Abstract
Craniofrontonasal syndrome (CFNS) is an X-linked disorder caused by EFNB1 mutations in which females are more severely affected than males. Severe male phenotypes are associated with mosaicism, supporting cellular interference for sex bias in this disease. Although many variants have been found in the coding region of EFNB1, only 2 pathogenic variants have been identified in the same nucleotide in 5′UTR, disrupting the stop codon of an upstream open reading frame (uORF). uORFs are known to be part of a wide range of post-transcriptional regulation processes, and just recently, their association with human diseases has come to light. In the present study, we analyzed EFNB1 in a female patient with typical features of CFNS. We identified a variant, located at c.-411, creating a new upstream ATG (uATG) in the 5′UTR of EFNB1, which is predicted to alter an existing uORF. Dual-luciferase reporter assays showed significant reduction in protein translation, but no difference in the mRNA levels. Our study demonstrates, for the first time, the regulatory impact of uATG formation on EFNB1 levels and suggests that this should be the target region in molecular diagnosis of CFNS cases without pathogenic variants in the coding and splice sites regions of EFNB1.
Key Words: Genetic counseling, Molecular diagnosis and testing, Protein translation, Regulatory variant, Upstream ORF and ATG
Craniofrontonasal syndrome (CFNS; OMIM 304110) is a rare X-linked dominant disorder caused by loss-of-function mutations in EFNB1 [Twigg et al., 2004; Wieland et al., 2004]. Greater severity is observed in heterozygous female patients compared with hemizygous male individuals. Female patients can present with severe hypertelorism, a central nasal groove, craniofacial asymmetry, and coronal craniosynostosis associated with extracranial features such as sloping shoulders, mild cutaneous syndactyly, grooved nails, duplication of the first digit, and wiry hair. On the other hand, hemizygous males present only with hypertelorism and occasional cleft lip [Twigg et al., 2004, 2013; van den Elzen et al., 2014]. However, severe phenotypes have been observed in male patients with mosaic EFNB1 mutations [Twigg et al., 2013], a finding that supports the hypothesis that cellular interference underlies clinical severity [Wieacker and Wieland, 2005; Twigg et al., 2013; Niethamer et al., 2017].
More than a hundred EFNB1 mutations have been described in association with CFNS. Almost all pathogenic variants are in the coding region, with the majority (>95%) leading to premature termination codons [Twigg et al., 2004, 2006, 2013; Wieland et al., 2004, 2005, 2007; Shotelersuk et al., 2006; Torii et al., 2007; Wallis et al., 2008; Hogue et al., 2010; Makarov et al., 2010; Grasso et al., 2011; Apostolopoulou et al., 2012; Cui et al., 2012; Ramirez-Garcia et al., 2013; Seven et al., 2013]. Only 2 mutations affecting the same nucleotide have been reported outside the coding and splice site sequences of EFNB1: c.-95T>G and c.-95T>C. These variants are present in the 5′UTR and abolish the stop codon of a small upstream open reading frame (uORF), leading to an overlapped and out-of-frame ORF with the main downstream ORF (dORF, corresponding to EFNB1), with reduced translation of EFNB1 as a consequence [Twigg et al., 2013].
uORFs regulate gene expression through multiple functional mechanisms, e.g., in a peptide-dependent manner, by consumption of functional pre-initiation complex, or triggering nonsense-mediated decay [Rahmani et al., 2009; Barbosa et al., 2013; Hurt et al., 2013; Ebina et al., 2015]. These regulatory sequences are mostly linked to repression of translation of the main dORF, although under specific conditions, uORFs can confer induction of protein translation [Wethmar, 2014]. uORF-altering variants have been described in association with the etiology of human disorders [Barbosa et al., 2013]. Such variants can introduce new uORFs, as found in gonadal dysgenesis and van der Woude syndrome [Poulat et al., 1998; Kondo et al., 2002; Calvo et al., 2009], as well as disrupt uORFs in diseases such as Marie Unna hereditary hypotrichosis [Wen et al., 2009] and thrombocythemia [Kondo et al., 1998].
In this study, we demonstrate the regulatory impact of a variant-created upstream ATG (uATG) on mRNA translation of EFNB1. Identification of such variants contributes to the expanding field of translation regulation and to the definition of critical noncoding sequences that can be targeted in genetic diagnosis.
Case Report
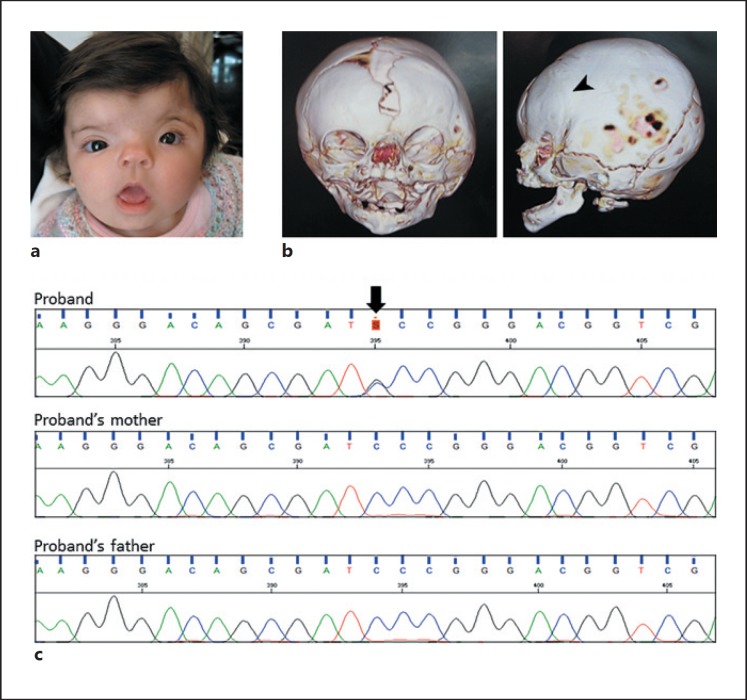
The female patient (referred to as F5162-1), first evaluated 10 days after birth, presented with typical features of CFNS: unilateral coronal craniosynostosis (left), hypertelorism, asymmetric craniofacial contour, widows peak, arched eyebrows, hypoplastic supraorbital ridges, telecanthus, downslanting palpebral fissures, short nose with depressed nasal bridge, bifid nasal tip, deep philtrum, tented upper lip, widened metopic suture, and a frontal bone defect (Fig. 1a,b). A fronto-orbital advancement surgery was performed at the age of 5 months. No clinical manifestations were observed in her parents, and there is no additional family history.
Fig. 1.
a Frontal view of the patient showing facial asymmetry, hypertelorism, telecanthus, downslanting palpebral fissures, short nose with bifid nasal tip, and tented mouth. b Cranial CT scans showing unilateral coronal craniosynostosis (arrowhead). c Dideoxy sequencing of the 5′UTR of EFNB1 in the patient and her parents. A de novo c.-411C>G (chrX:g.68049209C>G) variant was detected (arrow).
Materials and Methods
DNA Samples
Genomic DNA from the patient and her clinically unaffected parents was extracted from peripheral blood using Gentra Systems Autopure LS (AutoGen), according to the manufacturer's instructions.
DNA Sequencing
Standard genetic screening of the coding regions of EFNB1 was carried out with the patient's DNA using Sanger sequencing. Primers were designed with Primer3 [Untergasser et al., 2012] (primer sequences available upon request). Sequencing reactions were performed using the BigDye Terminator v3.1 Cycle Sequencing Kit, and the amplicons were sequenced using an ABI 3730 DNA Analyzer (Thermo Fisher Scientific). Data were analyzed using Sequencher v.5.1 software.
A more comprehensive analysis of the patient's DNA was performed by targeted next-generation sequencing. Specific probes were used to capture 5′ and 3′UTR sequences and all coding regions of EFNB1 (RefSeq NM_004429) as well as splice acceptor and donor sites (Nextera - custom target enrichment, Illumina), using MiSeq (Illumina) according to the manufacturer's protocols. Data were aligned to the human reference sequence (hg19) with the Burrows-Wheeler Aligner (BWA) [Li and Durbin, 2009], and PCR duplicates were removed with the Picard toolkit. Variants were called by the Unified Genotyper tool from Genome Analysis Toolkit [McKenna et al., 2010] and annotated with ANNOVAR [Wang et al., 2010]. Sanger sequencing was used to analyze the candidate variant in parental DNA (primer sequences available upon request).
Variant Analysis
Variants present in the 1000 Genomes database, dbSNP150, Genome Aggregation database (gnomAD), or in the Online Archive of Brazilian Mutations (ABraOM) [Naslavsky et al., 2017] were excluded from downstream analysis as unlikely to be pathogenic. Evolutionary sequence conservation was assessed using the Vertebrate Multiz Alignment & Conservation track (100 vertebrate species) and Genomic Evolutionary Rate Profiling (GERP) (35 species of mammals), both through the UCSC Genome Browser. For multiple vertebrate sequence alignment, 20 EFNB1 orthologues were extracted from the Ensembl genome browser along with the human gene. All sequences comprised the first coding exon plus the 5′UTR (when it was found annotated) or ~700 bp of the upstream sequence from the main ATG start codon. The sequences were aligned using MAFFT [Katoh et al., 2017] through the AliView v.1.18.1 program [Larsson, 2014]; consensus sequence for the alignment was obtained using Jalview v.2.10.2b2 [Waterhouse et al., 2009]. Open Reading Frame Finder (ORFfinder, NCBI) was utilized to search for uORFs in the sequences. NetStart 1.0 (http://www.cbs.dtu.dk/services/NetStart/) was taken to predict and score translation initiation sites.
Microsatellite Analysis
Microsatellite genotyping was conducted using fluorescently labeled PCR primer pairs from the Linkage Mapping Set v2.5, according to the manufacturer's protocol. PCR fragments were read in an ABI 3730 DNA Analyzer (Thermo Fisher Scientific) and analyzed with GeneMapper v5.0.
Plasmids
Site-directed mutagenesis was carried out to introduce specific nucleotide substitutions within the 5′UTR of EFNB1, using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies), following the manufacturer's instructions. The wild type (WT) 5′UTR of EFNB1 was inserted into a previously modified psiCHECK-2 vector [Calvo et al., 2009; referred to as WT construct] in which the ATG of Renilla was modified to TTG, so that Renilla expression was driven from the primary EFNB1 ATG codon. We used this vector as a template to generate a construct containing the c.-411C>G variant (referred to as F5162 construct) and a second construct containing c.-227G>T and c.-225G>A in addition to c.-411C>G (referred to as F5162SHORT; further explanation below). Sanger sequencing was used to confirm the incorporated modifications (primer sequences available upon request).
For comparative analysis in the luciferase assays, a previously described construct taken to investigate the c.-95T>G variant [Twigg et al., 2013] was also used (referred to as 1330).
Luciferase Assay
Human embryonic kidney 293T (HEK293T) cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin (all provided by Life Technologies). Cells were seeded at 6 × 103 cells/well in 96-well optical-bottom plates (Nunc™, Thermo Scientific) 24 h prior to transfection. Transient transfections were performed using 100 ng of the vector constructs, in triplicate, with TurboFectin 8.0 (OriGene), according to the manufacturer's instructions. Forty-eight hours after DNA transfection, Renilla activity was measured with the Dual-Glo Luciferase Assay System (Promega), in a GloMax-Multi Detection System (Promega). All constructs also contained the Firefly luciferase gene, used as a transfection control. Renilla luminescence results were normalized by Firefly luciferase. Statistical analysis was performed by one-way ANOVA with a post-hoc Bonferroni multiple comparison test using the GraphPad Prism 6.0. Data were presented relative to the WT construct as mean ± SEM.
Real-Time Quantitative PCR
HEK293T cells were seeded at 105 cells per well in 6-well culture plates 24 h prior to transfection. Cells were transfected with 1 μg of vector construct, in duplicate, with TurboFectin 8.0. Forty-eight hours after transfection, cells were washed with PBS, and total RNA was obtained using the NucleoSpin RNA II extraction kit (Macherey-Nagel), following the manufacturer's recommendations. cDNA synthesis was performed with the SuperScript IV First-Strand Synthesis System (Life Technologies) and oligo-dT primers using 1.5 μg of total RNA from each cell sample as starting material. RT-qPCR reactions were performed with 2X Fast SYBR Green PCR Master Mix (Applied Biosystems) and 200 nM of each primer. Primer pairs for Renilla luciferase 5′-TCCATGCTGAGAGTGTCGTG-3′ (forward) and 5′-CAAGCACCATTTTCTCGCCC-3′ (reverse) and Firefly luciferase 5′-TCTGGCGACATTGCCTACTG-3′ (forward) and 5′-CGGCGTCGAAAATGTTAGGG-3′ (reverse) were designed with the Primer-BLAST tool (NCBI) [Ye et al., 2012], and their amplification efficiencies (E) were determined by serial cDNA dilutions; primers were supplied by Exxtend. Fluorescence was detected using the QuantStudio 5 System (Applied Biosystems) under standard temperature protocol. The relative mRNA expression values were determined by dividing E-ΔCt of the target gene (Renilla luciferase) by E-ΔCt of the endogenous control (Firefly luciferase) [Pfaffl, 2001]. Statistical significance was assessed by one-way ANOVA with a post-hoc Bonferroni multiple comparison test using the GraphPad Prism 6.0. Data were presented relative to the WT construct as mean ± SEM.
Results
Detection of a uAUG Codon-Creating EFNB1 Variant
As there was a clinical diagnosis of CFNS, the patient was referred to our center for analysis of EFNB1. Sanger sequencing of the coding regions did not identify potential causative variants in EFNB1, and next-generation sequencing also confirmed no potential pathogenic variants in the coding regions or in splice sites. However, we identified a variant located within the 5′UTR of EFNB1, NM_004429.4:c.-411C>G. This variant (chrX:68049209C>G) was not inherited from the patient's parents (Fig. 1c), and the analysis of microsatellite markers was consistent with the correct biological relationships (correct segregation of 15/15 markers). The identified variant was novel and absent from 1000 Genomes, dbSNP150, ABraOM, and gnomAD.
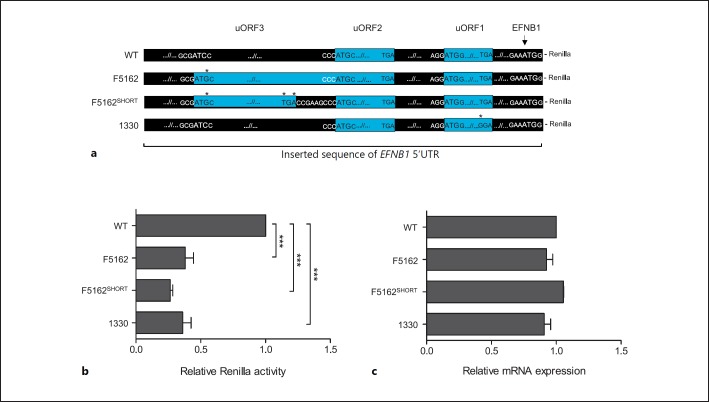
The WT 5′UTR sequence of EFNB1 contains 2 uORFs on the positive strand and downstream of the c.-411C>G variant: the first one is the functional 4 codon uORF (uORF1), studied by Twigg et al. [2013]; the second is a 10 codon uORF (uORF2) that was predicted by ORFfinder in silico analysis. Substitution of C by G at position c.-411 was predicted to introduce a uAUG start codon, which is in-frame with the uUGA stop codon of uORF2, establishing the creation of a new 76 codon uORF (hereafter referred to as uORF3) overlapping with uORF2 and sharing the same uUGA stop codon. All uORFs1-3 are out-of-frame with the AUG of the main dORF (Fig. 2a).
Fig. 2.
Functional analysis of the uORF3-creating variant. a Constructs generated to investigate the effect of uORFs on translation from the main dORF. WT, wild type 5′UTR sequence of EFNB1 containing the in silico predicted uORF2 and the functional validated uORF1 [Twigg et al., 2013]. F5162, the mutant allele found in the patient containing the new c.-411C>G uAUG codon; note the new uORF3 shares the same stop codon with uORF2 (blue bars). F5162SHORT, c.-411C>G variant along with an earlier stop codon in uORF3 which separates it from uORF2. 1330, c.-95T>G construct, found in a mosaic CFNS male patient [Twigg et al., 2013]; this variant disrupts the stop codon of uORF1 that is out-of-frame with and overlaps the main dORF. Asterisks indicate nucleotide changes, and blue bars represent uORFs. The AUG sequence context is shown for each uORF/ORF up to base −3 and +4. b Graph depicting the relative Renilla luciferase activity ([experimental sample ratio - negative control ratio] / [positive control ratio - negative control ratio]) for each construct. c RT-qPCR analysis of relative Renilla luciferase mRNA levels in transfected HEK293T cells. All values are relative to wild type as mean ± SEM of 3 experiments which were performed in triplicate. One-way ANOVA with a post-hoc Bonferroni multiple comparison test was done. * p < 0.05, **p < 0.01, ***p < 0.001.
Additionally, we investigated whether reported variants in gnomAD would alter, create, and/or delete some uORFs in the 5′UTR of EFNB1. Forty-seven point variants (allele frequency <0.09) were found. None of them were predicted to modify uORFs in the sequence, according to predefined parameters of the ORFfinder.
As the sequence around uAUGs has been associated with the likelihood of translation initiation, we evaluated the AUG context in uORFs1-3 and the main dORF. In the Kozak consensus, the most critical bases are a purine (A or G) at position −3 and a G at position +4 (the A of AUG, is designated as +1) [Kozak, 1987, 1989, 2002]. The uORF3 sequence has a match with the Kozak sequence at purine −3. The uORF2 sequence context lacks a purine at position −3 and the G base at position +4, while uORF1 and the main dORF do have a match at position −3 and +4 of the Kozak sequence. The AUGs of uORF1-3 and the main dORF were scored as probable translation sites (NetStart score > 0.5) (online suppl. Fig. 1; for suppl. material see www.karger.com/doi/10.1159/000490635).
The c.-411C site was found to be evolutionary conserved (GERP score = 2.48) and no uATG was identified in any species at this position. Analysis of EFNB1 orthologues in 20 vertebrates revealed a high degree of identity, most strongly among mammalian species. No uORFs were observed near the human c.-411C, with the exception of 24 and 16 codon uORFs found in zebra finch and zebrafish, respectively. The region comprising uORF2 shows a high degree of sequence homology in several species of mammals, while zebra finch and zebrafish also have a uORF just a few bases downstream the start codon of human uORF2. The multispecies alignment comprising uORF1-3 and the main ORF show more than 1 uORF in all species (online suppl. Fig. 1). Collectively, these observations suggest that the introduction of a uATG, by the c.-411C>G variant, could potentially interfere with EFNB1 translation.
c.-411C>G Reduces Translation of the Main EFNB1 ORF
We performed dual luciferase reporter assays to functionally evaluate the regulatory importance of the 5′UTR and to test the effect of c.-411C>G on protein expression from the main dORF, which could explain the observed CFNS phenotype. In the presence of c.-411C>G, translation of the main dORF was reduced by 62% when compared to WT (F5162 construct compared to WT; p < 0.001). Next, we investigated if the variant-created uORF3 would have an independent effect from uORF2 in the translation of the main uORF. To test this hypothesis, we built a construct with a premature stop codon, located 13 codons upstream of the UGA of uORF2 (F5162SHORT), thus containing 3 separate uORFS. We found a slightly stronger reduction in the main dORF translation in F5162SHORT compared to F5162 (the main dORF translation was about 11% less expressed when compared to F5162) (Fig. 2a,b).
To determine if the c.-411C>G variant in our patient has similar regulatory effects to c.-95T>G, previously described in CFNS by Twigg et al. [2013], we compared luciferase activity using the c.-95G construct. Both c.-411C>G (F5162) and c.-95T>G (1330) variants significantly reduced the main dORF translation on average 63% compared to WT.
As an RNA sample was not available from the patient to evaluate if the reduced protein levels could be a consequence of impaired EFNB1 transcription, we transfected HEK293T cells with each construct (F5162, F5162SHORT, 1330, and WT) and analyzed the Renilla luciferase RNA expression. No significant difference was observed between any of the constructs relative to WT (Fig. 2c), suggesting that the effect of the c.-411C>G variant is most likely at the level of translation.
Discussion
There are an increasing number of examples of diseases in which the underlying mechanisms are caused by variants at 5′UTR [Calvo et al., 2009; Barbosa et al., 2013]. However, pathogenicity interpretation of such variants is usually difficult, and detailed functional analyses are required to provide evidence of causality. Here, we add an additional, novel, and de novo c.-411C>G variant in EFNB1, to the still scarce list of pathogenic 5′UTR variants causative of CFNS.
The c.-411C>G change is a de novo variant located in a highly evolutionary conserved region, providing evidence that it may be functional. In silico analysis predicted the creation of a new uAUG, leading to a new uORF (uORF3) or representing a 5′ elongation of a presumed uORF2. Favoring this hypothesis, we observed that this new uAUG of uORF3 has a good Kozak consensus sequence that scored as a probable translation site. The potential functional effect of this variant was also supported by the observation that none of the 47 single nucleotide variants, annotated at the 5′UTR in gnomAD, seemed to disrupt, create, or alter uORFs. Therefore, all the in silico analysis supported a functional effect for the c.-411C>G variant, identified in a region usually not sequenced in CFNS patients.
Addressing this functional hypothesis, through in vitro assays of sequences containing uORF3, our findings suggest that this variant leads to translation dysregulation. Both the F5162 and F5162SHORT assays provided evidence that the c.-411C>G variant decreases protein translation of the main dORF. The F5162SHORT analysis showed a slightly greater reduction in protein expression of the main dORF (compared to F5162 construct), in agreement with the literature for an additional number of uORFs [Iacono et al., 2005; Calvo et al., 2009; Chew et al., 2016; Johnstone et al., 2016]. Furthermore, the c.-411C>G uORF3-creating variant and c.-95T>G uORF1-disrupting variant [Twigg et al., 2013] showed highly similar effects on the protein levels of the main dORF, despite their different positions and sizes. Taken together, these results strongly suggest that the c.-411C>G variant has a functional effect on translation. Although one of the mechanisms thought to be triggered by uORFs is the nonsense-mediated decay [Barbosa et al., 2013], this is not supported by our in vitro mRNA analysis.
To date, at least 24 diseases have been described to be caused by uORF-altering variants in the 5′UTR of different genes. It is interesting to note that the c.-411C>G variant is one of the most distantly located uORF variants compared with other pathogenic uORF-altering mutations in different disorders [Barbosa et al., 2013; Hornig et al., 2016]. Therefore, in CFNS patients negative for pathogenic variants in coding and splice sites of EFNB1, a careful analysis of the entire 5′UTR should be carried out as causative variants in the entire region could result in decreased protein levels and disease.
About 49% of human transcripts contain uORFs that are most likely to be physiologically relevant, with their functional importance supported by conservation in vertebrate evolution. The mean length of uORFs across vertebrates was found to be <60 nucleotides [Johnstone et al., 2016]. The novel 228 nucleotide uORF3 described in this work is much longer than the majority of uORFs present within the EFNB1 5′UTR of the evaluated species, except for 1 predicted uORF found in the mammal hyrax (>680 bp; data not shown in the alignment), in agreement with depletion of long ORFs in vertebrate 5′UTRs [Johnstone et al., 2016].
This study contributes an additional CFNS case with a regulatory variant in the EFNB1 5′UTR sequence and supports screening of this noncoding region in CFNS patients where other variants or gene copy number changes are absent. The 5′UTRs of disease-related genes are not routinely screened, raising the possibility that similar pathogenic variants contribute more widely to disease. In light of this, it is interesting to note that uORFs can be targeted by antisense oligonucleotides to reestablish normal levels of the main protein [Liang et al., 2016]. In this context, a CRISPR-Cas9 approach may be useful to generate different mutations in a disease-relevant cell type, such as iPSC-derived neuroepithelial cells, to provide further insights into pathophysiology of the disease along with investigation of antisense oligonucleotide strategies. In summary, this report adds to the understanding of the functional effects of uAUG/uORFs on protein expression and pathophysiological mechanisms in CFNS.
Statement of Ethics
Ethics approval for this study was provided by the Ethics Committee of the Instituto de Biociências at Universidade de São Paulo, São Paulo, SP, Brazil (Protocol 024/2004). Patients donated biological samples after providing signed informed consent.
Disclosure Statement
The authors declare no conflicts of interest.
Acknowledgments
The authors would like to thank Monica Varela, Suzana Ezquina, Melina Rodrigues, and Francine Favaro for technical support. We are also grateful to Carlos Menck and Diogo Meireles for helpful discussions. M.R.P.-B. is funded by CEPID/FAPESP (2013/08028-1) and CNPq (PQ10/2011). S.R.F.T. is funded by Action Medical Research (GN2483).
References
- Apostolopoulou D, Stratoudakis A, Hatzaki A, Kaxira OS, Panagopoulos KP, et al. A novel de novo mutation within EFNB1 gene in a young girl with craniofrontonasal syndrome. Cleft Palate Craniofac J. 2012;49:109–113. doi: 10.1597/10-247. [DOI] [PubMed] [Google Scholar]
- Barbosa C, Peixeiro I, Romão L. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet. 2013;9:e1003529. doi: 10.1371/journal.pgen.1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci USA. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew GL, Pauli A, Schier AF. Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish. Nat Commun. 2016;7:11663. doi: 10.1038/ncomms11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Zhao H, Liu Z, Liu C, Luan J, et al. A systematic review of genetic skeletal disorders reported in Chinese biomedical journals between 1978 and 2012. Orphanet J Rare Dis. 2012;7:55. doi: 10.1186/1750-1172-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina I, Takemoto-Tsutsumi M, Watanabe S, Koyama H, Endo Y, et al. Identification of novel Arabidopsis thaliana upstream open reading frames that control expression of the main coding sequences in a peptide sequence-dependent manner. Nucleic Acids Res. 2015;43:1562–1576. doi: 10.1093/nar/gkv018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso C, Licata F, Rossi C, Balistreri M, Sorge G. Craniofrontonasal syndrome: genetic aspects and description of a clinical case (in Italian) Minerva Pediatr. 2011;63:431–438. [PubMed] [Google Scholar]
- Hogue J, Shankar S, Perry H, Patel R, Vargervik K, Slavotinek A. A novel EFNB1 mutation (c.712delG) in a family with craniofrontonasal syndrome and diaphragmatic hernia. Am J Med Genet A. 2010;152A:2574–2577. doi: 10.1002/ajmg.a.33596. [DOI] [PubMed] [Google Scholar]
- Hornig NC, de Beaufort C, Denzer F, Cools M, Wabitsch M, et al. A recurrent germline mutation in the 5′UTR of the androgen receptor causes complete androgen insensitivity by activating aberrant uORF translation. Plos One. 2016;11:e0154158. doi: 10.1371/journal.pone.0154158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt JA, Robertson AD, Burge CB. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res. 2013;23:1636–1650. doi: 10.1101/gr.157354.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono M, Mignone F, Pesole G. uAUG and uORFs in human and rodent 5′untranslated mRNAs. Gene. 2005;349:97–105. doi: 10.1016/j.gene.2004.11.041. [DOI] [PubMed] [Google Scholar]
- Johnstone TG, Bazzini AA, Giraldez AJ. Upstream ORFs are prevalent translational repressors in vertebrates. EMBO J. 2016;35:706–723. doi: 10.15252/embj.201592759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform, E-pub ahead of print. 2017 doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, et al. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet. 2002;32:285–289. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Okabe M, Sanada M, Kurosawa M, Suzuki S, et al. Familial essential thrombocythemia associated with one-base deletion in the 5′-untranslated region of the thrombopoietin gene. Blood. 1998;92:1091–1096. [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 2014;30:3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Shen W, Sun H, Migawa MT, Vickers TA, et al. Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames. Nat Biotechnol. 2016;34:875–880. doi: 10.1038/nbt.3589. [DOI] [PubMed] [Google Scholar]
- Makarov R, Steiner B, Gucev Z, Tasic V, Wieacker P, Wieland I. The impact of CFNS-causing EFNB1 mutations on ephrin-B1 function. BMC Med Genet. 2010;11:98. doi: 10.1186/1471-2350-11-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naslavsky MS, Yamamoto GL, de Almeida TF, Ezquina SAM, Sunaga DY, et al. Exomic variants of an elderly cohort of Brazilians in the ABraOM database. Hum Mutat. 2017;38:751–763. doi: 10.1002/humu.23220. [DOI] [PubMed] [Google Scholar]
- Niethamer TK, Larson AR, O';Neill AK, Bershteyn M, Hsiao EC, et al. EPHRIN-B1 mosaicism drives cell segregation in craniofrontonasal syndrome hiPSC-derived neuroepithelial cells. Stem Cell Reports. 2017;8:529–537. doi: 10.1016/j.stemcr.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulat F, Desclozeaux M, Tuffery S, Jay P, Boizet B, Berta P. Mutation in the 5′ noncoding region of the SRY gene in an XY sex-reversed patient. Hum Mutat. 1998;(Suppl 1):S192–S194. doi: 10.1002/humu.1380110162. [DOI] [PubMed] [Google Scholar]
- Rahmani F, Hummel M, Schuurmans J, Wiese-Klinkenberg A, Smeekens S, Hanson J. Sucrose control of translation mediated by an upstream open reading frame-encoded peptide. Plant Physiol. 2009;150:1356–1367. doi: 10.1104/pp.109.136036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Garcia MA, Chacon-Camacho OF, Leyva-Hernandez C, Cardenas-Conejo A, Zenteno JC. A novel de novo EFNB1 gene mutation in a Mexican patient with craniofrontonasal syndrome. Case Rep Genet. 2013;2013:349725. doi: 10.1155/2013/349725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seven M, Gezdirici A, Ulucan H, Karatas OF, Yosunkaya E, et al. A novel EFNB1 mutation in a patient with craniofrontonasal syndrome and right hallux duplication. Gene. 2013;527:675–678. doi: 10.1016/j.gene.2013.06.038. [DOI] [PubMed] [Google Scholar]
- Shotelersuk V, Siriwan P, Ausavarat S. A novel mutation in EFNB1, probably with a dominant negative effect, underlying craniofrontonasal syndrome. Cleft Palate Craniofac J. 2006;43:152–154. doi: 10.1597/05-014.1. [DOI] [PubMed] [Google Scholar]
- Torii C, Izumi K, Nakajima H, Takahashi T, Kosaki K. EFNB1 mutation at the ephrin ligand-receptor dimerization interface in a patient with craniofrontonasal syndrome. Congenit Anom (Kyoto) 2007;47:49–52. doi: 10.1111/j.1741-4520.2006.00140.x. [DOI] [PubMed] [Google Scholar]
- Twigg SRF, Kan R, Babbs C, Bochukova EG, Robertson SP, et al. Mutations of ephrin-B1 (EFNB1), a marker of tissue boundary formation, cause craniofrontonasal syndrome. Proc Natl Acad Sci USA. 2004;101:8652–8657. doi: 10.1073/pnas.0402819101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg SRF, Matsumoto K, Kidd AMJ, Goriely A, Taylor IB, et al. The origin of EFNB1 mutations in craniofrontonasal syndrome: frequent somatic mosaicism and explanation of the paucity of carrier males. Am J Hum Genet. 2006;78:999–1010. doi: 10.1086/504440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg SRF, Babbs C, van den Elzen MEP, Goriely A, Taylor S, et al. Cellular interference in craniofrontonasal syndrome: males mosaic for mutations in the X-linked EFNB1 gene are more severely affected than true hemizygotes. Hum Mol Genet. 2013;22:1654–1662. doi: 10.1093/hmg/ddt015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, et al. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Elzen MEP, Twigg SRF, Goos JAC, Hoogeboom AJM, van den Ouweland AMW, et al. Phenotypes of craniofrontonasal syndrome in patients with a pathogenic mutation in EFNB1. Eur J Hum Genet. 2014;22:995–1001. doi: 10.1038/ejhg.2013.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis D, Lacbawan F, Jain M, Der Kaloustian VM, Steiner CE, et al. Additional EFNB1 mutations in craniofrontonasal syndrome. Am J Med Genet A. 2008;146A:2008–2012. doi: 10.1002/ajmg.a.32388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2 - a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Liu Y, Xu Y, Zhao Y, Hua R, et al. Loss-of-function mutations of an inhibitory upstream ORF in the human hairless transcript cause Marie Unna hereditary hypotrichosis. Nat Genet. 2009;41:228–233. doi: 10.1038/ng.276. [DOI] [PubMed] [Google Scholar]
- Wethmar K. The regulatory potential of upstream open reading frames in eukaryotic gene expression. Wiley Interdiscip Rev RNA. 2014;5:765–778. doi: 10.1002/wrna.1245. [DOI] [PubMed] [Google Scholar]
- Wieacker P, Wieland I. Clinical and genetic aspects of craniofrontonasal syndrome: towards resolving a genetic paradox. Mol Genet Metab. 2005;86:110–116. doi: 10.1016/j.ymgme.2005.07.017. [DOI] [PubMed] [Google Scholar]
- Wieland I, Jakubiczka S, Muschke P, Cohen M, Thiele H, et al. Mutations of the ephrin-B1 gene cause craniofrontonasal syndrome. Am J Hum Genet. 2004;74:1209–1215. doi: 10.1086/421532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland I, Reardon W, Jakubiczka S, Franco B, Kress W, et al. Twenty-six novel EFNB1 mutations in familial and sporadic craniofrontonasal syndrome (CFNS) Hum Mutat. 2005;26:113–118. doi: 10.1002/humu.20193. [DOI] [PubMed] [Google Scholar]
- Wieland I, Weidner C, Ciccone R, Lapi E, McDonald-McGinn D, et al. Contiguous gene deletions involving EFNB1OPHN1PJA1 and EDA in patients with craniofrontonasal syndrome. Clin Genet. 2007;72:506–516. doi: 10.1111/j.1399-0004.2007.00905.x. [DOI] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]