Abstract
Channelrhodopsins (ChRs) are light-gated ion channels in widespread use in neuroscience for mediating the genetically targetable optical control of neurons (optogenetics). ChRs pass multiple kinds of ions, and although nonspecific ChR-mediated conductance is not an issue in many neuroscience studies, conductance of calcium and protons, which can mediate diverse cellular signals, may be undesirable in some instances. Here, we turned our attention to the creation of ChRs that have high cation photocurrent but pass fewer calcium ions and protons. We developed an automated, time-resolved screening method capable of rapidly phenotyping channelrhodopsin-2 (ChR2) variants. We found substitution mutations throughout ChR2 that could boost current while altering ion selectivity and observed that the mutations that reduced calcium or proton conductance have additive effects. By combining four mutations, we obtained a ChR, ChromeQ, with improved photocurrent that possesses order-of-magnitude reductions in calcium and proton conductance and high fidelity in driving repetitive action potentials in neurons. The approach presented here offers a viable pathway toward customization of complex physiological properties of optogenetic tools. We propose that our screening method not only enables elucidation of new ChR variants that affect microbial opsin performance but may also reveal new principles of optogenetic protein engineering.
Keywords: optogenetics, calcium, protein engineering, physiology, neurobiology, channelrhodopsin, proton, screening
Introduction
Channelrhodopsins are light-gated ion channels in widespread use in neuroscience for mediating the genetically targetable optical control of neuron electrical potential. Cation-conducting channelrhodopsins naturally conduct multiple kinds of ions: sodium, protons, calcium, and potassium (1, 2). Although the nonspecific conductance of channelrhodopsins is not a problem for many neuroscience questions, especially when proper control experiments are done, there are some contexts in which the calcium or proton conductance may be undesirable. For example, recent studies have shown that the proton conductance of channelrhodopsins may be able to cause glial acidification and release of glutamate (3), and the calcium conductance of channelrhodopsins may cause release of calcium from intracellular stores (4, 5). Calcium and proton signaling pathways have also been implicated in many aspects of cellular physiology: mitochondrial calcium plays a role in synaptic plasticity (6), calcium signaling via various kinases and transcription factors causes changes in neural gene expression (7–10), calcium-dependent changes in receptor phosphorylation and membrane insertion result in altered synaptic strength (11), protons can activate specific ion channels and receptors (12), and so forth. Accordingly, we here turned our attention to the creation of channelrhodopsins that have high cation photocurrent but pass fewer calcium ions or protons, which can mediate diverse cellular signals.
We developed an automated microscopy screening platform that enables time-resolved phenotyping of mutants of optogenetic tools and applied it to the systematic protein-wide screening of mutants of the light-driven nonspecific cation channel channelrhodopsin-2 (ChR2).3 We found mutations throughout ChR2 that were capable of boosting current or altering ion selectivity. These results validate and extend homology- and structure-predicted optogenetic tool optimizations (13–15), revealing many novel regions of the protein, scattered throughout its backbone, that contribute to photocurrent amplitude and ion selectivity. Furthermore, we found that mutations that reduced calcium or proton conductance could be combined in a synergistic fashion. Thus, our novel multidimensional screening approach can be used to reveal new principles governing optogenetic tool design. We combined four mutations to result in a light-gated ion channel, ChromeQ, that possesses order-of-magnitude reductions in calcium and proton conductance while preserving sodium and potassium currents.
Results
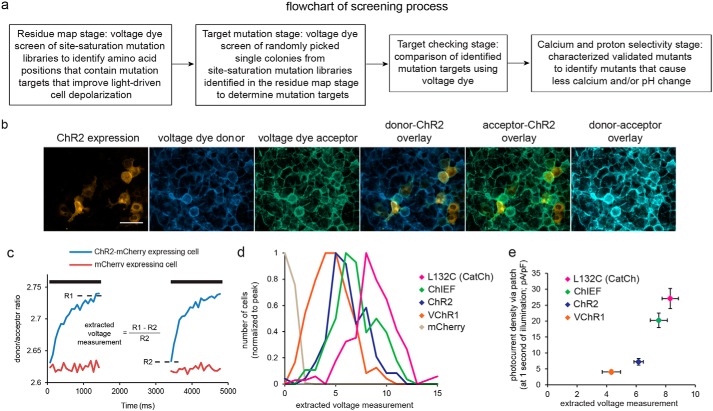
To systematically screen libraries of ChR2 mutants to select for molecules bearing three properties, high light-driven cell depolarization, low calcium photocurrent, and low proton photocurrent, we broke the screen down into four stages, each of which involved expressing a library of ChR2 point mutants into cultured HEK cells, loading the cells with a specific fluorescent dye that measures physiological responses, and then light activating the ChR2 point mutants while monitoring the dye responses. We first created saturation mutagenesis libraries of ChR2 (Fig. 1a, “residue map” stage), transfected the libraries into HEK cells, and identified amino acid positions for which one or more point mutants resulted in strongly increased light-driven responses using a voltage-sensitive dye reporter. For amino acid positions thus identified, we then screened the 19 possible individual mutants at each amino acid position for improved light-driven cell depolarization (Fig. 1a, “target mutation” stage), checking those mutants for improved light-driven cell depolarization (Fig. 1a, “target checking” stage) and then finally screened the identified mutants for reduced proton or calcium fluxes (Fig. 1a, “calcium and proton selectivity” stage). For each amino acid (positions 2–299) in ChR2 that we examined, we generated a library containing a physical mixture of the possible variants at that position, transfected each library into a well of a 96-well plate containing HEK cells in a fashion aiming for single-copy transfection (see “Experimental procedures”), and loaded cell membranes with an oxonol FRET voltage-sensitive dye (CC2-DMPE (donor); DiSBAC2(3) (acceptor) (16, 17)) (Fig. 1b). Delivering 405 nm light both excites the donor and activates ChR2 (1), causing the cell to depolarize and increasing the donor/acceptor fluorescence ratio (Fig. 1c). We illuminated cells twice, subtracting the fluorescence ratio at the beginning of the second pulse from that at the end of the first to compensate for dye photobleaching (see “Experimental procedures”).
Figure 1.
A screen for isolating mutations governing optogenetic protein operation. a, flowchart of the screening process. b, fluorescence images of live HEK293FT cells, highlighting from left to right ChR2-mCherry expression, CC2-DMPE (voltage dye donor), DiSBAC2(3) (voltage dye acceptor), overlaid donor and ChR2, overlaid acceptor and ChR2, and overlaid donor and acceptor. Scale bar, 50 μm. c, HEK cell voltage changes for ChR2-expressing (blue) and control (red) cells elicited by two 405 nm illumination pulses (1.5 s long, separated by 2 s, 6 milliwatts/mm2 irradiance; black bars) measured using the ratiometric voltage-sensitive FRET pair of b. Indicated by the dotted lines are the averages of donor-to-acceptor ratio of the last three frames at the end of the first light pulse (R1) and donor-to-acceptor ratio at the beginning of the second pulse (R2) used to calculate extracted voltage measurement. d, histograms of extracted voltage measurements, measured as in c, for four previously described channelrhodopsins (ChR2 L132C (CatCh) (18), ChIEF (20), ChR2 (1), and VChR1 (36)) and control cells (n = 94–224 cells each; histograms normalized to peak). e, steady-state photocurrent density measured at the end of a 1-s illumination using whole-cell voltage clamp (y axis) versus extracted voltage measurement (x axis) for the opsins shown in d (n = 10–12 HEK293FT cells each). Plotted are means and error bars representing S.E.
This “extracted voltage measurement,” calculated as shown in Fig. 1c, accurately reflected the performance of known channelrhodopsins (Fig. 1d; n = 94–224 HEK293FT cells for each opsin; F(4, 629) = 477.6, p < 0.0001, ANOVA of voltage measure with factor of opsin) and agreed with photocurrents measured via whole-cell patch clamp (Fig. 1e; n = 10–12 HEK293FT cells each opsin; F(3, 33) = 29.47, p < 0.0001, ANOVA). We found that 48 amino acid positions in ChR2 could be mutated to result in higher light-driven cell depolarization (Fig. 2, Fig. S1, Table S1, column 1; significance gauged by Bonferroni-corrected Kolmogorov–Smirnov (K-S) test comparing extracted voltage measurement of library versus ChR2, p < 0.05), some of which matched mutations known or previously hypothesized to enhance photocurrent (e.g. a full half of the residues in helix 2). We also found many sites in unpredicted places, including all other transmembrane domains, and in cytosolic and extracellular loops throughout the entire length of the protein (Fig. 2, Fig. S1, and Table S1, column 2). Thus, the screen suggests that parts of an opsin distributed throughout the backbone can, upon mutation, yield improved performance. In addition, two hits, at amino acids Asp-253 and Lys-257 (the latter forming the Schiff base with all-trans-retinal) showed no light-driven depolarization (Fig. S1a).
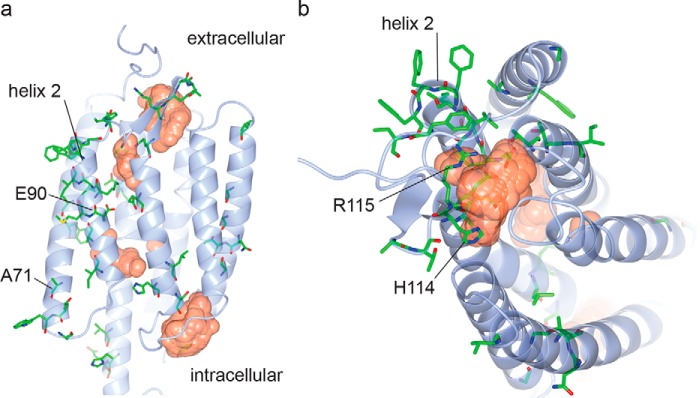
Figure 2.
Mutations mapped on the ChR2 structure. Amino acid positions of mutations identified in the residue mapping stage (colored by atom type; green, carbon; red, oxygen; blue, nitrogen; yellow, sulfur) are indicated on the side (a) and top (b) views of the crystal structure of ChR2 (shown in light blue) (44). Cavities within the protein are indicated as orange surfaces. Four positions mutated in ChromeQ are also additionally indicated with their residue numbers. The structure is based on Protein Data Bank (PDB) code 6EID, visualized using CCP4 (45). Cavities were calculated using HOLLOW (46).
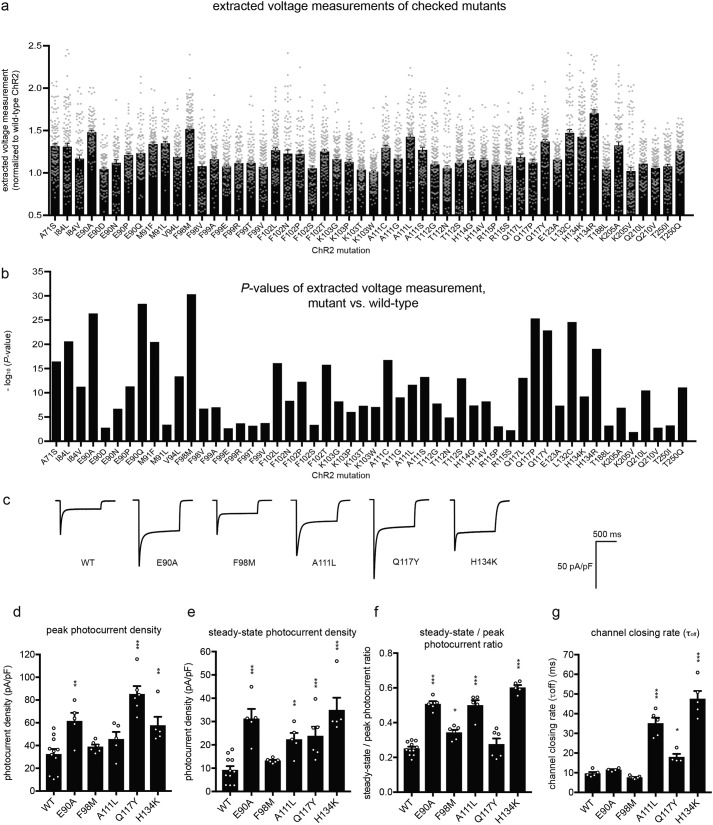
For each amino acid revealed in the residue map, we then screened the possible amino acids at each amino acid position for improved light-driven responses (target mutation stage). We began with the 27 amino acid positions from the residue map screen with the highest improvement (>1.5× increase in extracted voltage measurement) as well as those that were near those residues (Table S1, column 3) and identified 92 point mutants (at 24 of the 27 amino acid positions) with significantly higher extracted voltage measurement (n = 39–396 HEK293FT cells, Bonferroni-corrected K-S test comparing extracted voltage measurement with that of ChR2, p < 0.05). We checked these candidates with a second round of voltage imaging (target checking), confirming 52 single-point mutations from 21 of 24 amino acid positions (n = 71–164 HEK293FT cells, Bonferroni-corrected K-S test comparing extracted voltage measurement with that of WT ChR2, p < 0.05; Fig. 3, a and b), including two that were previously described to enhance photocurrents, L132C (18) and H134R (19) (Fig. 3, a and b). We examined five of the 52 mutations that exhibited the highest -fold change in the extracted voltage measurement (Fig. 3c) using patch clamp and 470 nm light at 10 milliwatts/mm2, and four of the five had enhancements in either peak (Fig. 3d), or steady-state (Fig. 3e) photocurrent density, or both (n = 4–11 HEK293FT cells, p < 0.05 non-Bonferroni–corrected t test comparing peak component or steady-state component versus that of ChR2). The fifth mutant, F98M, was not significantly enhanced over WT, although it was functional, exemplifying the fact that as with any screen, identified hits must be assessed through more detailed methodologies (in this case, using patch clamp). These mutants exhibited in general less run-down during illumination than the WT (Fig. 3f); we also measured the kinetics of channel closing postillumination (Fig. 3g).
Figure 3.
Mutations that improve light-driven cell depolarization. a, extracted voltage measurements (normalized to that of WT ChR2) for mutant opsins emerging from the screen (see Fig. S1 and Table S1 for residue-by-residue data from the residue map and target mutation stages, respectively, of the screen) that had mean >1 (normalized to WT ChR2) and p < 0.05 (Bonferroni-corrected K-S test against WT ChR2, n = 71–164 HEK293FT cells each). Gray dots indicate data from individual cells. b, K-S test p values (pre-Bonferroni correction) for the mutants in a. c, representative traces of photocurrent density (470 nm, 1-s illumination, 10 milliwatts/mm2 irradiance) measured using whole-cell voltage clamp for five mutants from f. d–g, population data (measured as in c) for peak photocurrent density (d), steady-state photocurrent density (measured at the end of 1-s illumination, 470 nm, 10 milliwatts/mm2 irradiance) (e), steady-state-to-peak photocurrent ratio (f), and channel closing rate (τoff) (measured using 470 nm, 2-ms illumination and 10 milliwatts/mm2 irradiance) (g) of five ChR2 mutants shown in c (n = 4–11 HEK293FT cells each). Plotted are means and error bars representing S.E. Circles indicate data for individual cells. Statistics for d–g, *, p < 0.05; **, p < 0.01, ***, p < 0.001, non-Bonferroni–corrected t test comparing mutant versus WT.
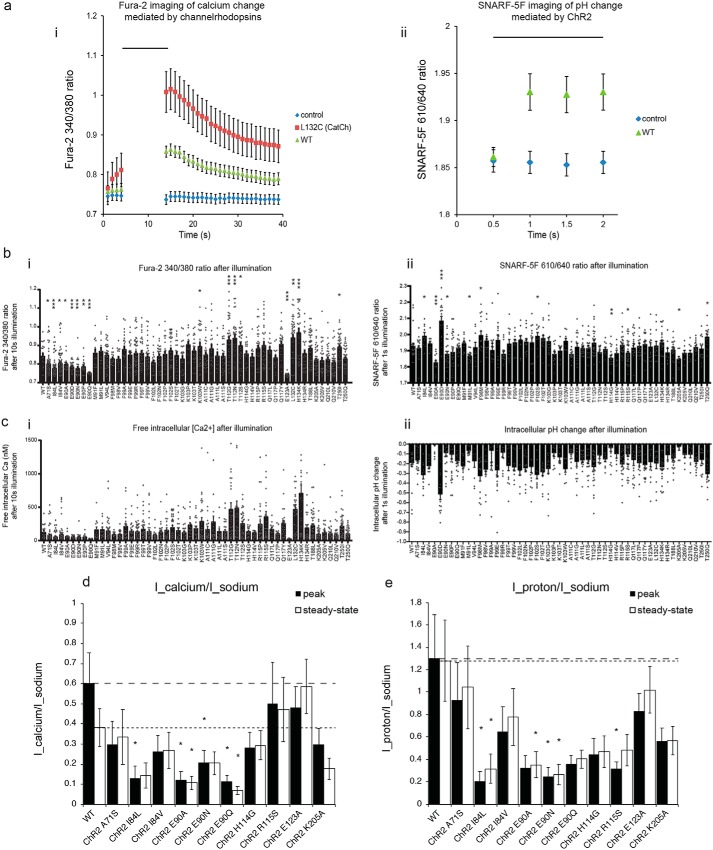
We screened the 52 mutants identified in Fig. 3a for reduced calcium and proton flux using Fura-2 (18, 20) to measure calcium flux (Fig. 4a, panel i) and SNARF-5F (20) to measure proton flux (Fig. 4a, panel ii). We screened mutants according to the appropriate calculated fluorescence ratios for Fura-2 (Fig. 4b, panel i) and SNARF-5F (Fig. 4b, panel ii) and calculated intracellular free calcium (Fig. 4c, panel i) and pH change (Fig. 4c, panel ii) in the screening conditions. HEK293 cells expressing WT ChR2 had resting calcium calculated to be 32.5 nm, which increased to 119 nm after illumination (n = 21 cells), and resting pH calculated to be 7.34, which decreased to 7.16 after illumination (n = 23 cells). Nine mutants (A71S, I84L, I84V, E90A, E90D, E90N, E90P, E90Q, and E123A) showed reduced calcium flux compared with ChR2 (Fig. 4b, panel i; n = 12–69 HEK293FT cells, p < 0.05 non-Bonferroni–corrected t test comparing 340/380 emission ratio after 10-s illumination of 470 nm at 4.6 milliwatts/mm2 with that of ChR2). Six mutants (E90A, E90N, M91L, H114G, R115S, and K205A) showed reduced proton flux (Fig. 4b, panel ii, n = 8–35 HEK293FT cells, p < 0.05 non-Bonferroni–corrected t test comparing pH change after 1-s illumination of 500 nm at 4 milliwatts/mm2 with that of WT ChR2). Moreover, a few had increased calcium or proton flux: in addition to the previously known mutant L132C, we found six mutants (K103W, T112G, T112N, T112S, H134K, and T250I) that showed increased calcium flux (Fig. 4b, panel i; n = 12–69 HEK293FT cells, p < 0.05 non-Bonferroni–corrected t test comparing 340/380 emission ratio after 10-s illumination with that of WT ChR2) and five that showed increased proton flux (I84L, E90D, F98M, F102S, and T250Q) (Fig. 4b, panel ii, n = 8–35 HEK293FT cells, p < 0.05 non-Bonferroni–corrected t test comparing pH change after 1-s illumination with that of WT ChR2). Although not the focus of this study, these latter mutants may provide residues for future constructs with greater calcium- or proton-conducting properties and thus may be of value for future optogenetic tools.
Figure 4.
Physiological screening for calcium and proton selectivity. a, change in Fura-2 340/380 nm emission ratio (panel i) and SNARF-5F 610/640 nm emission ratio (panel ii) mediated by previously described channelrhodopsins. Cells expressing fluorophores only were used as controls (mCherry in panel i and EGFP in panel ii) (n = 20–35 cells each). WT, WT ChR2. Black bar, illumination period, 470 nm for 10 s; irradiance, 4.6 milliwatts/mm2 (panel i); illumination period, 500 nm, 1.5 s; irradiance, 4 milliwatts/mm2 (panel ii). b and c, outcome of the calcium and proton selectivity of the screen for channelrhodopsins emerging from the screen of Fig. 1, showing the Fura-2 340/380 ratio (b, panel i) and calcium concentration (c, panel i) after blue light illumination (delivered as in a) in HEK cells expressing the indicated ChR2 mutants (n = 12–69 cells) and the SNARF-5F 610/640 ratio (b, panel ii) and pH (c, panel ii) after green light illumination (delivered as in a but with 1-s pulses) in HEK cells expressing the indicated ChR2 mutants (n = 8–35 cells). Gray dots indicate data from individual cells. d and e, population data for photocurrent density ratios measured using whole-cell patch clamp in HEK cells in ion-specific extracellular solutions (see “Experimental procedures” for details), calcium photocurrent (Icalcium) measured in 90 mm CaCl2, pH 7.4, divided by sodium photocurrent (Isodium) measured in 145 mm NaCl, pH 7.4 (d), and proton photocurrent (Iproton) measured in 135 mm NMDG, pH 6.4, divided by sodium photocurrent (Isodium) (e) of WT ChR2 and mutants with improved ion selectivity identified from b and c using illumination conditions 470 nm, 1 s, and 10 milliwatts/mm2 (n = 5–12 HEK293FT cells each). In d and e, peak and steady-state photocurrent ratios for WT ChR2 are indicated by dashed lines and dotted lines, respectively. Plotted are means and error bars representing S.E. Statistics for b, panels i and ii, d, and e, *, p < 0.05; **, p < 0.01; ***, p < 0.001, non-Bonferroni–corrected t test comparing mutant versus WT.
We characterized, in detail using whole-cell patch clamp, a subset of the mutants identified in the screen: 10 mutants with either reduced calcium (seven mutants; A71S, I84L, I84V, E90A, E90N, E90Q, and E123A) or proton (five mutants; E90A, E90N, H114G, R115S, and K205A) flux using ion-specific extracellular solutions to measure ion-specific photocurrents as done previously (14, 15, 18, 21, 22). Because ChR2 shows two apparent conducting states, exhibited by peak and steady-state components of its photocurrent (1, 23, 24), we characterized the ion selectivity for both components. For completeness, in addition to obtaining raw photocurrents (Fig. S4), we additionally characterized ion-specific reversal potentials (see supporting Results for data and a detailed discussion), although as discussed there, the interpretation of the reversal potentials in terms of calculated permeabilities may not be accurate due to the assumption of independent movement of multiple different ionic species in the Goldman–Hodgkin–Katz equation, which may not be met for channelrhodopsins (23, 25, 26).
Four of the seven calcium flux–reduced mutants identified in the imaging screen and chosen for further investigation were found to have either reduced peak or steady-state calcium photocurrent ratios (compared with sodium) as measured in patch clamp (Fig. 4d) with mutants E90A and E90Q having the largest reductions of ∼5-fold compared with WT; the mutation E123A did not have a reduced calcium flux when examined in the patch-clamp condition. Three of five proton flux–reduced mutants identified in the imaging screen and chosen for further investigation were similarly found to have reduced proton-to-sodium photocurrent ratios in patch clamp (normalized to sodium currents; Fig. 4e) with mutant I84L (chosen for its reduced calcium current despite its increased proton current, at least when measured with optical imaging) surprisingly having the largest reduction of ∼5-fold compared with WT. Interestingly, three of the seven calcium flux–reduced mutants identified in the imaging screen and chosen for further investigation had reduced proton-to-sodium photocurrent ratios, and two of the five proton flux–reduced mutants identified in the imaging screen and chosen for further investigation had reduced calcium-to-sodium photocurrent ratios. Indeed, the peak photocurrents of calcium and proton, normalized to sodium, appeared to be correlated to one another when we regressed across the various point mutants (Fig. S3). Thus, residues that alter ion selectivity for one ion may also alter ion selectivity for a second, independent ion.
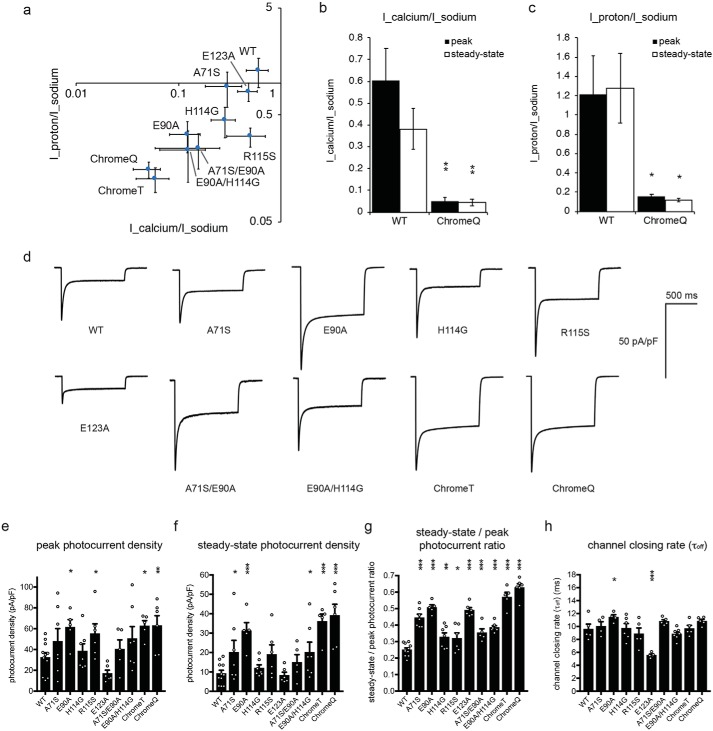
We next combined multiple mutations explored in Fig. 4 to see whether we could improve ion selectivity further, i.e. whether the mutations might be synergistically additive. We found through making double and triple mutants that we could progressively shift both the proton-to-sodium and calcium-to-sodium photocurrent ratios downward (Fig. 5a and raw photocurrents in Fig. S5) with the triple mutant A71S/E90A/H114G exhibiting a calcium-to-sodium photocurrent ratio reduced 10-fold (n = 5–12 HEK293FT cells, p < 0.001, t test comparing Icalcium/Isodium with that of WT ChR2; Fig. 5a) and the proton-to-sodium photocurrent ratio reduced 10-fold (n = 5–12 HEK293FT cells, p < 0.001, t test comparing Icalcium/Isodium with that of WT ChR2; Fig. 5a). A quadruple mutant, A71S/E90A/H114G/R115S, had calcium-to-sodium photocurrent and proton-to-sodium photocurrent ratios similar to those of the triple mutant (n = 5–7 HEK293FT cells, p > 0.2, t test comparing Icalcium/Isodium or Iproton/Isodium with that of triple mutant; Fig. 5, a–c). We nicknamed the triple and quadruple mutants ChromeT (Chr + omitting certain ions, triple mutant) and ChromeQ (quadruple mutant), respectively. Thus, we have created, through our screening methodology, new channelrhodopsins with order-of-magnitude improvement in their selectivity for two different ions, demonstrating the power of physiological screening.
Figure 5.
Combinatorial mutations leading to multidimensional molecular optimization and more monovalent ion–selective opsins. a, population data for proton-to-sodium photocurrent ratio versus calcium-to-sodium photocurrent ratio of ChR2 mutants measured using whole-cell patch clamp in HEK cells in ion-selective extracellular solutions (see “Experimental procedures” for details) using 470 nm, 1-s illumination and 10 milliwatts/mm2 irradiance (n = 5–12 HEK293FT cells each). b and c, population data for photocurrent density ratios measured using whole-cell patch clamp for peak (filled bars) and steady-state (open bars) calcium photocurrent (Icalcium) divided by sodium photocurrent (Isodium) (b) and peak (filled bars) and steady-state (open bars) proton photocurrent (Iproton) divided by sodium photocurrent (Isodium) (c) of WT ChR2 and ChromeQ (n = 7–9 HEK293FT cells each) using the same illumination as in a. d, representative traces of photocurrent density measured using whole-cell voltage clamp for the mutants in a using the same illumination conditions. e–h, population data for peak photocurrent density (e), steady-state photocurrent density (f), steady-state-to-peak photocurrent ratio (g) (measured using 470 nm, 1-s illumination and 10 milliwatts/mm2 irradiance) and channel closing rate (τoff) (h) (measured using 470 nm, 2-ms illumination and 10 milliwatts/mm2 irradiance) of mutants and mutant combinations in a (n = 4–11 HEK293FT cells each). Plotted are means and error bars representing S.E. Open circles indicate data from individual cells. Statistics for b, c, and e–h, *, p < 0.05; **, p < 0.01; ***, p < 0.001, non-Bonferroni–corrected t test comparing mutant versus WT. mW, milliwatt(s).
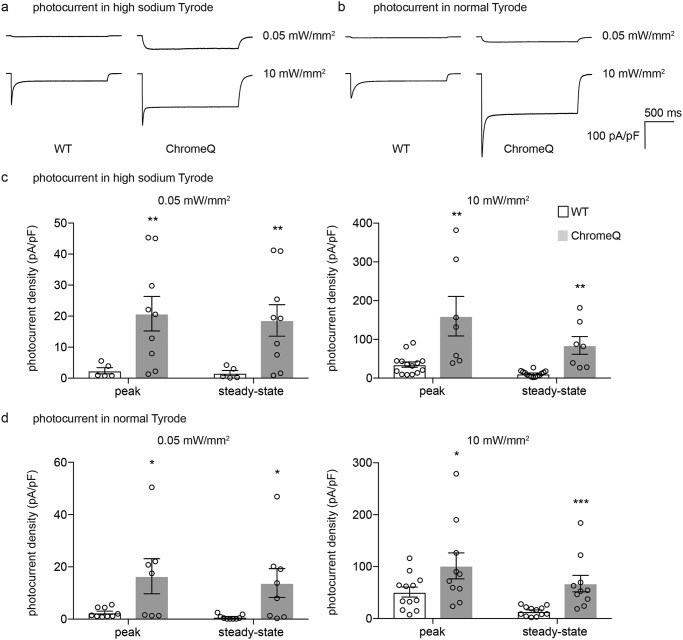
Mutations that reduce the conductance of an ion could, in principle, reduce overall ion flux through a channel. However, because we prefaced our ion selectivity screen with a screen for improved photocurrent, ChromeT and ChromeQ also possessed an increased peak as well as steady-state (Fig. 5, d–g) photocurrent density and unchanged channel closing kinetics (Fig. 5h). ChromeT and ChromeQ had greater overall photocurrent when normalized to the fluorescence of a fluorophore fused to the C terminus, a measure of expression level (Fig. S8a and supporting Results), suggesting but not proving an increased overall channel conductance. ChromeT and ChromeQ also had similar potassium selectivity, compared with sodium, versus WT (Fig. S8, b and c). When the sodium photocurrent (i.e. measured in high-sodium Tyrode's solution) and overall photocurrent (i.e. measured in conventional Tyrode's solution) of ChR2 and ChromeQ were compared under low (0.5 milliwatt/mm2) and high (10 milliwatts/mm2) light powers, ChromeQ consistently showed significantly larger sodium photocurrent (n = 5–14 HEK293FT cells, p < 0.01, t test comparing photocurrent of ChromeQ in high-sodium Tyrode's solution with that of WT ChR2; Fig. 6, a and c) and overall photocurrent (n = 5–14 HEK293FT cells, p < 0.05, t test comparing photocurrent of ChromeQ in normal Tyrode's solution with that of WT ChR2; Fig. 6, b and d) than ChR2. ChromeQ generated steady-state photocurrents under 0.05 milliwatt/mm2 similar to those generated by WT ChR2 under 10 milliwatts/mm2 when measured in normal Tyrode's solution (i.e. 14 ± 7 pA/pF for ChromeQ versus 14 ± 3 pA/pF for ChR2; n = 5–14 HEK293FT cells; Fig. 6, b and d). Therefore, if ChromeQ is illuminated with such low light powers, it could be useful for delivering steady-state currents similar to those conducted by ChR2 but with order-of-magnitude reductions in calcium and proton flux.
Figure 6.
Light power– and external sodium–dependent photocurrents in HEK293FT cells expressing WT ChR2 or ChromeQ. a and b, photocurrents were recorded in high-sodium (see “Experimental procedures”) Tyrode's solution (a) or in normal Tyrode's solution (b) in response to 470 nm light at 0.05 or 10 milliwatts/mm2 irradiance in HEK293FT cells expressing WT ChR2 or ChromeQ. c and d, population data for photocurrent density measured in high-sodium Tyrode's solution (c) or in normal Tyrode's solution (d) in response to 470 nm light at 0.05 or 10 milliwatts/mm2 irradiance (n = 5–14 HEK293FT cells each). Plotted are means and error bars representing S.E. Circles indicate data from individual cells. Statistics for c and d, *, p < 0.05; **, p < 0.01; ***, p < 0.001, Student's t test comparing ChR2 and ChromeQ.
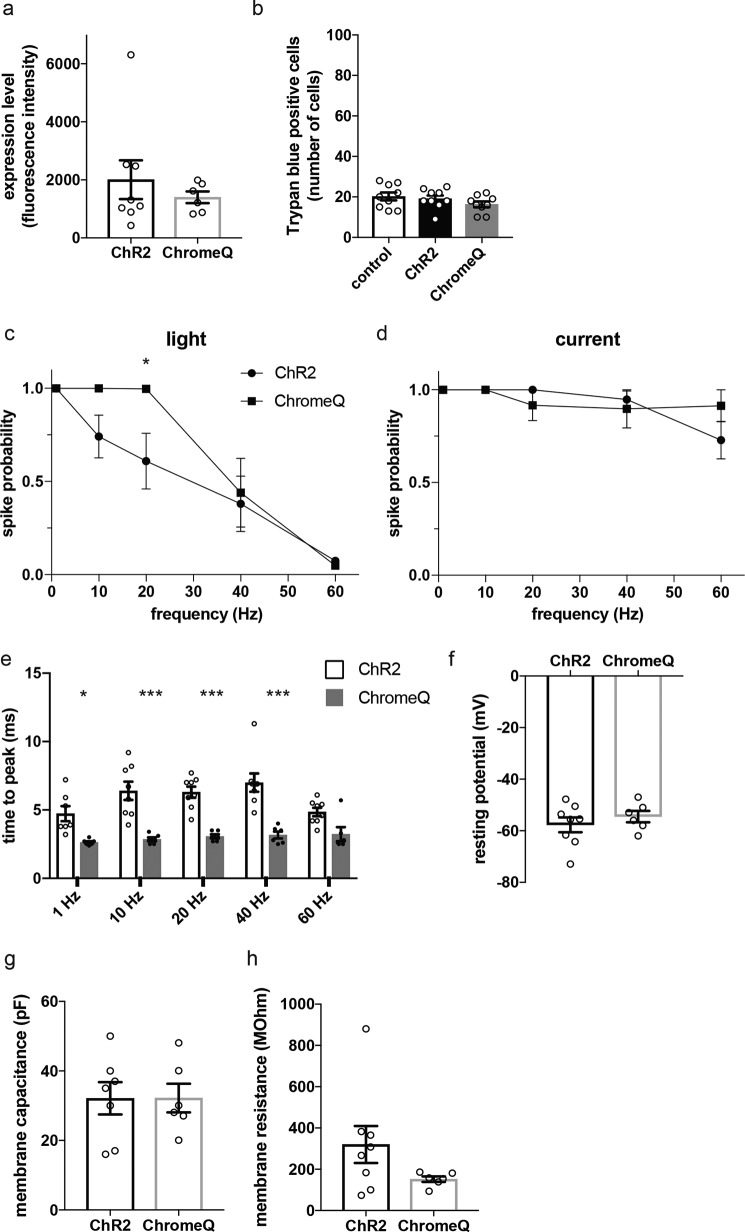
We next characterized ChromeQ in cultured primary mouse hippocampal neurons. When we expressed WT ChR2 and ChromeQ in primary mouse hippocampal neurons, they showed similar expression levels when analyzed by fluorescence of the fused genetically encoded fluorophore tdTomato (Fig. 7a; n = 6–8 neurons, p > 0.4, t test comparing expression level of ChromeQ with that of ChR2) and did not affect cell viability (Fig. 7b; n = 9 each condition, F(2, 24) = 1.406, p > 0.3, ANOVA). To assess the efficiency of light-driven action potential generation by ChromeQ, we compared ChR2 and ChromeQ in mediating responses to trains of photostimulation pulses of varying pulse frequency. Whole-cell current clamp was conducted on neurons expressing ChR2 or ChromeQ while 2-ms pulses of 470 nm light were delivered at an intensity of 4.9 milliwatts/mm2 and at frequencies of 1, 10, 20, 40, and 60 Hz (40 pulses each; Fig. 7, c–e). ChromeQ showed significantly higher success probability of light-driven action potentials at a frequency of 20 Hz compared with when ChR2 was used (Fig. 7c; n = 6–8 neurons, p < 0.05, repeated-measures ANOVA with Sidak's multiple comparisons test comparing spike probability with that of ChR2), whereas cells were similarly capable of electrically driven action potentials at this frequency (Fig. 7d; n = 6–8 neurons, p > 0.2, repeated-measures ANOVA with Sidak's multiple comparisons test comparing spike probability with that of ChR2). ChromeQ showed >98% success probability at frequencies between 1 and 20 Hz, whereas ChR2 showed reduced spike probability starting at 10 Hz (Fig. 7c). Latency of spike generation, as quantified by time to the peak of action potential after light onset, was significantly reduced in ChromeQ-expressing neurons at all frequencies tested except 60 Hz (Fig. 7e; n = 6–7 neurons, p < 0.05, repeated-measures ANOVA with Sidak's multiple comparisons test comparing time to the peak for ChromeQ versus ChR2). All other electrophysiological parameters such as resting potential, membrane capacitance, and membrane resistance were not significantly different in neurons expressing ChR2 and ChromeQ (Fig. 7, f–h; n = 6–8 neurons; p > 0.1, t tests comparing each parameter in ChromeQ- versus ChR2-expressing neurons).
Figure 7.
Characterization of cultured primary mouse hippocampal neurons expressing ChR2 or ChromeQ. a, expression level quantified by whole-cell fluorescence of cells transfected with ChR2 or ChromeQ (tdTomato fusions, n = 6–8 neurons each). b, number of trypan blue–positive cells in neurons transfected with ChR2 or ChromeQ. Control cells were untransfected (n = 9 neurons each). c–e, population data for spike probability of neurons transfected with ChR2 or ChromeQ under repetitive stimulation (40 trials for each frequency) using light (c) (using 470 nm, 2-ms illumination and 4.9 milliwatts/mm2 irradiance) or current injection (d) and spike latencies quantified by time between light onset and the spike peak (e) (n = 6–8 neurons each). f–h, population data for resting potential (f), membrane capacitance (g), and membrane resistance (h) of cells transfected with ChR2 or ChromeQ (n = 6–8 neurons each). Plotted are means and error bars representing S.E. Statistics for c–e, *, p < 0.05; **, p < 0.01; ***, p < 0.001, repeated-measures ANOVA with Sidak's multiple comparisons test comparing ChR2 and ChromeQ.
Discussion
Here, we present a novel screening method capable of evaluating libraries of microbial opsin mutants for altered and improved optogenetic physiological characteristics. We devised a pipeline capable of rapidly homing in on key residues, starting with libraries of mixed mutants and converging upon mutants of interest, as indicated by optical imaging of responses (i.e. voltage change, calcium flux, and proton flux) to optogenetic perturbation mediated by individual mutants. Although this strategy in principle can miss mutants because it randomly samples mutations, it results in efficient and inexpensive library preparation and screening. From the results of our screen, we discovered insights into how microbial opsin protein engineering can take place. For example, we found that mutants that improve photocurrent are distributed throughout significant fractions of the opsin, including practically all transmembrane and extracellular or intracellular loop regions, in contrast to previous studies that focused on key subdomains of the protein highlighted by homology or structure. In addition, we found that mutants that improve ion conductance can have additive and synergistic effects upon combination. As a result, we identified triple and quadruple mutants of channelrhodopsin-2, here named ChromeT and ChromeQ, with order-of-magnitude reduction in calcium and proton photocurrents compared with WT but preserved overall photocurrent amplitude and kinetics. Although many users of channelrhodopsins are focusing on short-term optical activation to drive spiking activity and many behavioral experiments with appropriate control experiments are being conducted, the calcium ions and protons that permeate the channelrhodopsin may be undesired in specific scientific contexts. Thus, ChromeT and ChromeQ may be of interest for a variety of experimental paradigms in the biosciences. For example, in experiments where extended illumination may result in significant permeation of calcium ions and protons, ChromeQ will significantly reduce the permeation of calcium ions and protons by using less light power to drive photocurrent similar to that of WT ChR2.
Earlier reports had identified several point mutations that altered cation-specific photocurrents (14, 15, 18, 21, 26–29), finding mutations that altered cation photocurrents typically by 2–4-fold. For example, several earlier reports have shown that mutations in channelrhodopsin residue Glu-90 alter proton selectivity (14, 15, 27) as found in our study as well. We also discovered many mutations unpredicted in structure-guided studies as well as new findings that may suggest rules governing optogenetic protein engineering (e.g. the observed correlation between calcium current and proton current across many point mutants of ChR2). Our screening approach may be applied to engineer channel selectivity to anions such as chloride, which has been achieved primarily through structure-guided mutations (30–32) and homology analysis (33–35). Thus, our screening method not only allows for elucidation of new mutants that affect microbial opsin performance but may reveal new principles of optogenetic protein engineering.
Experimental procedures
Molecular cloning and library construction
Mammalian codon-optimized ChR2(1–311) that constitutes the light-activated channel function (1, 36) from Chlamydomonas reinhardtii was cloned into pEGFP-N3 vector (Clontech) as a BamHI-AgeI fragment with mCherry as the fluorophore (mCherry was chosen among other red fluorophores because in HEK293FT cells the photocurrent amplitude of ChR2-mCherry was similar to ChR2-EGFP) to avoid spectral overlap with the dyes used in our screen. For the residue map stage of the screen, 298 site-saturation mutation libraries were generated, one for each amino acid between residues 2 and 299 of ChR2 (to focus on the membrane-spanning and intrahelical domains) using site-saturation mutagenesis reactions with primers containing NNK degenerate codons (Integrated DNA Technologies, Inc.). The site-saturation reaction mixture contained 50 ng of template plasmid mixed with 10 pmol of forward and reverse degenerate primers each, 0.2 mm dNTP mixture (New England Biolabs), and 1 unit of Phusion high-fidelity polymerase (New England Biolabs) in 50 μl. The thermocycle reaction consisted of a denaturation step at 95 °C for 60 s followed by 18 cycles of 95 °C for 50 s, 58 °C for 50 s, 72 °C for 3 min, and an extension step at 72 °C for 5 min. The reaction product was digested with 20 units of DpnI (New England Biolabs) for 6–12 h to completely remove the WT template plasmid. Each library was transformed in XL10-Gold competent Escherichia coli (Agilent) and purified using a QIAprep miniprep kit (Qiagen) run on an automated EVO75 liquid-handling robot (Tecan). The mutant codon frequency was checked (occurrences of 1 in 32 for amino acids Cys, Asp, Glu, Phe, His, Ile, Lys, Met, Asn, Gln, Trp, and Tyr; 2 in 32 for Ala, Gly, Pro, Thr, and Val; 3/32 for Leu, Arg, and Ser) (37) by sequencing 123 colonies from three libraries (amino acid positions Ile-84, Arg-120, and Leu-132), and the quality of library plasmids was checked for the presence of multiple peaks at the NNK degenerate codon sites (all four peaks for N; G and T peaks for K).
In the target mutation stage of the screen, plasmids containing point mutants were isolated to identify target mutations from site-saturation libraries by transforming XL10-Gold competent E. coli (Agilent) with each library, plating on a LB agar plate, and picking 96 single colonies (∼5-fold coverage of the library size) grown subsequently in 2-ml deep-well plates (Greiner). Plasmid from each colony was purified by an automated EVO100 liquid-handling robot (Tecan) using a CosMCprep kit (Beckman Coulter Genomics) following the manufacturer's instructions.
Culture and transfection of HEK293FT cells
HEK293FT cells (Invitrogen) were cultured in phenol red–free high-glucose DMEM (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (HyClone) and 1 mm sodium pyruvate, maintained in a 5% CO2 humidified incubator. The cells were plated on optical-bottom 24- or 96-well plates (Greiner Bio-One) coated with 2% growth factor–reduced Matrigel (BD Biosciences) in DMEM for 1 h at 37 °C at a density of 20,000 or 6,000 cells/well, respectively, which resulted in a confluent layer of cells that allowed us to consistently image the center of the wells. 24 h after plating cells, the medium was replaced with fresh medium (500 μl for 24-well and 50 μl for 96-well plates), and cells were placed in the incubator for 1–2 h at 37 °C before transfection.
For screening site-saturation mutation libraries in the residue map stage, each library plasmid was diluted in pUC18 plasmid that does not replicate in mammalian cells at a mass ratio of 1:100 to reduce uptake of multiple plasmids by a cell as described previously (38); having on average a single copy of a library plasmid per cell allows the detection of activity from each mutant. 625 ng of total plasmid DNA mixed with 1.563 μl of 2 m CaCl2 (250 mm final concentration) in 12.5 μl of double distilled H2O was mixed rapidly and thoroughly with 12.5 μl of 2× HEPES-buffered saline (50 mm HEPES, 280 mm NaCl, 1.5 mm Na2HPO4, pH 7.0) to obtain the transfection mixture (for a 4-well transfection; for larger or smaller numbers of wells, the recipes were scaled accordingly). The transfection mixture was incubated for 30 s at room temperature, and 5 μl was added to each well. The plates were placed in the incubator for 16–18 h, and the medium in each well was replaced with 100 μl of prewarmed culture medium containing 2 μm all-trans-retinal (Sigma). Although the addition of all-trans-retinal is not required for ChR2 to achieve photocurrent in HEK cells, we wanted to prototype a screen that could be of general use in screening for opsins, even those that have smaller affinities for all-trans-retinal. For transfecting cells in 24-well plates, all steps were identical to those above except the DNA mixture for each well contained 1.25 μg of total plasmid DNA and 3.125 μl of 2 m CaCl2 in 25 μl of double distilled H2O and was mixed with 25 μl of 2× HEPES-buffered saline.
Voltage-sensitive FRET imaging
HEK293FT cells were imaged in Tyrode's solution (125 mm NaCl, 2 mm KCl, 3 mm CaCl2, 1 mm MgCl2, 10 mm HEPES, 30 mm glucose, pH 7.3 (NaOH-adjusted), 305 mosm) 48 h after transfection. Cells in each well of 96-well plates were washed once with 50 μl of Tyrode's solution and incubated in 40 μl of voltage dye staining solution containing 40 μm CC2-DMPE (Invitrogen), 5 μm DiSBAC2(3) (Invitrogen), and 0.01% (w/v) Pluronic F-127 (Sigma) in Tyrode's solution for 40 min at room temperature. After incubation, cells were washed three times with 50 μl of Tyrode's solution and imaged.
The imaging was performed on an inverted microscope (Leica DMI6000B) equipped with an automated stage with a CMOS camera (Hamamatsu Orca Flash 2.8). The donor dye was excited using a 405 nm LED (Thorlabs) and detected using a dual-edge dichroic filter (440/520 Semrock) paired with an emission filter (448/20 Semrock). The acceptor was detected with the same dichroic mirror paired with an emission filter (572/28). mCherry was excited using a 590 nm LED filtered with an emission filter (590/20 Semrock) and detected with a dichroic filter (600 LP Chroma) paired with an emission filter (640/50 Chroma). The 405 nm LED and 590 nm LED were coupled using a dichroic mirror (409 nm long-pass Semrock). All the imaging components were controlled using MetaMorph (Molecular Devices).
To quantify the voltage changes mediated by opsins, the following imaging scheme was used. First, the cells were focused by imaging mCherry using a 590 nm LED (Thorlabs) at 3.5 milliwatts/mm2 accompanied by the autofocus function of MetaMorph, which allowed accurate focusing without activating ChR2 or photobleaching the dye. Next, each well was illuminated with a 405 nm LED (Thorlabs) at 6 milliwatts/mm2, which excites the voltage dye donor at its maximum absorption (16) and simultaneously activates ChR2 (1). The cells were illuminated with a 1.5-s pulse, during which the cell depolarizes, followed by a 2-s delay that allows recovery of membrane potential and a second 1.5-s pulse. Background fluorescence of donor and acceptor channels was determined by averaging signals from images recorded from six empty wells in the same plate under the same illumination conditions. In quantifying the light-driven depolarization of ChR2 using voltage-sensitive FRET imaging, we first attempted to quantify the voltage change by calculating the difference in the acceptor-to-donor emission ratio during an illumination period. However, we occasionally noticed false-positive signals (i.e. an increase in donor-to-acceptor ratio over time during illumination) for control cells not expressing any opsin caused by significantly larger photobleaching of the acceptor image compared with the donor image, perhaps due to sensitized photobleaching of the acceptor (39). Therefore, instead of measuring the change in donor-to-acceptor ratio during illumination, we calculated the difference in the acceptor-to-donor ratio before and after the dark period between illuminations (which we termed “extracted voltage measurement”), during which the membrane potential recovers from the depolarized state driven by channelrhodopsins and no photobleaching occurs. This imaging scheme allowed us to detect light-driven depolarization of ChR2 without any false-positive signals from non-opsin–expressing control cells (Fig. 1c). The extracted voltage measurement is defined as follows,
| (Eq. 1) |
where R1 is the average of background-subtracted donor-to-acceptor emission ratio of the last three frames at the end of the first 405 nm light pulse and R2 is the background-subtracted donor-to-acceptor emission ratio at the beginning of the second 405 nm light pulse. Note that this means that any mutants that stay open for longer than 2 s may be missed in this screen design, which focuses on obtaining fast, high-amplitude, novel ion conductance mutants. (Detecting slow mutants could be achieved, in principle, using our screen design and a longer interval or by driving ChRs to a closed state by illuminating with a 560 nm light (40). Between each pair of pulses at a given light power, cells were illuminated with the 590 nm LED at 3.5 milliwatts/mm2, which facilitates ChR2 to reach its dark-adapted closed state (40), for 1 s while the stream of images is saved to disk (which typically takes 5–6 s).
To calculate the extracted voltage measurement from opsin-expressing cells, binary masks were created. Binary masks of ChR-expressing cells were created using the mCherry image of each well using CellProfiler (modules “CorrectIlluminationCalculation” with “Regular” setting and “Fit Polynomial” smoothing, “ApplyThreshold” with “Otsu Global” two-class thresholding, “IdentifyPrimaryObjects” with objects larger than 20 pixels, and “RobustBackground Global” thresholding) (41). CellProfiler generally separates even touching cells into separate masks (in the IdentifyPrimaryObjects function, there is an option called “Method to Distinguish Clumped Objects”). The “Shape” option was used to distinguish clumps.
Using the binary masks, donor and acceptor fluorescence from each cell or set of contiguous cells, masked as described above, was averaged over each cell for each frame, and then the extracted voltage measurement was computed using a custom MATLAB script (MathWorks). For each construct, the extracted voltage measurements from cells across wells were pooled and compared with the similarly pooled measurements from WT ChR2 in the same plate using the K-S test. The mean of the extracted voltage measurement for each library was also computed and compared with that of the WT ChR2.
We used ChR2 L132C (CatCh) (18) as a control for cell health. This was necessary because during experiments to optimize the voltage-sensitive dye imaging protocol, we noticed occasional day-to-day variations in the results. For example, the difference in extracted voltage measurement between WT ChR2 and ChR2 L132C, known to have a larger steady-state component of photocurrent (18), was much less, at 1.1 or less, about 25% of the time, compared with the normal range of 1.4–1.5. We found that the main reason for such difference is caused by positively shifted membrane potential of cells, potentially due to poor cell health. To exclude data under such abnormal conditions, we added ChR2 L132C in the first plate of each day of screening and repeated experiments when the normalized extracted voltage measurement of ChR2 L132C was less than 1.3.
In the residue map stage, each site-saturation mutagenesis library was transfected to 4 wells of a 96-well plate, which resulted in imaging of on average 160 cells per library (159 ± 30, n = 298 libraries), allowing the 20 possible mutants in each library to be imaged roughly eight times. A typical layout of the plate used is shown in Table S2. The difference between WT and mutant was assessed using the K-S test with a cutoff of p < 0.05 (Bonferroni-corrected by multiplying by 298). As noted above, each experiment used both WT ChR2 and ChR2 L132C as controls, and experiments for 9 plates of a total of 36 plates were repeated because the ratio of extracted voltage measurement of ChR2 L132C to WT ChR2 was below 1.3.
Toward the end of the residue map stage, we noticed a strong column effect that was not present before (previously the hits were scattered in all columns). Specifically, two columns on the right-hand side of plate columns 11 and 12 showed large extracted voltage measurement values. These libraries were from residues located near the N and C termini of ChR2 (residues 2–17 and 289–299), and we initially thought these could be affecting trafficking, but we noticed that the voltage dye staining level was much brighter in these wells and found that the medium evaporated faster in these wells in the incubator during culture, potentially due to lower humidity in dry weather in later parts of the screen (done in the winter). Therefore, we did not use these data and repeated these experiments with the columns reversed in each row with the edge of the plates filled with water (similar to the layout shown below) and found that indeed there was a column effect. The problem subsided when we added water around the edge of the plates as shown below. Use of a humidified stage incubator might in principle eliminate this problem as well.
In the target mutation stage to identify single-point mutations from site-saturation mutagenesis libraries, 96 colonies were picked from each library and screened. The layout of the plate used is shown in Table S3. As in the previous experiment, each experiment used both WT and ChR2 L132C as a positive control (extracted voltage measurement of ChR2 L132C to WT ChR2 was below 1.3 as explained above, and then the experiments were repeated; a total of 33 plates of 146 were repeated). In this format, typically 70 cells (73 ± 17) were imaged per mutant construct. The difference between WT and mutant was assessed using the K-S test with a cutoff of p < 0.05 (Bonferroni-corrected by multiplying by 96). This screen yielded 92 point mutant hits from 24 of the 27 libraries. Libraries Phe-100, Asn-187, and Gly-206 did not yield hits.
In the target checking stage, the 92 mutants were screened with a larger number of cells (106 ± 17). Here, the mutants were also recloned into the pN3-mCherry backbone to eliminate any possible mutations in the backbone such as in the promoter. Each mutant was transfected in 4 wells of a 96-well plate and imaged in the same manner as the single-point mutant screen. The plate layout used is shown in Table S4.
As in previous rounds of the screen, WT and ChR2 L132C were used as controls (three of 12 plates repeated). The difference between WT ChR2 and mutant was assessed using the K-S test with a cutoff of p < 0.05 (non-Bonferroni–corrected). This screen yielded 52 point mutants with improved light-driven cell depolarization from 92 mutants screened.
Intracellular calcium and pH imaging
The image acquisition protocols (filter cubes, exposure times, etc.) were largely based on a previous publication that quantified intracellular calcium and pH change due to channelrhodopsin photocurrent using Fura-2 and SNARF 5-AM (20) in addition to our experience in imaging pH change in Arch-expressing cells (42).
For intracellular calcium imaging in HEK293FT cells, 15,000 cells/well were plated in glass-bottom 24-well plates (Greiner Bio-One) and transfected using the same procedure as in the voltage imaging. The cells were washed with Tyrode's solution and stained with 5 mm Fura-2 AM (Invitrogen) in Tyrode's solution at room temperature for 30 min. After dye loading, the cells were incubated in Tyrode's solution for 10 min for recovery. The cells were washed twice and imaged in an extracellular solution composed of 80 mm CaCl2, 20 mm glucose, 23 mm N-methyl-d-glucamine, 5 mm NaCl, 3 mm KCl, 1 mm MgCl2, and 10 mm HEPES, pH 7.3.
Channelrhodopsin expression was detected by mCherry fluorescence using a Sutter DG-4 (Semrock 575/15) and imaged using a dichroic mirror (600 LP Chroma) paired with an emission filter (640/50 Chroma). Fura-2 was excited with a DG-4 using two emission filters (Semrock 340/26 at 0.25 milliwatt/mm2 and 387/11 at 0.40 milliwatt/mm2; light powers at this level did not result in significant activation of WT ChR2) and imaged using a filter cube (Chroma, 495 nm dichroic mirror paired with 500 nm long-pass emission filter) with a CMOS camera (Hamamatsu Orca Flash 2.8) with 250 and 300 ms of exposure time for 340 and 380 nm emission, respectively. Images were taken with a 1-s interval for 4 s to measure baseline calcium levels followed by a 10-s illumination with the DG-4 (Chroma 470/22 at 6.8 milliwatts/mm2) to activate channelrhodopsins. Following channelrhodopsin activation, images were taken every 1 s for 30 s. The ratio of fluorescence at 340/380 was used as an indicator of intracellular calcium concentration. The 340/380 fluorescence ratio was calibrated to a known intracellular calcium concentration using a calcium calibration buffer kit (Invitrogen) and ionomycin (Invitrogen) following the manufacturer's instructions with solutions provided (free calcium concentrations between 0 and 1.35 μm) containing Fura-2 and fitted to the following calibration curve (R2 = 0.986)
| (Eq. 2) |
where R indicates the ratio of 340/380 nm emission, F380 indicates 380 nm emission, and subscripts min and max indicates measurements at [Ca2+]free of 0 and 1.35 μm, respectively.
For intracellular pH imaging, channelrhodopsin constructs were cloned into the pN3-EGFP vector (Clontech) as EGFP fusions to avoid spectral overlap between fluorophore and dye emission. HEK293FT cells transfected in glass-bottom 24-well plates (Nunc) were stained with 5 mm SNARF-5F AM (Invitrogen) at room temperature for 30 min. After loading, the cells were washed with Tyrode's solution and incubated for 10 min for recovery. The cells were washed twice and imaged in an extracellular solution that consisted of 1 mm CaCl2, 20 mm glucose, 145 mm NaCl, 3 mm KCl, 1 mm MgCl2, and 10 mm HEPES, pH 7.35. SNARF-5F and channelrhodopsins were simultaneously excited with the DG-4 (Semrock 500/15 nm at 4 milliwatts/mm2), and then SNARF-5F emission was imaged at 640 nm (Chroma, 600 LP dichroic, 640/50 nm filter; 200-ms exposure) and 610 nm (Chroma, 565 LP dichroic, 610/70 nm filter; 300-ms exposure) for a total illumination period of 1 s. Finally, EGFP expression was imaged (Chroma, 535/30 nm filter; 100-ms exposure using 470/22 nm at 2.5 milliwatts/mm2). The 610/640 nm emission ratio after 1-s illumination was used as the parameter to monitor intracellular pH change. A small amount of residual bleed-through of EGFP fluorescence in the 610 and 640 nm channels was corrected by quantifying images of cytosolic EGFP-expressing cells acquired under the same setting, and plotting the bleed-through of EGFP versus the EGFP emission measured at 535 nm, which yielded a linear curve (R2 = 0.897). The ratio of 610/640 nm emission to intracellular pH was calibrated according to the manufacturer's protocol with solutions, pH 6.5–8, containing 110 mm potassium gluconate, 10 mm NaCl, 25 mm KCl, 1 mm MgCl2, 10 mm HEPES, and 20 mm nigericin (Invitrogen) and fitted to the following calibration curve (R2 = 0.984),
| (Eq. 3) |
where R indicates the ratio of 610/640 nm emission, F640 indicates 640 nm emission, and subscripts A and B indicates measurements at pH 6.5 and 8, respectively.
Primary neuron culture
All procedures involving animals were in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the Massachusetts Institute of Technology Committee on Animal Care. Hippocampal neurons were prepared from postnatal day 0 or 1 Swiss Webster (Taconic) mice as described previously (2, 42) but with the following modifications. Dissected hippocampal tissue was digested with 50 units of papain (Worthington Biochemical) for 8 min, and the digestion was stopped with ovomucoid trypsin inhibitor (Worthington Biochemical). Cells were plated at a density of 20,000–50,000/glass coverslip coated with Matrigel (BD Biosciences). Neurons were seeded in 100 μl of plating medium containing MEM (Life Technologies), glucose (33 mm; Sigma), transferrin (0.01%; Sigma), HEPES (10 mm), Glutagro (2 mm; Corning), insulin (0.13%; Millipore), B27 supplement (2%; Gibco), and heat-inactivated fetal bovine serum (7.5%; Corning). After cell adhesion, additional plating medium was added. Ara-C (0.002 mm; Sigma) was added when glial density reached 50–70%. Neurons were grown at 37 °C in 5% CO2 in a humidified atmosphere.
Primary neuron transfection
Cultured neurons were transfected at 5–6 days in vitro with a commercial calcium phosphate kit (Invitrogen). We added an additional washing step with acidic MEM buffer, pH 6.8–6.9, after calcium phosphate precipitate incubation to completely resuspend residual precipitates (2, 43). pUC DNA (1.2 μg) was used as a cotransfectant DNA reagent with 0.3 μg of channelrhodopsin DNA (AAV-syn-ChR2-tdTomato or AAV-syn-ChromeQ-tdTomato). The viability of cultured neurons transfected with AAV-syn-ChR2-GFP or AAV-syn-ChromeQ-GFP was assessed at 22–25 days after transfection by staining with trypan blue (0.2% solution in PBS; Thermo Fisher) at room temperature for 3 min.
Electrophysiology recording
Whole-cell patch-clamp recordings were performed in isolated HEK293FT cells and neurons. All recordings were performed on an inverted microscope (Leica DMI6000B) or an upright microscope (Olympus BX51WI) using an Axopatch 200B or Multiclamp 700B amplifier and Digidata 1440 or 1550A digitizer (Molecular Devices) at room temperature. To allow isolated cell recording, HEK293FT cells were plated at a density of 15,000/per well in 24-well plates that contained round glass coverslips (0.15 mm thick, 25 mm in diameter, coated with 2% growth factor–reduced Matrigel in DMEM for 1 h at 37 °C). To make accurate measurements, data from cells with access resistance less than 25 megaohms and holding current less than ±50 pA were used. Typical membrane resistance was between 500 megaohms and 2 gigaohms and pipette resistance was between 4 and 10 megaohms.
For comparing (peak and steady-state) photocurrents and channel closing rates, recordings were performed using Tyrode's solution as the extracellular solution and an intracellular solution consisting of 125 or 135 mm potassium gluconate, 8 mm NaCl, 0.1 mm CaCl2, 0.6 mm MgCl2, 1 mm EGTA, 10 mm HEPES, 4 mm MgATP, and 0.4 mm NaGTP, pH 7.3 (KOH-adjusted), 295–300 mosm (sucrose-adjusted). The composition of high-sodium Tyrode's solution was 145 mm NaCl, 5 mm KCl, 1 mm CaCl2,10 mm HEPES, 5 mm glucose, and 2 mm MgCl2, pH 7.4.
For assessing ion selectivity in channelrhodopsins, extracellular and intracellular solutions listed in Table S5 with extracellular osmolarity of 290–300 mosm (sucrose-adjusted) and intracellular osmolarity of 285 mosm (sucrose-adjusted) were used. Data from different extracellular buffers were normalized to the average of photocurrent measured from 145 mm NaCl extracellular buffer (Table S5, second row) in separate cells. We attempted to measure them in the same cells by exchanging the extracellular solutions; however, we found that the order of buffer exchange greatly influenced the result. Therefore, we decided to measure them in separate cells. To calculate the standard deviation of the photocurrent ratios, we used the following formula (adapted from Stuart and Ord 47),
| (Eq. 4) |
where R and S indicate two variables, Var indicates variance of R, Cov indicates covariance, and μ indicates mean.
Whole-cell patch-clamp recordings in primary mouse neurons were made using a Multiclamp 700B amplifier, a Digidata 1550A digitizer, and a PC running pClamp (Molecular Devices). For in vitro current clamp recordings, neurons were patched 21–23 days in vitro (15–17 days post-transfection). Neurons were bathed in Tyrode's solution containing 125 mm NaCl, 2 mm KCl, 3 mm CaCl2, 1 mm MgCl2, 10 mm HEPES, 30 mm glucose, 0.01 mm 1,2,3,4-tetrahydro-6-nitro-2,3-dioxo-benzo[f]quinoxaline-7-sulfonamide (NBQX), and 0.01 mm GABAzine, pH 7.3. Borosilicate glass pipettes (Warner Instruments) with an outer diameter of 1.2 mm and a wall thickness of 0.255 mm were pulled to a resistance of 4–10 megaohms with a P-97 Flaming/Brown micropipette puller (Sutter Instruments) and filled with a solution containing 135 mm potassium gluconate, 8 mm NaCl, 0.1 mm CaCl2, 0.6 mm MgCl2, 1 mm EGTA, 10 mm HEPES, 4 mm MgATP, and 0.4 mm NaGTP, pH 7.3, 290 mosm. To ensure accurate measurements, we used cells with access resistance between 5 and 35 megaohms and holding current within ±100 pA (at −65 mV in voltage-clamp mode) (2).
Photostimulation of patch-clamped cells was conducted with a 470 nm LED (Thorlabs or Lumencor) at 10 milliwatts/mm2 unless otherwise stated. 1-s illumination was delivered for measuring peak and steady-state components of the photocurrent. 2-ms illumination was delivered, and the decay of photocurrent was fitted to a monoexponential curve using Clampfit (Molecular Devices) to determine the channel closing rate (τoff).
For testing repetitive trains of action potentials in cultured neurons, 40 pulses of 2-ms photostimulation (470 nm LED, Lumencor; 4.9 milliwatts/mm2) were delivered at various frequencies using a wave generator (Agilent, 33250A). There was a 1-min wait between photostimulations. To assess excitability, current injection of 700–1200 pA with 5-ms duration was conducted in the same cells.
Spikes were defined as depolarizing above 0 mV and then repolarizing below −30 mV within each stimulation interval. Spike peak time and spike amplitude were measured using macros written in IgorPro (Wavemetrics). Measured peak times were subtracted from photostimulation (light onset) times to calculate spike peak latency.
Fluorescence from patch-clamped cells was quantified by drawing a line around the patched cell manually using ImageJ and calculating the average intensity over the entire area. Background fluorescence was calculated by averaging the intensity in an area within the same image without any cell and subtracted from the cell fluorescence. All statistical comparisons were performed using JMP (SAS Institute Inc.) unless mentioned otherwise.
Author contributions
Y. K. C., N. C. K., and E. S. B. conceptualization; Y. K. C., D. P., A. Y., and A. S. C. data curation; Y. K. C., D. P., A. Y., F. C., and A. S. C. formal analysis; Y. K. C., D. P., A. Y., F. C., A. S. C., N. C. K., and E. S. B. investigation; Y. K. C., A. S. C., N. C. K., and E. S. B. methodology; Y. K. C. and E. S. B. writing-original draft; Y. K. C., A. S. C., N. C. K., and E. S. B. writing-review and editing; E. S. B. supervision; E. S. B. funding acquisition; E. S. B. project administration.
Supplementary Material
This work was supported by the AT&T Foundation, the Benesse Foundation, Jerry and Marge Burnett, Defense Advanced Research Projects Agency (DARPA) Living Foundries Program Grant HR0011-12-C-0068, the Harvard/Massachusetts Institute of Technology (MIT) Joint Grants Program in Basic Neuroscience, the Human Frontiers Science Program, the MIT McGovern Institute and McGovern Institute Neurotechnology (MINT) Program, the MIT Media Lab and Media Lab Consortia, the MIT Neurotechnology Fund (and its generous donors), the New York Stem Cell Foundation-Robertson Investigator Award, National Institutes of Health (NIH) Director's New Innovator Award 1DP2OD002002, NIH EUREKA Award 1R01NS075421, NIH Grant 1R01DA029639, National Science Foundation (NSF) CAREER Award CBET 1053233, SkTech, and the Wallace H. Coulter Foundation (all to E. S. B.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
This article contains supporting Methods and Results, Figs. S1–S9, and Tables S1–S5.
- ChR
- channelrhodopsin
- HEK
- human embryonic kidney
- ANOVA
- analysis of variance
- K-S
- Kolmogorov–Smirnov
- EGFP
- enhanced GFP
- DMEM
- Dulbecco's modified Eagle's medium
- CMOS
- complementary metal-oxide semiconductor
- LED
- light-emitting diode
- MEM
- minimum Eagle's medium
- AAV-syn
- adeno-associated virus–synapsin
- pF
- picofarad
- DiSBAC2(3)
- bis-(1,3-dialkyl-2-thiobarbiturate)-trimethineoxonol
- CC2-DMPE
- chlorocoumarin-2-dimyristoyl phosphatidylethanolamine.
References
- 1. Nagel G., Szellas T., Huhn W., Kateriya S., Adeishvili N., Berthold P., Ollig D., Hegemann P., and Bamberg E. (2003) Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. U.S.A. 100, 13940–13945 10.1073/pnas.1936192100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Klapoetke N. C., Murata Y., Kim S. S., Pulver S. R., Birdsey-Benson A., Cho Y. K., Morimoto T. K., Chuong A. S., Carpenter E. J., Tian Z., Wang J., Xie Y., Yan Z., Zhang Y., Chow B. Y., et al. (2014) Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 10.1038/nmeth.2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beppu K., Sasaki T., Tanaka K. F., Yamanaka A., Fukazawa Y., Shigemoto R., and Matsui K. (2014) Optogenetic countering of glial acidosis suppresses glial glutamate release and ischemic brain damage. Neuron 81, 314–320 10.1016/j.neuron.2013.11.011 [DOI] [PubMed] [Google Scholar]
- 4. Perea G., Yang A., Boyden E. S., and Sur M. (2014) Optogenetic astrocyte activation modulates response selectivity of visual cortex neurons in vivo. Nat. Commun. 5, 3262 10.1038/ncomms4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Figueiredo M., Lane S., Stout R. F. Jr., Liu B., Parpura V., Teschemacher A. G., and Kasparov S. (2014) Comparative analysis of optogenetic actuators in cultured astrocytes. Cell Calcium 56, 208–214 10.1016/j.ceca.2014.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zucker R. S. (1999) Calcium- and activity-dependent synaptic plasticity. Curr. Opin. Neurobiol. 9, 305–313 10.1016/S0959-4388(99)80045-2 [DOI] [PubMed] [Google Scholar]
- 7. Bito H., Deisseroth K., and Tsien R. W. (1996) CREB phosphorylation and dephosphorylation: a Ca2+- and stimulus duration-dependent switch for hippocampal gene expression. Cell 87, 1203–1214 10.1016/S0092-8674(00)81816-4 [DOI] [PubMed] [Google Scholar]
- 8. Bading H., Ginty D. D., and Greenberg M. E. (1993) Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science 260, 181–186 10.1126/science.8097060 [DOI] [PubMed] [Google Scholar]
- 9. Graef I. A., Mermelstein P. G., Stankunas K., Neilson J. R., Deisseroth K., Tsien R. W., and Crabtree G. R. (1999) L-type calcium channels and GSK-3 regulate the activity of NF-ATc4 in hippocampal neurons. Nature 401, 703–708 10.1038/44378 [DOI] [PubMed] [Google Scholar]
- 10. Sheng M., McFadden G., and Greenberg M. E. (1990) Membrane depolarization and calcium induce c-fos transcription via phosphorylation of transcription factor CREB. Neuron 4, 571–582 10.1016/0896-6273(90)90115-V [DOI] [PubMed] [Google Scholar]
- 11. Malenka R. C., and Bear M. F. (2004) LTP and LTD: an embarrassment of riches. Neuron 44, 5–21 10.1016/j.neuron.2004.09.012 [DOI] [PubMed] [Google Scholar]
- 12. Krishtal O. (2003) The ASICs: signaling molecules? Modulators? Trends Neurosci. 26, 477–483 10.1016/S0166-2236(03)00210-8 [DOI] [PubMed] [Google Scholar]
- 13. Sugiyama Y., Wang H., Hikima T., Sato M., Kuroda J., Takahashi T., Ishizuka T., and Yawo H. (2009) Photocurrent attenuation by a single polar-to-nonpolar point mutation of channelrhodopsin-2. Photochem. Photobiol. Sci. 8, 328–336 10.1039/b815762f [DOI] [PubMed] [Google Scholar]
- 14. Ruffert K., Himmel B., Lall D., Bamann C., Bamberg E., Betz H., and Eulenburg V. (2011) Glutamate residue 90 in the predicted transmembrane domain 2 is crucial for cation flux through channelrhodopsin 2. Biochem. Biophys. Res. Commun. 410, 737–743 10.1016/j.bbrc.2011.06.024 [DOI] [PubMed] [Google Scholar]
- 15. Kato H. E., Zhang F., Yizhar O., Ramakrishnan C., Nishizawa T., Hirata K., Ito J., Aita Y., Tsukazaki T., Hayashi S., Hegemann P., Maturana A. D., Ishitani R., Deisseroth K., and Nureki O. (2012) Crystal structure of the channelrhodopsin light-gated cation channel. Nature 482, 369–374 10.1038/nature10870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. González J. E., and Maher M. P. (2002) Cellular fluorescent indicators and voltage/ion probe reader (VIPR) tools for ion channel and receptor drug discovery. Receptors Channels 8, 283–295 [PubMed] [Google Scholar]
- 17. González J. E., and Tsien R. Y. (1995) Voltage sensing by fluorescence resonance energy transfer in single cells. Biophys. J. 69, 1272–1280 10.1016/S0006-3495(95)80029-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kleinlogel S., Feldbauer K., Dempski R. E., Fotis H., Wood P. G., Bamann C., and Bamberg E. (2011) Ultra light-sensitive and fast neuronal activation with the Ca2+-permeable channelrhodopsin CatCh. Nat. Neurosci. 14, 513–518 10.1038/nn.2776 [DOI] [PubMed] [Google Scholar]
- 19. Nagel G., Brauner M., Liewald J. F., Adeishvili N., Bamberg E., and Gottschalk A. (2005) Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr. Biol. 15, 2279–2284 10.1016/j.cub.2005.11.032 [DOI] [PubMed] [Google Scholar]
- 20. Lin J. Y., Lin M. Z., Steinbach P., and Tsien R. Y. (2009) Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys. J. 96, 1803–1814 10.1016/j.bpj.2008.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Plazzo A. P., De Franceschi N., Da Broi F., Zonta F., Sanasi M. F., Filippini F., and Mongillo M. (2012) Bioinformatic and mutational analysis of channelrhodopsin-2 protein cation-conducting pathway. J. Biol. Chem. 287, 4818–4825 10.1074/jbc.M111.326207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Prigge M., Schneider F., Tsunoda S. P., Shilyansky C., Wietek J., Deisseroth K., and Hegemann P. (2012) Color-tuned channelrhodopsins for multiwavelength optogenetics. J. Biol. Chem. 287, 31804–31812 10.1074/jbc.M112.391185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Berndt A., Prigge M., Gradmann D., and Hegemann P. (2010) Two open states with progressive proton selectivities in the branched channelrhodopsin-2 photocycle. Biophys. J. 98, 753–761 10.1016/j.bpj.2009.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nikolic K., Grossman N., Grubb M. S., Burrone J., Toumazou C., and Degenaar P. (2009) Photocycles of channelrhodopsin-2. Photochem. Photobiol. 85, 400–411 10.1111/j.1751-1097.2008.00460.x [DOI] [PubMed] [Google Scholar]
- 25. Gradmann D., Ehlenbeck S., and Hegemann P. (2002) Modeling light-induced currents in the eye of Chlamydomonas reinhardtii. J. Membr. Biol. 189, 93–104 10.1007/s00232-002-1006-8 [DOI] [PubMed] [Google Scholar]
- 26. Schneider F., Gradmann D., and Hegemann P. (2013) Ion selectivity and competition in channelrhodopsins. Biophys. J. 105, 91–100 10.1016/j.bpj.2013.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eisenhauer K., Kuhne J., Ritter E., Berndt A., Wolf S., Freier E., Bartl F., Hegemann P., and Gerwert K. (2012) In channelrhodopsin-2 Glu-90 is crucial for ion selectivity and is deprotonated during the photocycle. J. Biol. Chem. 287, 6904–6911 10.1074/jbc.M111.327700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gradmann D., Berndt A., Schneider F., and Hegemann P. (2011) Rectification of the channelrhodopsin early conductance. Biophys. J. 101, 1057–1068 10.1016/j.bpj.2011.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Richards R., and Dempski R. E. (2017) Adjacent channelrhodopsin-2 residues within transmembranes 2 and 7 regulate cation selectivity and distribution of the two open states. J. Biol. Chem. 292, 7314–7326 10.1074/jbc.M116.770321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Berndt A., Lee S. Y., Ramakrishnan C., and Deisseroth K. (2014) Structure-guided transformation of channelrhodopsin into a light-activated chloride channel. Science 344, 420–424 10.1126/science.1252367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wietek J., Wiegert J. S., Adeishvili N., Schneider F., Watanabe H., Tsunoda S. P., Vogt A., Elstner M., Oertner T. G., and Hegemann P. (2014) Conversion of channelrhodopsin into a light-gated chloride channel. Science 344, 409–412 10.1126/science.1249375 [DOI] [PubMed] [Google Scholar]
- 32. Berndt A., Lee S. Y., Wietek J., Ramakrishnan C., Steinberg E. E., Rashid A. J., Kim H., Park S., Santoro A., Frankland P. W., Iyer S. M., Pak S., Ährlund-Richter S., Delp S. L., Malenka R. C., et al. (2016) Structural foundations of optogenetics: determinants of channelrhodopsin ion selectivity. Proc. Natl. Acad. Sci. U.S.A. 113, 822–829 10.1073/pnas.1523341113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wietek J., Broser M., Krause B. S., and Hegemann P. (2016) Identification of a natural green light absorbing chloride conducting channelrhodopsin from Proteomonas sulcata. J. Biol. Chem. 291, 4121–4127 10.1074/jbc.M115.699637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Govorunova E. G., Sineshchekov O. A., and Spudich J. L. (2016) Proteomonas sulcata ACR1: a fast anion channelrhodopsin. Photochem. Photobiol. 92, 257–263 10.1111/php.12558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Govorunova E. G., Sineshchekov O. A., Rodarte E. M., Janz R., Morelle O., Melkonian M., Wong G. K., and Spudich J. L. (2017) The expanding family of natural anion channelrhodopsins reveals large variations in kinetics, conductance, and spectral sensitivity. Sci. Rep. 7, 43358 10.1038/srep43358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang F., Prigge M., Beyrière F., Tsunoda S. P., Mattis J., Yizhar O., Hegemann P., and Deisseroth K. (2008) Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri. Nat. Neurosci. 11, 631–633 10.1038/nn.2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cwirla S. E., Peters E. A., Barrett R. W., and Dower W. J. (1990) Peptides on phage: a vast library of peptides for identifying ligands. Proc. Natl. Acad. Sci. U.S.A. 87, 6378–6382 10.1073/pnas.87.16.6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Agarwal N., and Shusta E. V. (2009) Multiplex expression cloning of blood-brain barrier membrane proteins. Proteomics 9, 1099–1108 10.1002/pmic.200800368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zal T., and Gascoigne N. R. (2004) Photobleaching-corrected FRET efficiency imaging of live cells. Biophys. J. 86, 3923–3939 10.1529/biophysj.103.022087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stehfest K., Ritter E., Berndt A., Bartl F., and Hegemann P. (2010) The branched photocycle of the slow-cycling channelrhodopsin-2 mutant C128T. J. Mol. Biol. 398, 690–702 10.1016/j.jmb.2010.03.031 [DOI] [PubMed] [Google Scholar]
- 41. Carpenter A. E., Jones T. R., Lamprecht M. R., Clarke C., Kang I. H., Friman O., Guertin D. A., Chang J. H., Lindquist R. A., Moffat J., Golland P., and Sabatini D. M. (2006) CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chow B. Y., Han X., Dobry A. S., Qian X., Chuong A. S., Li M., Henninger M. A., Belfort G. M., Lin Y., Monahan P. E., and Boyden E. S. (2010) High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature 463, 98–102 10.1038/nature08652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang S., and Cho Y. K. (2016) An optimized calcium-phosphate transfection method for characterizing genetically encoded tools in primary neurons. Methods Mol. Biol. 1408, 243–249 10.1007/978-1-4939-3512-3_16 [DOI] [PubMed] [Google Scholar]
- 44. Volkov O., Kovalev K., Polovinkin V., Borshchevskiy V., Bamann C., Astashkin R., Marin E., Popov A., Balandin T., Willbold D., Büldt G., Bamberg E., and Gordeliy V. (2017) Structural insights into ion conduction by channelrhodopsin 2. Science 358, eaan8862 10.1126/science.aan8862 [DOI] [PubMed] [Google Scholar]
- 45. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ho B. K., and Gruswitz F. (2008) HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct. Biol. 8, 49 10.1186/1472-6807-8-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stuart A., and Ord K. (2010) Kendall's Advanced Theory of Statistics: Distribution Theory, Vol. 1, 6th Ed., Wiley, Hoboken, NJ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.