Abstract
N-Nitroso compounds (NOCs) are common DNA-alkylating agents, are abundantly present in food and tobacco, and can also be generated endogenously. Metabolic activation of some NOCs can give rise to carboxymethylation and pyridyloxobutylation/pyridylhydroxybutylation of DNA, which are known to be carcinogenic and can lead to gastrointestinal and lung cancer, respectively. Herein, using the competitive replication and adduct bypass (CRAB) assay, along with MS- and NMR-based approaches, we assessed the cytotoxic and mutagenic properties of three O6-alkyl-2′-deoxyguanosine (O6-alkyl-dG) adducts, i.e. O6-pyridyloxobutyl-dG (O6-POB-dG) and O6-pyridylhydroxybutyl-dG (O6-PHB-dG), derived from tobacco-specific nitrosamines, and O6-carboxymethyl-dG (O6-CM-dG), induced by endogenous N-nitroso compounds. We also investigated two neutral analogs of O6-CM-dG, i.e. O6-aminocarbonylmethyl-dG (O6-ACM-dG) and O6-hydroxyethyl-dG (O6-HOEt-dG). We found that, in Escherichia coli cells, these lesions mildly (O6-POB-dG), moderately (O6-PHB-dG), or strongly (O6-CM-dG, O6-ACM-dG, and O6-HOEt-dG) impede DNA replication. The strong blockage effects of the last three lesions were attributable to the presence of hydrogen-bonding donor(s) located on the alkyl functionality of these lesions. Except for O6-POB-dG, which also induced a low frequency of G → T transversions, all other lesions exclusively stimulated G → A transitions. SOS-induced DNA polymerases played redundant roles in bypassing all the O6-alkyl-dG lesions investigated. DNA polymerase IV (Pol IV) and Pol V, however, were uniquely required for inducing the G → A transition for O6-CM-dG exposure. Together, our study expands our knowledge about the recognition of important NOC-derived O6-alkyl-dG lesions by the E. coli DNA replication machinery.
Keywords: DNA polymerase, DNA replication, DNA damage, mutagenesis, mass spectrometry (MS), DNA alkylation, N-nitroso compounds, translesion synthesis
Introduction
Environmental exposure and endogenous metabolism can give rise to a variety of DNA adducts (1). N-Nitroso compounds (NOCs),2 a common type of alkylating agents, are capable of inducing cancer in ∼40 different animal species, including higher primates, and are carcinogenic in multiple organs in animals (2). Humans are exposed to NOCs through dietary consumption, tobacco smoking, and endogenous metabolism (3, 4). In this vein, two potential types of carcinogenic adducts formed from NOC exposure are of emerging concern, i.e. the pyridyloxobutyl (POB) and pyridylhydroxybutyl (PHB) adducts derived from tobacco-specific N-nitrosamines and carboxymethyl adducts arising from endogenous NOCs (5, 6).
Tobacco-specific nitrosamines 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and N′-nitrosonornicotine (NNN) are well-documented rodent carcinogens and classified as group I carcinogens by the International Agency for Research on Cancer (7). Metabolic activation of NNK and NNN via cytochrome P450 enzymes can produce reactive intermediates that can react with DNA to yield POB and/or PHB adducts along with other DNA lesions, which may induce mutations during DNA replication (5, 8–15). Another type of extensively investigated promutagenic lesions, carboxymethyl (CM) adducts, can be induced in DNA through metabolic activation of endogenous NOCs, e.g. N-nitroso bile acid conjugate or N-nitrosoglycine (16–18). It was suggested that the mutations in the TP53 gene found in gastrointestinal cancer emanate from carboxymethylation rather than methylation of DNA elicited by NOCs (19).
Previous studies have led to the identification and quantification of a series of POB and PHB derivatives on nucleobases in DNA. O2-POB-dT, O2-PHB-dT, N7-POB-guanine, N7-PHB-guanine, O6-POB-dG, O6-PHB-dG, N6-POB-dA, and N6-PHB-dA were detected in lung and liver tissues of NNK-treated rats (8, 9, 13, 14); O2-POB-dT, O6-POB-dG, N7-POB-guanine, and O2-POB-cytosine were found in esophageal tissues of rats treated with NNN (10, 20). In addition, it was reported that appreciable levels of O4-POB-dT along with the above-mentioned O2-POB-dT and O6-POB-dG could be formed in cultured mammalian cells upon exposure to 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone, a model pyridyloxobutylating agent (21, 22). Carboxymethylated DNA lesions, such as N6-CM-dA, N3-CM-dT, O4-CM-dT, O6-CM-dG, and N4-CM-dC were also identified in vitro (16, 17, 23, 24). Furthermore, O6-CM-dG could be induced in cultured human cells upon treatment with azaserine, a pancreatic carcinogen that can be converted to diazoacetate by cellular esterases (25).
Multiple lines of evidence suggested that the O6 position of 2′-deoxyguanosine is the most facile exocyclic atom to be alkylated on nucleobases (26), and the resulting lesions alter the hydrogen bonding property of the guanine base (6, 27–30). In this vein, O6-methyl-dG (O6-Me-dG) and O6-POB-dG have been implicated in lung tumorigenesis mediated by NNK (12, 31–33), and O6-CM-dG may contribute to the mutations found in TP53 tumor suppressor gene of human gastrointestinal tumors (19) and is correlated with colorectal cancer (6). Thus, it is important to understand how the O6-alkyl-2′-deoxyguanosine (O6-alkyl-dG) lesions derived from the above-mentioned NOCs affect the efficiency and fidelity of DNA replication.
Several replication studies have been performed to assess the impact of O6-POB-dG and O6-CM-dG lesions on the flow of genetic information. O6-POB-dG was found to be highly mutagenic, and it induces exclusively G → A transition in Escherichia coli, whereas it elicits both G → A transition and G → T transversion in mammalian cells (15, 33, 34). In addition, multiple translesion synthesis DNA polymerases, particularly Pol η, were found to be involved in the efficient bypass of O6-POB-dG (15), which is consistent with in vitro results showing that Pol η exhibits much higher efficiency in bypassing O6-POB-dG than Pol ι, Pol κ, and Rev1 (35, 36). O6-CM-dG is able to form Watson—Crick–type bp with thymidine, thereby inducing G → A transition mutation (37). In mammalian cells, Pol κ and Pol ζ are required for the efficient bypass of this lesion, and only G → A transition was observed (38). However, it remains unknown how O6-PHB-dG compromises the efficiency and accuracy of DNA replication.
Herein, we incorporated O6-POB-dG, O6-PHB-dG, O6-CM-dG, and its two analogs, i.e. O6-aminocarbonylmethyl-dG (O6-AMC-dG) and O6-hydroxyethyl-dG (O6-HOEt-dG), into single-stranded plasmids and examined how these lesions impede DNA replication and elicit mutation(s) in E. coli cells. We also explored the roles of SOS-induced DNA polymerases in bypassing these lesions.
Results
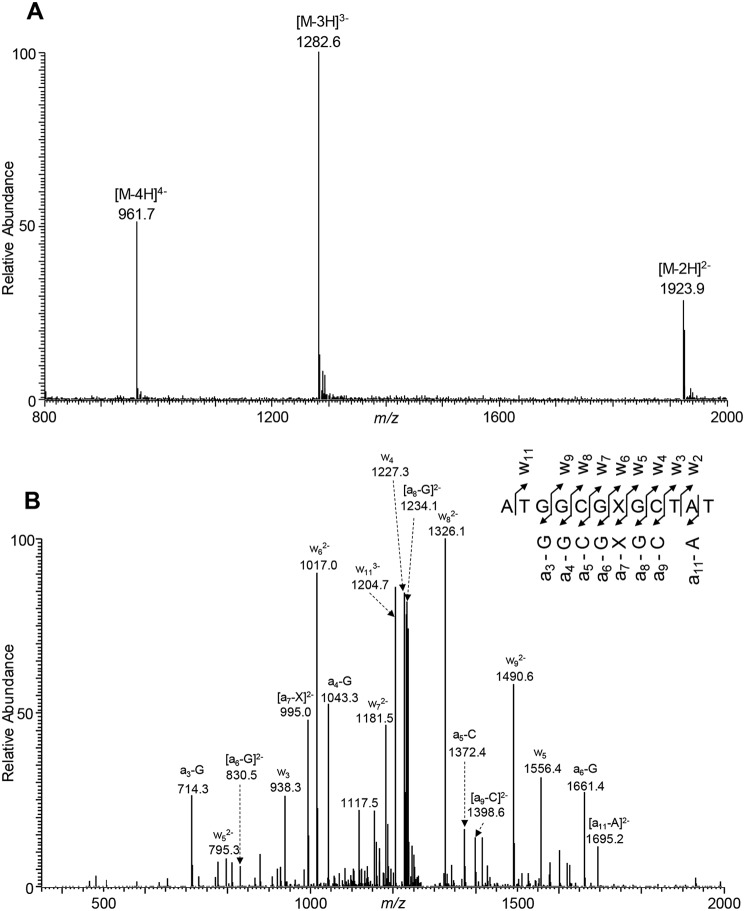
The primary objective of this project was to elucidate how the O6-substituted guanine lesions arising from tobacco-derived and endogenous NOCs (Fig. 1) compromise the efficiency and fidelity of DNA replication in E. coli cells. To this end, ODNs harboring a series of O6-alkyl-dG lesions at a specific site were synthesized and confirmed by ESI-MS and MS/MS analyses (Fig. 2 and Figs. S1–S7). A previously described CRAB assay (Fig. 3 and Fig. S8) was employed to determine the impacts of these lesions on DNA replication in E. coli cells and to determine how replication across these lesions is influenced by SOS-induced DNA polymerases (39, 40). After cellular replication, the region of interest in the progeny genomes was amplified by PCR, the ensuing products were digested with two restriction endonucleases, i.e. BbSI and MluCI, and the restriction fragments were analyzed by LC-MS/MS and native PAGE (Fig. 4 and Figs. S9–S14).
Figure 1.
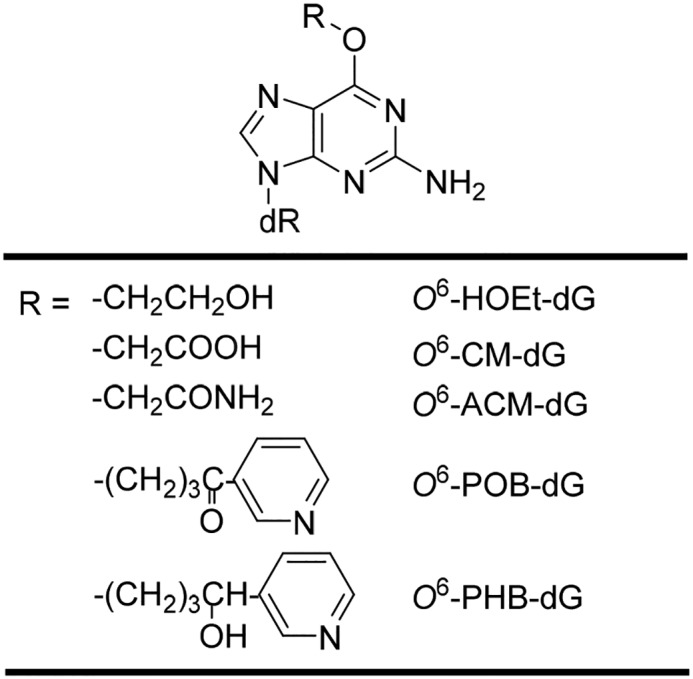
The O6-substituted dG lesions considered in this study. dR represents 2-deoxyribose.
Figure 2.
ESI-MS and MS/MS characterizations of d(ATGGCGXGCTAT), where X = O6-PHB-dG. A, negative-ion ESI-MS. B, product-ion spectrum of the [M − 3H]3− ion (m/z 1282.6).
Figure 3.
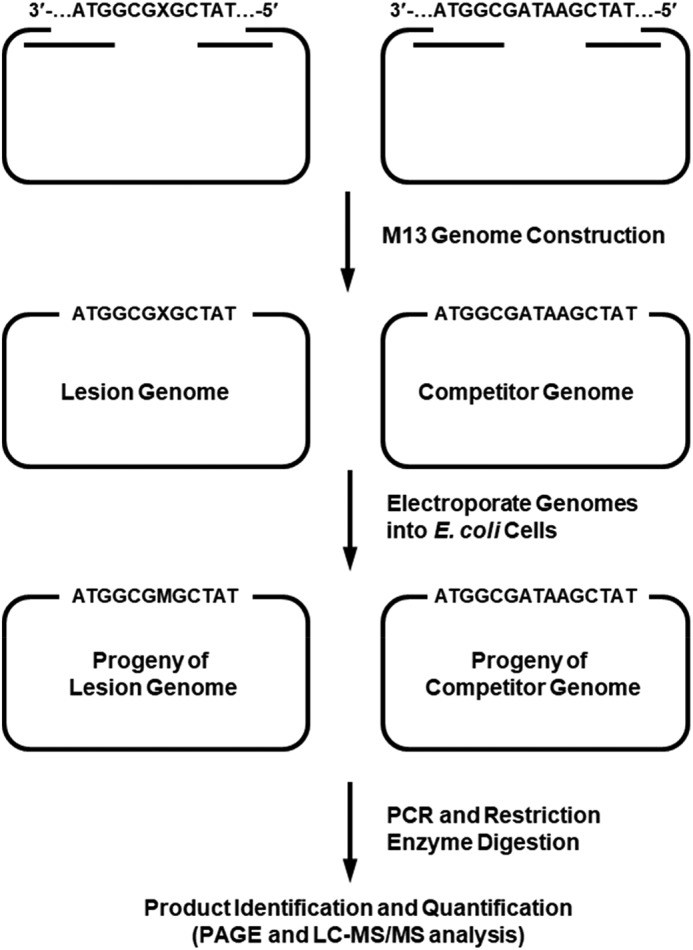
A schematic diagram illustrating the experimental procedures of the modified CRAB assay.
Figure 4.
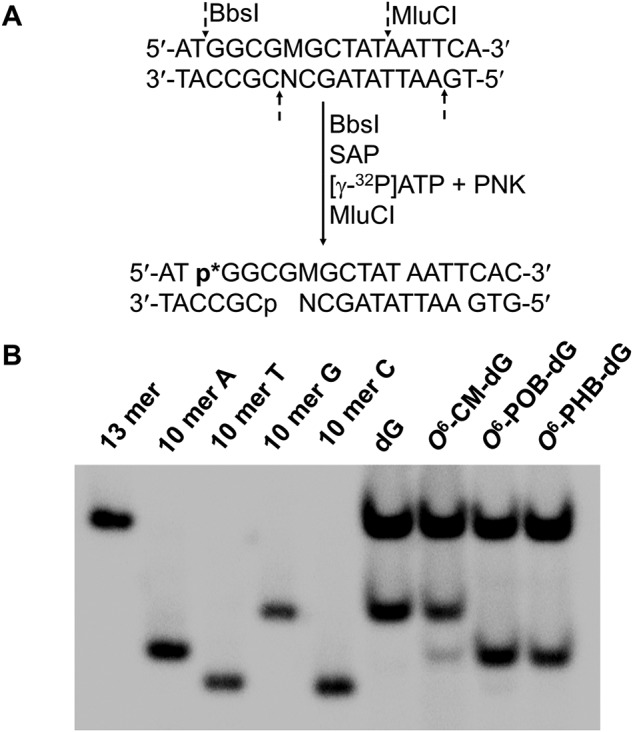
Native PAGE (30%) for monitoring the bypass efficiencies and mutation frequencies of O6-CM-dG, O6-POB-dG, and O6-PHB-dG in SOS-induced WT AB1157 E. coli cells. A, sequential restriction enzyme digestion and selective radiolabeling of the strand initially bearing the lesion. SAP and PNK designate shrimp alkaline phosphatase and T4 polynucleotide kinase, respectively. B, gel image showing 10-mer radiolabeled restriction fragments released from the original lesion-containing strand of the PCR products of the progeny of the control or lesion-carrying genome, where 10-mer A, 10-mer C, 10-mer G, and 10-mer T represent the 5′-32P-labeled standard ODNs 5′-GGCGMGCTAT-3′, where M is A, C, G, and T, respectively. Both O6-POB-dG and O6-PHB-dG are highly mutagenic and mainly induce G → A transition (10-mer A), although a low rate of G → T mutation (10-mer T) was also observed for O6-POB-dG. O6-CM-dG induced a much lower frequency of G → A mutation. The 13-mer radiolabeled band is the corresponding fragment liberated from the PCR product of the competitor genome.
The impacts of O6-POB-dG and O6-PHB-dG on the efficiency and fidelity of DNA replication
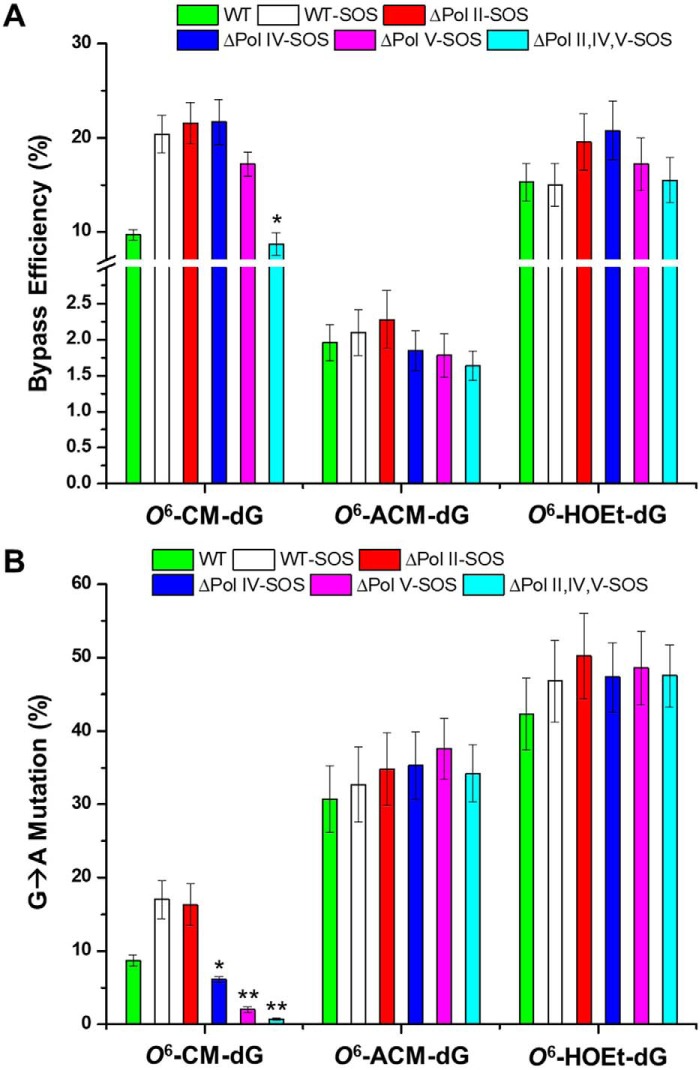
Our results from PAGE and LC-MS/MS analyses revealed the lack of insertion or deletion mutations (Fig. 4 and Fig. S9–S14). Hence, we calculated the bypass efficiencies of O6-POB-dG and O6-PHB-dG from the ratio of the 10-mer products emanating from lesion and control genomes, over the 13-mer product, generated from the competitor plasmid (Fig. 4 and Figs. S11–S14), by taking into account the molar ratios of the lesion and control over the competitor genomes employed at the initial transfection. It turned out that these two lesions were moderate impediments to DNA replication in WT AB1157 cells, with O6-PHB-dG exerting a stronger blockage effect (Fig. 5A).
Figure 5.
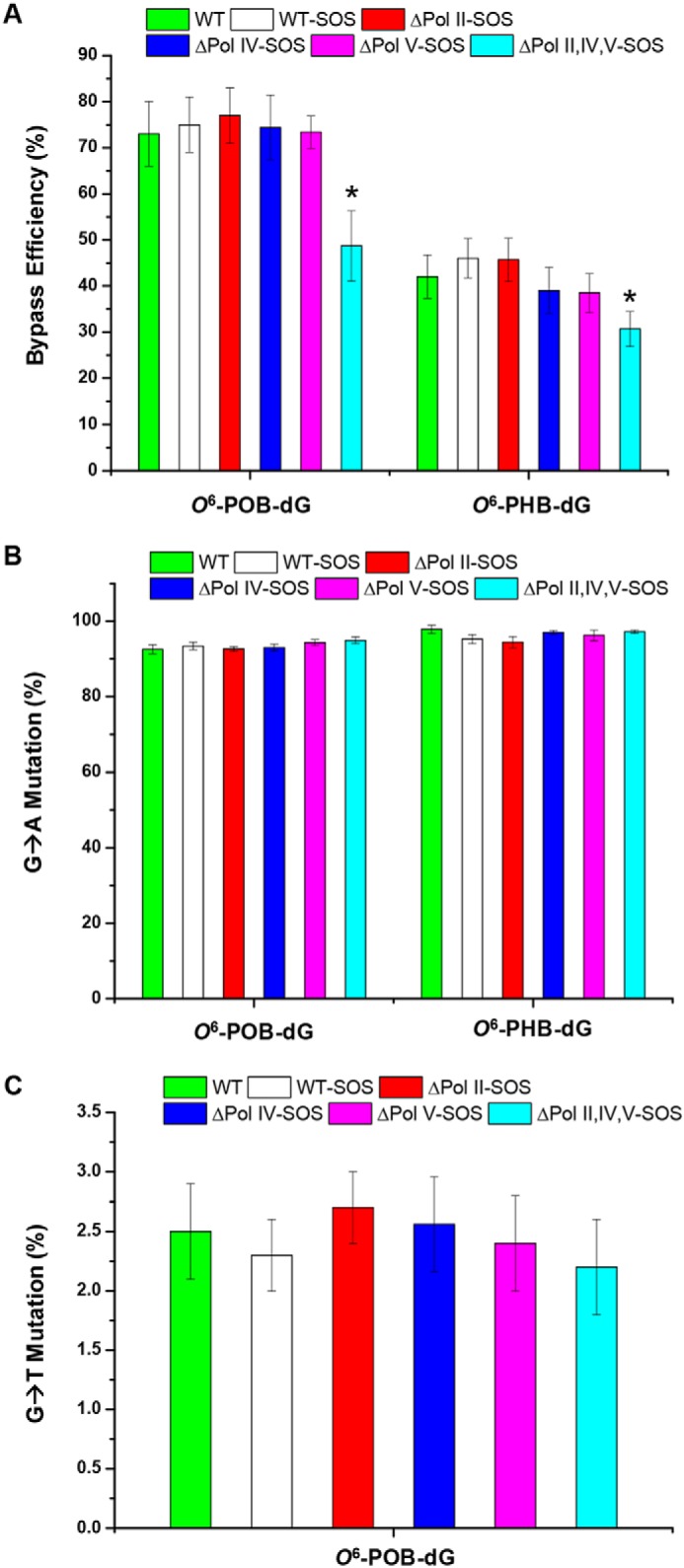
The bypass efficiencies (A) and G → A (B) or G → T (C) mutation frequencies of O6-POB-dG and O6-PHB-dG in AB1157 E. coli strains that are proficient in translesion synthesis or deficient in Pol II, Pol IV, and Pol V, alone or all three together. The data represent the means and standard deviation of results from three independent replication experiments. The p values were calculated using two-tailed, unpaired Student's t test. *, 0.01 < p < 0.05.
The roles of SOS-induced DNA polymerases in bypassing the O6-POB-dG and O6-PHB-dG were also investigated by performing the assay in E. coli strains deficient in Pol II, Pol IV, Pol V, or all three in combination. It turned out that a substantial diminution in bypass efficiency was observed only when all three SOS-induced DNA polymerases were depleted, which is consistent with the observations made previously for other O6-alkyl-dG lesions (30). No significant differences were observed for the bypass efficiencies of O6-POB-dG and O6-PHB-dG lesions in AB1157 cells and the isogenic cells with a single depletion of any of the three polymerases (Fig. 5A). This result supports that the three SOS-induced DNA polymerases assume somewhat redundant roles in bypassing O6-POB-dG and O6-PHB-dG.
The native PAGE analysis also facilitated the quantification of the mutation frequencies of O6-POB-dG and O6-PHB-dG. In agreement with previous findings (30), the major type of mutation was G → A transition for both O6-POB-dG and O6-PHB-dG in all E. coli strains (Fig. 5B). A low frequency of G → T transversion, however, was also detected for O6-POB-dG (Fig. 5C), which differs from the previously reported results (34). We also found that the frequencies of the mutations conferred by O6-POB-dG and O6-PHB-dG were not affected by depletion of any of the three SOS-induced DNA polymerases, individually or all three together (Fig. 5, B and C). This result underscores that the observed mutations can be attributed to the intrinsic hydrogen-bonding properties of the O6-subsituted guanine, which preferentially forms bp with thymine.
The impacts of O6-CM-dG, O6-ACM-dG, and O6-HOEt-dG on the efficiency and fidelity of DNA replication
Different from O6-POB-dG, O6-PHB-dG, and the previously reported O6-alkyl-dG lesions (30), O6-CM-dG displayed strong blockage to DNA replication machinery in WT AB1157 cells, as manifested by a ∼10% bypass efficiency (Fig. 6A). Interestingly, this bypass efficiency was augmented by almost 2-fold upon SOS induction.
Figure 6.
The bypass efficiencies (A) and mutation frequencies (B) of O6-CM-dG, O6-HOEt-dG, and O6-ACM-dG in AB1157 E. coli strains that are proficient in translesion synthesis or deficient in Pol II, Pol IV, and Pol V, alone or all three in combination. The data represent the means and standard deviation of results from three independent replication experiments. The p values were calculated using two-tailed, unpaired Student's t test. *, 0.01 ≤ p < 0.05; **, 0.001 ≤ p < 0.01.
The unique strong blockage effect of O6-CM-dG on cellular DNA replication prompted us to ask whether the negative charge located on the carboxymethyl functionality modulates the bypass efficiency of the lesion. To test this, we synthesized two other O6-modified derivatives with the carboxymethyl moiety being replaced with an aminocarbonylmethyl or hydroxyethyl group (Fig. 1), which are neutral under cellular pH. It turned out that O6-ACM-dG and O6-HOEt-dG also exhibited very low bypass efficiencies (Fig. 6A). Hence, we attribute the low bypass efficiencies of O6-CM-dG and the other two derivatives to the terminal hydrogen bond donor (i.e. hydroxyl or amino group) conjugated with the alkyl functionality adducted to the O6 position of guanine. It is worth noting that, similar to what we observed for O6-POB-dG and O6-PHB-dG, the bypass efficiencies of these three lesions were not affected by individual depletion of Pol II, Pol IV, or Pol V (Fig. 6A).
The mutation frequencies were also quantified, where only G → A transition was observed for all three lesions in all E. coli strains studied (Fig. 6B). In addition, the frequencies of G → A transition for O6-ACM-dG and O6-HOEt-dG were not influenced by depletion of any of the SOS-induced DNA polymerases, alone or all three in combination. However, O6-CM-dG distinguishes from other lesions, where Pol V and, to a lesser extent, Pol IV were required for the G → A transition mutation. Thus, the negative charge possessed by the carboxymethyl functionality affects the involvement of SOS-induced DNA polymerases in dTMP misincorporation opposite the lesion (Fig. 6B).
Discussion
There is established evidence for the association of tobacco use with lung cancer, and lung cancer is the second most common type of cancer diagnosed in both men and women (41). In addition, red meat–containing diet, especially processed meat, are risk factors for gastrointestinal cancer, which constitutes one of the leading causes of cancer-related deaths worldwide (42, 43). Previous studies revealed that the POB, PHB, and CM adducts of DNA derived from the NOCs in tobacco and red meat may be important factors contributing to the development of lung and colorectal cancer.
We investigated the replicative bypass of O6-POB-dG, O6-PHB-dG, and O6-CM-dG in E. coli cells, where O6-PHB-dG was studied here for the first time. Our results revealed several novel findings. First, we observed that O6-POB-dG and O6-PHB-dG were slight and moderate impediments to DNA replication, respectively, whereas replication past O6-CM-dG was strongly inhibited (Figs. 5A and 6A). Second, we found that the three SOS-induced DNA polymerases, i.e. Pol II, Pol IV, and Pol V, assume redundant roles in bypassing the three O6-alkyl-dG lesions (Fig. 5A and 4A). Third, our results demonstrated that these lesions induce G → A transition exclusively, except that G → T transversion was also observed for O6-POB-dG (Fig. 5, B and C, and 6B). Fourth, we revealed that the mutation frequencies of O6-POB-dG and O6-PHB-dG were not affected by depletion of Pol II, Pol IV, Pol V, or all three polymerases; however, Pol V and, to a lesser extent, Pol IV play important roles in eliciting G → A transition for O6-CM-dG (Fig. 5, B and C, and 6B).
Previously, it was found that O6-alkyl-dG lesions, with the alkyl group ranging from methyl to sec-butyl, do not inhibit DNA replication in E. coli cells and the three SOS-induced DNA polymerases play somewhat redundant roles in bypassing these lesions (30). Here, we found that O6-POB-dG, O6-PHB-dG, and O6-CM-dG exhibited slight, moderate, and strong blockage to DNA replication, respectively.
We further explored the mechanism underlying the very low bypass efficiency of O6-CM-dG. On the grounds that the carboxylic acid functionality in O6-CM-dG would be deprotonated and carry a negative charge in cellular pH, we initially hypothesized that the negative charge may contribute to the diminished replicative bypass of O6-CM-dG. To test that hypothesis, we assessed how two neutral analogs of O6-CM-dG, i.e. O6-ACM-dG and O6-HOEt-dG, impede DNA replication. It turned out that the latter two lesions strongly block DNA replication, with the bypass efficiency of O6-ACM-dG being merely ∼2% (Fig. 6A). These results unveiled that the poor replicative bypass of O6-CM-dG is not due to the presence of a negative charge situated on the carboxymethyl group of the lesion. Instead a hydrogen bond-donating hydroxyl or amino group at the terminus of the alkyl chain significantly impedes the replicative bypass (Fig. 6A), which is perhaps due to the disruption of the interaction between the lesions and the active site of polymerases (44, 45). Similar mechanism is likely at play for the substantially lower bypass efficiency of O6-PHB-dG than O6-POB-dG (Fig. 5A). However, future structural studies are required for understanding the detailed molecular mechanism underlying the strong blockage effects of these lesions. In addition, it should be noted that, consistent with the previously reported results for simple O6-alkyl-dG lesions (30), genetic ablation of Pol II, Pol IV, or Pol V did not confer any significant difference in replicative bypass of any of the five O6-alkyl-dG lesions examined in the present study (Fig. 6A).
It was previously shown that in E. coli cells, simple aliphatic O6-alkyl-dG lesions only induce G → A transition, which also holds true for the complex O6-POB-dG lesion (30, 34). In mammalian cells, the primary type of mutation elicited by O6-alkyl-dG lesions is the G → A transition, although a low frequency of G → T transversion was also detected for O6-POB-dG (15, 34). In line with these previous observations, the five lesions examined in the present study directed primarily G → A transition (Figs. 5, B and C, and 6B). However, different from the results obtained from replication experiments employing colony picking and Sanger sequencing analysis (34), a low frequency of G → T transversion (∼2%) was also observed for O6-POB-dG in E. coli cells (Fig. 5C). This result underscores that the modified CRAB assay is more sensitive, thereby allowing for the detection of low frequencies of mutations better than the previously reported method (34).
The mutation frequencies for O6-POB-dG, O6-PHB-dG, O6-ACM-dG, and O6-HOEt-dG were not modulated by SOS-induced DNA polymerases, where genetic ablation of Pol II, Pol IV, and Pol V, alone or all three in conjunction, did not confer any appreciable changes in frequencies of G → A transition, or, for O6-POB-dG, G → T transversion mutation (Figs. 5, B and C, and 6B), suggesting that the mutagenic properties of the O6-alkyl-dG lesions are independent of the size or shape of the alkyl groups and arise from the intrinsic chemical property of the lesions, i.e. their capabilities in hydrogen bonding and in mispairing with thymidine.
Intriguingly, different from the above four O6-alkyl-dG lesions, two SOS-induced DNA polymerases, i.e. Pol IV and Pol V, play important roles in inducing G → A transition for O6-CM-dG (Fig. 6B), and the frequency of this mutation was increased by 2-fold upon SOS induction, substantiating that the mutagenic properties of O6-alkyl-dG lesions can be modulated by the negative charge situated on the O6 position of guanine. For all the neutral substituents, from simple methyl to complex POB or PHB, the nucleotide insertion opposite the lesion was not affected by SOS-induced DNA polymerases. The mutagenic nucleotide insertion opposite the negatively charged O6-CM-dG is, however, influenced by Pol IV and Pol V (Fig. 6B). It should be noted that, in non–SOS-induced E. coli cells, Pol V could not be detected biochemically, and its expression level would increase by ∼100-fold upon SOS induction (46–48). In addition, Pol V plays crucial roles in replicative bypass of multiple lesions, such as oxidized guanine lesions and O2-alkylated thymidine lesions, where it promotes error-prone replication under many circumstances (40, 48, 49).
It is worth comparing the findings made in E. coli cells with previously reported results in mammalian cells. It was found that O6-POB-dG directed primarily the G → A transition with a low frequency of G → T transversion in HEK293T cells (15), which is in agreement with what we observed in E. coli cells. However, translesion synthesis polymerases, i.e. Pol η, Pol ι, Pol κ, and Pol ζ, are all required for the efficient bypass of this lesion in human cells (15). Likewise, O6-CM-dG confers exclusively G → A transition in HEK293T cells, and it moderately impedes DNA replication (with a 38% bypass efficiency), where Pol κ and Pol ζ are involved in bypassing this lesion (38). On the other hand, O6-CM-dG displays only a ∼10% bypass efficiency in WT E. coli cells, and the three SOS-induced polymerases play redundant roles in bypassing this lesion.
It is also important to compare the results obtained for the major-groove O6-alkyl-dG lesions with previous findings made for minor-groove N2-alkyl-dG lesions. In this vein, the minor-groove N2-alkyl-dG lesions exert moderate blocking effects on DNA replication (50). For instance, a site-specifically incorporated N2-(1-carboxyethyl)-2′-deoxyguanosine (N2-CE-dG) was not a strong impediment to DNA replication in WT AB1157 cells, with the bypass efficiencies of S- and R-N2-CE-dG being ∼80 and ∼40%, respectively (50). However, the efficient and accurate replication across N2-alkyl-dG lesions in E. coli requires Pol IV, and their accurate bypass in human cells necessitates both Pol κ (an ortholog of Pol IV) and Pol ι (50–54). These differences perhaps can be attributed to the differences in replication machineries of E. coli and human cells.
In summary, our shuttle vector-based study on a group of NOC-derived O6-alkyl-dG lesions provided important and novel insights into the impact of carboxymethyl and pyridyloxobutyl/pyridylhydroxybutyl DNA lesions on the efficiency and fidelity of DNA replication in cells. Our results revealed strong miscoding potentials of the O6-POB-dG and O6-PHB-dG derived from tobacco-specific nitrosamines. In addition, our results revealed the hydrogen-bonding donor and negative charge situated on the O6-alkyl group as important determinants of DNA replication in E. coli cells. It will be important to assess how these O6-alkyl-dG lesions compromise DNA replication and transcription in mammalian cells in the future.
Experimental procedures
All chemicals, unless otherwise specified, were from Sigma–Aldrich or Thermo Fisher Scientific. 1,1,1,3,3,3-Hexafluoro-2-propanol was obtained from Oakwood Products, Inc. (West Columbia, SC). Reagents for solid-phase DNA synthesis were purchased from Glen Research Co. (Sterling, VA) and unmodified oligodeoxyribonucleotides (ODNs) were from Integrated DNA Technologies (Coralville, IA). All enzymes were obtained from New England Biolabs (Ipswich, MA), and [γ-32P]ATP was purchased from PerkinElmer Life Sciences.
M13mp7(L2) and WT AB1157 E. coli strain were kindly provided by Prof. John M. Essigmann. Polymerase-deficient AB1157 strains (Δpol B1::spec (Pol II–deficient), ΔdinB (Pol IV–deficient), ΔumuC::kan (Pol V–deficient), ΔumuC::kanΔdinB (Pol IV, Pol V double knockout), and Δpol B1::specΔdinB ▵umuC::kan (Pol II, Pol IV, Pol V triple knockout)) were generously provided by Prof. Graham C. Walker (49).
MS and NMR
ESI-MS and MS/MS experiments were carried out on an LCQ Deca XP ion-trap mass spectrometer (Thermo Fisher Scientific). A mixture of acetonitrile and water (50:50, v/v) was used as the solvent for electrospray. The spray voltage was 3.0 kV, and the temperature of the ion transport tube was maintained at 275 °C. High-resolution mass spectra were acquired on an Agilent 6510 Q-TOF LC/MS mass spectrometer (Agilent Technologies, Palo Alto, CA) equipped with an ESI source. 1H and 31P NMR spectra were recorded at 400 and 80 MHz on an Avance NEO 400 NMR spectrometer (Bruker, Billerica, MA) at 25 °C, respectively.
Preparation of the lesion-carrying ODNs
The 12-mer lesion-containing ODNs, 5′-ATGGCGXGCTAT-3′ (X = O6-alkyl-dG; Fig. 1), were synthesized via automated solid-phase DNA synthesis on a Beckman Oligo 1000M DNA synthesizer (Fullerton, CA). Commercially available ultramild phosphoramidites were employed for the incorporation of unmodified nucleotides (Glen Research Inc., Sterling, VA) following the standard ODN assembly protocol. The phosphoramidite building block for O6-HOEt-dG was prepared following published procedures (Scheme S1) (30, 55, 56); the yields and spectroscopic characterizations of the synthetic products are provided in the supporting materials (Figs. S1–S4).
O6-Aminocarbonylmethyl-dG (O6-AMC-dG)-containing ODN was deprotected by treating the solid support of O6-CM-dG containing ODN with concentrated ammonium hydroxide (57). All other 12-mer lesion-containing ODNs were cleaved from the solid support and deprotected with 0.25 m NaOH at room temperature overnight, and the resulting solution was subsequently neutralized with 0.5 m HCl. The O6-POB-dG–containing ODN was synthesized following previously published procedures (58), and the O6-PHB-dG–containing ODN was obtained by incubation of the O6-POB-dG–containing ODN with sodium borohydride (5 eq) in H2O for 5 min in an ice-water bath.
The solvents were removed under vacuum, and the solid residues were redissolved in water and purified via reversed-phase HPLC, with the use of a Kinetex XB-C18 column (4.6 × 150 mm, 5 μm in particle size, and 100 Å in pore size; Phenomenex Inc., Torrance, CA), as described elsewhere (39, 40). The HPLC traces for the purification of the 12-mer lesion-carrying ODNs are displayed in Fig. S5, and the mass spectrometric characterizations of the purified lesion-containing ODNs are shown in Fig. 2 and supporting Figs. S6 and S7. The purified 12-mer O6-alkyl-dG–containing ODNs were then ligated individually with a 10-mer ODN (5′-AGTGGAAGAC-3′) with T4 DNA ligase in the presence of a template ODN, and the desired 22-mer lesion-bearing ODNs were purified using denaturing PAGE, as described previously (39, 40).
Construction of single-stranded lesion-containing and lesion-free competitor M13 genomes
The lesion-containing and lesion-free M13mp7(L2) genomes were prepared following previously reported procedures (Fig. S8) (59). Briefly, 20 pmol of single-stranded M13 genome was first linearized via digestion with 40 units of EcoRI at 23 °C for 8 h. The ensuing linearized vector was then annealed with two scaffolds: 5′-CTTCCACTCACTGAATCATGGTCATAGCTTTC-3′ and 5′-AAAACGACGGCCAGTGAATTATAGC-3′ (25 pmol). To the resulting mixture was subsequently added 30 pmol of the 5′-phosphorylated 22-mer O6-alkyl-dG-bearing ODN or the competitor ODN (25-mer, 5′-AGTGGAAGACATGGCGATAAGCTAT-3′). The mixture was then treated with T4 DNA ligase at 16 °C for 8 h. After the ligation, excess scaffolds and the unligated vector were degraded by incubating with T4 DNA polymerase (22.5 U) at 37 °C for 4 h. The desired lesion-containing and the lesion-free M13 genomes were purified from the resultant mixture using a Cycle Pure kit (Omega). The constructed lesion-containing or control genomes were normalized against the lesion-free competitor genome following published procedures to determine accurately the relative concentrations of the constructed genomes (59).
Transfection of lesion-containing and competitor M13 genomes into E. coli cells
The control lesion-free M13 genome was mixed with the competitor genome at a molar ratio of 1:1. The O6-POB-dG- and O6-PHB-dG–containing M13 genomes were mixed individually with the competitor genome at a molar ratio of 2:1. The O6-CM-dG–, O6-ACM-dG–, and O6-HOEt-dG–carrying M13 genomes were mixed with the competitor genome at a molar ratio of 5:1. The mixtures were transfected into SOS-induced, electrocompetent WT AB1157 E. coli cells and the isogenic E. coli cells that are deficient in pol II, pol IV, pol V, or all three polymerases (59). The SOS response was induced by irradiating E. coli cells with 254 nm light at a dose of 45 J/m2, as previously described (49).
The E. coli cells were subsequently grown in lysogeny broth culture medium at 37 °C for 6 h. The phage was recovered from the supernatant by centrifugation at 13,000 r.p.m. for 5 min and further amplified in SCS110 E. coli cells to increase the progeny/lesion-genome ratio. The amplified phage was finally purified using the QIAprep Spin M13 kit (Qiagen) to obtain the ssM13 DNA template for PCR amplification.
Quantification of bypass efficiencies and mutation frequencies
A modified version of the CRAB assay was employed to assess the bypass efficiencies of the O6-alkyl-dG lesions in vivo (Fig. 3) (50, 59). The region of interest in the purified ssM13 DNA template was amplified by PCR with Phusion high-fidelity DNA polymerase (NEB) using two primers, 5′-YCAGCTATGACCATGATTCAGTGAGTGGA-3′ and 5′-YTCGGTGCGGGCCTCTTCGCTATTAC-3′ (where Y is an amino group). PCR amplification was conducted for 30 cycles, each of which consisted of 10 s at 98 °C, 30 s at 65 °C, and 15 s at 72 °C, with a final extension at 72 °C for 5 min.
The PCR products were then subjected to sequential restriction endonuclease digestion and native PAGE analysis for determination of bypass efficiency and mutation frequency (Fig. 4 and Figs. S11–S14). Briefly, 80 ng of the PCR products were treated with BbsI (10 units) restriction endonuclease and shrimp alkaline phosphatase (SAP, 1 unit) in 10 μl of CutSmart buffer (NEB) at 37 °C for 30 min. The SAP was deactivated by heating at 80 °C for 10 min, and the mixture was incubated with T4 polynucleotide kinase (PNK, 10 units), DTT (5 mm), and [γ-32P]ATP at 37 °C for 30 min to radiolabel the newly released 5′-termini of the restriction fragments. After deactivation of T4 PNK via heating at 80 °C for 10 min, the resultant mixture was digested with MluCI (10 μl) at 37 °C for 30 min. Under these conditions, the 10-mer ODNs emanating from initial lesion-situated strand were radiolabeled as d(p*GGCGMGCTAT), where M indicates A, T, G, and C, and p* designates the 5′-32P-labeled phosphate (Fig. 4A).
The digestion was subsequently quenched with 15 μl of formamide gel-loading buffer (2×), and the labeled fragments were separated using 30% native PAGE, as previously described (Fig. 4 and Figs. S11–S14) (39, 40). The intensities of the radiolabeled DNA bands were measured using a Typhoon 9410 variable mode imager, and the bypass efficiency was derived using the following equation: bypass efficiency (%) = (lesion signal/competitor signal)/(control signal/competitor signal) × 100%, where the competitor signal was employed as the internal standard.
The mutation frequencies were determined according to previously published procedures (39, 40). As shown in Fig. 4B, the two products with M being an A or G could be well-resolved by native PAGE analysis; hence, native PAGE analysis of the initial lesion-bearing strand was sufficient for quantifying the G → A transition frequency. In addition, the G → T transversion elicited by O6-POB-dG was further confirmed via PAGE analysis of the strand that is complementary to the original lesion-containing strand (Fig. S14).
Identification of mutagenic products by LC-MS/MS
The PCR products were digested with 50 units of BbsI restriction endonuclease and 20 units of shrimp alkaline phosphatase in 250 μl of NEB CutSmart buffer at 37 °C for 2 h, followed by deactivation of the enzymes through heating at 80 °C for 20 min. To the mixture was added 50 units of MluCI, and the solution was incubated at 37 °C for 1 h. The resulting solution was extracted once with phenol/chloroform/isoamyl alcohol (25:24:1, v/v). The aqueous layer was subsequently dried in a SpeedVac, desalted with HPLC, and dried again. The resultant solid residues were reconstituted in 20 μl of water, and a 10-μl aliquot was injected for LC-MS/MS analysis using an Agilent Zorbax SB-C18 column (0.5 × 250 mm, 5 μm in particle size). The gradient for LC-MS/MS analysis was 5 min of 5–20% methanol followed by 35 min of 20–50% methanol in 400 mm 1,1,1,3,3,3-hexafluoro-2-propanol. The temperature for the ion-transport tube was maintained at 300 °C. The LTQ linear ion trap mass spectrometer (Thermo Electron, San Jose, CA) was set up for monitoring the fragmentation of the [M-3H]3− ions of the 10-mer d(GGCGMGCTAT), where M represents A, T, C, or G. The fragment ions detected in the MS/MS were manually assigned (Figs. S9 and S10).
Author contributions
P. W. data curation; P. W. formal analysis; P. W. and J. L. investigation; P. W. methodology; P. W. writing-original draft; P. W., J. L., and Y. W. writing-review and editing; Y. W. conceptualization; Y. W. supervision; Y. W. funding acquisition.
Supplementary Material
This work was supported by National Institutes of Health Grant R01 ES025121. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Scheme S1 and Figs. S1–S14.
- NOC
- N-nitroso compound
- CRAB
- competitive replication and adduct bypass
- POB
- pyridyloxobutyl
- PHB
- pyridylhydroxybutyl
- NNK
- 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- NNN
- N′-nitrosonornicotine
- CM
- carboxymethyl
- Pol
- polymerase
- ESI
- electrospray ionization
- ODN
- oligodeoxyribonucleotide
- PNK
- polynucleotide kinase.
References
- 1. Fu D., Calvo J. A., and Samson L. D. (2012) Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer 12, 104–120 10.1038/nrc3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bogovski P., and Bogovski S. (1981) Animal species in which N-nitroso compounds induce cancer. Int. J. Cancer 27, 471–474 10.1002/ijc.2910270408 [DOI] [PubMed] [Google Scholar]
- 3. Mirvish S. S. (1995) Role of N-nitroso compounds (NOC) and N-nitrosation in etiology of gastric, esophageal, nasopharyngeal and bladder cancer and contribution to cancer of known exposures to NOC. Cancer Lett. 93, 17–48 10.1016/0304-3835(95)03786-V [DOI] [PubMed] [Google Scholar]
- 4. Tricker A. R. (1997) N-nitroso compounds and man: sources of exposure, endogenous formation and occurrence in body fluids. Eur. J. Cancer Prev. 6, 226–268 10.1097/00008469-199706000-00003 [DOI] [PubMed] [Google Scholar]
- 5. Hecht S. S. (1998) Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem. Res. Toxicol. 11, 559–603 10.1021/tx980005y [DOI] [PubMed] [Google Scholar]
- 6. Lewin M. H., Bailey N., Bandaletova T., Bowman R., Cross A. J., Pollock J., Shuker D. E., and Bingham S. A. (2006) Red meat enhances the colonic formation of the DNA adduct O6-carboxymethyl guanine: Implications for colorectal cancer. Cancer Res. 66, 1859–1865 10.1158/0008-5472.CAN-05-2237 [DOI] [PubMed] [Google Scholar]
- 7. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans (2007) Smokeless tobacco and some tobacco-specific N-nitrosamines. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans 89, 1–592 [PMC free article] [PubMed] [Google Scholar]
- 8. Balbo S., Johnson C. S., Kovi R. C., James-Yi S. A., O'Sullivan M. G., Wang M., Le C. T., Khariwala S. S., Upadhyaya P., and Hecht S. S. (2014) Carcinogenicity and DNA adduct formation of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol in F-344 rats. Carcinogenesis 35, 2798–2806 10.1093/carcin/bgu204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carlson E. S., Upadhyaya P., Villalta P. W., Ma B., and Hecht S. S. (2018) Analysis and identification of 2′-deoxyadenosine-derived adducts in lung and liver DNA of F-344 rats treated with the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyI)-1-butanone and enantiomers of its metabolite 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem. Res. Toxicol. 31, 358–370 10.1021/acs.chemrestox.8b00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lao Y., Yu N., Kassie F., Villalta P. W., and Hecht S. S. (2007) Analysis of pyridyloxobutyl DNA adducts in F344 rats chronically treated with (R)- and (S)-N′-nitrosonornicotine. Chem. Res. Toxicol. 20, 246–256 10.1021/tx060208j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lao Y., Villalta P. W., Sturla S. J., Wang M., and Hecht S. S. (2006) Quantitation of pyridyloxobutyl DNA adducts of tobacco-specific nitrosamines in rat tissue DNA by high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry. Chem. Res. Toxicol. 19, 674–682 10.1021/tx050351x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peterson L. A., and Hecht S. S. (1991) O6-Methylguanine is a critical determinant of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone tumorigenesis in A/J mouse lung. Cancer Res. 51, 5557–5564 [PubMed] [Google Scholar]
- 13. Upadhyaya P., Kalscheuer S., Hochalter J. B., Villalta P. W., and Hecht S. S. (2008) Quantitation of pyridylhydroxybutyl-DNA adducts in liver and lung of F-344 rats treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem. Res. Toxicol. 21, 1468–1476 10.1021/tx8001109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang S., Wang M., Villalta P. W., Lindgren B. R., Upadhyaya P., Lao Y., and Hecht S. S. (2009) Analysis of pyridyloxobutyl and pyridylhydroxybutyl DNA adducts in extrahepatic tissues of F344 rats treated chronically with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem. Res. Toxicol. 22, 926–936 10.1021/tx900015d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Du H., Leng J., Wang P., Li L., and Wang Y. (2018) Impact of tobacco-specific nitrosamine-derived DNA adducts on the efficiency and fidelity of DNA replication in human cells. J. Biol. Chem. 293, 11100–11108 10.1074/jbc.RA118.003477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harrison K. L., Fairhurst N., Challis B. C., and Shuker D. E. (1997) Synthesis, characterization, and immunochemical detection of O6-(carboxymethyl)-2′-deoxyguanosine: a DNA adduct formed by nitrosated glycine derivatives. Chem. Res. Toxicol. 10, 652–659 10.1021/tx960203u [DOI] [PubMed] [Google Scholar]
- 17. Harrison K. L., Jukes R., Cooper D. P., and Shuker D. E. (1999) Detection of concomitant formation of O6-carboxymethyl- and O6-methyl-2′-deoxyguanosine in DNA exposed to nitrosated glycine derivatives using a combined immunoaffinity/HPLC method. Chem. Res. Toxicol. 12, 106–111 10.1021/tx980057n [DOI] [PubMed] [Google Scholar]
- 18. Busby W. F. Jr, Shuker D. E., Charnley G., Newberne P. M., Tannenbaum S. R., and Wogan G. N. (1985) Carcinogenicity in rats of the nitrosated bile acid conjugates N-nitrosoglycocholic acid and N-nitrosotaurocholic acid. Cancer Res. 45, 1367–1371 [PubMed] [Google Scholar]
- 19. Gottschalg E., Scott G. B., Burns P. A., and Shuker D. E. (2007) Potassium diazoacetate-induced p53 mutations in vitro in relation to formation of O6-carboxymethyl- and O6-methyl-2′-deoxyguanosine DNA adducts: relevance for gastrointestinal cancer. Carcinogenesis 28, 356–362 [DOI] [PubMed] [Google Scholar]
- 20. Yang J., Villalta P. W., Upadhyaya P., and Hecht S. S. (2016) Analysis of O6-4-(3-pyridyl)-4-oxobut-1-yl-2′-deoxyguanosine and other DNA adducts in rats treated with enantiomeric or racemic N'-nitrosonornicotine. Chem. Res. Toxicol. 29, 87–95 10.1021/acs.chemrestox.5b00425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leng J., and Wang Y. (2017) Liquid chromatography-tandem mass spectrometry for the quantification of tobacco-specific nitrosamine-induced DNA adducts in mammalian cells. Anal. Chem. 89, 9124–9130 10.1021/acs.analchem.7b01857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li L., Perdigao J., Pegg A. E., Lao Y., Hecht S. S., Lindgren B. R., Reardon J. T., Sancar A., Wattenberg E. V., and Peterson L. A. (2009) The influence of repair pathways on the cytotoxicity and mutagenicity induced by the pyridyloxobutylation pathway of tobacco-specific nitrosamines. Chem. Res. Toxicol. 22, 1464–1472 10.1021/tx9001572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang J., and Wang Y. (2009) Chemical synthesis of oligodeoxyribonucleotides containing N3- and O4-carboxymethylthymidine and their formation in DNA. Nucleic Acids Res. 37, 336–345 10.1093/nar/gkn946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang J., and Wang Y. (2010) Synthesis and characterization of oligodeoxyribonucleotides containing a site-specifically incorporated N6-carboxymethyl-2′-deoxyadenosine or N4-carboxymethyl-2′-deoxycytidine. Nucleic Acids Res. 38, 6774–6784 10.1093/nar/gkq458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu Y., Wang J., Wang P., and Wang Y. (2016) Quantification of azaserine-induced carboxymethylated and methylated DNA lesions in cells by nanoflow liquid chromatography-nanoelectrospray ionization tandem mass spectrometry coupled with the stable isotope-dilution method. Anal. Chem. 88, 8036–8042 10.1021/acs.analchem.6b01349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sedgwick B. (2004) Repairing DNA-methylation damage. Nat. Rev. Mol. Cell Biol. 5, 148–157 10.1038/nrm1312 [DOI] [PubMed] [Google Scholar]
- 27. Shrivastav N., Li D., and Essigmann J. M. (2010) Chemical biology of mutagenesis and DNA repair: cellular responses to DNA alkylation. Carcinogenesis 31, 59–70 10.1093/carcin/bgp262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Swann P. F. (1990) Why do O6-alkylguanine and O4-alkylthymine miscode: the relationship between the structure of DNA containing O6-alkylguanine and O4-alkylthymine and the mutagenic properties of these bases. Mutat. Res. 233, 81–94 10.1016/0027-5107(90)90153-U [DOI] [PubMed] [Google Scholar]
- 29. Meikrantz W., Bergom M. A., Memisoglu A., and Samson L. (1998) O6-Alkylguanine DNA lesions trigger apoptosis. Carcinogenesis 19, 369–372 10.1093/carcin/19.2.369 [DOI] [PubMed] [Google Scholar]
- 30. Wang P., and Wang Y. (2018) Cytotoxic and mutagenic properties of O6-alkyl-2′-deoxyguanosine lesions in Escherichia coli cells. J. Biol. Chem. 293, 15033–15042 10.1074/jbc.RA118.004676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Peterson L. A., Thomson N. M., Crankshaw D. L., Donaldson E. E., and Kenney P. J. (2001) Interactions between methylating and pyridyloxobutylating agents in A/J mouse lungs: implications for 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced lung tumorigenesis. Cancer Res. 61, 5757–5763 [PubMed] [Google Scholar]
- 32. Urban A. M., Upadhyaya P., Cao Q., and Peterson L. A. (2012) Formation and repair of pyridyloxobutyl DNA adducts and their relationship to tumor yield in A/J mice. Chem. Res. Toxicol. 25, 2167–2178 10.1021/tx300245w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ronai Z. A., Gradia S., Peterson L. A., and Hecht S. S. (1993) G to A transitions and G to T transversions in codon 12 of the Ki-ras oncogene isolated from mouse lung tumors induced by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and related DNA methylating and pyridyloxobutylating agents. Carcinogenesis 14, 2419–2422 10.1093/carcin/14.11.2419 [DOI] [PubMed] [Google Scholar]
- 34. Pauly G. T., Peterson L. A., and Moschel R. C. (2002) Mutagenesis by O6-4-oxo-4-(3-pyridyl)butyl guanine in Escherichia coli and human cells. Chem. Res. Toxicol. 15, 165–169 10.1021/tx0101245 [DOI] [PubMed] [Google Scholar]
- 35. Choi J.-Y., Chowdhury G., Zang H., Angel K. C., Vu C. C., Peterson L. A., and Guengerich F. P. (2006) Translesion synthesis across O6-alkylguanine DNA adducts by recombinant human DNA polymerases. J. Biol. Chem. 281, 38244–38256 10.1074/jbc.M608369200 [DOI] [PubMed] [Google Scholar]
- 36. Choi J.-Y., and Guengerich F. P. (2008) Kinetic analysis of translesion synthesis opposite bulky N2- and O6-alkylguanine DNA adducts by human DNA polymerase REV1. J. Biol. Chem. 283, 23645–23655 10.1074/jbc.M801686200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang F., Tsunoda M., Suzuki K., Kikuchi Y., Wilkinson O., Millington C. L., Margison G. P., Williams D. M., CzarinaMorishita E., and Takénaka A. (2013) Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing pyrimidine transition mutations. Nucleic Acids Res. 41, 5524–5532 10.1093/nar/gkt198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu J., Wang P., Li L., Williams N. L., Ji D., Zahurancik W. J., You C., Wang J., Suo Z., and Wang Y. (2017) Replication studies of carboxymethylated DNA lesions in human cells. Nucleic Acids Res. 45, 7276–7284 10.1093/nar/gkx442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang P., Amato N. J., Zhai Q., and Wang Y. (2015) Cytotoxic and mutagenic properties of O4-alkylthymidine lesions in Escherichia coli cells. Nucleic Acids Res. 43, 10795–10803 10.1093/nar/gkv941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhai Q., Wang P., Cai Q., and Wang Y. (2014) Syntheses and characterizations of the in vivo replicative bypass and mutagenic properties of the minor-groove O2-alkylthymidine lesions. Nucleic Acids Res. 42, 10529–10537 10.1093/nar/gku748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. de Groot P. M., Wu C. C., Carter B. W., and Munden R. F. (2018) The epidemiology of lung cancer. Trans. Lung Cancer Res. 7, 220–233 10.21037/tlcr.2018.05.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jakszyn P., Bingham S., Pera G., Agudo A., Luben R., Welch A., Boeing H., Del Giudice G., Palli D., Saieva C., Krogh V., Sacerdote C., Tumino R., Panico S., Berglund G., et al. (2006) Endogenous versus exogenous exposure to N-nitroso compounds and gastric cancer risk in the European Prospective Investigation into Cancer and Nutrition (EPIC-EURGAST) study. Carcinogenesis 27, 1497–1501 10.1093/carcin/bgl019 [DOI] [PubMed] [Google Scholar]
- 43. Cross A. J., Pollock J. R., and Bingham S. A. (2003) Haem, not protein or inorganic iron, is responsible for endogenous intestinal N-nitrosation arising from red meat. Cancer Res. 63, 2358–2360 [PubMed] [Google Scholar]
- 44. Warren J. J., Forsberg L. J., and Beese L. S. (2006) The structural basis for the mutagenicity of O6-methyl-guanine lesions. Proc. Natl. Acad. Sci. U.S.A. 103, 19701–19706 10.1073/pnas.0609580103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Johnson S. J., and Beese L. S. (2004) Structures of mismatch replication errors observed in a DNA polymerase. Cell 116, 803–816 10.1016/S0092-8674(04)00252-1 [DOI] [PubMed] [Google Scholar]
- 46. Janion C. (2008) Inducible SOS response system of DNA repair and mutagenesis in Escherichia coli. Int. J. Biol. Sci. 4, 338–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fuchs R. P., and Fujii S. (2013) Translesion DNA synthesis and mutagenesis in prokaryotes. Cold Spring Harb. Perspect. Biol. 5, a012682 10.1101/cshperspect.a012682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vaisman A., McDonald J., and Woodgate R. (2012) Translesion DNA synthesis. EcoSal Plus 5, doi: 10.1128/ecosalplus.7.2.2 10.1128/ecosalplus.7.2.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Neeley W. L., Delaney S., Alekseyev Y. O., Jarosz D. F., Delaney J. C., Walker G. C., and Essigmann J. M. (2007) DNA polymerase V allows bypass of toxic guanine oxidation products in vivo. J. Biol. Chem. 282, 12741–12748 10.1074/jbc.M700575200 [DOI] [PubMed] [Google Scholar]
- 50. Yuan B., Cao H., Jiang Y., Hong H., and Wang Y. (2008) Efficient and accurate bypass of N2-(1-carboxyethyl)-2′-deoxyguanosine by DinB DNA polyrnerase in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 105, 8679–8684 10.1073/pnas.0711546105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Minko I. G., Yamanaka K., Kozekov I. D., Kozekova A., Indiani C., O'Donnell M. E., Jiang Q., Goodman M. F., Rizzo C. J., and Lloyd R. S. (2008) Replication bypass of the acrolein-mediated deoxyguanine DNA-peptide cross-links by DNA polymerases of the DinB family. Chem. Res. Toxicol. 21, 1983–1990 10.1021/tx800174a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Choi J. Y., Angel K. C., and Guengerich F. P. (2006) Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase κ. J. Biol. Chem. 281, 21062–21072 10.1074/jbc.M602246200 [DOI] [PubMed] [Google Scholar]
- 53. Yuan B. F., You C., Andersen N., Jiang Y., Moriya M., O'Connor T. R., and Wang Y. (2011) The roles of DNA polymerases κ and ι in the error-free bypass of N2-carboxyalkyl-2′-deoxyguanosine lesions in mammalian cells. J. Biol. Chem. 286, 17503–17511 10.1074/jbc.M111.232835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Washington M. T., Minko I. G., Johnson R. E., Wolfle W. T., Harris T. M., Lloyd R. S., Prakash S., and Prakash L. (2004) Efficient and error-free replication past a minor-groove DNA adduct by the sequential action of human DNA polymerases ι and κ. Mol. Cell Biol. 24, 5687–5693 10.1128/MCB.24.13.5687-5693.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li B. F. L., and Swann P. F. (1989) Synthesis and characterization of oligodeoxynucleotides containing O6-methyl-, O6-ethyl-, and O6-isopropylguanine. Biochemistry 28, 5779–5786 10.1021/bi00440a012 [DOI] [PubMed] [Google Scholar]
- 56. Smith C. A., Xu Y. Z., and Swann P. F. (1990) Solid-phase synthesis of oligodeoxynucleotides containing O6-alkylguanine. Carcinogenesis 11, 811–816 10.1093/carcin/11.5.811 [DOI] [PubMed] [Google Scholar]
- 57. Xu Y. Z. (2000) Synthesis and characterization of DNA containing O6-carboxymethylguanine. Tetrahedron 56, 6075–6081 10.1016/S0040-4020(00)00556-1 [DOI] [Google Scholar]
- 58. Wang L., Spratt T. E., Pegg A. E., and Peterson L. A. (1999) Synthesis of DNA oligonucleotides containing site-specifically incorporated O6-4-oxo-4-(3-pyridyl)butyl guanine and their reaction with O6-alkylguanine-DNA alkyltransferase. Chem. Res. Toxicol. 12, 127–131 10.1021/tx980251+ [DOI] [PubMed] [Google Scholar]
- 59. Delaney J. C., and Essigmann J. M. (2006) Assays for determining lesion bypass efficiency and mutagenicity of site-specific DNA lesions in vivo. Methods Enzymol. 408, 1–15 10.1016/S0076-6879(06)08001-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.