Abstract
Advances in sequencing technology and bioinformatics have greatly enhanced our ability to understand the human microbiome. Over the last decade, a growing body of literature has linked nutrition and the environment to the microbiome and is now thought to be an important contributor to overall health. This paper reviews the literature from the past 10 years to highlight the influence of environmental factors such as diet, early life adversity and stress in shaping and modifying our microbiome towards health and disease. The review shows that many factors such as the mode of delivery, breast milk, stress, diet and medications can greatly influence the development of our gut microbiome and potentially make us more prone to certain diseases. By incorporating environmental factors into models that study the microbiome in the setting of health and disease, may provide a better understanding of disease and potentially new areas of treatment. To highlight this, we will additionally explore the role of the environment and the microbiome in the development of obesity and functional bowel disorders.
Keywords: gut microbiome, environment, diet, early life, stress, obesity, irritable bowel syndrome
INTRODUCTION
In the last decade, advances in sequencing technology and bioinformatics have enhanced our ability to understand the human microbiome, and how the environment contributes to shifts in these complex systems over time.1,2 The human microbiome represents a microbial community that encompasses 10 times more cells and approximately 100 times more genes than contained in the human body alone.3 While the major function of the gut microbiome is to aid in the fermentation and energy extraction of indigestible dietary fiber, multiple studies have linked the microbiome to energy homeostasis, immune function, and the development of certain diseases.4 An increased understanding of the relationship between humans, their microbes, and the environment can help us better understand the maintenance of health and the development of disease.5 This review explores the recent literature related to the influence of environmental factors such as early life events, diet, pathogens, social factors, and stress on the complex host-microbe interactions and how these interactions contribute to or are protective against disease. Example disease models such as obesity and irritable bowel syndrome (IBS) will be discussed in order to highlight how environmental perturbations in the human microbiome contribute to disease.
ENVIRONMENTAL FACTORS
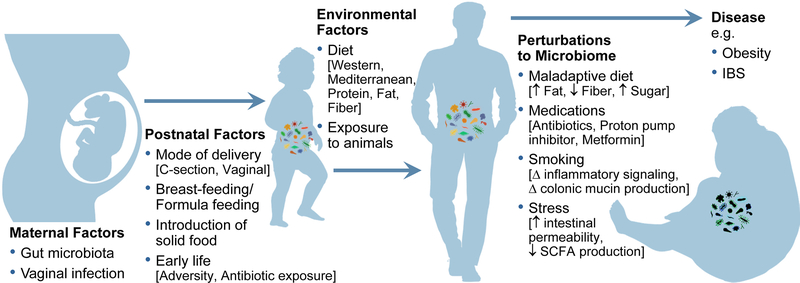
The environment plays a critical part in the composition of the human microbiome (Figure 1). In fact, 22–36% of the inter-person microbiome variability is associated with environmental factors and only 1.9 – 9% by genetics.2 Environmental factors start in the early days of life and extend well into adulthood. Below, we highlight how environmental factors such as the mode of delivery, breast feeding, and introductions of foods are critical steps in the development of a mature adult microbiome. We later show how such environmental factors as diet, smoking, home life, and stress can induce shifts in our microbiome during the lifespan and make us more prone to certain diseases.
Figure 1: Environmental Factors Shape and Change the Microbiome Over Time and Perturbations can lead to disease.
Maternal Factors: Vaginal infections and gut microbiome can lead to bacterial translocation into the uterus.
Postnatal Factors: Mode of delivery, breast feeding vs. formula feeding, introduction of solid food, and early life adversity and antibiotic exposure can shape the developing microbiome in early childhood.
Environmental Factors Across the Lifespan: Long-term diet and exposure to animals can modify the microbiome throughout childhood and adulthood.
Perturbations to the Microbiome: Medications such as antibiotics, proton pump inhibitors, and metformin and a variety of different diets can make individuals more prone to disease like inflammatory bowel disease (IBD), irritable bowel syndrome (IBS), and obesity. Stress can lead to changes in the microbiome that affects intestinal permeability and SCFA production. Smoking can cause microbial shifts that changes inflammatory signaling and colonic mucin production, all of which can be mechanisms that leads to the development of disease.
Early Life Events
Early life events are critical to the development of the human microbiome because they can shape the sequence of microbial community establishment and ultimately the final composition of our mature adult microbiome.6 In this section, we summarize how the microbiome matures during the transition from inside the womb, which is a relatively sterile environment, into the external environment after birth when ingestion of milk and solid food are introduced. Microbial community differences during each key early life process are summarized in Table 1.
Table 1:
Microbial Communities Described by Type of Early Life Environmental Factors
| Environmental Factors | Bacterial Community | Reference #s |
|---|---|---|
| Vaginal Delivery vs Cesarean Section Delivery | ↑Lactobacillus ↑Prevotella ↑Sneathia ↑Bifidobacterium ↑Bacteroides ↓Staphylococcus ↓Propionibacterium ↓Corynebacterium |
19,20 |
| Breast Feeding vs Formula Feeding | ↑Bifidobacteria ↑Lactobacillus ↓Clostridiales ↓Proteobacteria |
32,33 |
| Introduction of Solid Food | ↑Lachnospiraceae ↑Ruminococcaceae ↑Bacteroidaceae ↓Lactobacillaceae ↓Bifidobacterium ↓Enterococcaceae ↓Enterobacteriaceae |
36,37 |
The table summarizes the bacterial shifts during major early life events such as delivery, breast feeding, and introduction to solid food.
Prenatal.
Studies have suggested that the introduction of microbes can occur as early as during the prenatal period. While certain intrauterine infections and bacteria from such groups as Burkholderia, Actinomycetales and Alphaproteobacteria are associated with preterm delivery, it has been shown that a variety of other microbes may be present in the placenta, umbilical cord, amniotic fluid, and meconium of normal pregnancy.7–9 The maternal microbiome is likely translocated into the uterus via the bloodstream, an idea supported by the detection of labeled Enterococcus faecium in the meconium of inoculated pregnant mice.7 In a recent population-based study, researchers found the most abundant phyla isolated from first-pass meconium were Firmicutes, Proteobacteria, and Bacteriodetes.10 All bacteria isolated from the umbilical cord blood of healthy neonates belonged to the genus Enterococcus, Streptococcus, Staphylococcus, or Propionibacterium.11 In the placenta, the genus Bifidobacterium and Lactobacillus were identified.8
However, the role and function of these microbes in human health or disease during the prenatal period remains unclear. Due to the possibility of maternal contamination, it is difficult to definitively establish the presence of a prenatal microbiome.12 Further studies will be required to confirm the existence of a viable intrauterine-resident microbiota with the use of adequate controls (such as maternal blood or sampling at a site nearby delivery) in order to determine if the existence of such a microbiome might affect the future development of the newborn.
Delivery.
While the existence of a prenatal microbiome may be controversial, many agree that the first major introduction of a microbial community to a newborn is through delivery.13 As the newborn is passing through the vaginal canal, it is ultimately introduced to the commensal vaginal and fecal microbiome of the mother.14 This community of microbes seems to be distinct from the community of non-pregnant women as the vaginal microbiome changes during pregnancy.15 For example, healthy pregnant women, when compared to non-pregnant women, had lower vaginal bacterial diversity with higher levels of Lactobaccillus, Clostridiales, Bacteroidales, and Actinomycetales; these levels were associated with gestational age.16 Beyond the vaginal microbiome, there is also evidence that the community of the maternal gut also changes during the course of pregnancy. For example, a Finnish cohort of 91 healthy pregnant women demonstrated decreased bacterial diversity as evidenced by increased levels of high-energy-yielding fecal microbiota with increasing gestational age.17 From the first to the third trimester, the proportion of Proteobacteria, including species of the Enterobacteriaceae family and Streptococcus genus, decreased while the proportion of Faecalibacterium prausnitzii increased. These changes in the microbiome were independent of pre-pregnancy body weight, gestational diabetes, diet, and antibiotic use, suggesting that they were due to the changes of normal pregnancy. These changes in the microbiome have a beneficial role for both the mother and neonate by protecting against certain infections such as Neisseria gonorrhea and bacterial vaginosis and also by permitting greater efficiency for energy harvest to support the growth of the mother and fetus.17,18
With these specific pregnancy-related changes effecting the vaginal and fecal microbiome of the mother, it is unsurprising that the mode of delivery also greatly affects how the newborn microbiome develops. The differences seen between Cesarean section (C-section) and vaginally delivered babies are drastic. Compared to vaginally born babies, those that are born by C-section without membrane rupture have no vaginal microbes such as Lactobacillus, Prevotella, and Sneathia. Instead, babies born by C-section are colonized with skin microbes such as Staphylococcus, Propionibacterium, and Corynebacterium.19 These babies have a delayed colonization of intestinal microbes such as Bacteroides and Bifidobacterium.20 While the exact length of time these differences exist is unknown, microbial differences between C-section and vaginally delivered babies have been observed to as far out as 7 years of age.21 The deficits in the human microbiome associated with C-section deliveries have been implicated in certain childhood autoimmune disease like celiac disease, asthma, and type I diabetes.22 These studies also suggest that restoration of a more “normal” microbiome after C-section deliveries may therefore be beneficial. “Vaginal seeding,” or the process by which vaginal fluids are applied to a newborn child delivered by C-section has been a method used to restore the human microbiome. Although a small pilot study of 4 babies, demonstrated the feasibility of restoring the early microbiome of babies born by C-section,23 the long-term health consequence of such a procedure remains unknown and may even increase the risk of transmittable diseases to the newborn. Therefore, further prospective studies are needed to determine the safety and potential benefits, if any, of these methods used to restore the human microbiome.
Even though the mode of delivery is important to microbial seeding, it may not be the only mode for vertical transmission. Recent human studies have highlighted maternal vertical transmission from multiple different sources such as the skin, mouth, and GI track.24,25 By examining strain level data, these studies demonstrated that the direct mother to infant transmission would change over time as different floras were introduced through processes such as skin contact and breast feeding.24,25
Breast Feeding and Introduction of Solid Food.
The other major early life events that affect the development and maturation of the newborn microbiome are breastfeeding and the introduction of solid food. Breast milk bacteria such as Corynebacterium and Rothia can seed the infant gut and influence the bacteria that follows, affecting the communities even through adulthood.26 These early seeding events may be the mechanism by which breast milk can protect children against such autoimmune diseases like asthma and type 1 diabetes.27
Similar to the vaginal microbiome, it has been shown that the microbiome of breast milk also varies with increasing gestational age, and is related to maternal health and mode of delivery.27,28 The breast-milk microbial community is dominated by Corynebacterium, Ralstonia, Staphylococcus, Streptococcus, Serratia, Pseudomonas, Propionibacterium, Sphingomonas, and Bradyrhizobiaceae.27 In a study of 107 healthy women who were breastfeeding their infants for the first 30 days of life, the gut microbiome changed in a dose-dependent manner with 27.7% of the mean bacteria being derived from breast milk and 10.3 % from areola skin.27
Breast milk also contains many important prebiotic compounds such as human milk oligosaccharides. These sugar polymers are almost exclusively metabolized by the gut microbiome29 and they can promote the growth of key communities including Bifidobacterium spp.30 Bifidobacteria has been shown to inhibit the growth of pathogenic organisms and improve barrier function in the infant gut.31 In a mouse model, human milk oligosaccharides were found to be protective in the development of autoimmune disease and obesity.32,33
There are clear differences in the composition of the microbiome in infants who are breastfed versus those who are formula fed. Infants who are breastfed have a higher proportion of Bifidobacteria and Lactobacillus spp, while infants who are formula-fed have a higher proportion of Clostridiales and Proteobacteria.34,35 Formula-fed infants also have lower diversity and richness even after the first year of life as compared to their breastfed counterparts.34 In a study of 30 preterm infants, breastfeeding was found to be protective against gut immaturity and possibly necrotizing enterocolitis.36 Several other epidemiological studies have provided support for the beneficial role of breastfeeding in the development of disease. Formula feeding has been associated with various inflammatory and autoimmune diseases.37 In contrast, breastfeeding, through its effects on the microbiome, has been associated with a protective role against asthma, autism spectrum disorder, and type 1 diabetes.27,37
One of the last major events in early life affecting microbial development is the introduction of solid food. While breastmilk keeps the microbiome in a state that is characterized by low diversity and Bifidobacterium predominance, the introduction of solid food and the cessation of breastfeeding increases adult-associated microbes such as Lachnospiraceae and Ruminococcaceae.38 In a Danish study of 330 children between 9 and 36 months of age, Lactobacillaceae, Bifidobacteriaceae, Enterococcaceae and Enterobacteriaceae decreased while Lachnospiraceae, Ruminococcaceae and Bacteroidaceae increased during the period when solid foods was being introduced.39 Another study of 531 children born in 5 different countries showed similar results independent of location, use of antibiotics, mode of delivery or milk feeding practices, suggesting that these changes were typical of the normal developing microbiome as solid foods are being introduced.40 This transition is both necessary and beneficial. It allows for a microbial community that is better equipped to extract energy and process a diet that is no longer dependent on milk to a diet that is higher in fiber and protein, similar to the diet of a mature adult.
Early Life Adversity.
Recently, researchers are discovering that early life adverse events can manipulate our microbial community in significant ways. In rats, limited nesting stress during post-natal days 2 to 10 led to a delayed maturation of the hypothalamic-pituitary-adrenal axis that was associated with decreased microbial diversity, an increase of gram positive cocci, and a reduction of fiber-degrading bacteria.41 Similar findings were demonstrated in mice and rhesus monkeys when exposed to stress at an early age.42,43 Finally, in patients with IBS, those that had a microbiome profile distinct from healthy controls were more likely to have a history of early adverse events and trauma than those with a microbiome that was more similar to healthy controls.44 While these studies are predominantly associations, exploring how adverse events and early gut dysbiosis can cause such diseases as IBS is an active research area.
Antibiotic use can also play a significant role during early life. The average US child receives about one to three antibiotic courses by the age of two years old.45 Several studies have highlighted how antibiotic exposure in children can be associated with an increased risk for obesity, diabetes, allergies, asthma, IBS and inflammatory bowel disease (IBD).34,46 Children exposed to antibiotics have delayed maturation of their microbiome as compared to their respective controls, but whether this is the exact mechanism by which early antibiotics predisposes children to disease is still unclear.34 In animal models, peri-partum antibiotic exposure in the mother can lead to persistent gut dysbiosis in the offspring and colitis in susceptible individuals.47,48 Although these studies do not provide an exact mechanistic explanation of the effect of antibiotic use on microbiome development or on disease susceptibility, they do highlight that early antibiotic exposure is linked in some way to the normal development of microbial community and to disease development.
Diet
During the early stages of life, we see that breastmilk and the introduction of solid food are critical events in the development of the human microbiome. Therefore, it is not surprising that the introduction of diet throughout life has large effects on the human microbiome. The gut microbiota can change within days of a new diet, but to what extent these changes are permanent once the new diet has terminated remains uncertain.49 In this section, we review the current literature, focusing on popular diets and how the microbiome is connected (summarized in Table 2).
Table 2:
Influence of Diet on the Gut Microbiome
| Diet | Species Richness/ Diversity | Microbes Altered | Associated Physiological Effect | Associated Disease State | References |
|---|---|---|---|---|---|
| Western Diet | ↓ | ↑Bacteroides ↑ Enterobacteria ↓ Bifidobacteria ↓ Lactobacilli ↓Eubacteria |
Reduced SCFA Higher LPS levels Higher inflammation Decrease gut barrier |
Obesity Colon Cancer Type 2 Diabetes |
47–49 |
| Mediterranean Diet | ↑ | ↑Bifidobacteria ↑Lactobacilli ↑Eubacteria ↑Bacteroides ↑Prevotella ↑Roseburia ↓Clostridium |
Increase SCFA Decrease inflammation |
Decrease risk of CVD and Obesity | 47,52,54 |
| Protein | |||||
| Plant Protein | ↑ | ↑Bifidobacteria ↑Lactobacilli ↓Bacteroides ↓Clostridium perfringes |
Increase SCFA's Increase gut barrier, Reduce Inflammation |
47,57 | |
| Animal Protein | ↑ | ↑Alistipes ↑Bilophila ↑Clostridia ↓Roseburia |
Increase TMAO Reduce SCFA Increase amines and sulfides |
CVD IBD |
47,58,59 |
| Fats | |||||
| Unsaturated Fats | ↑ | ↑ Lactobacillus ↑ Lachnospiraceae ↑ Streptococcus ↑ Akkermansia muciniphila |
Reduce TLR activation Reduce white adipose tissue inflammation |
Decrease risk for IBD, obesity, psoriatic arthritis | 47,64,65 |
| Saturated Fats | ↓ | ↑Bacteroides ↑Bilophila ↑Faecalibacterium prausnitzii |
TLR activation Promote pro-inflammatory TH1 |
CVD Obesity Diabetes |
47,63 |
| Dietary Fiber | ↑ | ↑Lactobacilli ↑Bifidobacteria ↑Clostridia ↑Prevotella ↑Treponema |
SCFA production Anti-inflammatory Anti-cancer activities |
Decrease risk for CVD, obesity, diabetes, colon cancer | 47,72 |
The table summarizes bacterial richness, microbial community changes, physiological effects, and diseases associated with specific diets (Western, Mediterranean, Plant Protein, Animal Protein, Unsaturated Fats, Saturated Fats, and Dietary Fiber).
Abbreviations: CVD: Cardiovascular Disease, IBD: Inflammatory Bowel Disease, SCFA: Short chain fatty acid, LPS: Lipopolysaccharides, TMAO: trimethylamine N-oxide; TLR: Toll-like receptor
Western Diet.
The Western diet or standard American diet is a diet that is characterized by high-fat, high-sugar, high level of red and processed meat, high levels of refined grains and a lower level of fiber.49 Many studies have linked the Western diet to inflammation, diabetes, cardiovascular risks, obesity, and metabolic syndrome.49,50 While a Western diet affects many different cell types such as adipocytes, immune cells, and endocrine cells, there is also a strong link that connects the deleterious effects of a Western diet to shifts in the microbiome.49 Compared to other indigenous diets, the microbiome on a Western diet is characterized by a significantly lower microbial diversity and species richness.51 The Western diet microbial composition is classically characterized by an overabundance of the phyla Firmicutes and a decrease in Bacteroidetes.52 On a genus level, a Western diet shows a decrease in Bifidobacteria and Lactobacilli, while being high in Enterobacteria.53 Consequently, the Western diet has been linked to an increase in endotoxemia, a state characterized by decreased intestinal barrier function and increased levels of bacterial lipopolysaccharides and inflammatory signaling.54,55 Furthermore, a Western diet can possibly lead to permanent microbiome changes that may be responsible for post-dieting weight regain or the common concept of yo-yo dieting, which has been linked to higher long-term weight gain, increased obesity-related risk factors, and increased difficulty reducing weight.56
Mediterranean Diet.
In contrast to a Western diet, a Mediterranean diet is considered a healthier diet. It is characterized by a beneficial fatty acid profile, higher intake of fiber, vegetables, and fruits, and with lower intake of sugar and red meat.57 A recent study demonstrated that out of 153 participants, those who were more adherent to a Mediterranean diet had an increased level of short chain fatty acids (SCFA), Prevotella, and certain Firmicutes, which have all been associated with decreased cardiovascular events.58 Additionally, they also showed that low adherence to the Mediterranean diet led to decreases in urinary trimethylamine oxide levels, which is associated with higher cardiovascular risk.58,59 Several studies have shown that consumption of foods encompassing the typical Mediterranean diet improved obesity, inflammation, and lipid profile and were associated with an increases in Lactobacillus, Bifidobacterium, and Prevotella, but with decreases in Clostridium levels.53,60
Protein.
While diets like the Western diet and the Mediterranean diet have varying compositions of protein, fat and fiber, studies have found that the gut microbiome can also be affected by these individual components. High protein consumption can have a large effect on human health and the microbiome. Overall, studies have shown that a diet that is high in protein correlates positively with microbial diversity, which is important in intestinal health and barrier function.61,62 However, not all protein is the same as the source of the protein, whether from animals or plants, can have very different effects on the microbiome.
Diets high in plant-based protein can increase Bifidobacterium and Lactobacillus, while decreasing pathogenic species such as Bacteroides fragilis and Clostridium perfringens.63 These shifts are associated with higher levels of SCFA and possibly improved gut barrier function.63 On the other hand, diets high in animal protein have consistently been associated with an increase in Bacteroides, Alistipes, Bilophila, and intestinal inflammation.64,65 Although, diets high in animal-protein can have immediate effects associated with weight loss, these shifts can be detrimental to colonic health in the long-term.66 Animal protein is associated with an increased level of microbial derived toxic metabolites, such as amines and sulfides, within the colon.67 The pro-inflammatory state induced by animal protein is one of the potential mechanisms that explained the 3.3-fold increased risk of IBD in a prospective cohort of 67,581 participants.64 This observation was corroborated in a recent animal study demonstrating how dietary protein can change the density of the fecal microbiota while increasing intestinal permeability and the severity of dextran sulfate sodium induced colitis.68
Fats.
Similar to the changes seen in the microbiome with proteins, fats are also not all equal. A Western diet that is high in saturated and trans-fat has been linked to obesity and cardiovascular disease while a diet rich in mono and polyunsaturated fats can be protective.69 To study this, Fava et al. examined 88 subjects at risk for metabolic syndrome and fed them varying amounts and types of fats. They found that diets high in saturated fats led to an increase of Faecalibacterium prausnitzii and reduced bacterial richness, while a diet high in monounsaturated fats did not have any significant shifts in any bacterial genera.70 In another larger, cross-sectional study with 876 women, Menni et al demonstrated that polyunsaturated fats were associated with an increase in microbial diversity and an increase in members of the Lachnospiraceae family.71
Mouse studies have also offered insights regarding differences in microbial composition related to type and amount of fat intake. For example, a recent study found that mice fed lard fat had a higher abundance of Bacteroides and Bilophila, while mice fed fish oil-derived fat had higher levels of Akkermansia and Lactobaccillus.72 The mice on a lard fat diet also had higher toll-like receptor activation and white adipose tissue inflammation, in addition to a decrease in insulin sensitivity relative to mice fed on the fish oil-derived fat. Transplantation of the microbiome of the lard fat-fed mice into germ-free mice successfully replicated the donor’s inflammatory and metabolic phenotypes, suggesting that these pathways were at least in part mediated by the gut microbiome.72 Another potential mechanism by which fat intake can affect the host’s microbiome is through alterations of the host circadian rhythm.73 Several recent studies have shown that the microbiome has a diurnal variation in both structure and function and that these variations can modify the host circadian clock genes.74,75 A diet high in saturated fats disturbs normal diurnal microbial patterns leading to host dysregulation of circadian rhythm and metabolism, potentially promoting diet-induced obesity.73
Dietary Fiber.
Unlike protein and fat, the gut microbiome is absolutely necessary for the metabolism of dietary fiber. Carbohydrates, in particular those with dietary fiber, are a principal source of energy for colonic microbes. The recommended daily intake of fiber per day is 25–30 grams. The average American only ingests about 15 grams per day, highlighting the substantial deficit in fiber intake needed for the healthy function of the colon.76 Evolutionarily, the human diet was very heavy in dietary fiber as compared to fiber content in the current more modern diets. The diet and microbiome of indigenous populations in Papua New Guinea, Tanzania, and the Amazon rainforest are markedly different from those in the industrialized societies.77 With the advent of farming and industrialization, modern diets have become heavier in protein and fat while lighter in dietary fiber. This dietary shift has led to a reduction in microbial diversity that has been linked to a higher susceptibility of more western diseases such as diabetes, obesity, and inflammatory bowel disease.77 Several studies have shown fiber to be protective against such diseases as type II diabetes, cardiovascular disease, colon cancer and obesity.78,79 One of the main mechanism by which fiber and the microbiome impact health is through the production of short chain fatty acids (SCFAs). SCFAs are the major product of fiber fermentation and represent a major substrate for energy for colonic cells. Acetate, propionate and butyrate are the major SCFAs, accounting for 90% of the total, with ratios approximating 65:20:15.80 SCFA can affect gene regulation and colonocyte proliferation and inflammation.81 In diversion colitis, a disease state characterized by distal inflammation, the role of butyrate enemas as a treatment highlights the importance of SCFA to colonic health. The capacity of SCFA to regulate colonocyte differentiation and apoptosis further underscores its potential to protect against colon cancer.81 In mouse models of obesity, fiber supplementation prevented inflammation and metabolic syndrome by restoring IL-22 production within the colon.82 Diets high in fiber tend to have higher microbial richness and diversity with an abundance in such genera as Prevotella and Treponema.83 These shifts have been linked to a decreases in inflammatory signaling, protection against obesity, and possibly decreases in the presence of colorectal cancer.83
OTHER ENVIRONMENTAL FACTORS
Even though early life events and diet can shape the way our microbiome is formed, there are many other environmental exposures throughout the lifespan into adulthood that can lead to changes in our microbial composition and ultimately our health (Figure 1).
Pharmaceuticals
One of the most significant ways we can affect our health and our microbiome is through the use of drugs. Antibiotics are the most well-known class of medications to cause shifts in the microbiome. While the microbial community mostly returns to its pre-antibiotic state, studies have suggested that post-antibiotic exposure can lead to a new steady state that is different from the original pre-antibiotic community.84 This new steady state that emerges after antibiotic exposure increases the host’s susceptibility to infection, atopic diseases, and metabolic syndrome.85 Long-term studies, have shown that these effects of antibiotics can be long lasting, as far out as 4 years post-exposure.86 Antibiotics may also expand antibiotic-resistant strains which can act as a reservoir for resistance genes in the gut microbiome.87
The effect of antibiotics on the microbiome is best highlighted by the research regarding Clostridium difficile. Changes in the gut microenvironment after the exposure of antibiotics creates a metabolic environment that favors C. difficile germination and colonization.88 C. difficile can be a debilitating disease for patients and costly to the medical system. While the mainstay of C. difficile treatment is further antibiotic therapy, fecal microbiota transplantation (FMT) has gained substantial credibility for the treatment of recurrent C. difficile.89 The first randomized trial of FMT demonstrated higher rates of resolution of C. difficile infection when compared to placebo (81% vs 31%).90 Since then, several other studies have shown the efficacy and safety of FMT for patients with C. difficile.89
Medications other than antibiotics have also been associated with changes in the human microbiome. Proton pump inhibitors (PPIs) and metformin are two notable common examples, with over 100 million users of either medication in the US alone.91,92 Through reduced acid production and hence higher luminal pH, PPIs have been shown to alter the flora of the stomach in chronic users as well as increase the incidence of small bacterial overgrowth.93 In a study examining individuals with type 2 diabetes, Wu et al. demonstrated that the introduction of metformin strongly alters the gut microbiome.94 They further demonstrated that the beneficial effects of metformin can be transferred to germ-free mice through fecal transplantation, providing support that the alteration of the gut microbiome mediates some of metformin’s antidiabetic effects.94 The idea of non-antibiotic drugs effecting the microbiome led to Maier et al. to screen more than 1,000 marketed drugs highlighting that 24% of them could affect bacterial growth.95 Although, these studies demonstrate dramatic effects on the gut microbiome, further research is needed to see how these drugs can affect microbial communities and how these changes are related to drug efficacy.
Family Life
Pets.
Other than pharmaceuticals, household pets and animal exposure can also influence our microbiome. In a study of 60 families with or without pets, household members’ skin flora was more similar to each other and to their pets than to other households without pets.96 This highlights the theory that pets play a critical part as a conduit to the environment and their human co-inhabitants. Many studies have reported that the exposure during early life to pets or to farm animals is associated with less pediatric allergy.97,98 The theory is related to the idea that pets and farm animals introduce environmental allergens that sensitize the developing immune system of the child.99 The fact that these exposures are mediated through microbes is supported by studies that demonstrate how certain bacterial species like Acinetobacter lwofii and Lactococcus lactis which are isolated from farming communities can reduce allergic responses in mouse models.100 In a study of 24 healthy, full term infants, microbial richness and diversity of fecal samples were higher in infants living with pets.101 Infants living with pets showed a reduction of Bifidobacteriaceae and an increase in Peptostreptococcaceae. Another study found that Bifidobacterium longum levels were higher in fecal samples of infants with pet exposure compared to those without and that the abundance of B. longum was inversely associated with the onset of wheezy bronchitis. However, the role of animal exposure on the adult gut microbiome still remains unclear.
Smoking.
Another important environmental exposure is smoking. While the link between smoking and disease has been well established, several studies have emerged over the last several years that explore the influence of smoking on the oral, esophageal, and gastric microbiome.102 Within the mouth, smokers tend to have an increase in anaerobic bacteria leading to shifts in a community that possibly favors pathogenic microbes.102 In regards to the intestinal microbiome, both mouse models and human studies have indicated that smoking can affect microbial composition and intestinal inflammation. A recent study, which used side-stream smoke on wild type C57BL/6 mice, demonstrated that smoke can increase the abundance of Clostridium clostridiforme and decrease Lactococcus, Ruminococcus albus, and the family of Enterobacteriacae.103 These shifts were associated with a change in tight junction proteins and a inflammatory signaling.
In humans, smoking has been associated with a decrease in Firmicutes and Actinobacteria, in addition to an increase in Bacteroidetes and Proteobacteria.104 An observational study demonstrated that after smoking cessation, the levels of Firmicutes and Actinobacteria reversed and microbial diversity increases.104,105 Similar findings were found in patients with active Crohn’s disease. In a study of 103 smoking and non-smoking patients with active Crohn’s disease compared to 66 smoking and non-smoking healthy controls, active smoking was independently associated with higher abundance of Bacteroides and Prevotella, genera commonly associated with colonic inflammation.106,107 The exact mechanism on how smoking affects the microbiome is an active area of research; however, it may be linked to alterations of intestinal tight junction, immune signaling, and/or mucin production.103,108 Whether these changes in the intestinal microbiome mediate some of the deleterious effects of smoking remains unclear.
Stress.
In recent years there is growing evidence that there are bidirectional communication pathways between the gut microbiome and the brain, and recent studies have highlighted that this communication takes place through various processes including the vagus nerve, gut hormone signaling, inflammatory processes, and neurotransmitter production, just to name a few.109,110 Therefore, real or perceived stress can modulate this bidirectional communication in a way that increases dysbiosis and increases an individual’s propensity to develop disease.111,112 For example, stress can trigger the flight or fight response, increasing the production of corticotropin-releasing-hormone and catecholamine production from the central nervous system (CNS) which then modulates gut microbiome function.112,113 On the other hand, bottom up processes involving release of microbial products such as tryptophan or serotonin during stressful events can contribute to the enteric dysbiosis, increased intestinal permeability and the release of certain neurotransmitters associated with certain diseases.114 Studies have also shown that in the absence of stress and during stress reducing practices such as meditation, the microbiota increase production of SCFAs and anti-inflammatory processes, further highlighting the negative effects of stress on gut function and health.112,115 These studies highlight the importance of considering the modulation of the brain-gut-microbiome axis as an effective strategy for the development of treatments for various disorders.
CLINICAL IMPLICATIONS OF ALTERED HUMAN MICROBIOME
While previous sections have shown how environmental exposures such as diet, stress, pets, and medications can affect the microbiome, perturbations in the microbiome can also be a potential cause for disease (Figure 1). While there are many diseases that are associated with microbial dysbiosis, we will review the diseases with the best causal relationship between the environment and the microbiome: obesity and IBS.
Obesity.
In the previous section, we underscored the notable shifts in the microbiome that occurs with a Western diet and a high fat diet. These diets are tightly linked to obesity and many believe that the microbiome is a key mediator. One of the early seminal papers that linked obesity and diet to the gut microbiome came from Turnbaugh et al. in 2006.116 They used human and mice16s rRNA analyses to highlight how obesity and a high fat diet was related to an increased ratio of Firmicutes to Bacteroidetes and that colonization of this obesity-related microbial profile could recapitulate the obese phenotype in germ-free mice. Since then, animal studies have shown that gut microbes can influence weight gain and adiposity by affecting host gene expression, metabolic pathways, and the gut-brain-axis.117 Moreover, weight loss interventions have also been associated with distinct shifts in the gut microbiome. A variety of surgical weight loss intervention in humans and in animals have shown distinct and long lasting microbial profile differences.118 A study utilizing roux-en-y gastric bypass in mice lead to a to sustained increase in Escherichia and Akkermansia and that the transfer of fecal material of post-bariatric surgery mice into non-operated germ-free mice can successfully transfer the lean phenotype.119 Mechanisms by which microbes can cause host metabolic changes includes shifts in short-chain fatty acid production, changes in energy extraction, decrease gut hormones like glucagon-like peptide and peptide YY, and changes in toll-like receptor signaling.120
Irritable Bowel Syndrome.
Much like obesity, IBS is one of the most common diseases seen by primary care doctors and gastroenterologists. Factors linked to the pathogenesis of IBS includes a history of enteric infection, alteration in the gut-brain axis, changes in visceral sensitivity, and modifications in the gut microbiome.44,121 Studies have linked early life stress and adverse events to microbial shifts that could potentially be the cause of visceral sensitivity and subsequent IBS development.44,109,122 IBS patients generally had less Lactobacillus, Bifidobacterium, and Faecalibacterium prausnitzii than healthy controls.114 While it is unclear which changes are necessary and/or sufficient, studies in germ-free mice transplanted with fecal microbes from IBS patients causes alterations in gut permeability, motility, visceral perception, and food processing that ultimately triggers IBS symptoms.123 The treatment of IBS is varied and often personalized, but certain therapeutics for IBS with diarrhea often targets the gut microbiota. These includes the use of non-systemic antibiotic rifaximin, dietary modification with a low Fermentable Oligo-, Di-, Mono-saccharides And Polyols (FODMAP) diet, and certain probiotic formulations.123 Even though these treatments are promising, more research is required to elucidate how alterations in the gut microbiota can affect disease outcome in other subtypes of IBS. In this section, we have highlighted how certain microbial communities can lead to certain endpoints such as weight loss and patient symptomology. However, whether these microbial communities lead to similar endpoints outside of these specific disease states remains unknown and remains an area of active investigation.
LIMITATIONS OF CURRENT METHODS AND FUTURE DIRECTIONS
Obesity and IBS are two example diseases that highlights the significant gains microbiome research has undergone in the last decade. But despite those gains, there is still much more research to be done. To date, much of the current research in human samples have been predominantly association studies, with little insight into the underlying mechanisms associated with altered gut microbiome and disease. The limitation of these studies is that they are unable to clearly state if dysbiosis is a cause of disease or merely a byproduct. Currently, there is an important shift away from these types of analyses and a move towards investigations to define mechanisms by utilizing germ-free animals and multi’omics analysis.5 By starting with a multi’omics approach that integrates metabolomics, proteomics, and 16S rRNA analysis, for example, researchers can more clearly postulate microbial dependent mechanistic pathways that leads to disease. By recapitulating the disease phenotype through microbial transplantation in germ-free animal models, researchers can then clearly model out the direct causative effects of dysbiosis to disease development. Even though the majority of mechanistic studies have been done in animals, the observations seen in animal models and how they have correlated in humans have provided us with a deeper understanding of microbial-host signaling and pathogenesis of diseases. Future research may also focus on the interplay between the microbiome and epigenetics. There is an emerging body of literature on the role of transgenerational epigenetic inheritance on health and disease.124 As reviewed here, stress and the environment play a major role in the development and stability of the microbiome over time. But exactly how these signals can alter host epigenetics, particularly across generations, is an active area of research.
The other current limitation in the field is the lack of standardization across similar studies or even across same disease groups. Currently, there are not any definitive studies on how microbial analysis differ across the various sampling techniques and the many analytical pipelines. For example, it is difficult to compare studies of similar patients if sampling was done using differing methods such as via fresh or frozen stool collection, endoscopic aspiration, or via mucosal scrapings/biopsy. As the field matures, sample collection and processing will need to be standardized to help ensure reproducibility and to allow researchers to readily compare outcomes across different studies using different patient groups.
Another major shift will involve cheaper sequencing technology in order obtain strain-level data. 16S rRNA sequencing is currently the most commonly used method for microbiome analysis.5 However 16S rRNA sequencing is only able to obtain species level resolution. Due to horizontal gene transfer, bacterial genomes of strains even within the same species can be potentially diverse.125 While shotgun sequencing is available and able to provide strain-level resolution, it is out of reach for many due to its high costs. But with greater advances in sequencing technology, this cost may be reduced enough for widespread use in the future.126 Even though we may have a long way to go, improvements in sequencing throughput and accuracy, complemented by an upsurge of novel bioinformatics analysis of proteomes, metabolomes, and transcriptomes, has brought us closer to a better understanding of human disease and treatment.
CONCLUSIONS
The microbiome is a complex system that is at the intersection of our environment and our health. While many aspects of our environment can cause shifts in our microbiome, we see through the example of obesity and IBS that the perturbations of this intricate system can also be at the core of disease. Even though longitudinal studies are still required to determine the direct relationship between environmental factors and gut dysbiosis with disease development, improvements in sequencing technology and an ever-growing field of bioinformatics are providing us with new daily insights on how our environment, our heath, and our microbes are linked. This has implications for not only better understanding the underlying pathophysiology of disease but can also help better develop targeted treatments based on specific microbial molecules.
Acknowledgements:
Grant support DK106529 (AG) and DK 07180 (TD). We would like to thank Cathy Liu and Noelle Min for assistance with manuscript review, technical support and figure design and creation.
Abbreviations:
- CNS
central nervous system
- FMT
fecal microbiota transplantation
- IBS
irritable bowel syndrome
- IBD
inflammatory bowel disease
- PPIs
Proton pump inhibitors
- SCFA
short chain fatty acid
- FODMAP
Fermentable Oligo-, Di-, Mono-saccharides And Polyols
Footnotes
Disclosures: All authors have no disclosures or conflicts of interest
References
- 1.Lax S, Smith DP, Hampton-Marcell J, et al. Longitudinal analysis of micorbial interaction between humans and the indoor environment. Science (80- ). 2014;345(6200). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothschild D, Weissbrod O, Barkan E, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555(7695):210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 3.Ursell LK, Metcalf JL, Parfrey LW, Knight R. Defining the human microbiome. Nutr Rev. 2012;70 Suppl 1:S38–44. doi: 10.1111/j.1753-4887.2012.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shreiner AB, Kao JY, Young VB. The gut microbiome in health and in disease. Curr Opin Gastroenterol. 2015;31(1):69–75. doi: 10.1097/MOG.0000000000000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med. 2018;24(4):392–400. doi: 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamburini S, Shen N, Wu HC, Clemente JC. The microbiome in early life: Implications for health outcomes. Nat Med. 2016;22(7):713–722. doi: 10.1038/nm.4142. [DOI] [PubMed] [Google Scholar]
- 7.Jiménez E, Marín ML, Martín R, et al. Is meconium from healthy newborns actually sterile? Res Microbiol. 2008;159(3):187–193. doi: 10.1016/j.resmic.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Satokari R, Grönroos T, Laitinen K, Salminen S, Isolauri E. Bifidobacterium and Lactobacillus DNA in the human placenta. Lett Appl Microbiol. 2009;48(1):8–12. doi: 10.1111/j.1472-765X.2008.02475.x. [DOI] [PubMed] [Google Scholar]
- 9.Morimoto S, Usui H, Kobayashi T, et al. Bacterial culture-negative subclinical intra-amniotic infection can be detected by bacterial 16S ribosomal DNA-amplifying polymerase chain reaction. Jpn J Infect Dis. April 2018. doi: 10.7883/yoken.JJID.2017.468. [DOI] [PubMed] [Google Scholar]
- 10.Tapiainen T, Paalanne N, Tejesvi MV, et al. Maternal influence on the fetal microbiome in a population-based study of the first-pass meconium. Pediatr Res. May 2018. doi: 10.1038/pr.2018.29. [DOI] [PubMed] [Google Scholar]
- 11.Jiménez E, Fernández L, Marín ML, et al. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr Microbiol. 2005;51(4):270–274. doi: 10.1007/s00284-005-0020-3. [DOI] [PubMed] [Google Scholar]
- 12.Kliman HJ. Comment on “the placenta harbors a unique microbiome”. Sci Transl Med. 2014;6(254):254le4. doi: 10.1126/scitranslmed.3009864. [DOI] [PubMed] [Google Scholar]
- 13.Tamburini S, Shen N, Wu HC, Clemente JC. The microbiome in early life: implications for health outcomes. Nat Med. 2016;22(7):713–722. doi: 10.1038/nm.4142. [DOI] [PubMed] [Google Scholar]
- 14.Martin R, Makino H, Cetinyurek Yavuz A, et al. Early-Life Events, Including Mode of Delivery and Type of Feeding, Siblings and Gender, Shape the Developing Gut Microbiota. PLoS One. 2016;11(6):e0158498. doi: 10.1371/journal.pone.0158498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freitas AC, Chaban B, Bocking A, et al. The vaginal microbiome of pregnant women is less rich and diverse, with lower prevalence of Mollicutes, compared to non-pregnant women. Sci Rep. 2017;7(1):9212. doi: 10.1038/s41598-017-07790-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romero R, Dey SK, Fisher SJ. Preterm labor: one syndrome, many causes. Science. 2014;345(6198):760–765. doi: 10.1126/science.1251816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koren O, Goodrich JK, Cullender TC, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150(3):470–480. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mueller NT, Bakacs E, Combellick J, Grigoryan Z, Dominguez-Bello MG. The infant microbiome development: mom matters. Trends Mol Med. 2015;21(2):109–117. doi: 10.1016/j.molmed.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biasucci G, Benenati B, Morelli L, Bessi E, Boehm G. Cesarean delivery may affect the early biodiversity of intestinal bacteria. J Nutr. 2008;138(9):1796S–1800S. doi: 10.1093/jn/138.9.1796S. [DOI] [PubMed] [Google Scholar]
- 21.Salminen S, Gibson GR, McCartney AL, Isolauri E. Influence of mode of delivery on gut microbiota composition in seven year old children. Gut. 2004;53(9):1388–1389. doi: 10.1136/gut.2004.041640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neu J, Rushing J. Cesarean versus vaginal delivery: long-term infant outcomes and the hygiene hypothesis. Clin Perinatol. 2011;38(2):321–331. doi: 10.1016/j.clp.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med. 2016;22(3):250–253. doi: 10.1038/nm.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferretti P, Pasolli E, Tett A, et al. Mother-to-Infant Microbial Transmission from Different Body Sites Shapes the Developing Infant Gut Microbiome. Cell Host Microbe. 2018;24(1):133–145.e5. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Navarro LA, French DL, Zauscher S. Synthesis of Modular Brush Polymer-Protein Hybrids Using Diazotransfer and Copper Click Chemistry. Bioconjug Chem. 2018;29(8):2594–2605. doi: 10.1021/acs.bioconjchem.8b00309. [DOI] [PubMed] [Google Scholar]
- 26.Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509(7500):357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pannaraj PS, Li F, Cerini C, et al. Association Between Breast Milk Bacterial Communities and Establishment and Development of the Infant Gut Microbiome. JAMA Pediatr. 2017;171(7):647–654. doi: 10.1001/jamapediatrics.2017.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khodayar-Pardo P, Mira-Pascual L, Collado MC, Martínez-Costa C. Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. J Perinatol. 2014;34(8):599–605. doi: 10.1038/jp.2014.47. [DOI] [PubMed] [Google Scholar]
- 29.Rudloff S, Kunz C. Milk oligosaccharides and metabolism in infants. Adv Nutr. 2012;3(3):398S–405S. doi: 10.3945/an.111.001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coppa GV, Bruni S, Morelli L, Soldi S, Gabrielli O. The first prebiotics in humans: human milk oligosaccharides. J Clin Gastroenterol. 2004;38(6 Suppl):S80–3. http://www.ncbi.nlm.nih.gov/pubmed/15220665. [DOI] [PubMed] [Google Scholar]
- 31.Sudo N, Sawamura S, Tanaka K, Aiba Y, Kubo C, Koga Y. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J Immunol. 1997;159(4):1739–1745. http://www.ncbi.nlm.nih.gov/pubmed/9257835. [PubMed] [Google Scholar]
- 32.Xiao L, Van’t Land B, Engen PA, et al. Human milk oligosaccharides protect against the development of autoimmune diabetes in NOD-mice. Sci Rep. 2018;8(1):3829. doi: 10.1038/s41598-018-22052-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamilton MK, Ronveaux CC, Rust BM, et al. Prebiotic milk oligosaccharides prevent development of obese phenotype, impairment of gut permeability, and microbial dysbiosis in high fat-fed mice. Am J Physiol Gastrointest Liver Physiol. 2017;312(5):G474–G487. doi: 10.1152/ajpgi.00427.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bokulich NA, Chung J, Battaglia T, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8(343):343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bezirtzoglou E, Tsiotsias A, Welling GW. Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe. 2011;17(6):478–482. doi: 10.1016/j.anaerobe.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 36.Gregory KE, Samuel BS, Houghteling P, et al. Influence of maternal breast milk ingestion on acquisition of the intestinal microbiome in preterm infants. Microbiome. 2016;4(1):68. doi: 10.1186/s40168-016-0214-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brugman S, Visser JTJ, Hillebrands JL, Bos NA, Rozing J. Prolonged exclusive breastfeeding reduces autoimmune diabetes incidence and increases regulatory T-cell frequency in bio-breeding diabetes-prone rats. Diabetes Metab Res Rev. 2009;25(4):380–387. doi: 10.1002/dmrr.953. [DOI] [PubMed] [Google Scholar]
- 38.Laursen MF, Bahl MI, Michaelsen KF, Licht TR. First Foods and Gut Microbes. Front Microbiol. 2017;8:356. doi: 10.3389/fmicb.2017.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bergström A, Skov TH, Bahl MI, et al. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl Environ Microbiol. 2014;80(9):2889–2900. doi: 10.1128/AEM.00342-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fallani M, Amarri S, Uusijarvi A, et al. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology. 2011;157(Pt 5):1385–1392. doi: 10.1099/mic.0.042143-0. [DOI] [PubMed] [Google Scholar]
- 41.Moussaoui N, Jacobs JP, Larauche M, et al. Chronic Early-life Stress in Rat Pups Alters Basal Corticosterone, Intestinal Permeability, and Fecal Microbiota at Weaning: Influence of Sex. J Neurogastroenterol Motil. 2017;23(1):135–143. doi: 10.5056/jnm16105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bailey MT, Coe CL. Maternal separation disrupts the integrity of the intestinal microflora in infant rhesus monkeys. Dev Psychobiol. 1999;35(2):146–155. http://www.ncbi.nlm.nih.gov/pubmed/10461128. [PubMed] [Google Scholar]
- 43.Tannock GW, Savage DC. Influences of dietary and environmental stress on microbial populations in the murine gastrointestinal tract. Infect Immun. 1974;9(3):591–598. http://www.ncbi.nlm.nih.gov/pubmed/4593471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Labus JS, Hollister EB, Jacobs J, et al. Differences in gut microbial composition correlate with regional brain volumes in irritable bowel syndrome. Microbiome. 2017;5(1):49. doi: 10.1186/s40168-017-0260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hicks LA, Taylor TH, Hunkler RJ. U.S. outpatient antibiotic prescribing, 2010. N Engl J Med. 2013;368(15):1461–1462. doi: 10.1056/NEJMc1212055. [DOI] [PubMed] [Google Scholar]
- 46.Shao X, Ding X, Wang B, et al. Antibiotic Exposure in Early Life Increases Risk of Childhood Obesity: A Systematic Review and Meta-Analysis. Front Endocrinol (Lausanne). 2017;8:170. doi: 10.3389/fendo.2017.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miyoshi J, Bobe AM, Miyoshi S, et al. Peripartum Antibiotics Promote Gut Dysbiosis, Loss of Immune Tolerance, and Inflammatory Bowel Disease in Genetically Prone Offspring. Cell Rep. 2017;20(2):491–504. doi: 10.1016/j.celrep.2017.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schulfer AF, Battaglia T, Alvarez Y, et al. Intergenerational transfer of antibiotic-perturbed microbiota enhances colitis in susceptible mice. Nat Microbiol. 2018;3(2):234–242. doi: 10.1038/s41564-017-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1(6):6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zinöcker MK, Lindseth IA. The Western Diet-Microbiome-Host Interaction and Its Role in Metabolic Disease. Nutrients. 2018;10(3). doi: 10.3390/nu10030365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gupta VK, Paul S, Dutta C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front Microbiol. 2017;8(JUN). doi: 10.3389/fmicb.2017.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh RK, Chang H-W, Yan D, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15(1):73. doi: 10.1186/s12967-017-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rivera C a., Adegboyega P, van Rooijen N, Tagalicud A, Allman M, Wallace M. Toll-like receptor-4 signaling and Kupffer cells play pivotal roles in the pathogenesis of non-alcoholic steatohepatitis. J Hepatol. 2007;47(4):571–579. doi: 10.1016/j.jhep.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pendyala S, Walker JM, Holt PR. A high-fat diet is associated with endotoxemia that originates from the gut. Gastroenterology. 2012;142(5):1100–1101.e2. doi: 10.1053/j.gastro.2012.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thaiss CA, Itav S, Rothschild D, et al. Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature. November 2016. doi: 10.1038/nature20796. [DOI] [PubMed] [Google Scholar]
- 57.Willett WC, Sacks F, Trichopoulou A, et al. Mediterranean diet pyramid: a cultural model for healthy eating. Am J Clin Nutr. 1995;61(6 Suppl):1402S–1406S. doi: 10.1093/ajcn/61.6.1402S. [DOI] [PubMed] [Google Scholar]
- 58.De Filippis F, Pellegrini N, Vannini L, et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut. 2016;65(11):1812–1821. doi: 10.1136/gutjnl-2015-309957. [DOI] [PubMed] [Google Scholar]
- 59.Velasquez MT, Ramezani A, Manal A, Raj DS. Trimethylamine N-Oxide: The Good, the Bad and the Unknown. Toxins (Basel). 2016;8(11). doi: 10.3390/toxins8110326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koloverou E, Panagiotakos DB, Pitsavos C, et al. Adherence to Mediterranean diet and 10-year incidence (2002–2012) of diabetes: correlations with inflammatory and oxidative stress biomarkers in the ATTICA cohort study. Diabetes Metab Res Rev. 2016;32(1):73–81. doi: 10.1002/dmrr.2672. [DOI] [PubMed] [Google Scholar]
- 61.Clarke SF, Murphy EF, O’Sullivan O, et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut. 2014;63(12):1913–1920. doi: 10.1136/gutjnl-2013-306541. [DOI] [PubMed] [Google Scholar]
- 62.Cotillard A, Kennedy SP, Kong LC, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500(7464):585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 63.Świątecka D, Dominika Ś, Narbad A, et al. The study on the impact of glycated pea proteins on human intestinal bacteria. Int J Food Microbiol. 2011;145(1):267–272. doi: 10.1016/j.ijfoodmicro.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 64.Jantchou P, Morois S, Clavel-Chapelon F, Boutron-Ruault M-C, Carbonnel F. Animal protein intake and risk of inflammatory bowel disease: The E3N prospective study. Am J Gastroenterol. 2010;105(10):2195–2201. doi: 10.1038/ajg.2010.192. [DOI] [PubMed] [Google Scholar]
- 65.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Russell WR, Gratz SW, Duncan SH, et al. High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am J Clin Nutr. 2011;93(5):1062–1072. doi: 10.3945/ajcn.110.002188. [DOI] [PubMed] [Google Scholar]
- 67.Hughes R, Magee EA, Bingham S. Protein degradation in the large intestine: relevance to colorectal cancer. Curr Issues Intest Microbiol. 2000;1(2):51–58. http://www.ncbi.nlm.nih.gov/pubmed/11709869. [PubMed] [Google Scholar]
- 68.Llewellyn SR, Britton GJ, Contijoch EJ, et al. Interactions Between Diet and the Intestinal Microbiota Alter Intestinal Permeability and Colitis Severity in Mice. Gastroenterology. 2018;154(4):1037–1046.e2. doi: 10.1053/j.gastro.2017.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kris-Etherton PM, Harris WS, Appel LJ, American Heart Association. Nutrition Committee. Fish consumption, fish oil, omega-3 fatty acids, and cardiovascular disease. Circulation. 2002;106(21):2747–2757. http://www.ncbi.nlm.nih.gov/pubmed/12438303. [DOI] [PubMed] [Google Scholar]
- 70.Fava F, Gitau R, Griffin BA, Gibson GR, Tuohy KM, Lovegrove JA. The type and quantity of dietary fat and carbohydrate alter faecal microbiome and short-chain fatty acid excretion in a metabolic syndrome “at-risk” population. Int J Obes (Lond). 2013;37(2):216–223. doi: 10.1038/ijo.2012.33. [DOI] [PubMed] [Google Scholar]
- 71.Menni C, Zierer J, Pallister T, et al. Omega-3 fatty acids correlate with gut microbiome diversity and production of N-carbamylglutamate in middle aged and elderly women. Sci Rep. 2017;7(1):11079. doi: 10.1038/s41598-017-10382-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Caesar R, Tremaroli V, Kovatcheva-Datchary P, Cani PD, Bäckhed F. Crosstalk between Gut Microbiota and Dietary Lipids Aggravates WAT Inflammation through TLR Signaling. Cell Metab. 2015;22(4):658–668. doi: 10.1016/j.cmet.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leone V, Gibbons SM, Martinez K, et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. 2015;17(5):681–689. doi: 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zarrinpar A, Chaix A, Yooseph S, Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 2014;20(6):1006–1017. doi: 10.1016/j.cmet.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thaiss CA, Levy M, Korem T, et al. Microbiota Diurnal Rhythmicity Programs Host Transcriptome Oscillations. Cell. 2016;167(6):1495–1510.e12. doi: 10.1016/j.cell.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 76.NCHS. NATIONAL CENTER FOR HEALTH STATISTICS NCHS Fact Sheet | March 2017 NCHS Nutrition Data. 2017;(March):1–2. https://www.cdc.gov/nchs/data/factsheets/factsheet_nutrition.pdf.
- 77.Makki K, Deehan EC, Walter J, Bäckhed F. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe. 2018;23(6):705–715. doi: 10.1016/j.chom.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 78.Murphy N, Norat T, Ferrari P, et al. Dietary fibre intake and risks of cancers of the colon and rectum in the European prospective investigation into cancer and nutrition (EPIC). PLoS One. 2012;7(6):e39361. doi: 10.1371/journal.pone.0039361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bodinham CL, Smith L, Wright J, Frost GS, Robertson MD. Dietary fibre improves first-phase insulin secretion in overweight individuals. PLoS One. 2012;7(7):e40834. doi: 10.1371/journal.pone.0040834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Conlon MA, Bird AR. The impact of diet and lifestyle on gut microbiota and human health. Nutrients. 2014;7(1):17–44. doi: 10.3390/nu7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fung KYC, Cosgrove L, Lockett T, Head R, Topping DL. A review of the potential mechanisms for the lowering of colorectal oncogenesis by butyrate. Br J Nutr. 2012;108(5):820–831. doi: 10.1017/S0007114512001948. [DOI] [PubMed] [Google Scholar]
- 82.Zou J, Chassaing B, Singh V, et al. Fiber-Mediated Nourishment of Gut Microbiota Protects against Diet-Induced Obesity by Restoring IL-22-Mediated Colonic Health. Cell Host Microbe. 2018;23(1):41–53.e4. doi: 10.1016/j.chom.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sonnenburg ED, Sonnenburg JL. Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab. 2014;20(5):779–786. doi: 10.1016/j.cmet.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108 Suppl:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Francino MP. Antibiotics and the Human Gut Microbiome: Dysbioses and Accumulation of Resistances. Front Microbiol. 2015;6:1543. doi: 10.3389/fmicb.2015.01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jakobsson HE, Jernberg C, Andersson AF, Sjölund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010;5(3):e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Löfmark S, Jernberg C, Jansson JK, Edlund C. Clindamycin-induced enrichment and long-term persistence of resistant Bacteroides spp. and resistance genes. J Antimicrob Chemother. 2006;58(6):1160–1167. doi: 10.1093/jac/dkl420. [DOI] [PubMed] [Google Scholar]
- 88.Theriot CM, Koenigsknecht MJ, Carlson PE, et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gianotti RJ, Moss AC. Fecal Microbiota Transplantation: From Clostridium difficile to Inflammatory Bowel Disease. Gastroenterol Hepatol (N Y). 2017;13(4):209–213. http://www.ncbi.nlm.nih.gov/pubmed/28546791. [PMC free article] [PubMed] [Google Scholar]
- 90.van Nood E, Vrieze A, Nieuwdorp M, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368(5):407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 91.Rotman SR, Bishop TF. Proton pump inhibitor use in the U.S. ambulatory setting, 2002–2009. PLoS One. 2013;8(2):e56060. doi: 10.1371/journal.pone.0056060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Medical Expenditure Panel Survey (MEPS). Content last reviewed April 2018. Agency for Healthcare Research and Quality, Rockville, MD: http://www.ahrq.gov/research/data/meps/index.html. [Google Scholar]
- 93.Lombardo L, Foti M, Ruggia O, Chiecchio A. Increased incidence of small intestinal bacterial overgrowth during proton pump inhibitor therapy. Clin Gastroenterol Hepatol. 2010;8(6):504–508. doi: 10.1016/j.cgh.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 94.Wu H, Esteve E, Tremaroli V, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017;23(7):850–858. doi: 10.1038/nm.4345. [DOI] [PubMed] [Google Scholar]
- 95.Maier L, Pruteanu M, Kuhn M, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555(7698):623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Song SJ, Lauber C, Costello EK, et al. Cohabiting family members share microbiota with one another and with their dogs. Elife. 2013;2:e00458. doi: 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fujimura KE, Slusher NA, Cabana MD, Lynch SV. Role of the gut microbiota in defining human health. Expert Rev Anti Infect Ther. 2010;8(4):435–454. doi: 10.1586/eri.10.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pelucchi C, Galeone C, Bach J-F, La Vecchia C, Chatenoud L. Pet exposure and risk of atopic dermatitis at the pediatric age: a meta-analysis of birth cohort studies. J Allergy Clin Immunol. 2013;132(3):616–622.e7. doi: 10.1016/j.jaci.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 99.Wegienka G, Zoratti E, Johnson CC. The role of the early-life environment in the development of allergic disease. Immunol Allergy Clin North Am. 2015;35(1):1–17. doi: 10.1016/j.iac.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Debarry J, Garn H, Hanuszkiewicz A, et al. Acinetobacter lwoffii and Lactococcus lactis strains isolated from farm cowsheds possess strong allergy-protective properties. J Allergy Clin Immunol. 2007;119(6):1514–1521. doi: 10.1016/j.jaci.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 101.Azad MB, Konya T, Maughan H, et al. Infant gut microbiota and the hygiene hypothesis of allergic disease: impact of household pets and siblings on microbiota composition and diversity. Allergy Asthma Clin Immunol. 2013;9(1):15. doi: 10.1186/1710-1492-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vogtmann E, Flores R, Yu G, et al. Association between tobacco use and the upper gastrointestinal microbiome among Chinese men. Cancer Causes Control. 2015;26(4):581–588. doi: 10.1007/s10552-015-0535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang H, Zhao J-X, Hu N, Ren J, Du M, Zhu M-J. Side-stream smoking reduces intestinal inflammation and increases expression of tight junction proteins. World J Gastroenterol. 2012;18(18):2180–2187. doi: 10.3748/wjg.v18.i18.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Biedermann L, Zeitz J, Mwinyi J, et al. Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS One. 2013;8(3):e59260. doi: 10.1371/journal.pone.0059260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Biedermann L, Brülisauer K, Zeitz J, et al. Smoking cessation alters intestinal microbiota: insights from quantitative investigations on human fecal samples using FISH. Inflamm Bowel Dis. 2014;20(9):1496–1501. doi: 10.1097/MIB.0000000000000129. [DOI] [PubMed] [Google Scholar]
- 106.Benjamin JL, Hedin CRH, Koutsoumpas A, et al. Smokers with active Crohn’s disease have a clinically relevant dysbiosis of the gastrointestinal microbiota. Inflamm Bowel Dis. 2012;18(6):1092–1100. doi: 10.1002/ibd.21864. [DOI] [PubMed] [Google Scholar]
- 107.Lucke K, Miehlke S, Jacobs E, Schuppler M. Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J Med Microbiol. 2006;55(Pt 5):617–624. doi: 10.1099/jmm.0.46198-0. [DOI] [PubMed] [Google Scholar]
- 108.Allais L, Kerckhof F-M, Verschuere S, et al. Chronic cigarette smoke exposure induces microbial and inflammatory shifts and mucin changes in the murine gut. Environ Microbiol. 2016;18(5):1352–1363. doi: 10.1111/1462-2920.12934. [DOI] [PubMed] [Google Scholar]
- 109.Tillisch K, Labus JS. Neuroimaging the microbiome-gut-brain axis. Adv Exp Med Biol. 2014;817:405–416. doi: 10.1007/978-1-4939-0897-4_18. [DOI] [PubMed] [Google Scholar]
- 110.Mohajeri MH, La Fata G, Steinert RE, Weber P. Relationship between the gut microbiome and brain function. Nutr Rev. 2018;76(7):481–496. doi: 10.1093/nutrit/nuy009. [DOI] [PubMed] [Google Scholar]
- 111.Tetel MJ, de Vries GJ, Melcangi RC, Panzica G, O’Mahony SM. Steroids, stress and the gut microbiome-brain axis. J Neuroendocrinol. 2018;30(2). doi: 10.1111/jne.12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Foster JA, Rinaman L, Cryan JF. Stress & the gut-brain axis: Regulation by the microbiome. Neurobiol Stress. 2017;7:124–136. doi: 10.1016/j.ynstr.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mayer EA, Hsiao EY. The Gut and Its Microbiome as Related to Central Nervous System Functioning and Psychological Well-being: Introduction to the Special Issue of Psychosomatic Medicine. Psychosom Med. 2017;79(8):844–846. doi: 10.1097/PSY.0000000000000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu H-N, Wu H, Chen Y-Z, Chen Y-J, Shen X-Z, Liu T-T. Altered molecular signature of intestinal microbiota in irritable bowel syndrome patients compared with healthy controls: A systematic review and meta-analysis. Dig Liver Dis. 2017;49(4):331–337. doi: 10.1016/j.dld.2017.01.142. [DOI] [PubMed] [Google Scholar]
- 115.Rea K, Dinan TG, Cryan JF. The microbiome: A key regulator of stress and neuroinflammation. Neurobiol Stress. 2016;4:23–33. doi: 10.1016/j.ynstr.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 117.Maruvada P, Leone V, Kaplan LM, Chang EB. The Human Microbiome and Obesity: Moving beyond Associations. Cell Host Microbe. 2017;22(5):589–599. doi: 10.1016/j.chom.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 118.Tremaroli V, Karlsson F, Werling M, et al. Roux-en-Y Gastric Bypass and Vertical Banded Gastroplasty Induce Long-Term Changes on the Human Gut Microbiome Contributing to Fat Mass Regulation. Cell Metab. 2015;22(2):228–238. doi: 10.1016/j.cmet.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liou AP, Paziuk M, Luevano J-M, Machineni S, Turnbaugh PJ, Kaplan LM. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med. 2013;5(178):178ra41. doi: 10.1126/scitranslmed.3005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Parekh PJ, Balart LA, Johnson DA. The Influence of the Gut Microbiome on Obesity, Metabolic Syndrome and Gastrointestinal Disease. Clin Transl Gastroenterol. 2015;6:e91. doi: 10.1038/ctg.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ford AC, Lacy BE, Talley NJ. Irritable Bowel Syndrome. N Engl J Med. 2017;376(26):2566–2578. doi: 10.1056/NEJMra1607547. [DOI] [PubMed] [Google Scholar]
- 122.O’Mahony SM, Marchesi JR, Scully P, et al. Early life stress alters behavior, immunity, and microbiota in rats: implications for irritable bowel syndrome and psychiatric illnesses. Biol Psychiatry. 2009;65(3):263–267. doi: 10.1016/j.biopsych.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 123.Stern EK, Brenner DM. Gut Microbiota-Based Therapies for Irritable Bowel Syndrome. Clin Transl Gastroenterol. 2018;9(2):e134. doi: 10.1038/ctg.2018.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Heard E, Martienssen RA. Transgenerational epigenetic inheritance: myths and mechanisms. Cell. 2014;157(1):95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ranjan R, Rani A, Metwally A, McGee HS, Perkins DL. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem Biophys Res Commun. 2016;469(4):967–977. doi: 10.1016/j.bbrc.2015.12.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.de Muinck EJ, Trosvik P, Gilfillan GD, Hov JR, Sundaram AYM. A novel ultra high-throughput 16S rRNA gene amplicon sequencing library preparation method for the Illumina HiSeq platform. Microbiome. 2017;5(1):68. doi: 10.1186/s40168-017-0279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]