Abstract
Background
GNL3 has been reported to be up-regulated in cancers and function in tumor progression, whereas the role of GNL3 in the progression of osteosarcoma remains unclear.
Materials and methods
In this study, we blocked the expression of GNL3 by siRNA interference in osteosarcoma cell lines MG63 and U20S. CCK8, colony formation, wound-healing, Transwell, flow cytometry, and Hoechst/PI staining assays were used to examine the effects of GNL3 knockdown on cell proliferation, migration, invasion and apoptosis in MG63 and U20S cells. The relative activity of MMP9 was detected using Gelatin zymography assay. Western blot was performed to detect the expression of related proteins.
Results
We found that silencing of GNL3 reduced the growth, migration, and invasion abilities of MG63 and U20S cells. Moreover, silencing GNL3 triggered cell cycle arrest in MG63 and U20S cells, as well as promoted cell apoptosis. In addition, depletion of GNL3 was observed to reduce the activity of MMP9 and suppress the process of epithelial–mesenchymal transition (EMT) through up-regulation of E-cadherin and down-regulation of N-cadherin. Furthermore, we found that X-box-binding protein 1 (XBP1) could bind to GNL3 using dual-luciferase reporter assay, and XBP1 overexpression could restore the inhibitory effects on proliferation, invasion, and EMT in MG63 and U20S cells caused by GNL3 knockdown.
Conclusion
These data suggest that GNL3 functions as an oncogene in the progression of osteosarcoma by regulation of EMT, and XBP1 is also involved in its mechanism.
Keywords: G protein nucleolar 3, GNL3, osteosarcoma, EMT, XBP1
Introduction
As known, osteosarcoma, a bone tumor mainly occurring in children and adolescents, with an incidence rate of approximately 3–4 million in the world, results in a high rate of disability.1–4 Despite advances in the treatment technology of osteosarcoma, the prognosis of osteosarcoma patients remains poor, mainly due to pulmonary metastasis and recurrence, especially pulmonary metastasis, which is the leading cause of death.5–7 In order to develop a new osteosarcoma treatment strategy, novel available osteosarcoma metastasis–related genes and the underlying mechanism must be identified to provide effective therapeutic targets for osteosarcoma.
GNL3, originally named nucleostemin, is reported to be expressed in proliferating cells, including tumor cells and stem cells.8–10 Increasing evidence reveals that GNL3 is up-regulated in various types of cancer tissues and promotes cell proliferation via modulation of cell cycle, such as in prostate cancer,11 hepatocellular carcinoma,12 and ovarian cancer.13 It is also demonstrated that GNL3 is associated with the prognosis of patients with cancer.14–16 Moreover, recent studies found that GNL3 impacts the invasion ability of ovarian cancer13 and colon cancer cells.17 Furthermore, GNL3 has been shown to play important roles in various cellular physiological and pathological processes as a multi-functional protein, such as cellular self-renewal, apoptosis, and the maintenance of genome stability and telomere integrity.9,12,17,18 However, the role and mechanism of GNL3 in the progression of osteosarcoma remain unclear, which is the aim of this study.
In this study, we revealed an oncogenic role of GNL3 in the progression of osteosarcoma. Our data demonstrated that depletion of GNL3 inhibited the proliferation, migration, and invasion abilities of osteosarcoma cells, induced cell cycle arrest, and promoted apoptosis. Further research demonstrated that the transcription factor X-box-binding protein 1 (XBP1) could bind to GNL3 in osteosarcoma cells, which might be involved in the suppression of GNL3 knockdown in the proliferation and invasion of osteosarcoma.
Materials and methods
Cell culture
MG63, U20S, and normal chondrocyte cells (Cell Bank of Chinese Academy of Sciences, Shanghai, China) were cultured in DMEM supplemented with 10% FBS and antibiotics (penicillin, 100 U/mL; streptomycin, 0.1 mg/mL; Sigma-Aldrich, Hamburg, Germany). Cells were transfected with 50 nM siRNA-GNL3 (Oligobio, Beijing, China) using Lipofectamine 2000 to block the expression of GNL3, and scrambled siRNA was used as negative control (NC).
Real-time (RT-PCR)
After being transfected for 24 hours, the total RNA isolated from MG63 and U20S cells was reverse transcribed in complementary cDNA using a HiFiScript cDNA Synthesis Kit (CWBIO, Beijing, China). The obtained RT products were then used as templates to evaluate the expression of GNL3 mRNA using a SYBR Premix Ex Taq II kit (Takara, Shiga, Japan). The primers used in this study were as follows: GNL3, 5’-GCAGCAGAAACTTGACAGGC-3’ (forward), 5’-CGAATGGCTTTGCTGCAAGT-3’ (reverse); β-actin, 5’-CCCGAGCCGTGTTTCCT-3’ (forward), and 5’-GTCCCAGTTGGTGACGATGC-3’ (reverse).
Western blot assay
Cells transfected with siRNA for 48 hours were collected and lysed using RIPA Lysis Buffer (CWBIO) at 4°C for protein extraction; 20 µg of protein of each sample was electrophoresed by SDS-PAGE gel and then electrotransferred onto a polyvinylidene fluoride membrane (PVDF; Millipore, Billerica, MA, USA). Then 5% non-fat milk was used to block the PVDF membrane, followed by the incubation with primary antibodies (1:1000, Proteintech Group, IL, USA) at 4°C overnight. After incubation with secondary antibodies (1:3000, Proteintech Group) in blocking buffer, the bands were visualized using an enhanced chemiluminescence kit (CWBIO).
CCK8 assay
Cells transfected with siRNA for 24 hours were seeded into a 96-well plate at a density of 1×103 cells per well. Cell viability was measured every 24 hours, and CCK8 reagent was added to each well before the test. After 1.5 hours of incubation, the OD value of excitation light was detected using an enzyme standard instrument at a wavelength of 450 nm.
Colony formation assay
After being transfected for 24 hours, MG63 and U20S cells were grown in each plate and cultured at 37°C for about 1 week until visible colonies formed. After being fixed with 5 mL of 4% paraformaldehyde, the colonies were stained with 0.1% crystal violet. The colonies were counted and photographed.
Wound-healing assay
MG63 and U20S cells (5×105 cells per well) transfected with siRNA for 24 hours were grown in 6-well plates for a further 12 hours. After the wound was generated with a pipette tip, cells were incubated in a serum-free medium supplemented for a further 24 hours. After washing, cells were imaged and the wound closure was quantitated with ImageJ software.
Transwell invasion assay
First, the Transwell chambers (Millipore, MA, USA) were pre-covered with Matrigel (BD Bioscience, CA, USA). 1×105 of MG63 or U20S cells transfected with siRNA for 24 hours were grown into the upper compartment of Transwell chambers and 600 µL of DMEM with 20% FBS was then added into the lower compartment. After being cultured for a further 24 hours, the non-invaded cells were removed, and the invaded cells were stained with 0.1% crystal violet for 20 minutes after being fixed with 4% paraformaldehyde. After that, the invaded cells were captured (magnification, ×40) and counted under a light microscope.
Flow cytometry assay
Cell cycles of MG63 and U20S cells were analyzed by staining with propidium iodide (PI, BD Bioscience) by a flow cytometer. Briefly, cells transfected with siRNA for 24 hours were fixed in pre-cold 70% ethanol at −20°C overnight prior to stain with PI for 30 minutes at 37°C. Flow cytometry (BD FACSCanto II, BD Bioscience) was performed to analyze the cell cycle distribution, and the obtained data were determined using the BD FACSDiva software (BD Bioscience). Annexin V-FITC (BD Bioscience) and PI double staining were performed to detect the rate of apoptosis. The rate of apoptosis was analyzed by flow cytometer and calculated using BD FACSDiva software.
Hoechst/PI staining assay
To stain the cells with fluorescent dyes, MG63 and U20S cells transfected with siRNA for 24 hours were seeded into dishes followed by co-staining with Hoechst solution (10 µg/mL, Beyotime, Shanghai, China) and PI solution (5 µg/mL, Beyotime). Cells were captured and counted with a fluorescent microscope.
Gelatin zymography
After being transfected with siRNA for 24 hours, MG63 and U20S cells were grown into 6-cm dishes at a density of 2.5×106 cells per dish and incubated with serum-free medium for 24 hours. Culture supernatants were collected and electrophoresed by 10% SDS-PAGE containing 0.5 mg/mL gelatin. After washing, the gel was cultured at 37°C for a further 40 hours. After that, the gel was stained with Coomassie Blue R250 for 4 hours prior to decolorization at room temperature. The band was scanned by Image Scanner (Amersham, Piscataway, NJ, USA) and analyzed by ImageQuant TL V2003 software.
Dual-luciferase reporter assay
The pGL3.0-GNL3-promoter or pGL3.0 Basic vector was constructed to co-transfect into MG63 and U20S cells with pcDNA3.1-XBP1 vector using Lipofectamine 2000 (Invitrogen), and pGL3.0-GNL3-promoter vector was used as control. After 48 hours of co-transfection, the luciferase activity was evaluated using the Dual-Luciferase Reporter Assay System.
Statistical analysis
Data in this study was shown as mean±SD from triplicate independent experiments and analyzed using GraphPad Prism 7.0. The Student’s t-test or one-way ANOVA was used for comparison of the difference between groups, and a P-value lower than 0.05 was considered statistically significant.
Results
GNL3 is up-regulated in sarcoma tissues and its up-regulation is associated with poor prognosis of patients
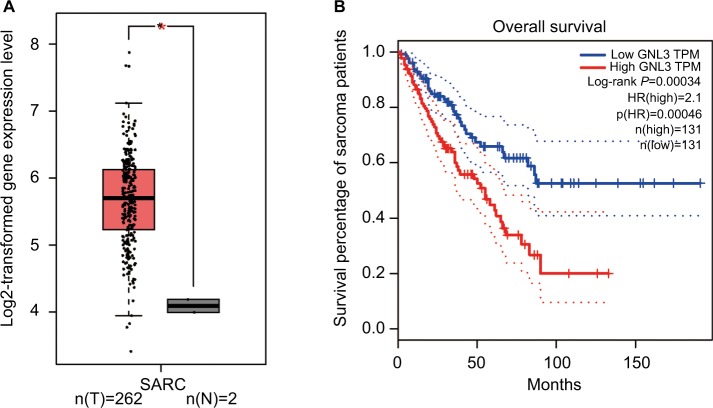
Analysis from Gene Expression Profiling Interactive Analysis (GEPIA) showed that the expression level of GNL3 mRNA in sarcoma (SARC) tissues was significantly higher than that in normal tissues (Figure 1A). Moreover, we observed that the expression level of GNL3 mRNA was associated with overall survival of patients with sarcoma (P<0.001, Figure 1B), and patients with high levels of GNL3 showed a shorter overall survival.
Figure 1.
GNL3 is up-regulated in SARCs and its up-regulation is associated with poor prognosis of patients with sarcoma.
Notes: (A) The expression of GNL3 mRNA in sarcoma tissues (red box) and normal tissues (black box) by GEPIA databases. The y-axis indicates the log2-transformed gene expression level. (B) Survival percentage of sarcoma patients with high or low expression of GNL3 mRNA by GEPIA databases. Blue: patients with low expression of GNL3; Red: patients with high expression of GNL3. *P<0.05.
Abbreviations: SARC, sarcoma tissue; TPM, trans per million.
GNL3 knockdown reduces cell viability and colony formation ability in osteosarcoma cells
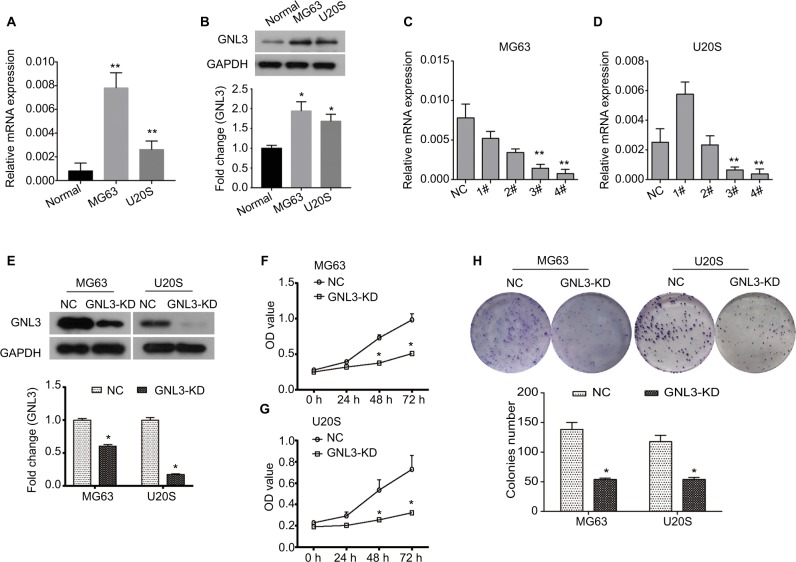
By RT-PCR and Western blot analysis, we found that the expression of GNL3 was significantly higher in osteosarcoma cell lines MG63 and U20S compared with the normal chondrocyte at mRNA and protein levels (Figure 2A and B). In order to investigate the role of GNL3, we knocked down its expression in MG63 and U20S cells using siRNA-GNL3 interference, and a scrambled siRNA was used as the NC. As shown in Figure 2C, the expression of GNL3 mRNA was significantly blocked in MG63 cells transfected with si-GNL3-3# or si-GNL3-4#, which was similarly observed in U20S cells (Figure 2D). Therefore, the si-GNL3-3# was used in all subsequent experiments due to its appropriate interference efficiency. As Western blot results indicate, the expression of GNL3 protein was also decreased in si-GNL3-transfected MG63 and U20S cells (Figure 2E). CCK8 assay was carried out to evaluate the effect of GNL3 expression on cell viability of osteosarcoma. As shown in Figure 2F and G, significant inhibition in cell viability was observed following transfection with si-GNL3 in MG63 and U20S cells. Moreover, the inhibitory effect of GNL3 knockdown on the growth of osteosarcoma cells was further confirmed by colony formation assay. GNL3 knockdown significantly inhibited the clonogenic abilities of MG63 and U20S cells (Figure 2H). Our investigations indicated that GNL3 knockdown inhibited the viability and colony formation ability of osteosarcoma cells.
Figure 2.
GNL3 knockdown inhibits cell viability and colony formation in osteosarcoma cells.
Notes: (A, B) The expression of GNL3 in normal chondrocyte and osteosarcoma cell lines MG63 and U20S was measured by Real-time PCR (A) and Western blot (B). Four siRNA-GNL3 sequences (1#-4#) were transfected into osteosarcoma cell lines MG63 and U20S cells as GNL3 silencing group (si-GNL3), respectively, scrambled siRNA was used as NC. After 24 hours of transfection (C, D). The relative expression of GNL3 mRNA in MG63 (A) and U20S (B) cells was examined by RT-PCR. (E) After 48 hours of transfection, the expression of GNL3 protein in MG63 and U20S cells was examined by Western blot assay. After transfection, cell viability in MG63 (F) and U20S (G) cells was determined by CCK8 assay. (H) Colony formation ability of MG63 and U20S cells was determined using colony formation assay. GNL3-KD, GNL3 knockdown, cells transfected with siRNA-GNL3. Data are expressed as the mean±SD (n=3). Results were obtained in three separate experiments. *P<0.05, **P<0.01 vs the control group.
Abbreviations: NC, negative control; RT-PCR, real-time PCR.
GNL3 knockdown suppresses migration and invasion abilities of osteosarcoma cells
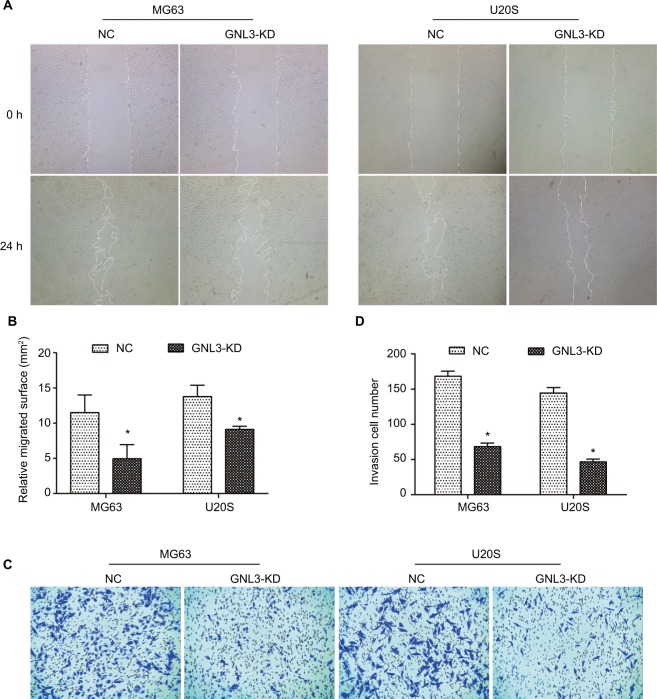
Metastasis is the main cause of poor prognosis of patients with osteosarcoma. Hence, a wound-healing assay was performed to assess the effect of GNL3 expression on the cell migration ability of osteosarcoma. As indicated in Figure 3A and B, MG63 cells transfected with si-GNL3 displayed a remarkable delay in the ability to migrate into the blank space compared with the NC group. Similarly, the migration ability of U20S cells was also inhibited by GNL3 deficiency (Figure 3A and B). In addition, in comparison with NC, the number of invasive cells in the si-GNL3 transfected MG63 group was decreased, which was also observed in U20S cells (Figure 3C and D).
Figure 3.
GNL3 knockdown inhibits migration and invasion in osteosarcoma cells.
Notes: (A, B) After 24 hours of transfection, a wound-healing assay was performed to detect the effect of si-GNL3 on cell migration in MG63 and U20S cells. Magnification ×40. (C, D) Transwell assay was employed to assess the effect of si-GNL3 on the cell invasion ability of MG63 and U20S cells. Magnification ×100. Data are presented as the mean±SD (n=3). Results were obtained in three replicates. *P<0.05 vs the control group.
Abbreviation: NC, negative control.
GNL3 knockdown triggers cell cycle arrest in osteosarcoma cells
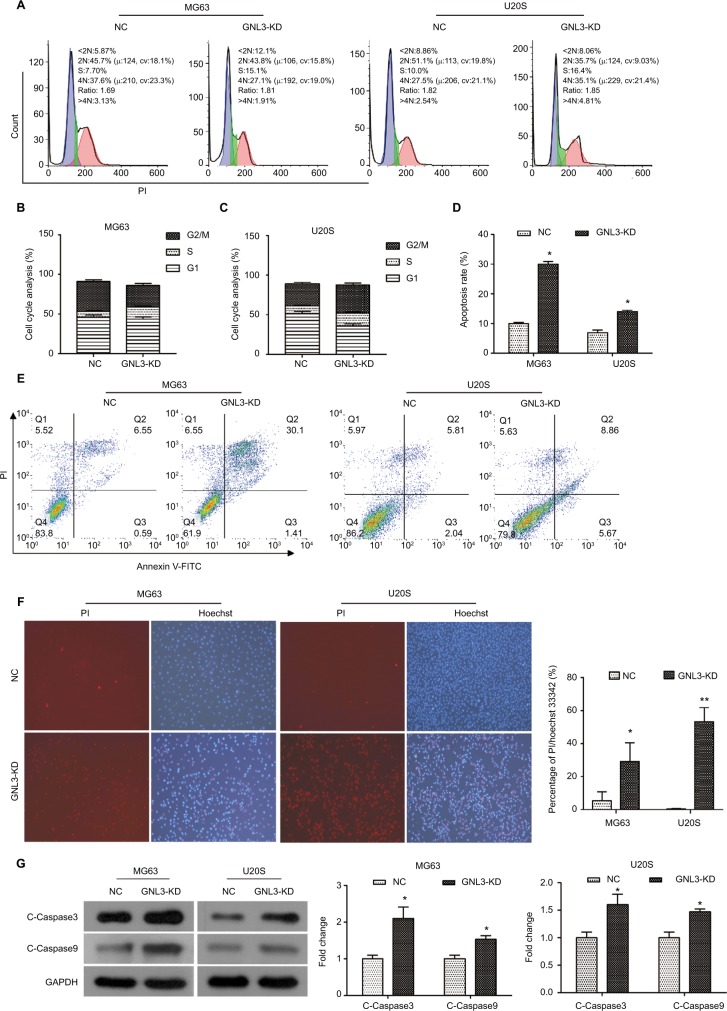
Flow cytometry was performed to assess whether GNL3 expression impaired the cell cycle distribution in osteosarcoma cells. As indicated in Figure 4A and B, compared with the NC group, the proportion of cells in the S phase was significantly increased in MG63 cells transfected with si-GNL3, and the proportions of cells in the G1 and G2/M phases were decreased. In U20S cells, in comparison with the NC, more cells were retained in S and G2/M phases, the proportion of cells staying in the G1 phase decreased following the transfection with si-GNL3 (Figure 4A and C). These findings indicated that GNL3 down-regulation induced cell cycle arrest in osteosarcoma cells, resulting in the inhibition of cell proliferation.
Figure 4.
GNL3 knockdown induces cell cycle arrest and promotes apoptosis in osteosarcoma cells.
Notes: (A, B, C) Flow cytometry analyses were applied to test the effects of si-GNL3 on cell cycle distribution in MG63 and U20S cells. (D, E) Cell apoptosis was also assessed by flow cytometry in MG63 and U20S cells. (F) Hoechst/PI staining assay was used to observe the effect of si-GNL3 on cell apoptosis in MG63 and U20S cells. Magnification ×100. (G) After 48 hours of transfection, Western blot assays of apoptosis-related proteins (cleaved Caspase3 and cleaved Caspase9) were conducted in MG63 and U20S cells. Data are presented as the mean±SD (n=3). Results were obtained in three replicates. *P<0.05, **P<0.01 vs the control group.
Abbreviations: NC, negative control; PI, propidium iodide.
GNL3 knockdown promotes apoptosis in osteosarcoma cells
We used the flow cytometry to assess the rate of apoptosis in MG63 and U20S cells transfected with siRNA-GNL3. Our data revealed that GNL3 knockdown significantly promoted the rate of apoptosis in MG63 cells in comparison with the NC (Figure 4D and E). Similarly, silencing GNL3 with siRNA also increased the rate of apoptosis in U20S cells (Figure 4D and E). The promotion of apoptosis in MG63 and U20S cells by loss-of-function of GNL3 was further substantiated by a Hoechst/PI staining assay (Figure 4F). Next, Western blot was performed to quantitate the expression of apoptosis-associated proteins to investigate the mechanism underlying the increased apoptosis caused by down-regulation of GNL3 in osteosarcoma cells. GNL3 knockdown up-regulated the expression level of Cleaved Caspase3 and Cleaved Caspase 9 in MG63 and U20S cells compared to the NC (Figure 4G). These results suggested that the increase in apoptosis induced by GNL3 knockdown in MG63 and U20S cells was mediated via blocking the Caspase cascade.
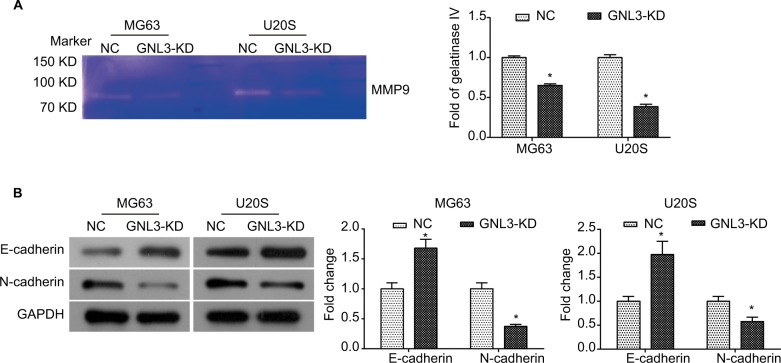
GNL3 knockdown diminishes the MMP9 and process of epithelial–mesenchymal transition (EMT) in osteosarcoma cells
Further study was conducted to explore the molecular mechanism underlying the suppression in cell migration and invasion caused by GNL3 knockdown in osteosarcoma cells. Due to the importance of MMP9 and EMT in tumor adhesion and invasion, the changes in MMP9 activity and EMT were examined in si-GNL3 transfected cells. Here, the gelatin zymography analysis demonstrated that GNL3 knockdown significantly decreased the activity of MMP9 in MG63 and U20S cells (Figure 5A). Besides, the expression level of N-cadherin was down-regulated in GNL3 knockdown cells in comparison with the NC, while the expression of E-cadherin was up-regulated (Figure 5B). Collectively, all these data shed light on the fact that the attenuation of MMP9 and EMT might be involved in the suppression in cell migration and invasion caused by silencing of GNL3 in osteosarcoma cells.
Figure 5.
GNL3 knockdown diminishes the MMP9 and EMT process in osteosarcoma cells.
Notes: After transfection, the MMP9 activity was examined by gelatin zymography enzyme activities assay (A). Changes in the expression of marker proteins (E-cadherin and N-cadherin) in the EMT process were detected by Western blot (B). Data are expressed as the mean±SD from three independent experiments. *P<0.05 vs the control group.
Abbreviations: EMT, epithelial–mesenchymal transition; NC, negative control.
Up-regulation of X-box-binding protein 1 (XBP1) rescues the GNL3 knockdown-induced suppression in osteosarcoma cells
GeneCards database (https://www.genecards.org/) analysis revealed that XBP1 was a possible transcription factor binding GNL3, which was further confirmed by dual-luciferase reporter assay. The pGL3.0-GNL3-promoter or pGL3.0 Basic vectors were constructed for co-transfection with pcDNA3.1-XBP1 vector into MG63 and U20S cells, and the pGL3.0-GNL3-promoter was used as control. After co-transfection, we observed that XBP1 greatly up-regulated the expression level of GNL3 promoter, suggesting that XBP1 could regulate the expression of GNL3 via binding its promoter (Figure 6A). Moreover, Western blot results also revealed that co-transfection with XBP1 and si-GNL3 significantly restored the expression of GNL3 protein compared to the si-GNL3 transfected cells (Figure 6B–D). Besides, the expression of N-cadherin and E-cadherin was also reversed by up-regulation of XBP1 in GNL3 knockdown cells (Figure 6B–D). As shown in Figure 6E and F, up-regulation of XBP1 restored the suppression in cell viability caused by GNL3 knockdown both in MG63 and U20S cells. Furthermore, the depression in cell invasion caused by GNL3 knockdown was significantly rescued by the up-regulation of XBP1 in MG63 cells (Figure 6G and H). Similar results were observed in U20S cells (Figure 6G and H). Collectively, these results suggested that XBP1 acted as a positive regulator of GLN3, and its overexpression could reverse the inhibition of GLN3 knockdown in cell proliferation and invasion in osteosarcoma cells.
Figure 6.
XBP1 binds to GNL3 in osteosarcoma cells.
Notes: (A) The pGL3.0-GNL3-promoter or pGL3.0 Basic vector was constructed to co-transfect with pcDNA3.1-XBP1 vector into MG63 and U20S cells, and pGL3.0-GNL3-promoter was used as control. After co-transfection, dual-luciferase reporter assay was used to assess the relative luciferase activity in MG63 and U20S cells. The si-GNL3 was co-transfected with pcDNA3.1-XBP1 into MG63 and U20S cells. After co-transfection, Western blot was used to detect the expression of GNL3, N-cadherin, and E-cadherin (B, C, and D). (E, F) CCK8 assay was used to assess the effect of XBP1 on cell viability in MG63 (E) and U20S (F). (G, H) Transwell assay was employed to assess the effect of XBP1 on cell invasion ability of MG63 and U20S cells. Magnification ×100. Data are presented as mean±SD (n=3). Results were obtained in three replicates. *P<0.05, **P<0.01 vs the control group.
Abbreviations: NC, negative control; XBP1, X-box-binding protein 1.
Discussion
GNL3 has been revealed to be up-regulated in some types of cancers and involved in tumor progression through regulating cell proliferation, cell cycle, and invasiveness. It is demonstrated that up-regulation of GNL3 increases the proliferation of prostate cancer PC-3 cells and inhibits cell migration and invasion.11 Tang et al17 revealed that GNL3 is up-regulated in colon cancer tissues, and GNL3 overexpression promotes the proliferation and invasion in HT-29 cells, whereas GNL3 knockdown reduces the proliferation and invasion in HCT-116 cells. With this study, we first demonstrated that silencing GNL3 in two osteosarcoma cell lines (MG63 and U20S) significantly reduced cell proliferation, migration, and invasion abilities of osteosarcoma cells. Additionally, this promoted the apoptosis through blocking the Caspase cascade, suggesting a pro-oncogenic role of GNL3 in the progression of osteosarcoma in vitro.
As is well known, cell cycle dysregulation is one of the most common characteristics of tumor cells. Considering the regulation of GNL3 on cell cycle distribution in tumor cells, we assessed the effect of GNL3 knockdown on the cell cycle of osteosarcoma cells. However, it is noteworthy that there are inconsistent reports regarding the cell cycle arrest caused by GNL3 dysregulation in different tumor cells. It is demonstrated that GNL3 knockdown triggers cell cycle arrest at the G1 phase in hepatocellular carcinoma HepG2 and Hep3B cells,12 ovarian cancer SKOV-3 cells,13 and glioma U251 and U373MG cells.19 Conversely, Meng et al20 claim that GNL3 knockdown triggers G2-M arrest in U2OS cells, while overexpression of GNL3 is revealed to induce G1 arrest through inhibiting MDM2 in U2OS cells.21 Herein, our data showed that si-GNL3 transfection triggered cell cycle arrest at G2/M in U20S cells, which was consistent with the results of the previous study.20 However, in MG63 cells transfected with si-GNL3, the proportion of cells in the S phase increased, and the cells in G1 and G2 phases decreased, indicating that the MG63 cell cycle was blocked in S phase by loss-of-function of GNL3. The different regulatory functions of GNL3 in different tumor cell cycles may be related to the diversity of tumor cells, which needs further study.
In respect to our research, our data demonstrated that loss-of-function of GNL3 inhibited migration and invasion in MG63 and U20S cells. Therefore, further study was needed to investigate the relevant mechanism of GNL3 underlying the regulation in cell invasion. MMP9 has been revealed to be up-regulated in cancers and involved in the process of tumor cell invasion and metastasis,22 including osteosarcoma.23 We found that GNL3 down-regulation significantly hindered the activity of MMP9 in MG63 and U20S cells. Moreover, we also observed that expression of E-cadherin was increased in GNL3 knockdown cells, while the expression of N-cadherin was down-regulated, indicating that the EMT process was diminished in GNL3 knockdown cells. EMT is the main process that promotes tumor metastasis and invasion.24 Wang et al13 claim that GNL3 knockdown inhibits the migration and invasion in SKOV3 cells via suppressing EMT process by the up-regulation of E-cadherin. In colon cancer, depletion of GNL3 is also revealed to suppress tumor metastasis through suppressing the EMT process by increasing expression of E-cadherin and decreasing expression of N-cadherin in HCT-116 cells.17 These results are similar to what our data revealed in this study. Collectively, these data suggest that GNL3 plays a pivotal role in tumor metastasis via regulating the MMP9 and EMT process in osteosarcoma.
Previous studies show that GNL3 regulates tumor growth and metastasis through different mechanisms, such as promoting EMT in colon cancer17 and glioma19 by activating the Wnt/β-catenin pathway. It is also revealed that GNL3 interacts with some proteins to modulate cell proliferation, such as P53, p27, and MDM2.20,21,25 Hu et al12 showed that GNL3 is a binding partner of p27 in hepatocellular carcinoma cells and promotes the progression of hepatocellular carcinoma by promoting the nucleolar polyubiquitination of p27kip1. In the current study, for the first time, we demonstrated that XBP1 could bind to the promoter of GNL3 in MG63 and U20S cells by GeneCards database analysis and dual-luciferase reporter assay. XBP1, a well-characterized transcription factor, plays a key role in response to endoplasmic reticulum stress26 and tumorigenesis via regulating target genes related to cell proliferation and survival.27–29 We further demonstrated that up-regulation of XBP1 could restore the inhibitory effects on proliferation, invasion, and EMT in MG63 and U20S cells caused by GNL3 knockdown. These findings indicate that XBP1 might be involved in the regulation of GNL3 in the growth and metastasis of osteosarcoma by binding to GNL3.
In summary, we found that depletion of GNL3 inhibited the progression and metastasis of osteosarcoma via triggering cell cycle arrest and apoptosis and down-regulating the MMP9 and EMT process, indicating an oncogenic role of GNL3 in tumor progression. In addition, we concluded that XBP1 was also involved in the regulation of GNL3 in tumor progression. These findings indicate that GNL3 may be a potential target for osteosarcoma treatment. However, further in vivo studies are needed to validate the results of this study and elucidate the specific mechanism.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 81501844) and the Natural Science Foundation of Shandong Province, China (No. BS2015YY009).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Berner K, Johannesen TB, Berner A, et al. Time-trends on incidence and survival in a nationwide and unselected cohort of patients with skeletal osteosarcoma. Acta Oncol. 2015;54(1):25–33. doi: 10.3109/0284186X.2014.923934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee L, Fei L, Pope J, Wagner LM. Early lymphocyte recovery and outcome in osteosarcoma. J Pediatr Hematol Oncol. 2017;39(3):179–183. doi: 10.1097/MPH.0000000000000717. [DOI] [PubMed] [Google Scholar]
- 3.Maximov VV, Aqeilan RI. Genetic factors conferring metastasis in osteosarcoma. Future Oncol. 2016;12(13):1623–1644. doi: 10.2217/fon-2016-0014. [DOI] [PubMed] [Google Scholar]
- 4.Anderson ME. Update on survival in osteosarcoma. Orthop Clin North Am. 2016;47(1):283–292. doi: 10.1016/j.ocl.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 5.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global Cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 6.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 7.Liu K, Huang J, Ni J, et al. MALAT1 promotes osteosarcoma development by regulation of HMGB1 via miR-142-3p and miR-129-5p. Cell Cycle. 2017;16(6):578–587. doi: 10.1080/15384101.2017.1288324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai RYL, Mckay RDG. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002;16(23):2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamashita M, Nitta E, Nagamatsu G, et al. Nucleostemin is indispensable for the maintenance and genetic stability of hematopoietic stem cells. Biochem Biophys Res Commun. 2013;441(1):196–201. doi: 10.1016/j.bbrc.2013.10.032. [DOI] [PubMed] [Google Scholar]
- 10.Wu H, Wang W, Tong S, Wu C. Nucleostemin regulates proliferation and migration of gastric cancer and correlates with its malignancy. Int J Clin Exp Med. 2015;8(10):17634. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Lee M, Williams KA, Hu Y, et al. GNL3 and SKA3 are novel prostate cancer metastasis susceptibility genes. Clin Exp Metastasis. 2015;32(8):769–782. doi: 10.1007/s10585-015-9745-y. [DOI] [PubMed] [Google Scholar]
- 12.Hu B, Hua L, Ni W, et al. Nucleostemin/GNL3 promotes nucleolar polyubiquitylation of p27kip1 to drive hepatocellular carcinoma progression. Cancer Lett. 2017;388:220–229. doi: 10.1016/j.canlet.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Wang L, Ji Q, Zhu H, Han S. Knockdown of Nucleostemin in an ovarian cancer SKOV-3 cell line and its effects on cell malignancy. Biochem Biophys Res Commun. 2017;487(2):262–267. doi: 10.1016/j.bbrc.2017.04.046. [DOI] [PubMed] [Google Scholar]
- 14.Nakajima TE, Yoshida H, Okamoto N, et al. Nucleostemin and twist as predictive markers for recurrence after neoadjuvant chemotherapy for esophageal carcinoma. Cancer Sci. 2012;103(2):233–238. doi: 10.1111/j.1349-7006.2011.02142.x. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi T, Masutomi K, Tamura K, et al. Nucleostemin expression in invasive breast cancer. BMC Cancer. 2014;14(1):215. doi: 10.1186/1471-2407-14-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yıldırım GD, Arık D, Yıldız B, Dinçer M, Yaşar NF, Paşaoğlu Ö. High nucleostemin expression has a favorable prognostic effect on gastric carcinomas. Turk J Med Sci. 2018;48(3):484–490. doi: 10.3906/sag-1705-165. [DOI] [PubMed] [Google Scholar]
- 17.Tang X, Zha L, Li H, et al. Upregulation of GNL3 expression promotes colon cancer cell proliferation, migration, invasion and epithelial-mesenchymal transition via the Wnt/β-catenin signaling pathway. Oncol Rep. 2017;38(4):2023–2032. doi: 10.3892/or.2017.5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng L, Lin T, Peng G, et al. Nucleostemin deletion reveals an essential mechanism that maintains the genomic stability of stem and progenitor cells. Proc Natl Acad Sci USA. 2013;110(28):11415–11420. doi: 10.1073/pnas.1301672110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bao Z, Wang Y, Yang L, et al. Nucleostemin promotes the proliferation of human glioma via Wnt/β-catenin pathway. Neuropathology. 2016;36(3):237–249. doi: 10.1111/neup.12265. [DOI] [PubMed] [Google Scholar]
- 20.Meng L, Lin T, Tsai RYL. Nucleoplasmic mobilization of nucleostemin stabilizes MDM2 and promotes G2-M progression and cell survival. J Cell Sci. 2008;121(Pt 24):4037–4046. doi: 10.1242/jcs.037952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai MS, Sun XX, Lu H. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol Cell Biol. 2008;28(13):4365–4376. doi: 10.1128/MCB.01662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dechow TN, Pedranzini L, Leitch A, et al. Requirement of matrix metalloproteinase-9 for the transformation of human mammary epithelial cells by Stat3-C. Proc Natl Acad Sci USA. 2004;101(29):10602–10607. doi: 10.1073/pnas.0404100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yi WR, Li ZH, Qi BW, Ernest MER, Hu X, Yu AX. Downregulation of IDH2 exacerbates the malignant progression of osteosarcoma cells via increased NF-κB and MMP-9 activation. Oncol Rep. 2016;35(4):2277–2285. doi: 10.3892/or.2016.4553. [DOI] [PubMed] [Google Scholar]
- 24.Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 25.Tsai RYL, Meng L. Nucleostemin: a latecomer with new tricks. Int J Biochem Cell Biol. 2009;41(11):2122–2124. doi: 10.1016/j.biocel.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Back SH, Schröder M, Lee K, Zhang K, Kaufman RJ. ER stress signaling by regulated splicing: IRE1/HAC1/XBP1. Methods. 2005;35(4):395–416. doi: 10.1016/j.ymeth.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Chen X, Iliopoulos D, Zhang Q, et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature. 2014;508(7494):103–107. doi: 10.1038/nature13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng L, Xiao Q, Chen M, et al. Vascular endothelial cell growth-activated XBP1 splicing in endothelial cells is crucial for angiogenesis. Circulation. 2013;127(16):1712–1722. doi: 10.1161/CIRCULATIONAHA.112.001337. [DOI] [PubMed] [Google Scholar]
- 29.Chae U, Park SJ, Kim B, et al. Critical role of XBP1 in cancer signalling is regulated by PIN1. Biochem J. 2016;473(17):2603–2610. doi: 10.1042/BCJ20160482. [DOI] [PubMed] [Google Scholar]