SUMMARY
Neurons exhibit a rich diversity of morphological phenotypes, electrophysiological properties and gene expression patterns. Understanding how these different characteristics are interrelated at the single-cell level has been difficult due to the lack of techniques for multimodal profiling of individual cells. We recently developed Patch-seq, a technique that combines whole-cell patch clamp recording, immunohistochemistry and single-cell RNA-sequencing (scRNA-seq) to comprehensively profile single neurons from mouse brain slices. Here we present a detailed step-by-step protocol including modifications to the patching mechanics and recording procedure, reagents and recipes, procedures for immunohistochemistry, and other tips to assist researchers in obtaining high-quality morphological, electrophysiological and transcriptomic data from single neurons. Successful implementation of Patch-seq allows researchers to explore the multidimensional phenotypic variability among neurons and to correlate gene expression with phenotype at the level of single cells. The entire procedure can be completed in approximately two weeks through the combined efforts of a skilled electrophysiologist, molecular biologist, and biostatistician.
Keywords: Patch-seq, electrophysiology, single-cell RNA-sequencing, RNA sequencing, single cell sequencing, transcriptomics, morphology, cell type, scRNA-seq, whole-cell patch clamp, patch clamp, immunohistochemistry, IHC, neuron, mouse, brain, tissue slice, morphology
EDITORIAL SUMMARY
This protocol describes how to integrate whole-cell patch clamp in single neurons from mouse brain tissue slices with single-cell RNA-sequencing and morphological recovery.
INTRODUCTION
Recent advances in single-cell RNA sequencing (1–4) have enabled unbiased molecular cell type classification in complex tissues (5, 6), including several brain regions (3, 7–11). However, linking these specific molecular cell types with their corresponding morphological, electrophysiological, and connectivity-level phenotypes has been difficult. While Cre lines provide a tremendous resource for further characterizing molecularly defined cell types, many morphological classes cannot be clearly defined by differential expression of a single gene (12, 13). New technologies that correlate transcriptomic and phenotypic data at the level of single cells may provide a powerful and complementary approach to better understand the molecular underpinnings of cell type diversity.
Many groups have attempted to combine whole-cell patch clamp recording, a classic technique used for studying the electrophysiological and morphological properties of individual neurons, with analysis of single-cell gene expression. Early studies performed RT-PCR of the patch pipette contents following whole-cell recording (14–17). However, these techniques require the prior selection of a handful of genes (up to a few dozen (18, 19)), which provides only a small glimpse of the full complexity of genes expressed by each cell and does not allow for identification of new genes that may be important in determining cellular phenotype or defining cell types. More recently, single-cell microarray has been used to assess genome-wide expression after patch recording (20), however microarray techniques have a limited dynamic range and poor sensitivity and specificity compared to sequencing-based approaches (1, 21), and cannot detect novel transcripts or splice variants. Initial attempts to perform single-cell RNA-sequencing (scRNA-seq) on patched neurons yielded poor quality sequencing data: one study sequenced in total three neurons from acute slices and detected <2,000 genes per cell with a mean correlation of ~0.25 across cells, suggesting high technical variability between samples (22). In parallel with another group (23), we recently demonstrated that high quality scRNA-seq data, comparable to that obtained from dissociated cells (7, 8), is indeed feasible from patch clamp-recorded neurons with a mean correlation ~0.6 and ~7,000 genes detected per cell in our dataset (24). We have since further refined our methodology to improve direct morphological recovery of cells after Patch-seq. Here we describe in detail the current protocol used in our laboratory, and highlight key steps and common pitfalls to help other groups incorporate Patch-seq into their research toolbox.
Overview of the protocol
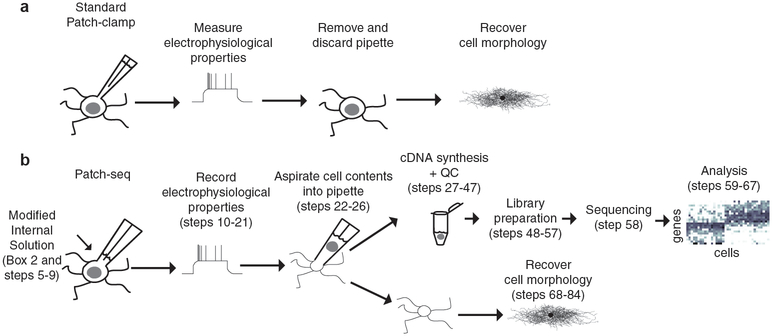
Our protocol, outlined in Fig. 1, consists of several stages including patch clamp recording, aspiration of cell contents, cDNA amplification, sequencing, data analysis, and immunohistochemistry. Several simple but critical modifications to the patching mechanics, internal solution, and downstream cDNA amplification steps are applied to improve cDNA yield. For example, when patching, we use large diameter pipettes (14, 15) loaded with a small volume of internal solution; we try to patch the cell with the pipette approaching from the side rather than the top (Fig. 2 and Supplementary Movie S1); and we continuously monitor the holding currents in voltage-clamp mode to ensure the cell remains healthy and the seal intact throughout the aspiration process. Previous studies suggested that silanization of the glass used for pulling patch pipettes may improve RNA recovery (15, 16), however we found that this actually decreased RNA yield in our hands. Therefore, we use autoclaved, nonsilanized glass to pull patch pipettes. Great caution is used to prepare solutions and collect samples in a strict RNAse-free manner (see also Box 1). Our internal solution has several modifications, some of which have been previously shown to facilitate isolation of mRNA from patched cells. For example glycogen, which serves as an RNA carrier (15, 16), and ethylene glycol-bis(β-aminoethyl ether)-N,N,N’,N’-tetraacetic acid (EGTA), which scavenges free calcium and reduces RNase activity (14), have been previously reported to increase RNA recovery from patched neurons and we included them in our internal solution. In addition to glycogen and EGTA, we also add recombinant RNase inhibitor directly to the internal solution which increased RNA yield approximately three-fold in our pilot studies. However, the addition of RNase inhibitor directly to the internal solution dramatically increases the osmolarity of the solution, interfering with standard techniques for immunohistochemistry (12). Therefore, we made additional modifications to develop and internal solution recipe that has a more physiologic osmolarity and is compatible with avidin-biotin immunohistochemistry. Our library preparation and sequencing protocol is very similar to that of Smart-seq2 (25), except that we use a slightly shorter template-switching oligonucleotide (TSO) and a lower ratio of beads:cDNA when purifying the amplified full-length cDNA. An oligo-dT primer is used to enrich for mRNAs. Due to the small amount of starting material, additional mRNA purification steps prior to reverse transcription are not practical, and samples should be assessed for quality (i.e. percent of reads mapped to exons) after sequencing.
Figure 1. Schematic of Patch-seq technique.
a) Standard whole-cell patch clamp recording technique is used to record the electrophysiological properties of a neuron and dialyze the cell with a dye (e.g. biocytin) for later histological staining and recovery of cell morphology. b) For Patch-seq, a modified internal solution and patching approach is used. Cell contents are aspirated into the pipette after recording the cell’s electrophysiological properties and used for cDNA synthesis, amplification, and sequencing. The collapsed soma and axodendritic processes remain embedded in the tissue slice and can still be stained for morphological recovery. Adapted from (24).
Figure 2. Ideal patching approach for aspiration of cell contents.
a) The target cell (Step 14) is approached from the side (Step 17–18) rather than the top to provide the most direct path for cell contents to enter the pipette. After obtaining a gigaseal, the cell membrane is quickly ruptured so that the patch pipette contents can freely diffuse into the cell (Step 19). If using a high osmolarity internal solution, the cell will gradually swell over the next 5–15 min and can be helpful to move the pipette back a few microns (Step 21) to release the tension on the membrane and prevent current leakage. At the end of the recording session (~30 min if aiming for morphological reconstruction), the content of the cell are aspirated into the pipette using gentle, negative pressure (Step 22). The cell is continuously monitored, both visually and electrophysiologically, to ensure that no extracellular contents are entering the pipette and the seal between the pipette and the membrane is intact (no large current leakage). Once aspiration is complete, the negative pressure is released and the pipette is slowly pulled away from the cell to separate the cell membrane from the pipette, leaving the cell body intact in the slice (Step 23). Once the membrane has released, the pipette can be quickly removed from the tissue and the single-cell contents collected in a 0.2 mL PCR tube. b) Examples of collapse of the cell body under two-photon imaging guidance (2PI) following aspiration of cell contents into the pipette. For these experiments, Alexa-488 was added to the internal solution to enable visualization of the pipette and patched neuron. Scale bars, 10 μm. Adapted from (24) c) Examples of collapse of the cell body after aspiration of cell contents, visualized using differential interference contrast (DIC) imaging. See Supplementary Movie S1 for video of sample collection from DIC Cell 2. Scale bar, 10 μm. All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at Baylor College of Medicine.
Box 1. Tips for reducing RNase contamination.
Ribonucleases (RNases) are ubiquitous in the natural environment and secreted by pancreatic, endothelial, and other cell types into the extracellular space presumably to protect against RNA viruses and other microorganisms (38). Due to the minute amount of RNA present in a single cell (~10 pg (39)), even a small amount of RNase contamination can easily degrade the sample RNA leading to truncated or completely absent cDNA. Therefore, utmost caution should be used to eliminate any potential sources of RNase contamination during sample collection and preparation of solutions that will be used prior to the RT (step 31) including the internal solution, lysis buffer, and reverse transcription mix.
Sample handling and preparation of solutions
In addition to using sterile techniques, certified RNase-free solutions, tubes, and pipette tips should be used whenever possible. Any reagents not available as certified RNase-free should be ordered as the highest purity available. Glassware, pipettes, counters and all other equipment should cleaned thoroughly with RNase Zap and autoclaved if possible. Any items that may have come into contact with previously amplified cDNA (counters, pipettes, glassware) should also be cleaned with DNA-off. Ideally, a dedicated RNase-free positive pressure room or dedicated RNase-free bench stocked with RNase-free equipment (pipettes, tips, glassware, tubes, vortex, tabletop centrifuge) should be used for all pre-RT sample handling steps. Following cDNA amplification (steps 32–33), samples should never return to the RNase-free work area to reduce the possibility of cross-contamination between experiments.
Sample collection
Additional steps during patching and sample collection can help reduce potential sources of contamination, including maintaining positive pressure on the pipette while navigating through ACSF and tissue, visually confirming that the pipette is clean (nothing is stuck on the inside or outside of the glass), and close visual and electrophysiological monitoring of the cell during aspiration to ensure the cell is healthy, the seal between the pipette and the cell membrane is intact and extracellular contents are not entering the pipette. As discussed under Experimental design, the inclusion of positive and negative controls in each experiment is crucial to detect RNase activity quickly so that the offending solution/equipment/habit can be identified and replaced.
Applications
Patch-seq is ideally suited to answer questions that require correlating gene expression with physiology and/or morphology at the level of single cells. For example, Patch-seq can be used to classify cell types by integrating information about each cell’s morphology, physiology and gene expression into a common framework. Along these lines, we recently demonstrated that it is feasible to identify molecular cell types using only the data generated from Patch-seq (24), and that these molecular cell types largely correlate with physiological and morphological phenotypes (24). To extend this approach to more complex brain regions several major challenges need to be overcome. First, the technique must be made high-throughput, as thousands of samples are likely to be required to characterize all of the cell types in more complex brain regions. Thus cost is an important factor and we attempt to use off-the-shelf reagents rather than commercially available kits whenever possible to achieve lower costs (see also “Limitations”). Second, our original paper took advantage of the high correlation between physiology and morphology for the particular cell types studied, however in many cases morphology cannot be inferred from physiology (13) and detailed morphological recovery of sequenced cells will be necessary. Here we provide additional modifications to the Patch-seq protocol that can improve morphological recovery of the sequenced cells, which represents a crucial improvement over our original protocol. These advances make Patch-seq a viable option for stand-alone, unbiased, multidimensional characterization to identify cell types in the nervous system.
Patch-seq can also be used as a complementary method to scRNA-seq of dissociated neurons to link molecular cell types with their corresponding morphology and physiology. In this approach, a large scRNA-seq dataset from dissociated neurons is first generated to identify all of the molecular cell types in a given brain region. Then Patch-seq is performed on a smaller number of targeted neurons to gather information about the cells’ morphology and/or physiology. For example, our Patch-seq data from layer 1 interneurons in mouse primary visual cortex (V1)(24) corresponds very well with the molecular subtypes identified by another group using scRNA-seq of dissociated V1 neurons (8), allowing the layering of phenotypic information onto the molecular cell taxonomy ((8) and unpublished data from the Allen Brain Institute)
In addition to cell type studies, Patch-seq can be applied to study a number of other scientific questions that require multimodal analysis of single cells. For example, combining Patch-seq with multiple simultaneous whole-cell recording techniques to study connectivity (12) could provide insights into the molecular mechanisms that determine synaptic specificity. We also envision many applications of Patch-seq in other cell types and organs outside of the nervous system, such as hormone-secreting pancreatic islet cells (26). In summary, we believe that Patch-seq is a powerful tool that can enhance many research programs and permit new avenues of investigation into the molecular underpinnings of cellular diversity.
Level of expertise needed to implement the protocol
While it is feasible for one person to carry out the entire experiment from start to finish, we have found that it is more efficient to have at least two people involved: one is primarily an electrophysiologist and the other is primarily a molecular biologist. In particular, when trying to preserve both the morphology and the RNA of a single cell there are two time-sensitive steps that need to be done essentially simultaneously: fixation of the tissue slice (Step 26) and ejection of the pipette contents into a PCR tube (Step 25). Thus, having two people available during sample collection is preferred. It is also highly recommended to enlist the assistance of a biostatistician who is experienced in single-cell RNA-sequencing analysis, as the large amount of highly complex data generated can be overwhelming and analysis tools are rapidly evolving (27–30).
Comparison with other methods
An alternative approach to characterize the morphology and physiology of molecularly specified neuronal cell types is through the use of transgenic mouse lines (e.g. Cre driver lines or tetracycline controlled transcription factors). By expressing a fluorescent reporter in the cell type of interest, standard patch-clamp techniques can then be applied to characterize the physiology and morphology of the targeted cell population. In practice, however, this approach is often limited by the availability and specificity of driver lines, which frequently label multiple distinct morphological cell types (12). Recent single-cell transcriptomic studies suggest that the combinatorial expression of two or more genes may be necessary to uniquely identify many molecular subtypes (3, 7, 8, 10), and the continued development of intersectional genetic tools may expand the utility of transgenic approaches to link transcriptomic cell types with corresponding phenotypes.
Another group developed a similar technique to combine patch-clamp recording with single-cell RNA-seq, which was published in parallel with ours (23). Both protocols call for the use of relatively large patch pipettes which are loaded with a small volume of internal solution, and the addition of EGTA to the internal solution. The protocols differ slightly in the composition of the internal solution, stimulation protocol and sequencing method used. As more groups adopt these techniques and continue to make iterative improvements to the protocol, we expect that the quality of single-cell transcriptomic data obtained from patched neurons will continue to improve.
Limitations
The main limitation of Patch-seq compared to other scRNA-seq methods is throughput. Since sample collection involves patching a cell in situ and aspirating the cell contents into a pipette, it is not compatible with droplet-based or microfluidic cell-sorting technologies. Patching neurons is a high-level skill that can take years to master and is difficult to automate, although some have tried (31, 32). In our laboratory with 2–3 people working together and optimal conditions, we can routinely collect 30–40 samples per day by targeted patching. This number can go as high as 50–60 samples per day if patching random neurons, and as low as 5–10 per day if we extend the recording times to better recover axonal morphology. Of the samples collected, approximately 80–90% will yield high-quality cDNA. While these numbers are sufficient to answer many important biological questions, they will never rival truly high-throughput techniques such as Drop-seq (3, 33, 34). Second, cost is a major limitation in scRNA-seq experiments and we have reduced costs to ~$21/library (excluding equipment and sequencing costs) by using in-house produced and off-the-shelf reagents over commercial kits whenever possible. It is not necessary to have dedicated electrophysiology rig for Patch-seq experiments, a shared rig that is thoroughly cleaned before the experiment will suffice. Finally, since our cDNA synthesis and sequencing protocol is based on Smart-seq2, it suffers from the same inherent limitations as that method such as only detecting polyadenylated RNA and not incorporating unique molecular identifiers (UMIs) (2, 25). However, the basic protocol we describe for collecting single-cell RNA from patched neurons could potentially be combined with other sequencing methods.
Experimental design
Controls The inclusion of appropriate positive and negative controls as early in the sample collection process as possible, and at various intermediate stages of processing, is critical to ensure sample quality and can help tremendously to localize problems if sample quality becomes compromised. Endogenous (within the tissue itself) or exogenous (from the environment or experimenter) RNase, RNA from cells other than the one targeted, and cross-contamination with amplified cDNA from prior experiments are all important potential sources of contamination that should be eliminated to the extent possible (see Box 1) and controlled for by experimental design. We recommend that labs attempting single-cell RNA-seq for the first time initially optimize their protocol using ~10pg of positive control RNA (isolated from whole brain and diluted to approximate the amount in a single cell) to ensure that the solutions and sample handling procedures are sufficiently RNase-free to allow amplification of single-cell RNA under optimal conditions. We also recommend including negative controls at each stage of the experiment (sample collection, first strand synthesis, PCR amplification, and so on) when initially setting up the protocol to identify any source of non-target RNA or previously amplified cDNA contamination. Even when the protocol has been well established in a lab, it is important to continue to include positive and negative controls in every experiment to monitor for new sources of contamination. As a positive control, we include ERCC spike-in RNA in every sample (in the lysis buffer that the sample is collected into) to monitor for new sources RNase contamination. As a negative control, for each experiment we include at least one sample without a cell (identical sample collection except that no cell is patched/aspirated) to rule out contaminations with non-target RNA or previously amplified cDNA.
Technical variability and bias In addition to including positive and negative controls at the time of sample collection, another important consideration in experimental design for any scRNA-seq experiment is technical variability and bias that can be introduced during the library preparation, PCR amplification and sequencing. Batch effects that arise due to day-to-day variations in library preparation and sequencing (different reagent lots, different experimenters, changes in humidity, etc.) can easily be misinterpreted as biological variability between treatment groups or cell types if, for example, all of cell type “A” is processed one day and all of cell type “B” is processed a different day. To control for this, it is important to collect and process samples in a randomized fashion if possible. We typically store the full length cDNA at −20°C, generate the sequencing library for all of the samples at the same time using unique cell-specific indices, and pool them to be sequenced together on the same lane(s).
MATERIALS
REAGENTS
CRITICAL All reagents for RNase-free solutions (see Reagents Setup) should be ordered at the highest purity available.
Wild type C57Bl/6 mice (obtained from the Center for Comparative Medicine at Baylor College of Medicine) or F1 double heterozygous offspring from PV-Cre (Jackson Laboratories stock #008069) × Ai9 (Jackson Laboratories Stock #007909) crosses.
CAUTION Any experiments involving live mice or other animals must conform to relevant Institutional and National guidelines. All experiments described in this protocol were approved by the Institutional Animal Care and Use Committee (IACUC) at Baylor College of Medicine.
Isoflurane, Piramal Healthcare (Patterson Veterinary, cat. no. 07-890-8115) CAUTION Causes mild skin irritation. Causes serious eye irritation. May cause respiratory irritation. May cause drowsiness or dizziness. Suspected of damaging fertility or the unborn child. May cause damage to organs on repeat exposure. Keep container tightly closed. Do not breathe vapors. Use in well-ventilated area.
Internal solution reagents
Potassium D-gluconate (K-gluconate; ≥99%; Sigma-Aldrich, cat. no. 64500)
Potassium chloride (KCl; for molecular biology; ≥99.0%; Sigma-Aldrich, cat. no. P9541)
HEPES solution (1 M; BioReagent; Sigma-Aldrich, cat.no. H3537)
Ethylene glycol-bis(2-aminoethylether)-N,N,N’,N’-tetraacetic acid (EGTA; for molecular biology; ≥97.0%; Sigma-Aldrich, cat. no. E3889)
Adenosine 5′-triphosphate magnesium salt (Mg-ATP; ≥95%; Sigma-Aldrich, cat. no. A9187)
Guanosine 5′-triphosphate sodium salt hydrate (Na-GTP; ≥90%; Sigma-Aldrich, cat. no. 51120)
Phosphocreatine disodium salt hydrate (Na2-phosphocreatine; ≥97%; Sigma-Aldrich, cat. no. P7936)
Glycogen (RNA grade; Thermo Fisher Scientific, cat. no. R0551)
Biocytin (≥98%, Sigma-Aldrich, cat. no. B4261)
Potassium hydroxide (KOH; ACS reagent; ≥85%; pellets; Sigma-Aldrich, cat. no. 221473) CAUTION Harmful if swallowed. Causes severe skin burns and eye damage. Wear protective gloves, protective clothing, eye and face protection.
Sterile water (molecular biology grade; endotoxin tested; VWR, cat. no. 95016–376)
RNA isolation and cDNA library preparation reagents
RNase Zap (Thermo Fisher Scientific, cat. no. AM9780) CAUTION Causes mild skin irritation. Wear protective gloves, eye protection, and face protection.
DNA-OFF (Clontech, cat. no. 9036)
Recombinant Ribonuclease Inhibitor (RRI; Clontech, cat. no. 2313A)
ERCC RNA spike-in mix (Thermo Fisher Scientific, cat. no. 4456740)
Tris-EDTA buffer solution (TE buffer; BioUltra; for molecular biology; Sigma-Aldrich, cat. no. 93283)
Triton X-100 (Sigma-Aldrich, cat. no. T8787) CAUTION Harmful if swallowed. Causes skin irritation. Causes serious eye damage. Very toxic to aquatic life with long lasting effects. Wear eye protection and face protection.
Betaine (BioUltra; ≥99.0%; Sigma-Aldrich, cat. no. 61962)
Ethanol (200 proof; Thermo Fisher Scientific, cat. no. 04355223) Caution Flammable. Keep away from flames and hot surfaces.
Sodium dodecyl sulfate solution (SDS; BioUltra; for molecular biology; 20% (wt/vol) in H2O; Sigma-Aldrich cat. no. 05030) CAUTION Causes skin irritation. Causes serious eye damage. May cause respiratory irritation. Wear eye and face protection.
Nextera XT index kit v2 set A for 96 indices, 384 samples (Illumina, cat. no. FC-131–2001)
Sodium hydroxide solution (1.0 N; BioReagent; for cell culture; Sigma-Aldrich, cat. no. S2770) CAUTION May be corrosive to metals. Causes severe skin burns and eye damage. Wear protective gloves, protective clothing, eye protection, and face protection.
dNTPs (25 mM each; ThermoFisher Scientific, cat. no. R1121)
Superscript II reverse transcriptase (SSIIRT; Thermo Fisher Scientific, cat. no. 18064014)
MgCl2 (1 M; molecular biology grade; Thermo Fisher Scientific, cat. no. AM9530G)
KAPA Biosystems HiFi HotStart Ready Mix (Thermo Fisher Scientific, cat. no. NC0295239)
Axygen AxyPrep mag PCR clean-up kit (Thermo Fisher Scientific, cat. no. 14223151)
Ethyl alcohol (pure; 200 proof; for molecular biology; Sigma-Aldrich, cat. no. E7023) CAUTION Flammable. Keep away from flames and hot surfaces.
Buffer EB (Qiagen, cat. no. 19086)
Nextera XT DNA sample preparation kit (Illumina, cat. no. FC-131–1096). Alternatively, in-house produced Tn5 and off-the-shelf reagents can be used for tagmentation (see (35))
TAPS (≥99.5%; Sigma-Aldrich, cat. no. T5130)
Polyethylene glycol solution (PEG-8000, 40 % (wt/vol); Sigma-Aldrich, cat. no. P1458)
KAPA HiFi PCR Kit (KAPA Biosystems, cat. no. KK2103)
Immunohistochemistry and other reagents
Sodium phosphate dibasic (Na2HPO4; BioXtra; ≥99.0%; Sigma-Aldrich, cat. no. S7907)
Sodium phosphate monobasic (NaH2PO4; BioPerformance certified; for cell culture; Sigma-Aldrich, cat. no. S5011)
Sodium chloride (NaCl; PharmaGrade; Sigma-Aldrich, cat. no. RES0926S-A7)
Trizma Base (BioPerformance certified; ≥99.9%; Sigma-Aldrich, cat. no. T6066)
Hydrochloric acid solution (HCl; 1.0 N; BioReagent; for cell culture; Sigma-Aldrich, cat. no. H9892) CAUTION May be corrosive to metals. Causes severe skin burns and eye damage. May cause respiratory irritation. Wear protective gloves, protective clothing, eye and face protection.
Glycerol (for molecular biology; ≥99%; Sigma-Aldrich, cat. no. G5516)
Mowiol 4–88 (Sigma-Aldrich, cat. no. 81381)
3,3′-diaminobenzidine (DAB; ≥99%; Sigma-Aldrich, cat. no. D8001) CAUTION Harmful if swallowed. Causes serious eye irritation. Suspected of causing genetic defects. May cause cancer. Wear eye and face protection.
Dimethyl sulfoxide (DMSO; sterile-filtered; BioPerformance certified; Sigma-Aldrich, cat. no. D2438)
Paraformaldehyde (PFA; reagent grade; Sigma-Aldrich, cat. no. P6148) CAUTION Flammable solid. Harmful if swallowed or inhaled. Causes skin irritation. May cause and allergic skin reaction. Causes serious eye damage. May cause respiratory irritation. Suspected of causing cancer. Keep away from heat, sparks, open flames, hot surfaces. Avoid breathing dust. Wear protective gloves, eye protection, and face protection.
Glutaraldehyde 25% (wt/vol) solution (EM grade; distillation purified; Electron Microscopy Sciences, cat. no. 16220) CAUTION May cause allergy or asthma symptoms or breathing difficulties if inhaled. Causes severe skin burns and eye damage. Causes serious eye damage. Harmful if swallowed. May cause an allergic skin reactions. Combustible liquid. Keep away from flames and hot surfaces. Do not breathe dusts or mists. In case of inadequate ventilation wear respiratory protection. Wear protective gloves, eye protection, and face protection.
Hydrogen peroxide solution (H2O2), with inhibitor, 30 % (vol/vol) in H2O (ACS reagent; Sigma-Aldrich, cat. no. 216763) CAUTION Corrosive to metal and eyes. Wear appropriate protective equipment, if necessary, to prevent eye contact.
Vectastain ABC HRP Kit (peroxidase; standard; Vector Laboratories, cat. no. PK-4000)
Sodium bicarbonate (NaHCO3; BioReagent; Sigma-Aldrich, cat. no. S5761)
D-(+)-glucose (glucose; ACS reagent; Sigma-Aldrich, cat. no. G5767)
Calcium chloride (CaCl2; anhydrous; Sigma-Aldrich, cat. no. C1016) CAUTION Causes serious eye irritation.
Oligonucleotides
Oligo-dT30VN (sequence: 5′- AAGCAGTGGTATCAACGCAGAGTACTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTVN-3′, where V represents A, C or G and N represents any nucleotide; HPLC purified; Biomers.net; see Reagent Setup for suspension and storage information)
ISPCR Oligo (sequence: 5′- AAGCAGTGGTATCAACGCAGAGT-3′, HPLC purified, Biomers.net, see Reagent Setup for suspension and storage information)
Template switching oligonucleotide (LNA-TSO; sequence: 5′- AAGCAGTGGTATCAACGCAGAGTACrGrG+G-3′, where rG indicates riboguanosines and +G indicates a locked nucleic acid (LNA)-modified guanosine; RNase-free; HPLC purified; Exiqon; see Reagent Setup for suspension and storage information)
EQUIPMENT
Fi-streem II still 2L/hr water distillation system (Barnstead Lab Water Products, cat. no. A74415–60)
MaxiMix II vortex mixer (Thermo Fisher Scientific, cat. no. 12-814-5Q)
Avanti J-E BioSafe Centrifuge System (Beckman Coulter, cat. no. A20698)
125 mL narrow mouth Erlenmeyer flask (Corning, cat. no. 4980–125)
Eppendorf research plus adjustable volume pipettes (0.5–10 μL, 10–100 μL, and 100–1000 μL; Thermo Fisher Scientific, cat. nos. 13-690-026, 13-690-029 and 13-690-032)
B10P benchtop pH meter with pH probe (VWR, cat. no. 89231–664)
Vapro pressure osmometer 5600 (Thermo Fisher Scientific, cat. no. NC0044806)
Qorpak industrial trigger spray bottle (Thermo Fisher Scientific, cat. no. 02-991-721)
50 mL narrow mouth Erlenmeyer flask (Corning, cat. no. 4980–50)
Flaming/Brown micropipette puller (Sutter Instrument, model no. P-1000)
3.0 mm square box filament (Sutter Instrument, cat. no. FB330B)
BIC classic lighter (Bic, http://www.shopbic.com, prod. no. LCP51DC) 5′ purifier logic class II, type A2 biological safety cabinet with 10” sash opening, UV light and service fixture (Labconco, cat. no. 3450001)
Mag touch 2 ice bucket with lid (Sigma-Aldrich, cat. no. BAM168072002)
Marker (VWR, cat. no. 52899–310)
PCR storage rack (VWR, cat. no. 83009–690)
Panasonic WV-BM1910 Black and White monitor (Broadcaststore.com, cat. no. MO21245)
Temperatur controller TC07 (Luigs-Neumann, cat. no 200–100 500 0145)
Remote control SM7 (Luigs-Neumann, cat. no. 200–100 900 9050)
EPC 10 USB Quadro (HEKA Elektronik, cat. no. 895003)
Patchmaster software (HEKA Elektronic, cat. no. 895040)
An upright light microscope with differential interference contrast (DIC) such as Zeiss Axioskop 2 FS Plus, equipped with Zeiss A-Plan 10×(0.2 NA) and Achroplan 40× (0.8 NA) objectives
Arclamp power supply XBO 75 HBO 100 (Zeiss, cat. no. 910426)
CCD camera (Hamamatsu, cat. no. C3077–70)
Camera controller (Hamamatsu, cat. no. C2741–63)
A platinum ring with mesh or other device to secure tissue slice in the recording chamber Dell Precision 690 Workstation
Style 5 super fine forceps (Miltex Premium, cat. no. 17–305)
Leica VT1200 semiautomatic vibrating blade microtome with Vibrocheck
Fiber-Lite MI-150 illuminator (Thomas Scientific, cat. no. 6625B38)
Dual light guide (Thomas Scientific, cat. no. 6625B47)
2L water bath (VWR, cat. no. 89501–460)
MyFuge 12 mini centrifuge (Benchmark Scientific, cat. no. C1012)
24-well tissue culture plates (sterile; VWR, cat. no. 10062–896)
Eppendorf mastercycler pro S and control panel (Thermo Fisher Scientific, cat. nos. E950030020 and E950030050)
Ambion magnetic stand-96 (Thermo Fisher Scientific, cat. no. AM10027)
2100 Bioanalyzer desktop bundle (Agilent, cat. no. G2940CA)
Qubit 3.0 quantitation starter kit (Thermo Fisher Scientific, cat. no. Q33217)
A compatible sequencing machine such as the Illumina HiSeq 2500
Staining chamber (Custom-made by Nason Machine, Fort Bragg, CA)
Clinical rotator (Thermo Fisher Scientific, cat. no. 11-402-12Q)
Light microscope for visualizing cell morphology.
Consumables
RNase-free 50 mL conical tubes (Thermo Fisher Scientific, cat. no. AM12502)
Nonstick, RNase-free 1.5 mL microfuge tubes (Thermo Fisher Scientific, cat. no. AM12450)
RNase-free filter pipette tips (10, 100, and 1000 μL; Thermo Fisher Scientific, cat. nos. AM12635, 12648 and 12665)
BD 10 and 60 mL disposable syringes, luer lock tip (Patterson Veterinary, cat. nos. 078032645 and 078676493)
0.45 μm cellulose acetate sterile syringe filter (VWR, cat. no. 28145–481)
Parafilm M (Sigma-Aldrich, cat. no. P7793)
Borosilicate glass (O.D. 2.0 mm; I.D. 1.16 mm; 10 cm length; without filament; Sutter Instrument, cat. no. B200-116-10)
BD 1 mL disposable syringe, tuberculin slip tip (Patterson Veterinary, cat. no. 078821438)
Thin-walled, RNase-free 0.2 mL PCR tubes (Thermo Fisher Scientific, cat. no. AM 12225)
13 mm syringe filter with 0.2 μm PTFE membrane (VWR, cat. no. 28145–491)
96-well polypropylene deep well plates (VWR, cat. no. 47743–996)
High Sensitivity DNA Kit (Agilent, cat. no. 5067–4626)
Superfrost plus gold microscope slides (Thermo Fisher Scientific, cat. no. FT-4981I-GL+)
Micro cover glass (VWR, cat. no. 48404–454)
REAGENT SETUP
CRITICAL All glassware, spatulas, stir bars, pipettes and pipette tips should certified RNase-free and/or be cleaned thoroughly with RNase Zap prior to preparing any RNase-free solution. RNase-free solutions are denoted by “(RNase-free)”.
0.2 M phosphate buffer (PB) Dissolve 23.004 g Na2HPO4 (142.0 g/mol) and 4.56 g NaH2PO4 (120.0 g/mol) in 1 L distilled water (dH2O). Solution can be stored at room temperature (25°C) for up to a month.
0.01 M phosphate buffered saline (PBS) Dissolve 9 g NaCl in 50 mL 0.2 M PB and 950 mL dH2O. Solution can be stored at room temperature for up to a month.
0.2 M Tris buffer Dissolve 2.4228 g Trizma base (121.14 g/mol) in 35–50 mL dH2O. Adjust pH to 8.5 using 1 M HCl. Add dH2O to total volume of 100 mL. Solution can be stored at room temperature for up to six months.
Mowiol mounting medium Weigh out 6 g glycerol in a 50 mL conical tube. Add 2.4 g mowiol 4–88 and vortex thoroughly. Add 6 mL dH2O, vortex, and leave at room temperature for 2 h. Add 12 mL 0.2 M Tris buffer and incubate at ~53°C, vortexing occasionally, until the mowiol dissolves. Clarify by centrifugation at 2,000–3,000 g for 20 min. Store 1 mL aliquots in 1.5 mL tubes at −20°C for up to one year. Once thawed, the solution is stable at room temperature for up to a month.
DAB stock solution Dissolve DAB powder at 20 mg/mL in DMSO. Store 50 μL aliquots at −20°C for up to one year.
ERCC spike-in stock solution (RNase-free) Thoroughly vortex ERCC RNA spike-in mix. Transfer 5 μL of the original ERCC mix to a new RNase-free 1.5 mL tube containing 45 μL RNase-free sterile water. Vortex well, this is now a 1:10 (vol:vol) dilution from the original ERCC spike-in. Transfer 10 μl of the 1:10 ERCC dilution to a new RNase-free 1.5 mL tube containing 990 μL RNase-free sterile water. Vortex well, this is now a 1:103 (vol:vol) dilution from the original ERCC spike-in. Transfer 25 μL of the 1:103 (vol:vol) ERCC dilution to a new RNase-free 1.5 mL tube containing 975 μL RNase-free sterile water. Vortex well, this is now a 1:4×104 (vol:vol) dilution from the original ERCC spike-in. Store 15 μL aliquots in RNase-free 1.5 mL tubes for up to 6 months at −20°C. Avoid freeze/thaw cycles by using a fresh aliquot for each experiment.
Oligo-dT30VN (RNase-free) Dissolve in TE buffer at 100 μM. Store 50 μL aliquots at −20°C for up to a year.
ISPCR Oligo (RNase-free) Dissolve in TE buffer at 100 μM. Store 50 μL aliquots at −20°C for up to a year.
LNA-TSO (RNase-free) Dissolve in TE buffer at 100 μM. Store 20 μL aliquots at −80°C for up to a year.
Triton X-100 stock (RNase-free) Make 33% (vol/vol) stock solution in RNase-free sterile water. Store at room temperature for up to a year. Dilute stock solution 1:100 to make working dilution 0.33% (vol/vol). Store working dilution at −20°C for up to 6 months.
5 M Betaine (RNase-free) Dissolve in RNase-free sterile water at 5M. Store at −20°C for up to 6 months.
0.5 M KOH (RNase-free) Dissolve in RNase-free sterile water at 0.5 M. Store at room temperature for up to a year.
Internal solutions (RNase-free) See Box 2 for detailed instructions on preparing and storing the internal solutions. The aliquots can be stored at −20C for up to six months.
Box 2. Preparation of internal solution TIMING 1 d.
Great care should be taken to prepare the internal solution under strict RNase-free conditions, since this will be the first solution to come into contact with the RNA sample.
Reagents:
Potassium D-gluconate (K-gluconate; ≥99%; Sigma-Aldrich, cat. no. 64500)
Potassium chloride (KCl; for molecular biology; ≥99.0%; Sigma-Aldrich, cat. no. P9541)
HEPES solution (1 M; BioReagent; Sigma-Aldrich, cat.no. H3537)
Ethylene glycol-bis(2-aminoethylether)-N,N,N’,N’-tetraacetic acid (EGTA; for molecular biology; ≥97.0%; Sigma-Aldrich, cat. no. E3889)
Adenosine 5’-triphosphate magnesium salt (Mg-ATP; ≥95%; Sigma-Aldrich, cat. no. A9187)
Guanosine 5’-triphosphate sodium salt hydrate (Na-GTP; ≥90%; Sigma-Aldrich, cat. no. 51120)
Phosphocreatine disodium salt hydrate (Na2-phosphocreatine; ≥97%; Sigma-Aldrich, cat. no. P7936)
Glycogen (RNA grade; Thermo Fisher Scientific, cat. no. R0551)
Biocytin (≥98%, Sigma-Aldrich, cat. no. B4261)
Potassium hydroxide (KOH; ACS reagent; ≥85%; pellets; Sigma-Aldrich, cat. no. 221473), dissolved in RNase-free water to a concentration of 0.5 M. CAUTION Harmful if swallowed. Causes severe skin burns and eye damage. Wear protective gloves, protective clothing, eye and face protection.
Sterile water (molecular biology grade; endotoxin tested; VWR, cat. no. 95016–376)
| Component | Amount | Final Concentration (assuming 50 mL final volume) |
|---|---|---|
| K-gluconate (234.25 g/mol) | 1.4406 g | 123 mM |
| KCl (74.55 g/mol) | 0.0447 g | 12 mM |
| HEPES (1 M) | 500 μL | 10 mM |
| EGTA (380.35 g/mol) | 0.0038 g | 0.2 mM |
| Mg-ATP (507.18 g/mol) | 0.1014 g | 4 mM |
| Na-GTP (523.18 g/mol) | 0.0078 g | 0.3 mM |
| Na2-phosphocreatine (255.08 g/mol) | 0.1275 g | 10 mM |
| Glycogen (20 mg/mL) | 50 μL | 20 μg/mL 13 mM |
| Biocytin (372.5 g/mol) | 0.2421 g |
Procedure:
-
Dissolve K-gluconate, KCl, HEPES and EGTA in ~40 mL RNase-free water in a 125 mL Erlenmeyer flask.
CRITICAL STEP Ensure that all glassware, spatulas, stir bars, counters and anything else that may come into contact with the reagents or solution is cleaned thoroughly with RNase Zap.
-
Adjust the pH to 7.25 with RNase-free 0.5 M KOH.
CRITICAL STEP Ensure to use a pH meter electrode that is cleaned with RNase Zap and RNase-free water prior to use.
-
Cover with aluminum foil and autoclave. Cool to room temperature before proceeding.
PAUSE POINT The solution can be stored at 4C overnight.
-
Add Mg-ATP, Na-GTP, Na2-phosphocreatine, glycogen, and biocytin. Stir until biocytin is completely dissolved.
CRITICAL STEP Ensure that all glassware, spatulas, stir bars, counters and anything else that may come into contact with the reagents or solution is cleaned thoroughly with RNase Zap.
-
Adjust osmolarity to ~35 mOsm lower than the osmolarity of the ACSF solution that will be used during the experiment using RNase-free sterile water.
CRITICAL STEP Let the ACSF and internal solution come to room temperature and alternate measuring the osmolarity of the two solutions to obtain the most accurate comparative measurements. Add small volumes of water to the internal and stir thoroughly before each measurement. Obtain at least three measurements when approaching the desired osmolarity (within ~10 mOsm of the goal osmolarity) before adding more water to ensure the readings are stable before adding more water.
CRITICAL STEP Be careful to keep the internal solution RNase-free by using RNase-free pipettes and pipette tips while adjusting the osmolarity.
Filter the internal solution using an RNase-free 60 mL syringe (cleaned with RNase Zap) and 0.45 μm syringe filter.
-
Aliquot into 1 mL samples in RNase-free 1.5 mL tubes.
PAUSE POINT The aliquots can be stored at −20C for up to six months.
70% (vol/vol) ethanol Dissolve the non-molecular biology grade ethanol in dH2O to a final concentration of 70% (vol/vol). Store in RNase Zap-treated spray bottle for up to a week.
0.2% (wt/vol) SDS Dilute 20% (wt/vol) SDS 1:100 (vol:vol) with sterile water to make a working diltion of 0.2% (wt/vol) SDS. The solution can be stored at room temperature for up to six months.
Nextera XT primers Dilute the primers from the Nextera XT v2 index kit 1:4 with sterile water. They can be stored at −20C for up to six months.
PFA solution See Box 3 for detailed instructions on preparing the PFA solution. The PFA solution should be prepared fresh for each experiment (same day).
Box 3. Preparation of paraformaldehyde TIMING 20 min.
If preservation of cellular morphology is desired, paraformaldehyde should be prepared fresh on the day of the experiment. This solution does not need to be RNase-free.
Reagents:
Sodium hydroxide solution (1.0 N; BioReagent; for cell culture; Sigma-Aldrich, cat. no. S2770) CAUTION May be corrosive to metals. Causes severe skin burns and eye damage. Wear protective gloves, protective clothing, eye protection, and face protection.
Distilled water.
Paraformaldehyde (PFA; reagent grade; Sigma-Aldrich, cat. no. P6148) CAUTION Flammable solid. Harmful if swallowed or inhaled. Causes skin irritation. May cause and allergic skin reaction. Causes serious eye damage. May cause respiratory irritation. Suspected of causing cancer. Keep away from heat, sparks, open flames, hot surfaces. Avoid breathing dust. Wear protective gloves, eye protection, and face protection.
Glutaraldehyde 25% (wt/vol) solution (EM grade; distillation purified; Electron Microscopy Sciences, cat. no. 16220) CAUTION May cause allergy or asthma symptoms or breathing difficulties if inhaled. Causes severe skin burns and eye damage. Causes serious eye damage. Harmful if swallowed. May cause an allergic skin reactions. Combustible liquid. Keep away from flames and hot surfaces. Do not breathe dusts or mists. In case of inadequate ventilation wear respiratory protection. Wear protective gloves, eye protection, and face protection.
0.2 M Phosphate buffer (see Reagent Setup)
Final concentrations:
4% (wt/vol) Paraformaldehyde
2.5% (vol/vol) Glutaraldheyde
in 0.1 M Phosphate Buffer
Procedure:
Add 40 μL of 1N NaOH to 7.5 mL distilled water in a 50 mL Erlenmeyer flask.
Add a stir bar, cover with parafilm, and stir on a heating stir plate set on low-medium heat for 5–10 min.
When the solution is warm, add 0.8 g paraformaldehyde (PFA) and re-cover with parafilm.
As soon as PFA dissolves remove the solution from heat and let cool to room temperature.
Add 2 mL of 25% (wt/vol) glutaraldehyde and 10 mL of 0.2 M phosphate buffer. Swirl gently to combine.
Store the PFA/glutaradehyde solution at 4 °C. For best morphological recovery, use it the same day.
Oxygenated artificial cerebrospinal fluid (ACSF) Dissolve the following salts in distilled water: NaCl 125 mM, KCl 2.5 mM, NaH2PO4 1.25 mM, NaHCO3 25 mM, MgCl2 1 mM, glucose 25mM, CaCl2 2 mM. Oxygenate for 30 min before adding the MgCl and CaCl2. The solution can be stored at 4 °C for up to two weeks.
200 mM TAPS-NaOH stock solution (optional) Dissolve TAPS in sterile molecular biology grade water and adjust pH to 8.5 (at room temperature). The solution can be stored at room temperature for up to six months. (Only prepare if using in-house Tn5 transposase, see “Reagents”).
10× Tagmentation buffer (optional) Dilute the 200 mM TAPS-NaOH stock solution and 1 M MgCl2 in sterile molecular biology grade water to final concentrations of 10 mM TAPS-NaOH and 50 mM MgCl2). The solution can be stored at room temperature for up to six months. (Only prepare if using in-house Tn5 Transposase, see “Reagents”).
EQUIPMENT SETUP
Agilent Bioanalyzer Set up according to manufacturer’s instructions.
Electrophysiology rig Set up according to manufacturer’s instructions.
Autoclave glass for pulling electrodes Autoclave borosilicate glass using a 20 min exposure time.
Pipette puller program Install the box filament in the pipette puller and measure the ramp temperature according to the manufacturer’s instructions. Set the velocity to 25 and the time to 250 for four pull cycles. Adjust the heat value as needed to obtain 4–6 MΩ resistance pipettes. For the best results, it is critical for the glass to break on the fourth pull cycle and to allow the jaw temp to cool to below 30 °C between pulls.
Syringe fitted with tapered pipette tip Use a handheld lighter to melt the end of a 200 μL pipette tip, focusing the heat approximately 2 cm from the very tip. When the plastic is soft, stretch it out to make a long thin tapered end. Allow to cool. Cut off the end, leaving approximately 5 cm of the thin tapered section, which will be used for backfilling pipettes with internal solution. Also cut of approximately 0.5 cm of the proximal end so that it will fit on the 1 mL syringe. See example syringe in Fig. 3. A new syringe should be prepared for each experiment and cleaned thoroughly with RNase Zap. Other devices for backfilling pipettes are also acceptable as long as they can be cleaned with RNase Zap.
Figure 3. Custom equipment.
a) 1 mL syringe with 0.2 μm syringe filter and tapered pipette tip, used for filtering internal solution and backfilling glass pipettes (Steps 8–9, 15). b) Patch pipette marked at approximately 0.3 μL volume, used as a reference while loading other pipettes (Step 15). c) Positive pressure device used to eject pipette contents into PCR tube after aspirating cell contents (Step 24). d) Staining chamber used for immunohistochemistry (Step 68).
Pipette volume model To gauge the volume of internal solution to load, it may be helpful to keep model glass pipette on hand to refer to during the experiment (Fig. 3b). First, eject about 0.3 μL of water onto a piece of parafilm. Pull a glass electrode using the program you plan to use the day of the experiment. Carefully place the tip of the glass pipette into the small drop of water and very lightly touch it to the parafilm. This should barely break the tip and allow the water to slowly backfill into the pipette. Once all of the water has been drawn into the tip of the glass pipette, mark the meniscus using an ultra-fine tip marker and keep as a reference. The model can be kept as long as the mark is visible.
Positive pressure device We built a custom device to hold the pipette and eject its contents into a sample tube (Fig. 3c). However, any device that can apply positive air pressure on the pipette while expelling the contents into a PCR tube will suffice (e.g. a syringe with rubber seal, or the pressure system on the electrophysiology rig). The same device can be used for up to six months but should be cleaned with RNase Zap prior to each experiment.
PROCEDURE
Prepare work surfaces and equipment, TIMING 1 h
1 Clean the following equipment with RNase Zap, rinsing with 70% (vol/vol) ethanol or distilled water after each cleaning step: Biosafety hood or other dedicated RNase-free bench; P10, P100 and P1000 pipettes; two ice buckets; marker; PCR tube rack; Glass pipette holder (disassembled) from electrophysiology rig; Micromanipulator nobs; Outside surfaces of pipette puller; All other surfaces, such as counters, that you will touch frequently during the experiment; Wire electrode on electrophysiology rig; Number 5 forceps for reassembling glass pipette holder; 1 mL syringe; melted and elongated 200 μl pipette tip, cut to fit a 0.2 μm syringe filter;
CRITICAL STEP Great care should be taken to reduce or eliminate any source of RNase contamination during sample collection and solution preparation (see Box 1 for additional tips). Any items that could come into contact with the samples, such as the recording electrode, must be thoroughly cleaned with RNase Zap and re-cleaned during the experiment if they may have become contaminated (for example, if the glass pipette breaks and the wire electrode comes into contact with ACSF it should be re-cleaned with RNase Zap).
CRITICAL STEP Prior to cleaning with RNase Zap, any equipment that may have come into contact with post-PCR products (tube holders, pipettes if not using a dedicated set) should also be cleaned thoroughly with DNA-OFF to prevent cross-contamination with amplified cDNA from previous experiments.
PAUSE POINT These items can be cleaned the day before the experiment and left in the hood overnight to dry.
Prepare lysis buffer, TIMING 30 min (for 30 tubes)
2 Prepare lysis buffer for all the samples (including the negative control) plus an extra 10–15% by combining the reagents listed in the table below.
| Component | Volume (μL) | Final Concentration |
|---|---|---|
| Nuclease-free sterile water | 1.75 | – |
| Triton X-100 (0.33% vol/vol) | 1.15 | 0.1% (vol/vol) |
| dNTPs (25 mM each) | 0.80 | 5 mM (each dNTP) |
| Oligo dT30VN (100 μM) | 0.10 | 2.5 μM |
| Recombinant RNase Inhibitor (40 U/μL) | 0.10 | 1 U/μL |
| ERCC spike-ins (1:4×10−4 dilution) | 0.10 | 1.6×10−5 dilution |
| Total Volume | 4.00 | – |
CRITICAL STEP Thaw all reagents ahead of time and vortex thoroughly before and after combining.
CRITICAL STEP If two people are working together, one can prepare the lysis buffer and internal solution (steps 2–9) while the other person cuts slices and makes the PFA (steps 10–13).
3 Label enough RNase-free 0.2 mL PCR tubes for the number of samples anticipated, plus one extra tube to serve as a negative control.
4 Distribute 4 μL lysis buffer into each PCR tube. Store tubes in the clean PCR tube holder (from step 1) on ice (using the cleaned ice bucket from step 1).
Prepare internal solution, TIMING 10 min
5 Thaw a 1 mL aliquot of internal solution on ice. Vortex well.
6 Prepare the high osmolarity or the physiologic osmolarity internal solution, depending on whether the cell needs to be patched for a long period of time (e.g. for detailed morphological recovery or long electrophysiological stimulation protocols). The high osmolarity internal solution (Option A) gives the highest concentration of RNase inhibitor and will best preserve the RNA of the cell, but long recordings >15–20 min are more difficult due to swelling of the cell body. The physiologic osmolarity internal solution (Option B) will have a lower concentration of RNase inhibitor, resulting in a modest decrease in cDNA yield (Fig. 4), but permits stable recordings for over an hour. Both the long recording time and the lower viscosity of this solution allow better filling of the cell processes with biocytin for subsequent morphological recovery.
Figure 4. cDNA yield using physiological and high osmolarity internal solution.
Amplified cDNA yield (a) and mean size (b) as a function of recording time for both types of internal solution used. The physiological internal results in slightly lower yield of cDNA compared to the high osmolarity internal solution (likely due to a reduction in the amount of RRI), however no further decrease in yield is noted when recording time is extended up to 45 min. The grey and black “X”‘s indicate negative controls (no cell patched) for the high osmolarity and physiologic osmolarity internal solutions, respectively. All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at Baylor College of Medicine.
- High osmolarity internal solution:
- Combine 487.5 μL internal and 12.5 μl RRI in an RNase-free 1.5 mL tube.
-
Vortex well.CRITICAL STEP Store the internal solution on ice until ready to begin patching.
- Physiologic osmolarity internal solution:
- Combine 496 μL internal and 4 μL RRI in an RNase-free 1.5 mL tube.
- Vortex well.
CRITICAL STEP Measure the osmolarity of the Physiologic osmolarity internal solution using an osmometer after adding the RRI. If necessary, dilute with RNase-free water until it is 10–15 mOSM higher osmolarity than the ACSF that will be used during the experiment.
CRITICAL STEP Store the internal solution on ice until ready to begin patching.
7 Draw the internal solution into an RNase-free 1 mL syringe (from step 1). Carefully tap to remove any air bubbles.
CRITICAL STEP If not removed, air bubbles will destroy the integrity of the syringe filter (step 8) and can lead to clogging of the pipette during patching.
8 Eject any remaining air from the syringe, place a 0.2 μm syringe filter on the end of the syringe, and slowly push the internal solution through the filter.
9 When internal solution begins to emerge from the filter, place the clean, melted 200 μL pipette tip (from step 1) on the other end of the filter. Continue to slowly push the solution through until it reaches the end of the melted pipette tip. See example of syringe assembly for internal solution in Fig. 3a.
Prepare acute brain slices, TIMING 1 h
CAUTION All experiments should be carried out in accordance with National Research Council’s “Guide for the Care and Use of Laboratory Animals” and with approval from the appropriate Institutional Animal Care and Use Committee. Permission for the experiments described here was provided by the Institutional Animal Care and Use Committee at Baylor College of Medicine.
10 Deeply anesthetize the mouse with isoflurane and cut acute brain slices according to standard protocols for the brain region of interest (12, 36).
11 Allow the slices to recover in oxygenated ACSF for 30–45 min at 37±0.5 °C.
Prepare fresh paraformaldehyde solution TIMING 20 min
12 Prepare fresh paraformaldehyde (PFA) for morphological recovery of the cells (as described in Box 2) and store at 4 °C.
Patching and sample collection, TIMING ~5 h (for 30 cells)
CRITICAL Sample tubes should be kept on ice on a clean counter in close proximity to the electrophysiology rig to facilitate sample collection. Having two people present during sample collection is highly recommended – one will patch, aspirate the cell contents, and fix the tissue slice while the other takes notes and collects the sample into a PCR tube.
CRITICAL Great care should be taken to maintain an RNase-free environment during sample collection by re-cleaning any items that become contaminated (such as the electrode wire if it comes into contact with ACSF) and changing gloves frequently.
13 Reassemble the cleaned glass pipette holder (from step 1) using the clean number 5 forceps (from step 1) and install it on the clean micromanipulator (from step 1).
14 Place a tissue slice in the recording chamber and select a cell to target using differential interference contrast (DIC) and/or wide-field fluorescence light microscopy. Center the target cell in the field of view at 40× magnification.
?Troubleshooting
15 Pull an autoclaved glass pipette and backfill with 0.1–0.3 μL of internal solution. Tap or shake to remove any bubbles. Compare to the marked model pipette (see equipment setup) to ensure less than 0.3 μL is loaded (Figure 3b).
CRITICAL STEP Using too much internal solution at this stage will inhibit the downstream reverse transcription reaction.
16 Secure the glass pipette in the clean pipette holder and lower it into the circulating ACSF. Immediately apply a small amount of positive pressure, push out any air bubbles (Supplementary Movie S1), and measure the resistance.
CRITICAL STEP If the pipette resistance is too high (>7 MΩ for most cortical neurons) try lowering the pull temperature by one or two degrees and pulling another pipette. In general, pipettes of 4–6 MΩ will give good cDNA yield. However, smaller pipettes may be sufficient to aspirate very small cells (e.g. certain cortical interneurons). Small pipettes (7–8 MΩ) will also make it easier to hold the cell for a long period of time if morphological recovery is a high priority for the experiment.
CRITICAL STEP Make sure the electrode wire is long enough to reach the internal solution with only 0.3 μL loaded into the pipette.
?Troubleshooting
17 Carefully but quickly advance the pipette up to the target cell until the cell membrane is observed to dimple under the tip and the resistance across the pipette slightly increases (Supplementary Movie S1).
CRITICAL STEP Maintain a small amount of positive pressure to prevent the inside of the pipette tip from getting dirty as it is pushed through tissue.
18 Immediately release the positive pressure and apply a small amount of suction if necessary to achieve a GΩ seal on the cell membrane (Supplementary Movie S1). Set the holding potential to −70 mV and compensate the fast capacitance.
CRITICAL STEP Reposition the pipette so that the cell membrane is not under tension, and the pipette approaches from the side of the cell rather than the top (Fig. 2, Supplementary Movie S1).
?Troubleshooting
19 Apply brisk suction while monitoring the pipette resistance until whole-cell access is achieved (Supplementary Movie S1). Compensate the slow capacitance.
CRITICAL STEP Discontinue suction immediately when the cell membrane opens to prevent losing the seal between the cell membrane and the pipette. Signs of leakage through a broken seal include an unstable resting membrane potential, a large current required to hold the cell at −70 mV (>200–300 pA), or blunted/absent action potentials when stimulated in current clamp. If the recording seems unstable or there is a large amount of current leakage, try adjusting the pipette position or waiting a few minutes to see if it improves.
?Troubleshooting
20 Run the desired electrophysiology protocol. We typically deliver a series of hyperpolarizing, subthreshold, and suprathreshold current steps to record the intrinsic electrophysiological properties and firing pattern of the cell and ensure it is healthy (Supplementary Movie S1). For example, if 50 pA of negative current are needed to hold the cell at −70 mV, we may start our currents steps at −100 pA and increase by 10–20 pA on each sweep until the cell is firing many action potentials.
21 If morphological recovery is important for the experimental question, hold the cell in current clamp mode for 30–60 min.
CRITICAL STEP Injecting the cell with large depolarizing currents (e.g. 15 × 100 ms pulses of 10–20 nA delivered at 1 Hz) may help to diffuse biocytin throughout the fine axonal processes and improve morphological recovery.
?Troubleshooting
22 Aspirate the cell contents into the patch pipette by gently applying negative pressure (0.7–1.5 psi is typically applied). Continuously monitor the holding currents in voltage-clamp mode and size of the cell body on DIC. If the cell becomes unstable, discontinue the suction and wait a few minutes to see if it recovers. Continue until the cell body has significantly decreased in size (typically 2–10 min) or the cell cannot tolerate more suction without becoming unstable as described in step 19 (Supplementary Movie S1).
CRITICAL STEP Be cautious to apply the suction very slowly and gradually. Use a large (20–30 mL), new syringe to ensure smooth contact between the rubber piston and syringe wall and change the syringe if necessary. If the cell is lost (i.e. sudden increase in the amount of current required to hold at −70 mV or any extracellular contents seen entering the pipette) immediately discontinue suction and remove the pipette. The RNA from the cell may still be salvageable, but endogenous RNases from the extracellular space may degrade it quickly. If a significant amount of extracellular material has entered the pipette the sample should be discarded.
CRITICAL STEP Sometimes when aspirating the cell contents there is a gradual increase in the measured resistance, which may correlate with the entry of the nucleus or other organelles into the pipette. If this is observed continue to apply constant gentle suction and monitor for a sudden drop in resistance, suggesting that the organelle(s) have entered the pipette. Once the nucleus is completely aspirated, the cell becomes extremely fragile and very little suction is needed to aspirate the remaining cell contents.
?Troubleshooting
23 When the cell contents have completely entered the pipette, or the cell has destabilized to the point that additional suction is not possible, release all suction and quickly remove the pipette (Supplementary Movie S1).
CRITICAL STEP If attempting to preserve cell morphology, first move the pipette back slowly from the cell to allow the membrane to slowly retract from the pipette before removing the pipette from the tissue (Supplementary Movie S1).
24 Immediately eject the contents of the pipette into a 0.2 mL PCR tube containing 4 μL lysis buffer (prepared in step 4) by applying a small amount of positive pressure (using device depicted in Fig. 3c, or something similar) and gently breaking the tip on the bottom of the tube. Avoid making any bubbles. Cap the PCR tube and store on wet ice.
CRITICAL STEP If bubbles occur during ejection, tap the tube gently to remove them and immediately spin down the sample (700 g for 10 s at 4 °C) before storing on ice.
25 Optional: If morphological recovery of the cell is desired, immediately remove the slice from the recording chamber and place it in one well of a 24-well culture dish. Remove the excess ACSF and add ~1mL of cold, fresh PFA (from step 12). Cover the plate using 2–3 layers of parafilm underneath the lid to form an airtight seal.
CRITICAL STEP To be most efficient, one experimenter can eject the cell contents into the PCR tube (step 24) while the other one is fixing the tissue slice (step 25). These steps are both time-sensitive and should be done as quickly as possible after the pipette is removed from the tissue.
PAUSE POINT The fixed tissue samples can be stored at 4 °C for up to 10–14 days before immunohistochemistry (Steps 67–84).
26 Repeat steps 14–25 until the desired number of samples have been collected. Be sure to collect at least one negative control at the end of the experiment, which is identical to the samples except that no cell is patched (the pipette is loaded with internal and inserted into the tissue, positive pressure is released, the pipette is removed from tissue and its contents are ejected into a PCR tube).
Denaturation and Reverse Transcription, TIMING 3h
27 Remove any remaining bubbles by gently tapping the side of each tube. Spin down the samples (700 g for 10 s at 4 °C) and place back on ice.
28 Denature the secondary structure of the RNA by incubating the samples in a thermal cycler using the following program.
| Cycle | Temperature (°C) | Time | Purpose |
|---|---|---|---|
| 1 | 72 | 3 min | Denaturation of RNA |
| 2 | 4 | Hold | Safe storage |
29 Assemble the reverse transcription (RT) mix for all reactions plus 10% extra by combining the reagents listed in the following table. Be sure that each component is completely thawed and thoroughly vortexed before adding to mix. The RT mix can be assembled while the samples are in the thermal cycler for denaturation (step 28).
| Component | Volume (μL) | Final Concentration (in 10 μL) |
|---|---|---|
| Superscript II first strand buffer (5×) | 2.00 | 1× |
| Betaine (5 M) | 2.00 | 1 M |
| Superscript II Reverse Transcriptase (200 U/μL) | 0.50 | 10 U/μL (100 U total) |
| DTT (0.1 M) | 0.50 | 5 mM |
| Nuclease-free sterile water | 0.29 | – |
| Recombinant RNase Inhibitor (40 U/μL) | 0.25 | 1 U/μL (10 U total) |
| LNA-TSO (100 μM) | 0.1 | 1 μM |
| MgCl2 (1 M) | 0.06 | 6 mM |
| Total Volume | 5.70 |
30 Add 5.7 μL of RT mix to each tube and pipette up and down four times to mix. Gently tap each tube as needed to remove bubbles and spin down at 700 g for 10 s at 4 °C. Immediately return to ice.
31 Incubate the samples in a thermal cycler according to the following program:
| Cycle | Temperature (°C) | Time | Purpose |
|---|---|---|---|
| 1 | 42 | 90 min | RT and template switching |
| 2–11 | 50 | 2 min | Unfolding RNA secondary structure |
| 42 | 2 min | Continuation of RT and template switching | |
| 12 | 70 | 15 min | Enzyme inactivation |
| 13 | 4 | Hold | Safe storage |
PAUSE POINT Samples can be left in the PCR machine overnight while running the RT reaction.
Amplification of full-length cDNA, TIMING 3 h
32 Prepare the PCR mix for all reactions, plus ~10% extra, by combining the reagents in the following table:
| Component | Volume (μL) | Final Concentration (in 25 μL) |
|---|---|---|
| KAPA HiFi HotStart ReadyMix (2×) | 12.50 | 1× |
| Nuclease-free sterile water | 2.25 | – |
| IS PCR primers (10 μM) | 0.25 | 0.1 μM |
| Total Volume | 15.00 |
33 Add 15 μL of the PCR mix to each tube. Vortex to mix, spin down at 700 g for 10 s at 4 °C, and run the PCR in a thermal cycler using the following program:
| Cycle | Temperature (°C) | Time | Purpose |
|---|---|---|---|
| 1 | 98 | 3 min | Denature |
| 2–19 | 98 | 20 s | Denature |
| 67 | 15 s | Anneal | |
| 72 | 6 min | Extend | |
| 20 | 72 | 5 min | Extend |
| 21 | 4 | Hold | Safe storage |
PAUSE POINT PCR Products can be stored at −20 or −80 °C for up to six months before proceeding with PCR purification.
PCR Purification, TIMING 3 h (for 30 samples)
CRITICAL From this point forward, the amplified cDNA should be handled and stored in a separate post-PCR area away from the pre-PCR samples to prevent cross-contamination of new samples with amplified cDNA.
34 Allow the Axygen AxyPrep mag PCR beads to come to room temperature for 15 min. Vortex well.
35 Add 17.5 μL of beads (0.7:1 (vol:voL) ratio) to each sample and mix thoroughly by pipetting up and down at least 10 times. Transfer to a 96-well V-bottom plate.
36 Cover the plate with a lid and incubate the mixture for 8 min at room temperature.
37 Place the V-bottom plate on the magnetic stand and incubate for an additional 5 min. The solution should be clear and the beads collected at one corner of each well.
38 With the plate on the magnetic stand, carefully remove the solution without touching the beads.
39 Wash the beads with freshly prepared 80% (vol/vol) ethanol (molecular biology grade ethyl alcohol in sterile water) for 30 s. Carefully remove the ethanol without touching the beads.
CRITICAL STEP Purify the samples in small batches (8–12 at a time) to allow sufficient time to remove the ethanol after 30 s.
40 Repeat step 39 once.
41 Carefully remove as much ethanol as possible using a P1000 followed by a P100 pipette. Rotate the plate 45 degrees to collect any residual ethanol on the side of the wells, being very careful not to disturb the beads.
42 Cover the plate with a lid and leave at room temperature until the beads are completely dry and small cracks appear on the surface (5–10 min depending on humidity). Do not over-dry, as this will make it difficult to resuspend the beads.
43 Take the plate off the magnetic stand, add 17.5 μL of Buffer EB to each well and pipette up and down ~10 times to resuspend the beads.
?Troubleshooting
44 Incubate the plate off the magnetic stand for 2 min.
45 Place the plate on the magnetic stand and incubate for an additional 2–3 min. The solution should be clear and the beads should be collected in the corner of each well.
46 Without touching the beads, collect 15 μL of the clear liquid from each well and transfer to new 0.2 mL PCR tubes.
PAUSE POINT Purified full-length cDNA can be stored at −20 or −80 °C for up to six months before proceeding with the quality check (QC).
Quality check of the full-length cDNA library, TIMING 3 h (for 30 samples)
47 Check the concentration and size distribution of the full-length cDNA library on an Agilent Bioanalyzer using the Agilent High Sensitivity DNA Kit according to the manufacturer’s instructions. A good quality library should have a broad peak at 1500–2000 bp and very few short fragments <500 bp (Fig. 5).
Figure 5. Quality check of full-length single-cell cDNA libraries.
Bioanalyzer profiles of single-cell amplified cDNA from several cells from a typical experiment (a) showing variable cDNA yield, as well as a negative control from the same experiment that consisted of ERCC spike-in RNA only (diluted 1:4×106 in the RT reaction). Profiles from degraded (b) or contaminated (c) samples are also shown. The low molecular weight fragments in (c) were traced back to bacterial contamination of a particular reagent. All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at Baylor College of Medicine.
PAUSE POINT Purified full-length cDNA can be stored after the QC step at −20 or −80 °C for up to six months before proceeding with tagmentation and sequencing.
?Troubleshooting
Tagmentation reaction, TIMING 1 h
48 Dilute each sample to 50 pg/μL in sterile water.
49 Proceed with tagmentation using either the Nextera XT DNA sample preparation kit according to the manufacturer’s instructions, or using in-house produced Tn5 transposase (35) according to the following table:
| Component | Volume (μL) | Final Concentration (in 25 μL) |
|---|---|---|
| 10× Tagmentation buffer | 1.00 | 1× |
| PEG-8000 (40% wt/vol) | 2.50 | 10% (wt/vol) |
| Tn5 Transposase (12.5 μM) | 0.10 | 1.25 μM |
| Nuclease-free sterile water | 0.40 | - |
| Sample DNA | 6.00 | 300 pg |
| Total Volume | 10.00 | - |
50 Incubate the samples in a thermal cycler at 55C for 5 min (if using Nextera XT DNA sample preparation kit) or 8 min (if using in-house Tn5 transposase (35).
51 Immediately strip the Tn5 transposase off of the tagmented DNA by 2.5 μL of either the NT reagent (if using Nextera XT DNA sample preparation kit) or 2.5 μL of 0.2% (wt/vol) SDS (if using in-house Tn5 (35)) to each sample and incubate the mixture at room temperature for 5 min. Immediately return the samples to ice.
Amplification of adapter-ligated fragments, TIMING 2 h
52 Add 2.5 μL each of index 1 (N7XX) and index 2 (S5XX) primers, diluted 1:4 (vol:vol) from the Nextera XT index kit, ensuring that each sample receives a unique combination of indices.
53 Assemble the enrichment PCR mix for all the samples plus ~10% extra, according to the following table:
| Component | Volume (μL) | Final Concentration (in 25 μL) |
|---|---|---|
| KAPA HiFi Buffer (5×) | 5.00 | 1× |
| KAPA dNTP mix (10 mM each) | 0.75 | 0.3 mM each |
| Nuclease-free sterile water | 1.25 | – |
| KAPA enzyme (1U/μL) | 0.50 | 0.02 U/μL (0.5 U total) |
| Total Volume | 7.50 |
54 Add 7.5 μL of the PCR mix to each tube. Spin down at 700 g for 10 s at 4 °C, and run the PCR in a thermal cycler using the following program:
| Cycle | Temperature (°C) | Time | Purpose |
|---|---|---|---|
| 1 | 72 | 3 min | Extend |
| 2 | 95 | 30 s | Denature |
| 3–14 | 95 | 10 s | Denature |
| 55 | 30 s | Anneal | |
| 72 | 30 s | Extend | |
| 15 | 72 | 5 min | Extend |
| 16 | 4 | Hold | Safe storage |
CRITICAL STEP We use 10–12 cycles of amplification for 300–500 pg of input cDNA. The number of cycles can be adjusted if more or less cDNA is used.
PAUSE POINT Amplified cDNA libraries can be stored at −20°C or −80°C for up to six months.
PCR Purification, TIMING 45 min
55 Pool 2.5 μL from each sample into a single 1.5 mL tube.
56 Repeat steps 34–46 to purify the pooled library, with the following modifications: use a 1:1 ratio of beads:cDNA, resuspend the beads in a volume of 0.875 μL × the number of samples pooled, and collect a final volume of 0.75 μL × the number of samples pooled.
PAUSE POINT Pooled cDNA libraries can be stored at −20°C or −80°C for up to 6 months before sequencing.
57 Determine the mean size and mean concentration of the final cDNA library using an Agilent Bioanalyzer and Qubit, respectively. Use these measurements to dilute the library to ~3nM.
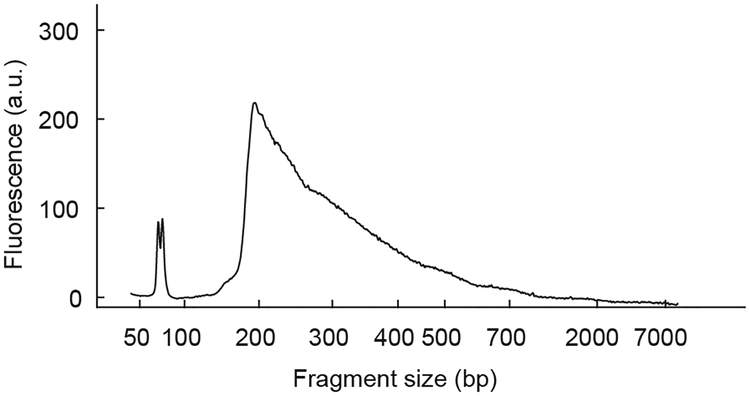
CRITICAL STEP The mean size of fragments in the final library should be <500 bp (see Fig. 6).
Figure 6. Bioanalyzer profile of pooled cDNA library.
Example of bioanalyzer profile showing the size distribution of the final pooled sequencing library for 224 Patch-seq neurons. The mean size of the cDNA fragments for this library (integrated from ~100 bp to 8 kb) is 336 bp and the concentration is ~3 ng/μL. All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at Baylor College of Medicine.
?Troubleshooting
Sequencing, TIMING 1–2 d
58 Sequence the library on a HiSeq 2500 or other compatible system according to the manufacturer’s instructions.
Preliminary Analysis, TIMING 2–14 d
59 Demultiplex the reads (i.e. assign reads to individual cells) using the unique indices assigned during library preparation (step 52).
60 Assemble the appropriate genome or transcriptome files and align the raw reads to the reference data using an alignment program such as STAR (37).
61 Examine the basic statistics of each cell such as total number of reads, percent of mapped reads aligned to intron/exons, total number of genes detected, and so on. Remove cells that appear to be outliers or do not meet quality control criteria from further analysis, as these likely reflect poor quality samples that may be contaminated with pre-mRNA, rRNA, and/or genomic DNA (See Anticipated Results and Fig. 7).
Figure 7. Initial Patch-seq data processing steps.
Quality of the samples is assessed by analyzing the sequencing depth (a), number of detected genes (b), maximum spearman correlation between each cell and all other cells (c), and fraction of reads mapped to exons, introns and intergenic segments (d). This example dataset originally contained of 224 neurons which were primarily excitatory (n = 218/224, ~97%). Twenty-eight cells were discarded based on QC criteria (dashed lines in a and b represent the QC cutoffs), leaving 196 for subsequent analyses. e) Histogram of mean normalized expression across cells for all genes in the example dataset. Genes with a mean expression less than one (dotted line) may be excluded to minimize the impact of gene dropout on further analyses. Panels a, b, and e were generated by following the tutorial described in (30). All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at Baylor College of Medicine.
62 Remove lowly expressed genes (i.e. mean read count across cells < 1) to de-noise the data. The most appropriate criteria will depend on the expected heterogeneity within the population, the sample size, and the goals of the experiment.
63 Normalize the data using reads per kilobase of transcript per million total reads (RPKM), transcripts per million (TPM), or an alternative method developed specifically for scRNA-seq data such as scran (30).
64 Identify genes with the highest biological variability across cells (27, 30).
65 Use dimensionality reduction techniques such as principal component analysis (PCA) and t-stochastic neighborhood embedding (t-SNE) to visualize the distribution of cells within the high-dimensional gene space (24).
66 Perform clustering analysis using an appropriate algorithm such as hierarchical clustering or affinity propagation (24).
67 Perform additional exploratory analyses as indicated, such as correlating transcriptomic classes with electrophysiological and morphological phenotypes (24) and identifying differentially expressed genes between cell types (24, 28, 30).
Optional: Immunohistochemistry and morphological recovery, TIMING 11–14 d
CRITICAL If morphological recovery of the cells is an important part of the experiment (i.e. for cell types that cannot be distinguished by known molecular markers), the physiological pH internal solution should be used, the recording time should be extended to 30–60 min, and the tissue should be immediately immersed in fresh PFA after the recordings for best immunohistochemical results.
68 After 10–14 days of fixation, transfer the tissue slices (from step 25) to a staining chamber that has mesh-bottom wells to facilitate changing solutions for all slices simultaneously (Fig. 3d). Alternatively, staining can be done in a 24-well culture dish by exchanging the solution in each well.
69 Carefully remove any trace of PFA and add wash the slices with 0.01 M PBS. Cover and leave on shaker for 10 min at room temperature.
70 Wash the tissue slices two more times with 0.01 M PBS as in step 69.
71 Prepare 50 mL of 3% (vol/vol) H2O2 by combining 45 mL of 0.01 M PBS with 5 mL of 30% (vol/vol) H 2O2 in a conical tube. Vortex briefly.
72 Remove the PBS and submerge slices in the 3% (vol/vol) H2O2. Leave on shaker at room temperature for 30 min.
73 Remove the 3% (vol/vol) H2O2 and wash the slices with 0.01 M PBS. Leave on shaker at room temperature for 10 min.
74 Wash the tissue slices two more times with 0.01 M PBS as in step 73.
75 Combine 10 mL of 0.01 M PBS, 100 μL Triton X-100, 4 drops of reagent A and 4 drops of reagent B (from ABC Elite Kit) in a 50 mL conical tube (makes enough for ten slices). Vortex briefly.
76 Remove the PBS from the staining chamber and add the PBS/Triton/A/B mixture (from step 75). Store overnight at 4°C.
77 Remove the PBS/Triton/A/B mixture and wash with 0.01 M PBS. Leave on shaker for 10 min at room temperature.
78 Wash the tissue slices three more times with 0.01 M PBS for 10 min at room temperature.
79 Combine 10 mL 0.01 M PBS, 0.25 mL DAB stock solution (5 aliquots) and 3.3 μL 30% (vol/vol) H2O2 in a 50 mL conical tube. Vortex briefly.
80 Remove the PBS from each well and the DAB mixture from step 79. Leave on shaker at room temperature for 7–8 min.
81 Remove the DAB mixture and wash with 0.01 M PBS. Leave on shaker at room temperature for 10 min.
82 Wash the tissue slices two more times with 0.01 M PBS as in step 81.
83 Mount the slices on microscope slides in Mowiol mounting medium and cover them with a glass coverslip
PAUSE POINT The mounted samples can be stored indefinitely at room temperature, protected from light
84 Visualize cell morphology under a light microscope (see Fig. 8).
Figure 8. Direct morphological recovery of Patch-seq cells.
a) cDNA yield as a function of recording time. Points are color coded according to the quality of the morphological recovery. b) Venn diagram summarizing success rate for obtaining electrophysiological, morphological and cDNA information from single cells. Examples of two excitatory (c,e) and two inhibitory (d,f) neurons demonstrating successful characterization of all three data modalities: morphology (left), electrophysiology (right, upper panel) and full-length cDNA profile assessed using an Agilent Bioanalyzer (right, lower panel). Scale bars for immunohistochemical staining, 50 μm. Scale bars for electrophysiological traces, 100 ms and 50 mV/500pA. All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at Baylor College of Medicine.
CRITICAL STEP Staining quality may continue to improve for 48–72 h after mounting on slides.
?Troubleshooting
TIMING
Day 1
Step 1, prepare work surfaces and equipment, 1 h
Day 2
Steps 2–9, prepare lysis buffer and internal solution, 40 min
Steps 10–12, cut brain slices, make fresh PFA solution, 1 h 20 min
Steps 13–26, patching and sample collection, 5 h
Steps 27–31, first strand synthesis, 3 h
Day 3
Steps 32–33, full-length cDNA amplification, 3 h
Steps 34–46, PCR purification, 3 h
Step 47, quality control check of full length cDNA library, 3 h
Day 4
Steps 48–51, tagmentation, 1 h
Steps 52–54, enrichment PCR, 2 h
Step 55–57, PCR purification, 45 min
Days 5–6
Step 58, sequencing of final cDNA library, 1–2 d
Days 6–14
Steps 59–67, preliminary analysis, 2–14 d
Days 10–14
Steps 68–84, immunohistochemistry, 11–14 d
TROUBLESHOOTING
Troubleshooting advice can be found in Table 1.
Table 1.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 14 | Difficult to visualize cells using DIC | Cells died during or shortly after slicing | Use solutions optimized for the age of the animal (12, 36) and adjust the slicing angle to avoid cutting through cellular processes (i.e. apical dendrites) |
| Cells are swollen due to hypo-osmolar ACSF | Check the osmolarity of the ACSF and re-make the solution if needed to achieve osmolarity ~290 mOsm | ||
| 16 | No internal solution is seen flowing out of pipette despite applying positive pressure | Air leak | Check all connections and tubing from pressure application to pipette holder for air leakage |
| Air bubble in pipette | Apply strong positive pressure to expel any air bubble from the pipette. If this does not work, remove the pipette from the holder and tap or shake it to remove the bubble | ||
| The pipette tip is clogged with debris | Discard the pipette and try another, it will be difficult to achieve a gigaseal if there is debris on the pipette tip. If debris is a recurrent problem it is likely the syringe filter integrity has been compromised. Re-make the internal solution and filter more carefully | ||
| The pipette resistance cannot be measured | The silver electrode wire is not submerged in the internal solution | Pull patch pipettes of a consistent length. Replace the silver electrode wire with a slightly longer one. Re-coat and clean with RNase Zap | |
| 18 | No gigaseal can be formed, or cell dies upon approach | Cells are unhealthy | Try a different tissue slice or slice another animal. |
| Problem with internal solution composition | Try another internal solution that has been successfully used before to determine if this is the problem. If so, re-make the RNase-free internal solution | ||
| The cell was approached too slowly | Improve patching technique so that fewer and faster approaches are required to achieve gigaseal | ||
| The pipette tip is too large for a particular cell | Patch a larger cell or use a smaller pipette | ||
| 19 | The cell is unstable immediately after break-in | Too much suction or a non-optimal pipette position when breaking into cell | Wait to see if cell stabilizes. Adjust the pipette position (typically move back slightly) to relieve tension on the cell membrane |
| 21 | The cell becomes unstable while holding in current clamp mode | The cell is gradually swelling leading to mechanical and biochemical stress | Move the pipette back slightly to ease any tension on the cell membrane. Apply a small amount of suction to offset cell swelling. Stimulate the cell with positive current injection to cause it to fire. Adjustments should be made on a cell-by-cell basis according to what the cell seems to ‘like’ |
| 22 | The cell is “lost”, i.e. it quickly destabilizes, requiring large negative current (>200 pA) to maintain a membrane potential of −60 to −70 mV | The cell died and/or the seal between the pipette and the cell membrane is broken | Quickly remove the pipette and collect the cell’s RNA into a 0.2 mL PCR tube. If extracellular material was seen to enter the pipette on DIC, discard the sample |
| 43 | It is difficult to resuspend the beads | The beads were over-dried | Resuspend the beads in the order they become ready, rather than waiting until all the samples are completely dry before beginning to resuspend |
| 47 | Poor yield of cDNA | RNase contamination | Test the RT and cDNA amplification protocol using 10 pg control RNA as input. Re-make RNase-free solutions and clean the lab space/equipment more thoroughly until 10 pg can be consistently amplified. Change gloves frequently during sample collection. |
| Poor sample quality | Take steps to improve quality of tissue slices (i.e. optimized buffers and slice angle). Store samples on ice and run the RT immediately after sample collection. If collecting samples over a long period of time, consider running interim RT reactions every 2–3 hours to prevent sample degradation | ||
| Too much internal solution added to reaction | Use less than 0.3 μL of internal solution in the patch pipette | ||
| Low molecular weight fragments on Bioanalyzer profile | <150 bp: Residual primer dimers, reaction components or ethanol | Completely remove the solutions during each wash step to prevent carry-over. Make sure ethanol is prepared fresh and has completely evaporated prior to resuspending the beads | |
| 300–600 bp, also present in negative control: Degraded/contaminated ERCC RNA spike-in (see examples Fig. 5c) | Run additional controls with/without ERCC RNA spike-ins to confirm. If ERCC has degraded, order new and make fresh aliquots of the working dilution | ||
| The mean size of full-length cDNA is <1500 bp: degradation of sample RNA prior to RT (see examples Fig. 5b) | See above under ‘Poor yield’ (step 47) | ||
| 57 | Mean size of final cDNA library >500 bp | Incomplete tagmentation | Use less input cDNA or more Tn5 transposase. Be careful to completely remove the SDS during PCR purification, thoroughly dry the beads before resuspending, and elute in a low-salinity buffer. |
| 84 | Poor staining results | Poor diffusion of biocytin into fine cellular processes | Hold the cell for 30–60 min in current-clamp mode. Use the low osmolarity internal solution, which is also less viscous, to promote diffusion of biocytin into the cell. |
| The soma is destroyed after aspiration of cell contents, biocytin leaked out into the extracellular space | Apply suction gently and avoid breaking the seal between the pipette and the membrane. After aspiration of the cell contents, slowly move the pipette back away from the cell and wait until the membrane retracts from the pipette tip before removing it completely from the slice | ||
| Tissue slice was poorly fixed | Use fresh, carefully prepared PFA solution and fix the tissue immediately after removing the pipette. Be sure to remove all ACSF before adding the PFA, and make sure the slice is floating so that the PFA can penetrate both sides. Allow to fix for at least 10 – 14 days. | ||
| Poor staining | Be sure to completely remove each solution during the staining procedure. Prepare reagents fresh. At each solution change, ensure the slice is floating so that the solution can penetrate both sides of the slice. |
ANTICIPATED RESULTS
Steps 13–26, Sample collection
The health of the tissue during sample collection is critical to the success of the experiment. Under DIC, many healthy cells should be visualized and the electrophysiological recording from the patched cell should be very stable (see Fig. 8 and Supplementary Movie S1). The resting membrane potential should be between −60 and −80 mV and the cell should not require a large amount of negative current (>200 pA) to maintain a physiological membrane potential. If the cell is unhealthy or dying, RNA degradation may occur. Healthy, stable cells will be able to tolerate a substantial amount of suction during aspiration of cell contents (Supplementary Movie S1).
For the cell types we have studied (24), the modified internal solution used for Patch-seq does not seem to substantially alter the intrinsic electrophysiological properties of the cells. However, given the addition of the calcium chelator EGTA there is potential to decrease the amplitude of calcium-dependent currents, which the experimenter should keep in mind. We recommend investigators perform a pilot study to compare the electrophysiological properties of cells using the modified Patch-seq internal solution and a standard internal solution to determine whether there are important changes in physiology in the cell population of interest.
When aspirating the cell contents into the patch pipette there should be a visible decrease in the size of the cell body (see Fig. 2 and Supplementary Movie S1). Ideally, the seal between the pipette and the cell should remain intact. If the seal is leaky, the recording may become unstable and the negative current required to hold the cell at a physiological resting potential may increase (Supplementary Movie S1). If the seal is completely lost, no additional suction should be applied as this may draw extracellular material into the pipette. Once the nucleus has entered the pipette very little, if any suction, is required to aspirate the remaining cell contents. Therefore, it is important to continuously monitor the cell visually under DIC as well as electrophysiologically while applying suction and the suction protocol should be adapted to each individual cell.
Step 47, QC on full-length cDNA
Due of the very small amount of RNA in a single cell (~10–30 pg), we do not assess the sample quality until after it has been reverse transcribed to cDNA, amplified and purified. We analyze the size profile of every full-length cDNA sample on an Agilent Bioanalyzer to quantify the yield and quality. High quality samples will have a broad peak of cDNA with a mean of ~2000 bp and few short fragments (<500 bp, see Fig. 5 for examples of good and bad samples). Degradation and contamination (by bacteria, genomic DNA) will result in an atypical size distribution. If ERCC spike-in RNA is added to the samples, which can vary from one experiment to another, this may affect quantitative measures of cDNA yield making it difficult to apply rigid cutoffs to all samples. Therefore, we visually inspect each sample to be certain that the size distribution is consistent with the presence of full-length cDNA from a single cell. There is a slight decrease in cDNA yield when using the physiologic pH internal solution (for morphological recovery) compared with our standard high osmolarity internal solution, however no further decrease in yield is seen with longer recording times (Fig. 4).
Individual samples within an experiment may vary substantially in total yield (Fig. 5) due to the nonlinearity of PCR amplification. We keep samples with as little as 300–500 pg total cDNA yield if the size profile looks good, and dilute the samples with higher yield so that the same amount of full-length cDNA from each sample is used to generate the final sequencing library. Screening for sample quality before sequencing also helps to control costs (i.e. money is not wasted sequencing poor quality samples).
Step 57, QC on sequencing library
The size distribution of the pooled sequencing library is analyzed using the Agilent Bioanalyzer, and the concentration is determined using Qubit. The mean size should be <500 bp (Fig. 6) and may have a small amount of primer dimers (peak <100 bp, see Fig. 6). A long tail of large fragments, or average size >500 bp, suggests incomplete tagmentation of the input cDNA (see Table 1).
Steps 59–67, Preliminary analysis of gene expression data
A pipeline for high-throughput processing of scRNA-seq data should be developed in collaboration with an experienced biostatistician. Algorithms for quality control of the raw reads, and demultiplexing to assign reads to cells, are mostly standardized and may even be performed by the sequencing core before the investigator receives the data. Several in-depth tutorials are available (30), and new methods are constantly being developed, to identify and remove poor quality samples and to normalize the data (which is necessary for all scRNA-seq experiments to account for the high technical variability across samples). At a minimum the investigator should examine basic statistics for each cell such as total number of reads, percent of mapped reads aligned to exons/introns, and number of genes detected (see Fig. 7a–d). Samples that seem poor quality (i.e. low number of reads, few genes detected, high percent mapped to intergenic regions) should be removed from further analysis. It may be helpful to compare the results to published datasets on the same cell type to help gauge the quality of the samples and determine thresholds for inclusion. However, in many cases it may not be possible to set thresholds a priori on QC measures such as number of detected genes if the ground truth is unknown for the cell type of interest. In this case, using a threshold such as two median absolute deviations below the median (30) is reasonable. We typically discard ~10–15% of the samples after sequencing.
After excluding poor quality cells, lowly expressed genes should also be excluded (Fig. 7e) and the data should be normalized. The list of top expressed genes should be examined to confirm that it contains the expected markers for the cell type being studied. The top differentially expressed may show bimodal distributions suggesting two discrete transcriptomic populations within the data. Dimensionality reduction techniques can be used to visualize the data in the high-dimensional gene-space spanned by these variable genes and clustering algorithms can be applied to identify transcriptomic cell types (24). The clusters should be externally validated by comparison with known physiological information about the cells (i.e. morphological cell type, firing pattern, brain region) or known molecular markers. Outliers should prompt further investigation into the sample inclusion criteria and pre-processing steps.
Step 84, Direct morphological recovery of Patch-seq neurons
After staining, the cell morphology can be visualized using a light microscope and the quality of the stain may slowly improve for 48–72 h after the slices are mounted on slides (Fig. 8). Many variables can contribute to poor staining (see Table 1).
Supplementary Material
Supplementary Movie S1. Example of Patch-seq sample collection visualized using differential interference contrast (DIC). The entire sample collection process including cell approach, pipette positioning, and aspiration of cell contents into the pipette is shown under DIC. The simultaneous electrophysiological recording is shown in the bottom left of the screen.
ACKNOWLEDGEMENTS
We thank Paul Fahey, Jacob Reimer, Qiaolin Deng, Per Johnsson and Bosiljka Tasic for helpful discussions and suggestions on improving protocol and the manuscript. This study was supported by grants R01MH103108, DP1EY023176, P30EY002520, T32EY07001, and DP1OD008301 from the National Institutes of Health (NIH) to A.S.T.; grants from the Swedish Research Council and the Vallee Foundation to R. S.; the McKnight Scholar Award to A.S.T.; and the Arnold and Mabel Beckman Foundation Young Investigator Award to A.S.T. X. J. was supported by BCM Faculty Startup Fund. C.R.C was supported by NIH grants F30MH095440, T32GM007330 and T32EB006350. Supported by the Intelligence Advanced Research Projects Activity (IARPA) via Department of Interior/Interior Business Center (DoI/IBC) contract number D16PC00003. The U.S. Government is authorized to reproduce and distribute reprints for Governmental purposes notwithstanding any copyright annotation thereon. Disclaimer: The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of IARPA, DoI/IBC, or the U.S. Government.
Footnotes
TWEET #PatchSeq for combined morphology, electrophysiology and gene expression in single neurons. @AToliasLab @sandberglab
COVER TEASER Single-cell RNAseq of patch-clamp recorded neurons
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Key References
- Cadwell CR, Palasantza A, Jiang X, Berens P, Deng Q, Yilmaz M, Reimer J, Shen S, Bethge M, Tolias KF, Sandberg R & Tolias AS (2016). Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat Biotechnol, 34 (2), 199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell CR & Tolias AS (2016) Correlating cellular morphology, physiology, and gene expression using Patch-Seq. In: Using single-cell genomics to analyze neurons, glia, and circuits. (McCarroll S, ed) pp. 41–51. San Diego, CA: Society for Neuroscience. [Google Scholar]
- Cadwell CR, Sandberg R, Jiang X & Tolias AS (2017). Q&A: using Patch-seq to profile single cells. BMC Biology, 15 (1), 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES
- 1.Tang F et al. , mRNA-Seq whole-transcriptome analysis of a single cell. Nature methods 6, 377–382 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Picelli S et al. , Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nature methods 10, 1096–1098 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Macosko EZ et al. , Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hashimshony T, Wagner F, Sher N, Yanai I, CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep 2, 666–673 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Jaitin DA et al. , Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Treutlein B et al. , Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509, 371–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeisel A et al. , Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Tasic B et al. , Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci 19, 335–346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.La Manno G et al. , Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell 167, 566–580 e519 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romanov RA et al. , Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat Neurosci 20, 176–188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Usoskin D et al. , Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 18, 145–153 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Jiang X et al. , Principles of connectivity among morphologically defined cell types in adult neocortex. Science 350, aac9462 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burkhalter A, Many specialists for suppressing cortical excitation. Frontiers in neuroscience 2, 155–167 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eberwine J et al. , Analysis of gene expression in single live neurons. Proc Natl Acad Sci U S A 89, 3010–3014 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sucher NJ, Deitcher DL, PCR and patch-clamp analysis of single neurons. Neuron 14, 1095–1100 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Sucher NJ, Deitcher DL, Baro DJ, Warrick RM, Guenther E, Genes and channels: patch/voltage-clamp analysis and single-cell RT-PCR. Cell Tissue Res 302, 295–307 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Simanski M, Köten B, Schröder JM, Gläser R, Harder J, Antimicrobial RNases in Cutaneous Defense. Journal of Innate Immunity 4, 241–247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toledo-Rodriguez M et al. , Correlation maps allow neuronal electrical properties to be predicted from single-cell gene expression profiles in rat neocortex. Cerebral cortex (New York, N.Y.: 1991) 14, 1310–1327 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Toledo-Rodriguez M, Markram H, Single-cell RT-PCR, a technique to decipher the electrical, anatomical, and genetic determinants of neuronal diversity. Methods in molecular biology (Clifton, N.J.) 1183, 143–158 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Subkhankulova T, Yano K, Robinson HP, Livesey FJ, Grouping and classifying electrophysiologically-defined classes of neocortical neurons by single cell, whole-genome expression profiling. Frontiers in molecular neuroscience 3, 10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang F et al. , RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat Protoc 5, 516–535 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu S et al. , Single-neuron RNA-Seq: technical feasibility and reproducibility. Frontiers in genetics 3, 124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuzik J et al. , Integration of electrophysiological recordings with single-cell RNA-seq data identifies neuronal subtypes. Nature biotechnology 34, 175–183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cadwell CR et al. , Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nature biotechnology 34, 199–203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Picelli S et al. , Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Briant LJ et al. , Functional identification of islet cell types by electrophysiological fingerprinting. J R Soc Interface 14, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brennecke P et al. , Accounting for technical noise in single-cell RNA-seq experiments. Nature methods, (2013). [DOI] [PubMed] [Google Scholar]
- 28.Kharchenko PV, Silberstein L, Scadden DT, Bayesian approach to single-cell differential expression analysis. Nature methods 11, 740–742 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herman J, paper presented at the Boston Single-Cell Network Single-Cell Genomics Workshop, Boston, MA, Nov 3–4 2015. [Google Scholar]
- 30.Lun A, McCarthy D, Marioni J, A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor [version 2; referees: 3 approved, 2 approved with reservations]. (2016), vol. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perin R, Markram H, A computer-assisted multi-electrode patch-clamp system. Journal of visualized experiments: JoVE, e50630 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kodandaramaiah SB, Franzesi GT, Chow BY, Boyden ES, Forest CR, Automated whole-cell patch-clamp electrophysiology of neurons in vivo. Nature methods 9, 585–587 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klein AM et al. , Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao J et al. , Comprehensive single cell transcriptional profiling of a multicellular organism by combinatorial indexing. bioRxiv, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Picelli S et al. , Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome research 24, 2033–2040 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ting JT, Daigle TL, Chen Q, Feng G, Acute brain slice methods for adult and aging animals: application of targeted patch clamp analysis and optogenetics. Methods in molecular biology (Clifton, N.J.) 1183, 221–242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dobin A et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eller CH, Lomax JE, Raines RT, Bovine Brain Ribonuclease Is the Functional Homolog of Human Ribonuclease 1. The Journal of Biological Chemistry 289, 25996–26006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dueck HR et al. , Assessing characteristics of RNA amplification methods for single cell RNA sequencing. BMC Genomics 17, 966 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie S1. Example of Patch-seq sample collection visualized using differential interference contrast (DIC). The entire sample collection process including cell approach, pipette positioning, and aspiration of cell contents into the pipette is shown under DIC. The simultaneous electrophysiological recording is shown in the bottom left of the screen.