Abstract
Background:
Variants in the cardiomyocyte-specific RNA splicing factor RBM20 have been linked to familial cardiomyopathy but the causative genetic architecture and clinical consequences of this disease are incompletely defined.
Methods and Results:
To define the genetic architecture of RBM20 cardiomyopathy, we first established a database of RBM20 variants associated with cardiomyopathy and compared these to variants observed in the general population with respect to their location in the RBM20 coding transcript. We identified two regions significantly enriched for cardiomyopathy-associated variants in exons 9 and 11. We then assembled a registry of 74 patients with RBM20 variants from 8 institutions across the world (44 index cases and 30 from cascade testing). This RBM20 patient registry revealed highly prevalent family history of sudden cardiac death (51%) and cardiomyopathy (72%) among index cases, and a high prevalence of composite arrhythmias (including AF, NSVT, ICD discharge and sudden cardiac arrest, 43%). Patients harboring variants in cardiomyopathy-enriched regions identified by our variant database analysis were enriched for these findings. Further, these characteristics were more prevalent in the RBM20 registry than in large cohorts of patients with DCM and titin (TTNtv) cardiomyopathy, and not significantly different from a cohort of patients with Lamin A/C associated (LMNA) cardiomyopathy.
Conclusions:
Our data establish RBM20 cardiomyopathy as a highly penetrant and arrhythmogenic cardiomyopathy. These findings underline the importance of arrhythmia surveillance and family screening in this disease and represent the first step in defining the genetic architecture of RBM20 disease causality on a population level.
Background
Autosomal dominant variants in a subset of cardiomyocyte-expressed genes represent a pathologically significant contributor to the etiology of cardiomyopathies. Variants in RBM20 represent an unusually complex genetic etiology of cardiomyopathy in that RBM20 controls post-transcriptional splicing of many sarcomeric and calcium handling genes in the cardiomyocyte 1–3. Loss of function in RBM20 leads to missplicing of such genes as TTN, LDB3, CAMK2D, and RYR2 in humans and murine models, all of which are independently implicated either in the cause or molecular propagation of ventricular dysfunction or arrhythmia 1,3,4. In vivo, Rbm20 −/− and Rbm20+/− rats develop both dilated cardiomyopathy and ventricular arrhythmias, and most recently, data in isolated cardiomyocytes from Rbm20 −/− mice demonstrate pro-arrhythmic calcium release from the sarcoplasmic reticulum 4,5.
As such, the study of RBM20 cardiomyopathy stands at the center of our understanding of disrupted alternative transcriptional splicing in the contractile dysfunction and arrhythmogenesis of heart failure. A small number of pathogenic variants in RBM20 have been reported from kindred studies 6–9, the majority of which lie within regions of RBM20 critical to spliceosome binding: the (RS-domain) and RNA recognition motif (RRM). However, a global understanding of variant pathogenicity across the coding transcript of this gene remains elusive. Further, though small cohort and kindred-based studies of RBM20 cardiomyopathy have been reported, a comprehensive clinical description of this patient population in the broader context of genetic cardiomyopathy has yet to be established.
Here, we compare the genetic distribution of clinically reported cardiomyopathy-associated RBM20 variants to RBM20 variants found in the general population, allowing us to define regions of the RBM20 transcript enriched for cardiomyopathy-associated variants, and therefore more prone to pathogenic variation. We go on to establish an international registry of patients harboring variants in RBM20 and characterize their clinical features. We compare this registry to large patient cohorts with dilated cardiomyopathy (DCM) and specifically cardiomyopathy due to truncating variants in TTN (TTNtv) and Lamin A/C (LMNA).
Methods
The data, analytic methods, and study materials will made available upon request to other researchers for purposes of reproducing the results or replicating the procedure.
RBM20 cardiomyopathy-associated variant database and comparison to population variant frequency
We compiled a database of 403 clinical genetic tests revealing variants in RBM20 from patients with cardiomyopathy. These variants were collated by the Laboratory for Molecular Medicine at Harvard Medical School and Invitae, Inc, and cross-referenced with reports in ClinVar (as of January 3, 2017). Variants reported by these testing providers in ClinVar were not double-counted. Synonymous coding variants and non-coding variants were eliminated from analysis to focus on those variants with the greatest potential impact on protein function. 171 missense, insertion/deletion and nonsense variants were included. The position of insertion/deletion and nonsense variants was taken as the position at the start of the insertion/deletion, and the position of termination for nonsense variants respectively. Variants were included regardless of their classification by the reporting institution (e.g., as benign, VUS or pathogenic). A full list of included variants is supplied in Supplemental Table 1.
Comparison of this cardiomyopathy-associated RBM20 variant database to general population variation in RBM20 was achieved by comparison to variants reported in 123,136 exomes and 15,496 genomes in the Genome Aggregation Database (gnomAD). The gnomAD database was used instead of the Exome Aggregation Consortium (ExAC) because mean coverage of RBM20 is greater in gnomAD (~40X) compared to ExAC (<10X) 10. We limited this analysis to the number of unique variants rather than patients (details included in Supplemental Methods). In all, we identified 171 unique cardiomyopathy-associated variants and 604 unique variants in the general population (Figure 1).
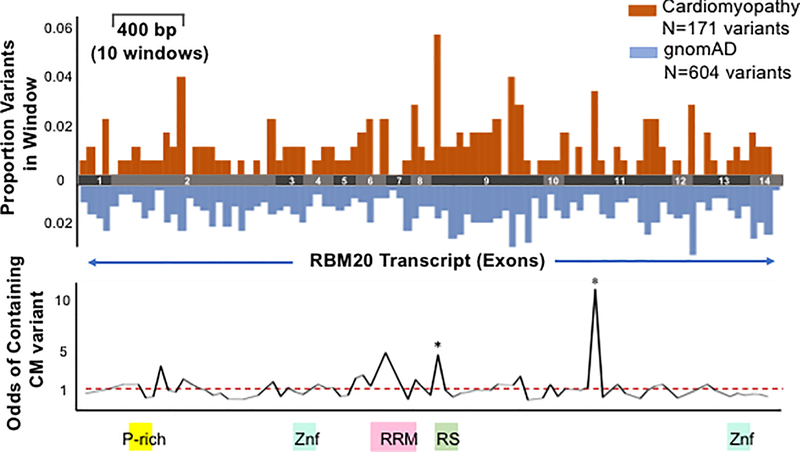
Figure 1. Regional likely pathogenic genetic variation in RBM20.
Comparison of regional frequencies of cardiomyopathy-associated variants found on genetic testing to those observed in the population (gnomAD) illuminates two regions enriched for cardiomyopathy-associated variants, and therefore more likely to contain pathogenic variation. The coding transcript of RBM20 is divided into ~100 windows of 40 bp each and the odds of cardiomyopathy-associated versus population variants lying within that window (versus any other window) are displayed. Orange bars denote percentage of cardiomyopathy-associated variants in each window, blue bars denote percentage of population variants per window. * denotes windows with statistically significant odds of containing cardiomyopathy-associated variants after adjustment for multiple testing. Red dashed line denotes OR of 1.
We divided the ~3700 bp coding transcript into windows of 40 bp each, creating 93 windows along the length of the transcript (for details of window size optimization, see Supplemental Methods). We then compared the fraction of unique variants within each of these windows (# unique variants falling in a window / # total unique variants in respective database) between the cardiomyopathy and general population groups. We then identified windows with high odds of containing cardiomyopathy-associated variants.
International RBM20 Patient Registry
An international registry of patients with RBM20 variants was assembled from pre-existing registries at 8 inherited cardiomyopathy centers (University of Sydney, Australia, University of Trieste, Italy, Johns Hopkins University, USA, University of Colorado, USA, Brigham and Women’s Hospital at Harvard University, USA, The Ohio State University, USA, University of Heidelberg, Germany and Stanford Center for Inherited Cardiovascular Disease, USA). Data were collected, stored and shared in a de-identified manner in accordance with the institutional review board (IRB) at each contributing center by retrospective chart review. (Please see Supplemental Methods for description of clinical characteristic definitions.) Where information could not be collected, it was marked as missing, and that patient was excluded from analysis for that variable.
The RBM20 patient registry was assembled independently of the RBM20 variant database described above, and the two datasets are not combined in our analyses. However, variants discovered in registry patients may have been included in the database due to the small number of cardiovascular genetic testing providers in the US and the ability of testing providers and care teams to post novel variants in ClinVar. These 74 patients were diagnosed independently by their respective centers of excellence for genetic cardiomyopathy, and therefore, specific sequencing tests used varied by center. In general, disease-specific panel-based testing was used for diagnosis in probands and variant specific sequencing was used for cascade screening.
Dilated Cardiomyopathy (DCM), Titin truncating variant (TTNtv), and LMNA cardiomyopathy cohorts
To better understand the clinical characteristics of RBM20 cardiomyopathy in the context of other causes of DCM, RBM20 registry data was compared with prevalence data from three large existing cohorts: one of all comers with DCM (N=633), one with TTNtv (N=83) cardiomyopathy patients,11 and one with LMNA cardiomyopathy patients (N=87)12 to serve as a comparison to known arrhythmogenic cardiomyopathy.
Statistical Analysis
RBM20 patient registry statistics were compared using the Fisher Exact test for discrete variables and Student’s T-test for continuous variables. Statistical significance for odds ratios in variant database analysis was determined using a Fisher Exact test due to sample size <100 per window and was corrected for multiple testing using a Benjamini-Hochberg false discovery rate (FDR) set at 20% 13–15. For multiple comparisons of clinical observations, a Bonferroni correction was used. These corrections for multiple testing were used separately for these respective purposes. All statistical comparisons were performed using Stata® 14.
Results
Comparison of RBM20 sequence architecture between cardiomyopathy patients and the general population identifies high confidence regions of pathogenicity in exons 9 and 11
We identified two windows enriched for cardiomyopathy-associated variants compared to population variants after correction for multiple testing. These windows occurred at positions c.2721– 2760 (exon 11), and c.1881–1920 (exon 9, including the RS-domain), where the odds of inclusion of a cardiomyopathy-population associated variant were higher than that of a variant from the general population (Figure 1). The c.1601–1640 window (in exon 7) is the third most confidently ranked pathogenic window, and encodes a section of the RRM domain. It also encompasses the V535I variant reported by Li et al. in a single individual with a strong family history of DCM 7. The significance value of this region (p=0.025) did not meet our gene-wide multiple testing criterion for statistical significance, but this was the third most highly ranked window. A full list of windows ranked by statistical probability of enrichment with cardiomyopathy variants is included in Supplemental Table 2.
Together, these three windows encode two previously characterized functional domains of the RBM20 protein (the RS domain and the RRM, Figure 1), as well as an area of exon 11 where a familial variant has previously been identified, but for which no function has been identified 8. Only one of the 8 previously reported cardiomyopathy variants was not encompassed in the three mostly confidently predicted windows (R716Q), but was found to be in a window where population and cardiomyopathy associated variants are equally prevalent. On further examination, the analysis window in which the corresponding variant (c.2147G>A) lies is enriched both for cardiomyopathy associated and general population variants, indicating that it may represent an area of high tolerance to genetic variation and not a site for pathogenic variation (purple indicator arrow, Figure 2). Prior evidence for the potential pathogenicity of this variant includes a reporter assay for splicing of the PEVK segment of TTN in which this variant displayed altered splicing similar to variants in the RS domain. 16 This large family of 43 individuals was originally described in 20107, at which time, 17 of its members were noted to carry the c.2147G>A variant. 10 of 17 of these family members also had DCM. One family member with mild disease (sick sinus syndrome and mild DCM easily reverse with medical therapy) did not carry the variant. Therefore it was felt to segregate with disease (though with low penetrance), and the familial DCM was attributed to this variant in RBM20. Of note, 8 members of this family also carried a previously reported variant in LMNA 7,17, 3 of whom also had DCM, and one of whom died suddenly. Taken together, our population-level genetic data and this family-level data cast some doubt on the pathogenicity of this variant and highlight the utility of the integrated model incorporating population and patient variation.
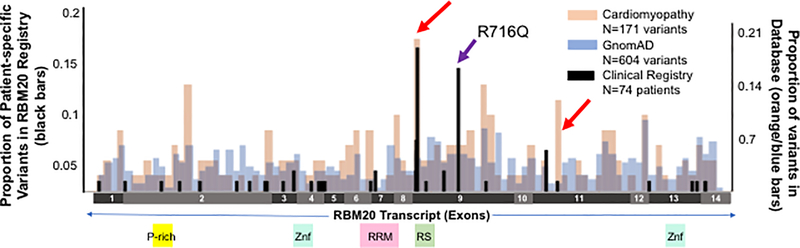
Figure 2. Distribution of variants observed in RBM20 registry patients.
Black bars represent proportion of RBM20 registry patients (N=74) with variants in each window (“patient-specific variants” left axis). Position for nonsense and insertion/deletion variants is represented as the first location of change in the transcript sequence. Orange and blue bars represent proportion of variants in each window in cardiomyopathy-associated and population (gnomAD) variant databases respectively (right axis). Red arrows indicate predicted pathogenic regions. Purple arrow indicates region containing previously reported p.R716Q variant lacking high level evidence for pathogenicity that lies outside highly predicted pathogenic regions.
Cardiomyopathy associated with RBM20 variants has an arrhythmogenic, highly penetrant phenotype
74 total patients are included in the RBM20 registry. 44 of these patients were index cases, while the remaining 30 patients were identified through cascade screening (Table 1). Patients in this registry harbored variants distributed across the length of the RBM20 transcript and included those inside (N=30) and outside (N=44) highly predicted windows in exons 9 and 11 from our variant database analysis (Figure 2).
Table 1.
Baseline Clinical Characteristics of the patients in the RBM20 registry
| All RBM20 Registry Cases (N=74) | Cases Carrying Variants in Predicted Windows (N=30) | Odds Ratio [CI] p-value | |
|---|---|---|---|
| Index Cases (vs. identified by cascade screening) (%) | 44/74 (59) | 14/30 (47) | 2.4 [0.9, 7.1] p=0.06 |
| Current Age (years, mean±SD) | 46±17 | 42±16 | p=0.17 |
| Age at diagnosis (years, mean±SD) (index cases only) | 37±15 40±15 |
34±14 40±12 |
p=0.14 p=0.98 |
| HCM phenotype (%) | 5/74 (7) | 0/30 (0) | 0.0 [0.0,1.1] p=0.07 |
| Family History SCD (%) (index cases only) | 22/43 (51) | 13/14 (93) | 11.4 [1.9, 117.2] p=0.002 |
| Family History DCM (%) (index cases only) | 31/43 (72) | 12/14 (86) | †p=0.06 |
| Any Arrhythmia (%) | 25/58 (43) | 16/24 (66) | 5.6 [1.6, 20.4] p=0.002 |
| NSVT (%) | 21/59 (36) | 14/26 (54) | 4.3 [1.2, 16.0] p=0.009 |
| Atrial Fibrillation (%) | 10/58 (17) | 7/23 (30) | 4.7 [0.9, 30.8] p=0.04 |
| SCA (%) | 5/60 (8) | 2/26 (8) | 0.9 [0.1, 8.2] p=1.00 |
| ICD implant (%) | 36/61 (59) | 15/25 (60) | 1.1 [0.3, 3.5] p=1.00 |
| Appropriate ICD discharge (%) (of patients with ICDs) | 9/32 (28) | 6/15 (40) | 2.9 [0.5, 21.9] p=0.25 |
| LVEF by Echocardiography (%) | 40±17 | 39±15 | p=0.67 |
| LVEDD (mm, mean±SD) | 56±12 | 55±13 | p=0.51 |
| Dilated (LVEDD>57mm) (%) | 33/69 (48) | 13/28 (46) | 0.9 [0.3, 2.7] p=1.00 |
| LVEF by CMR (%, mean±SD) | 45±17 (N=22) | 51±14 (N=7) | p=0.21 |
| LV Volume Index by CMR (ml/m2, mean±SD) | 117±50 (N=20) | 98±14 (N=7) | p=0.29 |
| RVEF by CMR (%, mean±SD) | 50±16 (N=11) | 53±9 (N=5) | p=0.52 |
| RV Volume Index by CMR (ml/m2, mean±SD) | 101±59 (N=11) | 90±22 (N=5) | p=0.60 |
| Delayed Gadolinium Enhancement (CMR) (%) | 11/22 (50) | 3/7 (42) | 0.7 [0.1, 5.6] p=0.64 |
| PR duration (ms, mean±SD) | 169±33 | 150±21 | p=0.0003 |
| QRS duration (ms, mean±SD) | 110±27 | 102±19 | p=0.09 |
| LVAD (%) | 1/74 (1) | 1/30 (3) | †p=0.45 |
| Transplant (%) | 5/74 (7) | 2/30 (7) | 1.0 [0.1, 9.1] p=0.97 |
| Death (%) | 3/74 (4) | 3/30 (10) | †p=0.06 |
HCM: Hypertrophic cardiomyopathy, DCM: Dilated Cardiomyopathy, SCD: Sudden cardiac death, SCA: sudden cardiac arrest, NSVT: Nonsustained Ventricular Tachycardia, LVEF: LV ejection fraction: LVEDD: LV end diastolic dimension, RVEF: RV ejection fraction, CMR: Cardiac Magnetic Resonance, LVAD: Left Ventricular Assist Device. Where sample size is less than stated registry size, this indicates lack of available data on a number of subjects.
indicates insufficient observations in a single group to accurately report odds ratio. Bolded p-vales indicate those that remain significant after correction for multiple comparisons (p-value<0.003).
We first examined the clinical characteristics of this registry as a whole (N=74, Table 1). Among index cases alone, was a high prevalence of family history of both cardiomyopathy and sudden cardiac death (72% and 51%, respectively). We also noted a young age at diagnosis among index cases (40±12 years). In the complete registry, there was also a high prevalence of sudden cardiac arrest (SCA, 8%), and appropriate implantable cardiac defibrillator (ICD) discharges among those patients with ICDs (28%). This was coupled with a high prevalence of nonsustained ventricular tachycardia (NSVT) and atrial fibrillation (AF) (36% and 17%, respectively). The mean PR interval was 169±33 ms (mean±SD) and 4 patients displayed first degree AV block (PR>220 ms, ranging between 230 ms and 260 ms). Mean left ventricular ejection fraction (LVEF) was 40±17% (mean±SD). Forty-eight percent of patients had an LV end diastolic dimension (LVEDD) greater that 5.7 cm. Cardiac magnetic resonance (CMR) imaging from the RBM20 registry reports delayed gadolinium enhancement in 11/22 patients (50%) who had undergone MRI. This represents a risk factor for arrhythmia in multiple types of cardiomyopathy 18–20. Five patients in the registry carried a diagnosis of hypertrophic (HCM) by chart review and 1 carried a diagnosis of left ventricular non-compaction in the absence of LV dilation or reduced LV systolic function (Table 1).
Arrhythmia and family history of sudden death are enriched in patients harboring RBM20 variants within high confidence likely-pathogenic regions
We sought to examine the clinical consequences of variation specific to the likely pathogenic regions identified from our population level analysis above. We hypothesized that patients harboring variants in the two statistically predicted pathogenic regions (c.2721– 2760 in exon 11 and c.1881–1920 in exon 9) would have higher prevalence of family history and arrhythmia outcomes compared to patients harboring variants outside of likely pathogenic regions. We identified 30 patients in the registry with variants lying inside these high confidence, likely pathogenic regions (high likelihood regions, Figure 2). We found that index patients with variants in high likelihood windows were more likely to have a family history of SCD (93% vs. 51% in the overall registry, p=0.002) than index patients with variants lying outside of these regions (Table 1). They were also more likely to personally have an arrhythmia (66% vs. 43% p=0.002), in particular non-sustained ventricular tachycardia (NSVT, p=0.009) and atrial fibrillation (p=0.04), though these did not remain statistically significant after correction for multiple comparisons (Bonferroni corrected α = 0.002, Table 1). Lastly, patients with variants in high likelihood regions had significantly shorter PR intervals (150±21 ms vs. 169±33 ms, p=0.0003).
That five patients in this registry expressed a hypertrophic cardiomyopathy phenotype is uncharacteristic of prior reports of a dilated phenotype associated with RBM20 pathogenic variants 7,8,21 (Table 1). Therefore, we examined whether these patients’ variants were associated with high confidence likely pathogenic regions. None of these HCM patients harbored variants in the top three regions of the gene predicted to be pathogenic by our analysis, which also encompasses the RS and RRM domains (these variants occurred at residues 374, 429, 545, 888 and 1089).
RBM20 cardiomyopathy has increased penetrance and higher prevalence of arrhythmias compared to other causes of cardiomyopathy
To better understand the clinical characteristics of RBM20 cardiomyopathy in the context of other causes of DCM, RBM20 registry data was compared with prevalence data from two large existing cohorts. To compare RBM20 cardiomyopathy patients at the time of evaluation to other patients with DCM, we first compared RBM20 index cases (N=43) to a large cohort of all-comers with DCM (N=633) 11. Further, as RBM20 is thought to control TTN splicing, we also compared these patients to a cohort of TTNtv cardiomyopathy patients, specifically (N=83) 11.
We found family history of DCM and SCD to be more common among RBM20 registry index patients (N=43) than in the DCM cohort (N=633, p<0.0001 for both comparisons) and the TTNtv cardiomyopathy cohort (N=83, p<0.0001 for both comparisons) (Table 2). A combined metric of evidence of sustained ventricular arrhythmia (VA) (reported personal history of SCA or appropriate ICD discharge) in the RBM20 registry was more common than reported sustained VT in the DCM population and TTNtv cardiomyopathy cohort (RBM20: 20.0%, DCM 2.2%, TTNtv 1.2%, p<0.0001 for both comparisons, Table 2). NSVT was also more common in the RBM20 registry (RBM20: 36.0%, DCM 10.6% (p<0.0001), TTNtv 19.3% (p=0.03) Table 2). Atrial fibrillation prevalence was not different between RBM20 and DCM or TTNtv cardiomyopathy groups. The average age of patients in the DCM cohort (54±14 years) and TTNtv cohort (49±13 years) was significantly older than the RBM20 registry (40±15 years, p<0.0001 vs. DCM and p=0.0008 vs TTNtv cardiomyopathy, Table 2).
Table 2.
RBM20 cardiomyopathy is highly penetrant and arrhythmogenic cardiomyopathy
| RBM20 CM (Index cases) (N=43) | DCM (N=633) | Odds Ratio [CI] p value | TTNtv CM (N=83) | Odds Ratio [CI] p value | LMNA CM (N=87) | Odds Ratio [CI], p value | |
|---|---|---|---|---|---|---|---|
| Age at evaluation (yrs, mean±SD) | 40±15 | 54±14 | p<0.0001 | 49±14 | p=0.0008 | 43±14 | p=0.35 |
| Family History of DCM (%) (RBM20 Index cases only) | 31/43 (72) | 87/633 (13.7) | 16.0 [8.0, 32.8] p=2.4×10−16 | 26/83 (31) | 5.6 [2.5,12.7] p=1.5×10−5 | 46/87 (52.9) | 1.3 [1.0,1.8] p=0.04 |
| Family History of SCA (%) (RBM20 Index cases only) | 22/43 (51) | 95/633 (15) | 5.9 [3.1,11.2] p=1.3×10−7 | 12/83 (15) | 6.2 [2.6,14.5] 2.0×10−5 | 38/87 (44) | 1.4 [0.6,2.8], p=0.46 |
| Atrial Fibrillation (%) | 9/41 (22) | 140/633 (22.1) | 1.0 [0.5,2.1] p=0.90 | 23/83 (28) | 0.7 [0.3,1.8] p=0.52 | 37/87 (43) | 0.4 [0.2,0.9], p=0.03 |
| Evidence of Sustained VA (%)* | 10/40 (25) | 14/633 (2.2) | 14.7 [6.0,36.0] p=1.9×10−7 | 1/83 (1) | 27.3 [3.4,223.0] p=5×10−5 | 18/87 (21) | 1.2 [0.6,2.4], p=0.65 |
| NSVT (%) | 17/39 (43) | 67/633 (10.6) | 6.5 [3.3,12.9] p=5.1×10−7 | 16/83 (19) | 3.4 [1.4,7.5] p=0.008 | 18/87 (21) | 2.1 [1.3,6.7], p=0.01 |
CM = Cardiomyopathy, SCA = Sudden Cardiac Arrest, ICD = Implantable Cardiac Defibrillator, NSVT = Nonsustained Ventricular Tachycardia, DCM = Dilated Cardiomyopathy, LVEF = Left ventricular ejection fraction, LVEDD = Left ventricular end diastolic dimension, SD = standard deviation. VA= ventricular arrhythmia Data from DCM and TTNtv cohorts have been previously reported (8). Where N is indicated as less that total cohort for a given variable, data was not available for some individuals.
Defined as sustained VT or VF on monitoring for DCM, TTNtv and LMNA and as SCA and/or ICD discharge for RBM20. Bolded p-values remain statistically significant after correction multiple comparisons (p<.0027).
As we found evidence of increased ventricular arrhythmia in the RBM20 cardiomyopathy index cases, we also compared them with Lamin A/C (LMNA) cardiomyopathy patients from a recently published cohort 12. LMNA cardiomyopathy is an inherited dilated cardiomyopathy with a high burden of ventricular and atrial arrhythmias, observed even in the absence of significant LV remodeling 12,17. We found that, at initial evaluation, RBM20 cardiomyopathy displayed similar rates of evidence of ventricular arrhythmia (VA) compared to documented VA in the LMNA cohort. There were trends toward less atrial fibrillation in the RBM20 index patients (OR=0.4 [0.2,0.9], p=0.03), and increased NSVT (OR= 2.1[1.3,6.7], p=0.01), but these were not statistically significant after correction for multiple comparisons (Bonferroni corrected α= 0.0027, Table 2). Family history of SCD and DCM in index cases was also similar between RBM20 and LMNA cardiomyopathy index cases (Table 2). We additionally hypothesized that arrhythmia in RBM20 cardiomyopathy might occur in the absence of significant LV remodeling, as has been observed with LMNA cardiomyopathy12,22. Indeed, of 12 RBM20 cardiomyopathy patients with evidence of sustained ventricular arrhythmia (appropriate ICD discharge and/or SCA), 4 patients (33%) had an LVEF≥45% at the time of evaluation.
Discussion
Here, we provide a comprehensive view of genetic variation and clinical characteristics of RBM20 cardiomyopathy, defining it as a highly penetrant, arrhythmogenic cardiomyopathy. We have: (i) used population and clinical genetics data to define the regional architecture of RBM20 variant pathogenicity, confirming the RS domain as a pathogenic domain and identifying a new likely pathogenic region in exon 11, (ii) assembled an international registry of patients with variants in RBM20 to define the clinical characteristics of this disease, (iii) demonstrated that patients within the RBM20 registry harboring variants in predicted pathogenic regions have an increased prevalence of family history of SCD, and increased personal history of arrhythmia compared to those with variants outside these regions and (iv) contextualized these findings in comparison to a large cohort of DCM patients, as well as TTNtv cardiomyopathies, finding a higher incidence of arrhythmias and more frequent family history of DCM and SCD in the RBM20 registry, similar to a large cohort of LMNA cardiomyopathy patients.
Identification of likely pathogenic regions of RBM20 highlights the power of population-level genetic data to illuminate structure-function relationships
We, and others, have previously demonstrated structure-function relationships in larger genes with frequent genetic variation 23,24. These analyses rely on the premise that areas of genes enriched with variants in diseased patients (compared to the general population) represent regions of low functional tolerance to genetic variants, and thus high probability of pathogenicity. Careful mechanistic validation of highly predicted pathogenic regions in genes of interest remains critical. Kindred-based studies of RBM20 variants in the US and Europe have harnessed the disease-modeling power of induced pluripotent stem cell derived cardiomyocytes to better understand the downstream effects of familial RBM20 variants on RNA splicing, myocyte contractility and calcium cycling, specifically in the RS domain of exon 9, independently identified by our analysis as well 8,9,25–27. The transcript-wide pathogenicity data for RBM20 generated by our study adds to this body of work because it incorporates population-scale data, which can improve diagnostic power in rare disease, as well as increase our understanding of structure-function relationships at the protein level.
Clinical characteristics of RBM20 registry underline arrhythmia risk for patients with pathogenic variants in RBM20
Our data draw attention to unique clinical characteristics and risks for patients with RBM20 variants as compared to other patients with a personal or family history of DCM. They are supported by a recent report showing deranged calcium handling in the Rbm20−/− mouse, coupled with results from a smaller cohort of RBM20 cardiomyopathy patients with increased ventricular arrhythmias compared to TTNtv cardiomyopathy 5. Our findings underline the imperative of family screening in this group, especially as some of the ICD discharge and sudden cardiac arrest was observed prior to the onset of severe LV dysfunction. Though current guidelines in DCM point to severe and irrecoverable LV dysfunction or personal history of arrest as indications for ICD placement 28, a small number of studies have shown that some genetic cardiomyopathies carry a significantly greater risk of malignant arrhythmia than the general population with DCM. Patients with LMNA cardiomyopathy display a highly progressive disease with high rates of significant ventricular arrhythmia. This prompted a recent revision to the ACC/AHA/HRS guidelines with a Class IIa recommendation for ICD implant in patients with LMNA variants and two or more risk factors (nonmissense variant, male sex, EF <45% or NSVT) 12,22. As we establish here, RBM20 cardiomyopathy is more highly penetrant with increased arrhythmogenicity than TTNtv cardiomyopathy and DCM without a genetic diagnosis, with ventricular arrhythmia rates at evaluation not significantly different from an LMNA cardiomyopathy cohort.
As we find that RBM20 cardiomyopathy is more similar to LMNA cardiomyopathy with respect to these characteristics, we suggest that these patients should be viewed with clinical concern similar to other arrhythmogenic cardiomyopathies or to catecholaminergic polymorphic ventricular tachycardia (caused by RYR2, whose splicing is affected by RBM20), in particular with respect to their capacity for early life-threatening arrhythmia 29. Longitudinal data will be required to make specific recommendations regarding risk stratification for SCD in RBM20 cardiomyopathy beyond depressed EF alone, and this is clearly of urgent importance. At present, we would suggest that early and aggressive arrhythmia monitoring in patients carrying likely pathogenic or pathogenic variants in RBM20 with or without severe ventricular remodeling is critical to identify patients at risk for life threatening arrhythmia.
Study limitations
Our study, focused on rare disease, is limited by small sample size and the inter-relatedness of study subjects. However, we believe this population represents a significant proportion of patients known to harbor RBM20 variants internationally, as it represents a culmination of data from 8 major inherited cardiovascular disease centers in the world.
Conclusion
Taken together, our findings define the clinical syndrome of RBM20 cardiomyopathy as a highly penetrant, arrhythmogenic cardiomyopathy. We have prioritized discrete regions of the RBM20 transcript that are likely critical to the pathogenesis of RBM20 cardiomyopathy and defined the clinical characteristics of the largest reported multicenter international cohort of these patients to date, including prominent arrhythmogenesis and high penetrance of cardiomyopathy and sudden death. These findings lay the foundation for examination of longitudinal outcomes in this registry, investigation of the novel putative RBM20 functional domain in exon 11, and, more broadly, for defining the role of RBM20 and alternative splicing in the biology of heart failure and arrhythmia.
Supplementary Material
Clinical Impact
- What is New?
- Largest international description of the clinical features of RBM20 cardiomyopathy to date
- Draws attention to unique clinical characteristics and arrhythmic risk for patients with RBM20 variants as compared to other patients with a personal or family history of DCM
- First population-based regional analysis of variant-disease association across the length of the RBM20 transcript
- What are the clinical implications?
- Immediately clinically impactful information about regional variant pathogenicity can be applied to variants of uncertain significance in a clinical setting
- Implications for arrhythmia monitoring and family screening in patients with RBM20 variants thought likely to contribute to disease
Acknowledgements
We would like to acknowledge the generous participation of our patients in this research.
Sources of Funding
NIH R01HG009117–01 (EAA), NIH F32HL134233–02 (VNP), NIH R01 GM111873 (LS), a National Health and Medical Research Council (NHMRC) Practitioner Fellowships (#105915) (CS), National Heart Foundation of Australia Future Leader Fellowship (#100833) (JI), NIH UL1 TR001082, R01 HL69071, R01 116906, and in part by a Trans-Atlantic Network of Excellence grant from the Leducq Foundation (14-CVD 03) (LM).
Footnotes
Disclosures: VP: None. CC: IP, royalties - genome interpretation technology, Stockholder – Personalis Advisor/Consultant – Phosphorus, Gene Matters, Myokardia. CR: None. LL: None. JI: None. JG: None. KM:None. TA: None. FSH: None. SK: None. SG: None. MG: None. DS: None. MDH: None. AYI: None. RN: None. BF: None. MTW: minor ownership interest in Personalis REH: None. SC: None. LS: ownership interest in Sophia Genetics and Levitas. NKL: None. MRGT: Consulting for: Array Biopharma, GeneDx, and Sanofi/Genzyme. LM: None. MM: None. GS: None. CS:None. BM:None. DPJ: None. EA: Founder Personalis and DeepCell, Inc. and Advisor for SequenceBio and Genome Medical.
References
- 1.Li S, Guo W, Dewey CN, Greaser ML. Rbm20 regulates titin alternative splicing as a splicing repressor. Nucleic Acids Res. 2013;41:2659–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, Maatz H, Schulz H, Li S, Parrish AM, Dauksaite V, Vakeel P, Klaassen S, Gerull B, Thierfelder L, Regitz-Zagrosek V, Hacker TA, Saupe KW, Dec GW, Ellinor PT, MacRae CA, Spallek B, Fischer R, Perrot A, Özcelik C, Saar K, Hubner N, Gotthardt M. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18:766–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maatz H, Jens M, Liss M, Schafer S, Heinig M, Kirchner M, Adami E, Rintisch C, Dauksaite V, Radke MH, Selbach M, Barton PJR, Cook SA, Rajewsky N, Gotthardt M, Landthaler M, Hubner N. RNA-binding protein RBM20 represses splicing to orchestrate cardiac pre-mRNA processing. J Clin Invest. 2014;124:3419–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, Maatz H, Schulz H, Li S, Parrish AM, Dauksaite V, Vakeel P, Klaassen S, Gerull B, Thierfelder L, Regitz-Zagrosek V, Hacker TA, Saupe KW, Dec GW, Ellinor PT, MacRae CA, Spallek B, Fischer R, Perrot A, Özcelik C, Saar K, Hubner N, Gotthardt M. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18:766–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van den Hoogenhof MMG, Beqqali A, Amin AS, van der Made I, Aufiero S, Khan MAF, Schumacher CA, Jansweijer JA, van Spaendonck-Zwarts KY, Remme CA, Backs J, Verkerk AO, Baartscheer A, Pinto YM, Creemers EE. RBM20 Mutations Induce an Arrhythmogenic Dilated Cardiomyopathy Related to Disturbed Calcium Handling. Circulation. 2018;138:1330–1342 [DOI] [PubMed] [Google Scholar]
- 6.Brauch KM, Karst ML, Herron KJ, de Andrade M, Pellikka PA, Rodeheffer RJ, Michels VV, Olson TM. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J Am Coll Cardiol. 2009;54:930–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li D, Morales A, Gonzalez-Quintana J, Norton N, Siegfried JD, Hofmeyer M, Hershberger RE. Identification of novel mutations in RBM20 in patients with dilated cardiomyopathy. Clin Transl Sci. 2010;3:90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beqqali A, Bollen IAE, Rasmussen TB, van den Hoogenhof MM, van Deutekom HWM, Schafer S, Haas J, Meder B, Sørensen KE, van Oort RJ, Mogensen J, Hubner N, Creemers EE, van der Velden J, Pinto YM. A mutation in the glutamate-rich region of RNA-binding motif protein 20 causes dilated cardiomyopathy through missplicing of titin and impaired Frank-Starling mechanism. Cardiovasc Res. 2016;112:452–463. [DOI] [PubMed] [Google Scholar]
- 9.Streckfuss-Bömeke K, Tiburcy M, Fomin A, Luo X, Li W, Fischer C, Özcelik C, Perrot A, Sossalla S, Haas J, Vidal RO, Rebs S, Khadjeh S, Meder B, Bonn S, Linke WA, Zimmermann W-H, Hasenfuss G, Guan K. Severe DCM phenotype of patient harboring RBM20 mutation S635A can be modeled by patient-specific induced pluripotent stem cell-derived cardiomyocytes. J Mol Cell Cardiol. 2017;113:9–21. [DOI] [PubMed] [Google Scholar]
- 10.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won H-H, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG, Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tayal U, Newsome S, Buchan R, Whiffin N, Halliday B, Lota A, Roberts A, Baksi AJ, Voges I, Midwinter W, Wilk A, Govind R, Walsh R, Daubeney P, Jarman JWE, Baruah R, Frenneaux M, Barton PJ, Pennell D, Ware JS, Prasad SK, Cook SA. Phenotype and Clinical Outcomes of Titin Cardiomyopathy. J Am Coll Cardiol. 2017;70:2264–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar S, Baldinger SH, Gandjbakhch E, Maury P, Sellal J-M, Androulakis AFA, Waintraub X, Charron P, Rollin A, Richard P, Stevenson WG, Macintyre CJ, Ho CY, Thompson T, Vohra JK, Kalman JM, Zeppenfeld K, Sacher F, Tedrow UB, Lakdawala NK. Long-Term Arrhythmic and Nonarrhythmic Outcomes of Lamin A/C Mutation Carriers. J Am Coll Cardiol. 2016;68:2299–2307. [DOI] [PubMed] [Google Scholar]
- 13.Li J, Witten DM, Johnstone IM, Tibshirani R. Normalization, testing, and false discovery rate estimation for RNA-sequencing data. Biostatistics. 2012;13:523–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benjamini Y Quantitative Trait Loci Analysis Using the False Discovery Rate. Genetics. 2005;171:783–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abramovich F, Benjamini Y. False Discovery Rate. In: Wiley StatsRef: Statistics Reference Online. 2014. [Google Scholar]
- 16.Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, Maatz H, Schulz H, Li S, Parrish AM, Dauksaite V, Vakeel P, Klaassen S, Gerull B, Thierfelder L, Regitz-Zagrosek V, Hacker TA, Saupe KW, Dec GW, Ellinor PT, MacRae CA, Spallek B, Fischer R, Perrot A, Özcelik C, Saar K, Hubner N, Gotthardt M. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18:766–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parks SB, Kushner JD, Nauman D, Burgess D, Ludwigsen S, Peterson A, Li D, Jakobs P, Litt M, Porter CB, Rahko PS, Hershberger RE. Lamin A/C mutation analysis in a cohort of 324 unrelated patients with idiopathic or familial dilated cardiomyopathy. Am Heart J. 2008;156:161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halliday BP, Cleland JGF, Goldberger JJ, Prasad SK. Personalizing Risk Stratification for Sudden Death in Dilated Cardiomyopathy: The Past, Present, and Future. Circulation. 2017;136:215–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moon JCC, McKenna WJ, McCrohon JA, Elliott PM, Smith GC, Pennell DJ. Toward clinical risk assessment in hypertrophic cardiomyopathy with gadolinium cardiovascular magnetic resonance. J Am Coll Cardiol. 2003;41:1561–1567. [DOI] [PubMed] [Google Scholar]
- 20.Gulati A, Jabbour A, Ismail TF, Guha K, Khwaja J, Raza S, Morarji K, Brown TDH, Ismail NA, Dweck MR, Di Pietro E, Roughton M, Wage R, Daryani Y, O’Hanlon R, Sheppard MN, Alpendurada F, Lyon AR, Cook SA, Cowie MR, Assomull RG, Pennell DJ, Prasad SK. Association of fibrosis with mortality and sudden cardiac death in patients with nonischemic dilated cardiomyopathy. JAMA. 2013;309:896–908. [DOI] [PubMed] [Google Scholar]
- 21.Brauch KM, Karst ML, Herron KJ, de Andrade M, Pellikka PA, Rodeheffer RJ, Michels VV, Olson TM. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J Am Coll Cardiol. 2009;54:930–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Khatib SM, Stevenson WG, Ackerman MJ, Gillis AM, Bryant WJ, Hlatky MA, Callans DJ, Granger CB, Curtis AB, Hammill SC, Deal BJ, Joglar JA, Dickfeld T, Kay GN, Field ME, Matlock DD, Fonarow GC, Myerburg RJ, Page RL. 2017 AHA/ACC/HRS Guideline for Management of Patients With Ventricular Arrhythmias and the Prevention of Sudden Cardiac Death: Executive Summary: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Rhythm Society. Heart Rhythm. 2017;15: e190–e252. [DOI] [PubMed] [Google Scholar]
- 23.Deo RC. Alternative Splicing, Internal Promoter, Nonsense-Mediated Decay, or All ThreeCLINICAL PERSPECTIVE: Explaining the Distribution of Truncation Variants in Titin. Circ Cardiovasc Genet. 2016;9:419–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Homburger JR, Green EM, Caleshu C, Sunitha MS, Taylor RE, Ruppel KM, Metpally RPR, Colan SD, Michels M, Day SM, Olivotto I, Bustamante CD, Dewey FE, Ho CY, Spudich JA, Ashley EA. Multidimensional structure-function relationships in human β-cardiac myosin from population-scale genetic variation. Proc Natl Acad Sci U S A. 2016;113:6701–6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wyles SP, Hrstka SC, Reyes S, Terzic A, Olson TM, Nelson TJ. Pharmacological Modulation of Calcium Homeostasis in Familial Dilated Cardiomyopathy: An In Vitro Analysis From an RBM20 Patient-Derived iPSC Model. Clin Transl Sci. 2016;9:158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wyles SP, Li X, Hrstka SC, Reyes S, Oommen S, Beraldi R, Edwards J, Terzic A, Olson TM, Nelson TJ. Modeling structural and functional deficiencies of RBM20 familial dilated cardiomyopathy using human induced pluripotent stem cells. Hum Mol Genet. 2016;25:254–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sedaghat-Hamedani F, Haas J, Zhu F, Geier C, Kayvanpour E, Liss M, Lai A, Frese K, Pribe-Wolferts R, Amr A, Li DT, Samani OS, Carstensen A, Bordalo DM, Müller M, Fischer C, Shao J, Wang J, Nie M, Yuan L, Haßfeld S, Schwartz C, Zhou M, Zhou Z, Shu Y, Wang M, Huang K, Zeng Q, Cheng L, Fehlmann T, Ehlermann P, Keller A, Dieterich C, Streckfuß-Bömeke K, Liao Y, Gotthardt M, Katus HA, Meder B. Clinical genetics and outcome of left ventricular non-compaction cardiomyopathy. Eur Heart J. 2017;38:3449–3460. [DOI] [PubMed] [Google Scholar]
- 28.Yancy CW, Jessup M, Bozkurt B, Butler J, Casey DE. 2013 ACCF/AHA guideline for the management of heart failure: executive summary. Circulation. 2013; 128:1810–1852. [DOI] [PubMed] [Google Scholar]
- 29.Broendberg AK, Nielsen JC, Bjerre J, Pedersen LN, Kristensen J, Henriksen FL, Bundgaard H, Jensen HK. Nationwide experience of catecholaminergic polymorphic ventricular tachycardia caused by RyR2 mutations. Heart. 2017;103:901–909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.