Abstract
The liver X receptors Lxrα/NR1H3 and Lxrβ/NR1H2 are ligand-dependent nuclear receptors critical for midbrain dopaminergic (mDA) neuron development. We found previously that 24(S),25-epoxycholesterol (24,25-EC), the most potent and abundant Lxr ligand in the developing mouse midbrain, promotes mDA neurogenesis in vitro. In this study, we demonstrate that 24,25-EC promotes mDA neurogenesis in an Lxr-dependent manner in the developing mouse midbrain in vivo and also prevents toxicity induced by the Lxr inhibitor geranylgeranyl pyrophosphate. Furthermore, using MS, we show that overexpression of human cholesterol 24S-hydroxylase (CYP46A1) increases the levels of both 24(S)-hydroxycholesterol (24-HC) and 24,25-EC in the developing midbrain, resulting in a specific increase in mDA neurogenesis in vitro and in vivo, but has no effect on oculomotor or red nucleus neurogenesis. 24-HC, unlike 24,25-EC, did not affect in vitro neurogenesis, indicating that the neurogenic effect of 24,25-EC on mDA neurons is specific. Combined, our results indicate that increased levels of 24,25-EC in vivo, by intracerebroventricular delivery in WT mice or by overexpression of its biosynthetic enzyme CYP46A1, specifically promote mDA neurogenesis. We propose that increasing the levels of 24,25-EC in vivo may be a useful strategy to combat the loss of mDA neurons in Parkinson's disease.
Keywords: development, lipid metabolism, MS, neurodegenerative disease, neurogenesis, CYP46A1, dopamine neuron, liver X receptor, midbrain, oxysterol
Introduction
The vertebrate central nervous system is composed of an extensive variety of neurons that are generated following tightly regulated developmental programs. Characterization of the function and specificity of molecules selectively controlling distinct neuronal populations is thus essential to enhance our understanding of how such complexity is achieved in the developing brain, how it is maintained in the adult brain, and how it can be used for therapeutic purposes. Specific nuclear hormone receptors and their ligands have been identified as crucial factors in these processes (1–3). We have shown previously that liver X receptors (Lxrα3 and Lxrβ, encoded by NR1H3 and NR1H2, respectively) and their endogenous brain ligands (oxidized derivatives of cholesterol and related molecules) regulate the development of midbrain dopamine (mDA) neurons (4–6), red nucleus neurons (5), as well as oculomotor neurons (7). Moreover, enzymes involved in the biosynthesis of cholesterol, oxysterols, and 24(S),25-epoxycholesterol (24,25-EC), such as 2,3-oxidosqualene-lanosterol cyclase, cytochrome P450 family 11 subfamily A member 1 (CYP11A1), and CYP46A1 (also known as cholesterol 24S-hydroxylase), are expressed in the developing mouse ventral midbrain (VM) during VM neurogenesis (4, 8, 9).4 The enzyme CYP46A1 oxidizes cholesterol to 24(S)-hydroxycholesterol (24-HC), the most abundant oxysterol in the adult brain, present at 20–40 ng/mg in the mouse and human (11). It has been shown, in vitro in human embryonic kidney 293 cells transfected with CYP46A1, that 24-HC can be further oxidized to 24,25-dihydroxycholesterol (24,25-diHC) and to 24,27-diHC (systematic name 24,26-diHC (12)) by CYP46A1 (13, 14). It has been suggested by these studies that 24,25-diHC could then be converted to 24,25-EC, but definite evidence for such a mechanism is lacking. Interestingly, it has been shown in vitro that CYP46A1 can also oxidize desmosterol to 24,25-EC and to 27-hydroxydesmosterol (systematic name 26-hydroxydesmosterol) (15), thereby providing a distinct 24,25-EC biosynthetic pathway via desmosterol in the brain. In agreement with this study, there is a reduction in both 24-HC and 24,25-EC levels in the Cyp46a1-knockout mouse adult brain compared with the WT brain (16). An alternative route to 24,25-EC formation is via a shunt of the mevalonate pathway, specifically the Bloch arm of the pathway, in which an extra oxygen atom is introduced by squalene epoxidase into 3S-squalene-2,3-epoxide to give squalene-2,3(S);22(S),23-diepoxide prior to cyclization by 2,3-oxidosqualene-lanosterol cyclase (17). This pathway is also expected to be impaired in the Cyp46a1-knockout mouse, as the necessary enzymes are down-regulated as a consequence of reduced cholesterol biosynthesis (18).
The functions of 24-HC and 24,25-EC in the central nervous system are diverse. 24-HC plays a role as a cholesterol transport molecule, crossing the blood–brain barrier and thus facilitating transport of cholesterol to the liver for further metabolism (8, 11, 19). 24-HC is also a ligand for Lxrα and Lxrβ in the brain (5) and binds to the endoplasmic reticulum–resident protein INSIG (insulin-induced gene) (20), modulating processing of SREBP-2 (sterol response element–binding protein-2) to its active form as the master transcription regulator for cholesterol biosynthesis. On the other hand, 24,25-EC is the most abundant Lxr ligand in the developing but not the adult brain (5, 21). Within the embryonic VM, 24,25-EC is present at a much higher concentration than 24-HC (Ref. 5 and this study). Moreover, we found previously that 24,25-EC is the most potent endogenous Lxr ligand to promote mDA neurogenesis in mouse progenitor VM cultures, embryonic stem cells in vitro, and zebrafish in vivo (5). However, the function of 24,25-EC in the developing mouse brain in vivo remains to be determined. In this study, we address this question by examining the midbrain of mouse embryos either injected intracerebroventricularly with 24,25-EC in utero or transgenic mice expressing CYP46A1 under the control of a hybrid β-actin promoter (22). We show that increases in 24,25-EC in the developing VM, by either of these two strategies, result in increased number of mDA neurons in vivo. Thus, our results identify a new function of CYP46A1 and 24,25-EC in the mammalian brain in vivo.
Results
CYP46A1-overexpressing mice exhibit elevated levels of 24-HC and 24,25-EC
To examine the role of CYP46A1 in the developing brain, we examined the VM of transgenic mice overexpressing this enzyme. We first analyzed the levels of several sterols, oxysterols, and related compounds in WT and CYP46A1-overexpressing mice. We found a 29.2-fold increase in 24-HC levels and a 3.9-fold increase in 24,25-EC levels in the developing VM of CYP46A1-overexpressing mice compared with WT mice at E11.5 (Fig. 1 and Table S1). We also found a 1.98-fold increase in cholesterol levels in the developing VM of CYP46A1-overexpressing mice compared with WT mice (Table S1). However, the level of desmosterol (266-fold higher than that of 24,25-EC in WT mice) did not change in CYP46A1-overexpressing mice. Furthermore, we did not find any alteration in 22(R)-HC, 25-HC, 27-HC (systematic name (25R)26-HC), 7α-HC, or 7α,24-dihydroxycholesterol in CYP46A1-overexpressing mice (Table S1), indicating that the increases in 24-HC and 24,25-EC levels are very specific.
Figure 1.
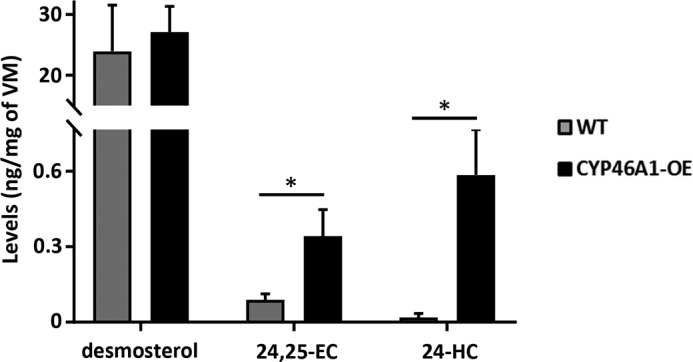
CYP46A1-overexpressing mice exhibit elevated levels of 24,25-EC and 24-HC. LC-MS(MSn) analysis demonstrated a significant increase in 24,25-EC and 24-HC but not desmosterol concentrations in the developing VM of CYP46A1-overexpressing mice compared with WT mice. Data are means ± S.E. (n = 4–6); *, p < 0.05 by Mann–Whitney test compared with the WT group.
To determine whether these changes were stable over time, we analyzed the levels of these compounds in the adult brain of CYP46A1-overexpressing mice. We found that, although the levels of cholesterol were not significantly different from WT mice, the levels of 24-HC and 24,25-EC increased by 22% and 25%, respectively, in CYP46A1-overexpressing mice (Table S2). Thus, our results portend CYP46A1 as a highly relevant enzyme in the biosynthesis of 24,25-EC in the developing and adult mouse brain.
Interestingly, our analysis of the developing mouse VM by single-cell RNA-Seq (9) indicates that Cyp46a1 is expressed at higher levels in two cell types lining the ventricle, ependymal and radial glia–like3 cells (Fig. S1), suggesting that these cell types may be the endogenous source of 24-HC and 24,25-EC in the developing VM.
Increased dopamine neuron number in midbrain cultures from CYP46A1-overexpressing mice
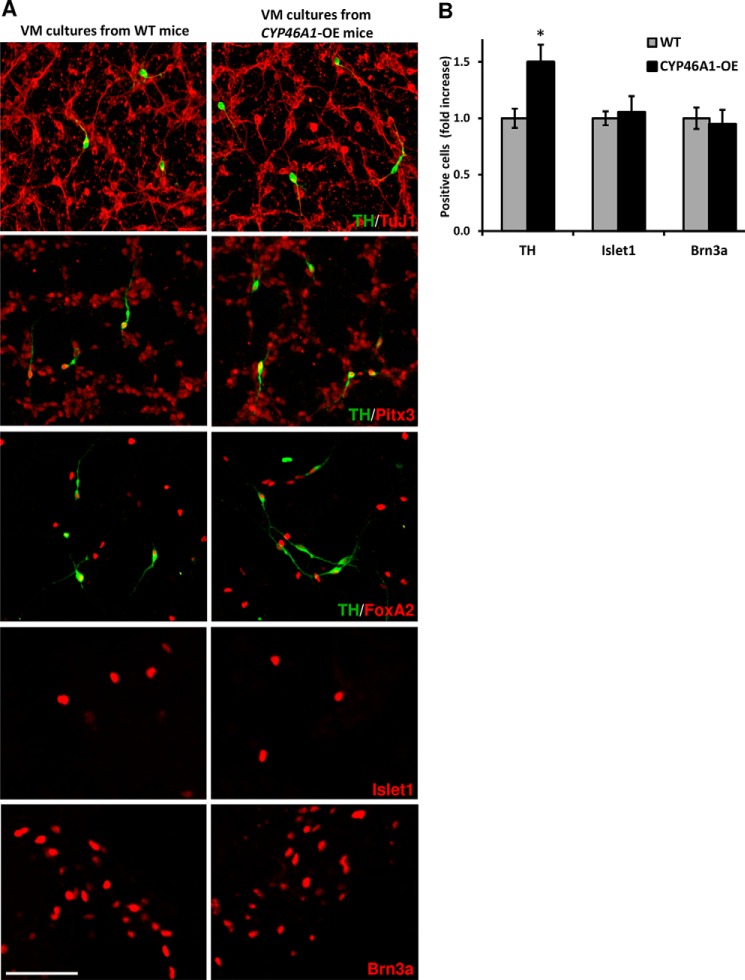
We next studied the impact of CYP46A1 overexpression on distinct neuronal populations in the developing VM. Notably, mouse VM progenitor cultures from CYP46A1-overexpressing mice exhibited a significant 49.8% increase in the number of mDA neurons compared with cultures from WT mice (Fig. 2, A and B). These neurons co-expressed the rate-limiting enzyme in the synthesis of dopamine tyrosine hydroxylase (TH), β III tubulin (TuJ1, a pan-neuronal marker), Forkhead box transcription factor (Foxa2, required for midbrain development regulation) (23), and pituitary homeobox 3 (Pitx3, a transcription factor required for the survival and maintenance of mDA neurons) (24) (Fig. 2A), thereby showing that they were true mDA neurons. Because mouse Cyp46a1 was also expressed in other cell types of the developing VM (Fig. S1), we examined adjacent neuronal populations. No significant change in the number of Islet1+ oculomotor neurons or Brn3a+ red nucleus neurons was detected (Fig. 2B), thereby demonstrating that the effect of CYP46A1 overexpression is specific to mDA neurons. We next examined whether 24-HC and 24,25-EC increase the number of mDA neurons when added to WT VM progenitor cultures. Although 24,25-EC enhances mDA neurogenesis (Ref. 5 and Fig. S2), we found that 24-HC had no significant effect on the number of TH+ mDA neurons (Fig. S3). Interestingly, the effect of 24,25-EC on TH+ mDA neurons was abolished in VM progenitor cultures from Lxrαβ double knockout mice (Fig. S2), thereby showing that LXR receptors are required for the increase in mDA neuron numbers by 24,25-EC. Combined, our results indicate that elevated levels of 24,25-EC lead to increased numbers of mDA neurons in CYP46A1-overexpressing mice.
Figure 2.
Increased dopamine neuron numbers in midbrain cultures from CYP46A1-overexpressing mice. A, representative images of TH+ and TuJ1+ neurons as well as Foxa2+, Pitx3+, Islet1+, and Brn3a+ neuron nuclei in VM cultures from WT and CYP46A1-overexpressing mice. Scale bar = 50 μm. B, quantification of TH+, Islet1+, and Brn3a+ neurons in VM cultures from WT and CYP46A1-overexpressing mice. Data are means ± S.E. (n = 3); *, p < 0.05 by Student's t test compared with the WT group.
CYP46A1 overexpression increases the number of mDA neurons in the developing brain in vivo
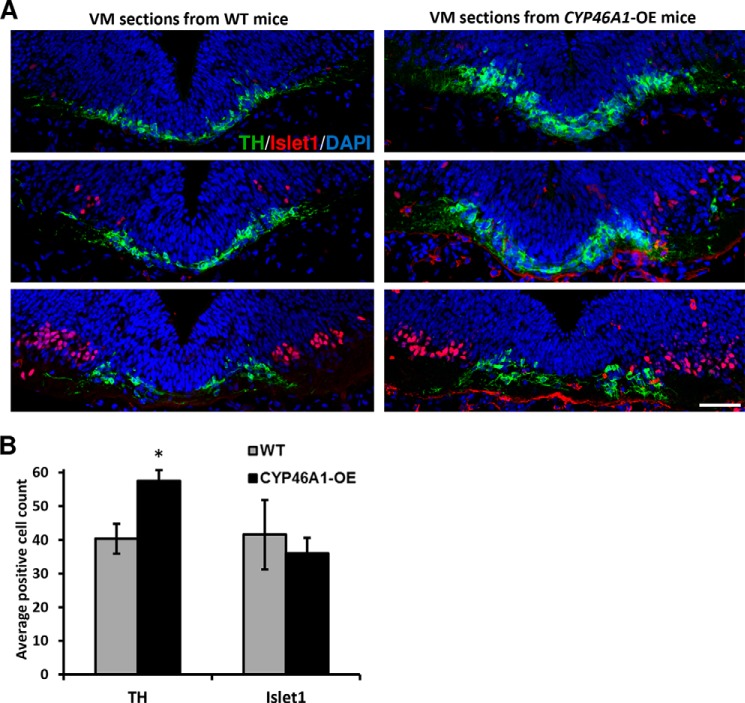
We also investigated whether CYP46A1-overexpression impacts VM development in vivo. We thus examined the number of mDA and oculomotor neurons in coronal sections through the VM of CYP46A1-overexpressing and WT mice at E11.5. We observed that the number of TH+ mDA neurons significantly increased by 42.6% in CYP46A1-overexpressing compared with WT mice (Fig. 3, A and B). However, the number of Islet1+ oculomotor neurons did not change, arguing for a specific effect of CYP46A1 overexpression on mDA neurons in vivo. Combined, our results indicate that elevated levels of 24,25-EC in CYP46A1-overexpressing mice lead to increased numbers of mDA neurons.
Figure 3.
Increased dopamine neuron numbers in the VM of CYP46A1-overexpressing mice. A, representative images of anterior-to-posterior coronal VM sections from E11.5 WT and CYP46A1-overexpressing mice showing TH+ neurons, Islet1+ neuron nuclei, and 4′,6-diamidino-2-phenylindole–stained nuclei. Scale bar = 50 μm. B, quantification of TH+ and Islet1+ neurons in VM sections from WT and CYP46A1-overexpressing mice. Data are means ± S.E. (n = 5–15); *, p < 0.05 by Mann–Whitney test compared with the WT group.
24,25-EC promotes mouse midbrain dopaminergic neurogenesis in vivo and prevents toxicity by GGPP
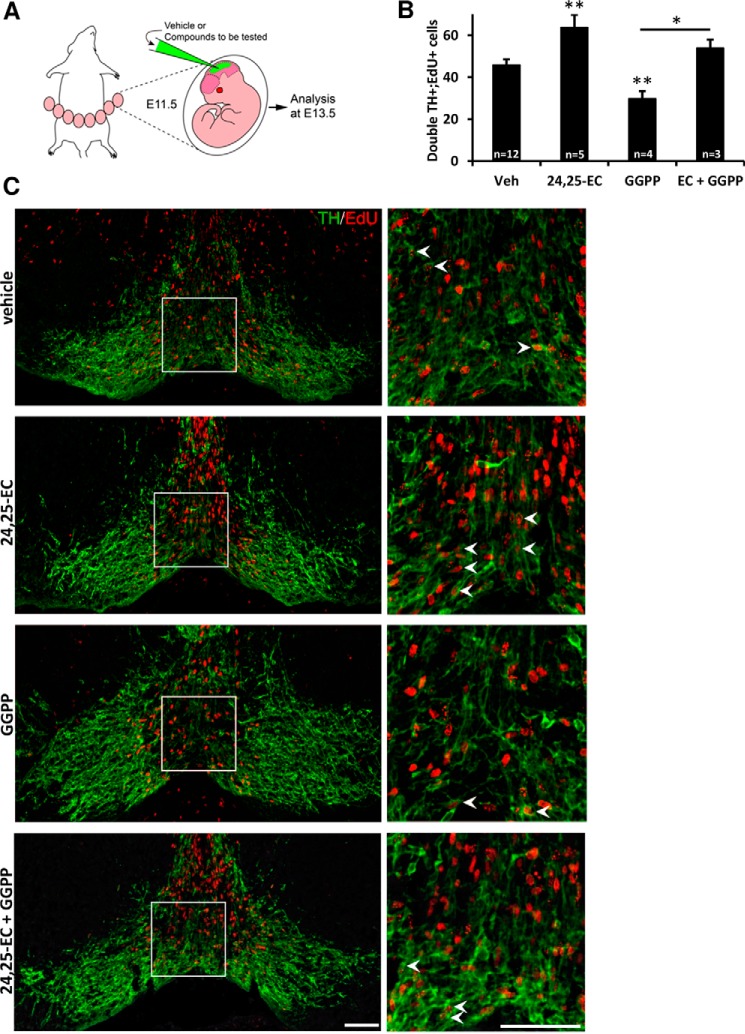
Finally, to directly examine the function of 24,25-EC in the developing mouse midbrain in vivo, we performed 24,25-EC injections into the cerebrospinal fluid, at the level of the aqueduct, in E11.5 WT mouse embryos in utero and analyzed brain sections at the midbrain level at E13.5 (Fig. 4A). Neurogenesis was examined by performing a pulse of EdU intraperitoneally at E11.5 to label proliferative progenitors and assess their capacity to undergo neurogenesis and give rise to mDA neurons that can be identified by the expression of tyrosine hydroxylase (Th). Upon injection of 24,25-EC, we found that the number of double EdU+;TH+ cells increased by 39% (Fig. 4, B and C), thereby demonstrating that 24,25-EC promotes mDA neurogenesis in vivo. In contrast, injection of the Lxr inhibitor geranylgeranyl pyrophosphate (GGPP), reduced the number of double EdU+;TH+ cells, indicating that LXR activity is required for mDA neuron development. Notably, the effect of GGPP was blocked by co-injection of 24,25-EC, indicating that 24,25-EC is not only required and sufficient to promote mDA neurogenesis in vivo but can also prevent the toxic effect of GGPP. Thus, our results demonstrate that elevated levels of 24,25-EC promote mDA neurogenesis in vivo.
Figure 4.
24,25-EC promotes neurogenesis of mouse midbrain dopamine neurons in vivo and prevents toxicity by GGPP. A, vehicle or 24,25-EC and/or GGPP was injected into the mesencephalic ventricle of E11.5 mouse embryos in utero, and embryos were collected at E13.5. Dopamine neurogenesis was examined by EdU intraperitoneal injection and by assessing the acquisition of TH expression. B, quantification of double EdU+;TH+ cell numbers (mean ± S.E.) for the indicated conditions: vehicle, 24,25-EC-, GGPP-, and 24,25-EC + GGPP–injected embryos. Data are means ± S.E. (n = 3–12); *, p < 0.05; **, p < 0.01 by Mann–Whitney test compared with the vehicle group or as indicated. C, microphotographs of midbrain coronal sections showing TH+ dopamine neurons (green) and EdU+ cells (red) (left panels) and higher-magnification pictures of the boxed region (right panels) for the indicated conditions. Arrowheads indicate double EdU+;TH+ cells. Scale bars = 50 μm.
Discussion
In this study, we show that overexpression of CYP46A1 in transgenic mice increases the levels of 24-HC and 24,25-EC in the VM but does not alter desmosterol or other oxysterol levels, which remain at a similar level as in WT mice. Our results, together with previous findings showing a reduction in both 24-HC and 24,25-EC levels in Cyp46a1-knockout mice (16), lend support to the hypothesis that CYP46A1 is highly relevant in the biosynthesis of 24,25-EC. This could be achieved either by increased biosynthesis of 24,25-EC from desmosterol by CYP46A1 (as suggested in Ref. 15) or by increased biosynthesis of 24,25-EC from cholesterol by CYP46A1 via 24-HC and 24,25-diHC (as suggested in Refs. 13, 14). In either case, our results demonstrate the importance of CYP46A1 in 24,25-EC biosynthesis in the developing mammalian VM. We also found that 24-HC does not affect the number of mDA neurons, whereas 24,25-EC strongly promotes mDA neurogenesis, a finding consistent with our previous results showing Lxr ligand–specific activities in the developing mouse VM (5, 7). These results also indicate that the observed increase in mDA neurogenesis in vitro and in vivo in CYP46A1-overexpressing mice is indeed associated with the increase in 24,25-EC in these mice. Notably, the effect of CYP46A1 was specific to mDA neurons, as neighboring cell populations in the developing basal plate of the VM, such as oculomotor neurons or red nucleus neurons, were not affected in CYP46A1-overexpressing mice. These results show that increased levels of Lxr ligands do not alter their cell type specificity, which is conferred by LXRs, as we described previously (5, 7). Mechanistically, we found that intracerebroventricular injection of the LXR agonist 24,25-EC or the LXR antagonist GGPP was capable of, respectively, promoting or inhibiting mDA neurogenesis in vivo. These effects were specific because 24,25-EC had no effect on red nucleus, serotonin+ neurons, oculomotor neurons, or GABA+ neurons in vitro and in vivo (Ref. 5 and this work). In sum, our results demonstrate a clear role of LXR receptors and 24,25-EC in mDA neurogenesis in vivo.
Several studies have associated a reduced level of CYP46A1 with neurodegeneration and neuronal dysfunction as well as restoration of normal CYP46A1 levels with functional recovery and neuroprotection. For instance, knockdown of Cyp46a1 in mice results in deficits in spatial, associative, and motor learning and in hippocampal long-term potentiation (25). In addition, reduced Cyp46a1 levels result in cognitive deficits, elevated production of β-amyloid peptides, and abnormal phosphorylation of tau (26) as well as in progressive loss of hippocampal neurons and an Alzheimer's disease–like phenotype (27). Conversely, increased expression of CYP46A1 improves spatial memory retention in aged female mice (28) and reduces cognitive decline and amyloid burden in several mouse models of Alzheimer's disease (29–31). Furthermore, similar results have been obtained by enhancing CYP46A1 activity with the reverse transcriptase inhibitor efavirenz (32), arguing for the feasibility of using a pharmacological treatment to reduce neurodegeneration.
With regard to neurodegeneration in the basal ganglia, it has been reported that the level of CYP46A1 is decreased in the putamen of patients with Huntington's disease (33). Notably, CYP46A1 knockdown in the mouse striatum induced spontaneous striatal neurodegeneration associated with abnormal balance and motor coordination. Conversely, increased levels of CYP46A1 in the R6/2 Huntington's disease mouse model decreased striatal neuron atrophy, protein aggregates, and motor deficits.
Much less is known about the role of CYP46A1 in Parkinson's disease. For instance, it remains to be determined whether the level and functionality of CYP46A1 are conserved or altered in Parkinson's disease models or patients. The results in this study provide first evidence that 24,25-EC can rescue a defect in mDA neurogenesis induced by GGPP in vivo, suggesting a potential application of CYP46A1 and 24,25-EC in regenerating mDA neurons in vivo. Previous studies have also shown that a synthetic LXR ligand can prevent the degeneration of mDA neurons in an animal model of Parkinson's disease (34). Thus combined, our results and data in the literature suggest that 24,25-EC or pharmacological tools capable of activating LXR receptors or enhancing the function and/or levels of CYP46A1 could be used to enhance mDA neurogenesis, limit neurodegeneration, and advance cell replacement strategies for the treatment of Parkinson's disease.
Experimental procedures
Extraction of sterols
Sterols were extracted from mouse adult brain and mouse embryonic VM into ethanol and fractionated by reverse-phase solid phase extraction (SPE) to give an oxysterol-rich fraction devoid of cholesterol (5, 7, 16, 36).
Charge-tagging of sterols
The sterols were charge-tagged with GP-hydrazine as described previously (5, 7, 16, 36). This greatly enhances their response when analyzed by LC-electrospray ionization-MS (LC-ESI-MS) and MS with multistage fragmentation (MSn).
Reagents
HPLC-grade water and solvents were from Fisher Scientific or Sigma-Aldrich. Authentic sterols and oxysterols were from Avanti Polar Lipids. Girard P (GP) reagent (1-[carboxymethyl]pyridinium chloride hydrazide, [2H0]GP) was from TCI Europe or synthesized in-house ([2H5]GP) as in earlier studies (37), and cholesterol oxidase from Streptomyces sp. was from Sigma-Aldrich. Certified Sep-Pak C18 200-mg (SPE1) and OASIS HLB 60-mg (SPE2) columns were from Waters.
LC-ESI-MSn on the Orbitrap ELITE
LC-ESI-MS and LC-ESI-MSn were performed using an Ultimate 3000 HPLC system (Dionex, now Thermo Fisher Scientific) linked to the ESI source of an Orbitrap ELITE (Thermo Fisher Scientific) mass spectrometer as described previously (5, 7, 16, 35, 36).
WT mice
Mice were housed, bred, and treated according to the guidelines of the European Communities Council (directive 86/609/EEC) and the Society for Neuroscience. Ethics approval for mouse experimentation was granted by Stockholm Norra Djurförsöksetisks Nämnd N154/06, N145/09, N370/09, N273/11, and N486/12.
Mice overexpressing human CYP46A1
Human CYP46A1-overexpressing transgenic mice were generated as described before (22, 28). All animal experiments received full approval from the local Animal Experimentation Ethics Committee. Tissue sampling from these mice was performed under the aegis of the UK Scientific Procedures (Animals) Act, 1986.
Primary midbrain cultures
Brains from E11.5 mice were obtained. The ventral midbrain region was dissected, mechanically dissociated, plated on poly-d-lysine (150,000 cells/cm2), and grown in serum-free N2 media consisting of a 1:1 mixture of F12 and Dulbecco's modified Eagle's medium with 10 ng/ml insulin, 100 μg/ml apo-transferrin, 100 μm putrescine, 20 nm progesterone, 30 nm selenium, 6 mg/ml glucose, and 1 mg/ml BSA. Cells were treated for 3 days in vitro with the compounds of interest, fixed with 4% PFA, and processed for staining using appropriate antibodies. Hoechst staining was performed by permeabilizing cells with a 0.3% Triton X-100/PBS solution for 5 min, followed by incubation with Hoechst 33258 (Sigma) for 10 min.
Immunocytochemical analysis
Cells were fixed in 4% paraformaldehyde (PFA), washed in PBS, and blocked in 5% normal goat serum/PBS for 1 h at room temperature. Primary antibodies were diluted in PBS (pH 7.4), 0.3% Triton X-100, and 1% BSA, and incubations were carried out overnight at +4 °C or at room temperature for 2 h. The antibodies used were anti-TH (1:1000, Pel-Freeze), anti-Islet1 (1:100, Developmental Studies Hybridoma Bank), anti-Brn3a (1:250, Millipore), anti-TuJ1 (1:1000, Promega), anti-FoxA2 (1:400, Cell Signaling Technology), anti-Pitx3 (1:400, Invitrogen) and appropriate secondary antibodies (Jackson ImmunoResearch Laboratories or Alexa). Cells positive for the corresponding marker were counted directly at the microscope at a magnification of ×20. Cells were counted in every well, in eight consecutive fields (going from one side of the well to the other, passing through the center), in three different wells per experiment, and in three different experiments per condition. Positive cell counts were normalized to the total number of cells (counted utilizing Hoechst-stained nuclei) and presented as -fold increase over WT or vehicle. Random pictures of the wells were taken for every condition to document the result, and representative pictures were subsequently selected to represent the quantitative data. Photos were acquired with a Zeiss Axioplan microscope and a Hamamatsu camera (C4742-95) using the Openlab software.
In utero intraventricular injections
Mouse in utero injections were performed as described previously (7, 38). Female WT CD-1 mice (25–35 g, Charles River Breeding Laboratories) were used for these experiments. Ethics approval was granted by Stockholm Norra Djurförsöksetisks Nämnd N273/11 and N486/12. For embryo analyses, WT CD-1 mice were mated overnight, and noon of the day the plug was considered E0.5. E11.5 pregnant females were deeply anesthetized using isoflurane (IsoFlo®, Abbott Labs), and the uterine horns were accessed through an abdominal incision. 1 μl of 24,25-EC (5 mm), GGPP (5 mm), or vehicle solution (methanol/PBS, 50% v/v) was injected into the cerebral aqueduct. The uterine horns were replaced into the abdominal cavity, which was then closed with sutures. For EdU pulse-chase experiments, EdU (50 mg/kg of body weight) was injected by intraperitoneal injection 30 min after the injections to the embryo. Embryos were analyzed 48 h later, at E13.5. The concentration and volume of the compounds utilized in these experiments were chosen for the compounds to be in a physiological range because the cerebrospinal fluid volume in the E11.5 mouse embryo is ∼40 μl, the cerebrospinal fluid is replaced at a speed of 3.3 × 10−4 ml/min in mice (10), and mouse embryos were analyzed 48 h after injection.
Mouse VM coronal sections and immunohistochemical analysis
Embryos were dissected out of the uterine horns in ice-cold PBS, fixed in 4% PFA for 4 h to overnight, cryoprotected in 15–30% sucrose, frozen in Tissue-Tek Optimum Cutting Temperature compound (Sakura Fine-Tek) on dry ice, and stored at −80 °C until use. 14-μm serial coronal sections through the E11.5 or E13.5 midbrain region were cut on a cryostat and placed serially on 10 slides. Slides 1 and 6 were subjected to immunohistochemistry. Sections were preincubated for 1 h in blocking solution, followed by incubation at 4 °C overnight with the following primary antibodies: sheep anti-TH (1:500, Novus Biologicals), rabbit anti-TH (1:750, Pel-Freeze), and mouse anti-Islet-1 (1:100, Developmental Studies Hybridoma Bank). After washing, slides were incubated for 1 h at room temperature with the appropriate fluorophore-conjugated (Cy2, Cy3, and Cy5, 1:300, Jackson ImmunoResearch Laboratories; Alexa 488, 555, and 647, 1:1000, Invitrogen) secondary antibodies. The EdU click reaction was performed according to the instructions of the manufacturer (Life Technologies). Confocal pictures were taken on a Zeiss LSM700 microscope. TH+ cells, Islet1+ cells, and double EdU+;TH+ cells were counted on three sections covering the rostral to caudal midbrain for each embryo.
Statistical analysis
Statistical analyses (Mann–Whitney test and Student's t test) were performed using Prism 4 (GraphPad Software, La Jolla, CA). A p value less than 0.05 was considered significant; *, p < 0.05; **, p < 0.01. Data represent mean ± S.E.
Animal studies approval statement
Ethics approval for WT and CYP46A1-overexpressing transgenic mouse experimentation was granted by the local Animal Experimentation Ethics Committee (Stockholm Djurförsöksetisks Nämnd N154/06, N145/09, N370/09, N273/11, and N486/12).
Author contributions
S. T., I. B., Y. W., W. J. G., and E. A. conceptualization; S. T., W. A. A. d. O., S. Y., E. Y., A. S., J. A.-K., and A. U. data curation; S. T., W. A. A. d. O., S. Y., E. Y., A. S., J. A.-K., I. B., Y. W., W. J. G., and E. A. formal analysis; S. T., I. B., Y. W., W. J. G., and E. A. supervision; S. T., I. B., Y. W., W. J. G., and E. A. funding acquisition; S. T., W. A. A. d. O., S. Y., E. Y., A. S., J. A.-K., A. U., A. C.-M., I. B., Y. W., W. J. G., and E. A. investigation; S. T. and W. A. A. d. O. visualization; S. T. methodology; S. T. writing-original draft; S. T., I. B., Y. W., W. J. G., and E. A. project administration; S. T., I. B., Y. W., W. J. G., and E. A. writing-review and editing; A. C.-M., I. B., Y. W., W. J. G., and E. A. resources.
Supplementary Material
Acknowledgments
We thank members of the European Network for Oxysterol Research for informative discussions.
This work was supported by funding from the Swedish Research Council (VR projects: DBRM, 2011-3116, 2011-3318, and 2016-01526), the European Union (NeuroStemcellRepair and DDPD), the Swedish Foundation for Strategic Research (DBRM, SRL program, and SB16-0065), Hjärnfonden (FO2015:0202 and FO2017-0059), Cancerfonden (CAN 2016/572), and Karolinska Institutet (SFO Strat Regen, senior grant 2018) (to E. A.); by a Sêr Cymru II Rising Stars grant from the Welsh Government (to S. T.); by the Biotechnology and Biological Sciences Research Council (BB/I001735/1 and B/N015932/1 to W. J. G. and BB/L001942/1 to Y. W.); by a PhD studentship from Imperial College Healthcare Charities (to J. A.-K.); and by Hjärnfonden, the Stockholm County Council (ALF), and Stiftelsen för Gamla Tjänarinnor (to I. B.). The derivatization method used for the LC-MS analysis of sterols has been licensed to Avanti Polar Lipids Inc. and Cayman Chemical by Swansea Innovations Ltd., a wholly owned subsidiary of Swansea University.
This article contains Figs. S1–S3 and Tables S1 and S2.
The array data have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO accession number GSE76381 (https://github.com/linnarsson-lab/ipynb-lamanno2016).
- Lxr
- liver X receptor
- mDA
- midbrain dopamine
- 24,25-EC
- 24(S),25-epoxycholesterol
- VM
- ventral midbrain
- 24-HC
- 24(S)-hydroxycholesterol
- 24,25-diHC
- 24,25-dihydroxycholesterol
- E11.5
- embryonic day 11.5
- TH
- tyrosine hydroxylase
- GGPP
- geranylgeranyl pyrophosphate
- SPE
- solid phase extraction
- GP
- Girard P
- ESI
- electrospray ionization
- MSn
- MS with multistage fragmentation
- PFA
- paraformaldehyde
- EdU
- 5-ethynyl-2'-deoxyuridine.
References
- 1. Förthmann B., Aletta J. M., Lee Y. W., Terranova C., Birkaya B., Stachowiak E. K., Stachowiak M. K., and Claus P. (2015) Coalition of nuclear receptors in the nervous system. J. Cell Physiol. 230, 2875–2880 10.1002/jcp.25036 [DOI] [PubMed] [Google Scholar]
- 2. Katayama K., Wada K., Nakajima A., Kamisaki Y., and Mayumi T. (2005) Nuclear receptors as targets for drug development: the role of nuclear receptors during neural stem cell proliferation and differentiation. J. Pharmacol. Sci. 97, 171–176 10.1254/jphs.FMJ04008X3 [DOI] [PubMed] [Google Scholar]
- 3. Chawla A., Repa J. J., Evans R. M., and Mangelsdorf D. J. (2001) Nuclear receptors and lipid physiology: opening the X-files. Science 294, 1866–1870 10.1126/science.294.5548.1866 [DOI] [PubMed] [Google Scholar]
- 4. Sacchetti P., Sousa K. M., Hall A. C., Liste I., Steffensen K. R., Theofilopoulos S., Parish C. L., Hazenberg C., Richter L. A., Hovatta O., Gustafsson J. A., and Arenas E. (2009) Liver X receptors and oxysterols promote ventral midbrain neurogenesis in vivo and in human embryonic stem cells. Cell Stem Cell 5, 409–419 10.1016/j.stem.2009.08.019 [DOI] [PubMed] [Google Scholar]
- 5. Theofilopoulos S., Wang Y., Kitambi S. S., Sacchetti P., Sousa K. M., Bodin K., Kirk J., Saltó C., Gustafsson M., Toledo E. M., Karu K., Gustafsson J. Å., Steffensen K. R., Ernfors P., Sjövall J., et al. (2013) Brain endogenous liver X receptor ligands selectively promote midbrain neurogenesis. Nat. Chem. Biol. 9, 126–133 10.1038/nchembio.1156 [DOI] [PubMed] [Google Scholar]
- 6. Theofilopoulos S., and Arenas E. (2015) Liver X receptors and cholesterol metabolism: role in ventral midbrain development and neurodegeneration. F1000Prime Rep. 7, 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Theofilopoulos S., Griffiths W. J., Crick P. J., Yang S., Meljon A., Ogundare M., Kitambi S. S., Lockhart A., Tuschl K., Clayton P. T., Morris A. A., Martinez A., Reddy M. A., Martinuzzi A., Bassi M. T., et al. (2014) Cholestenoic acids regulate motor neuron survival via liver X receptors. J. Clin. Invest. 124, 4829–4842 10.1172/JCI68506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lund E. G., Xie C., Kotti T., Turley S. D., Dietschy J. M., Russell D. W. (2003) Knockout of the cholesterol 24-hydroxylase gene in mice reveals a brain-specific mechanism of cholesterol turnover. J. Biol. Chem. 278, 22980–22988 10.1074/jbc.M303415200 [DOI] [PubMed] [Google Scholar]
- 9. La Manno G., Gyllborg D., Codeluppi S., Nishimura K., Salto C., Zeisel A., Borm L. E., Stott S. R. W., Toledo E. M., Villaescusa J. C., Lönnerberg P., Ryge J., Barker R. A., Arenas E., and Linnarsson S. (2016) Molecular diversity of midbrain development in mouse, human, and stem cells. Cell 167, 566–580.e19 10.1016/j.cell.2016.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smith J. S., Angel T. E., Chavkin C., Orton D. J., Moore R. J., and Smith R. D. (2014) Characterization of individual mouse cerebrospinal fluid proteomes. Proteomics 14, 1102–1106 10.1002/pmic.201300241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lütjohann D., Breuer O., Ahlborg G., Nennesmo I., Sidén A., Diczfalusy U., and Björkhem I. (1996) Cholesterol homeostasis in human brain: evidence for an age-dependent flux of 24S-hydroxycholesterol from the brain into the circulation. Proc. Natl. Acad. Sci. U.S.A. 93, 9799–9804 10.1073/pnas.93.18.9799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fakheri R. J., and Javitt N. B. (2012) 27-Hydroxycholesterol, does it exist? On the nomenclature and stereochemistry of 26-hydroxylated sterols. Steroids 77, 575–577 10.1016/j.steroids.2012.02.006 [DOI] [PubMed] [Google Scholar]
- 13. Mast N., Norcross R., Andersson U., Shou M., Nakayama K., Bjorkhem I., and Pikuleva I. A. (2003) Broad substrate specificity of human cytochrome P450 46A1 which initiates cholesterol degradation in the brain. Biochemistry 42, 14284–14292 10.1021/bi035512f [DOI] [PubMed] [Google Scholar]
- 14. Pikuleva I. A. (2006) Cholesterol-metabolizing cytochromes P450. Drug Metab. Dispos. 34, 513–520 10.1124/dmd.105.008789 [DOI] [PubMed] [Google Scholar]
- 15. Goyal S., Xiao Y., Porter N. A., Xu L., and Guengerich F. P. (2014) Oxidation of 7-dehydrocholesterol and desmosterol by human cytochrome P450 46A1. J. Lipid Res. 55, 1933–1943 10.1194/jlr.M051508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meljon A., Wang Y., and Griffiths W. J. (2014) Oxysterols in the brain of the cholesterol 24-hydroxylase knockout mouse. Biochem. Biophys. Res. Commun. 446, 768–774 10.1016/j.bbrc.2014.01.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nelson J. A., Steckbeck S. R., and Spencer T. A. (1981) Biosynthesis of 24,25-epoxycholesterol from squalene 2,3;22,23-dioxide. J. Biol. Chem. 256, 1067–1068 [PubMed] [Google Scholar]
- 18. Russell D. W., Halford R. W., Ramirez D. M., Shah R., and Kotti T. (2009) Cholesterol 24-hydroxylase: an enzyme of cholesterol turnover in the brain. Annu. Rev. Biochem. 78, 1017–1040 10.1146/annurev.biochem.78.072407.103859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Griffiths W. J., Abdel-Khalik J., Hearn T., Yutuc E., Morgan A. H., and Wang Y. (2016) Current trends in oxysterol research. Biochem. Soc. Trans. 44, 652–658 10.1042/BST20150255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Radhakrishnan A., Ikeda Y., Kwon H. J., Brown M. S., and Goldstein J. L. (2007) Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: oxysterols block transport by binding to Insig. Proc. Natl. Acad. Sci. U.S.A. 104, 6511–6518 10.1073/pnas.0700899104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Y., Sousa K. M., Bodin K., Theofilopoulos S., Sacchetti P., Hornshaw M., Woffendin G., Karu K., Sjövall J., Arenas E., and Griffiths W. J. (2009) Targeted lipidomic analysis of oxysterols in the embryonic central nervous system. Mol. Biosyst. 5, 529–541 10.1039/b819502a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shafaati M., Olin M., Båvner A., Pettersson H., Rozell B., Meaney S., Parini P., and Björkhem I. (2011) Enhanced production of 24S-hydroxycholesterol is not sufficient to drive liver X receptor target genes in vivo. J. Intern. Med. 270, 377–387 10.1111/j.1365-2796.2011.02389.x [DOI] [PubMed] [Google Scholar]
- 23. Ferri A. L., Lin W., Mavromatakis Y. E., Wang J. C., Sasaki H., Whitsett J. A., and Ang S. L. (2007) Foxa1 and Foxa2 regulate multiple phases of midbrain dopaminergic neuron development in a dosage-dependent manner. Development 134, 2761–2769 10.1242/dev.000141 [DOI] [PubMed] [Google Scholar]
- 24. van den Munckhof P., Luk K. C., Ste-Marie L., Montgomery J., Blanchet P. J., Sadikot A. F., and Drouin J. (2003) Pitx3 is required for motor activity and for survival of a subset of midbrain dopaminergic neurons. Development 130, 2535–2542 10.1242/dev.00464 [DOI] [PubMed] [Google Scholar]
- 25. Kotti T. J., Ramirez D. M., Pfeiffer B. E., Huber K. M., and Russell D. W. (2006) Brain cholesterol turnover required for geranylgeraniol production and learning in mice. Proc. Natl. Acad. Sci. U.S.A. 103, 3869–3874 10.1073/pnas.0600316103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Djelti F., Braudeau J., Hudry E., Dhenain M., Varin J., Bièche I., Marquer C., Chali F., Ayciriex S., Auzeil N., Alves S., Langui D., Potier M. C., Laprevote O., Vidaud M., et al. (2015) CYP46A1 inhibition, brain cholesterol accumulation and neurodegeneration pave the way for Alzheimer's disease. Brain 138, 2383–2398 10.1093/brain/awv166 [DOI] [PubMed] [Google Scholar]
- 27. Ayciriex S., Djelti F., Alves S., Regazzetti A., Gaudin M., Varin J., Langui D., Bièche I., Hudry E., Dargère D., Aubourg P., Auzeil N., Laprévote O., and Cartier N. (2017) Neuronal cholesterol accumulation induced by Cyp46a1 down-regulation in mouse hippocampus disrupts brain lipid homeostasis. Front. Mol. Neurosci. 10, 211 10.3389/fnmol.2017.00211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maioli S., Båvner A., Ali Z., Heverin M., Ismail M. A., Puerta E., Olin M., Saeed A., Shafaati M., Parini P., Cedazo-Minguez A., and Björkhem I. (2013) Is it possible to improve memory function by upregulation of the cholesterol 24S-hydroxylase (CYP46A1) in the brain? PLoS ONE 8, e68534 10.1371/journal.pone.0068534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bryleva E. Y., Rogers M. A., Chang C. C., Buen F., Harris B. T., Rousselet E., Seidah N. G., Oddo S., LaFerla F. M., Spencer T. A., Hickey W. F., and Chang T. Y. (2010) ACAT1 gene ablation increases 24(S)-hydroxycholesterol content in the brain and ameliorates amyloid pathology in mice with AD. Proc. Natl. Acad. Sci. U.S.A. 107, 3081–3086 10.1073/pnas.0913828107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Burlot M. A., Braudeau J., Michaelsen-Preusse K., Potier B., Ayciriex S., Varin J., Gautier B., Djelti F., Audrain M., Dauphinot L., Fernandez-Gomez F. J., Caillierez R., Laprévote O., Bièche I., Auzeil N., et al. (2015) Cholesterol 24-hydroxylase defect is implicated in memory impairments associated with Alzheimer-like Tau pathology. Hum. Mol. Genet. 24, 5965–5976 10.1093/hmg/ddv268 [DOI] [PubMed] [Google Scholar]
- 31. Hudry E., Van Dam D., Kulik W., De Deyn P. P., Stet F. S., Ahouansou O., Benraiss A., Delacourte A., Bougnères P., Aubourg P., and Cartier N. (2010) Adeno-associated virus gene therapy with cholesterol 24-hydroxylase reduces the amyloid pathology before or after the onset of amyloid plaques in mouse models of Alzheimer's disease. Mol. Ther. 18, 44–53 10.1038/mt.2009.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mast N., Saadane A., Valencia-Olvera A., Constans J., Maxfield E., Arakawa H., Li Y., Landreth G., and Pikuleva I. A. (2017) Cholesterol-metabolizing enzyme cytochrome P450 46A1 as a pharmacologic target for Alzheimer's disease. Neuropharmacology 123, 465–476 10.1016/j.neuropharm.2017.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boussicault L., Alves S., Lamazière A., Planques A., Heck N., Moumné L., Despres G., Bolte S., Hu A., Pagès C., Galvan L., Piguet F., Aubourg P., Cartier N., Caboche J., and Betuing S. (2016) CYP46A1, the rate-limiting enzyme for cholesterol degradation, is neuroprotective in Huntington's disease. Brain 139, 953–970 10.1093/brain/awv384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dai Y. B., Tan X. J., Wu W. F., Warner M., and Gustafsson J. Å. (2012) Liver X receptor β protects dopaminergic neurons in a mouse model of Parkinson disease. Proc. Natl. Acad. Sci. U.S.A. 109, 13112–13117 10.1073/pnas.1210833109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ogundare M., Theofilopoulos S., Lockhart A., Hall L. J., Arenas E., Sjövall J., Brenton A. G., Wang Y., and Griffiths W. J. (2010) Cerebrospinal fluid steroidomics: are bioactive bile acids present in brain? J. Biol. Chem. 285, 4666–4679 10.1074/jbc.M109.086678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Griffiths W. J., and Wang Y. (2011) Analysis of oxysterol metabolomes. Biochim. Biophys. Acta 1811, 784–799 10.1016/j.bbalip.2011.05.012 [DOI] [PubMed] [Google Scholar]
- 37. Crick P. J., William Bentley T., Abdel-Khalik J., Matthews I., Clayton P. T., Morris A. A., Bigger B. W., Zerbinati C., Tritapepe L., Iuliano L., Wang Y., and Griffiths W. J. (2015) Quantitative charge-tags for sterol and oxysterol analysis. Clin. Chem. 61, 400–411 10.1373/clinchem.2014.231332 [DOI] [PubMed] [Google Scholar]
- 38. Yang S., Edman L. C., Sánchez-Alcañiz J. A., Fritz N., Bonilla S., Hecht J., Uhlén P., Pleasure S. J., Villaescusa J. C., Marín O., and Arenas E. (2013) Cxcl12/Cxcr4 signaling controls the migration and process orientation of A9-A10 dopaminergic neurons. Development 140, 4554–4564 10.1242/dev.098145 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.