 832–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Larocca RA, et al. , Vaccine protection against Zika virus from Brazil. Nature, 2016. 536(7617): p. 474–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Abbink P, et al. , Protective efficacy of multiple vaccine platforms against Zika virus challenge in rhesus monkeys. Science, 2016. 353(6304): p. 1129–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Plotkin SA, Correlates of protection induced by vaccination. Clin Vaccine Immunol, 2010. 17(7): p. 1055–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thomas SJ, et al. , Fast-Track Zika Vaccine Development - Is It Possible? N Engl J Med, 2016. 375(13): p. 1212–6. [DOI] [PubMed] [Google Scholar]
- 87.Xiao X, et al. , Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem Biophys Res Commun, 2009. 390(3): p. 404–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bonsignori M, et al. , Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody. Cell, 2016. 165(2): p. 449–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McGuire AT, et al. , Specifically modified Env immunogens activate B-cell precursors of broadly neutralizing HIV-1 antibodies in transgenic mice. Nat Commun, 2016. 7: p. 10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kumar S, Stecher G, and Tamura K, MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol, 2016. 33(7): p. 1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]