Abstract
Phycoerythrin (PE) is a green light–absorbing protein present in the light-harvesting complex of cyanobacteria and red algae. The spectral characteristics of PE are due to its prosthetic groups, or phycoerythrobilins (PEBs), that are covalently attached to the protein chain by specific bilin lyases. Only two PE lyases have been identified and characterized so far, and the other bilin lyases are unknown. Here, using in silico analyses, markerless deletion, biochemical assays with purified and recombinant proteins, and site-directed mutagenesis, we examined the role of a putative lyase-encoding gene, cpeF, in the cyanobacterium Fremyella diplosiphon. Analyzing the phenotype of the cpeF deletion, we found that cpeF is required for proper PE biogenesis, specifically for ligation of the doubly linked PEB to Cys-48/Cys-59 residues of the CpeB subunit of PE. We also show that in a heterologous host, CpeF can attach PEB to Cys-48/Cys-59 of CpeB, but only in the presence of the chaperone-like protein CpeZ. Additionally, we report that CpeF likely ligates the A ring of PEB to Cys-48 prior to the attachment of the D ring to Cys-59. We conclude that CpeF is the bilin lyase responsible for attachment of the doubly ligated PEB to Cys-48/Cys-59 of CpeB and together with other specific bilin lyases contributes to the post-translational modification and assembly of PE into mature light-harvesting complexes.
Keywords: cyanobacteria, post-translational modification (PTM), light-harvesting complex (antenna complex), mass spectrometry (MS), fluorescence, photosynthesis, photosynthetic pigment, bilin lyase, phycobiliprotein, phycobilisome
Introduction
Cyanobacteria, often referred to as “blue-green algae,” are photosynthetic prokaryotes capable of absorbing light energy from their environment using a light-harvesting complex called the phycobilisome (PBS5; see Fig. 1A). The PBS comprises homologous, pigmented phycobiliproteins that are composed of α- and β-subunits in heterohexameric forms (αβ)6, assembled in disklike stacked structures with the aid of specific linker proteins. In the freshwater cyanobacterium Fremyella diplosiphon (Tolypothrix sp. PCC 7601), the PBS consists of an allophycocyanin (AP; λmax = 650–655 nm) core and both phycocyanin (PC; λmax = 615–640 nm) and phycoerythrin (PE; λmax = 495–575 nm) rods with PE being core-distal (1).
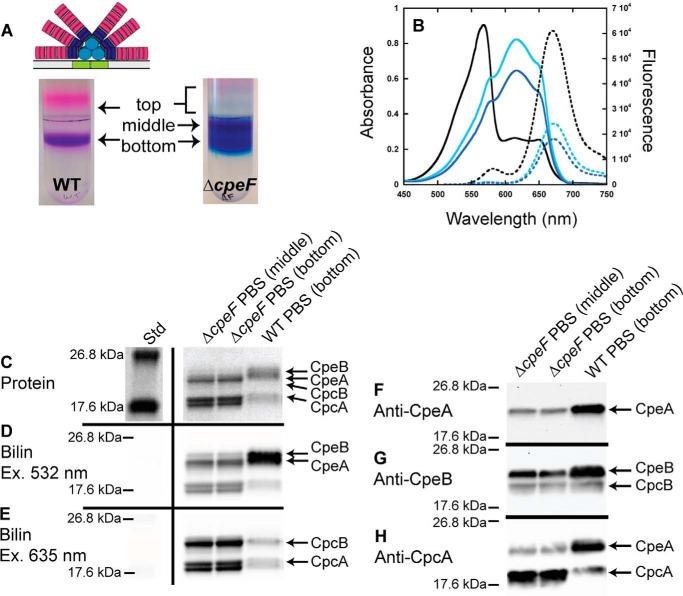
Figure 1.
Analyses of PBSs purified from WT and ΔcpeF. The WT PBS structure is illustrated above the sucrose gradient (30). A, sucrose gradients of WT (left) and ΔcpeF (right) cellular extracts from cells grown in green light. Fractions are labeled top, middle, and bottom. B, absorbance (solid lines) and fluorescence emission (dashed lines; excitation set at 490 nm) spectra of PBS fractions purified from WT (bottom fraction, black lines) and ΔcpeF (middle fraction, cyan lines; bottom fraction, blue lines) F. diplosiphon cells grown in green light. C, the Coomassie-stained SDS-polyacrylamide gel for PBSs purified from WT (bottom fraction) and ΔcpeF (middle and bottom fractions). The gel was loaded with 5 μg of total protein per sample. D and E, the Zn-enhanced fluorescence of the gel in C excited at 532 (D) and 635 nm (E). Lane Std indicates the molecular mass standard. F–H, Western blotting analyses of purified PBS samples from WT (bottom fraction) and ΔcpeF (bottom and middle fractions) using anti-CpeA (F), anti-CpeB (G), or anti-CpcA (H) antibodies. The gel was loaded with 5 μg of total protein per sample. These results are representative of three independent replicates.
Many cyanobacteria are capable of altering their PBS rod compositions in the presence of a changing photoenvironment in a process called chromatic acclimation (2). F. diplosiphon is classified as a Type III member in which the distal portions of the PBS rods are made of PE under green light conditions, but under red light conditions they are made of PC (3). This is a photoreversible process that is regulated both at transcriptional and post-transcriptional levels (2, 4) to control the expression of PE and PC subunits, lyases, chromophore biosynthesis enzymes, and linker proteins necessary for the assembly of the appropriate PBS (2). The AP core and the rod-proximal portions remain unaltered under both light conditions and are vital for the final energy transfer to the photosynthetic complexes embedded in the thylakoid membrane.
The brilliant color and light-absorbing properties of phycobiliproteins are due to the presence of tetrapyrrole phycobilins or bilin chromophores that are covalently attached by thioether linkages to highly conserved cysteine residues (5). The two bilins found in F. diplosiphon, which differ in double bond conjugation, are phycocyanobilin (PCB; λmax = 620–650 nm) attached to AP and PC and phycoerythrobilin (PEB; λmax = 540–565 nm) attached to PE (6). PCB and PEB are derived from heme (hence the name of linear tetrapyrroles with rings A–D; see Fig. S1A), which is first converted to biliverdin-IXα and then reduced by ferredoxin-dependent bilin reductases within cyanobacteria (7). Chromophore ligation to phycobiliprotein α- and β-subunits occurs post-translationally through bilin lyase–catalyzed reactions. Each bilin attachment site in the AP and PC subunits is chromophorylated by a specific bilin lyase (8). PE in F. diplosiphon contains five covalently attached PEB chromophores for efficient photosynthetic light capture, two on CpeA (α-subunit of PE) at Cys-82 and Cys-139 and three on CpeB (β-subunit of PE), one each at Cys-80 and Cys-165 and one that is doubly attached at Cys-48/Cys-59 (see Fig. S1A).
There are three major families of bilin lyases that have been characterized thus far in cyanobacteria: the CpcS/CpcU type, the CpcT type, and the CpcE/CpcF type (9). Members of the CpcS/CpcU family of lyases have an antiparallel β-barrel structure (Protein Data Bank (PDB) code 3BDR) and are involved in the ligation of PEB or PCB to the Cys-82 equivalent on β-PC, β-PE, α-AP, and β-AP subunits (8, 11–13). The CpcT family of lyases are related to CpcS/CpcU lyases (13) and have high chromophorylation site specificity for the Cys-153–equivalent position on β-PC (9, 14). The crystal structures of CpcT from Nostoc (Anabaena) sp. PCC 7120 (15) and the homolog ΦCpeT from the P-HM1 cyanophage (16) were recently elucidated and are similar to CpcS/CpcU lyases; both CpcT and ΦCpeT proteins form a homodimer with a β-barrel fold. The third family of lyases is the CpcE/CpcF (E/F) type, which is unrelated in sequence and structure to the other lyase families. Members of this E/F family that have been characterized are generally involved in ligating chromophores to the α-subunits of phycobiliproteins (17–21). Some members of this lyase family have the ability to perform chaperone-like functions (21, 22), whereas others have been shown to function as bilin lyase/isomerases (23–25). All E/F-type lyases contain multiple HEAT-repeat motifs that facilitate protein–protein interactions (23, 25, 26). The crystal structure of CpcE/CpcF from Nostoc sp. PCC 7120 was determined, and the structure is primarily α-helical consisting of crescent-shaped elongated superhelices or solenoids (27).
It has been suggested that the five PEB-binding sites on PE in F. diplosiphon are chromophorylated by a minimum of five bilin lyases (28, 29). Thus far, only two PE lyases have been characterized, the CpcS/CpcU-type homolog CpeS (attaches PEB to CpeB-Cys-80 (29)) and the E/F-type homolog CpeY (attaches PEB to CpeA-Cys-82 with the aid of the CpeZ chaperone (29)).6 The other bilin lyases for the remaining Cys sites are unknown, although it is hypothesized to include lyases from each of the three lyase families (28), i.e. CpeT (14, 31), CpeU (11, 31), and CpeF (31, 32). Here, we characterize the functional role of CpeF, a member of the E/F-type lyase family in F. diplosiphon, by using a cpeF deletion mutant and recombinant protein studies. We found that it is involved in the chromophorylation of CpeB at position Cys-48/Cys-59, and we show that attachment at ring A of PEB to Cys-48 likely occurs prior to ring D ligation at Cys-59. This is the first report of an E/F-type lyase being responsible for chromophore ligation to a β-subunit of a phycobiliprotein.
Results
In silico analysis of CpeF from F. diplosiphon
In F. diplosiphon, the 912-bp cpeF gene is located ∼400 bp downstream from the pebAB operon (Fig. S1B), whose transcript abundance increases in cells grown under green light (33) and which encodes the ferredoxin-dependent bilin reductases PebA and PebB. These two proteins, in addition to heme oxygenase (encoded elsewhere in the genome by hoI), are responsible for producing PEB from cellular heme (7).
To gain insight into the potential role of CpeF, we analyzed its sequence (32.4 kDa predicted, residues 11–313 from GenBankTM accession number EKF00796.1) by BLASTp 2.3.1 (34, 35) and found high sequence similarity to phycobiliprotein lyases (41–74% similarity), phycoerythrin II biosynthesis protein MpeU from cyanobacteria (43–54% similarity), glycosyltransferase family 2 (37–87% similarity), and hypothetical proteins. CpeF showed high similarity to MpeV and MpeU from the marine Synechococcus sp. WH 8020 (54 and 50% similarity, respectively), both of which were originally identified within the phycoerythrin II operon and are members of the E/F family of lyases (32, 36, 37). CpeF was 36% similar to CpcE and 28% similar to CpcF from Synechococcus sp. PCC 7002, the first E/F-type bilin lyase to be characterized (20). CpeF was also 30% similar to MpeZ, an E/F-type lyase/isomerase characterized from Synechococcus sp. RS 9916 responsible for attaching PEB to Cys-83 of PEII α-subunit and subsequently isomerizing it to a different type of bilin called phycourobilin (PUB; λmax = 490–495 nm) (25). We further analyzed the CpeF sequence using the Phyre2 modeling program (38) to predict the secondary and 3D structures. Using PDB code 5N3U as a template (27), Phyre2 predicted that the CpeF protein has an α-solenoid type structure (Fig. S1C), consistent with the structure of the only E/F-type lyase crystallized to date, CpcE/CpcF from Nostoc sp. PCC 7120 (27). 74% of CpeF's secondary structure was predicted with very high confidence to form α-helices, and only 9% of the protein was disordered. The protein CpeF appears to have six HEAT-repeat domains (39), which are featured in all members of the E/F lyase family (23, 25, 26). For the predicted 3D structure (Fig. S1C), 243 residues of CpeF (303 residues total; 80%) were modeled with 99.89% confidence by the template PDB code 5N3U (27). After taking into account multiple factors, including its location in the F. diplosiphon genome, the six HEAT-repeat motifs within the sequence, the amino acid homology to other known E/F-type lyases, and the predicted secondary and 3D structure, we hypothesized that cpeF encodes an E/F-type bilin lyase that is responsible for attaching PEB to one or both subunits of PE.
Phenotypes and whole-cell analyses of WT F. diplosiphon and ΔcpeF
To determine the function of cpeF in cyanobacteria, we generated a markerless deletion (40–42) of cpeF in F. diplosiphon (ΔcpeF; Fig. S1B) and observed that these cells manifested a PE-deficient phenotype (Fig. S2). This phenotype was readily observable for cells grown in green light, which normally induces the red-colored PE in wildtype (WT; hence the brick red–colored cultures; Fig. S2A). In the ΔcpeF cells, green light failed to induce a high level of PE (hence the green-colored cultures; Fig. S2A). Whole-cell absorbance spectra confirmed that ΔcpeF mutant had reduced levels of PE when compared with WT (Fig. S2B). We also found that the ΔcpeF mutant grew slower than WT in green light (Fig. S2C), but in red light, there was no difference in growth between the mutant and WT cells (data not shown). These results indicate that deleting cpeF has an adverse effect on the expression of PE in F. diplosiphon during growth in green light.
Analyses of PBS assembly in ΔcpeF versus WT cells
To determine the effect of deleting cpeF on PBS composition, PBSs from WT and ΔcpeF were purified using sucrose density gradients and analyzed using absorbance and fluorescence emission spectroscopy. Following ultracentrifugation, two colored fractions were observed in the WT sucrose density gradient: a minor “top” fraction consisting of small amounts of dissociated phycobiliproteins and a major “bottom” purple fraction containing fully assembled PBSs (Fig. 1A). The absorption spectra of the WT complete PBSs showed a large amount of PE (λmax = 568 nm) compared with the amount of PC (λmax = 615 nm) and AP (λmax = 649 nm; Fig. 1B and Table S1). When excited at 490 nm, WT PBSs emit strongly at 669 nm with a small shoulder at 582 nm, indicating a transfer of energy from PE to PC to AP with some emission from PE (582 nm; Fig. 1B and Table S1).
In the ΔcpeF mutant, three fractions were collected from the sucrose gradients (which purified PBSs of various sizes and composition) and labeled as top, middle, and bottom with the bulk of the PBSs migrating to the middle and bottom fractions (Fig. 1A). As in WT, the top fractions consisted of dissociated proteins and was not analyzed further due to the small amounts present. The migration of the ΔcpeF PBSs to the middle fraction indicates a smaller molecular mass for the PBSs compared with those in the bottom fractions. Nevertheless, both the middle and bottom fractions isolated from ΔcpeF were blue in color, indicating a reduction in PE and a predominance of PC and AP. The absorbance spectra confirmed severely reduced levels of PE within the PBSs (Fig. 1B). However, when exciting the PBSs at 490 nm, energy was transferred from PE to PC to AP and exhibited fluorescence emission at 673 nm, indicating the presence of some PE within both middle and bottom fractions of the PBSs, albeit less than that found in WT (Fig. 1B and Table S1). The maximal absorbance by PE within the PBSs from the mutant cells in both middle and bottom fractions was at 577 nm (shoulder peak in Fig. 1B), which is red-shifted when compared with WT peak (λmax = 568 nm; Fig. 1B).
PBS samples from WT and ΔcpeF were further analyzed by SDS-PAGE for protein and bilin content. Covalent bilin attachment to proteins was examined by fluorescence emission of the gel after incubation with ZnSO4 with excitation at 532 nm (Fig. 1D), which detects both PEB and PCB, or at 635 nm (Fig. 1E), which detects PCB only (43). The protein stain indicated that, compared with WT, both CpeA and CpeB levels were reduced in the ΔcpeF mutant PBS samples with CpeB being less abundant than CpeA (Fig. 1C). PEB fluorescence was reduced as measured by Zn-enhanced fluorescence when excited at 532 nm (Fig. 1D) with CpeB fluorescing less than CpeA. This reduced Zn-enhanced fluorescence may be attributed to a reduced amount of PE produced in the mutant. Western blot analysis confirmed the presence of both CpeA and CpeB in the ΔcpeF samples, but the quantities of both proteins were drastically reduced compared with WT (Fig. 1, F and G). The relative amount of PC also apparently increased in the ΔcpeF mutant compared with WT (Fig. 1, C, E, and H). Together, these data show that when the putative lyase-encoding gene cpeF was deleted, PE synthesis was severely reduced, normal levels of PE being assembled into the PBSs was hindered, and the PE absorption peak was significantly red-shifted.
Analyses of PE in ΔcpeF versus WT cells
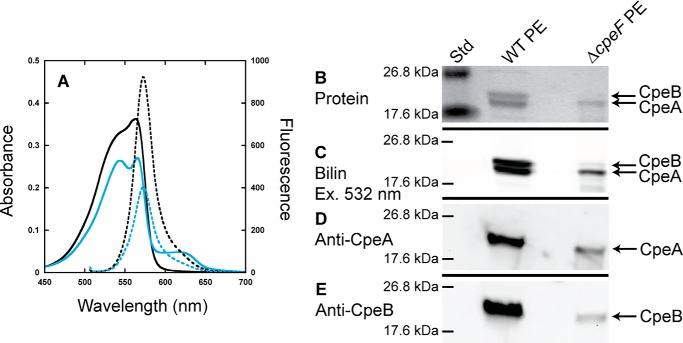
To determine how PE alone (i.e. free and assembled in PBSs) was being affected in the ΔcpeF mutant, total PE was isolated from WT and ΔcpeF cells and subsequently analyzed using spectroscopy and SDS-PAGE (Fig. 2). WT PE had a maximum absorbance at 563 nm with a shoulder at 545 nm, and the ΔcpeF PE had a maximum absorbance at 565 nm, which was lower relative to the shoulder at 544 nm, but both PE samples had a maximum fluorescence at 573 nm when excited at 490 nm (Fig. 2A). The small absorbance peak at 625 nm present in the mutant sample was caused from PC/AP contamination (data not shown). Zn-enhanced fluorescence (excitation at 532 nm) of WT PE showed a high fluorescence signal for both CpeB and CpeA (Fig. 2C). However, in the ΔcpeF sample, CpeB protein levels and Zn-enhanced fluorescence of CpeB were severely reduced when compared with CpeA (Fig. 2, B and C). Western blotting analyses using both anti-CpeA and anti-CpeB antibodies (Fig. 2, D and E, respectively) confirmed that CpeB levels in the mutant PE sample were severely reduced when compared with CpeA.
Figure 2.
Analyses of PE purified from WT and ΔcpeF. A, absorbance (solid lines) and fluorescence emission (dashed lines; excitation set at 490 nm) spectra of PE purified from WT (black lines) and ΔcpeF (cyan lines) F. diplosiphon cells grown in green light. B, the Coomassie-stained SDS-polyacrylamide gel for PE purified from WT and ΔcpeF. Lane Std indicates the molecular mass standard. The gel was loaded with 10 μl of each sample. C, the Zn-enhanced fluorescence of the gel in B excited at 532 nm. D and E, Western blotting analyses of PE purified from WT and ΔcpeF using anti-CpeA (D) and anti-CpeB (E) antibodies. These results are representative of three independent replicates.
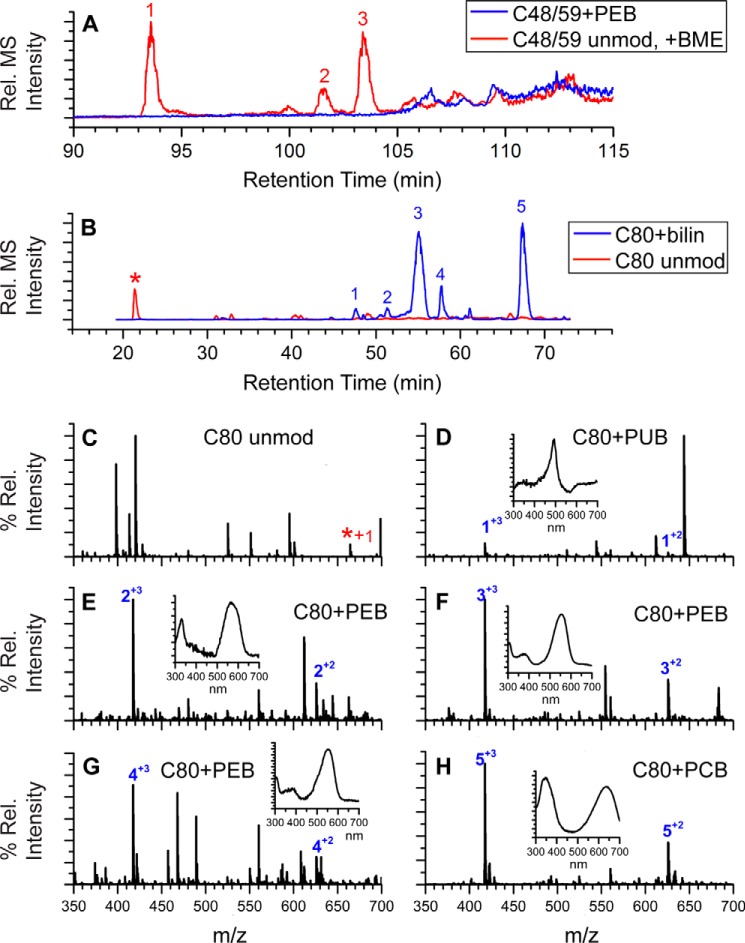
To identify potential changes in the post-translational modifications of PE, we used LC-MS/MS to analyze PE samples purified from both WT and ΔcpeF after trypsin digestion (Table 1). As expected, five peptides6 containing PEB were detected from the WT PE sample, two from CpeA and three from CpeB (Table 1). Peptides were separated by HPLC, and ligated chromophores were detected using visible absorbance in line with the mass spectrometer. The nonchromophorylated peptides containing the five Cys sites were also detected but in very low amounts within the peptide mixtures.6 In contrast, PE purified from the ΔcpeF mutant only had four observable peaks with bound PEB (Table 1). The m/z 1200.06 peak, corresponding to the (M + 4H)4+ ion of CpeB-Cys-48/Cys-59 peptide with the doubly ligated PEB, was not detected (Fig. 3A). Generally, a peptide with such a large m/z is more difficult to detect, especially given its size and the low abundance of CpeB in the sample. However, the same CpeB peptide was detected without a bilin (Table 1 and Fig. 3A; m/z 1404.323+; PEB molecular mass, 587 Da). The unmodified version of this peptide (no bound PEB) was modified by 2-mercaptoethanol (BME) present in the buffer during the protein purification and digestion processes (Fig. 3A). Interestingly, the CpeB-Cys-80 peptide mixtures from the ΔcpeF mutant PE sample had PEB, PCB, or PUB attached, as identified by their UV-visible spectra (Fig. 3, B–H; PEB, λmax = 540–565 nm; PCB, λmax = 620–650 nm; PUB, λmax = 490–495 nm). However, the ratio of peptides with PEB to peptides with PCB attached was 3:2 within the peptide mixture, and very low amounts of PUB-bound peptides were detected. Based on these observations we conclude that in vivo, cpeF is required for Cys-48/Cys-59 PEB chromophorylation on CpeB. Abrogation of this chromophorylation induces loss of CpeB and overall PE reduction in the cell. PEB addition at the Cys-48/Cys-59 sites may also enhance PEB addition at subsequent sites, such as Cys-80 in cyanobacteria.
Table 1.
Observed LC-MS/MS peaks of trypsin-digested PE peptides
| Strain | α-82 (m/z)1+ | α-139 (m/z)1+ | β-80 (m/z)1+ | β-165 (m/z)1+ | β-48/59 (m/z)1+ |
|---|---|---|---|---|---|
| WT | 936 | 1361 | 1251 | 2105 | 4797 |
| ΔcpeF | 936 | 1361 | 1251a | 2105 | 4210b |
a Peptide mixtures contained peptides with PEB, PCB, or PUB bound at that specific Cys residue.
b Only the peptide without bound PEB (m/z 4210; PEB molecular mass, 587 Da) was observed.
Figure 3.
Extracted ion chromatograms and LC-MS spectra for trypsin-digested peptides from ΔcpeF PE. A, combined extracted ion chromatograms for the peptides RLDAVNAIASNASC48MVSDAVAGMIC59ENQGLIQAGGNCYPNR at m/z 1200. 06 ((M + 4H)4+, PEB-modified; blue line) and m/z 1404.32 ((M + 3H)3+, unmodified (unmod) + one BME; red line) of CpeB isolated from ΔcpeF cells grown in green light. Numbers 1–3 indicate three versions of the unmodified peptide observed (one BME on Cys-48, -59, or -71). B, extracted ion chromatograms for the peptides MAAC80LR at m/z 625.81 ((M + 2H)2+, bilin-modified; blue line) and m/z 332.67 ((M + 2H)2+, unmodified; red line and asterisk) of CpeB isolated from ΔcpeF cells grown in green light. Numbers 1–5 indicate the MAAC80LR peptides that have a bilin bound. C, MS of unmodified MAAC80LR. D–H, MS for the five modified versions of the peptide MAAC80LR containing Cys-80 as specified in B. Inset graphs are the UV-visible absorbance spectra for the peaks at the retention times that are numbered with a + charge. The type of bilin bound to Cys-80 is indicated per panel. These results are representative of two independent replicates.
Recombinant CpeF protein coexpressions
To better define the role of CpeF in PE chromophorylation, we examined its role in a heterologous system where both CpeF and CpeB were coexpressed in Escherichia coli that were engineered to produce PEB (44).6 Hexahistidine-tagged (HT-) CpeB was purified using nickel-nitrilotriacetic acid affinity chromatography after cell lysis and analyzed using fluorescence emission spectroscopy and SDS-PAGE (Fig. 4). Recombinant CpeF was coproduced with HT-CpeB and the PEB synthesis enzymes PebS/HoI (45) to detect any potential lyase activity. This expression combination resulted in very low HT-CpeB solubility (Fig. 4B) with no detectable CpeB fluorescence (Fig. 4, A and C). It has been previously observed that very little soluble CpeB accumulates within E. coli when it is expressed without a PEB chromophore post-translationally attached to Cys-80 (29). Therefore, we used the lyase CpeS (CpeB-Cys-80 lyase (29)) to enhance the solubility of HT-CpeB both alone and in the presence of CpeF. In these experiments, CpeF was expressed after all others using a sequential induction system to chromophorylate Cys-80 prior to CpeF expression (see “Experimental procedures”). Isolated HT-CpeB showed an increase in fluorescence (Fig. 4A) both in the presence and absence of CpeF due to the covalent attachment of PEB to Cys-80 verified by Zn-enhanced fluorescence (Fig. 4C); however, the solubility of HT-CpeB remained very low (Fig. 4B), suggesting that even partially chromophorylated CpeB does not accumulate in an E. coli system very efficiently.
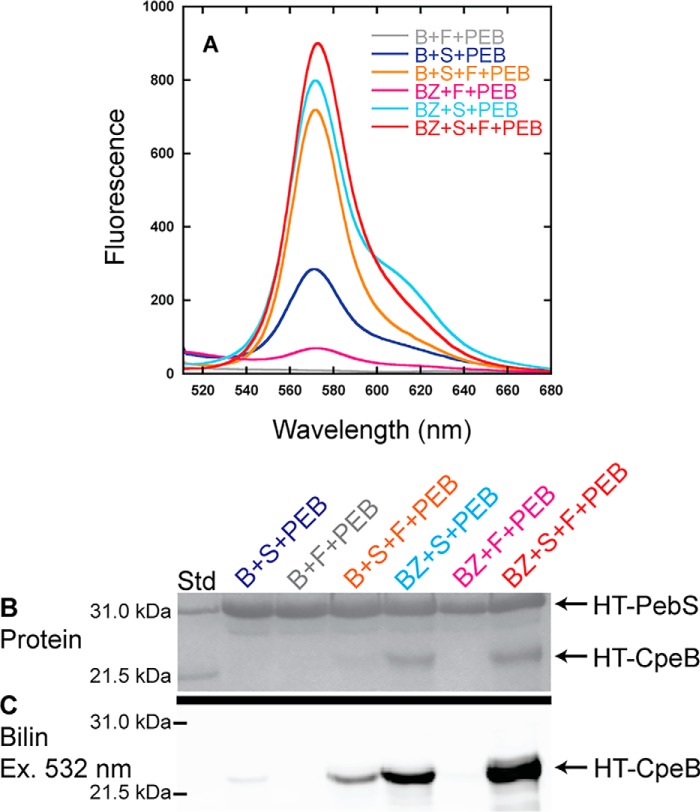
Figure 4.
Recombinant CpeF coexpressions with HT-CpeB. A, fluorescence emission (excitation set at 490 nm) spectra of purified HT-CpeB obtained from E. coli cells expressing pPebS in addition to pCpeB2 and pCpeS (B+S+PEB; blue); pCpeB2 and pHT-CpeF2 (B+F+PEB; gray); pCpeB2, pCpeS, and pHT-CpeF2 (B+S+F+PEB; orange); pCpeBZ and pCpeS (BZ+S+PEB; cyan); pCpeBZ and pHT-CpeF2 (BZ+F+PEB; magenta); or pCpeBZ, pCpeS, and pHT-CpeF2 (BZ+S+F+PEB; red). B, the Coomassie-stained SDS-polyacrylamide gel for purified HT-CpeB samples from A. Lane Std indicates the molecular mass standard. C, the Zn-enhanced fluorescence of the gel in B excited at 532 nm. These results are representative of three independent replicates.
To further increase the amount of soluble HT-CpeB and potentially boost the lyase activity of CpeF, we asked whether the chaperone-like protein CpeZ could enhance these processes (29).6 Coexpressing HT-CpeB, the lyase CpeS, the chaperone-like protein CpeZ, and PebS/HoI, with subsequent expression of CpeF, dramatically increased both HT-CpeB protein solubility (Fig. 4B) and PEB fluorescence (Fig. 4, A and C) when compared with the same coexpressions without CpeS. When HT-CpeB was coproduced with CpeS, CpeZ, and PebS/HoI, a fluorescent product was detected, but the signal was less intense than when CpeF was present. Because CpeS is known to attach PEB to Cys-80 of CpeB (29) and CpeZ acts as a chaperone only,6 we reasoned that CpeF was directly involved in PEB attachment to one or both of the remaining Cys sites in CpeB (Cys-165 and Cys-48/Cys-59).
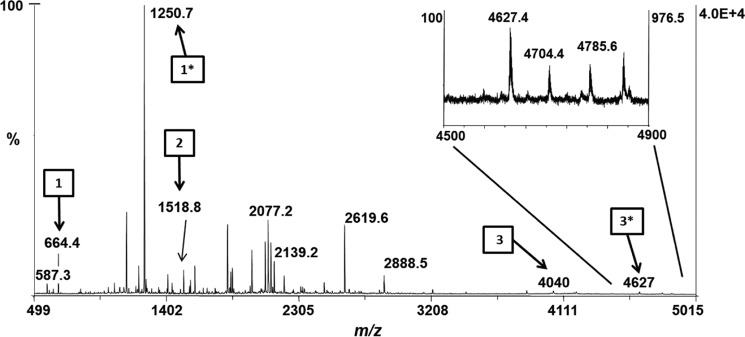
To further examine PEB attachment on CpeB at these cysteine sites, the recombinant protein samples were digested with trypsin and analyzed by MALDI-MS. As predicted, any sample including the lyase CpeS had a PEB attached to Cys-80 (m/z 1250.71+; Table S2). When HT-CpeB was expressed alone with CpeF or with CpeF and CpeZ, no PEB chromophore was detected on any HT-CpeB peptide by MS (Table S2). However, when HT-CpeB was expressed with CpeZ, CpeS, and CpeF, a PEB chromophore was detected on Cys-48/Cys-59 (m/z 4627.41+) as well as Cys-80 (m/z 1250.71+; Table S2 and Fig. 5). The peptide with m/z 4627.41+ was detected in low quantities (Fig. 5, arrow 3*); however, its detection by MS indicated that when recombinant CpeB was partially chromophorylated by CpeS and in the presence of the chaperone-like protein CpeZ, then CpeF functioned as a lyase by attaching one PEB to CpeB at Cys-48/Cys-59.
Figure 5.
MALDI-MS spectrum of recombinant HT-CpeB peptides. The MALDI-MS spectrum of the peptide mixture resulting from the trypsin digestion of HT-CpeB purified from E. coli cells expressing pCpeBZ, pCpeS, pHT-CpeF2, and pPebS (BZ+S+F+PEB sample) is shown. Arrows bearing numbers indicate peaks that were identified as tryptic peptides containing a Cys residue without PEB bound: Cys-80 (arrow 1, m/z 664.41+), Cys-165 (arrow 2, m/z 1518.81+), and Cys-48/Cys-59 (arrow 3, m/z 40401+). Arrows bearing numbers with asterisks indicate peaks that were identified as tryptic peptides containing a PEB chromophore. Attachment was found to occur at Cys-80 (arrow 1*, m/z 1250.71+) and double attachment at Cys-48/Cys-59 (arrow 3*, m/z 4627.41+). No attachment to Cys-165 was detected. The inset graph is a close-up view of the m/z 4500–4900 range. These results are representative of two independent replicates.
Given the previous observations that recombinant phycobiliprotein production is enhanced when both α- and β-subunits are coexpressed (13, 14), we sought to test whether coexpression with CpeA would improve the yield of soluble CpeB. For this we made a new construct, HT-CpeBA, and analyzed CpeB as before. To account for chromophorylation of CpeA-Cys-82, we also coexpressed the CpeY lyase (29). Although CpeA was not His-tagged, small amounts copurified with HT-CpeB, its natural interacting partner, in these expressions. When compared with the previous coexpressions, a greater amount of soluble HT-CpeB protein accumulated in the presence of CpeA and PebS/HoI (Fig. S3B), and very little nonenzymatic PEB addition occurred (Fig. S3C). No fluorescence was observed when the sample was excited at 490 nm (Fig. S3A). The control experiments with CpeF or CpeZ alone in the presence of HT-CpeBA and PebS/HoI resulted in low soluble protein yields and little to no fluorescence (Fig. S3, A–C) as seen previously when CpeA was not present (see Fig. 4). However, when both CpeF and CpeZ were expressed together with HT-CpeBA and PebS/HoI, a slightly fluorescent product was produced, indicating the presence of a covalently bound PEB (Fig. S3, A–C). Furthermore, increasing the number of covalently bound PEB molecules to either CpeB (by CpeS (29)) or CpeA (by CpeY (29)) helped increase the accumulation (Fig. S3E) and the fluorescence (Fig. S3, D and F) of CpeB.
To determine which Cys residues had a PEB covalently bound, all fluorescent protein samples were digested with trypsin and analyzed by LC-MS/MS. As expected, in every sample that had CpeS expressed, a PEB was covalently bound to CpeB at Cys-80 (m/z 417.543+; Table 2). In the control sample where only CpeS was expressed, no other Cys residues were chromophorylated in the peptide mixtures (Table 2 and Fig. 6A). Also, as expected, in every sample that had CpeY expressed, a PEB was bound to CpeA at Cys-82 (m/z 468.232+) even though only small amounts copurified with HT-CpeB. When CpeF was expressed alone with HT-CpeBA, no fluorescence was detected and therefore was not analyzed by MS. However, when both CpeF and CpeZ were expressed together, a small amount of fluorescence was detected, and MS revealed that a very small amount of CpeB-Cys-48/Cys-59 peptides was chromophorylated with PEB (Table 2 and Fig. 6B). This chromophorylated peptide was detected in every sample where CpeF was coexpressed (Table 2). The amount of chromophorylated CpeB-Cys-48/Cys-59 peptides appeared to increase as more proteins were expressed in the system, such as the lyases CpeS and CpeY and the chaperone-like protein CpeZ (Table 2 and Fig. 6, C and D). Overall, these results indicate that CpeF acts as a bilin lyase responsible for attaching PEB to CpeB at Cys-48/Cys-59, and this activity is increased in the presence of the CpeZ chaperone and when either CpeA (if present) or CpeB is partially chromophorylated.
Table 2.
Observed LC-MS/MS peaks of trypsin-digested recombinant HT-CpeBA peptides
ND represents peptides that were not detected, and Unmod represents unmodified peptides that lack a PEB chromophore. BA, pCpeBA; S, pCpeS; SF, pCpeSF2; F, pCpeF2; Z, pNT-CpeZ3; YZ, pNT-CpeYZ.
| Samplea | α-82 | α-139 | β-80 | β-165 | β-48/59 (% PEB)b |
|---|---|---|---|---|---|
| BA+S+PEB | ND | Unmod | PEB | Unmod | Unmod (0) |
| BA+SF+PEB | Unmod | Unmod | PEB | Unmod | PEB (14.1) |
| BA+F+Z+PEB | Unmod | Unmod | Unmod | ND | PEB (12.5) |
| BA+F+YZ+PEB | PEB | Unmod | Unmod | Unmod | PEB (2) |
| BA+SF+Z+PEB | Unmod | Unmod | PEB | Unmod | PEB (18.1) |
| BA+SF+YZ+PEB | PEB | Unmod | PEB | Unmod | PEB (35.6) |
a See Fig. S3 for sample descriptions.
b Values in parentheses represent the estimated percentage of β-48/59 peptides chromophorylated with PEB relative to non-chromophorylated β-48/59 peptides based upon the integrated intensities for the various charge states and missed cleavages or non-enzymatic modifications (e.g. addition of BME, oxidation of methionine, and internal disulfide bond).
Figure 6.
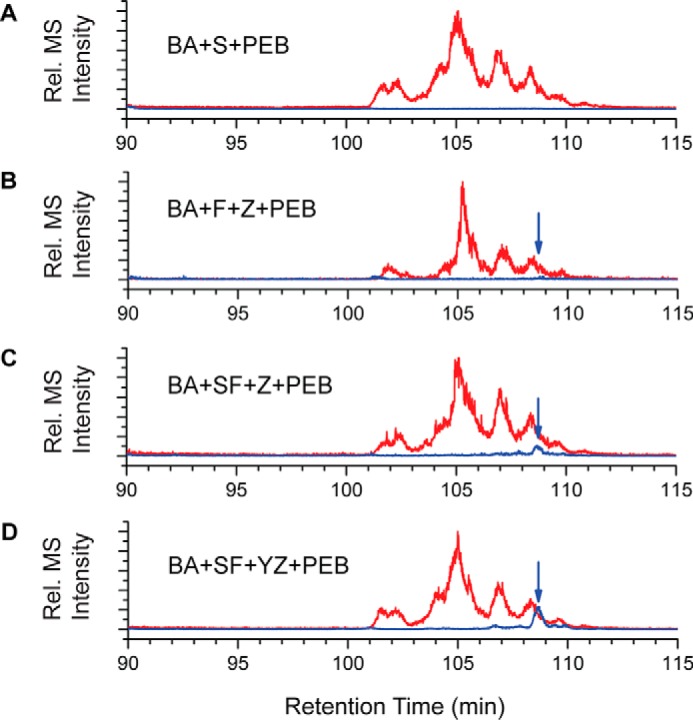
Extracted ion chromatograms for trypsin-digested peptides from HT-CpeBA coexpressions. Combined extracted ion chromatograms for unmodified Cys-48/Cys-59 peptides (red) and PEB-cross-linked versions of Cys-48/Cys-59 peptides (blue and indicated with arrows) from purified HT-CpeBA obtained from E. coli cells expressing pCpeBA and pNT-PebS in addition to pCpeS (BA+S+PEB) (A), pCpeF2 and pNT-CpeZ3 (BA+F+Z+PEB) (B), pCpeSF2 and pNT-CpeZ3 (BA+SF+Z+PEB) (C), or pCpeSF2 and pNT-CpeYZ (BA+SF+YZ+PEB) (D) are shown. These results are representative of two independent replicates.
Analyses of CpeF mechanism of PEB attachment using site-directed variants
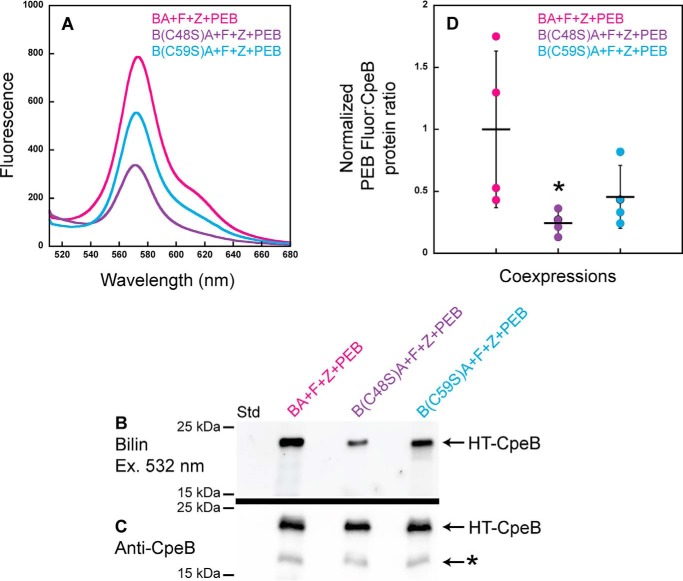
Crystal structures of PE from various species of cyanobacteria reveal that the bilin doubly bound to CpeB is attached to the Cys-48– and Cys-59–equivalent residues at the A ring and D ring of the bilin, respectively (46–48); however, the order of PEB attachment to these sites has not been examined. To identify the order of this attachment and, thus, gain insight into the mechanism of CpeF PEB ligation, site-specific variants within CpeB were produced (C48S and C59S) in our HT-CpeBA construct. Recombinant CpeF was coproduced in E. coli with HT-CpeBA or C48S or C59S variant with the CpeZ chaperone and the PEB synthesis enzymes PebS/HoI (45). His-tagged proteins were purified and analyzed by fluorescence emission spectroscopy, SDS-PAGE, and immunoblotting (Fig. 7). All three coexpressions produced fluorescent products due to the attachment of PEB to CpeB (Fig. 7, A and B); however, the fluorescence produced from the mutant variants was reduced (Fig. 7, A and B) when compared with the WT HT-CpeBA. The amount of soluble HT-CpeB in all samples (Fig. 7, A and B) was relatively similar as measured by Western blotting analyses (Fig. 7C). We quantitated the PEB fluorescence (Fig. 7B) and the amount of soluble CpeB protein band intensities (Fig. 7C) from four independent replicates. Fig. 7D represents how much CpeB was chromophorylated in each protein sample (e.g. bilin content:protein amount) after normalization (see “Experimental procedures”). The average bilin content:CpeB ratio (n = 4) of C59S variant was 46% of WT, whereas that of the C48S variant was 24% of WT, indicating that the C48S variant had a more severe reduction of chromophorylation, which was significantly different from that of WT (p = 0.0277; Fig. 7D). The chromophore conformation in the binding pocket of CpeB is such that Cys-48 will ligate to ring A followed by ligation at ring D to Cys-59 (46). Together, these data indicate that when either Cys-48 or Cys-59 is mutated to Ser, PEB addition to CpeB is still possible; however, there is a more severe reduction in binding to the C48S variant, indicating that this is likely the first site of attachment for PEB followed by thioether formation with ring D at Cys-59.
Figure 7.
Recombinant CpeF coexpressions with HT-CpeB variants show the order of PEB ring A versus ring D ligation. A, fluorescence emission (excitation set at 490 nm) spectra of purified HT-CpeBA variants obtained from E. coli cells expressing pCpeF2, pNT-CpeZ3, and pNT-PebS in addition to pCpeBA (BA+F+Z+PEB; magenta); pCpeB(C48S)CpeA (B(C48S)A+F+Z+PEB; purple); or pCpeB(C59S)CpeA (B(C59S)A+F+Z+PEB; cyan). All protein samples were diluted to 0.75 μg μl−1 prior to obtaining the fluorescence spectra. B, the Zn-stained SDS-polyacrylamide gel for purified HT-CpeBA variant samples from A. Lane Std indicates the molecular mass standard with size markers shown at left. The gel was loaded with 2 μg of total protein per lane. C, Western blot analysis of purified HT-CpeBA variant samples using anti-CpeB antibodies. The gel was loaded with 2 μg of total protein per sample. The asterisk indicates degraded CpeB; this degradation product was not observed in B because it did not have PEB covalently attached. D, scatter plot representing the normalized ratio of Zn fluorescence from PEB (B):CpeB intensities as obtained from the Western blotting in C for the HT-CpeB samples. Error bars represent the S.D. of four independent replicates (mean values are indicated with a horizontal line for each sample set). The average BA+F+Z+PEB ratio was set to 1, and the mutants are represented relative to this value in D. Normalized fluorescence data were analyzed by a two-way analysis of variance (α = 0.05) with experimental replicate and strain (or mutant) as factors. Fluorescence of the strains was significantly different (p = 0.0292). Of note, B(C48S)A+F+Z+PEB showed significantly lower fluorescence compared with BA+F+Z+PEB (Tukey's honestly, p = 0.0277, noted with asterisk). Strain B(C59S)A+F+Z+PEB was intermediate between B(C48S)A+F+Z+PEB and BA+F+Z+PEB (p = 0.0941 versus B(C48S)A+F+Z+PEB and p = 0.6054 versus BA+F+Z+PEB).
Discussion
This study investigated the bilin lyase function of CpeF. The deletion of cpeF caused a reduced PE phenotype, indicating that this protein plays a role in PE biosynthesis and/or stability. The remaining and minor PE amount that was incorporated into the PBSs of the ΔcpeF mutant had a 9-nm red-shifted absorbance when compared with WT PE. This red shift might be the result of the presence of a noncovalently attached PEB within the bilin-binding pocket of PE. This noncovalent PEB would have an extra double bond in conjugation (11, 18, 19, 49). Each bilin within the structure of the PBS contributes uniquely to the overall absorbance spectrum (1, 50). Another possibility is the complete absence of PEB from one or more Cys sites, which may also cause a shift in the absorbance spectrum as previously demonstrated in other lyase mutants when bilins were missing from phycobiliprotein subunits (11, 17, 18). CpeF's role as a bilin lyase is consistent with this phenotype. The absence of PEB at CpeB-Cys-48/Cys-59 in our ΔcpeF mutant is likely the cause of the decrease in the peak at 565 nm relative to the shoulder in the absorbance spectrum for the PE.
An unexpected result was that the MS analysis of PE purified from ΔcpeF mutant cells also showed that PCB (Fig. 3H) and PUB (Fig. 3D) were covalently attached to CpeB peptides at Cys-80 (in addition to PEB). This Cys-80–binding site is normally chromophorylated by CpeS (29). Because atypical bilins are covalently attaching to this site in the ΔcpeF mutant, it is possible that CpeF assists in the activity of CpeS in vivo to facilitate the interaction among CpeS, PEB, CpeZ, and/or CpeB; however, no interactions between CpeF and CpeS proteins were detected in pulldown assays (data not shown). Another possibility is that in cyanobacteria, CpeF acts before CpeS in PEB ligation, and that without CpeF, less efficient PEB ligation at Cys-80 by CpeS occurs. Bilins naturally have a high affinity for their binding pockets in phycobiliproteins. Nonenzymatic bilin addition has been seen in PE subunits where PEB was ligated at very slow reaction rates and, in some cases, resulted in the attachment of oxidized or isomerized bilin adducts (51). Because these analyses were performed on PE purified from whole cells, it is possible that these CpeB variants are not incorporated into mature PBSs. Phycobiliproteins with large defects in folding or chromophorylation are prevented from assembling into PBSs (52), and these partially chromophorylated subunits may be targeted for turnover.6
The lack of bilins on phycobiliproteins (due to a mutation of the Cys ligation site of the phycobiliprotein or in the gene encoding the bilin lyases) affects stability, turnover rates of the protein, the assembly of phycobiliproteins into the PBS, and energy transfer efficiency (1, 50, 52, 53). Once apophycobiliproteins are chromophorylated, they form monomers (αβ), trimers (αβ)3, and then hexamers (αβ)6, which are assembled into the PBSs. If PE is missing one or more PEB chromophores, this could result in instability of the PE subunits, leading to protein degradation, preventing proper assembly into the PBS, and thereby decreasing PBS light capturing and energy transfer efficiency (1, 53, 54). In the absence of bilin lyases and bilin ligation, there is a marked decrease in phycobiliprotein content (11, 14, 17, 55–57), consistent with our results when cpeF was deleted in F. diplosiphon.
Our heterologous expression system in E. coli demonstrated that recombinant CpeF was able to attach PEB to Cys-48/Cys-59 on CpeB only after CpeB was already chromophorylated by CpeS on the central Cys-80 site and stabilized by CpeZ (29). This sequential induction system was useful for detecting the lyase function of CpeF. CpeZ showed a chaperone-like function and helped stabilize the conformation of recombinant CpeB.6 It is possible that the stabilized conformation of CpeB is necessary for CpeF to recognize the Cys-48/Cys-59 site for the attachment of PEB. It is likely that an ordered ligation of each PEB to PE subunits is necessary in vivo to ensure that holo-PE is properly synthesized in cyanobacteria. Biswas et al. (29) suggested that addition to the central bilin at CpeB-Cys-80 occurs first to produce the appropriate conformational stability for CpeB, which allows the other lyases to act on the other two sites at Cys-165 and Cys-48/Cys-59. However, Zhao et al. (58) investigated the order of bilin addition in the homologous protein CpcB in Nostoc PCC 7120 and demonstrated that the primary addition of a bilin to the CpcB-Cys-155 site allowed for the subsequent chromophorylation of the CpcB-Cys-84 site, whereas the inverse scenario inhibited the addition of the second bilin (58). CpeB from F. diplosiphon is more complex than CpcB in terms of bilin-binding sites (CpcB has two; CpeB has three). Further experimentation is required to determine the actual sequence of PEB addition to CpeB.
The mechanism of E/F-type lyases is believed to be complex and has been investigated using the heterodimeric CpcE/CpcF and PecE/PecF (21, 22, 27). Both subunits are necessary to efficiently attach a bilin to the corresponding α-subunit. For example, PecE first binds PecA and is able to slowly attach PCB, which is accelerated if PecE and PecA form a complex prior to the addition of PCB, indicating that protein–protein interaction is the rate-limiting step (22). When the isomerizing subunit PecF is bound to the complex, the PCB on PecA is isomerized to phycoviolobilin (22). In F. diplosiphon, CpeF and CpeB interaction may be efficiently facilitated by CpeZ, which stabilizes CpeB's conformation. Ligation of ring A of PEB to Cys-48 and then ring D to Cys-59 likely occurs because the two proximal Cys residues are oriented appropriately in relation to PEB by CpeF. The site-specific variants showed that although single thioether linkages to either ring A (C59S) or ring D (C48S) were possible, the amount of PEB attached when only attachment via ring D was possible was about half that seen when only attachment at ring A was possible. This suggests a mechanism whereby ring A ligates first followed by ring D; however, another interpretation is that ligation at ring D is more contingent on ligation at ring A than the reverse scenario. If CpeB is not stabilized or does not adopt the appropriate structure, the Cys-binding site(s) may not be accessible for CpeF to recognize and bind to CpeB. The modeled structure of CpeF (Fig. S1C) is most similar to the crystal structure for CpcE (27); we did not investigate whether CpeF is active as a monomer or a dimer, but binding of the PEB might occur in the groove of CpeF (Fig. S1C, red arrow).
In conclusion, we have shown that CpeF plays a role in the biosynthesis of PE in cyanobacteria by functioning as the bilin lyase that attaches PEB to Cys-48/Cys-59 of CpeB. This double ligation mechanism likely occurs by the attachment of ring A to Cys-48 first, and then ring D is ligated to Cys-59. This study provides insight into the complicated process of PE biosynthesis in cyanobacteria and the mechanism of how this CpeF lyase facilitates the double ligation of PEB to CpeB.
Experimental procedures
Generation of cpeF deletion strain and culture conditions
SF33 (59, 60), a shortened filament mutant of F. diplosiphon UTEX 481 (also called Tolypothrix sp. PCC 7601) was used as WT and was cultured as described previously (61). To avoid polar effects on neighboring genes, the cpeF knockout mutant was generated as a clean deletion as described previously (40–42) with a few alterations. The cpeF deletion construct made by fusing two DNA fragments corresponding to the upstream and downstream regions of cpeF was cloned in the NcoI site of pJCF276 suicide plasmid (41) and used to transform F. diplosiphon as described previously.6 The site of cpeF deletion was designed to leave a short chimeric 28-amino acid–long polypeptide. The primers used to clone the cpeF deletion construct are provided in Table S3. Clean deletion of cpeF gene was confirmed by PCR polymorphism with primers flanking the region of interest. Growth curves were generated from two independent replicates of cells grown in ∼15 μmol m−2 s−1 of green light monitored by absorbance at 750 nm every 24 h for 12 days.
Genome and sequence analysis
The nucleotide sequence for cpeF from F. diplosiphon UTEX 481 was retrieved from GenBank (accession number AGCR01000053.1; locus tag FDUTEX481_08608) (10). Amino acid sequences (CpeF (residues 11–313), GenBank accession number EKF00796.1) were analyzed using the ClustalW alignment tool from MacVector software version 12.7.5 (MacVector Inc., Apex, NC), BLASTp 2.3.1 from NCBI (34, 35), and the Phyre2 prediction system (38).
Construction of expression plasmids
Plasmids used in this study are listed in Table S4. Some expression constructs were described previously, but the newly produced constructs were sequenced at the W. M. Keck Conservation and Molecular Genetics Laboratory (University of New Orleans). Each gene was amplified by PCR from F. diplosiphon chromosomal DNA using the primers listed in Table S3. Each resulting amplicon was cloned into a Duet-1 vector (Novagen/EMD Millipore Corp., Darmstadt, Germany) or pBAD/Myc-His A vector (Thermo Fisher Scientific, Waltham, MA) as listed in Table S4 after digestion with restriction enzymes (whose recognition sites were embedded into the primers; see Table S3). The pCpeBA plasmid was used as a template to produce the site-specific variants pCpeB(C48S)CpeA and pCpeB(C59S)CpeA using standard methods as described previously (29) and using the primers listed in Table S3.
Heterologous expression and purification of recombinant proteins
Recombinant proteins were expressed and purified from E. coli BL21 (DE3) cells as described previously (29). For cells containing both Duet-1 vectors and pBAD/Myc-His A vector, cultures were sequentially induced first with 1 mm isopropyl 1-thio-β-d-galactopyranoside (for expression of Duet-1 vectors) at 18 °C for 2–3 h and then with 0.2 mg ml−1 l-arabinose (for expression of the pBAD/Myc-His A vector) at 18 °C for an additional 16 h before being harvested by centrifugation. After purification, total protein concentrations were quantified using the Quick Start Bradford Protein Assay kit (Bio-Rad).
Isolation of cyanobacterial proteins
All protocols to isolate and purify F. diplosiphon proteins, including PBSs and PE, were described previously.6 Total protein concentrations were quantified using the Quick Start Bradford Protein Assay kit.
Tryptic digestion and MS analysis
Tryptic digestion of protein samples containing HT-CpeB expressed from the plasmids pCpeB2 and pCpeBZ was conducted as described earlier (29). All other digestions were conducted using the altered protocol as described previously.6 After digestion, samples were analyzed as described using MALDI MS and tandem MALDI MS/MS (29) or LC-MS/MS using a Waters Synapt HDMS quadrupole-TOF mass spectrometer (25).6
Protein analysis by spectroscopy and gel electrophoresis
Spectroscopy and gel electrophoresis were conducted as described previously.6 Protein purification was verified using SDS-PAGE. Protein band intensities and Zn-enhanced bilin fluorescence band intensities were quantified and analyzed using Quantity One 1-D Analysis Software and Image Lab Software version 5.2.1 (Bio-Rad). Ratios of the fluorescence from PEB:CpeB intensities from four independent replicates were analyzed by a two-way analysis of variance (α = 0.05) with experimental replicate and strain (or mutant) as factors. The WT ratio was set to 1, and the mutants were represented relative to this value.
Immunoblotting analysis
Immunoblotting analysis was performed as described (29) using the following primary antisera generated in rabbits: anti-CpeA (1:5,000 dilution (29)), anti-CpeB (1:20,000 dilution), anti-CpcA (1:5,000 dilution (11)), and anti-CpcS (1:5,000 (13)). Anti-CpeB antibodies were generated in rabbits against holo-CpeB purified from F. diplosiphon as described6 (YenZym Antibodies, San Francisco, CA). Protein band intensities were quantified and analyzed using Quantity One 1-D Analysis Software and Image Lab Software version 5.2.1.
Author contributions
C. M. K. and W. M. S. conceptualization; C. M. K. and W. M. S. data curation; C. M. K., J. P. F., J. A. K., M. N. B., R. B. C., and W. M. S. formal analysis; C. M. K., D. M. K., R. B. C., and W. M. S. supervision; C. M. K. and W. M. S. validation; C. M. K., C. V. H., J. P. F., L. S. H., A. G., J. A. K., M. N. B., and R. B. C. investigation; C. M. K., C. V. H., J. P. F., L. S. H., J. A. K., and M. N. B. visualization; C. M. K., J. A. K., M. N. B., D. M. K., R. B. C., and W. M. S. methodology; C. M. K. writing-original draft; C. M. K. and W. M. S. project administration; C. M. K., C. V. H., J. P. F., L. S. H., A. G., J. A. K., M. N. B., D. M. K., R. B. C., and W. M. S. writing-review and editing; D. M. K. and W. M. S. funding acquisition; W. M. S. resources.
Supplementary Material
Acknowledgments
We thank Dr. Tumulesh Solanky, Dr. Bernard Rees, Dr. Avijit Biswas, and Tyler Blensdorf for technical assistance on this work.
This work was supported by National Science Foundation Grants MCB-1029414 and MCB-1818187 (to D. M. K.) and MCB-1244339 and MCB-0843664 (to W. M. S.) and by a University of New Orleans Graduate School performance and accountability fellowship (to C. M. K.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3 and Tables S1–S4.
C. M. Kronfel, A. Biswas, J. P. Frick, A. Gutu, T. Blensdorf, J. A. Karty, D. M. Kehoe, and W. M. Schluchter, submitted for publication.
- PBS
- phycobilisome(s)
- AP
- allophycocyanin
- BME
- 2-mercaptoethanol
- HT-
- hexahistidine-tagged
- NT-
- non-tagged
- PC
- phycocyanin
- PCB
- phycocyanobilin
- PE
- phycoerythrin
- PEB
- phycoerythrobilin
- PUB
- phycourobilin
- E/F
- CpcE/CpcF.
References
- 1. Ong L. J., and Glazer A. N. (1991) Phycoerythrins of marine unicellular cyanobacteria. I. Bilin types and locations and energy transfer pathways in Synechococcus spp. phycoerythrins. J. Biol. Chem. 266, 9515–9527 [PubMed] [Google Scholar]
- 2. Gutu A., and Kehoe D. M. (2012) Emerging perspectives on the mechanisms, regulation, and distribution of light color acclimation in cyanobacteria. Mol. Plant 5, 1–13 10.1093/mp/ssr054 [DOI] [PubMed] [Google Scholar]
- 3. Tandeau de Marsac N. (1977) Occurrence and nature of chromatic adaptation in cyanobacteria. J. Bacteriol. 130, 82–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bezy R. P., Wiltbank L., and Kehoe D. M. (2011) Light-dependent attenuation of phycoerythrin gene expression reveals convergent evolution of green light sensing in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 108, 18542–18547 10.1073/pnas.1107427108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sidler W. A. (1994) Phycobilisome and phycobiliprotein structure, in The Molecular Biology of Cyanobacteria (Bryant D. A., ed.) pp. 139–216, Kluwer Academic, Dordrecht, The Netherlands [Google Scholar]
- 6. Glazer A. N. (1994) Phycobiliproteins—a family of valuable, widely used fluorophores. J. Appl. Phycol. 6, 105–112 10.1007/BF02186064 [DOI] [Google Scholar]
- 7. Dammeyer T., and Frankenberg-Dinkel N. (2006) Insights into phycoerythrobilin biosynthesis point toward metabolic channeling. J. Biol. Chem. 281, 27081–27089 10.1074/jbc.M605154200 [DOI] [PubMed] [Google Scholar]
- 8. Biswas A., Vasquez Y. M., Dragomani T. M., Kronfel M. L., Williams S. R., Alvey R. M., Bryant D. A., and Schluchter W. M. (2010) Biosynthesis of cyanobacterial phycobiliproteins in Escherichia coli: chromophorylation efficiency and specificity of all bilin lyases from Synechococcus sp. strain PCC 7002. Appl. Environ. Microbiol. 76, 2729–2739 10.1128/AEM.03100-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Scheer H., and Zhao K. H. (2008) Biliprotein maturation: the chromophore attachment. Mol. Microbiol. 68, 263–276 10.1111/j.1365-2958.2008.06160.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yerrapragada S., Shukla A., Hallsworth-Pepin K., Choi K., Wollam A., Clifton S., Qin X., Muzny D., Raghuraman S., Ashki H., Uzman A., Highlander S. K., Fryszczyn B. G., Fox G. E., Tirumalai M. R., et al. (2015) Extreme sensory complexity encoded in the 10-megabase draft genome sequence of the chromatically acclimating cyanobacterium Tolypothrix sp. PCC 7601. Genome Announc. 3, e00355–15 10.1128/genomeA.00355-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shen G., Schluchter W. M., and Bryant D. A. (2008) Biogenesis of phycobiliproteins. I. cpcS-I and cpcU mutants of the cyanobacterium Synechococcus sp. PCC 7002 define a heterodimeric phycocyanobilin lyase specific for β-phycocyanin and allophycocyanin subunits. J. Biol. Chem. 283, 7503–7512 10.1074/jbc.M708164200 [DOI] [PubMed] [Google Scholar]
- 12. Zhao K. H., Su P., Tu J M., Wang X., Liu H., Plöscher M., Eichacker L., Yang B., Zhou M., and Scheer H. (2007) Phycobilin:cystein-84 biliprotein lyase, a near-universal lyase for cysteine-84-binding sites in cyanobacterial phycobiliproteins. Proc. Natl. Acad. Sci. U.S.A. 104, 14300–14305 10.1073/pnas.0706209104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saunée N. A., Williams S. R., Bryant D. A., and Schluchter W. M. (2008) Biogenesis of phycobiliproteins. II. CpcS-I and CpcU comprise the heterodimeric bilin lyase that attaches phycocyanobilin to Cys-82 of β-phycocyanin and Cys-81 of allophycocyanin subunits in Synechococcus sp. PCC 7002. J. Biol. Chem. 283, 7513–7522 10.1074/jbc.M708165200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shen G., Saunée N. A., Williams S. R., Gallo E. F., Schluchter W. M., and Bryant D. A. (2006) Identification and characterization of a new class of bilin lyase: the cpcT gene encodes a bilin lyase responsible for attachment of phycocyanobilin to Cys-153 on the β subunit of phycocyanin in Synechococcus sp. PCC 7002. J. Biol. Chem. 281, 17768–17778 10.1074/jbc.M602563200 [DOI] [PubMed] [Google Scholar]
- 15. Zhou W., Ding W.-L., Zeng X.-L., Dong L.-L., Zhao B., Zhou M., Scheer H., Zhao K.-H., and Yang X. (2014) Structure and mechanism of the phycobiliprotein lyase CpcT. J. Biol. Chem. 289, 26677–26689 10.1074/jbc.M114.586743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gasper R., Schwach J., Hartmann J., Holtkamp A., Wiethaus J., Riedel N., Hofmann E., and Frankenberg-Dinkel N. (2017) Distinct features of cyanophage-encoded T-type phycobiliprotein lyase ΦCpeT: the role of auxiliary metabolic genes. J. Biol. Chem. 292, 3089–3098 10.1074/jbc.M116.769703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Swanson R. V., Zhou J., Leary J. A., Williams T., de Lorimier R., Bryant D. A., and Glazer A. N. (1992) Characterization of phycocyanin produced by cpcE and cpcF mutants and identification of an intergenic suppressor of the defect in bilin attachment. J. Biol. Chem. 267, 16146–16154 [PubMed] [Google Scholar]
- 18. Zhou J., Gasparich G. E., Stirewalt V. L., de Lorimier R., and Bryant D. A. (1992) The cpcE and cpcF genes of Synechococcus sp. PCC 7002: construction and phenotypic characterization of interposon mutants. J. Biol. Chem. 267, 16138–16145 [PubMed] [Google Scholar]
- 19. Zhou J. (1992) Mutational Analysis of the Genes Encoding Phycobilisome Components in the Cyanobacterium Synechococcus sp. PCC 7002. Ph.D. dissertation, Pennsylvania State University [Google Scholar]
- 20. Fairchild C. D., Zhao J., Zhou J., Colson S. E., Bryant D. A., and Glazer A. N. (1992) Phycocyanin α subunit phycocyanobilin lyase. Proc. Natl. Acad. Sci. U.S.A. 89, 7017–7021 10.1073/pnas.89.15.7017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fairchild C. D., and Glazer A. N. (1994) Oligomeric structure, enzyme kinetics, and substrate specificity of the phycocyanin α subunit phycocyanobilin lyase. J. Biol. Chem. 269, 8686–8694 [PubMed] [Google Scholar]
- 22. Böhm S., Endres S., Scheer H., and Zhao K. H. (2007) Biliprotein chromophore attachment: chaperone-like function of the PecE subunit of α-phycoerythrocyanin lyase. J. Biol. Chem. 282, 25357–25366 10.1074/jbc.M702669200 [DOI] [PubMed] [Google Scholar]
- 23. Storf M., Parbel A., Meyer M., Strohmann B., Scheer H., Deng M. G., Zheng M., Zhou M., and Zhao K. H. (2001) Chromophore attachment to biliproteins: specificity of PecE/PecF, a lyase-isomerase for the photoactive 3(1)-Cys-α84-phycoviolobilin chromophore of phycoerythrocyanin. Biochemistry 40, 12444–12456 10.1021/bi010776s [DOI] [PubMed] [Google Scholar]
- 24. Blot N., Wu X. J., Thomas J. C., Zhang J., Garczarek L., Böhm S., Tu J. M., Zhou M., Plöscher M., Eichacker L., Partensky F., Scheer H., and Zhao K. H. (2009) Phycourobilin in trichromatic phycocyanin from oceanic cyanobacteria is formed post-translationally by a phycoerythrobilin lyase-isomerase. J. Biol. Chem. 284, 9290–9298 10.1074/jbc.M809784200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shukla A., Biswas A., Blot N., Partensky F., Karty J. A., Hammad L. A., Garczarek L., Gutu A., Schluchter W. M., and Kehoe D. M. (2012) Phycoerythrin-specific bilin lyase-isomerase controls blue-green chromatic acclimation in marine Synechococcus. Proc. Natl. Acad. Sci. U.S.A. 109, 20136–20141 10.1073/pnas.1211777109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao K. H., Wu D., Zhou M., Zhang L., Böhm S., Bubenzer C., and Scheer H. (2005) Amino acid residues associated with enzymatic activities of the isomerizing phycoviolobilin-lyase PecE/F. Biochemistry 44, 8126–8137 10.1021/bi0500168 [DOI] [PubMed] [Google Scholar]
- 27. Zhao C., Höppner A., Xu Q.-Z., Gärtner W., Scheer H., Zhou M., and Zhao K.-H. (2017) Structures and enzymatic mechanisms of phycobiliprotein lyases CpcE/F and PecE/F. Proc. Natl. Acad. Sci. U.S.A. 114, 13170–13175 10.1073/pnas.1715495114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schluchter W. M., Shen G., Alvey R. M., Biswas A., Saunée N. A., Williams S. R., Miller C. A., and Bryant D. A. (2010) Phycobiliprotein biosynthesis in cyanobacteria: structure and function of enzymes involved in post-translational modification, in Recent Advances in Phototrophic Prokaryotes (Hallenbeck P. C., ed) pp. 211–228, Springer, New York: [DOI] [PubMed] [Google Scholar]
- 29. Biswas A., Boutaghou M. N., Alvey R. M., Kronfel C. M., Cole R. B., Bryant D. A., and Schluchter W. M. (2011) Characterization of the activities of the CpeY, CpeZ, and CpeS bilin lyases in phycoerythrin biosynthesis in Fremyella diplosiphon strain UTEX 481. J. Biol. Chem. 286, 35509–35521 10.1074/jbc.M111.284281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kehoe D. M., and Gutu A. (2006) Responding to color: the regulation of complementary chromatic adaptation. Annu. Rev. Plant Biol. 57, 127–150 10.1146/annurev.arplant.57.032905.105215 [DOI] [PubMed] [Google Scholar]
- 31. Bretaudeau A., Coste F., Humily F., Garczarek L., Le Corguillé G., Six C., Ratin M., Collin O., Schluchter W. M., and Partensky F. (2013) CyanoLyase: a database of phycobilin lyase sequences, motifs and functions. Nucleic Acids Res. 41, D396–D401 10.1093/nar/gks1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Six C., Thomas J.-C., Garczarek L., Ostrowski M., Dufresne A., Blot N., Scanlan D. J., and Partensky F. (2007) Diversity and evolution of phycobilisomes in marine Synechococcus spp.: a comparative genomics study. Genome Biology 8, R259 10.1186/gb-2007-8-12-r259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alvey R. M., Karty J. A., Roos E., Reilly J. P., and Kehoe D. M. (2003) Lesions in phycoerythrin chromophore biosynthesis in Fremyella diplosiphon reveal coordinated light regulation of apoprotein and pigment biosynthetic enzyme gene expression. Plant Cell 15, 2448–2463 10.1105/tpc.015016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., and Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Altschul S. F., Wootton J. C., Gertz E. M., Agarwala R., Morgulis A., Schäffer A. A., and Yu Y.-K. (2005) Protein database searches using compositionally adjusted substitution matrices. FEBS J. 272, 5101–5109 10.1111/j.1742-4658.2005.04945.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wilbanks S. M., and Glazer A. N. (1993) Rod structure of a phycoerythrin II-containing phycobilisome. I. Organization and sequence of the gene cluster encoding the major phycobiliprotein rod components in the genome of marine Synechococcus sp. WH8020. J. Biol. Chem. 268, 1226–1235 [PubMed] [Google Scholar]
- 37. de Lorimier R., Wilbanks S. M., and Glazer A. N. (1993) Genes of the R-phycocyanin II locus of marine Synechococcus spp., and comparison of protein-chromophore interactions in phycocyanins differing in bilin composition. Plant Mol. Biol. 21, 225–237 10.1007/BF00019939 [DOI] [PubMed] [Google Scholar]
- 38. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Andrade M. A., Petosa C., O'Donoghue S. I., Müller C. W., and Bork P. (2001) Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 309, 1–18 10.1006/jmbi.2001.4624 [DOI] [PubMed] [Google Scholar]
- 40. Noubir S., Luque I., Ochoa de Alda J. A., Perewoska I., Tandeau de Marsac N., Cobley J. G., and Houmard J. (2002) Co-ordinated expression of phycobiliprotein operons in the chromatically adapting cyanobacterium Calothrix PCC 7601: a role for RcaD and RcaG. Mol. Microbiol. 43, 749–762 10.1046/j.1365-2958.2002.02783.x [DOI] [PubMed] [Google Scholar]
- 41. Cobley J. G., Clark A. C., Weerasurya S., Queseda F. A., Xiao J. Y., Bandrapali N., D'Silva I., Thounaojam M., Oda J. F., Sumiyoshi T., and Chu M. H. (2002) CpeR is an activator required for expression of the phycoerythrin operon (cpeBA) in the cyanobacterium Fremyella diplosiphon and is encoded in the phycoerythrin linker-polypeptide operon (cpeCDESTR). Mol. Microbiol. 44, 1517–1531 10.1046/j.1365-2958.2002.02966.x [DOI] [PubMed] [Google Scholar]
- 42. Gutu A., Nesbit A. D., Alverson A. J., Palmer J. D., and Kehoe D. M. (2013) Unique role for translation initiation factor 3 in the light color regulation of photosynthetic gene expression. Proc. Natl. Acad. Sci. U.S.A. 110, 16253–16258 10.1073/pnas.1306332110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kronfel C. M., Kuzin A. P., Forouhar F., Biswas A., Su M., Lew S., Seetharaman J., Xiao R., Everett J. K., Ma L.-C., Acton T. B., Montelione G. T., Hunt J. F., Paul C. E., Dragomani T. M., et al. (2013) Structural and biochemical characterization of the bilin lyase CpcS from Thermosynechococcus elongatus. Biochemistry 52, 8663–8676 10.1021/bi401192z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dammeyer T., Hofmann E., and Frankenberg-Dinkel N. (2008) Phycoerythrobilin synthase (PebS) of a marine virus: crystal Structures of the biliverdin complex and the substrate-free form. J. Biol. Chem. 283, 27547–27554 10.1074/jbc.M803765200 [DOI] [PubMed] [Google Scholar]
- 45. Dammeyer T., Bagby S. C., Sullivan M. B., Chisholm S. W., and Frankenberg-Dinkel N. (2008) Efficient phage-mediated pigment biosynthesis in oceanic cyanobacteria. Curr. Biol. 18, 442–448 10.1016/j.cub.2008.02.067 [DOI] [PubMed] [Google Scholar]
- 46. Ficner R., Lobeck K., Schmidt G., and Huber R. (1992) Isolation, crystallization, crystal structure analysis and refinement of B-phycoerythrin from the red alga Porphyridium sordidum at 2.2 Å resolution. J. Mol. Biol. 228, 935–950 10.1016/0022-2836(92)90876-L [DOI] [PubMed] [Google Scholar]
- 47. Chang W. R., Jiang T., Wan Z. L., Zhang J. P., Yang Z. X., and Liang D. C. (1996) Crystal structure of R-phycoerythrin from Polysiphonia urceolata at 2.8 Å resolution. J. Mol. Biol. 262, 721–731 10.1006/jmbi.1996.0547 [DOI] [PubMed] [Google Scholar]
- 48. Ritter S., Hiller R. G., Wrench P. M., Welte W., and Diederichs K. (1999) Crystal structure of a phycourobilin-containing phycoerythrin at 1.9 Å resolution. J. Struct. Biol. 126, 86–97 10.1006/jsbi.1999.4106 [DOI] [PubMed] [Google Scholar]
- 49. Gindt Y. M., Zhou J., Bryant D. A., and Sauer K. (1994) Spectroscopic studies of phycobilisome subcore preparations lacking key core chromophores: assignment of excited state energies to the Lcm, β18 and αAP-B chromophores. Biochim. Biophys. Acta 1186, 153–162 10.1016/0005-2728(94)90174-0 [DOI] [PubMed] [Google Scholar]
- 50. Glazer A. N. (1989) Light guides: directional energy transfer in a photosynthetic antenna. J. Biol. Chem. 264, 1–4 [PubMed] [Google Scholar]
- 51. Fairchild C. D., and Glazer A. N. (1994) Nonenzymatic bilin addition to the α subunit of an apophycoerythrin. J. Biol. Chem. 269, 28988–28996 [PubMed] [Google Scholar]
- 52. Toole C. M., Plank T. L., Grossman A. R., and Anderson L. K. (1998) Bilin deletions and subunit stability in cyanobacterial light-harvesting proteins. Mol. Microbiol. 30, 475–486 10.1046/j.1365-2958.1998.01082.x [DOI] [PubMed] [Google Scholar]
- 53. Anderson L. K., and Toole C. M. (1998) A model for early events in the assembly pathway of cyanobacterial phycobilisomes. Mol. Microbiol. 30, 467–474 10.1046/j.1365-2958.1998.01081.x [DOI] [PubMed] [Google Scholar]
- 54. Martínez-Oyanedel J., Contreras-Martel C., Bruna C., and Bunster M. (2004) Structural-functional analysis of the oligomeric protein R-phycoerythrin. Biol. Res. 37, 733–745 [DOI] [PubMed] [Google Scholar]
- 55. Jung L. J., Chan C. F., and Glazer A. N. (1995) Candidate genes for the phycoerythrocyanin α subunit lyase: biochemical analysis of pecE and pecF interposon mutants. J. Biol. Chem. 270, 12877–12884 10.1074/jbc.270.21.12877 [DOI] [PubMed] [Google Scholar]
- 56. Bhalerao R. P., Lind L. K., and Gustafsson P. (1994) Cloning of the cpcE and cpcF genes from Synechococcus sp. PCC 6301 and their inactivation in Synechococcus sp. PCC 7942. Plant Mol. Biol. 26, 313–326 10.1007/BF00039542 [DOI] [PubMed] [Google Scholar]
- 57. Bolte K., Kawach O., Prechtl J., Gruenheit N., Nyalwidhe J., and Maier U. G. (2008) Complementation of a phycocyanin-bilin lyase from Synechocystis sp PCC 6803 with a nucleomorph-encoded open reading frame from the cryptophyte Guillardia theta. BMC Plant Biol. 8, 56 10.1186/1471-2229-8-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhao K. H., Zhang J., Tu J. M., Böhm S., Plöscher M., Eichacker L., Bubenzer C., Scheer H., Wang X., and Zhou M. (2007) Lyase activities of CpcS- and CpcT-like proteins from Nostoc PCC7120 and sequential reconstitution of binding sites of phycoerythrocyanin and phycocyanin β-subunits. J. Biol. Chem. 282, 34093–34103 10.1074/jbc.M703038200 [DOI] [PubMed] [Google Scholar]
- 59. Cobley J. G., and Miranda R. D. (1983) Mutations affecting chromatic adaptation in the cyanobacterium Fremyella diplosiphon. J. Bacteriol. 153, 1486–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cobley J. G., Zerweck E., Reyes R., Mody A., Seludo-Unson J. R., Jaeger H., Weerasuriya S., and Navankasattusas S. (1993) Construction of shuttle plasmids which can be efficiently mobilized from Escherichia coli into the chromatically adapting cyanobacterium, Fremyella diplosiphon. Plasmid 30, 90–105 10.1006/plas.1993.1037 [DOI] [PubMed] [Google Scholar]
- 61. Seib L. O., and Kehoe D. M. (2002) A turquoise mutant genetically separates expression of genes encoding phycoerythrin and its associated linker peptides. J. Bacteriol. 184, 962–970 10.1128/jb.184.4.962-970.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.