Abstract
Objectives
This study aimed to determine the in vitro susceptibility of commonly encountered Gram-negative bacilli (GNB) recovered from patients admitted to intensive care units (ICUs) in Taiwan against colistin, carbapenems, and other comparative agents.
Methods
In total, 758 nonduplicate GNB isolates were obtained from clinical specimens of ICU patients at seven medical centers in 2016. Minimum inhibitory concentrations (MICs) were determined using the Vitek 2 susceptibility system. The reference broth-microdilution method was performed to determine MICs of colistin. Five main carbapenemase genes among carbapenem-non-susceptible GNB and mcr-1–mcr5 genes among colistin non-wild-type or -resistant isolates were determined.
Results
After exclusion 38 Proteus mirabilis and 13 Morganella morganii spp. among 361 Enterobacteriaceae isolates, 34 (9.4%) isolates were carbapenem-insusceptible, 91.1% (n=31) were colistin wild type, and three and one Klebsiella pneumoniae isolates carried blaKPC and blaOXA48-like, respectively. Carbapenem-insusceptible isolates were found in 23.4% (30 of 128) and 63.0% (87 of 138) of isolates of the Pseudomonas aeruginosa and Acinetobacter baumannii complex, respectively. mcr-1 was detected in two (1.8%) Enterobacter cloacae isolates. Very major errors between two methods of susceptibility to colistin were found in 1.5% of K. pneumoniae, 27.5% of E. cloacae, 4.7% of P. aeruginosa, and 10.1% of A. baumannii complex isolates.
Conclusion
In this study, 8.7% of Enterobacteriaceae isolates from ICUs were not susceptible to carbapenem, and blaKPC and blaOXA48-like were found among three and one carbapenem-insusceptible K. pneumoniae isolates, respectively. Colistin MICs determined by Vitek 2 were not reliable, especially for E. cloacae and A. baumannii complex isolates.
Keywords: colistin, carbapenems, susceptibility, carbapenemase, mcr-1, intensive care units, SMART, P. aeruginosa, A. baumannii
Introduction
Intensive care units (ICUs) cater to saving the lives of critically ill patients, and their use is rapidly growing worldwide.1,2 However, the ICU is also a common place for acquiring nosocomial infections, due to the increasing number of immunocompromised patients and the frequent use of catheters, such as endotracheal tubes, central venous catheters, and Foley catheters.3,4 Moreover, the increasing number of multidrug-resistant organisms (MDROs) that cause health-care-acquired infections in the ICU complicates this condition further.5–7 In 2007, the World Health Organization highlighted in particular the threat of MDR Gram-negative bacteria (GNB), including carbapenem-resistant Acinetobacter baumannii complex, Pseudomonas aeruginosa, and Enterobacteriaceae as critical priority pathogens. There was no exception for Taiwan.8–10 In addition to carbapenem, MDR GNB can also develop resistance to colistin, which is one of the limited antibiotic choices for MDRO infections.11 Several resistance mechanisms, including extended-spectrum β-lactamases, such as the ampC gene, carbapenemase genes, and mcr genes, are reported to be responsible for carbapenem and colistin resistance.11,12 To overcome this life-threatening condition, active infection-control programs including infection surveillance and implementation of prevention guidelines should be a priority.
Therapeutic options for MDROs are limited, and carbapenems and colistin are the last drugs of choice. However, the threat of colistin and carbapenem resistance has become another serious concern globally.13–15 There is an urgent need to address these conditions of MDROs in ICUs. The Surveillance of Multicenter Antimicrobial Resistance in Taiwan (SMART), launched in 2000, is designed to monitor longitudinally the in vitro susceptibility profiles of clinical pathogens to promising antibiotic agents, particularly pathogens isolated from ICUs over time throughout Taiwan.5,6,16–19
This study aimed to determine the in vitro susceptibilities of commonly encountered GNB, including Enterobacteriaceae, and nonfermentative GNB (NFGNB) isolated from patients admitted to ICUs at different locations in Taiwan against colistin, carbapenems, and other comparative agents. It also investigated the prevalence of carbapenemase genes and mcr genes among carbapenem-insusceptible and colistin non-wild-type (NWT) isolates, respectively.
Methods
Bacterial isolates
We analyzed 758 nonduplicate isolates of GNB collected from various specimens of patients admitted to ICUs at seven medical centers from January to December 2016. One participating hospital submitted only 57 clinical isolates, whereas the other six hospitals submitted more than 100 each. These clinical isolates included A. baumannii complex (n=138), Klebsiella pneumoniae (n=137), P. aeruginosa (n=128), Escherichia coli (n=121), Stenotrophomonas maltophilia (n=61), Enterobacter cloacae (n=51), Serratia marcescens (n=42), Proteus mirabilis (n=38), Burkholderia cepacia (n=19), Morganella morganii (n=13), and Citrobacter freundii (n=10) (Table 1). Sputum/endotracheal aspirates were the most common source of isolates (n=495, 65.3%), followed by blood (n=93, 12.3%), urine (n=90, 11.9%), pus/abscess (n=42, 5.5%), and ascites (n=13, 1.7%) (Table 1). All the isolates were stored at –70°C in trypticase soy broth (BD, Franklin Lakes, NJ, USA) supplemented with 15% glycerol prior to testing. The isolates were then transported to National Taiwan University Hospital, Taipei for further identification using the Phoenix PMIC/ID-30 identification system (BD). The institutional review board of National Taiwan University Hospital (201512064RSB) approved this study and waived the requirement for written informed consent.
Table 1.
Clinical isolates obtained from patients admitted to the intensive care units of seven main teaching hospitals in Taiwan in 2016
| Sources | Isolates | Isolates (total 758), n (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli (n=121) | K. pneumoniae (n=137) | P. aeruginosa (n=128) | A. baumannii complex (n=138) | C. freundii (n=10) | E. cloacae (n=51) | S. marcescens (n=42) | P. mirabilis (n=38) | M. morganii (n=13) | B. cepacia (n=19) | S. maltophilia (n=61) | |||
| Hospital (location within Taiwan) | |||||||||||||
| NTUH (N) | 20 | 20 | 20 | 20 | 5 | 10 | 10 | 10 | 4 | 10 | 10 | 139 (18.3) | |
| TMWFH (N) | 1 | 18 | 8 | 20 | 2 | 1 | 1 | 0 | 1 | 1 | 4 | 57 (7.5) | |
| VGH Taichung (M) | 20 | 20 | 20 | 20 | 0 | 10 | 5 | 1 | 0 | 1 | 9 | 106 (14.0) | |
| CMMC (S) | 20 | 20 | 20 | 20 | 2 | 9 | 10 | 10 | 4 | 0 | 10 | 125 (16.5) | |
| NCKUH (S) | 20 | 20 | 20 | 20 | 1 | 10 | 4 | 3 | 3 | 0 | 10 | 111 (14.6) | |
| KMUH (S) | 20 | 20 | 20 | 18 | 0 | 3 | 4 | 9 | 0 | 5 | 9 | 108 (14.2) | |
| VGH Kaohsiung (S) | 20 | 19 | 20 | 20 | 0 | 8 | 8 | 5 | 1 | 2 | 9 | 112 (14.8) | |
| Clinical sources | |||||||||||||
| Sputum/endotracheal aspirates | 40 | 93 | 99 | 115 | 5 | 34 | 29 | 17 | 3 | 9 | 51 | 495 (65.3) | |
| Blood | 17 | 16 | 13 | 9 | 2 | 5 | 8 | 4 | 1 | 10 | 8 | 93 (12.3) | |
| Urine | 44 | 19 | 7 | 6 | 1 | 3 | 2 | 6 | 1 | — | 1 | 90 (11.9) | |
| Pus/wound | 7 | 3 | 3 | 5 | 2 | 8 | 3 | 7 | 4 | — | — | 42 (5.5) | |
| Ascites | 7 | — | 1 | 1 | — | — | — | 1 | 2 | — | 1 | 13 (1.7) | |
| Abscess fluids | 2 | 2 | 1 | 1 | — | — | — | 1 | — | — | — | 7 (0.9) | |
| Bile | 2 | 1 | — | — | — | 1 | — | — | — | — | — | 4 (0.5) | |
| Cerebrospinal fluid | — | 1 | — | — | — | — | — | — | — | — | — | 1 (0.1) | |
| Othersa | 2 | 2 | 4 | 1 | — | — | — | 2 | 2 | — | — | 13 (1.7) | |
Note:
Vaginal discharges (n=7), intravenous catheters (n=3), and tissue biopsy (n=3).
Abbreviations: CMMC, Chi Mei Medical Center; KMUH, Kaohsiung Medical University Hospital; M, middle; N, Northern; NCKUH, National Cheng Kung University Hospital; NTUH, National Taiwan University Hospital; S, Southern; TMWFH, Taipei Municipal Wan-Fang Hospital; VGH, Veterans General Hospital; E. coli; Escherichia coli; K. pneumoniae; Klebsiella pneumoniae; P. aeruginosa; Pseudomonas aeruginosa; A. baumannii; Acinetobacter baumannii; C. freundii; Citrobacter freundii; E. cloacae; Enterobacter cloacae; S. marcescens, Serratia marcescens; P. mirabilis, Proteus mirabilis; M. morganii; Morganella morganii; B. cepacia, Burkholderia cepacia; S. maltophilia; Stenotrophomonas maltophilia.
Antimicrobial-susceptibility testing
Minimum inhibitory concentrations (MICs) of 17 antimicrobial agents to the isolates, including colistin, were determined using the commercial Vitek 2 antimicrobial-susceptibility system (AST-NB card; BioMérieux, Marcy l’Etoile, France). The MICs of colistin were also determined using the reference broth microdilution (BMD) method recommended by the Clinical and Laboratory Standards Institute (CLSI).20 Ampicillin–sulbactam testing was performed with a 2:1 ratio and piperacillin–tazobactam testing with a fixed concentration (4 mg/L) of tazobactam. Interpretations of all MIC results were in accordance with the CLSI guidelines.20 E. coli ATCC 25922 and P. aeruginosa (ATCC 27853) were used as quality-control strains for each run of the MIC tests.
In addition to Proteus spp. and Morganella morganii, which have intrinsically elevated MICs to imipenem, carbapenem-insusceptible isolates were defined as clinical isolates exhibiting insusceptibility to any of the carbapenems, including ertapenem, imipenem, and meropenem. As per the CLSI, the E. coli, K. pneumoniae, and E. cloacae isolates are known as wild type (WT; MICs ≤2 mg/L) and non-WT (NWT; MICs ≥4 mg/L) based on their susceptibility to colistin. For P. aeruginosa and A. baumannii complex isolates, MICs of ≤2 and ≥4 mg/L for colistin are identified as susceptible and resistant, respectively.17 For the six other isolates tested in this study — P. mirabilis, M. morganii, C. freundii, S. marcescens, B. cepacia, and S. maltophilia — there were no CLSI MIC-interpretation criteria for defining susceptibility.20
To examine intertest agreement between the two methods for determining susceptibility to colistin, essential and categorical agreement and very major error (VME) were evaluated. Essential agreement between BMD and Vitek 2 susceptibility testing was measured as the difference between MICs of ±1 log2 dilution or less using BMD as a reference standard. Categorical agreement between the two susceptibility-testing methods was measured as the percentage of isolates that had concordant test results when determining susceptible or WT and resistant or NWT to colistin. A VME for the Vitek 2 was defined as discrepancy in MICs between the methods when a colistin-resistant or NWT isolate determined using the reference BMD method was interpreted as a colistin-susceptible or WT isolate by the Vitek 2.
Determination of carbapenemase-encoding genes among carbapenem-insusceptible Enterobacteriaceae
The Xpert Carba-R assay (Cepheid, Sunnyvale, CA, USA) was used to detect carbapenemase-encoding alleles, including blaKPC, blaNDM, blaIMP, blaVIM, and blaOXA48-like, among the carbapenem-insusceptible Enterobacteriaceae.15,16
Determination of mcr-1–mcr5 genes
PCR amplification of whole-cell DNA of isolates showing colistin MICs >2 mg/L was performed using previously described primers for mcr-1, mcr2, mcr3, mcr4, and mcr5, and PCR products were sequenced.21
Statistical analysis
We compared rates of WT susceptibility to colistin among selected isolates from patients admitted to ICUs of seven main teaching hospitals in Taiwan in this study and those from data reported in 2007.6 Analyses were performed using Excel 2013 (Microsoft, Redmond, WA, USA), and P<0.05 was considered statistically significant.
Results
Antimicrobial susceptibility
Table 2 shows the antibiotic susceptibility of 758 clinical isolates. Amikacin showed the highest in vitro activity against both Enterobacteriaceae and P. aeruginosa, and resistance rates were <8%. Among Enterobacteriaceae isolates, all three carbapenems — ertapenem, imipenem, and meropenem — exhibited good activity, with a resistance rate <10%, except E. cloacae (ertapenem-resistance rate:17.6%) and C. freundii (ertapenem-, imipenem-, and meropenem-resistance rates 10%, 20%, and 20%, respectively). Among the 128 P. aeruginosa isolates, imipenem- and meropenem-resistance rates were 20.3% and 19.5%, respectively. Among the 138 A. baumannii complex isolates, imipenem- and meropenem-resistance rates were 62.3% and 61.6%, respectively. Among the 61 S. maltophilia isolates, resistance rates of levofloxacin and trimethoprim–sulfamethoxazole were 16.4% and 29.5%, respectively. For B. cepacia, the susceptibility rate of both ceftazidime and meropenem was 100%.
Table 2.
Antimicrobial susceptibility of Gram-negative bacteria isolated from patients admitted to intensive care units of seven main teaching hospitals in Taiwan in 2016
| Organism and agents tested | MIC (mg/L) | Isolates, n (%) | ||||
|---|---|---|---|---|---|---|
| Range | MIC50 | MIC90 | S (%) | I (%) | R (%) | |
| Escherichia coli (121) | ||||||
| Ampicillin–sulbactam | ≤2–≥32 | ≥32 | ≥32 | 35 (28.9) | 12 (9.9) | 74 (61.2) |
| Cefazolin | ≤4–≥64 | ≥64 | ≥64 | 51 (42.1) | — | 70 (57.9) |
| Cefmetazole | ≤1–≥64 | ≤1 | 32 | 101 (83.5) | 10 (8.3) | 10 (8.3) |
| Cefotaxime | ≤1–≥64 | ≤1 | ≥64 | 64 (52.9) | 1 (0.8) | 56 (46.3) |
| Ceftazidime | ≤1–≥64 | ≤1 | ≥64 | 84 (69.4) | 1 (0.8) | 36 (29.8) |
| Cefepime | ≤1–≥64 | ≤1 | ≥64 | 101 (83.5) | 5 (4.1) | 15 (12.4) |
| Piperacillin–tazobactam | ≤4–≥128 | ≤4 | ≥128 | 93 (76.9) | 14 (11.6) | 14 (11.6) |
| Ertapenem | ≤0.5–≥8 | ≤0.5 | ≤0.5 | 118 (97.5) | 1 (0.8) | 2 (1.7) |
| Imipenem | ≤0.25–4 | ≤0.25 | ≤0.25 | 120 (99.2) | 0 | 1 (0.8) |
| Meropenem | ≤0.25–4 | ≤0.25 | ≤0.25 | 120 (99.2) | 0 | 1 (0.8) |
| Ciprofloxacin | ≤0.25–≥4 | 0.5 | ≥4 | 74 (61.2) | 1 (0.8) | 46 (38.0) |
| Levofloxacin | ≤0.12–≥8 | 1 | ≥8 | 75 (62.0) | 1 (0.8) | 45 (37.2) |
| Gentamicin | ≤1–≥16 | ≤1 | ≥16 | 91 (75.2) | 1 (0.8) | 29 (24.0) |
| Amikacin | ≤2–16 | ≤2 | 4 | 121 (100) | 0 | 0 |
| Trimethoprim–sulfamethoxazole | ≤1–≥16 | ≤1 | ≥16 | 73 (60.3) | — | 48 (39.7) |
| Tigecycline | ≤0.5–4 | ≤0.5 | ≤0.5 | NA | NA | NA |
| Colistin | ≤0.5 | ≤0.5 | ≤0.5 | NA | NA | NA |
| Colistin-BMD | 0.5–2 | 1 | 1 | NA | NA | NA |
| Klebsiella pneumoniae (137) | ||||||
| Ampicillin–sulbactam | ≤2–≥32 | 8 | ≥32 | 77 (56.2) | 5 (3.6) | 55 (40.1) |
| Cefazolin | ≤4–≥64 | ≤4 | ≥64 | 0 | 84 (61.3) | 53 (38.7) |
| Cefmetazole | ≤1–≥64 | ≤1 | ≥64 | 102 (74.5) | 16 (11.7) | 19 (13.9) |
| Cefotaxime | ≤1–≥64 | ≤1 | ≥64 | 96 (70.1) | 6 (4.4) | 35 (25.5) |
| Ceftazidime | ≤1–≥64 | ≤1 | ≥64 | 98 (71.5) | 6 (4.4) | 33 (24.1) |
| Cefepime | ≤1–≥64 | ≤1 | 32 | 117 (85.4) | 2 (1.5) | 18 (13.1) |
| Piperacillin–tazobactam | ≤4–≥128 | ≤4 | ≥128 | 97 (70.8) | 13 (9.5) | 27 (19.7) |
| Ertapenem | ≤0.5–≥8 | ≤0.5 | ≤0.5 | 124 (90.5) | 5 (3.6) | 8 (5.8) |
| Imipenem | ≤0.25–≥16 | ≤0.25 | 1 | 125 (91.2) | 8 (5.8) | 4 (2.9) |
| Meropenem | ≤0.25–≥16 | ≤0.25 | ≤0.25 | 130 (94.9) | 0 | 7 (5.1) |
| Ciprofloxacin | ≤0.25–≥4 | ≤0.25 | ≥4 | 107 (78.1) | 1 (0.7) | 29 (21.2) |
| Levofloxacin | ≤0.12–≥8 | ≤0.12 | ≥8 | 105 (76.6) | 4 (2.9) | 28 (20.4) |
| Gentamicin | ≤1–≥16 | ≤1 | ≥16 | 97 (70.8) | 9 (6.6) | 31 (22.6) |
| Amikacin | ≤2–≥64 | ≤2 | ≤2 | 132 (96.4) | 0 | 5 (3.6) |
| Trimethoprim–sulfamethoxazole | ≤1–≥16 | ≤1 | ≥16 | 89 (65.0) | — | 48 (35.0) |
| Tigecycline | ≤0.5–≥8 | ≤0.5 | ≥8 | NA | NA | NA |
| Colistin | ≤0.5–≥16 | ≤0.5 | ≤0.5 | NA | NA | NA |
| Colistin BMD | 0.5–16 | 1 | 1 | NA | NA | NA |
| Enterobacter cloacae (51) | ||||||
| Cefazolin | ≤4–≥64 | ≥64 | ≥64 | 1 (2.0) | 50 (98.0) | |
| Cefmetazole | 2–≥64 | ≥64 | ≥64 | 2 (3.9) | 3 (5.9) | 46 (90.2) |
| Cefotaxime | ≤1–≥64 | ≤1 | ≥64 | 30 (58.8) | 0 | 21 (41.2) |
| Ceftazidime | ≤1–≥64 | ≤1 | ≥64 | 31 (60.8) | 0 | 20 (39.2) |
| Cefepime | ≤1–≥64 | ≤1 | 16 | 39 (76.5) | 5 (9.8) | 7 (13.7) |
| Ertapenem | ≤0.5–≥8 | ≤0.5 | 4 | 40 (78.4) | 2 (3.9) | 9 (17.6) |
| Imipenem | ≤0.25–≥16 | 0.5 | 2 | 42 (82.4) | 4 (7.8) | 5 (9.8) |
| Meropenem | ≤0.25–≥16 | ≤0.25 | 1 | 47 (92.2) | 1 (2.0) | 3 (5.9) |
| Ciprofloxacin | ≤0.25–≥4 | ≤0.25 | ≥4 | 41 (80.4) | 2 (3.9) | 8 (15.7) |
| Levofloxacin | ≤0.12–≥8 | ≤0.12 | ≥8 | 41 (80.4) | 2 (3.9) | 8 (15.7) |
| Gentamicin | ≤1–≥16 | ≤1 | ≥16 | 43 (84.3) | 0 | 8 (15.7) |
| Amikacin | ≤2–32 | ≤2 | 4 | 49 (96.1) | 2 (3.9) | 0 |
| Trimethoprim–sulfamethoxazole | ≤1–≥16 | ≤1 | ≥16 | 38 (74.5) | — | 13 (25.5) |
| Tigecycline | ≤0.5–≥8 | 1 | ≥8 | NA | NA | NA |
| Colistin | ≤0.5–≥16 | ≤0.5 | ≤0.5 | NA | NA | NA |
| Colistin BMD | 0.5–>32 | 1 | >32 | NA | NA | NA |
| Serratia marcescens (42) | ||||||
| Cefazolin | ≥64–≥64 | ≥64 | ≥64 | 0 | 0 | 42 (100) |
| Cefmetazole | 4–≥64 | 8 | ≥64 | 35 (83.3) | 1 (2.4) | 6 (14.3) |
| Cefotaxime | ≤1–≥64 | ≤1 | 32 | 28 (66.7) | 2 (4.8) | 12 (28.6) |
| Ceftazidime | ≤1–≥64 | ≤1 | ≥64 | 37 (88.1) | 0 | 5 (11.9) |
| Cefepime | ≤1–32 | ≤1 | 8 | 35 (83.3) | 5 (11.9) | 2 (4.8) |
| Piperacillin–tazobactam | ≤4–≥128 | ≤4 | 16 | 39 (92.9) | 0 | 3 (7.1) |
| Ertapenem | ≤0.5 | ≤0.5 | ≤0.5 | 42 (100) | 0 | 0 |
| Meropenem | ≤0.25 | ≤0.25 | ≤0.25 | 42 (100) | 0 | 0 |
| Ciprofloxacin | ≤0.25–≥4 | ≤0.25 | ≥4 | 36 (85.7) | 0 | 6 (14.3) |
| Levofloxacin | ≤0.12–≥8 | ≤0.12 | ≥8 | 34 (81.0) | 3 (7.1) | 5 (11.9) |
| Gentamicin | ≤1–≥16 | ≤1 | 8 | 35 (83.3) | 3 (7.1) | 4 (9.5) |
| Amikacin | ≤2–≥64 | ≤2 | 8 | 40 (95.2) | 0 | 2 (4.8) |
| Trimethoprim–sulfamethoxazole | ≤1–≥16 | ≤1 | ≤1 | 40 (95.2) | — | 2 (4.8) |
| Tigecycline | ≤0.5–≥8 | 1 | ≥8 | NA | NA | NA |
| Colistin BMD | 1–>32 | >32 | >32 | NA | NA | NA |
| Proteus mirabilis (38) | ||||||
| Ampicillin–sulbactam | ≤2–≥32 | 4 | ≥32 | 20 (52.6) | 7 (18.4) | 11 (28.9) |
| Cefazolin | ≤4–≥64 | ≤4 | ≥64 | 19 (50) | 19 (50) | |
| Cefmetazole | ≤1–≥64 | 2 | 4 | 37 (97.4) | 0 | 1 (2.6) |
| Cefotaxime | ≤1–≥64 | ≤1 | 8 | 30 (78.9) | 0 | 8 (21.1) |
| Ceftazidime | ≤1–≥64 | ≤1 | 4 | 35 (92.1) | 0 | 3 (7.9) |
| Cefepime | ≤1–≥64 | ≤1 | 4 | 33 (86.8) | 3 (7.9) | 2 (5.3) |
| Piperacillin–tazobactam | ≤4–16 | ≤4 | ≤4 | 38 (100) | 0 | 0 |
| Ertapenem | ≤0.5–≥8 | ≤0.5 | ≤0.5 | 36 (94.7) | 1 (2.6) | 1 (2.6) |
| Imipenem | ≤0.25–≥16 | 4 | 8 | 2 (5.3) | 14 (36.8) | 22 (57.9) |
| Meropenem | ≤0.25–8 | ≤0.25 | 1 | 37 (97.4) | 0 | 1 (2.6) |
| Ciprofloxacin | ≤0.25–≥4 | ≤0.25 | ≥4 | 24 (63.2) | 5 (13.2) | 9 (23.7) |
| Levofloxacin | ≤0.12–≥8 | 0.5 | ≥8 | 26 (68.4) | 6 (15.8) | 6 (15.8) |
| Gentamicin | ≤1–≥16 | ≤1 | ≥16 | 20 (52.6) | 6 (15.8) | 12 (31.6) |
| Amikacin | ≤2–≥64 | ≤2 | 8 | 34 (89.5) | 1 (2.6) | 3 (7.9) |
| Trimethoprim–sulfamethoxazole | ≤1–≥16 | ≥16 | ≥16 | 11 (28.9) | — | 27 (71.1) |
| Tigecycline | 1–≥8 | 4 | 4 | NA | NA | NA |
| Colistin BMD | >32 | >32 | >32 | NA | NA | NA |
| Morganella morganii (13) | ||||||
| Ampicillin–sulbactam | 16–≥32 | ≥32 | ≥32 | 0 | 1 (7.7) | 12 (92.3) |
| Cefazolin | ≥64–≥64 | ≥64 | ≥64 | 0 | 0 | 13 (100) |
| Cefmetazole (12) | 8–≥64 | 8 | 32 | 10 (83.3) | 1 (8.3) | 1 (8.3) |
| Cefotaxime | ≤1–≥64 | ≤1 | ≥64 | 8 (61.5) | 0 | 5 (38.5) |
| Ceftazidime | ≤1–≥64 | ≤1 | ≥64 | 9 (69.2) | 0 | 4 (30.8) |
| Cefepime | ≤1–≥64 | ≤1 | 8 | 11 (84.6) | 1 (7.7) | 1 (7.7) |
| Piperacillin–tazobactam | ≤4–≥128 | ≤4 | ≤4 | 12 (92.3) | 0 | 1 (7.7) |
| Ertapenem | ≤0.5–≤0.5 | ≤0.5 | ≤0.5 | 13 (100) | 0 | 0 |
| Imipenem | ≤0.25–8 | 2 | 8 | 3 (23.1) | 4 (30.8) | 6 (46.2) |
| Meropenem | ≤0.25–1 | ≤0.25 | 1 | 13 (100) | 0 | 0 |
| Ciprofloxacin | ≤0.25–≥4 | ≤0.25 | ≥4 | 11 (84.6) | 0 | 2 (15.4) |
| Levofloxacin | ≤0.12–≥8 | ≤0.12 | ≥8 | 11 (84.6) | 0 | 2 (15.4) |
| Gentamicin | ≤1–≥16 | ≤1 | ≥16 | 9 (69.2) | 1 (7.7) | 3 (23.1) |
| Amikacin | ≤2–≥64 | ≤2 | 4 | 12 (92.3) | 0 | 1 (7.7) |
| Trimethoprim–sulfamethoxazole | ≤1–≥16 | ≤20 | ≥320 | 9 (69.2) | — | 4 (30.8) |
| Tigecycline | ≤0.5–≥8 | 1 | 4 | NA | NA | NA |
| Colistin | ≥16 | ≥16 | ≥16 | NA | NA | NA |
| Colistin BMD | >32 | >32 | >32 | NA | NA | NA |
| Citrobacter freundii (10) | ||||||
| Cefazolin | ≥64 | ≥64 | ≥64 | 0 | 0 | 10 (100) |
| Cefmetazole | 32–≥64 | 32 | ≥64 | 0 | 5 (50) | 5 (50) |
| Cefotaxime | ≤1–≥64 | ≤1 | ≥64 | 5 (50) | 0 | 5 (50) |
| Ceftazidime | ≤1–≥64 | 2 | ≥64 | 5 (50) | 0 | 5 (50) |
| Cefepime | ≤1–4 | ≤1 | 2 | 9 (90) | 1 (10) | 0 |
| Piperacillin–tazobactam | ≤4–≥128 | 32 | ≥128 | 4 (40) | 3 (30) | 3 (30) |
| Ertapenem | ≤0.5–4 | ≤0.5 | ≤0.5 | 9 (90) | 0 | 1 (10) |
| Imipenem | ≤0.25–≥16 | 0.5 | ≥16 | 8 (80) | 0 | 2 (20) |
| Meropenem | ≤0.25–≥16 | ≤0.25 | ≥16 | 8 (80) | 0 | 2 (20) |
| Ciprofloxacin | ≤0.25–2 | ≤0.25 | 1 | 9 (90) | 1 (10) | 0 |
| Levofloxacin | ≤0.12–4 | 0.5 | 4 | 8 (80) | 2 (20) | 0 |
| Gentamicin | ≤1–≥16 | ≤1 | ≥16 | 8 (80) | 0 | 2 (20) |
| Amikacin | ≤2 | ≤2 | ≤2 | 10 (100) | 0 | 0 |
| Trimethoprim–sulfamethoxazole | ≤1 | ≤1 | ≤1 | 10 (100) | — | 0 |
| Tigecycline | ≤0.5–1 | ≤0.5 | 1 | NA | NA | NA |
| Colistin | ≤0.5 | ≤0.5 | ≤0.5 | NA | NA | NA |
| Colistin BMD | 0.5–1 | 1 | 1 | NA | NA | NA |
| Pseudomonas aeruginosa (128) | ||||||
| Ceftazidime | ≤1–≥64 | 4 | 32 | 89 (69.5) | 23 (18.0) | 16 (12.5) |
| Cefepime | ≤1–≥64 | 2 | 16 | 107 (83.6) | 11 (8.6) | 10 (7.8) |
| Piperacillin–tazobactam | ≤4–≥128 | 8 | ≥128 | 74 (57.8) | 18 (14.1) | 36 (28.1) |
| Imipenem | ≤0.25–≥16 | 2 | ≥16 | 102 (79.7) | 0 | 26 (20.3) |
| Meropenem | ≤0.25–≥16 | 0.5 | ≥16 | 98 (76.6) | 5 (3.9) | 25 (19.5) |
| Ciprofloxacin | ≤0.25–≥4 | ≤0.25 | ≥4 | 105 (82.0) | 4 (3.1) | 19 (14.8) |
| Levofloxacin | ≤0.12–≥8 | 1 | ≥8 | 97 (75.8) | 10 (7.8) | 21 (16.4) |
| Gentamicin | ≤1–≥16 | ≤1 | 4 | 117 (91.4) | 5 (3.9) | 6 (4.7) |
| Amikacin | ≤2–≥64 | ≤2 | 4 | 127 (99.2) | 0 | 1 (0.8) |
| Trimethoprim–sulfamethoxazole | ≤1–≥16 | 8 | ≥16 | NA | NA | NA |
| Tigecycline | ≤0.5–≥8 | ≥8 | ≥8 | NA | NA | NA |
| Colistin | ≤0.5 | ≤0.5 | ≤0.5 | 128 (100) | — | 0 |
| Colistin BMD | 1–8 | 2 | 2 | 122 (95.3) | — | 6 (4.7) |
| Acinetobacter baumannii complex (138) | ||||||
| Ampicillin–sulbactam | ≤2–≥32 | 16 | ≥32 | 61 (44.2) | 18 (13.0) | 59 (42.8) |
| Cefotaxime | ≤1–≥64 | ≥64 | ≥64 | 42 (30.4) | 22 (15.9) | 74 (53.6) |
| Ceftazidime | ≤1–≥64 | ≥64 | ≥64 | 47 (34.1) | 18 (13.0) | 73 (52.9) |
| Cefepime | ≤1–≥64 | ≥64 | ≥64 | 48 (34.8) | 2 (1.4) | 88 (63.8) |
| Piperacillin–tazobactam | ≤4–≥128 | ≥128 | ≥128 | 44 (31.9) | 0 | 94 (68.1) |
| Imipenem | ≤0.25–≥16 | ≥16 | ≥16 | 51 (37.0) | 1 (0.7) | 86 (62.3) |
| Meropenem | ≤0.25–≥16 | ≥16 | ≥16 | 51 (37.0) | 2 (1.4) | 85 (61.6) |
| Ciprofloxacin | ≤0.25–≥4 | ≥4 | ≥4 | 44 (31.9) | 0 | 94 (68.1) |
| Levofloxacin | ≤0.12–≥8 | 4 | ≥8 | 45 (32.6) | 30 (21.7) | 63 (45.7) |
| Gentamicin | ≤1–≥16 | ≥16 | ≥16 | 65 (47.1) | 3 (2.2) | 70 (50.7) |
| Trimethoprim–sulfamethoxazole | ≤1–≥16 | 8 | ≥16 | 56 (40.6) | — | 82 (59.4) |
| Tigecycline | ≤0.5–≥8 | 1 | 4 | NA | NA | NA |
| Colistin | ≤0.5–2 | 0.5 | 0.5 | 138 (100) | — | 0 |
| Colistin BMD | 0.5–16 | 2 | 2 | 124 (89.9) | — | 14 (10.1) |
| Stenotrophomonas maltophilia (61) | ||||||
| Levofloxacin | 0.25–≥8 | 1 | ≥8 | 47 (77.0) | 4 (6.6) | 10 (16.4) |
| Trimethoprim–sulfamethoxazole | ≤1–≥16 | ≤1 | ≥16 | 43 (70.5) | — | 18 (29.5) |
| Colistin BMD (60) | 2–>32 | >32 | >32 | NA | NA | NA |
| Burkholderia cepacia (19) | ||||||
| Ceftazidime | 2–4 | 4 | 4 | 19 (100) | 0 | 0 |
| Cefepime | 2–32 | 8 | 32 | NA | NA | NA |
| Piperacillin–tazobactam | ≥128 | ≥128 | ≥128 | NA | NA | NA |
| Imipenem | ≥16 | ≥16 | ≥16 | NA | NA | NA |
| Meropenem | 1–4 | 4 | 4 | 19 (100) | 0 | 0 |
| Ciprofloxacin | 1–≥4 | 2 | ≥4 | NA | NA | NA |
| Levofloxacin | 1–≥8 | 4 | 4 | 6 (31.6) | 11 (57.9) | 2 (10.5) |
| Gentamicin | ≥16 | ≥16 | ≥16 | NA | NA | NA |
| Amikacin | ≥64 | ≥64 | ≥64 | NA | NA | NA |
| Trimethoprim–sulfamethoxazole | ≤1–4 | ≤1 | ≤1 | 18 (94.7) | — | 1 (5.3) |
| Tigecycline | 2–≥8 | ≥8 | ≥8 | NA | NA | NA |
| Colistin | ≥16 | ≥16 | ≥16 | NA | NA | NA |
| Colistin BMD | >32 | >32 | >32 | NA | NA | NA |
Abbreviations: BMD, broth microdilution; MIC, minimum inhibitory concentration; NA, not available; I, intermediate; R, resistant; S, susceptible.
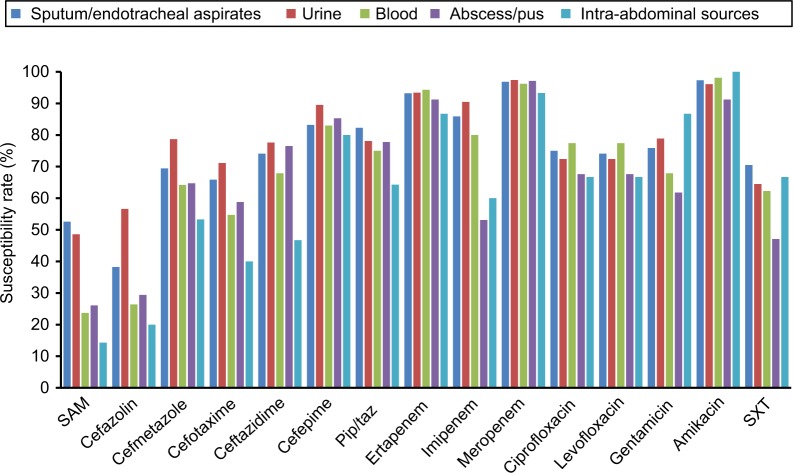
We further analyzed the antimicrobial-susceptibility pattern of Enterobacteriaceae isolated from different sources (Figure 1). Among the pathogens isolated from sputum/endotracheal aspirates, carbapenems had the best activity, with susceptibility >85%, and other commonly used antibiotics — ceftazidime, cefepime, piperacillin–tazobactam, ciprofloxacin, and levofloxacin — exhibited good activity, with susceptibility >70%. Similar patterns were noted for isolates from urinary and blood specimens. However, for the isolates from abscess/pus and intra-abdominal sources, imipenem-resistance rates were 21.9% (n=7) and 33.3% (n=5), respectively, which were much higher than the other two carbapenems, ertapenem and meropenem, which exhibited <10% resistance.
Figure 1.
Antibiotic-susceptibility rate of Enterobacteriaceae according to source of isolation.
Abbreviations: SAM, sulbactam–ampicillin; Pip/taz, piperacillin–tazobactam; SXT, sulfamethoxazole–trimethoprim.
Carbapenem-resistant Enterobacteriaceae, P. aeruginosa, A. baumannii complex, and carbapenemases
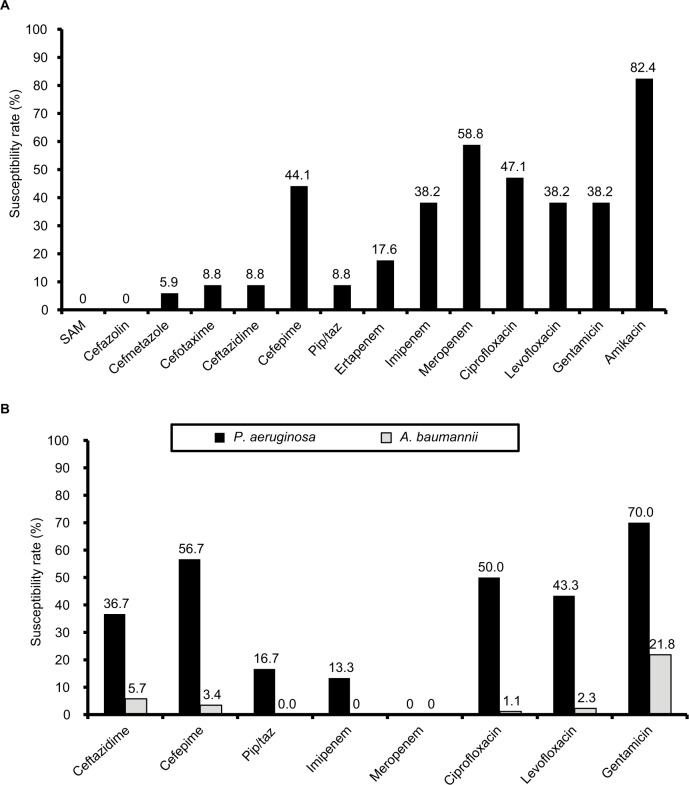
After excluding 38 P. mirabilis and 13 M. morganii isolates among the remaining 361 Enterobacteriaceae isolates, 34 (9.4%) were classified as carbapenem-insusceptible, including K. pneumoniae (n=18), E. cloacae (n=11), E. coli (n=3), and C. freundii (n=2). The prevalence of carbapenem-insusceptible Enterobacteriaceae among each Enterobacteriaceae species was highest for E. cloacae (21.6%), followed by C. freundii (20.0%), K. pneumoniae (13.1%), and E. coli (2.5%). Only amikacin showed good in vitro activity, with a susceptibility rate of 82.4% (Figure 2A). The susceptibility rates of imipenem and meropenem against 28 ertapenem-insusceptible Enterobacteriaceae were 46.4% (n=13) and 57.1% (n=16), respectively. Susceptibility rates of ertapenem and meropenem against 21 imipenem-insusceptible Enterobacteriaceae were 28.5% (n=6) and 42.9% (n=9), respectively. Susceptibility rates of ertapenem and imipenem against 15 meropenem-insusceptible Enterobacteriaceae were only 6.7% (n=1) and 13.3% (n=2), respectively. In total, 30 (23.4%) and 87 (63.0%) carbapenem-insusceptible P. aeruginosa and A. baumannii complex isolates, respectively, were identified, and their susceptibility patterns are shown in Figure 2B. For carbapenem-insusceptible A. baumannii complex isolates, all antibiotics tested showed poor in vitro activity. For carbapenem-insusceptible P. aeruginosa isolates, gentamicin exhibited good in vitro activity. In addition, 91.1% (n=31) of carbapenem-insusceptible Enterobacteriaceae exhibited WT susceptibility to colistin. All carbapenem-insusceptible Enterobacteriaceae isolates were screened for carbapenemases using the Xpert Carba-R. Furthermore, three and one K. pneumoniae isolates were found to carry the resistance genes blaKPC and blaOXA48, respectively. The three K. pneumoniae isolates carrying blaKPC were resistant to most of the other antibiotics, but the K. pneumoniae isolate carrying blaOXA48 was susceptible to most of the antibiotics (Table 3). All these four clinical isolates were colistin WT.
Figure 2.
Antibiotic-susceptibility rates.
Notes: (A) Carbapenem-insusceptible Enterobacteriaceae; (B) Pseudomonas aeruginosa and Acinetobacter baumannii complex. Abbreviations: SAM, sulbactam–ampicillin; Pip/taz, piperacillin–tazobactam.
Table 3.
Antimicrobial susceptibility of four carbapenem-insusceptible Klebsiella pneumoniae isolates harboring carbapenemase genes, and two mcr-1-carrying Enterobacter cloacae isolates to selected agents
| K. pneumoniae isolates (MIC, mg/L) | E. cloacae isolates (MIC, mg/L) | |||||
|---|---|---|---|---|---|---|
| KP1 | KP2 | KP3 | KP4 | EC1 | EC2 | |
| Source | Blood | Drainage | CSF | Surgical wound | Sputum | Urine |
| Resistant gene | blaKPC | blaKPC | blaKPC | blaOXA48 | mcr-1 | mcr-1 |
| Ampicillin–sulbactam | ≥32 (R) | ≥32 (R) | ≥32 (R) | ≥32 (R) | — | — |
| Cefazolin | ≥64 (R) | ≥64 (R) | ≥64 (R) | 4 (I) | ≥64 (R) | ≥64 (R) |
| Cefmetazole | ≥64 (R) | ≥64 (R) | ≥64 (R) | 1 (S) | ≥64 (R) | ≥64 (R) |
| Cefotaxime | ≥64 (R) | ≥64 (R) | ≥64 (R) | 1 (S) | ≤1 (S) | ≤1 (S) |
| Ceftazidime | ≥64 (R) | ≥64 (R) | ≥64 (R) | 1 (S) | ≤1 (S) | ≤1 (S) |
| Cefepime | ≥64 (R) | ≥64 (R) | ≥64 (R) | 1 (S) | ≤1 (S) | ≤1 (S) |
| Piperacillin–tazobactam | ≥128 (R) | ≥128 (R) | ≥128 (R) | ≥128 (R) | — | — |
| Ertapenem | ≥8 (R) | ≥8 (R) | ≥8 (R) | ≤0.5 (S) | ≤0.5 (S) | ≤0.5 (S) |
| Imipenem | ≥16 (R) | ≥16 (R) | ≥16 (R) | 2 (R) | 1 (S) | ≤0.25 (S) |
| Meropenem | ≥16 (R) | ≥16 (R) | ≥16 (R) | ≤0.25 (S) | ≤0.25 (S) | ≤0.25 (S) |
| Ciprofloxacin | ≥4 (R) | ≥4 (R) | ≥4 (R) | 1 (S) | ≤0.25 (S) | ≤0.25 (S) |
| Levofloxacin | ≥8 (R) | ≥8 (R) | ≥8 (R) | 1 (S) | ≤0.12 (S) | ≤0.12 (S) |
| Gentamicin | 1 (S) | ≥16 (R) | ≥16 (R) | ≤1 (S) | ≤1 (S) | ≤1 (S) |
| Amikacin | 2 (S) | ≥64 (R) | 4 (S) | ≤2 (S) | ≤2 (S) | ≤2 (S) |
| Tigecycline | 4 (S) | ≥8 (R) | 1 (S) | ≤0.5 (S) | 2 (S) | 1 (S) |
| Colistin | ≤0.5 (S) | ≤0.5 (S) | ≤0.5 (S) | ≤0.5 (S) | ≤0.5 (S) | ≤0.5 (S) |
| Colistin BMD | 1 (S) | 0.5 (S) | 1 (S) | 1 (S) | 32 (R) | 32 (R) |
Abbreviations: MIC, minimum inhibitory concentration; I, intermediate; R, resistant; S, susceptible; BMD, broth microdilution; CSF, cerebrospinal fluid.
MIC distribution of colistin determined by two susceptibility methods
Distribution of colistin MICs for isolates tested using BMD Vitek 2 are shown in Table 4. For some species (P. mirabilis, S. marcescens, and S. maltophilia), data retrieved from Vitek 2 are not available. In general, MIC values determined using BMD were higher than those obtained by the Vitek 2. When colistin MIC was measured using the Vitek 2 and not by BMD, MICs ≤0.5 were noted for colistin NWT among 14 (87.5%) of 16 E. cloacae and two (66.7%) of the three K. pneumoniae isolates (Table 4). Two E. cloacae and one K. pneumoniae isolate showed colistin MIC >16 using the Vitek 2 (Table 4). VMEs between the two methods were found to be 0 for E. coli, 66.7% for K. pneumoniae, 87.5% for E. cloacae, 100% for P. aeruginosa, and 100% for A. baumannii complex isolates (Table 4).
Table 4.
Distribution of MICs for isolates tested against colistin using the reference BMD method (recommended by the CLSI) and Vitek 2
| Organism (total isolates) | Method | Isolates with indicated colistin MICs (mg/L) determined by two methods, n (%) | Agreementa | Isolates with VME,b n (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ″0.5 | 1 | 2 | 4 | 8 | 16 | ≥32 | Essential | Categorical | ||||
| Escherichia coli (121) | BMD | 14 (11.7) | 103 (85.8) | 3 (2.5) | 0 | 0 | 0 | 0 | 118 (97.5) | 121 (100) | 0 | |
| Vitek 2 | 120 (100) | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Klebsiella pneumoniae (137) | BMD | 6 (4.4) | 124 (90.5) | 4 (2.9) | 1 (0.7) | 0 | 2 (1.5) | 0 | 131 (95.6) | 135 (98.5) | 2 (66.7) | |
| Vitek 2 | 136 (99.3) | 0 | 0 | 0 | 0 | 1 (0.7) | 0 | |||||
| Proteus mirabilis (38) | BMD | 0 | 0 | 0 | 0 | 0 | 0 | 38 (100) | NA | NA | NA | |
| Morganella morganii (12) | BMD | 0 | 0 | 0 | 0 | 0 | 0 | 12 (100) | 12 (100) | NA | 0 | |
| Vitek 2 | 0 | 0 | 0 | 0 | 0 | 12 (100) | 0 | |||||
| Citrobacter freundii (10) | BMD | 2 (20.0) | 8 (80.0) | 0 | 0 | 0 | 0 | 0 | 10 (100) | NA | 0 | |
| Vitek 2 | 10 (100) | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Enterobacter cloacae (51) | BMD | 1 (2.0) | 32 (62.7) | 2 (3.9) | 0 | 1 (2.0) | 1 (2.0) | 14 (27.5) | 35 (68.6) | 37 (72.5) | 14 (87.5) | |
| Vitek 2 | 49 (96.1) | 0 | 0 | 0 | 0 | 2 (3.9) | 0 | |||||
| Serratia marcescens (42) | BMD | 0 | 1 (2.4) | 1 (2.4) | 0 | 0 | 0 | 40 (95.2) | NA | NA | NA | |
| Pseudomonas aeruginosa (128) | BMD | 0 | 14 (10.9) | 108 (84.4) | 4 (3.1) | 2 (1.6) | 0 | 0 | 14 (10.9) | 122 (95.3) | 6 (100) | |
| Vitek 2 | 128 (100) | 0 | 0 | 0 | 0 | 0 | 0 | |||||
| Acinetobacter baumannii complex (138) | BMD | 2 (1.4) | 60 (43.5) | 62 (44.9) | 7 (5.1) | 5 (3.6) | 2 (1.4) | 0 | 62 (44.9) | 124 (89.9) | 14 (100) | |
| Vitek 2 | 133 (96.4) | 3 (2.2) | 2 (1.4) | 0 | 0 | 0 | 0 | |||||
| Burkholderia cepacia (19) | BMD | 0 | 0 | 0 | 0 | 0 | 0 | 19 (100) | 19 (100) | NA | 0 | |
| Vitek 2 | 0 | 0 | 0 | 0 | 0 | 19 (100) | 0 | |||||
| Stenotrophomonas maltophilia (61) | BMD | 0 | 0 | 2 (3.3) | 3 (4.9) | 2 (3.3) | 8 (13.1) | 45 (73.8) | NA | NA | NA | |
Notes:
Categorical agreement between susceptibility-testing methods measured as percentage of isolates with concordant test results when determining susceptible or WT and resistant or NWT to colistin;
major errors for Vitek 2 defined as colistin-resistant or NWT isolate determined using BMD method interpreted as colistin-susceptible or WT isolate.
Abbreviations: MICs, minimum inhibitory concentrations; BMD, broth microdilution; CLSI, Clinical and Laboratory Standards Institute; VME, very major error; NA, not available; WT, wild type; NWT, non-WT.
mcr-1 genes among colistin-NWT isolates
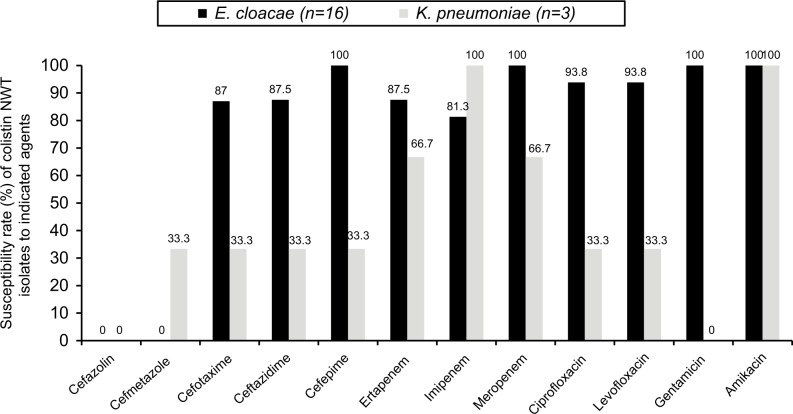
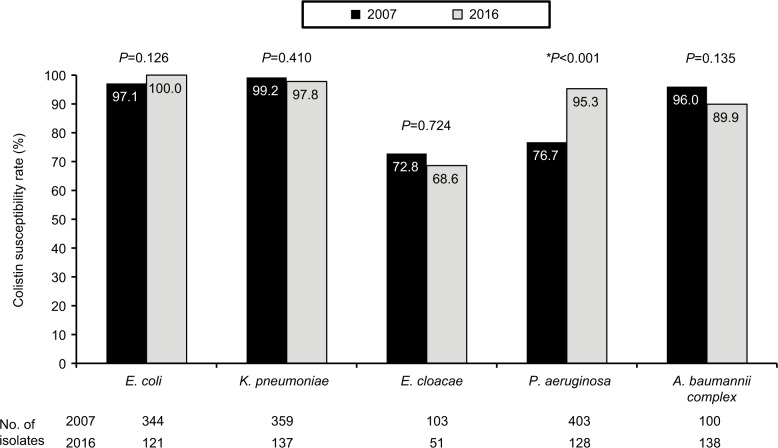
In total, 111 (25.7%) Enterobacteriaceae isolates were identified as colistin NWT (MIC ≥4 mg/L). In addition to bacterial species with inherent colistin resistance, such as P. mirabilis (100%, 38 of 38), M. morganii (100%, 14 of 14), and S. marcescens (95.2%, 40 of 42), colistin NWT isolates were found in 31.4% (16 of 51) of E. cloacae and 2.2% (three of 137) of K. pneumoniae. In contrast, all the E. coli (n=121) isolates were WT. Except cefazolin and cefmetazole, most antibiotics exhibited good in vitro activity against the 16 E. cloacae isolates that belonged to colistin NWT (Figure 3). We further compared rates of WT susceptibility to colistin among selected isolates from patients admitted to ICUs of the hospitals in a previous study6 in 2007 and the present work in 2016 (Figure 4). The colistin-susceptibility rate of P. aeruginosa was higher in 2016 than that in 2007 (P<0.001), but no significant difference was found among other pathogens: E. coli, K. pneumoniae, E. cloacae and A. baumannii complex (all P>0.05, Figure 4). The gene mcr-1 was detected only in two (1.8%) E. cloacae isolates among the colistin NWT isolates that had a colistin MIC of 32 mg/L. In addition to cefazolin and cefmetazole, these two E. cloacae isolates harboring mcr-1 were susceptible to all other antibiotics (Table 3).
Figure 3.
Antibiotic-susceptibility rates of colistin non-wild-type (NWT) Enterobacter cloacae and Klebsiella pneumoniae.
Figure 4.
Rates of wild-type susceptibility to colistin among selected isolates from patients admitted to intensive care units of seven major teaching hospitals in Taiwan in 20076 and 2016.
Notes: *Significant difference in susceptibility.
With regard to NFGNB isolates, the colistin-resistance rate was highest for B. cepacia (100%, 19 of 19) and S. maltophilia (96.7%, 58 of 60), followed by A. baumannii complex isolates (10.1%, 14 of 138) and P. aeruginosa (4.7%, six of 128).
Discussion
Based on the surveillance of common GNB in the ICU, including Enterobacteriaceae and NFGNB, we have several significant findings. First, the prevalence of carbapenem-insusceptible Enterobacteriaceae was found to be 9.4%. For P. aeruginosa and A. baumannii complex, carbapenem-insusceptibility rates were ~23% and 63%, respectively. Furthermore, we found that the carbapenem-resistance rate of Enterobacteriaceae varied depending on the site of infection. The imipenem-resistance rate was much higher for isolates from abscess/pus and intra-abdominal sources than from other sources. Moreover, the in vitro activity of the three carbapenems against carbapenem-insusceptible Enterobacteriaceae was different: meropenem demonstrated greater potency compared to ertapenem and imipenem. More than 40% ertapenem- or imipenem-insusceptible Enterobacteriaceae remained susceptible to meropenem. In addition to carbapenem, most of the other antibiotics exhibited poor in vitro activity against carbapenem-insusceptible Enterobacteriaceae. The only exception was amikacin, which demonstrated a susceptibility rate >80% against carbapenem-insusceptible Enterobacteriaceae. All these findings helped us recognize the resistance burden and antibiotic-resistance patterns of carbapenem-insusceptible Enterobacteriaceae. These data will provide clinicians with useful information regarding the use of empiric antibiotics in ICUs.
Colistin may be the drug of last resort for MDROs, including carbapenem-resistant organisms. However, the emergence of colistin resistance and mcr-1-encoding plasmid-mediated colistin resistance has been found among Enterobacteriaceae worldwide.22–25 In this study, rates of NWT-colistin susceptibility were 31.4% and 2.2% for E. cloacae and K. pneumoniae, respectively. In contrast, all the E. coli isolates belonged to WT colistin. Moreover, two isolates of E. cloacae were found to harbor the mcr-1 gene. For these colistin NWT Enterobacteriaceae, including the two carrying the mcr-1 gene, other broad-spectrum antibiotics showed good in vitro activities.22,26,27 Although the susceptibility profiles of colistin NWT Enterobacteriaceae to the other classes of antibiotics were favorable, regular surveillance is warranted to monitor the development of other resistance mechanisms among colistin NWT Enterobacteriaceae, which may help in determining resistance to broad-spectrum antibiotics.22,27
In this study, NWT Enterobacteriaceae were confirmed by BMD and compared with the results of the Vitek 2. We found that 87.5% (14 of 16) of NWT E. cloacae and 66.7% (two of three) of NWT K. pneumoniae isolates had MICs ≤0.5 mg/L using the Vitek 2. For colistin, a VME between the two susceptibility methods, ie, Vitek 2 and BMD, was found to be 87.5% for E. cloacae and 100% for both P. aeruginosa and A. baumannii complex isolates. All these findings are consistent with previous studies,28 and indicate that the Vitek 2 is a low-sensitivity tool to identify NWT.
We also found that the colistin-insusceptibility rates of P. aeruginosa and A. baumannii complex were 4.7% and 10.1%, respectively. This finding is consistent with a previous study in European hospitals between 2009 and 2011, where colistin exhibited good activity against P. aeruginosa strains, with 99.4% susceptibility rate in ICU patients.29 In contrast, the colistin-insusceptibility rate of P. aeruginosa in this surveillance was significantly lower compared to that observed in the previous SMART in 2007, where the colistin-insusceptibility rate of P. aeruginosa was 23.3% (94 of 403, Figure 4).6 As for other bacterial species tested, differences in insusceptibility (A. baumannii complex) or NWT to colistin and susceptibility to the carbapenems tested were not statistically significant (Figure 4).6 This comparison suggests that the resistance rate of P. aeruginosa seems to decrease with time in Taiwan. However, continuous monitoring of colistin resistance is still needed to investigate the secular trend.
In this surveillance, three K. pneumoniae isolates harbored blaKPC, and their prevalence was 16.7% among the 18 carbapenem-insusceptible K. pneumoniae isolates. This prevalence was similar to that found in the previous study,30 where 13 (21.7%) of the 60 carbapenem-insusceptible K. pneumoniae in ICUs in 2012 harbored blaKPC. In fact, no blaKPC gene was detected in Enterobacteriaceae isolates from the previous SMART from ICUs in Taiwan before 2011.5 This suggests the emergence of blaKPC among carbapenem-insusceptible K. pneumoniae in the ICUs of Taiwan.
Conclusion
Although carbapenem demonstrated good in vitro activity against most of the Enterobacteriaceae isolates from ICUs, 8.7% of Enterobacteriaceae isolates were not susceptible to carbapenem. Among these carbapenem-insusceptible Enterobacteriaceae isolates, the carbapenemase genes blaKPC and blaOXA48 were found in K. pneumoniae isolates. About a quarter of Enterobacteriaceae isolates were identified as colistin NWT, but the gene mcr-1 was detected in only two E. cloacae isolates among the colistin-NWT isolates. Colistin MICs determined by the Vitek 2 were not reliable, especially for the E. cloacae and A. baumannii complex isolates.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Halpern NA, Goldman DA, Tan KS, Pastores SM. Trends in critical care beds and use among population groups and Medicare and Medicaid beneficiaries in the United States: 2000–2010. Crit Care Med. 2016;44(8):1490–1499. doi: 10.1097/CCM.0000000000001722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai CC, Ho CH, Chang CL, et al. Critical care medicine in Taiwan from 1997 to 2013 under national health insurance. J Thorac Dis. 2018;10(8):4957–4965. doi: 10.21037/jtd.2018.07.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Navoa-Ng JA, Berba R, Galapia YA, et al. Device-associated infections rates in adult, pediatric, and neonatal intensive care units of hospitals in the Philippines: international nosocomial infection control Consortium (INICC) findings. Am J Infect Control. 2011;39(7):548–554. doi: 10.1016/j.ajic.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 4.Tao L, Hu B, Rosenthal VD, Gao X, He L. Device-associated infection rates in 398 intensive care units in Shanghai, China: international nosocomial infection control Consortium (INICC) findings. Int J Infect Dis. 2011;15(11):e774–e780. doi: 10.1016/j.ijid.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Jean SS, Hsueh PR, Lee WS, et al. Carbapenem susceptibilities and non-susceptibility concordance to different carbapenems amongst clinically important gram-negative bacteria isolated from intensive care units in Taiwan: results from the surveillance of multicentre antimicrobial resistance in Taiwan (SMART) in 2009. Int J Antimicrob Agents. 2013;41(5):457–462. doi: 10.1016/j.ijantimicag.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Jean SS, Lee WS, Yu KW, et al. Rates of susceptibility of carbapenems, ceftobiprole, and colistin against clinically important bacteria collected from intensive care units in 2007: results from the surveillance of multicenter antimicrobial resistance in Taiwan (SMART) J Microbiol Immunol Infect. 2016;49(6):969–976. doi: 10.1016/j.jmii.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Lee YL, Chen YS, Toh HS, et al. Antimicrobial susceptibility of pathogens isolated from patients with complicated intra-abdominal infections at five medical centers in Taiwan that continuously participated in the study for monitoring antimicrobial resistance trends (SMART) from 2006 to 2010. Int J Antimicrob Agents. 2012;40(Suppl):S29–S36. doi: 10.1016/S0924-8579(12)70007-9. [DOI] [PubMed] [Google Scholar]
- 8.Tsao LH, Hsin CY, Liu HY, et al. Risk factors for healthcare-associated infection caused by carbapenem-resistant Pseudomonas aeruginosa. J Microbiol Immunol Infect. 2018;51(3):359–366. doi: 10.1016/j.jmii.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 9.Chen CH, Kuo HY, Hsu PJ, et al. Clonal spread of carbapenem-resistant Acinetobacter baumannii across a community hospital and its affiliated long-term care facilities: A cross sectional study. J Microbiol Immunol Infect. 2018;51(3):377–384. doi: 10.1016/j.jmii.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Lee CM, Lai CC, Chiang HT, et al. Presence of multidrug-resistant organisms in the residents and environments of long-term care facilities in Taiwan. J Microbiol Immunol Infect. 2017;50(2):133–144. doi: 10.1016/j.jmii.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 12.Rodríguez-Baño J, Gutiérrez-Gutiérrez B, Machuca I, Pascual A. Treatment of infections caused by extended-spectrum-beta-lLactamase-, AmpC-, and carbapenemase-producing Enterobacteriaceae. Clin Microbiol Rev. 2018;31(2):e00079–17. doi: 10.1128/CMR.00079-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Can F, Menekse S, Ispir P, et al. Impact of the ST101 clone on fatality among patients with colistin-resistant Klebsiella pneumoniae infection. J Antimicrob Chemother. 2018;73(5):1235–1241. doi: 10.1093/jac/dkx532. [DOI] [PubMed] [Google Scholar]
- 14.Ripabelli G, Tamburro M, Guerrizio G, et al. Tracking multidrug-resistant Klebsiella pneumoniae from an Italian hospital: molecular epidemiology and surveillance by PFGE, RAPD and PCR-Based resistance genes prevalence. Nat Microbiol. 2018;75(8):977–987. doi: 10.1007/s00284-018-1475-3. [DOI] [PubMed] [Google Scholar]
- 15.Shen Y, Zhou H, Xu J, et al. Anthropogenic and environmental factors associated with high incidence of mcr-1 carriage in humans across China. Nat Microbiol. 2018;3(9):1054–1062. doi: 10.1038/s41564-018-0205-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hung MN, Hsueh PR, Chang HT, et al. In vitro activities of various piperacillin and sulbactam combinations against bacterial pathogens isolated from Intensive Care Units in Taiwan: SMART 2004 programme data. Int J Antimicrob Agents. 2007;29(2):145–152. doi: 10.1016/j.ijantimicag.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 17.Jean SS, Hsueh PR, Lee WS, et al. Nationwide surveillance of antimicrobial resistance among non-fermentative gram-negative bacteria in intensive care units in Taiwan: SMART programme data 2005. Int J Antimicrob Agents. 2009;33(3):266–271. doi: 10.1016/j.ijantimicag.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 18.Jean SS, Hsueh PR, Lee WS, et al. In vitro activities of doripenem and other carbapenems against clinically important bacteria isolated in intensive care units: nationwide data from the SMART programme. Eur J Clin Microbiol Infect Dis. 2010;29(4):471–475. doi: 10.1007/s10096-009-0866-6. [DOI] [PubMed] [Google Scholar]
- 19.Jean SS, Lee WS, Bai KJ, et al. Relationship between the distribution of cefepime minimum inhibitory concentrations and detection of extended-spectrum β-lactamase production among clinically important Enterobacteriaceae isolates obtained from patients in intensive care units in Taiwan: results from the surveillance of multicenter antimicrobial resistance in Taiwan (SMART) in 2007. J Microbiol Immunol Infect. 2015;48(1):85–91. doi: 10.1016/j.jmii.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing. 28th Informational Supplement. Wayne, PA: CLSI; 2018. pp. M100–ED28. [Google Scholar]
- 21.Rebelo AR, Bortolaia V, Kjeldgaard JS, et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 2018;23(6) doi: 10.2807/1560-7917.ES.2018.23.6.17-00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai CC, Lin YT, Lin YT, et al. Clinical characteristics of patients with bacteraemia due to the emergence of mcr-1-harboring Enterobacteriaceae in humans and pigs in Taiwan. Int J Antimicrob Agents. 2018;52(5):651–657. doi: 10.1016/j.ijantimicag.2018.08.015. [DOI] [PubMed] [Google Scholar]
- 23.Quan J, Li X, Chen Y, et al. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis. 2017;17(4):400–410. doi: 10.1016/S1473-3099(16)30528-X. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Tian GB, Zhang R, et al. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect Dis. 2017;17(4):390–399. doi: 10.1016/S1473-3099(16)30527-8. [DOI] [PubMed] [Google Scholar]
- 25.Kocsis B, Kádár B, Tóth Ákos, Fullár A, Szabó D. MgrB variants in colistin-susceptible and colistin-resistant Klebsiella pneumoniae ST258. J Microbiol Immunol Infect. 2017;50(5):735–736. doi: 10.1016/j.jmii.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 26.Corbella M, Mariani B, Ferrari C, et al. Three cases of mcr-1-positive colistin-resistant Escherichia coli bloodstream infections in Italy, August 2016 to January 2017. Euro Surveill. 2017;22(16):30517. doi: 10.2807/1560-7917.ES.2017.22.16.30517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nordmann P, Lienhard R, Kieffer N, Clerc O, Poirel L. Plasmid-Mediated Colistin-Resistant Escherichia coli in Bacteremia in Switzerland. Clin Infect Dis. 2016;62(10):1322–1323. doi: 10.1093/cid/ciw124. [DOI] [PubMed] [Google Scholar]
- 28.Lo-Ten-Foe JR, de Smet AM, Diederen BM, Kluytmans JA, van Keulen PH. Comparative evaluation of the VITEK 2, disk diffusion, etest, broth microdilution, and agar dilution susceptibility testing methods for colistin in clinical isolates, including heteroresistant Enterobacter cloacae and Acinetobacter baumannii strains. Antimicrob Agents Chemother. 2007;51(10):3726–3730. doi: 10.1128/AAC.01406-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sader HS, Farrell DJ, Flamm RK, Jones RN. Antimicrobial susceptibility of gram-negative organisms isolated from patients hospitalized in intensive care units in United States and European hospitals (2009–2011) Diagn Microbiol Infect Dis. 2014;78(4):443–448. doi: 10.1016/j.diagmicrobio.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 30.Chang YY, Chuang YC, Siu LK, et al. Clinical features of patients with carbapenem nonsusceptible Klebsiella pneumoniae and Escherichia coli in intensive care units: a nationwide multicenter study in Taiwan. J Microbiol Immunol Infect. 2015;48(2):219–225. doi: 10.1016/j.jmii.2014.05.010. [DOI] [PubMed] [Google Scholar]