Abstract
Toca 511, a retroviral replicating vector (RRV), uses an internal ribosomal entry site (IRES) to express an optimized yeast cytosine deaminase (yCD2), which converts 5-fluorocytosine to 5-fluorouracil. This configuration is genetically stable in both preclinical mouse models and human clinical trials. However, the use of IRES (∼600 bp) restricts choices of therapeutic transgenes due to limits in RRV genome size. This study replaced IRES with 2A peptides derived from picornaviruses with or without a GSG linker. The data show that GSG-linked 2A (g2A) peptide resulted in higher polyprotein separation efficiency than non-GSG linked 2A peptide. The study also shows that RRV can tolerate insertion of two separate 2A peptides to allow expression of two transgenes without compromising the assembly and function of the virus in addition to insertion of a single 2A peptide to confirm genetic stability with yCD2, green fluorescent protein, and HSV-1 thymidine kinase. In a parallel comparison of the RRV-IRES-yCD2 and RRV-g2A-yCD2 configurations, the study shows the yCD2 protein expressed from RRV-g2A-yCD2 has higher activity, resulting in a higher survival benefit in an intracranial tumor mouse model. These data enable a wider range of potential product candidates that could be developed using the RRV platform.
Keywords: : retroviral replicating vector, cancer gene therapy, 2A peptide
Introduction
Toca 511 (vocimagene amiretrorepvec) is a novel retroviral replicating vector (RRV) in late-stage clinical development. Toca 511 has been shown to infect cancer cells selectively and deliver a prodrug activator gene (yCD2) encoding an optimized yeast cytosine deaminase enzyme. The yCD2 produced in the infected cancer cells catalyzes the conversion of 5-fluorocytosine (5-FC) to the potent anticancer agent, 5-fluorouracil (5-FU), which kills cancer cells and immune-suppressive myeloid cells, resulting in immune activation against the cancer. This strategy leads to eradication of tumors and prolonged survival in mouse models.1–3 The preclinical data also demonstrate that Toca 511 with 5-FC treatment elicits a prolonged antitumor immunity in mouse glioma models.2,4 Clinically, favorable safety and tolerability profiles were reported in addition to durable complete responses and extended survival compared to historical controls.5 Toca 511 in combination with Toca FC (extended-release 5-fluorocytosine) is currently in Phase I and Phase II/III clinical trials for treating solid tumors and recurrent brain tumors (NCT02576665 and NCT02414165, respectively).
Viral genome stability during replication is a key criterion for maintaining sustained therapeutic gene expression in targeted cells resulting from cancer-selective gene therapy. In general, increasing the viral genome length beyond the native size can reduce replication efficiency leading to retroviral vector genome instability.6,7 Toca 511, which utilizes an internal ribosomal entry site (IRES) derived from encephalomyocarditis virus, is reported to be genetically stable in both preclinical mouse models and human clinical trials.5,8 However, use of the IRES (∼600 bp) in RRV imposes a restriction on choices of therapeutic genes for developing future gene therapy products, as instability can occur around 1,200–1,500 bp of total insertion size, depending on the transgene.7 This study investigated the use of relatively short 2A peptides derived from picornaviruses for expression of a single or multiple transgenes. The 2A peptides (∼80 bp) are markedly shorter in length than the IRES utilized in Toca 511, and most importantly, they efficiently co-express multiple proteins in an equal molar ratio manner from a single multi-cistronic transcript.9–11
Although expression of a transgene mediated by 2A peptide in a gammaretrovirus has been reported previously,12,13 it has not been well characterized in terms of its impact on viral replication, viral envelope protein processing, long-term viral genome stability, optimized coding sequences, as well as its potential applications in gene therapy. This study designed RRV with the 2A peptide in-frame with the viral envelope protein to express transgenes of various sizes and cellular compartments. It was demonstrated that replacement of the IRES with the GSG-linked 2A (g2A) configuration results in high efficiency of polyprotein separation and does not compromise assembly and function of the virus or the transgene expression. Furthermore, the replacement of the IRES with a gT2A peptide facilitates insertion of a transgene larger in size, as well as the insertion of multiple transgenes. The data suggest that the g2A configuration is an alternative RRV design for cancer gene therapy and may expand transgene selections for RRV product candidates.
Materials and Methods
Construction of RRV-2A plasmid DNAs
pAC3-T2A-GFP plasmid DNA was first generated by the Gibson assembly method and used as the template for additional constructs. It contains the 2A peptide sequence from Thosea asigna virus (T2A). The T2A-GFP cassette was inserted into pAC3 at the BstBI and NotI restriction enzyme sites so that the T2A-GFP coding sequence is in the same reading frame as the env gene. The T2A-GFP cassette consisted of two DNA fragments. The first fragment was generated by gene synthesis (Integrated DNA Technologies), and it consists of sequences spanning the BstBI site in the 3′ end of the env region (with removal of the stop codon in the env gene) followed by the T2A sequence and sequence of the AscI restriction enzyme site with an extra “T” nucleotide added to maintain the Env-2A-GFP coding sequence in-frame and partial 5′ region of the GFP sequence. The second fragment was amplified from pAC3-GFP8 using primers spanning the GFP sequence. Three-piece assembly (pAC3-GFP backbone digested with BstBI and NotI; two DNA fragments consist of the T2A-GFP cassette) was performed using Gibson Assembly Kit (New England Biolabs; #E2611). For RRV-2A constructs with the 2A peptides derived from other picornaviruses, the sequence of Env-2A was synthesized and replaced by subcloning at the BstBI and AscI restriction enzyme sites.
For the generation of the set of RRVs that utilized T2A, GSG-T2A, P2A, or GSG-P2A peptide sequences for yCD2, HSV-1 human codon optimized HSV-1 thymidine kinase gene (TKO), yCD2-2A-hGMCSF, or hGMCSF-2A-yCD2 expression, information is provided in the Supplementary Data (Supplementary Data are available online at www.liebertpub.com/hum).
Cell culture
Human 293T cells (obtained through a materials transfer agreement with the Indiana University Vector Production Facility; Stanford University deposited the cells with ATCC, SD-3515; lot 2634366), human glioma cell line U87-MG (ATCC; #HTB-14), mouse glioma cell line Tu2449,3 and Tu2449SC2 were cultured in complete Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (HyClone), sodium pyruvate (Cellgro), Glutamax, and penicillin-streptomycin (Invitrogen).
Virus production from 293T cells and virus titer
Twenty micrograms of plasmid DNA encoding the RRVs were used for transient transfection using the calcium phosphate method, as previously described.14 The viral titers of all RRVs were determined by quantitative polymerase chain reaction (qPCR) and viral titers, reported in transduction units per milliliter (TU/mL).8,15
Virus replication kinetics of RRVs in U87-MG, Tu2449, and Tu2449SC cells
U87-MG cells were infected with RRVs expressing the green fluorescent protein (GFP) transgene at a multiplicity of infection (MOI) of 0.01 in a six-well plate, and the percentage of GFP-positive cells at each cell passage during the course of infection was determined by flow cytometry using proper gating to exclude GFP-negative cells.16 To determine replication kinetics of the RRVs that do not express the GFP marker gene, RNA particle titer was assessed using reverse transcriptase qPCR (RT-qPCR) on viral RNAs extracted from U87-MG cell supernatants collected every 2 days for a period of 10 days post infection following an initial infection at a MOI of 0.01. Twenty microliters of collected supernatant from each time point was obtained to extract viral RNA as described.17 Maximally (>95% infectivity) infected U87-MG cells were expanded and cultured in larger tissue culture vessels. At approximate 90% confluency, media was replaced with fresh media, followed by the collection of virus containing supernatant and 0.45 μm filtration at 18–24 h post media replacement. The collected cell supernatant was aliquoted and stored at −80°C for immunoblotting, and the cell pellets were washed with phosphate-buffered saline (PBS) and stored at −20°C for cell lysates preparation or for assessment of viral genome stability.
In vitro superinfection resistant assay
For assessing the ability of maximally infected cells to resist subsequent infection by a second RRV utilizing the same receptor, Tu2449/RRV-IRES-yCD2 cells were mixed with Tu2449/RRV-IRES-GFP infected cells at ratios of 3–97% cells, respectively. The percentage of GFP infected cells were measured by flow cytometric analysis (BD Biosciences; FACS Canto II) at the time of cell mixing (day 0), 4, 8, and 11 days post mixing.
5-FC LD50 assay
Maximally infected U87-MG cells, Tu2449 cells (with RRV variants containing yCD2 in the transgene cassette), and naive cells were seeded at 1 × 103 cells/100 μL/well in triplicate into three 96-well plates, one plate per time point. The cells were monitored over a 7-day period in the presence of 5-FC concentrations (Sigma–Aldrich; #F7129) ranging from 0.00001 to 1 mM (10-fold dilutions). Naive U87-MG cells were included to determine cytotoxic effect of 5-FC. Cell viability was measured every 2 days using the CellTiter 96® Aqueous One Solution Cell Proliferation Assay System (Promega; #G3581), and detection at an OD of 490 nm was acquired using the Infinite M200 plate reader (Tecan Group Ltd.). All time points for each cell line and each concentration were performed in triplicate. The percentage of cell viability relative to 5-FC untreated but RRV-yCD2 infected cells was calculated and plotted for nonlinear four-parameter fit to determine the LD50 values using GraphPad Prism v7 (GraphPad Software, Inc.).
GCV LD50 assay
Maximally infected U87-MG cells with RRV-P2A-TKO, RRV-gP2A-TKO, RRV-T2A-TKO, and RRV-gT2A-TKO were used to determine its GCV LD50 dose via MTS assay, as described above, with the following exceptions. Treatment with ganciclovir (GCV; EMD Millipore; #345700-50MG) was performed in series of 1:10 dilutions ranging from GCV concentrations of 0.001 to 1.0 μM. GCV was added 1 day after seeding and then replenished with complete medium plus GCV every 2 days. No GCV treatment was included as a control, and naive U87-MG cells were included for cytotoxic effect of GCV. The cell viability was monitored over a 7-day incubation time, and cell viability was measured every 2 days as described. All time points were performed in triplicate, and the average OD values from triplicates of each sample were converted to percentage of cell survival relative to GCV untreated but RRV-infected cells. The percentage values were plotted against GCV concentrations in log scale using GraphPad Prism v7 to generate LD50 graphs.
In vitro 5-FC bystander cell killing assay
Tu2449 maximally infected by RRV-IRES-yCD2, RRV-gT2A-yCD2, and RRV-IRES-GFP that demonstrated block of superinfection (Supplementary Fig. S3, additional data not shown) were mixed with Tu2449/RRV-IRES-yCD2 (or Tu2449/RRV-gT2A-yCD2) to Tu2449/RRV-IRES-GFP at fixed ratios of 3/97, 15/85, 30/70, and 50/50, respectively. Total cells were seeded at 1 × 103 cells/200 μL/well in triplicate into 96-well plates. The cells were monitored over a 7-day period following treatment with 0.1 mM of 5-FC and without 5-FC. Cells infected with RRV-IRES-yCD2 and RRV-gT2A-yCD2 were included as positive and negative controls, respectively. Cell viability was measured every 2 days as described. All time points for each cell line were performed in triplicate, and the average OD values from triplicates of each sample were converted to percentage of cell survival relative to untreated, 100% Tu2449/RRV-IRES-yCD2, or Tu2449/RRV-GSG-T2A-yCD2 infected cells. As a control, 100% Tu2449/RRV-IRES-GFP cells were treated with 0.1 mM 5-FC to determine the cytotoxic effect of 5-FC.
Subcutaneous tumor model and survival study of orthotopic glioma model
All animal protocols and experiments were approved by the IRB/IACUC of Explora BioLabs. For the subcutaneous tumor model, 8-week-old female B6C3F1 mice (Harlan Sprague Dawley, Inc.) were assigned to one of six groups: groups 1 and 2 received subcutaneous implants of 1 × 106 2% Tu2449SC/RRV-IRES-yCD2 + 98% Tu-2249SC tumor cells; groups 3 and 4 received subcutaneous implants of 1 × 106 2% Tu2449SC/RRV-gT2A-yCD2 + 98% Tu-2249SC tumor cells on the right flank. Once tumors reached an average of 150 mm3, treatment was initiated. 5-FC was administered (500 mg/kg intraperitoneally once daily) for 5 days consecutively, followed by 2 days off drug. Control animals received PBS. Tumor sizes were monitored three times a week until tumor volumes reach sizes >2,000 mm3. Selective tumors were collected at termination.
For the orthotopic glioma model, female B6C3F1 mice underwent surgical implantation of the tumor cells by Hamilton syringe. The stereotaxic coordinates were anteroposterior (AP), 0.5 mm; mediolateral (ML), 1.8 mm; and dorso-ventral (DV), 3.5 mm (from bregma). The syngeneic cell line Tu2449 was used as an orthotopic brain tumor model. On day 0, animals received intracranial implants of 1.4 × 104 Tu2449 cells maximally infected by RRV-IRES-yCD2 or RRV-gT2A-yCD2 were mixed with Tu2449/RRV-IRES-GFP (or Tu2449/RRV-gT2A-GFP) at fixed ratios of 3%, 15%, 30%, and 100% cells, respectively. Starting on day 10, mice were treated with either PBS or 5-FC (500 mg/kg/dose intraperitoneally twice daily) for 4 days consecutively, followed by 10 days without drug. Cycles of PBS or 5-FC treatment were repeated a further three times.
Survival data were plotted by the Kaplan–Meier method, and were compared by the log-rank test or Student's t-test as noted. Tumor volumes were assessed for significance using two-way analysis of variance. p-Values of <0.05 were considered statistically significant in all analyses, which were done with GraphPad Prism v7 statistical software (GraphPad Software, Inc.).
Measurement of 5-FU production by competitive enzyme-linked immunosorbent assay
Cells were plated into a flat-bottom 96-well plate (Falcon; #353072) with 7,000 cells/well for 23–24 h. The cells were a mixture of Tu2449/IRES-yCD2 and Tu2449/IRES-GFP or Tu2449/gT2A-yCD2 and Tu2449/gT2A-GFP at 0/100, 3/97, 15/85, 30/70, 50/50, and 100/0 ratios in triplicate. The supernatant was aspirated off the cells, and 200 μL of 2.6 mg/mL 5-FC was added to the wells followed by incubation at 37°C/5% CO2 for 4 h. At the end of the incubation time, the supernatant was transferred into a round-bottom 96-well plate and sealed with aluminum film for long-term storage at ≤–65°C until the enzyme-linked immunosorbent assay (ELISA) was performed.
A competitive ELISA was performed to measure 5-FU concentrations generated from the conversion of 5-FC to 5-FU by the yCD2 proteins. 5-FU standards were prepared from a 1 mg/mL stock in PBS followed by a serial dilution in fresh culture medium. 5-FU conjugated to bovine serum albumin (BSA; 5-FU-BSA) was custom-made by Genscript and re-suspended in PBS to reach a stock concentration of 0.1 mg/mL, and a volume of 100 μL of 1:100 diluted 5-FU-BSA was added into each well to coat a medium-binding plate (Costar; #3591). The 5-FU-BSA-coated plate was then washed six times with 1 × Tris-buffered saline/Tween 20 (Thermo Fisher Scientific; #28360) then blocked with SuperBlock (300 μL/well; Thermo Fisher Scientific; #37536) for 1 h at 37°C/5% CO2 followed by aspiration. During the blocking, in a separate non-tissue culture–treated 96-well plate (USA Scientific; #1830-9610), the collected supernatant as well as the 5-FU standards were mixed with a 1:100 diluted anti-5-FU antibody (stock solution at 0.95 mg/mL; custom-made by Genscript) at a 1:1 ratio (55 μL each) to capture the 5-FU present in the mixture and immediately transferred to the 5-FU-BSA-coated plate (100 μL/well) followed by incubation at 37°C/5% CO2 for half an hour. The plate was then washed six times with 1 × Tris-buffered saline/Tween 20. The 5-FU-BSA/anti-5-FU antibody complex was detected with a goat anti-mouse HRP-conjugated antibody (1:2,000; Southern Biotech; #1010-05) after incubation at 37°C/5% CO2 for half an hour followed by six washes with 1 × Tris-buffered saline/Tween 20. A volume of 100 μL of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate solution (Southern Biotech; #0410-01) was added to each well and incubated at room temperature for 15 min, and the reaction was terminated by adding 100 μL of TMB STOP solution (Southern Biotech; #0412-01). Optical density was read by a plate reader (Spectra Max 190; Molecular Devices) at 450 nm using the Soft Max Pro software. The amount of 5-FU for each sample (mg/mL) was calculated from the standard curve.
Vector genome stability
The genomic DNA was extracted from maximally infected cells using the Maxwell 16 Cell DNA Purification Kit (Promega; #AS1020) as described.8
hGMCSF ELISA
Expression of hGMCSF produced from RRV-IRES-hGMCSF, RRV-gT2A-hGMCSF-gP2A-yCD2, and RRV-gT2A-yCD2-gP2A-hGMCSF maximally infected U87-MG human glioma cell supernatants were quantified using the Quantikine® ELISA to Human GM-CSF Immunoassay (R&D Systems, Inc.; DGM00), as recommended by the manufacturer. Briefly, maximally infected cells were grown to confluency in T75 flasks and replaced with fresh medium. Supernatants were collected 18–24 h later and filtered through a 0.45 μm syringe filter. Filtered supernatants were stored at −80°C prior to determination of hGMCSF by ELISA.
Immunoblots
For the assessment of the transgene protein expression and the separation efficiency of the polyproteins, cell lysates were generated from maximally infected cells. The whole cell lysates were then assayed for their protein concentration by BCA protein assay kit (Thermo Fisher Scientific; #23227). For assessment of the viral envelope protein processing and virion incorporation, 1 mL of viral supernatant from maximally infected U87-MG cells was pelleted through a 20% sucrose gradient at 19,502 × g for 30 min at 4°C. Cell lysate samples (20 μg of total protein) or viral particles were resolved on Criterion XT Precast 4–12% Bis-Tris gels (Bio-Rad; #345-0124). Subsequently, proteins were transferred to PVDF membranes (iblot 2; Invitrogen; #IB24001) and blotted to detect the expression of the viral envelope proteins (anti-Env, clone 83A25; 1:500 dilution; licensed from NIH), GFP (clone 4B10; 1:1,000; Cell Signaling Technology; #2955S), yCD2 (1:1,000; Tocagen, Inc.), HSV-1 TK (1:200; Santa Cruz Biotechnology; #sc28037), 2A Peptide (1:1,000; Millipore; #ABS31), and GAPDH (clone 6C5; Millipore; #MAB374). Protein expression was detected using the corresponding secondary antibody conjugated to horseradish peroxidase and detected by chemiluminescence (Bio-Rad, Clarity Western ECL Substrate; #170-5060).
Results
RRVs utilizing 2A peptides for transgene expression exhibited different rates of viral replication and levels of efficiency in polyprotein separation
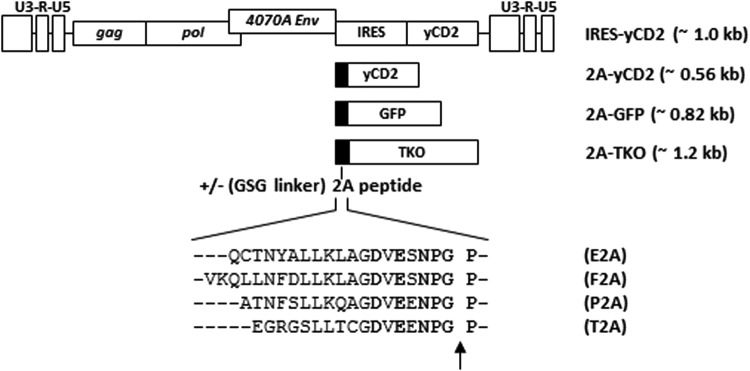
The length of the 2A peptide has been reported to influence separation efficiency of polyprotein linked by the 2A peptide.9,11,18 This study selected several picornavirus 2A peptides or 2A-peptide-like sequences from three mammalian viruses (foot-and-mouth disease [F2A], equine rhinitis A virus [E2A], and porcine teschovirus-1 [P2A]) and one insect virus (Thosea asigna virus [T2A]) used for the expression of multi-cistronic proteins.11,19 A second set of RRVs was also generated for all the 2A peptides to include a glycine–serine–glycine (GSG) linker at the N-terminus11 (Fig. 1).
Figure 1.
Schematic diagram of the retroviral replicating vector (RRV) configurations encoding different transgenes. RRV configurations with either an internal ribosomal entry site (IRES) or 2A peptide for transgene expression. The approximate size of the IRES-transgene and 2A-transgene cassette is shown in parentheses. The amino acid sequence of the 2A peptides from the equine rhinitis A virus (E2A), foot-and-mouth disease virus (F2A), porcine teschovirus-1 (P2A), and Thosea asigna virus (T2A) are shown. For each 2A peptide sequence, a glycine–serine–glycine peptide linker (GSG linker) was also included or excluded on the N-terminus. The conserved amino acid residues of the 2A peptides are shown in bold. The arrow indicates the site where the viral envelope protein at the N-terminus of the polyprotein is released from the ribosome in the 2A sequence.
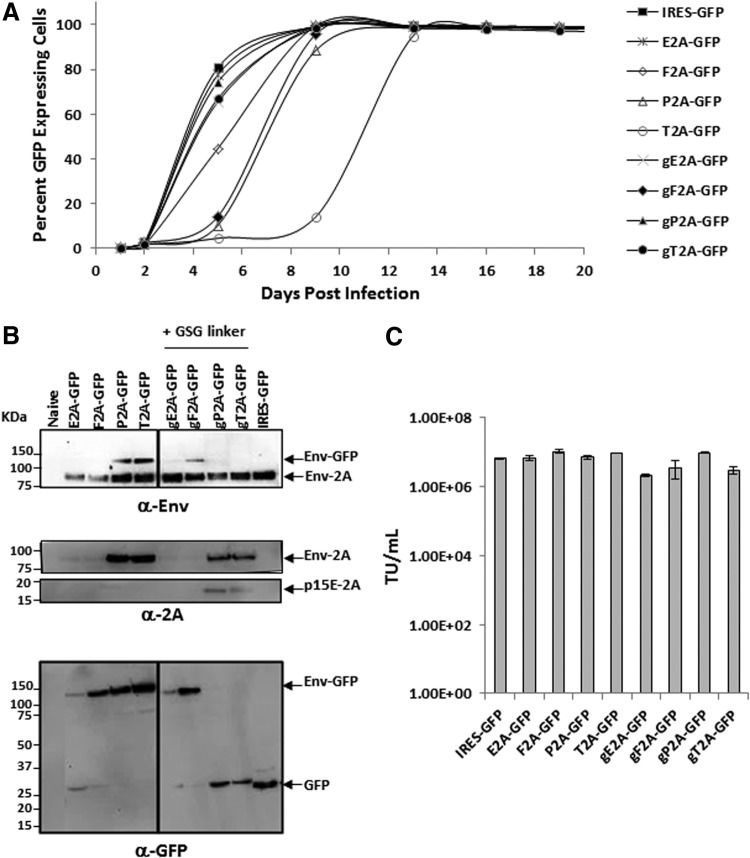
Since the strategy for transgene expression mediated by a 2A peptide encodes a polyprotein containing the viral envelope protein and therapeutic protein, and the viral envelope protein is known to play an important role in viral entry, RRV-2A variants expressing GFP were first generated so the viral replication during the course of infection could be easily monitored by measuring the percentage of GFP+ cells. U87-MG glioma cells were infected with RRV-2A-GFP variants generated from transient transfection in 293T cells at a MOI of 0.01. Viral spread of the RRV-2A-GFP variants were compared to RRV-IRES-GFP, which exhibited rapid viral spread and reached maximal infectivity by 7–8 days post infection (Fig. 2A). Among the four variants with GSG-linked 2A (g2A-GFP), gE2A-GFP, gP2A-GFP, and gT2A-GFP replicated at a similar rate as RRV-IRES-GFP, whereas gF2A-GFP showed a moderate delay in viral spread. Notably, viral replication of E2A-GFP with or without the GSG linker was comparable to RRV-IRES-GFP. In contrast, three out of the four variants without GSG-linked 2A showed a moderate (F2A and P2A) or significant (T2A) delay in viral spread compared to RRV-IRES-GFP. However, all variants were able to replicate and reached nearly 100% infectivity by day 14 (Fig. 2A).
Figure 2.
Vector characterization of RRV-2A-GFP in human U87-MG glioma cells. (A) Replication kinetics of RRV-2A-GFP variants. Percentage of green fluorescent protein (GFP)-expressing U87-MG cells were quantified by flow cytometric analysis at each cell passage following initial RRV-2A-GFP infection at a multiplicity of infection (MOI) of 0.01. RRV-IRES-GFP was included as a positive control and as a reference. Data shown represent one of the two independent experiments. Variants with GSG-linked 2A peptide are denoted as g2A. (B) Western blot analysis of viral envelope proteins produced by U87-MG cells maximally infected with the RRV-2A-GFP variants. Twenty micrograms of total protein lysates were loaded per well. Membranes were incubated with anti-Env antibody 83A25, which detects the viral envelope-GFP polyprotein (Env-GFP) and the viral envelope protein (Env-2A; top panel); anti-2A peptide antibody that detects unprocessed viral precursor envelope protein (Env-2A) and processed viral envelope protein tagged with the 2A peptide at the C-terminus, p15E-2A (middle panels); and anti-GFP antibody which detects both Env-GFP polyprotein and GFP (bottom panel). Protein standards are indicated in kDa. (C) Viral transduction titers of RRV-2A-GFP variants from RRVs collected from maximally infected U87-MG cells. RRV-IRES-GFP was included as a control and as a reference.
As proper viral envelope processing is an important determinant for viral replication,20 the separation efficiency of the viral envelope-GFP polyprotein mediated by the 2A peptide and its correlation with viral replication were examined. Separation efficiency of the GFP from the viral envelope protein in cell lysates was evaluated by immunoblotting using antibodies against the viral envelope protein, 2A peptide, and GFP. Consistent with the viral replication observed (Fig. 2A), gP2A and gT2A variants both showed efficient separation of GFP from the viral envelope protein (Fig. 2B, top and bottom panel). In addition, proper processing of the precursor viral envelope was also detected in gP2A and gT2A variants (indicated as p15E-2A; Fig. 2B, middle panel).
In contrast, variants that have low efficiency of separation, as indicated by a low signal of GFP (∼27 kDa) and a strong signal of the viral envelope-GFP polyprotein (Env-GFP ∼112 kDa), showed a delay in viral replication. Despite the separation observed between the GFP and the viral envelope protein in E2A-GFP and gE2A-GFP variants (and F2A-GFP and gF2A-GFP to a lesser extent), their overall polyprotein and GFP expression levels are lower than others. This reduced polyprotein expression level may be due to the lack of continuous protein translation of the GFP protein, as indicated by the differential detection of viral envelope protein by the anti-Env and anti-GFP antibodies (Fig. 2B).
It was also noticed that the P2A-GFP and T2A-GFP variants, which showed a delay in viral replication possibly due the inefficient separation of the Env-GFP polyprotein and thus improper processing of the viral envelope protein, also had the least amount of viral envelope protein incorporated into the viral particles (Supplementary Fig. S1). However, the level of viral envelope incorporation appears not to affect viral titer produced from the maximally infected cells (Fig. 2C). It is also noteworthy that there was no significant difference in titers produced among the variants from transient transfection of 293T cells (Supplementary Table S1). Together, it was discovered that among the four 2A peptides tested, gP2A and gT2A bearing a GSG linker are the most favorable configurations for transgene expression in RRV.
RRV-gT2A can tolerate insertion of a large transgene
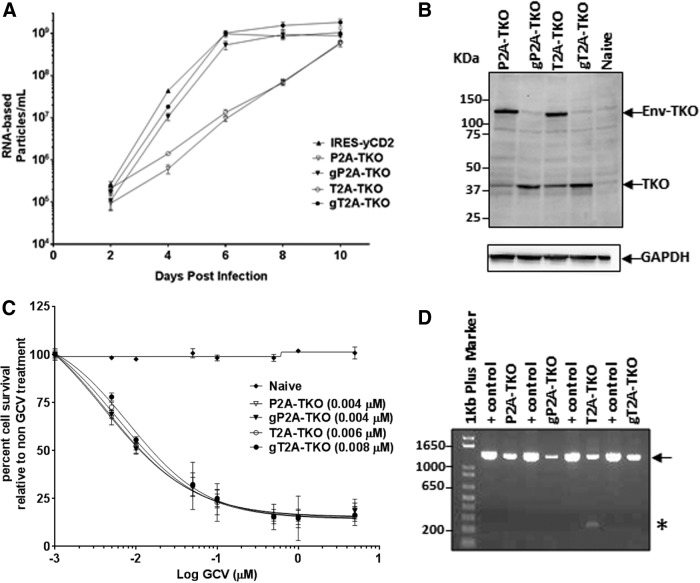
To test whether the observation made with the GFP described above is not a transgene-specific phenomenon and whether utilizing the 2A peptide for transgene expression in RRV allows insertion of a large gene while maintaining viral genome stability,7 gP2A, gT2A, and their matching pair without the GSG linker were selected to generate RRV expressing a human codon optimized HSV-1 thymidine kinase gene (TKO; Inagaki et al., manuscript submitted) with a cDNA size of approximately 1.1 kb (Fig. 1). Importantly, the insertion of the TKO gene in the RRV-2A configuration keeps the RRV genome size within the limitations of heterologous sequences, which would not be stable with the RRV-IRES configuration7 (pers. commun. Dr. Noriyuki Kasahara).
To monitor viral replication of RRV-2A-TKO variants, U87-MG cells were infected with RRV-2A-TKO variants produced from transiently transfected 293T cells (Supplementary Table S2) at a MOI of 0.01. Due to the lack of a GFP marker gene, the viral replication was assessed by quantifying the viral RNA levels in the particles during the course of infection using the RT-qPCR method. Consistent with data observed with RRV-2A-GFP variants, gP2A-TKO and gT2A-TKO variants both exhibited replication kinetics comparable to the control vector, RRV-IRES-yCD2. In contrast, a significant delay in replication was observed for P2A-TKO and T2A-TKO (Fig. 3A). The earlier time point reached to maximal infectivity seen in this set of vectors in comparison to RRV-IRES-GFP can be explained by the high sensitivity of the RT-qPCR assay compared to a cell-based GFP protein expression method, which requires protein production. Similar to the RRV-2A-GFP variants, efficient TKO protein separation from the viral envelope protein was observed with gP2A and gT2A but not with P2A and T2A (Fig. 3B). Although the TKO protein is normally expressed as a cytoplasmic protein, the TKO protein in the design can be expressed as a polyprotein with the TKO protein linked to the C-terminus of the viral envelope protein, as seen with the P2A-GFP and T2A-GFP variants (Fig. 3B). Next, we asked if there is a difference in the biological activity among the four 2A-TKO variants in maximally infected U87-MG cells by treating the cells with GCV. Unexpectedly, the results showed that all four variants exhibited similar sensitivity to GCV, despite the states (separated or polyprotein) of the TKO protein (Fig. 3C). Both the P2A-TKO and T2A-TKO variants, where the TKO protein was inefficiently separated from the viral envelope protein, have LD50 values comparable to that of the gP2A-TKO and gT2A-TKO variants. The data suggest that both forms of TKO proteins may be biologically functional in converting GCV to cytotoxic agent and kill tumor cells, or there was sufficient amount of biologically active separated TKO expressed to exert GCV-mediated cytotoxicity. In addition, when the proviral DNA was evaluated for viral genome stability, the PCR data showed that the viral genome remained stable during infection. In addition, vector genome stability from maximally infected U87-MG cells revealed that gP2A-TKO and gT2A-TKO transgene cassettes are more stable than the ones without the GSG linker (Fig. 3D). Altogether, data from both 2A-GFP and 2A-TKO vector sets are consistent and showed that the GSG-linked 2A is a more favorable configuration for transgene expression from the RRV.
Figure 3.
Vector characterization of RRV-2A encoding the TKO transgene in U87-MG cells. (A) Replication kinetics of RRV-2A-TKO variants. Viral genomes isolated from the viral supernatant of infected U87-MG cells were quantified by reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) using primers spanning the viral envelope region. Infected cells were passaged every other day for 10 days post initial infection, and supernatants were collected at each indicated time point. Data shown represent one of three independent experiments. RRV-IRES-yCD2 vector was included as control. (B) Western blot analysis of viral proteins produced by U87-MG cells maximally infected with RRV-2A-TKO variants. Twenty micrograms of total protein lysates was loaded per well. Membranes were incubated with anti-HSV-1-TK antibody (top panel). Detection of GAPDH using the anti-GAPDH antibody was included as a loading control. Protein standards are indicated in kDa. (C) LD50 of ganciclovir (GCV)-mediated killing of maximally infected U87-MG cells with RRV-2A-TKO variants. Cells were cultured in the presence of GCV in different concentrations for 7 days. Naïve cells were included as a control for GCV cytotoxicity at each concentration. Cell viability was quantified by using MTS assay at 7 days post GCV addition, and the percentage of cell survival was calculated relative to RRV-infected cells not treated with GCV. The data set represents one of the three independent experiments. Error bars indicate the standard deviation of the data set. Numbers in parentheses indicate LD50 values. (D) Stability 2A-TKO transgene cassette from proviral DNA in maximally infected U87-MG cells (15 days post infection). The arrow indicates the size of the PCR product expected for the intact 2A-TKO transgene cassette. The asterisk indicates deletion in the transgene cassette of RRV-T2A-TKO; + controls are PCR product amplified from plasmid DNA corresponding to each RRV-2A-TKO variant. DNA molecular ladder (1 kb plus; Life Technologies) is shown in the first lane of the gel; numbers indicate the size of the ladders in base pair.
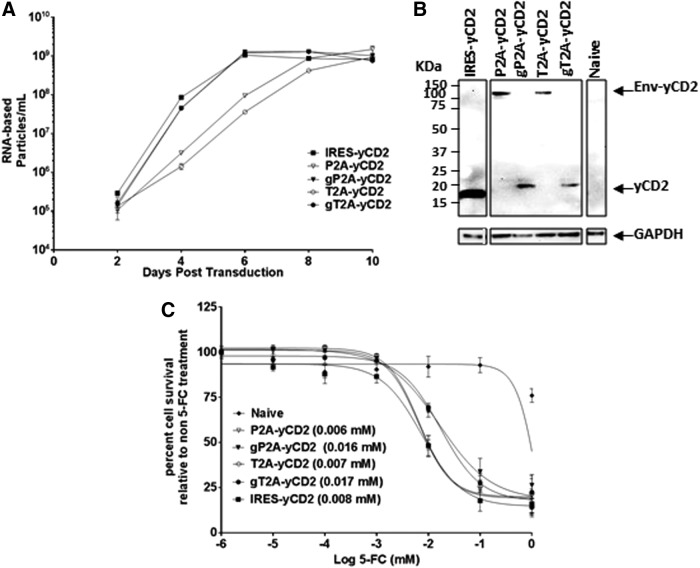
Efficient viral replication and polyprotein separation are also observed with RRV-gP2A-yCD2 and RRV-gT2A-yCD2 in human and mouse glioma cell lines
In this set of RRVs, the 2A-yCD2 variants were evaluated in comparison to RRV-IRES-yCD2 (i.e., Toca 511).8 Consistent with data observed with RRV-2A-GFP and RRV-2A-TKO sets, gP2A-yCD2 and gT2A-yCD2 variants in U87-MG cells both exhibited replication kinetics comparable to RRV-IRES-yCD2, and a delay in replication was observed for the P2A-yCD2 and T2A-yCD2 variants, as seen for P2A-GFP and T2A-GFP (Fig. 4A). In addition, efficient yCD2 protein separation from the viral envelope protein was observed with gP2A and gT2A but not with P2A and T2A (Fig. 4B). With respect to the separation efficiency of the yCD2 protein from the viral envelope protein and its effect on the incorporation of properly processed viral envelope protein, as well as titer produced from the maximally infected U87-MG cells, the data are consistent with what were observed in the corresponding RRV-2A-GFP variants (Supplementary Fig. S2A and B).
Figure 4.
Vector characterization of RRV-2A encoding the yCD2 transgene in U87-MG cells. (A) Replication kinetics of RRV-2A-yCD2 variants. Viral genomes isolated from the viral supernatant of infected U87-MG cells were quantified by RT-qPCR using primers spanning the viral envelope region. Infected cells were passaged every other day through 10 days post infection, and supernatants were collected at each indicated time point. Data shown represent one of three independent experiments. RRV-IRES-yCD2 vector was included as control. (B) Western blot analysis of viral proteins produced by U87-MG cells maximally infected with RRV-2A-yCD2 variants. Twenty micrograms of total protein lysates was loaded per well. Membranes were incubated with anti-yCD2 antibody, which detects the viral envelope-yCD2 polyprotein (Env-yCD2). Detection of GAPDH using the anti-GAPDH antibody was included as a loading control. Protein standards are indicated in kDa. (C) LD50 of 5-FC-mediated killing of maximally infected U87-MG cells with RRV-2A-yCD2 variants. Cells were cultured in the presence of 5-FC in different concentrations for 7 days. Naïve cells were included as a control for 5-FC cytotoxicity at each concentration. Cell viability was quantified by using MTS assay at 7 days post 5-FC addition, and the percentage of cell survival was calculated relative to RRV-infected cells not treated with5-FC. The data set represents one of the three independent experiments. Error bars indicate the standard deviation of the data set. Numbers in parentheses indicate LD50 values. Statistical significance was determined by two-way ANOVA. The p-value for P2A-yCD2 versus gP2A-yCD2 is <0.0079; the p-value for T2A-yCD2 vs. gT2A-yCD2 is <0.0001.
Due to the extensive characterizations for RRV-IRES-yCD2 (Toca 511) and its superior long-term viral genome stability,8 we sought to evaluate the genome stability of the 2A-yCD2 variants and compare to RRV-IRES-yCD2 in vitro. The viral genome stability was assessed by endpoint PCR using primers that span the IRES-transgene or 2A-transgene cassette from each cycle of infection. RRV-IRES-yCD28 was included as a reference. Supplementary Figure S2C shows that viral genomes of RRV-gP2A-yCD2 and RRV-gT2A-yCD2 are both stable, with no emergence of a deletion mutant in RRV-gP2A-yCD2 until infection cycle 13. In sharp contrast, deletion mutants emerged as early as infection cycles 2 and 4 for RRV-P2A-yCD2 and RRV-T2A-yCD2, respectively. As expected, the deletion mutants that emerged in RRV-P2A-yCD2 became predominant, as their genomes likely provide replicative advantages over time. Interestingly, deletion mutants that emerged in RRV-T2A-yCD2 appear to co-exist with RRV-T2A-yCD2 at equal proportion through the remaining infection cycles (Supplementary Fig. S2C).
Tumor-selective Toca 511 encoding the yCD2 therapeutic transgene converts the prodrug 5-fluorocytosine (5-FC) to cytotoxic 5-fluorouracil (5-FU) and kills rapidly dividing tumor cells and myeloid derived suppressor cells, as well as tumor-associated macrophages.2,3,8 We next asked if there is a difference in the biological activity of the yCD2 among the four 2A-yCD2 variants in maximally infected U87-MG cells due their different efficiency in polyprotein separation. Unexpectedly, as seen with the 2A-TKO variants, the results of LD50 assay showed that both P2A-yCD2 and T2A-yCD2 variants, where the yCD2 protein was inefficiently separated from the viral envelope protein, have LD50 values that are comparable to that of RRV-IRES-yCD2. The gP2A-yCD2 and gT2A-yCD2 variants, where the yCD2 protein was efficiently separated from the viral envelope protein, have LD50 values that are slightly higher than P2A-yCD2 and T2A-yCD2 variants and RRV-IRES-yCD2 (Fig. 4C; p < 0.0001). The data suggest that both forms of yCD2 proteins may be biologically functional in converting 5-FC to 5-FU to kill tumor cells.
Tumor-bearing mice transduced with RRV-gT2A-yCD2 exhibit superior survival benefit than RRV-IRES-yCD2 in orthotopic syngeneic mouse models
The in vitro characterization of the 2A-yCD2 variants was further extended in two mouse glioma cell lines, Tu2449 and Tu2449SC, in which Toca 511 demonstrated its cytotoxic and long-term antitumor immune effect in animal models.2,3,21 As described in U87-MG cells, both the Tu2449 and Tu2449SC cell lines support viral replication with stable viral genomes and express biologically active yCD2 (Supplementary Fig. S3). Consistently, separation of the viral envelope-yCD2 polyprotein is more efficient in gP2A-yCD2 and gT2A variants than in non-GSG linked P2A-yCD2 and T2A-yCD2.
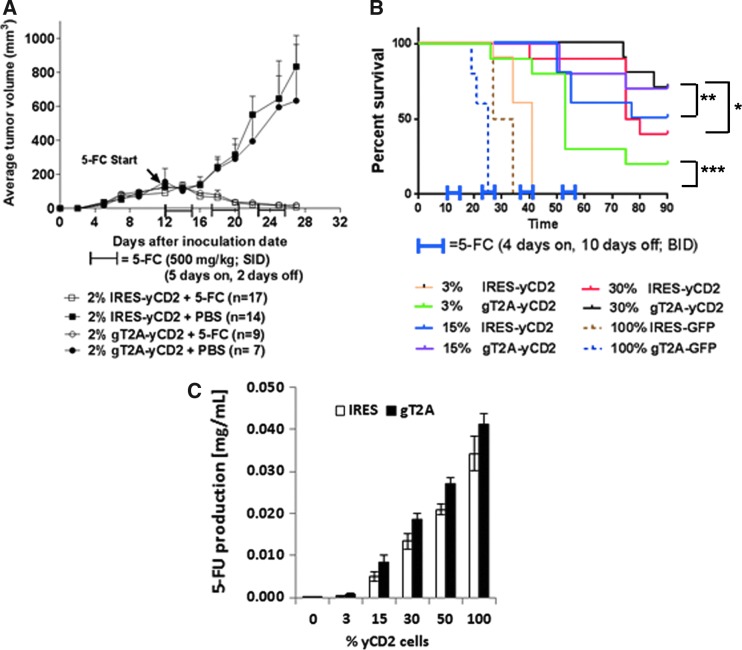
The high degree of genome stability of RRV-gT2A-yCD2 led to the feasibility of using the 2A peptides for expression of a therapeutic transgene delivered by RRV. RRV-gT2A-yCD2 was selected for evaluating its antitumor effect in comparison to RRV-IRES-yCD2 of which its antitumor effect has been demonstrated in both intracranial and subcutaneous syngeneic mouse models.2,3 The antitumor effect was first examined in a subcutaneous model. In this experiment, administration of RRVs (RRV-gT2A-yCD2 and RRV-IRES-yCD2) and subsequent viral replication in tumor was performed by mixing 2% RRV-infected cells with 98% naïve cells at initial tumor implantation followed by in situ viral spread. In this subcutaneous model, the 5-FC treatment started at day 12 post tumor implantation when tumors reached an average size of 100–200 mm3. Tumor burden for each group was assessed by averaging tumor size of all mice from each group over a period of approximately 30 days. Figure 5A shows that tumor growth in the RRV-IRES-yCD2 or RRV-gT2A-yCD2 infected Tu2449SC tumors in PBS-treated groups continued to grow, whereas tumors infected with RRV-IRES-yCD2 or RRV-gT2A-yCD2 were eradicated after three cycles of 5-FC treatment.
Figure 5.
Tumor burden and survival analysis of Tu2449 tumor mouse models. (A) Tumor burden of female B6C3F1 mice subcutaneously implanted with 1 × 106 cells mixed with 2% Tu2449SC/RRV-IRES-yCD2 + 98% Tu2449SC tumor cells or 2% Tu2449SC/RRV-gT2A-yCD2 + 98% Tu2449SC tumor cells on the right flank. On day 12, 5-FC treatment (arrow, 500 mg/kg in 800 μL intraperitoneally [i.p.], once daily [SID]) was administered in three cycles: 5 days on followed by 2 days off; or PBS treatment (800 μL i.p., SID) in parallel, through 30 days post tumor inoculation. Tumor sizes were monitored two to three times a week until tumor volumes reached >2000 mm3. Statistical significance was determined by two-way ANOVA of the following data sets: p < 0.0001 for g2A-yCD2 + 5-FC vs. g2A-yCD2 + PBS; p < 0.0001 for IRES-yCD2 + 5-FC vs. IRES-yCD2 + PBS. (B) Kaplan–Meier survival analysis of orthotopic glioma mouse model. B6C3F1 mice were implanted intracranially (i.c.) with the different ratios of Tu2449/RRV-IRES-yCD2 to Tu2449/RRV-IRES-GFP or Tu2449/RRV-gT2A-yCD2 to Tu2449/RRV-2A-GFP cells (3/97, 15/85, and 30/70, respectively) then dosed i.c. with vehicle (control, PBS) or i.c. with 5-FC, starting at 10 days post tumor implant over four cycles of 4 days on 5-FC followed by10 days off, treated twice daily. Survival analysis was monitored for 90 days. Statistical significance of survival between mice treated with RRV-IRES-yCD2 and RRV-gT2A-yCD2 for each subgroup was determined by the log-rank (Mantel–Cox) test and is indicated by brackets: *p = 0.1416; **p = 0.3755; ***p = 0.0013. (C) Measurement of extracellular 5-FU concentrations generated from the conversion of 5-FC to 5-FU by the yCD2 proteins. Error bars indicate the standard deviation of the data set.
The yCD2 protein in the RRV-gT2A-yCD2 is efficiently separated from the polyprotein. There is no remarkable difference in its LD50 values in vitro (Supplementary Fig. S3) and antitumor effect in vivo (Fig. 5A) compared to RRV-IRES-yCD2 in which tumor cells are at least 50–75% transduced (data not shown) in the subcutaneous setting at the time the 5-FC treatment initiated. It is difficult to differentiate the antitumor effect between two vectors due to excessive amount of 5-FU being generated.22 Thus, the animal experimental design was refined to address a question that is more clinically relevant. It was hypothesized that the level of transduction and/or level of yCD2 protein expression could greatly influence the 5-FU bystander and cytotoxic effect, as well as the long-term antitumor immune effect (unpublished data; Yagiz et al., manuscript in preparation). This experiment utilized retroviral superinfection resistance23 via receptor downregulation to generate tumor cells consisting of defined percentages of the cells transduced with RRV expressing the therapeutic transgene (i.e., RRV-IRES-yCD2 or RRV-gT2A-yCD2) with the remaining cells transduced with a non-therapeutic control RRV (i.e., RRV-IRES-GFP or RRV-gT2A-GFP). This artificial setting allowed the dose-dependent therapeutic effect of RRV expressing the therapeutic transgene to be investigated in a more controlled manner than dosing by intra-tumoral injection of RRV.3 In vitro, it was shown that the ratio (3/97) of RRV-GFP to non-GFP RRV infected Tu2449 cells remains constant over time (Supplementary Fig. S4). Importantly, it was shown that the bystander effect of 5-FU leading to cytotoxicity is greatly reduced under the condition in which <15% of cells are transduced with RRV-IRES-yCD2 (Supplementary Fig. S5A) or RRV-gT2A-yCD2 (Supplementary Fig. S5B). To demonstrate that the survival benefit can be influenced by the percentage of cells expressing the therapeutic yCD2 protein and to compare the antitumor effect between RRV-IRES-yCD2 and RRV-gT2A-yCD2, matching pairs of RRV-transduced Tu2449 cells infected with RRV-IRES-yCD2/RRV-IRES-GFP and RRV-gT2A-yCD2/RRV-gT2A-GFP were generated at ratios of 3/97, 15/85, and 30/70. In an orthotopic syngeneic mouse glioma model, mice bearing tumors of 100% transduced RRV-IRES-GFP and 100% transduced RRV-gT2A-GFP tumors were included as control groups for RRV-IRES-yCD2/RRV-IRES-GFP and RRV-gT2A-yCD2/RRV-gT2A-GFP, respectively, and showed no significant difference in their survival benefit when treated with 5-FC due to the absence of the yCD2 transgene (Fig. 5B). Expectedly, a dose-dependent survival benefit was observed within each subgroup of RRV-IRES-yCD2/RRV-IRES-GFP and RRV-gT2A-yCD2/RRV-gT2A-GFP. When comparing between the RRV-IRES-yCD2 and RRV-gT2A-yCD2 groups at the 3/97 ratio, the survival data over a 90-day period indicated that mice bearing tumors transduced with RRV-gT2A-yCD2/RRV-gT2A-GFP have a statistically significant higher survival benefit than mice bearing the tumor transduced with RRV-IRES-yCD2/RRV-IRES-GFP (Fig. 5B; p = 0.0013). A similar trend in survival benefit between the two groups at 15/85 and 30/70 ratios was also observed, but this was not statistically significant (Fig. 5B; p = 0.3755 and p = 0.1416, respectively).
The in vitro bystander data at the ratio of 3/97 setting (Supplementary Fig. S5B) and vector stability data (Supplementary Fig. S2C) did not indicate that RRV-gT2A-yCD2 is more stable and/or has more bystander-mediated cytotoxic effect than RRV-IRES-yCD2. To delineate the underlying mechanism for RRV-gT2A-yCD2 leading to a better survival benefit than RRV-IRES-yCD2 in tumor-bearing mice, the enzymatic activity of the yCD2 protein from the two vectors was evaluated by measuring the amount of 5-FU produced from the infected cells using the matching ratios conducted in the in vivo study. Figure 5C shows that RRV-gT2A-yCD2 produced more 5-FU than RRV-IRES-yCD2 at all ratios tested. The continuous increase in the 5-FU production with increasing percentage of yCD2-expressing cells suggests that the 5-FC administered is not limiting. It is also important to point out that the higher yCD2 potency observed with RRV-gT2A-yCD2 is not due to a higher level of protein expression (Supplementary Fig. S3D). In line with a recent publication on immune-mediated antitumor response,2 the data also indirectly suggest that the higher 5-FU production from the RRV-gT2A-yCD2/RRV-gT2A-GFP cells at the transduction level <30% in vivo may reflect an antitumor response dominated by an immune-mediated response leading to longer survival.
RRV-2A configuration can tolerate insertion of multiple transgenes
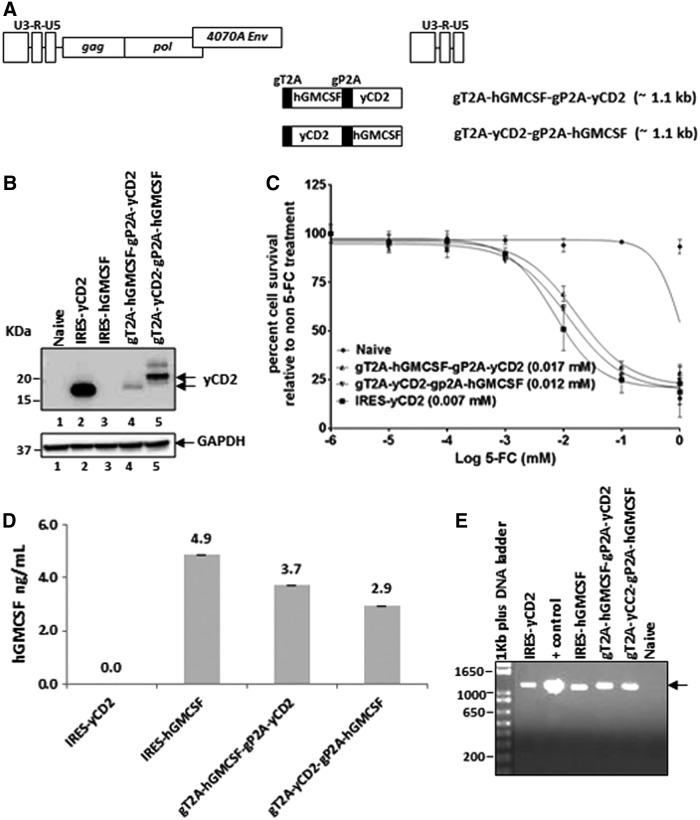
Since multiple 2A peptides have been successfully used to encode multiple genes in various expression vectors,11,24–26 the study asked whether RRV-2A configuration can be utilized for multiple gene expression. To explore this, the gP2A and gT2A were used to express yCD2 and human GM-CSF (hGMCSF) protein in one vector. Other than its known immune functions and clinical implications,27,28 hGMCSF was chosen mainly due to its small size (cDNA length of 435 bp) and its expression as a secreted protein, which is in a different cellular compartment than the yCD2 and TKO protein. Two versions of RRVs were made for multiple-transgene expression: RRV-gT2A-hGMCSF-gP2A-yCD2 and RRV-gT2A-yCD2-gP2A-hGMCSF (Fig. 6A). Briefly, viral titer produced from maximally infected U87-MG cells showed comparable titer values compared to that of RRV-IRES-yCD2 (Supplementary Table S3). Immunoblotting with anti-yCD2 antibody showed that the yCD2 protein is efficiently separated from the viral envelope protein in RRV-gT2A-yCD2-gP2A-hGMCSF and from the hGMCSF in RRV-gT2A-hGMCSF-gP2A-yCD2, but the overall yCD2 protein expression is reduced compared to that of RRV-IRES-yCD2 (Fig. 6B). In addition, the drastically reduced yCD2 protein expression by RRV-gT2A-yCD2-gP2A-hGMCSF is associated with aberrant yCD2 isoforms (Fig. 6B). Despite the presence of the reduced protein expression level and aberrant yCD2 isoforms, the LD50 data indicated that U87-MG cells maximally infected with RRV-gT2A-hGMCSF-gP2A-yCD2 or RRV-gT2A-yCD2-gP2A-yCD2 are comparable but slightly higher than those infected with RRV-IRES-yCD2 (Fig. 6C). However, the observed differences are not statistically significant. Furthermore, the level of hGMCSF secreted into cultured medium was also measured by ELISA using U87-MG cells infected with RRV-IRES-hGMCSF as a control. Figure 6D shows that all three vectors express hGMCSF in the ng/mL range and that the levels secreted from RRV-gT2A-hGMCSF-gP2A-yCD2 and RRV-gT2A-yCD2-gP2A-hGMCSF were within less than a twofold difference compared to RRV-IRES-hGMCSF. Most importantly, vector stability from maximally infected U87-MG cells indicated that the vectors remained stable during the course of infection (Fig. 6E). Altogether, the data suggest that it is feasible to utilize multiple 2A peptides to express more than one therapeutic transgenes with different cellular compartmentations in RRV.
Figure 6.
Vector characterization of RRVs expressing multiple transgenes from 2A peptides in U87-MG cells. (A) Schematic diagram of the RRV constructs encoding multiple-transgene cassette of gT2A-hGMCSF-gP2A-yCD2 and gT2A-yCD2-gP2A-hGMCSF. The approximate size of the multiple transgene cassette is shown in parentheses. (B) Western blot analysis of yCD2 proteins produced by U87-MG cells maximally infected with RRV-gT2A-hGMCSF-gP2A-yCD2 and RRV-gT2A-yCD2-gP2A-hGMCSF. RRV-IRES-yCD2 and RRV-IRES-hGMCSF were included as controls. Twenty micrograms of total protein lysates was loaded per well. Membranes were incubated with anti-yCD2 antibody. Detection of GAPDH using the anti-GAPDH antibody was included as a loading control. (C) LD50 of 5-FC-mediated killing of maximally infected U87-MG cells with RRV-2A-yCD2 variants. Cells were cultured in the presence of 5-FC in different concentrations for 7 days. Naïve cells were included as a control for 5-FC cytotoxicity at each concentration. Cell viability was quantified by using MTS assay at 7 days post 5-FC addition, and the percentage of cell survival was calculated relative to RRV-infected cells not treated with 5-FC. The data set represents one of the three independent experiments. Error bars indicate the standard deviation of the data set. Numbers in parentheses indicate LD50 values. Statistical significance was determined by two-way ANOVA. (D) hGMCSF in the supernatant produced from maximally infected U87-MG cells was quantified by ELISA. Error bars indicate the standard deviation of the data set. (E) Stability of yCD2-hGMCSF and hGMCSF-yCD2 transgene cassettes from proviral DNA in maximally infected U87-MG cells (16 days post infection). The arrow indicates the size of the PCR product expected for the intact yCD2-hGMCSF and hGMCSF-yCD2 transgene cassette + controls are PCR product amplified from plasmid DNA corresponding to each RRV-2A-TKO variants. DNA molecular ladder (1 kb plus; Life Technologies) is shown in the first lane of the gel; numbers indicate the size of the ladders in base pairs.
Discussion
Retroviral replicating vectors are attractive for cancer gene therapy due to their high selectivity for cancer cells, non-lytic property, and integration into the host genome. As a virus continues to replicate in a tumor, the stability of the therapeutic transgene incorporated into the viral genome is necessary to maintain therapeutic transgene expression.
The use of IRES to design a bi-cistronic messenger RNA has been widely used for expressing transgenes in various vector systems.29,30 RRV with the IRES configuration has a high degree of genome stability.8 However, to date, the choice of therapeutic genes may be restricted due to the size limitation of the viral genome. Although the 2A peptide has been utilized in F-MLV to express a single transgene, a full characterization of the virus and transgene expression was lacking, preventing insights into the feasibility of this configuration for clinical applications. This study shows that the 2A peptide configuration allows insertion of therapeutic transgene(s) up to approximately 1.2 kb compared to so far approximately 0.7 kb with the IRES configuration, and it demonstrated that the RRV-2A configuration consisting of a GSG linker allows high efficiency of polyprotein separation and tolerates single and multiple therapeutic transgene insertion without compromising the viral genome stability, infectivity, and transgene expression.
An interesting finding from this study is the higher separation efficiency of the viral envelope-transgene polyprotein bridged by a GSG-linked 2A. Although GSG linker has not been described as a necessary component in transgene protein expression and particularly not in the context of murine leukemia retrovirus genome,12 its presence in facilitating polyprotein separation has been described in some expression vectors and cell lines of different species.11,31 In the RRV system, the enhancement of polyprotein separation with the GSG linker upstream of the 2A peptide is confirmed with three transgenes tested in this study. The reasons for this improvement are unknown. This may result from the flexibility of the GSG linker or via creating more space from distal inhibitory residues that may interfere with the separation.9,31 It is possible that the requirement for the GSG linker to achieve high efficiency of polyprotein separation may depend on the tertiary structure of the protein N-terminus to the 2A peptide. Intriguingly, the biological activity of the polyproteins in both Env-TKO and Env-yCD2 without the GSG linker appeared to be equally functional as TKO and yCD2 expressed from the IRES, respectively. As the TKO and yCD2 protein in the RRV-2A configuration are translated in-frame with the transmembrane domain of the viral envelope protein, the Env-TKO and Env-yCD2 polyprotein may exist in a form that is membrane associated and could remain biological active intracellularly but not be incorporated into viral particles. However, because maximally infected cells were used to determine the LD50 values, we cannot rule out the possibility that there was sufficient level of biologically active TKO expressed to exert the 5-FC- or GCV-mediated cytotoxicity. Another interesting finding from this study is the markedly improved viral genome stability with the inclusion of the GSG linker in the RRV-2A-yCD2 variants (Supplementary Fig. S2C). It will be of a great interest to see if this observation also holds true in general in the RRV system.
In a parallel comparison of the RRV-IRES-yCD2 and RRV-2A-yCD2 configuration, the yCD2 protein translation in the IRES-yCD2 configuration is expected to occur in both the spliced and unspliced viral transcripts, whereas the yCD2 protein translation only occurs in the spliced viral transcript. Nevertheless, a similar level of transgene expression from both configurations is evident by the comparable yCD2 protein expression observed in RRV-IRES-yCD2 and RRV-gT2A-yCD2 in multiple cell lines. This observation raises the question that the protein translation mediated by the IRES from the unspliced viral transcript may not be as efficient as previously anticipated. Reports of gene expression downstream of IRES at a lower level than the transgene upstream of IRES in bi-cistronic expression vectors also support the finding.32–34
An intriguing finding from this study is the survival benefit of tumor-bearing mice pre-transduced with the RRV-IRES-yCD2 and RRV-gT2A-yCD2. The 2A configuration was observed to be superior to the IRES configuration under limited transduction setting in two independent in vivo studies (one set of data shown in Fig. 5B). The preliminary in vitro data show that the higher survival benefit observed with RRV-gT2A-yCD2 is partly due to higher potency of the yCD2 protein derived from the 2A configuration and not due to higher level of protein expression. It is speculated that the higher survival benefit observed in tumor cells infected with RRV-gT2A-yCD2 is dominated by an immune-mediated antitumor bystander effect by 5-FU elimination of myeloid-derived suppressor cells, as described in a recent publication.2 The immune-mediated antitumor effect in vivo may explain why a higher bystander-mediated cytotoxic effect of RRV-gT2A-yCD2 was not observed when compared to that of RRV-IRES-yCD2 in vitro (Supplementary Fig. S5A and B). The attribute of higher potency of the yCD2 protein derived from the gT2A-yCD2 configuration is undefined and remains to be elucidated.
In summary, the data demonstrate that placing a transgene cassette downstream of the viral envelope protein separated by a GSG-linked 2A peptide in RRV is a feasible strategy to allow long-term viral genome stability and transgene expression with potentially increased drug activity. In addition, the additional space allowed in the 2A configuration may broaden the selection of therapeutic genes for future single- or multiple-agent cancer gene therapy.
Supplementary Material
Acknowledgments
We thank Asha Das, John Wood, Marty J. Duvall, Nicholas A. Boyle, and Victoria Cohan for critical reading of the manuscript. The original RRV system was developed with support from NIH grants U01-NS59821 and R01-CA105171 to N.K. The work was also supported by Accelerate Brain Cancer Cure, American Brain Tumor Association (Washington, DC), Musella Foundation (Hewlett, NY), National Brain Tumor Society (Watertown, MA), Voices Against Brain Cancer (New York, NY), and the Department of Health and Human Services US (Washington, DC).
Author Disclosure
A.H., K.Y., D.M., F.L., A.W.M., C.B., H.E.G., D.J.J., and A.H.L. are full-time employees of Tocagen and hold stock or stock options of Tocagen.
References
- 1.Huang TT, Parab S, Burnett R, et al. Intravenous administration of retroviral replicating vector, Toca 511, demonstrates therapeutic efficacy in orthotopic immune-competent mouse glioma model. Hum Gene Ther 2015;26:82–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell LA, Lopez Espinoza F, Mendoza D, et al. Toca 511 gene transfer and treatment with the prodrug, 5-fluorocytosine, promotes durable antitumor immunity in a mouse glioma model. Neuro Oncol 2017;19:930–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ostertag D, Amundson KK, Lopez Espinoza F, et al. Brain tumor eradication and prolonged survival from intratumoral conversion of 5-fluorocytosine to 5-fluorouracil using a nonlytic retroviral replicating vector. Neuro Oncol 2012;14:145–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiraoka K, Inagaki A, Kato Y, et al. Retroviral replicating vector-mediated gene therapy achieves long-term control of tumor recurrence and leads to durable anticancer immunity. Neuro Oncol 2017;19:918–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cloughesy TF, Landolfi J, Hogan DJ, et al. Phase 1 trial of vocimagene amiretrorepvec and 5-fluorocytosine for recurrent high-grade glioma. Sci Transl Med 2016;8:341ra75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paar M, Klein D, Salmons B, et al. Influence of vector design and host cell on the mechanism of recombination and emergence of mutant subpopulations of replicating retroviral vectors. BMC Mol Biol 2009;10:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Logg CR, Tai CK, Logg A, et al. A uniquely stable replication-competent retrovirus vector achieves efficient gene delivery in vitro and in solid tumors. Hum Gene Ther 2001;12:921–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perez OD, Logg CR, Hiraoka K, et al. Design and selection of Toca 511 for clinical use: modified retroviral replicating vector with improved stability and gene expression. Mol Ther 2012;20:1689–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Felipe P, Luke GA, Hughes LE, et al. E unum pluribus: multiple proteins from a self-processing polyprotein. Trends Biotechnol 2006;24:68–75 [DOI] [PubMed] [Google Scholar]
- 10.Osborn MJ, Panoskaltsis-Mortari A, McElmurry RT, et al. A picornaviral 2A-like sequence-based tricistronic vector allowing for high-level therapeutic gene expression coupled to a dual-reporter system. Mol Ther 2005;12:569–574 [DOI] [PubMed] [Google Scholar]
- 11.Szymczak AL, Workman CJ, Wang Y, et al. Correction of multi-gene deficiency in vivo using a single “self-cleaving” 2A peptide-based retroviral vector. Nat Biotechnol 2004;22:589–594 [DOI] [PubMed] [Google Scholar]
- 12.Browne EP. An interleukin-1 beta-encoding retrovirus exhibits enhanced replication in vivo. J Virol 2015;89:155–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stavrou S, Crawford D, Blouch K, et al. Different modes of retrovirus restriction by human APOBEC3A and APOBEC3G in vivo. PLoS Pathogens 2014;10:e1004145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin AH, Liu Y, Burrascano C, et al. Extensive replication of a retroviral replicating vector can expand the A bulge in the encephalomyocarditis virus internal ribosome entry site and change translation efficiency of the downstream transgene. Hum Gene Ther Methods 2016;27:59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin AH, Burrascano C, Pettersson PL, et al. Blockade of type I interferon (IFN) production by retroviral replicating vectors and reduced tumor cell responses to IFN likely contribute to tumor selectivity. J Virol 2014;88:10066–10077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin AH, Timberlake N, Logg CR, et al. MicroRNA 142-3p attenuates spread of replicating retroviral vector in hematopoietic lineage-derived cells while maintaining an antiviral immune response. Hum Gene Ther 2014;25:759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin AH, Twitty CG, Burnett R, et al. Retroviral replicating vector delivery of miR-PDL1 inhibits immune checkpoint PDL1 and enhances immune responses in vitro. Mol Ther Nucleic Acids 2017;6:221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minskaia E, Ryan MD. Protein coexpression using FMDV 2A: effect of “linker” residues. Biomed Res Int 2013;2013:291730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JH, Lee SR, Li LH, et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PloS One 2011;6:e18556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murakami T. Retroviral env glycoprotein trafficking and incorporation into virions. Mol Biol Int 2012;2012:682850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hlavaty J, Jandl G, Liszt M, et al. Comparative evaluation of preclinical in vivo models for the assessment of replicating retroviral vectors for the treatment of glioblastoma. J Neurooncol 2011;102:59–69 [DOI] [PubMed] [Google Scholar]
- 22.Twitty CG, Diago OR, Hogan DJ, et al. Retroviral replicating vectors deliver cytosine deaminase leading to targeted 5-fluorouracil-mediated cytotoxicity in multiple human cancer types. Hum Gene Ther Methods 2016;27:17–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nethe M, Berkhout B, van der Kuyl AC. Retroviral superinfection resistance. Retrovirology 2005;2:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahier A, Jarriault S. Simultaneous expression of multiple proteins under a single promoter in Caenorhabditis elegans via a versatile 2A-based toolkit. Genetics 2014;196:605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Felipe P. Skipping the co-expression problem: the new 2A “CHYSEL” technology. Genet Vaccines Ther 2004;2:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Z, Chen O, Wall JBJ, et al. Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci Rep 2017;7:2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arellano M, Lonial S. Clinical uses of GM-CSF, a critical appraisal and update. Biologics 2008;2:13–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiomi A, Usui T. Pivotal roles of GM-CSF in autoimmunity and inflammation. Mediators Inflamm 2015;2015:568543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgan RA, Couture L, Elroy-Stein O, et al. Retroviral vectors containing putative internal ribosome entry sites: development of a polycistronic gene transfer system and applications to human gene therapy. Nucleic Acids Res 1992;20:1293–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Renaud-Gabardos E, Hantelys F, Morfoisse F, et al. Internal ribosome entry site-based vectors for combined gene therapy. World J Exp Med 2015;5:11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Wang F, Wang R, et al. 2A self-cleaving peptide-based multi-gene expression system in the silkworm Bombyx mori. Sci Rep 2015;5:16273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaufman RJ, Davies MV, Wasley LC, et al. Improved vectors for stable expression of foreign genes in mammalian cells by use of the untranslated leader sequence from EMC virus. Nucleic Acids Res 1991;19:4485–4490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizuguchi H, Xu Z, Ishii-Watabe A, et al. IRES-dependent second gene expression is significantly lower than cap-dependent first gene expression in a bicistronic vector. Mol Ther 2000;1:376–382 [DOI] [PubMed] [Google Scholar]
- 34.Zhou Y, Aran J, Gottesman MM, et al. Co-expression of human adenosine deaminase and multidrug resistance using a bicistronic retroviral vector. Hum Gene Ther 1998;9:287–293 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.