Abstract
Hypoxia-inducible factor 2α (HIF2α) directly regulates a battery of genes essential for intestinal iron absorption. Interestingly, iron deficiency and overload disorders do not result in increased intestinal expression of glycolytic or angiogenic HIF2α target genes. Similarly, inflammatory and tumor foci can induce a distinct subset of HIF2α target genes in vivo. These observations indicate that different stimuli activate distinct subsets of HIF2α target genes via mechanisms that remain unclear. Here, we conducted a high-throughput siRNA-based screen to identify genes that regulate HIF2α's transcriptional activity on the promoter of the iron transporter gene divalent metal transporter-1 (DMT1). SMAD family member 3 (SMAD3) and SMAD4 were identified as potential transcriptional repressors. Further analysis revealed that SMAD4 signaling selectively represses iron-absorptive gene promoters but not the inflammatory or glycolytic HIF2α or HIF1α target genes. Moreover, the highly homologous SMAD2 did not alter HIF2α transcriptional activity. During iron deficiency, SMAD3 and SMAD4 expression was significantly decreased via proteasomal degradation, allowing for derepression of iron target genes. Several iron-regulatory genes contain a SMAD-binding element (SBE) in their proximal promoters; however, mutation of the putative SBE on the DMT1 promoter did not alter the repressive function of SMAD3 or SMAD4. Importantly, the transcription factor forkhead box protein A1 (FOXA1) was critical in SMAD4-induced DMT1 repression, and DNA binding of SMAD4 was essential for the repression of HIF2α activity, suggesting an indirect repressive mechanism through DNA binding. These results provide mechanistic clues to how HIF signaling can be regulated by different cellular cues.
Keywords: hypoxia, iron, iron metabolism, gene expression, SMAD transcription factor, DMT1, HIF, Iron, SMAD
Introduction
Hypoxia plays a fundamental role in the pathophysiology of common causes of mortality, including ischemic heart disease, stroke, cancer, chronic lung disease, and congestive heart failure (1). In cancer, hypoxia is considered a hallmark for solid tumors. In response to hypoxia, tumor cells activate genes that are critical in angiogenesis, cell survival, cell proliferation, and glucose metabolism (2–4). Hypoxia signaling is mediated by the central transcriptional factor hypoxia-inducible factor (HIF),3 a family of per–ARNT–Sim of basic helix–loop–helix proteins, which includes three isoforms HIF1α, HIF2α, and HIF3α (5–9). HIF2α signaling is activated rapidly and is important for tumor inflammation and colon cancer progression (10–13). In addition to hypoxic regulation of HIF2α, HIF2α is stabilized by low iron. In response to low iron, HIF2α regulates iron absorption (14). Several iron-absorptive genes are direct HIF2α target genes in vivo (15). However, iron deficiency and iron overload disorders do not up-regulate any hypoxic HIF2α target genes in the intestine. Similarly, inflammatory and tumor foci can induce a distinct subset of HIF2α targets genes that are not regulated by iron demand. These observations demonstrate that different HIF2α stimuli activate different subsets of HIF2α target genes.
Mothers against decapentaplegic homolog (SMAD) 3 and SMAD4 are ligand-stimulated transcription factors, which are similar to HIF2α, and play essential role in inflammation, colon cancer progression, and iron regulation (16–19). Bone morphogenetic protein (BMP) and transforming growth factor (TGF) β are canonical ligands that activate SMAD signaling. The TGFβ superfamily signals exert growth inhibition effect on normal epithelial cells, and the loss of function promotes tumorigenesis (20). Moreover, BMP signaling is essential in regulating the hepatic master iron-regulatory hormone hepcidin. Upon ligand binding to type I and type II, TGFβ and BMP receptors lead to phosphorylation of receptor-activated SMADs (Smad2 and Smad3) at conserved C-terminal Ser-Ser-Xaa-Ser motifs (19, 21). The receptor-activated SMADs partner with common SMAD (SMAD4), translocate to the nucleus and drive transcription (22–24). The structure of SMAD protein is highly homologous, consisting of an N-terminal Mad homology domain 1 (MH1), a linker region, and a C-terminal Mad homology domain 2 (MH2) (23). MH1 domain is DNA-binding domain, which facilitate the nucleus import, whereas MH2 domain is important for protein–protein binding (25, 26).
Using a high-throughput siRNA screen for genes that modulate HIF2α activity, SMAD3 and SMAD4 were identified as selective repressors for HIF2α iron-regulatory genes but not angiogenic and glycolytic genes (27). Our data demonstrate that SMAD3 and SMAD4 are iron-regulated transcription factors that are decreased following iron deficiency, leading to a derepression and optimization of HIF2α-dependent iron absorption. Moreover, it provides a mechanistic insight into how a single transcription factor can regulate different target genes depending on the upstream stimuli.
Results
SMAD4 was an essential repressor of HIF2α-dependent DMT1 activation
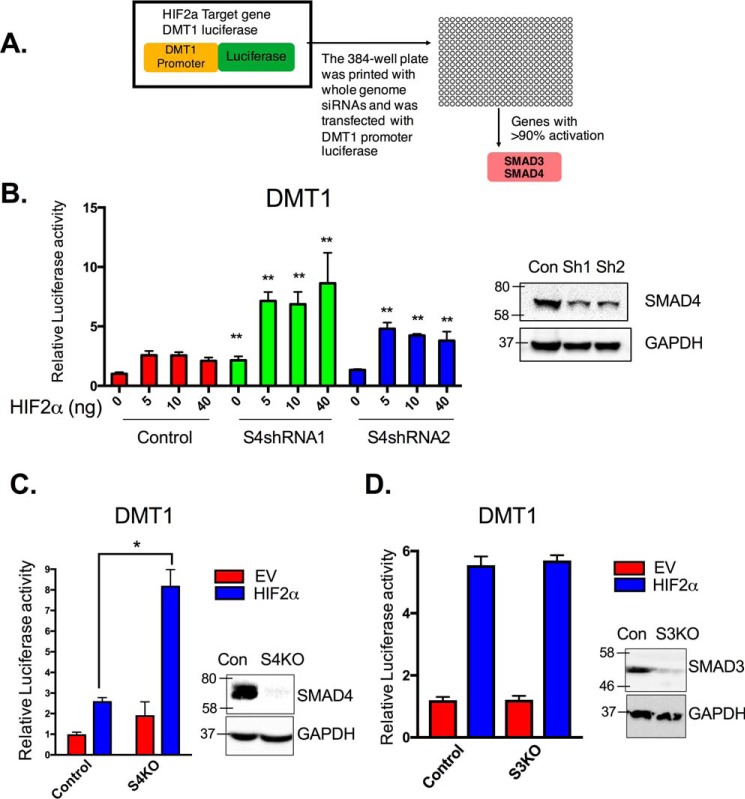
HIF2α modulators were assessed using a high-throughput siRNA screen for genes that regulate the Dmt1 promoter (a HIF2α-selective promoter) (15, 27). DMT1 has four isoforms because of the combination of two 5′ processing (transcribed from two distinct regulatory regions) and two 3′-UTR (presence or absence of an iron-response element (IRE)) variants: DMT1A, DMT1A-IRE, DMT1B, and DMT1B-IRE. Previous studies have shown that DMT1A is the most abundant isoform in the duodenum (28) and HIF2α specifically regulates DMT1A (± IRE), but not DMT1B (± IRE) (15). Therefore, to assess the role of SMADs in intestinal HIF2α regulation, the DMT1A promoter was utilized (a schematic representation and full promoter sequence are shown in Fig. S1). In brief, in HCT116 cells overexpressing HIF2α, DMT1 promoter luciferase activity was assessed using siRNA-based screen with a druggable target library (Fig. 1A). Previously we identified a battery of genes that were shown to be essential activators for HIF2α (27). In the present work, the data were assessed for genes that repressed HIF2α function. Through this analysis, we identified 37 genes that repressed HIF2α activity on the Dmt1 promoter (Table S1). HIF2α-induced DMT1 luciferase activity was significantly potentiated following siRNAs specific for SMAD3 or SMAD4 (Fig. 1A). To confirm the role of SMAD3 and SMAD4 in HIF2α regulation, SMAD4 stable knockdown in HCT116 cells were generated using shRNAs. SMAD4 protein level was confirmed with Western blotting analysis (Fig. 1B), and the cells were assessed using the Dmt1 promoter luciferase assay. Consistent with the siRNA screen, knockdown of SMAD4 potentiated HIF2α activity (Fig. 1B). These data were further confirmed using CRISPR/CAS9-mediated knockout of SMAD4 (Fig. 1C), but knockout of SMAD3 did not result in potentiation of HIF2α activity (Fig. 1D).
Figure 1.
SMAD4 is essential for the suppression of HIF2α iron target genes. A, schematic of the high-throughput siRNA screening assessing HIF2α-induced Dmt1 promoter luciferase assay in HCT116 cells. B, DMT1 promoter luciferase in HCT116 expressing scrambled shRNA (control, Con), or two different shRNAs for SMAD4 (S4shRNA1 or S4shRNA2) cell lines when transfected with different amounts of HIF2α overexpression construct. Right panel, SMAD4 Western blotting analysis. C and D, DMT1 promoter luciferase with either empty vector (EV) control or HIF2α-overexpressing construct (HIF2α) in HCT116 control cells or HCT116 SMAD4 knockout (C, S4KO) or SMAD3 knockout (D, S3KO) cell lines. The right panels are SMAD3 and SMAD4 Western blotting analysis. Luciferase data were normalized to β-galactosidase, and Western blots were normalized to GAPDH. *, p < 0.01; **, p < 0.001 compared with control or as indicated on the graph.
SMAD3 and SMAD4 were sufficient to selectively suppress HIF2α-dependent iron-regulatory promoters
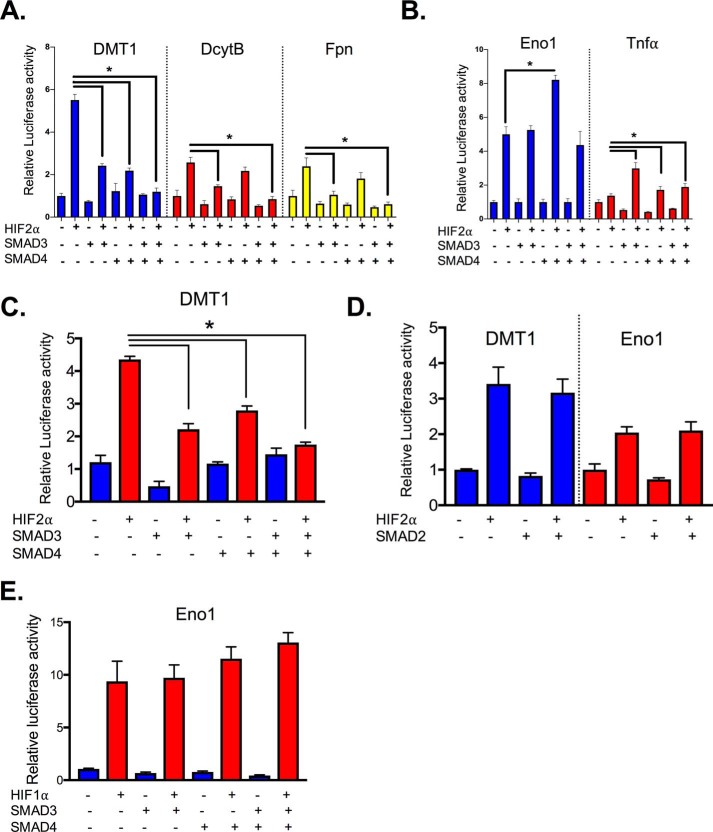
To further assess the role of SMADs, SMAD3 and SMAD4 were overexpressed, and HIF2α activity was evaluated. Dmt1, Dcytb, and Fpn promoter luciferase activities were significantly increased in HCT116 cells following HIF2α overexpression. SMAD3 and SMAD4, alone or combination, significantly inhibited HIF2α-induced activity (Fig. 2A). However, SMAD3 and SMAD4 exerted opposite effects on HIF2α glycolytic and inflammatory genes (Fig. 2B). Consistent with the data in HCT116 cells, the repressive function of SMAD3 and SMAD4 was also demonstrated in SW480 human colon cancer–derived cell line (Fig. 2C). Interestingly, the highly homologous SMAD2 did not repress HIF2α activity (Fig. 2D), and SMAD3 and SMAD4 did not alter HIF1α activity (Fig. 2E). Together, these data suggest that both SMAD3 and SMAD4 are sufficient to repress HIF2α activity; however, only SMAD4 is essential.
Figure 2.
SMAD3 and SMAD4 are sufficient and selective to decrease HIF2α transcriptional activity in iron-regulated target genes. A–C, HIF2α activity on the Dmt1, Dcytb, and Fpn (A), and Eno1 and Tnfa promoter luciferase in HCT116 (B), or SW480 cells transfected with SMAD3, SMAD4, and/or HIF2α (C). D, Dmt1 and Eno1 promoter luciferase in HCT116 cells transfected with SMAD2, and/or HIF-2α. E, Eno1 promoter luciferase in HCT116 cells transfected with SMAD3, SMAD4, and/or HIF1α. *, p < 0.01 compared as indicated on the graph.
Low-iron decreased SMAD3 and SMAD4 protein in vitro and in vivo
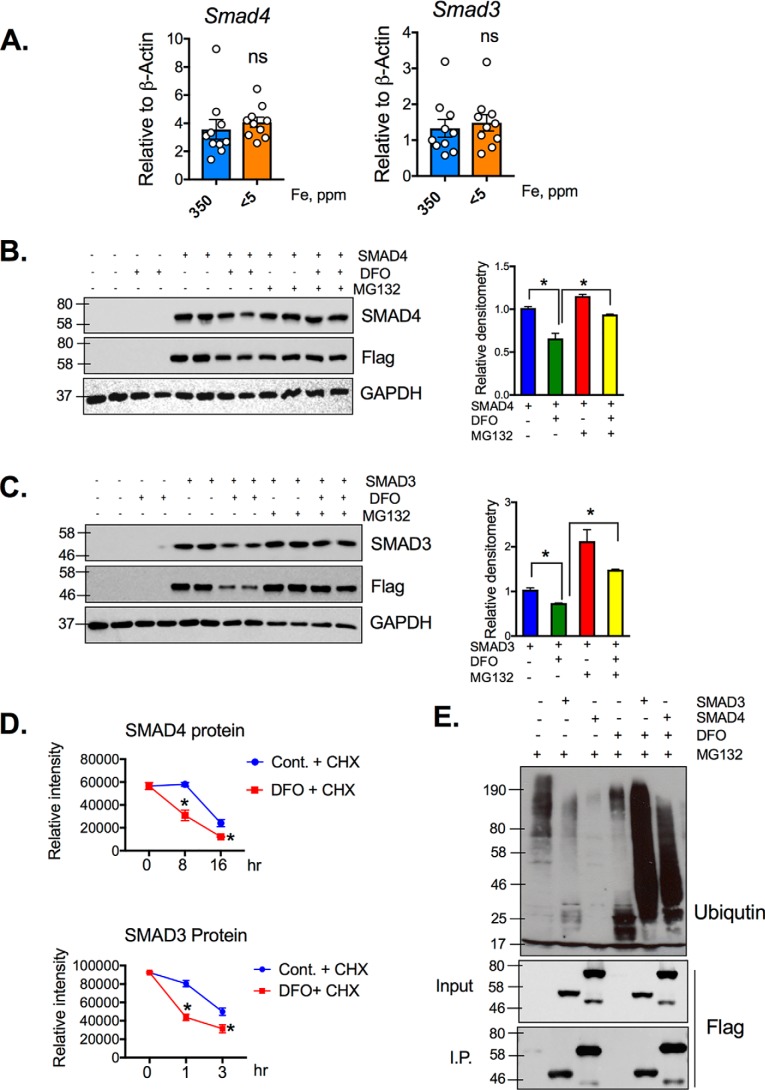
To understand whether SMAD signaling was integrated into cellular iron content, SMAD3 and SMAD4 levels were assessed following changes in cellular iron levels. Deferoxamine (DFO), an iron chelator, significantly decreased SMAD3 and SMAD4 protein levels (Fig. 3A). The iron chelation was confirmed by assessing iron storage protein ferritin (FtnH) expression. Interestingly, this decreased SMAD3 and SMAD4 protein levels were reversed once iron was restored by adding back ferric ammonium citrate (FAC) (Fig. 3A). In vivo experiments also confirmed the negative feedback regulation between iron and SMAD protein levels. The duodenum from the mice that were on 2 weeks of iron-enriched diet (350 ppm), iron-replete (35 ppm), or low-iron (<5 ppm) diet demonstrated a dose-dependent decrease of SMAD3 and SMAD4 levels (Fig. 3B). SMAD signaling is initiated via diverse BMP and TGFβ ligands; therefore it is unclear whether intestines have basal SMAD3 and SMAD4 signaling. To examine the distribution of SMAD3 and SMAD4, a cellular fractionation was performed. At the basal level, SMAD3 is present in cytosol, nucleus including both chromatin-bound and non–chromatin-bound fractions in HCT116 cells and mouse duodenal tissue (Fig. 3C), whereas SMAD4 was mainly localized to the nucleus and enriched in chromatin-associated fraction (Fig. 3C). These data demonstrate that SMAD3 and SMAD4 are responsive to the changes of cellular iron, and SMAD signaling is highly active in normal intestine.
Figure 3.
SMAD3 and SMAD4 protein expression is iron-regulated. A, Western blotting (left panel) and densitometric analysis (right panel) from HCT116 cells, treated with 100 μm FAC, 100 μm or 150 μm DFO for overnight, or 150 μm DFO for 16 h followed by 100 μm FAC for 8 h. B, Western blotting analysis from small intestines of WT mice (4–6 weeks old, males and females) on an iron-enriched (350 ppm), iron-replete (35 ppm), or low-iron (<5 ppm) diet for 1 week. C, SMAD3 and SMAD4 protein localization in HCT116 cells or mouse small intestines. *, p < 0.01; ***, p < 0.001 compared with control or as indicated on the graph.
Iron starvation degraded SMAD3 and SMAD4 via the proteasome
To understand the mechanism of decreased SMAD3 and SMAD4 protein levels following iron deprivation, gene expression was assessed in the small intestine from WT mice on iron-enriched or low-iron diet for 2 weeks. There was no difference in either SMAD3 or SMAD4 expression in the iron-deficient mice, compared with the mice fed with iron-enriched diet (Fig. 4A). To demonstrate that the loss of SMAD3 and SMAD4 upon iron deprivation was via post-transcriptional mechanisms, expression constructs of FLAG-tagged SMAD3 and SMAD4 were overexpressed in HCT116 cell line. The cells were treated with DFO, and SMAD3, SMAD4, or FLAG Western blots were performed. A decrease in SMAD3, SMAD4, and FLAG was observed following DFO treatment (Fig. 4, B and C). To investigate whether this was mediated by proteasomal degradation, the cells were also treated with the proteasomal inhibitor Mg132. Mg132 completely rescued SMAD3 and SMAD4 expression following DFO treatment (Fig. 4, B and C). SMAD3 and SMAD4 stability during iron deprivation were assessed by cycloheximide chase assays in DFO-treated or untreated HEK293T cells. DFO treatment reduced SMAD3 half-life from 3 h (29) to 1 h and that of SMAD4 from 16 h (30) to 8 h (Fig. 4D). To further study whether iron deprivation induced the proteasomal degradation of SMAD3 and SMAD4, an immunoprecipitation assay was performed in HCT116 cells transfected with FLAG-SMAD3 or FLAG-SMAD4 plasmid, followed by 1 h of Mg132 pretreatment and overnight DFO treatment. Following immunoprecipitation with anti-FLAG magnetic beads, ubiquitin Western blotting analysis showed dramatic increase of ubiquitin conjugation with SMAD3 and SMAD4 (Fig. 4E), suggesting that the low iron–induced decrease in SMAD3 and SAMD4 was through the ubiquitin proteasome pathway (Fig. 4E). These data illustrate that low-iron levels result in SMAD3 and SMAD4 protein degradation through ubiquitin proteasome pathway.
Figure 4.
Low-iron levels induce SMAD3 and SMAD4 proteasomal degradation. A, SMAD3 and SMAD4 gene expression in small intestine from WT mice on iron-enriched (350 ppm iron) or low-iron diet (<5 ppm iron). B and C, HCT116 cells transfected with SMAD4 plasmid (B) or SMAD3 (C) and pretreated with 10 μm Mg132 for 1 h followed by overnight DFO (150 μm) treatment. Quantification of respective Western blots is shown in the lower panels. D, HCT116 cells were treated with or without 150 μm DFO in presence of cycloheximide (CHX, 50 mg/ml), and stability of SMAD3 and SMAD4 protein was assessed by densitrometric analysis relative to GAPDH Western blots. E, HCT116 cells transfected with FLAG-SMAD3 or FLAG-SMAD4 plasmid and pretreated with 10 μm Mg132 for 1 h and then treated with 150 μm DFO for overnight, followed by immunoprecipitation with anti-FLAG magnetic beads. Ubiquitin and FLAG Western blotting analysis is shown. *, p < 0.05 compared as indicated on the graph. Cont., control; I.P., immunoprecipitation; ns, not significant.
SMAD3 and SMAD4 repressive function was not via phosphoactivation or direct protein–protein interaction
To examine the mechanism by which SMAD3 and SMAD4 repress HIF2α, direct protein–protein interaction was assessed. Co-immunoprecipitation was performed in HEK293T cells following transfection of HIF2α, SMAD3-FLAG, or SMAD4-FLAG or the co-transfection of HIF2α and SMAD3-FLAG or HIF2α and SMAD4-FLAG. Immunoprecipitation with FLAG and then HIF2α and FLAG immunoblotting were performed. HIF2α was not observed after FLAG pulldown, suggesting that there was no direct protein–protein interaction between HIF2α and SMAD3 or SMAD4 (Fig. 5A). Next, we assessed whether SMAD3 phosphorylation is required; three essential phosphoserine sites were mutated to alanine (S33A) (31). Interestingly, S33A still significantly suppressed HIF2α-induced DMT1 luciferase activity to the same extent as WT SMAD3. This indicated that the cross-talk between HIF2α and SMADs was not through SMAD3 activation (Fig. 5B). Interestingly, a consensus SBE (32) in the DMT1 promoter region was identified. To examine whether the suppression was through direct binding of SMAD3 or SMAD4 to DMT1 promoter, the consensus SBE sequence was deleted (DMT1ΔSBE). SMAD3 and SMAD4 were still able to suppress HIF2α-induced DMT1 activity (Fig. 5C). To further identify the region of SMAD-mediated repression, DMT1 promoter luciferase activity was performed in 5′-truncated mutants of the DMT1 promoter. Both SMAD3 and SMAD4 significantly suppressed HIF2α-induced DMT1 promoter activity that contained 0.2 kb of the proximal promoter. This suggests the possibility of novel interaction(s) with transcription factors that bind in this 0.2-kb region (Fig. 5D). Using publicly available databases JASPAR and GTRD, three overlapping transcription factor–binding sites in the 0.2-kb region were identified, namely FOXA1, KLF4, and Stat3. Upon deletion of these binding sites on DMT1 luciferase promoter (ΔFOXA1, ΔKLF4, and ΔStat3), ΔFOXA1 completely rescued the SMAD4-induced suppression (Fig. 5E). Taken together, SMAD3 and SMAD4 suppressive function on DMT1 activity was indirect and possibly through the regulation of FOXA1.
Figure 5.
SMAD3 and SMAD4 did not directly repress HIF2α transcriptional activity. A, co-immunoprecipitation for SMAD3 or SMAD4 and HIF2α in HEK-293T cells. B, Dmt1 promoter luciferase assay in HCT116 cells transfected with empty vector, SMAD3, SMAD3 phosphorylation site mutant (SMAD33A), and HIF2α. C, Dmt1 or Dmt1 SMAD-binding element deletion (DMT1ΔSBE) promoter luciferase in HCT116 cells transfected with SMAD3, SMAD4, and HIF2α. D, luciferase assay in HCT116 cells transfected with SMAD3, SMAD4, HIF2α, and truncated Dmt1 promoter constructs. E, Dmt1 promoter was mutated for binding sites of transcription factors FOXA1, KLF4, and Stat3 (ΔFOXA1, ΔKLF4, and ΔStat3); WT and mutant Dmt1 luciferase assay in HCT116 cells transfected with SMAD4, and HIF2α. *, p < 0.01 compared as indicated on the graph. IB, immunoblotting; IP, immunoprecipitation; ns, not significant.
SMAD4 DNA binding was critical for the suppression of HIF2α iron target genes
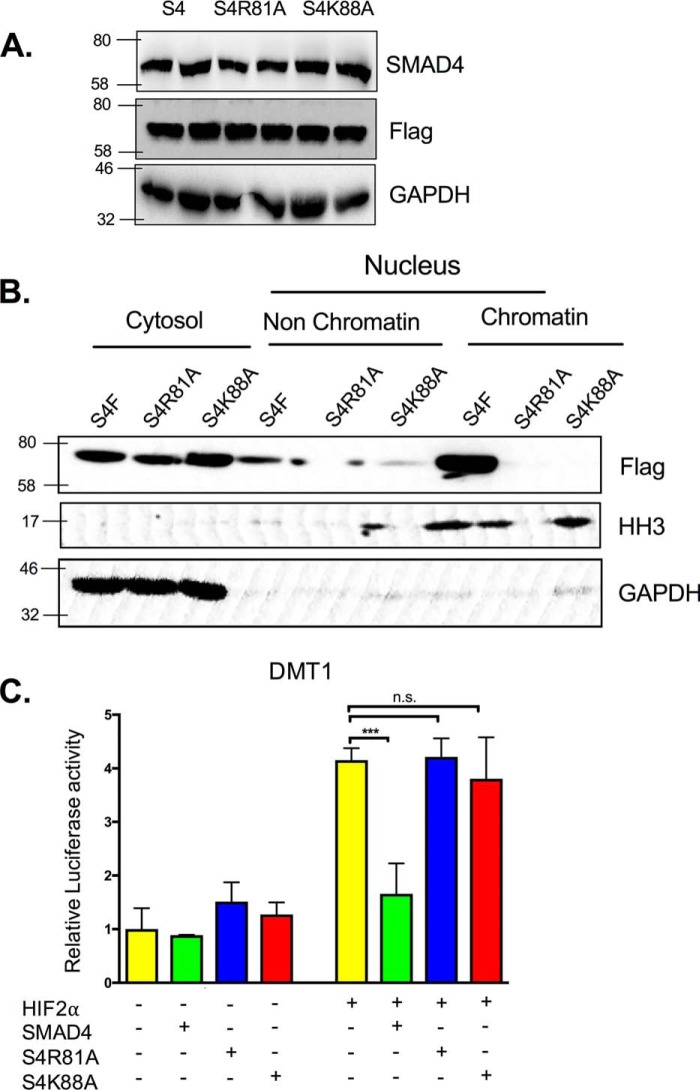
To understand whether SMAD4-mediated repression is indirect and is due to the activation of SMAD4-dependent target genes, DNA-binding mutants of SMAD4 were generated. Mutations of two critical residues (Arg-81 and Lys-88) in the MH1 dramatically reduce the ability of the SMAD4 to bind chromatin (33), which our data further confirmed (Fig. 6B). In addition, both mutations S4R81A and S4K88A did not alter SMAD4 expression (Fig. 6A). Strikingly, the suppression of DMT1 activity by SMAD4 was completely abolished by S4R81A and S4K88A mutants (Fig. 6C). These data indicate that the SMAD4 DNA binding is essential for the suppression of HIF2α iron target genes.
Figure 6.
SMAD4 DNA-binding domain is critical for the suppression of HIF2α iron target gene DMT1. A, HCT116 cells were transfected with WT SMAD4 or SMAD4 DNA-binding site mutants S4R81A and S4K88A. B, HCT116 cellular fractionations including cytosol, nucleus non–chromatin-bound protein, or nucleus chromatin-bound protein in cells transfected with WT SMAD4 or DNA-binding site mutants. C, Dmt1 promoter luciferase in HCT116 cells transfected with S4, S4R81A, S4K88A, and/or HIF2α. ***, p < 0.001 compared as indicated on the graph.
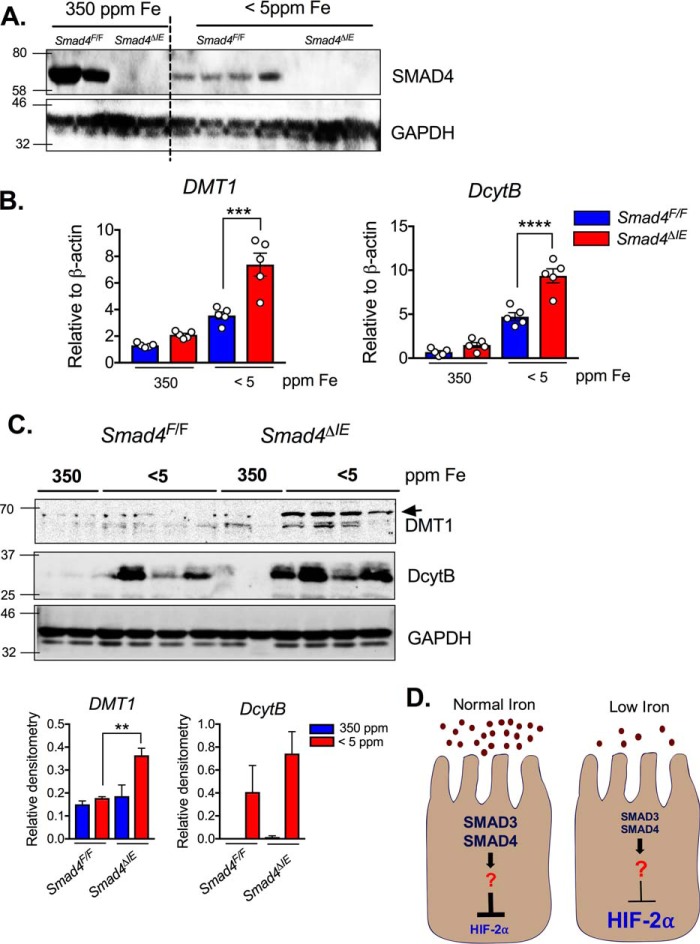
Intestine-specific SMAD4 knockout mice had increased expression of iron transporters
Mice with an intestinal SMAD4 deletion (Smad4ΔIE) were generated by crossing the Smad4F/F mice to the villin-Cre mice. SMAD4 Western blotting analysis showed successful SMAD4 disruption in the intestine of the Smad4ΔIE mice compared with the littermate controls (Smad4F/F) (Fig. 7A). Expression of two canonical HIF2α–iron target gene Dmt1 and Dcytb were unchanged in the Smad4ΔIE mice on an iron-enriched diet. However, Dmt1 and Dcytb were robustly potentiated following 1 week of low-iron diet (Fig. 7B). These data were consistent with protein-level changes, DMT1 was significantly elevated in the Smad4ΔIE mice, and DCYTB, although not significant, demonstrated an increase in expression compared with littermate controls (Fig. 7C).
Figure 7.
Intestine-specific SMAD4 knockout mice increase HIF2 iron target genes. A–C, SMAD4 Western blotting (A), gene expression analysis (B), and (C) DMT1 and DcytB Western blotting (C, upper panel) and quantification (C, lower panel) on small intestines from wild-type (Smad4F/F) and intestine-specific SMAD4 knockout (Smad4ΔIE) mice on iron-enriched (350 ppm) or low-iron diet (<5ppm) for 1 week. D, schematic representation of the SMAD3 and SMAD4 crosstalk to HIF2 signaling under normal iron (left panel) or low-iron environment (right panel). The left panel shows that constitutively expressed and highly active SMAD proteins restrict hyperinduction of iron-related genes by HIF2 under iron deficiency, whereas the right panel shows that decreased iron level results in SMAD protein degradation and a release of the molecular brake to increase HIF2 iron target gene. **, p <0.01; ***, p <0.001; ****, p <0.0001 compared as indicated on the graph.
Discussion
Intestinal HIF2α is a master regulator for iron absorption, glycolysis, angiogenesis, and inflammation (34–39). Interestingly, different HIF2α stimuli activate a distinct subset of HIF2α target genes. In the present manuscript, we provide a mechanism that is essential for the regulation of the iron response genes by HIF2α. We show that SMAD3 and SMAD4 are iron-regulated transcription factors that limit HIF2α transcriptional activity under normal cellular iron. Under low-iron conditions, SMAD3 and SMAD4 are degraded by the proteasome, leading to a derepression of HIF2α activity and an increase in iron-absorptive genes (Fig. 7D).
Our data demonstrate that SMAD3 and SMAD4 do not directly bind to HIF2α, and the consensus SBE is not required for SMAD3 or SMAD4 repression. Although it is possible that noncanonical SBEs are present in the proximal promoters of iron transporters, an in-depth search for cryptic SBEs still did not identify any additional interactions. Interestingly, a 0.2-kb DMT1 promoter luciferase construct was repressed by SMAD4, indicating a noncanonical and novel SMAD4 regulatory site. Data mining identified forkhead box protein A1 (FOXA1), which is known as a pioneer transcription factor and is essential in providing chromatin access to SMAD4 and its subsequent DNA binding (40). The deletion of FOXA1 on DMT1 promoter luciferase completely abolished SMAD4 induced DMT1 activity inhibition, suggesting a SMAD4–FOXA1–DMT1 interaction. Furthermore, the DNA-binding mutants of SMAD4 completely abolished the repressive activity of SMAD4. Taken together, these data suggest that SMAD signaling via indirect mechanisms and possibly through FOXA1 transcriptional activation of a repressor may lead to HIF2α inhibition. Further work is being done to identify the precise mechanism.
Our data indicated that SMAD3 and SMAD4 are constitutively expressed and highly active in tissues under basal iron conditions. Currently the mechanism for driving SMAD3 and SMAD4 activity in normal intestine is not known; however, BMP signaling is constitutively high in the villus. Moreover, other signals that are necessary for cell differentiation and intestinal stem cell homeostasis may cross-talk with SMAD3 and SMAD4 (41). Under low-iron diet, SMAD3 and SMAD4 are degraded via a proteasome-mediated mechanism. The best-characterized regulation of SMAD3 and SMAD4 signaling is via ligands such as TGFβ and BMPs. However, Smurf2 is a specific ubiquitin-protein isopeptide ligase specific for SMADs, and future work will focus on understanding whether the ligases are activated in low-iron conditions.
In conclusion our study reports evidence of a novel cross-talk between HIF2α and SMAD3 and SMAD4. The data demonstrate that this repressive mechanism is an important molecular switch to specifically shift HIF2α regulation to iron gene targets.
Experimental procedures
Animals and treatments
Intestine-specific SMAD4 knockout mice (SmadΔIE) were generated by crossing Smad4F/F (stock no. 017462, Jackson Laboratories) to the villin-cre mice, which have been described previously (10, 42). Smad4ΔIE mice, littermate controls (Smad4F/F), and WT C57BL/6 mice (6 weeks of age; both male and female) were maintained in standard cages in a light- and temperature-controlled room and were fed with iron-enriched (350 ppm iron; catalog no. 115180), iron-replete (35 ppm iron; catalog no. 115270), or low-iron (<5 ppm iron; catalog no. 115072) chow for 1–2 weeks (Dyets Inc., Bethlehem, PA). The mice had access to food and water ad libitum. All the animal studies were carried out in accordance with the Institute of Laboratory Animal Resources guidelines and approved by the University Committee on the Use and Care of Animals at the University of Michigan.
Cell lines and cell culture
Human colon cancer cell lines HCT116 and SW480 and human embryonic kidney cell line HEK293T were used. These cell lines express SMAD proteins basally and HIF2α-dependent iron-regulatory gene promoters are responsive in these cell lines. SMAD4shRNA are commercially available from Dharmacon (Lafayette, CO). HCT116 SMAD3 and SMAD4 knockout cell lines were generated by using CRISPR-Cas9 technology according to the Zhang lab protocol (43); the sgRNA sequences used for SMAD3 and SMAD4 are in Table 1. All cells were maintained in complete Dulbecco's modified Eagle's medium (supplemented with 10% fetal bovine serum and 1% antibiotic/antimycotic agent) at 37 °C in 5% CO2 and 21% O2.
Table 1.
Primer sequences
| Forward (5′ → 3′) | Reverse (5′ → 3′) | |
|---|---|---|
| Mouse quantitative PCR primers | ||
| β-actin | TGAAGCAGGCATCTGAGGG | CGAAGGTGGAAGAGTGGGAG |
| Dmt1 | TGTTTGATTGCATTGGGTCTG | CGCTCAGCAGGACTTTCGAG |
| DcytB | CATCCTCGCCATCATCTC | GGCATTGCCTCCATTTAGCTG |
| SMAD3 | AACGTGAACACCAAGTGCA | ACAGGCGGCAGTAGATAACG |
| SMAD4 | AGTTAAGCCACCTGCCCCTT | CACGTGAGCACAGTGCGTTTA |
| Cloning primers | ||
| SMAD4R81A | accagccacctgaagcgccccatccaatgttctc | gagaacattggatggggcgcttcaggtggctggt |
| SMAD4K88A | atcacatgaggaaatcctgcccgaccagccacctgaag | cttcaggtggctggtcgggcaggatttcctcatgtgat |
| DMT1ΔSBE | ctgcttccaactgtggtagctatttgcctaaggcaa | ttgccttaggcaaatagctaccacagttggaagcag |
| Dmt1ΔFOXA1 | tggctagagagaacacttaagctgagggaaaaatttcaaacc | ggtttgaaatttttccctcagcttaagtgttctctctagcca |
| Dmt1ΔKLF4 | ggtcaacactgagggaaaaatttcaatcaaagcataatcaaatttcatcttt | aaagatgaaatttgattatgctttgattgaaatttttccctcagtgttgacc |
| Dmt1 ΔStat3 | ttcaaaccgcaccaatcaaatttcatctttgaccggcc | ggccggtcaaagatgaaatttgattggtgcggtttgaa |
| Guide RNA | ||
| SMAD3 | TTCACGATCGGGGGAGTGAA | |
| SMAD4 | TTCTTCCTAAGGTTGCACAT | |
Luciferase assay
The cells were seeded into a 24-well plate at a cell density of 5 × 104 cells/well. The promoter luciferase constructs for divalent metal transporter 1 (Dmt1) (1 kb unless otherwise indicated as 0.4 or 0.2kb), duodenal cytochrome B (Dcytb), ferroportin (Fpn), enolase (eno1), or tumor necrosis factor (Tnf) α were previously described (15, 27, 42, 44). A 1-kb Dmt1 promoter luciferase construct was used as a template to mutate the binding sites for transcription factors FOXA1, KLF4, and Stat3 (ΔFOXA1, ΔKLF4, and ΔStat3) located in the 0.2-kb region. The primer sequences were shown in Table 1. The luciferase constructs were co-transfected with oxygen stable HIF2α, pCS2-FLAG-SMAD2 (catalog no. 14042, Addgene), pCS2-FLAG-SMAD3 (catalog no. 14052, Addgene), pcDNA3-FLAG-SMAD4 (catalog no. 80888, Addgene), pCS2-FLAG-SMAD3 mutation (SMAD33A) (three Ser-to-Ala mutations in the phosphorylation sites), SMAD4 DNA-binding site mutants (S4R81A and S4K88A), or empty vector into cells with polyethylenimine (Polysciences Inc., Warrington, PA). Cells were lysed in reporter lysis buffer (Promega, Madison, WI), and firefly luciferase activity was measured and normalized to β-galactosidase activity 48 h after transfection.
Western blotting analysis and immunoprecipitation
Western blotting analysis was performed as previously described (10, 45). Co-immunoprecipitation was carried out according to previous publications (10, 46). The blots were probed with antibodies against SMAD3 (catalog no. 9523, Cell Signaling Technology, Danvers, MA), SMAD4 (catalog no. 9101, Cell Signaling Technology), FtnH (catalog no. 3998, Cell Signaling Technology), HIF2α (catalog no. NB100–122, Novus Biologicals, Littleton, CO), GAPDH (sc-25778, Santa Cruz Biotechnology, Santa Cruz, CA), FLAG (catalog no. F1804, Sigma), and ubiquitin (catalog no. sc-8017, Santa Cruz Biotechnology) followed by incubation with appropriate horseradish peroxidase–conjugated secondary antibodies (Cell Signaling Technology). Immunoreactive protein species were detected using chemiluminescence methods.
SMAD3 and SMAD4 protein half-life
HCT116 cells were pretreated with cycloheximide (50 mg/ml) for 1 h and then treated with or without DFO (150 μm) for indicated time points. SMAD3 and SMAD4 half-lives were determined by relative densitometric analysis from SMAD3, SMAD4, and GAPDH Western blots.
Chromatin-associated protein isolation
Cytosol, nucleus, chromatin-bound, and non–chromatin-bound proteins were isolated from cell lines or mouse duodenal tissue using previously described methods (47). In brief, 1 × 107 to 2 × 107 cells were collected and washed with 2 ml of PBS. For tissues, mucosal duodenal scrapes were collected. The samples pellets were resuspended in 100–200 μl of buffer A (10 mm HEPES, pH 7.9, 10 mm KCl, 0.34 m sucrose, 10% glycerol, 1 mm DTT, 0.1% Triton X-100, 1.5 mm MgCl2, and protease inhibitor mixture). The pellet was homogenized with a glass Dounce homogenizer (20–30 strokes), then transferred to 1.5-ml centrifuge tubes, and incubated on ice for 8 min. The supernatant and nuclei pellet were separated by centrifugation (5 min at 1,300 × g in 4 °C). For the cytosolic fraction the supernatants were transferred to a new tube and spun at high speed (5 min at 20,000 × g in 4 °C). The nuclei were washed once in buffer A and processed in a Dounce homogenizer for an additional 20–30 times. Following centrifugation (5 min at 1,300 × g in 4 °C) the supernatants were discarded. The nuclei were lysed for 30 min in 50–80 μl of buffer B (3 mm EDTA, 0.2 mm EGTA, 1 mm DTT, and protease inhibitor mixture), and insoluble chromatin (chromatin bound protein) and soluble (non–chromatin-bound protein) fractions were separated by centrifugation (5 min, 1,700 × g, 4 °C). The pellet was washed with 0.5–1 ml of buffer B and resuspended in nucleus digestion buffer (radioimmune precipitation assay + phosphatase inhibitor + protease inhibitor mixture).
Quantitative real-time RT-PCR
RNA extraction, reverse transcription, and quantitative PCR were described previously (48, 49). The primers used in the study were listed in Table 1. The data were normalized to β-actin and expressed as fold difference from controls.
Statistical analysis
The results were expressed as means ± S.D. Significance between two groups were calculated by independent t test, and the significance among different groups was tested using one-way analysis of variance followed by Dunnett's post hoc comparisons.
Author contributions
X. M., M. P. V., and Y. M. S. conceptualization; X. M., N. K. D., C. C., A. G., A. O. P., M. P. V., and Y. M. S. formal analysis; X. M., N. K. D., M. P. V., and Y. M. S. investigation; X. M., N. K. D., C. C., and Y. M. S. methodology; X. M., N. K. D., and Y. M. S. writing-original draft; M. P. V. and Y. M. S. writing-review and editing; Y. M. S. project administration.
Supplementary Material
This work was supported by National Institutes of Health Grants CA148828 and DK095201 (to Y. M. S.) and CA190558 (to M. P. V.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1 and Fig. S1.
- HIF
- hypoxia-inducible factor
- SBE
- SMAD-binding element
- BMP
- bone morphogenetic protein
- TGF
- transforming growth factor
- MH
- Mad homology domain
- IRE
- iron-response element
- DFO
- deferoxamine
- FAC
- ferric ammonium citrate
- FOXA1
- forkhead box protein A1
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
References
- 1. Brahimi-Horn M. C., and Pouysségur J. (2007) Harnessing the hypoxia-inducible factor in cancer and ischemic disease. Biochem. Pharmacol. 73, 450–457 10.1016/j.bcp.2006.10.013 [DOI] [PubMed] [Google Scholar]
- 2. Bertout J. A., Patel S. A., and Simon M. C. (2008) The impact of O2 availability on human cancer. Nat. Rev. Cancer 8, 967–975 10.1038/nrc2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Höckel M., and Vaupel P. (2001) Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J. Natl. Cancer Inst. 93, 266–276 10.1093/jnci/93.4.266 [DOI] [PubMed] [Google Scholar]
- 4. Muz B., de la Puente P., Azab F., and Azab A. K. (2015) The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia 3, 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Semenza G. L., and Wang G. L. (1992) A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell Biol. 12, 5447–5454 10.1128/MCB.12.12.5447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tian H., McKnight S. L., and Russell D. W. (1997) Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 11, 72–82 10.1101/gad.11.1.72 [DOI] [PubMed] [Google Scholar]
- 7. Wang G. L., Jiang B. H., Rue E. A., and Semenza G. L. (1995) Hypoxia-inducible factor 1 is a basic–helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U.S.A. 92, 5510–5514 10.1073/pnas.92.12.5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang G. L., and Semenza G. L. (1993) Characterization of hypoxia-inducible factor 1 and regulation of DNA binding activity by hypoxia. J. Biol. Chem. 268, 21513–21518 [PubMed] [Google Scholar]
- 9. Wang G. L., and Semenza G. L. (1993) General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc. Natl. Acad. Sci. U.S.A. 90, 4304–4308 10.1073/pnas.90.9.4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ma X., Zhang H., Xue X., and Shah Y. M. (2017) Hypoxia-inducible factor 2α (HIF-2α) promotes colon cancer growth by potentiating Yes-associated protein 1 (YAP1) activity. J. Biol. Chem. 292, 17046–17056 10.1074/jbc.M117.805655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Triner D., and Shah Y. M. (2016) Hypoxia-inducible factors: a central link between inflammation and cancer. J. Clin. Invest. 126, 3689–3698 10.1172/JCI84430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xue X., and Shah Y. M. (2013) Hypoxia-inducible factor-2α is essential in activating the COX2/mPGES-1/PGE2 signaling axis in colon cancer. Carcinogenesis 34, 163–169 10.1093/carcin/bgs313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xue X., Taylor M., Anderson E., Hao C., Qu A., Greenson J. K., Zimmermann E. M., Gonzalez F. J., and Shah Y. M. (2012) Hypoxia-inducible factor-2α activation promotes colorectal cancer progression by dysregulating iron homeostasis. Cancer Res. 72, 2285–2293 10.1158/1538-7445.AM2012-2285,10.1158/0008-5472.CAN-11-3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shah Y. M., and Xie L. (2014) Hypoxia-inducible factors link iron homeostasis and erythropoiesis. Gastroenterology 146, 630–642 10.1053/j.gastro.2013.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shah Y. M., Matsubara T., Ito S., Yim S. H., and Gonzalez F. J. (2009) Intestinal hypoxia-inducible transcription factors are essential for iron absorption following iron deficiency. Cell Metab. 9, 152–164 10.1016/j.cmet.2008.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Friedl W., Kruse R., Uhlhaas S., Stolte M., Schartmann B., Keller K. M., Jungck M., Stern M., Loff S., Back W., Propping P., and Jenne D. E. (1999) Frequent 4-bp deletion in exon 9 of the SMAD4/MADH4 gene in familial juvenile polyposis patients. Genes Chromosomes Cancer 25, 403–406 [DOI] [PubMed] [Google Scholar]
- 17. Maesawa C., Tamura G., Nishizuka S., Iwaya T., Ogasawara S., Ishida K., Sakata K., Sato N., Ikeda K., Kimura Y., Saito K., and Satodate R. (1997) MAD-related genes on 18q21.1, Smad2 and Smad4, are altered infrequently in esophageal squamous cell carcinoma. Jpn. J. Cancer Res. 88, 340–343 10.1111/j.1349-7006.1997.tb00386.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Uchida K., Nagatake M., Osada H., Yatabe Y., Kondo M., Mitsudomi T., Masuda A., Takahashi T., and Takahashi T. (1996) Somatic in vivo alterations of the JV18–1 gene at 18q21 in human lung cancers. Cancer Res. 56, 5583–5585 [PubMed] [Google Scholar]
- 19. Wang R. H., Li C., Xu X., Zheng Y., Xiao C., Zerfas P., Cooperman S., Eckhaus M., Rouault T., Mishra L., and Deng C. X. (2005) A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2, 399–409 10.1016/j.cmet.2005.10.010 [DOI] [PubMed] [Google Scholar]
- 20. Massagué J. (2008) TGFβ in cancer. Cell 134, 215–230 10.1016/j.cell.2008.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Souchelnytskyi S., Tamaki K., Engström U., Wernstedt C., ten Dijke P., and Heldin C. H. (1997) Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with Smad4 and is required for transforming growth factor-β signaling. J. Biol. Chem. 272, 28107–28115 10.1074/jbc.272.44.28107 [DOI] [PubMed] [Google Scholar]
- 22. Chacko B. M., Qin B., Correia J. J., Lam S. S., de Caestecker M. P., and Lin K. (2001) The L3 loop and C-terminal phosphorylation jointly define Smad protein trimerization. Nat. Struct. Biol. 8, 248–253 10.1038/84995 [DOI] [PubMed] [Google Scholar]
- 23. Chacko B. M., Qin B. Y., Tiwari A., Shi G., Lam S., Hayward L. J., De Caestecker M., and Lin K. (2004) Structural basis of heteromeric smad protein assembly in TGF-β signaling. Mol. Cell 15, 813–823 10.1016/j.molcel.2004.07.016 [DOI] [PubMed] [Google Scholar]
- 24. Shi Y., Hata A., Lo R. S., Massagué J., and Pavletich N. P. (1997) A structural basis for mutational inactivation of the tumour suppressor Smad4. Nature 388, 87–93 10.1038/40431 [DOI] [PubMed] [Google Scholar]
- 25. Hata A., Lo R. S., Wotton D., Lagna G., and Massagué J. (1997) Mutations increasing autoinhibition inactivate tumour suppressors Smad2 and Smad4. Nature 388, 82–87 10.1038/40424 [DOI] [PubMed] [Google Scholar]
- 26. Kuang C., and Chen Y. (2004) Tumor-derived C-terminal mutations of Smad4 with decreased DNA binding activity and enhanced intramolecular interaction. Oncogene 23, 1021–1029 10.1038/sj.onc.1207219 [DOI] [PubMed] [Google Scholar]
- 27. Xue X., Ramakrishnan S. K., Weisz K., Triner D., Xie L., Attili D., Pant A., Gyorffy B., Zhan M., Carter-Su C., Hardiman K. M., Wang T. D., Dame M. K., Varani J., Brenner D., et al. (2016) Iron uptake via DMT1 integrates cell cycle with JAK–STAT3 signaling to promote colorectal tumorigenesis. Cell Metab. 24, 447–461 10.1016/j.cmet.2016.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hubert N., and Hentze M. W. (2002) Previously uncharacterized isoforms of divalent metal transporter (DMT)-1: implications for regulation and cellular function. Proc. Natl. Acad. Sci. U.S.A. 99, 12345–12350 10.1073/pnas.192423399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shuttleworth V. G., Gaughan L., Nawafa L., Mooney C. A., Cobb S. L., Sheerin N. S., and Logan I. R. (2018) The methyltransferase SET9 regulates TGFB1 activation of renal fibroblasts via interaction with SMAD3. J. Cell Sci. 131 [DOI] [PubMed] [Google Scholar]
- 30. Wan M., Tang Y., Tytler E. M., Lu C., Jin B., Vickers S. M., Yang L., Shi X., and Cao X. (2004) Smad4 protein stability is regulated by ubiquitin ligase SCFβ-TrCP1. J. Biol. Chem. 279, 14484–14487 10.1074/jbc.C400005200 [DOI] [PubMed] [Google Scholar]
- 31. Kretzschmar M., Doody J., Timokhina I., and Massagué J. (1999) A mechanism of repression of TGFβ/Smad signaling by oncogenic Ras. Genes Dev. 13, 804–816 10.1101/gad.13.7.804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jonk L. J., Itoh S., Heldin C. H., ten Dijke P., and Kruijer W. (1998) Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-β, activin, and bone morphogenetic protein-inducible enhancer. J. Biol. Chem. 273, 21145–21152 10.1074/jbc.273.33.21145 [DOI] [PubMed] [Google Scholar]
- 33. Jones J. B., and Kern S. E. (2000) Functional mapping of the MH1 DNA-binding domain of DPC4/SMAD4. Nucleic Acids Res. 28, 2363–2368 10.1093/nar/28.12.2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Endo H., Okuyama H., Ohue M., and Inoue M. (2014) Dormancy of cancer cells with suppression of AKT activity contributes to survival in chronic hypoxia. PLoS One 9, e98858 10.1371/journal.pone.0098858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mimeault M., and Batra S. K. (2013) Hypoxia-inducing factors as master regulators of stemness properties and altered metabolism of cancer- and metastasis-initiating cells. J. Cell Mol. Med. 17, 30–54 10.1111/jcmm.12004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pugh C. W., and Ratcliffe P. J. (2003) Regulation of angiogenesis by hypoxia: role of the HIF system. Nat. Med. 9, 677–684 10.1038/nm0603-677 [DOI] [PubMed] [Google Scholar]
- 37. Qiu G. Z., Jin M. Z., Dai J. X., Sun W., Feng J. H., and Jin W. L. (2017) Reprogramming of the tumor in the hypoxic niche: the emerging concept and associated therapeutic strategies. Trends Pharmacol. Sci. 38, 669–686 10.1016/j.tips.2017.05.002 [DOI] [PubMed] [Google Scholar]
- 38. Barsoum I. B., Smallwood C. A., Siemens D. R., and Graham C. H. (2014) A mechanism of hypoxia-mediated escape from adaptive immunity in cancer cells. Cancer Res. 74, 665–674 10.1158/1538-7445.AM2014-665,10.1158/0008-5472.CAN-13-0992 [DOI] [PubMed] [Google Scholar]
- 39. Yang Y., Yang X., Yang Y., Zhu H., Chen X., Zhang H., Wang F., Qin Q., Cheng H., and Sun X. (2015) Exosomes: a promising factor involved in cancer hypoxic microenvironments. Curr. Med. Chem. 22, 4189–4195 10.2174/0929867322666150825163318 [DOI] [PubMed] [Google Scholar]
- 40. Taube J. H., Allton K., Duncan S. A., Shen L., and Barton M. C. (2010) Foxa1 functions as a pioneer transcription factor at transposable elements to activate Afp during differentiation of embryonic stem cells. J. Biol. Chem. 285, 16135–16144 10.1074/jbc.M109.088096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen Y. G., Li Z., and Wang X. F. (2012) Where PI3K/Akt meets Smads: the crosstalk determines human embryonic stem cell fate. Cell Stem. Cell 10, 231–232 10.1016/j.stem.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xie L., Xue X., Taylor M., Ramakrishnan S. K., Nagaoka K., Hao C., Gonzalez F. J., and Shah Y. M. (2014) Hypoxia-inducible factor/MAZ-dependent induction of caveolin-1 regulates colon permeability through suppression of occludin, leading to hypoxia-induced inflammation. Mol. Cell Biol. 34, 3013–3023 10.1128/MCB.00324-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shalem O., Sanjana N. E., Hartenian E., Shi X., Scott D. A., Mikkelson T., Heckl D., Ebert B. L., Root D. E., Doench J. G., and Zhang F. (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Taylor M., Qu A., Anderson E. R., Matsubara T., Martin A., Gonzalez F. J., and Shah Y. M. (2011) Hypoxia-inducible factor-2α mediates the adaptive increase of intestinal ferroportin during iron deficiency in mice. Gastroenterology 140, 2044–2055 10.1053/j.gastro.2011.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xue X., Ramakrishnan S., Anderson E., Taylor M., Zimmermann E. M., Spence J. R., Huang S., Greenson J. K., and Shah Y. M. (2013) Endothelial PAS domain protein 1 activates the inflammatory response in the intestinal epithelium to promote colitis in mice. Gastroenterology 145, 831–841 10.1053/j.gastro.2013.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Anderson E. R., Taylor M., Xue X., Martin A., Moons D. S., Omary M. B., and Shah Y. M. (2012) The hypoxia-inducible factor-C/EBPα axis controls ethanol-mediated hepcidin repression. Mol. Cell. Biol. 32, 4068–4077 10.1128/MCB.00723-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wysocka J., Reilly P. T., and Herr W. (2001) Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol. Cell Biol. 21, 3820–3829 10.1128/MCB.21.11.3820-3829.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ma X., Patterson K. J., Gieschen K. M., and Bodary P. F. (2013) Are serum hepcidin levels chronically elevated in collegiate female distance runners? Int. J. Sport Nutr. Exerc. Metab. 23, 513–521 10.1123/ijsnem.23.5.513 [DOI] [PubMed] [Google Scholar]
- 49. Ma X., Pham V. T., Mori H., MacDougald O. A., Shah Y. M., and Bodary P. F. (2017) Iron elevation and adipose tissue remodeling in the epididymal depot of a mouse model of polygenic obesity. PLoS One 12, e0179889 10.1371/journal.pone.0179889 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.