Abstract
Mutations in isocitrate dehydrogenase 1 (IDH1) drive most low-grade gliomas and secondary glioblastomas and many chondrosarcomas and acute myeloid leukemia cases. Most tumor-relevant IDH1 mutations are deficient in the normal oxidization of isocitrate to α-ketoglutarate (αKG), but gain the neomorphic activity of reducing αKG to D-2-hydroxyglutarate (D2HG), which drives tumorigenesis. We found previously that IDH1 mutants exhibit one of two reactivities: deficient αKG and moderate D2HG production (including commonly observed R132H and R132C) or moderate αKG and high D2HG production (R132Q). Here, we identify a third type of reactivity, deficient αKG and high D2HG production (R132L). We show that R132Q IDH1 has unique structural features and distinct reactivities towards mutant IDH1 inhibitors. Biochemical and cell-based assays demonstrate that while most tumor-relevant mutations were effectively inhibited by mutant IDH1 inhibitors, R132Q IDH1 had up to a 16 300-fold increase in IC50 versus R132H IDH1. Only compounds that inhibited wild-type (WT) IDH1 were effective against R132Q. This suggests that patients with a R132Q mutation may have a poor response to mutant IDH1 therapies. Molecular dynamics simulations revealed that near the NADP+/NADPH-binding site in R132Q IDH1, a pair of α-helices switches between conformations that are more wild-type-like or more mutant-like, highlighting mechanisms for preserved WT activity. Dihedral angle changes in the dimer interface and buried surface area charges highlight possible mechanisms for loss of inhibitor affinity against R132Q. This work provides a platform for predicting a patient’s therapeutic response and identifies a potential resistance mutation that may arise upon treatment with mutant IDH inhibitors.
Introduction
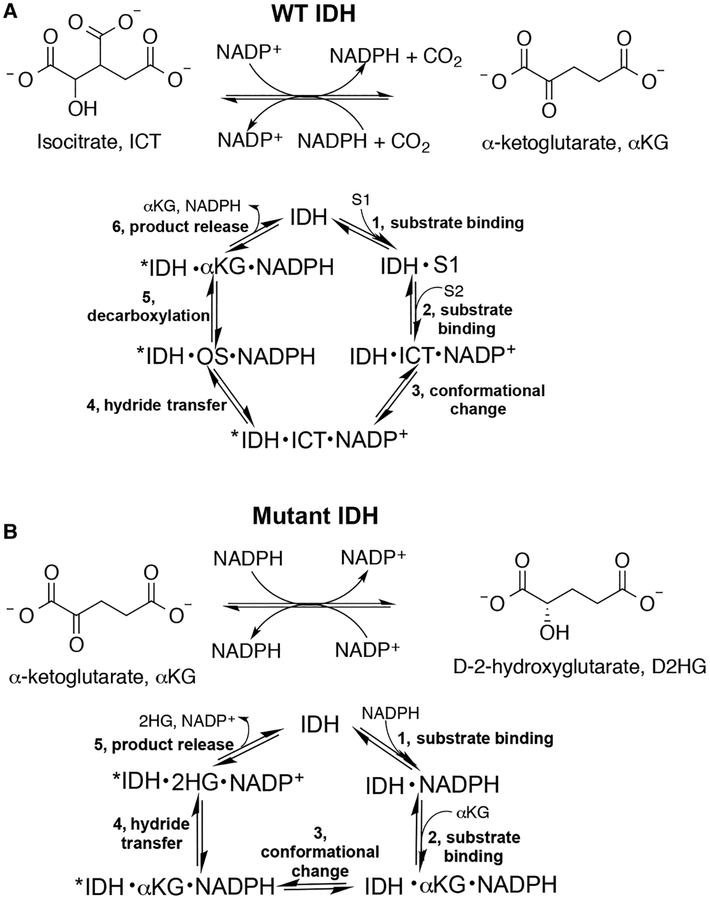
Although alterations in tumor metabolism have been first described nearly a century ago [1,2], it is only relatively recently that dysfunction in metabolic enzymes have been shown to drive tumor formation and growth [3]. Mutations in isocitrate dehydrogenase 1 (IDH1) or isocitrate dehydrogenase 2 (IDH2) are commonly implicated in cancers including gliomas, chondrosarcomas, and acute myeloid leukemia [4–8]. Typically, these mutants have two catalytic features. First, their ability to catalyze the normal reaction, the reversible β-nicotinamide adenine dinucleotide phosphate (NADP+)-dependent conversion of isocitrate (ICT) to α-ketoglutarate (αKG), is ablated [5,9]. Second, most mutants acquire a neomorphic activity, the β-nicotinamide adenine dinucleotide phosphate reduced (NADPH)-dependent synthesis of D-2-hydroxyglutarate (D2HG) from αKG (Figure 1) [10]. D2HG serves as an oncometabolite [10], competitively inhibiting αKG-dependent enzymes such as ten-eleven translocation (TET) methylcytosine dioxygenases and JmjC lysine demethylases, leading to genome hypermethylation [11,12].
Figure 1. Reactions catalyzed by WT and mutant IDH1.
(A) WT IDH1 catalyzes the reversible, NADP+-dependent oxidation of ICT to αKG (OS, oxalosuccinate). (B) Mutant IDH1 is typically deficient in the normal reaction shown in (A) and instead acquires a neomorphic reaction, the NADPH-dependent reduction in αKG to generate the oncometabolite, D2HG [10]. For both reactions, the predicted catalytic cycle is also shown.
Though most IDH1 mutations affect residue 132, which is involved in substrate coordination, there is wide structural and chemical diversity among the tumor-relevant mutations found at this residue [13]. By far, the most common mutation in gliomas is R132H (90% of cases), but R132C, R132G, R132Q, R132L, R132S and R132V IDH1 have been observed at ≤5% frequencies [14]. All but the R132V IDH1 mutation can occur through a single base mutation. Mutation frequencies are different in chondrosarcomas; in a small study on 43 chondrosarcoma tumor samples, 20 had IDH1 mutations, including 5% mutation frequencies each for R132L, R132Q, and R132H, with the remaining tumors having R132C (60%) or R132G (25%) IDH1 mutations [15]. Based in part on this, a mouse model containing the R132Q IDH1 mutation knocked-in was generated and shown to lead to tumor formation [15,16]. We have recently reported that several tumor-relevant IDH1 mutations have wide variation in catalytic efficiency for both the normal and neomorphic reactions [17]. While some trends in van der Waals volume and hydrophobicity were observed [17], the mechanistic details of these variations are still under investigation.
IDH1 and IDH2 mutants have emerged as successful therapeutic targets in the treatment of mutant IDH-driven cancers (reviewed recently in ref. [18]), with many groups developing small molecule inhibitors that have proved to be extremely selective for R132H and R132C versus wild-type (WT) IDH1. Agios Pharmaceuticals has recently received FDA approval for mutant IDH1 [19,20] and IDH2 inhibitors [21,22]. Crystal structures of inhibitors bound to R132H IDH1 show that the inhibitor-binding site is located between the α9 and α10 helices in both monomers, or at the dimer interface (Supplementary Figure S1). Recent work has suggested that the local conformation of the α10 regulatory domain may help drive selectivity over WT IDH1 [23], as this domain is a loop in a structure of WT IDH1 bound to NADP+ [23,24], and is disordered with electron density unresolved in R132H IDH1 in complex with NADP+ [23,25] (Supplementary Figure S1). To date, structural characterization of tumor-relevant mutations is limited to R132H, R132C, and WT IDH1, and inhibitors are typically designed for and assessed in primarily the R132H and R132C IDH1 backgrounds. While such inhibitors should benefit most patients with IDH1-driven tumors, the wide variation in catalytic behavior among IDH1 mutants suggests that there may be differences in inhibitor affinity and efficacy. Thus, it is possible that some patients with less common IDH mutations may not respond well to targeted IDH therapies. Furthermore, studying less common IDH1 mutations that are still capable of D2HG production may highlight possible resistance mutations that may arise following targeted treatment, an issue that has been recently highlighted with the FDA-approved mutant IDH2 inhibitor AG-221 (enasidenib) [26]. Thus, a detailed mechanistic study of a broad series of IDH1 mutants is needed to elucidate fundamental catalytic features, predict patient response to therapy, and identify possible therapeutic resistance mutations.
In this work, we have characterized the widely varying kinetic, inhibitory, and structural features of tumor-relevant IDH1 mutations affecting residue 132 and show that R132Q IDH1 shows unique properties in all three of these features. We report for the first time how a large panel of IDH1 mutants interacts with a series of mutant IDH1 inhibitors and show that D2HG production by R132Q IDH1 is robust despite treatment with these inhibitors. Using explicit-solvent molecular dynamics (MD) simulations, we provide atomic-level models for the local breaking and quaking protein motions around the NADP+ and inhibitor-binding sites that may drive the activity and inhibitor selectivity characteristics of R132Q IDH1 relative to other IDH1 mutants. These kinetic and atomic analyses are important for predicting which mutations are likely to be responsive to mutant IDH1 therapeutic inhibitors, for informing future drug design, and for identifying possible resistance mutations that may develop over the course of treatment.
Experimental procedures
Chemicals and reagents
Tris–hydrochloride, sodium chloride, magnesium chloride hexahydrate, αKG (sodium salt), and resazurin were purchased from Fisher (Hampton, NH). The pH of αKG stocks was adjusted to 7.0 before use. dl-isocitric acid trisodium salt hydrate was purchased from MP Biomedicals (Santa Ana, CA). NADPH (tetrasodium salt) and NADP+ (disodium salt) were purchased from Calbiochem (San Diego, CA). Diaphorase from Clostridium kluyveri and dimethyl sulfoxide (DMSO) were purchased from Sigma–Aldrich (St. Louis, MO). Bovine serum albumin (BSA) was purchased from SeraCare Lifescience (Milford, MA). ML309 was purchased from Sigma, but the manufacturer unable to confirm if the compound is stereospecific or racemic (Sigma–Aldrich, St. Louis, MO). AGI-5198 was purchased from Selleckchem (Houston, TX). GSK864 was purchased from Cayman Chemical Company (Ann Arbor, MI).
Protein expression and purification
WT and mutant homodimers were expressed and purified as described recently [17]. Briefly, after BL21 gold (DE3) Escherichia coli cells transformed with the IDH1 construct reached an A600 of 1.8–2.0 following incubation at 37°C, the culture was induced with 1 mM IPTG (isopropyl β-d-1-thiogalactopyranoside; final concentration) and incubated for an additional 18–20 h. IDH1 protein was purified using nickel–nitrilotriacetic acid column chromatography (Qiagen, Valencia, CA), concentrated, and flash frozen in liquid nitrogen for storage at −80°C for ≤2 months. All enzymes were assessed using small-angle X-ray scattering (SAXS) analysis as described in Supplementary Figure S2 and Table S1.
Steady-state parameter measurements
The activity of IDH1 homodimers was assessed at 37°C using steady-state methods described recently [17]. To quantify the amount of product formed, the molar extinction coefficient for NADPH (6.22 cm−1 mM−1) (absorbance measurements) or a standard curve of known NADPH concentrations (fluorescence measurements) was used. A plot of product versus time yielded an initial linear range that was fit to obtain kobs values (nM NADPH/nM enzyme s−1). These kobs values were then plotted against substrate concentration using Graphpad Prism (RRID:SCR_002798) and fit to a hyperbolic Michaelis–Menten function in order to obtain kcat and KM parameters, which were reported as ±SE. At least two different protein preparations were used in these assays.
MD simulations
We used Schrödinger PrepWizard [27–30] (RRID:SCR_014879) to prepare IDH1 apo structures (PDB: 1T09; [24]) for MD simulation and CHARMMGUI (RRID:SCR_014892) to prepare IDH1 holo NADP+-bound and NADP+/Ca2+-bound structures (PDB: 4KZO; [31]) (Supplementary Tables S2 and S3). Within Bioluminate [32], to prepare these structures for simulations, the crystallographic waters from the PDB structure were maintained. The systems were protonated at pH ~7.4, with the pKa of titratable residues determined with the Maestro integrated program PROPKA [33]. Convergence of protein dynamics was verified through root-mean-squared deviation (RMSD) analysis (Supplementary Figure S3). We solvated each system in a TIP3P model [33]. Five Na+ ions were added to balance the charge of IDH1, and the ion concentration was set at 0.15 M NaCl to mimic the salt concentration in cells. For MD system parameterization performed with the AMBER16 package (NADP+-bound system), system co-ordinates from the preparation (above) were parameterized in CHARMMGUI using the charmm36m force field parameters [34], and periodic boundary conditions were implemented. The Ca2+ bound to the IDH1 protein was parameterized as has been previously discussed [35]. NADP+ was parameterized using the in-house parameters in CHARMM.
GPU-enabled MD simulations were performed with NVIDIA GK110 (GeForce GTX Titan) GPUs using the CUDA version of PMEMD in AMBER16 [36] for the NADP+-bound systems. The simulation systems above were minimized with the CPU version of AMBER16 and equilibrated using the GPU version of AMBER16 [36,37]. We minimized in one step with 2500 steps of steepest-descent algorithm and the remaining 2500 steps with conjugate-gradient algorithm. In the following minimization, five steps of equilibration were performed, including a stage of heating: (1) 25 ps of hydrogen only minimization with a restraint weight of 250 kcal/mol on the protein and solvent, (2) 4 ps of water minimization with a restraint weight of 20 kcal/mol on the protein and salt atoms, (3) 20 ps of water heating with a restraint weight of 20 kcal/mol on the protein and salt atoms, and (4) 40 ps of full equilibration. We equilibrated the system using harmonic equilibration at 298.15 K over four sequential 500-ps runs, decreasing the restraint potential on the backbone on each step, starting at 4.0 kcal/mol and ending at 1.0 kcal/mol.
GPU-enabled AMBER16 production runs were carried out as an NPT ensemble at 298.15 K and 1 bar with a 2-fs time step and particle mesh Ewald [38] electrostatic approximation and non-bonding cutoff of 12.0 Å. For each of the systems simulated, five copies of the simulation were performed for 110 ns each. For simulation analysis, MD trajectories were processed within an ipython jupyter notebook environment [39] using PYTRAJ [40], MDTRAJ [41], and numpy, as well as with cpptraj for principal component analysis (PCA) and visual molecular dynamics (VMD) [42] for visualization. All frames were aligned using backbone atoms. Pytraj was used to perform dihedral, distance, buried surface area, root-mean-squared fluctuation (RMSF), and hydrogen bond-based measurements. Images were taken with VMD [42] and Maestro [43]. RMSD analysis was performed to ensure convergence over the simulations (Supplementary Figure S3).
VolMap Tool, a VMD plugin [42], was used to generate the images of the average buried cavity within each of the mutant and WT IDH1 simulations. The volume was made using the selection of ‘protein and within 10 of residues 277 and 702’, which is residue Q277 that is buried in the dimer interface. The volume around the S124 pocket opening was made using the selection of ‘protein and within 10 of residues 277 and 124’, which are residues W124 and Q277. SiteMap [44] was used to map the volume and size of the binding pockets between the dimers. Site 1 and Site 2 were identified in PDB: 4KZO and 3INM and the volumes from Sites 1 and 2 are outlined in Supplementary Table S4 and Figure S4. Canvas [44] was used to perform hierarchical clustering on binary fingerprints on the IDH1-bound ligands and inhibitors used in this study (Supplementary Figure S5).
Biochemical IDH1 inhibition assays
Methods for inhibition assays were performed as previously described [19]. Briefly, concentrations of AGI-5198 and ML309 ranged from 5 nM to 50 μM for all mutants, except WT and R132Q IDH1, where 5 nM to 500 μM was used. For GSK864, concentrations for WT, R132Q, and R132H IDH1 ranged from 5 nM to 50 μM. For mutant IDH1 studies, inhibitors were preincubated with IDH1 for 30 min on ice, and then 20 mM Tri–HCl (pH 7.5 at ambient temperature), 10 mM MgCl2, 150 mM NaCl, 0.05% (w/v) BSA, 4 μM NADPH, and 4 nM mutant IDH1 were added to a flat-bottom black 96-well plate (Corning Incorporated, Kennebunck, ME). Reactions were initiated by the addition of αKG, with concentrations selected based on the measured KM [17]. Each well had a final volume of 100 ml, and each series was run in triplicate. DMSO concentration was held constant at 1% (final, v/v). After incubation at ambient temperature for 1 h, 50 μl of diaphorase–resazurin solution (final concentrations of 12 μg/ml and 33 μM, respectively) was added to each well for 10 min to consume any remaining NADPH during the production of resofurin. Resofurin was measured at 544 nm excitation and 590 nm emission using an InfiniteM200 Tecan plate reader (Männedorf, Switzerland). For WT IDH1 studies, concentrations of ICT and NADP+ were used at the KM previously measured [17]. Due to high concentrations of NADPH produced during WT IDH1 catalysis, the resazurin–diaphorase solution was not used. Instead, data were collected directly with an InfiniteM200 Tecan instrument using an excitation at 340 nm and emission at 450 nm. Two different enzyme preparations were used for these inhibition studies, except for AGI-5198 and GSK864 activity against R132Q IDH1, where three different enzyme preparations were used. The data were plotted as % activity versus log inhibitor concentration. Biochemical IC50 measurements were calculated by fitting the data to the following equation using GraphPad software (RRID:SCR_002798): Y = 100/(1 + 10^((X − log(IC50)). Deviation of the data from the fit were indicated using 95% confidence intervals.
Transient cell line generation
To construct plasmids for mammalian cell expression, pcDNA3-Flag-IDH1-R132H [9] was purchased (Addgene, Cambridge, MA, plasmid #62907). The R132Q IDH1 construct was generated via a single point mutation with standard site-directed mutagenesis techniques (Kapa Biosystems, Wilmington, MA) using the following primers: R132Q-F: AAAACCTATCATCATAGGTCAGCATGCTTATGGGGATCAATAC; R132Q-R: GTATTGATCCCCATAAGCATGCTGACCTATGATGATAGGTTTT. The WT (R132) IDH1 construct was generated via the same method using the following primers: R132-F: GTAAAACCTATCATCATAGGTCGTCATGCTTATGGGGATCAATAC. R132-R: GTATTGATCCCCATAAGCATGACGACCTATGATGATAGGTTTTAC. Sequences for all plasmids were confirmed (Retrogen, San Diego, CA).
To generate cell lines transiently expressing IDH1, HEK293T cells (ATCC Cat# CRL-3016, RRID: CVCL_0063) and HeLa cells (ATCC Cat# CCL-2) were first maintained in Dulbecco’s Modified Eagle Medium (DMEM) (Thermo Fisher, Waltham, MA) augmented with 10% fetal bovine serum (FBS) at 5% CO2 and 37°C. Cells were purchased from ATCC just prior to use and were tested for mycoplasma by ATCC before use. Cells were passaged 1–2 times between thaw and use. Cell transfection when cells were at 60% confluency was achieved with polyethylenimine (PEI) according to manufacturer’s instructions. WT, R132H, and R132Q IDH1 constructs were used for HEK2937T cells, and the R132Q IDH1 construct was used for HeLa cells. Briefly, 3 × 106 cells were seeded in a 10 cm plate. After 24 h, 20 μg of plasmid DNA was incubated for 10 min at room temperature with 30 μg of PEI in DMEM. This solution was added dropwise to the cell media. After 24 h of incubation, the transfection solution was replaced with fresh media. Protein expression was confirmed via Western immunoblot analysis using anti-Flag (Thermo Fisher Scientific; Cat# PA1–984B, RRID:AB_347227) or anti-Actin primary antibodies (Santa Cruz Biotechnology; CAT# sc-47778) and ECL Plex goat-α-rabbit IgG-Cy5 secondary antibodies (GE Healthcare; Cat# PA45011, RRID:AB_772205) (Supplementary Figure S6).
Cell inhibition assays
Cellular inhibition assays were performed using HEK293T cells transiently expressing R132Q, R132H, or WT IDH1 or HeLa cells transiently expressing R132Q IDH1 that were seeded at 3 × 105 cells/well in six-well plates. Based on Western immunoblot analysis of optimal IDH1 expression, mutant IDH1 inhibition was initiated 72 h post-transfection with the addition of 0, 20, 40, 90, 180, 500, or 3000 nM AGI-5198 (0.2% DMSO final). An abbreviated set of concentrations was used for the HeLa cells. Non-transfected HEK293T and HeLa cells were used as controls. Cells were incubated with inhibitor for 48 h before metabolites were extracted with 50% methanol containing l-norvaline to serve as an internal standard. Derivatization and quantitation via gas chromatography–mass spectrometry (GC–MS) were performed as previously described [17,45].
Results
IDH1 mutations identified in tumors show different catalytic properties that can be broadly grouped into three types
Previously, we reported on the steady-state catalytic rates of several IDH1 mutants observed in patients [17]. We built upon this work by establishing kinetic, inhibitory, and structural features of these mutations. Here, we report catalytic efficiencies of additional IDH1 tumor-relevant mutations affecting residue 132 (Table 1) for the normal and neomorphic reactions, namely R132V, R132L, and R132S IDH1. We show that these mutants show very low catalytic efficiency when converting ICT to αKG, consistent with most mutants we have studied to date [17]. Thus, these mutations were unable to preserve normal activity, unlike R132Q IDH1. Interestingly, R132L IDH1 is nearly as catalytically efficient at D2HG production as R132Q IDH1, though this is driven through a low KM value as the measured kcat is the lowest among the mutants (Table 1). All mutants are more catalytically efficient at D2HG production than the most common IDH1 mutant, R132H IDH1. Thus, these kinetic analyses highlight the unique properties of R132Q IDH1 and allowed us to identify three distinct mutation types. While most tumor-relevant mutations explored to date are deficient in normal catalysis but have moderate rates of D2HG production (R132G, R132V, R132S, R132H, and R132C IDH1), or are deficient in normal catalysis but are highly efficient at D2HG production (R132L IDH1), only R132Q IDH1 is uniquely capable of highly efficient αKG and D2HG production.
Table 1. Steady-state kinetic parameters for the normal reaction catalyzed by IDH1, conversion of ICT to αKG, and for the neomorphic reaction catalyzed by IDH1, conversion of αKG to D2HG.
At least two enzyme preparations were used to obtain kobs rates (at 37°C), which were plotted against substrate concentration and fit to a hyperbolic function. The SE shown is determined from the deviation resulting from hyperbolic fits of plots of kobs versus substrate concentration in order to calculate catalytic efficiency (kcat/KM, mM−1 s−1). In some cases, maximal rates were still achieved at concentrations of NADPH that approached the limit of detection, and thus, KM values are listed as ≤ or < than the lowest concentration tested.
| IDH1 | kcat (s−1), ICT → αKG | KM, ICT (mM) ICT → αKG | KM, NADP+ (mM) ICT → αKG | kcat/KM (mM−1 s−1) ICT → αKG | kcat (s−1) αKG → D2HG | KM, αKG (mM) αKG → D2HG | KM, NADPH (mM) αKG → D2HG | kcat/KM (mM−1 s−1) αKG → D2HG |
|---|---|---|---|---|---|---|---|---|
| WT [17] | 85 ± 4 | 0.22 ± 0.02 | 0.08 ± 0.03 | 3.9 × 102 ± 0.4 × 102 | 0.019 ± 0.001 | 0.5 ± 0.3 | <0.010 | 0.04 ± 0.02 |
| R132H [17] | 2.4 ± 0.1 | 4.2 ± 0.6 | 1.6 ± 0.5 | 0.57 ± 0.08 | 1.44 ± 0.051 | 1.5 ± 0.21 | <0.025 | 1.0 ± 0.11 |
| R132C [17] | 4.4 ± 0.1 | 8.2 ± 0.8 | 0.75 ± 0.07 | 0.54 ± 0.05 | 1.60 ± 0.07 | 0.36 ± 0.05 | 0.010 ± 0.009 | 4.4 ± 0.6 |
| R132G [17] | 9.3 ±0.6 | 7 ± 1 | 0.067 ± 0.007 | 1.3 ± 0.2 | 1.59 ± 0.09 | 0.34 ± 0.08 | <0.025 | 5 ± 1 |
| R132Q [17] | 9.2 ±0.3 | 0.8 ± 0.2 | 0.22 ± 0.052 | 12 ± 3 | 4.7 ± 0.2 | 0.26 ± 0.04 | <0.0052 | 18 ± 3 |
| R132L | 4.0 ± 0.1 | 2.2 ± 0.2 | 0.055 ± 0.009 | 1.8 ± 0.2 | 0.79 ± 0.05 | 0.05 ± 0.01 | <0.005 | 16 ± 3 |
| R132S | 5.6 ± 0.2 | 3.9 ± 0.7 | 0.059 ± 0.009 | 1.4 ± 0.3 | 1.32 ± 0.07 | 0.20 ± 0.04 | <0.005 | 7 ± 1 |
| R132V | 1.38 ± 0.09 | 1.6 ± 0.4 | 0.073 ± 0.008 | 0.9 ± 0.2 | 1.10 ± 0.01 | 0.134 ± 0.006 | <0.005 | 8.2 ± 0.4 |
Additional measurements have led to slight adjustment in previously reported values.
Not reported previously.
Molecular dynamics simulations show residue 132 helps dictate features of the NADP+-binding pocket
Kinetic experiments show that R132Q IDH1 maintains WT-like and mutant-like catalytic activities. To gain insights into the short-timescale atomic-level changes associated with WT-like reactivity among R132Q IDH1 versus other mutants, we performed four replicates each of MD simulations on the R132H, R132Q, R132L, and WT IDH1 homodimers with NADP+ and Ca2+ bound and then with only NADP+ bound. With these simulations, we aimed to uncover local loop rearrangements and pocket breathing motions that are associated with the WT-like and mutant-like reactivities, specifically by investigating the MD starting from a crystal state that would be most similar to the transition states of both the IDH1 neomorphic and normal reactions.
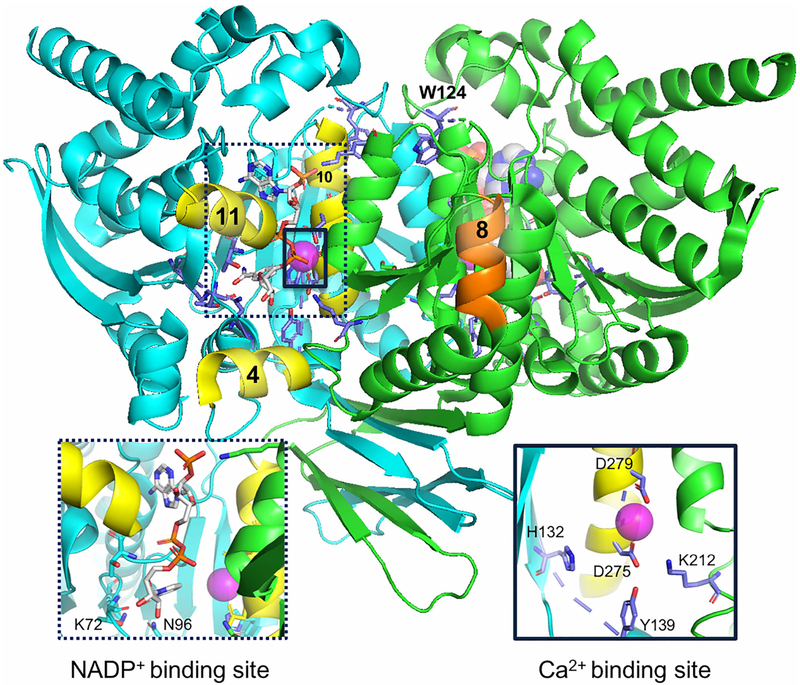
As an array of crystal structures are available for IDH1, we performed preliminary assessments on these crystal structures to determine which would be ideal for MD analyses (Supplementary Tables S2 and S3). Crystal structures were categorized based on substrates bound, surface area of buried ligands (including NADP+, αKG, and ICT), the distances between the center of mass of the α4 and α11 helices, and the center of mass of the α10 helix (a measure of the open and closed state), residue 132-D275 distances, the presence or absence of α10 helix density, the volume of buried cavity from SiteMap [46,47], resolution, and number of missing side chains. All crystal structures with missing loops were excluded as possibilities for simulation. We chose to model the structures of IDH1 bound to NADP+, Ca2+, and αKG with the rationale that a complex with both ligands and metal would most closely reflect a transition state-like or catalytically active complex of the IDH1 neomorphic and normal reactions. Of the crystal structures with these ligands and ion bound, 4KZO [31] was in the most closed state and had well-resolved α10 helix density (Figure 2).
Figure 2. IDH1 dimeric structure (PDB: 4KZO [31]).
α-Helices 4, 8, 10, and 11 are labeled for reference and colored in yellow (monomer A, helices 4, 10, and 11) and orange (monomer B and helix 8). The NADP+- and Ca2+-binding sites are expanded in the labeled sub-panels. Calcium is shown as a pink sphere. NADP+ is colored based on atoms, with nitrogen (blue), oxygen (red), phosphorus (orange), and carbon (gray) shown. Image was generated with PyMol [63].
Simulations were performed in the absence of αKG and in both the presence and absence of Ca2+. Here, the aim was to measure the impact of Ca2+ binding on mutant IDH1 stability and to observe breathing and quaking motions around the NADP+-binding site (sites are highlighted in Figure 2). The RMSF of the NADP+ atoms in the binding site of both monomers in each enzyme was measured after aligning the simulations to the backbone of the entire IDH1 dimer. RMSF analysis is a proxy for understanding how stably NADP+ is bound to the active site; we assume that NADP+ will be more stably bound to the active site of proteins that maintain WT activity, which is dependent upon binding NADP+. Conversely, NADP+ should bind less stably (higher RMSF) to the active site of mutants for which WT activity is abrogated (R132L and R132H IDH1), especially for the nicotinamide moiety. While KM values do not accurately reflect the thermodynamic affinity of an enzyme for its substrate, we do show that far less ICT is required to yield saturating rates of ICT to αKG conversion for WT and R132Q IDH1 versus the high concentrations of ICT needed for R132L and R132H IDH1 (Table 1).
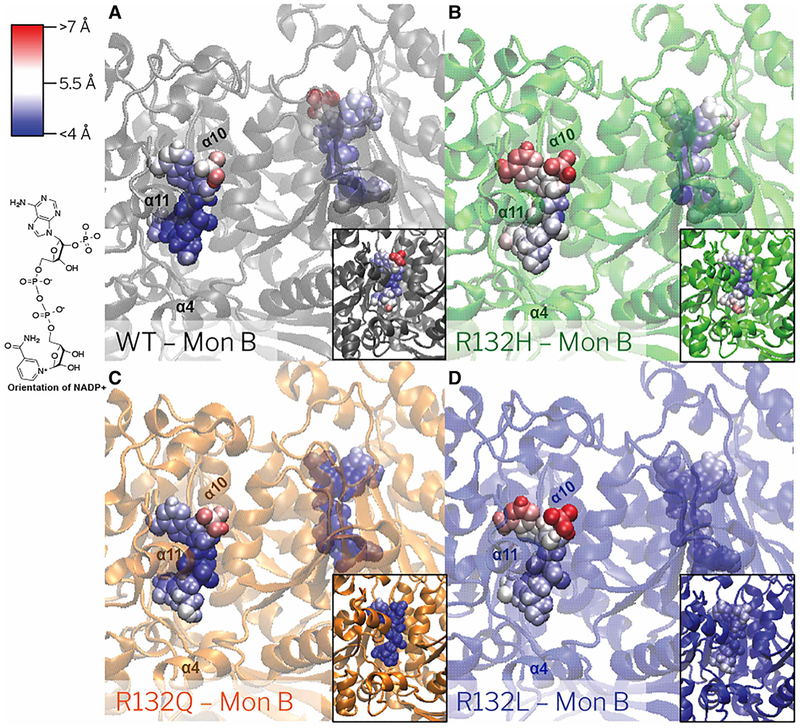
MD simulations reveal that in the B monomers, the atomic-level RMSF of NADP+ bound to R132L and R132H IDH1 is greater than the atomic-level RMSF of NADP+ bound to R132Q and WT IDH1 (Figure 3 and Supplementary Table S5). The atomic-level RMSF of R132Q IDH1 is also slightly greater than the RMSF of WT IDH1 for monomer B, but much less than that for monomer A. Especially when averaged over the atoms in the nicotinamide moiety, the average RMSF for R132L IDH1 per monomer (4.2 ± 0.4 Å and 4.7 ± 0.4 Å) and R132H IDH1 per monomer (5.0 ± 0.4 Å and 5.4 ± 0.3 Å) trend greater than WT IDH1 per monomer (4.3 ±0.6 Å and 4.2 ± 0.7 Å) and R132Q IDH1 (3.6 ± 0.2 Å and 4.3 ± 0.4 Å). The fluctuations seen in WT, R132L, and R132H IDH1 are within a single standard deviation between the monomers of the IDH1 dimer. Yet, simulations of R132Q IDH1 with NADP+/Ca2+ reveal large atomic-level RMSF for NADP+ for monomer B compared with NADP+ bound to monomer A. This suggests a deviation in the dynamics communicated across the dimer interface for mutants relative to WT and other IDH1 mutants. Measurements of IDH1 homodimer crystal structures also demonstrate monomer-to-monomer structural variability in NADP+ and NADP+/Ca2+/αKG-bound states (PDB: 1T09 [24] and 4KZO [31]) (Supplementary Table S2).
Figure 3. NADP+ RMSF in WT, R132Q, R132H, and R132L IDH1 simulations with Ca2+ bound.
The RMSF per atom is colored onto each atom for (A) WT IDH1 (gray), (B) R132H IDH1 (green), (C) R132Q IDH1 (orange), and (D) R132L IDH1 (blue). The NADP+ atoms that fluctuate more than 7 Å are in red, while the atoms which fluctuation less than 4 Å are in blue. Gradations between the two are modulated with white. The orientation of NADP+ in the crystal structures is shown.
Variable hydrogen bonding patterns are also seen for NADP+ in each of the mutants, especially for atoms within the nicotinamide moiety that are involved in hydrogen bonding (Supplementary Figures S7 and S8). Hydrogen bonding formation was monitored in two regions of the NADP+ moiety: first, the ester oxygen of the nicotinamide riboside and T311 (Supplementary Figure S7), and second, the amide oxygen of the nicotinamide and residues T75, N96, and K72 (Supplementary Figure S8). When comparing crystal structures of R132H IDH1 bound to NADP+ versus NADPH, the only difference in the binding pose is within the hydrogen bonding of the amide oxygen of the nicotinamide (atom name O7N; Supplementary Table S5). Thus, we predict that this region may be a better differentiator between features associated with NADP+ binding, which is required for the WT forward reaction, versus NADPH binding, which is required for the neomorphic reaction. In a structure of NADP+-bound R132H IDH1 [31], there are three hydrogen bonds between the amide oxygen of the nicotinamide and IDH1, while in its NADPH-bound form there are none [23]. From all-atom MD, WT IDH1 makes the largest number of hydrogen bond interactions with the nicotinamide ribose in the NADP+/Ca2+-bound simulations, while all mutants show fewer interactions at this site (Supplementary Figure S7). However, WT and R132Q IDH1 are more likely to form stable hydrogen bonds with the amide oxygen of the nicotinamide of NADP+ than R132H and R132L IDH1 (Supplementary Figure S8). This may be an indication that R132Q IDH1 is simulated to bind NADP+ with a more WT-like affinity, suggesting some qualitative agreement between these simulations and our experimental findings.
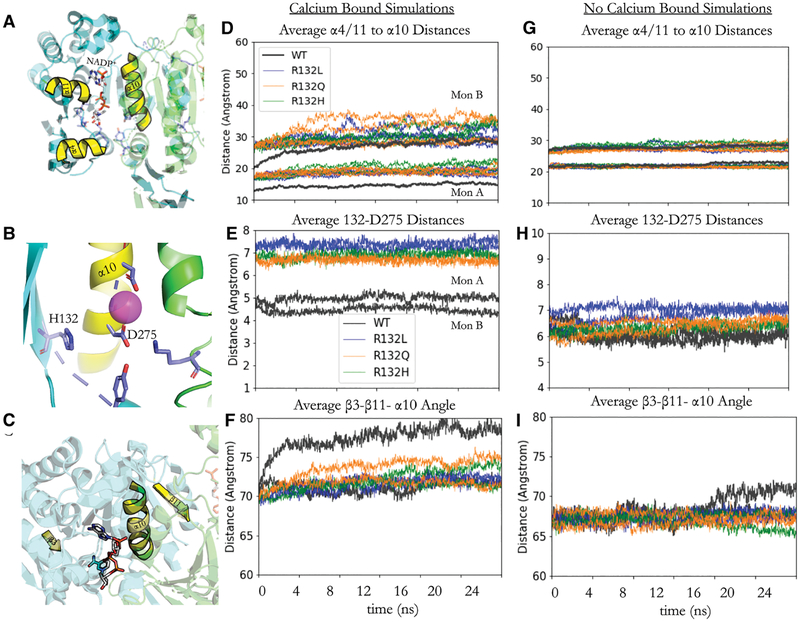
Results from all-atom MD also reveal that the R132 residue is critical for positioning the orientation of the β-sheets in both monomers of IDH1. The distance between the center of mass of the β carbons of residues 132 and the center of mass of the carboxyl carbon of D275 for the WT, R132H, R132Q, and R132L IDH1 simulations were measured in the Ca2+-bound and the Ca2+-unbound simulations (Figure 4). R132 from one monomer co-ordinates to D275 in the α10 helix of the other monomer. The trend in the distances between these two residues in the mutant and WT IDH1 simulations for Ca2+-bound simulations is as follows: R132L > R132H > R132Q >> WT IDH1. In Ca2+-unbound simulations, the trend in distances between these two residues in the mutant and WT IDH1 simulations is: WT > R132L > R132Q > R132H IDH1. This modeled difference in interaction distance between R132 and D275 in turn appears to cause a change in the positioning of the β-sheet relative to the dimer interface (Figure 4C). This tilt in the β-sheet is also seen in the crystal structure of R132H IDH1 bound to inhibitors [23] (Supplementary Figure S1C,E). In turn, this tilt of the β-sheet shifts the positions of the α4 and α11 helices. Helices α4 and α11 neighbor the NADP+-binding site and are connected in sequence through loops to the β3 and β14 sheets, respectively (Figure 4). This rearrangement in the β-sheets and α-helices opens the active site of IDH1. This opening suggests a possible explanation for the RMSF and hydrogen bond distances seen, which could be measured by the change in the angle between the center of mass of β3, β11, and α10 (Figure 4C). This measurement also reveals that one of the WT IDH1 monomers opens greater over the simulation than the other (Figure 4F). This same difference in openness over the simulation occurs with the following trend: WT > R132Q > R132H > R132L IDH1, the same trend that would be expected given the trend in the distance between R132 and D275 and the anchoring role of this residue interaction in the closed state. Interestingly, in the Ca2+-unbound simulations, the residue 132 to D275 distances are longer for WT IDH1 compared with the Ca2+-bound simulations, suggesting a role for divalent cations in the structures of the WT active site compared with the mutants. In all mutants, the α4/11 to α10 distances remain more stable over the MD trajectory in the Ca2+-unbound simulations than in the Ca2+-bound simulations.
Figure 4. NADP+-binding site conformational changes in R132Q, R132H, and R132L IDH1 simulations.
(A) The relative positions of the α-helices 4 and 11 (α4/11) and α10 near the NADP+-binding site (Figure 2). (B) Zoomed-in view of H132, D275 on α10, and Ca2+ (pink sphere). (C) The positions of β3, β11, and α10 are highlighted to give context for the center of mass angles measured in F/I. (D and G) The average distances between the α4/11 residues and the α10 residues on the other side of the NADP+-binding site are shown for Ca2+-bound simulations (D) and Ca2+-unbound simulations (G). The distances for each of the four replicate simulations for the R132L (blue), R132Q (orange), and R132H (green) IDH1 simulations are displayed while the average of the WT simulations is shown in gray for clarity. The standard deviation in the WT IDH1 simulations for monomer B (Mon B) is 2.23 Å, whereas the standard deviation for the WT IDH1 simulations for monomer A (Mon A) is 1.91 Å. (E and H) The average distances between the residue 132 center of mass and D275 center of mass over all four replicates of each mutant/WT trajectory for Ca2+-bound simulations (E) and Ca2+-unbound simulations (H). (F and I) The angles between the center of mass for β3, β11, and α10 are averaged over all four replicates of each mutant/WT trajectory for Ca2+-bound simulations (F) and Ca2+-unbound simulations (H).
In the Ca2+-bound R132Q IDH1 simulations, the openness of the monomers and the R132Q-D275 distances demonstrate both WT-like characteristics in two out of four replicate simulations, as well as more R132H/R132L IDH1-like characteristics in the other two remaining simulations for one of the monomers (Figure 4). These dynamics motions, which stem from R132 coordination with D275, may allow R132Q IDH1 to adopt either WT or R132H-like conformations and thus promote αKG or D2HG production activity. This trend is not seen in the simulations where Ca2+ is absent, suggesting that the structural change in the β-sheets and α4/11 led by coordination between residue 132 and D275 is also dependent upon the coordination of D275 with Ca2+.
R132Q IDH1 is refractory to inhibition in biochemical assays
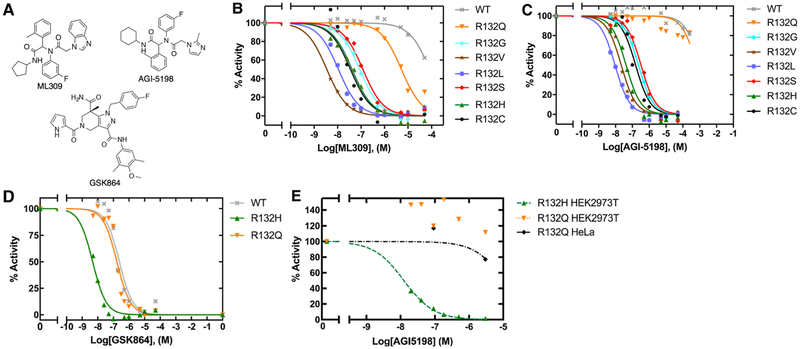
Since the kinetic features of IDH1 mutants varied widely, we sought to compare how these mutants interacted with three commercially available mutant IDH1 inhibitors, ML309 [48], AGI-5198 [19], and GSK864 [49] (Figure 5A). These compounds have been tested previously against both R132H and R132C IDH1, with nM IC50 values in both cellular [19,48–50] and biochemical [48–51] assays, but have not been tested against a wider range of IDH1 mutants. Additionally, these inhibitors are found within a similar cluster when compared among other crystalized inhibitors in a hierarchical clustering based on binary linear fingerprints (Supplementary Figure S5). Figure 5 and Table 2 show results of biochemical IC50 measurements of the selective mutant IDH1 inhibitors (ML309 and AGI-5198) against mutant IDH1 homodimers in an assay using an established resazurin/diaphorase system to quantify NADPH levels [19]. We show that all IDH1 mutants yield low to mid-nM range biochemical IC50 values for both ML309 and AGI-5198 except for WT IDH1 as expected, and except for R132Q IDH1. For ML309, the IC50 is 125-fold higher for R132Q IDH1 relative to R132H IDH1, and only 16-fold lower than WT IDH1. The IC50 value for AGI-5198 is >16 000-fold higher for R132Q IDH1 compared with R132H IDH1, and only 1.5-fold lower than WT IDH1. Importantly, since R132L IDH1 was effectively inhibited by both compounds, this suggests that it is the preservation of the normal activity by R132Q IDH1 that drives this loss of affinity, rather than highly efficient D2HG production.
Figure 5. Biochemical and cellular inhibition by ML309, AGI-5198, and GSK864.
(A) Selective mutant IDH1 inhibitors (ML309 and AGI-5198) designed to target R132H and R132C IDH1, and the pan-inhibitor GSK864 are commercially available. The loss of catalytic conversion of αKG to D2HG in the case of mutant IDH1, or conversion of ICT to αKG in WT IDH1, was measured upon inhibition by (B) ML309, (C) AGI-5198, and (D) GSK864 to obtain biochemical (protein-based) IC50 values. (E) The ability of AGI-5198 to inhibit mutant IDH1 in HEK293T cells transiently expressing R132H IDH1 or R132Q IDH1 and HeLa cells transiently expressing R132Q IDH1 was determined by quantifying cellular D2HG levels using GC–MS. An IC50 value of 0.013 μM with a confidence interval of [0.011–0.014] for AGI-5198 was calculated for R132H IDH1 in HEK293T cells based upon a fit of the plot shown. An IC50 value could not be determined for R132Q IDH1 as concentrations of AGI-5198 required are beyond solubility limits of the compound. Expression of R132Q IDH1 in both HEK2973 and HeLa cells support this finding. Additional information is shown in Supplementary Figure S9.
Table 2. Biochemical IC50 measurements for selective inhibitors of IDH1 mutants.
Percent activity of an incubation of IDH1, NADPH, and αKG with increasing concentrations of inhibitor were determined and plotted against log[inhibitor] and fit to a sigmoidal function. The 95% confidence intervals are shown as determined from the deviation from these fits.
| IDH1 | ML309 IC50 (μ-M) [confidence interval] | AGI-5198 IC50 (μM) [confidence interval] |
|---|---|---|
| WT | 80 [60–100] | 1000 [480–2100] |
| R132H | 0.04 [0.03–0.05] | 0.04 [0.03–0.06] |
| R132Q | 5 [4–7] | 650 [330–1300] |
| R132C | 0.04 [0.02–0.07] | 0.2 [0.07–0.4] |
| R132G | 0.09 [0.07–0.1] | 0.23 [0.20–0.27] |
| R132L | 0.01 [0.009–0.14] | 0.009 [0.008–0.011] |
| R132S | 0.1 [0.096–0.15] | 0.3 [0.2–0.5] |
| R132V | 0.0035 [0.0027–0.0044] | 0.02 [0.02–0.03] |
R132H, R132Q, and WT IDH1 were further evaluated with GSK864, which has been described as a pan-inhibitor [52], with high nM biochemical IC50 values reported for WT IDH1 (~470 nM) versus 9–17 nM IC50 values for R132H, R132C, and R132G IDH1 [49]. Thus, we sought to test if GSK864 also showed nM-level inhibition against R132Q IDH1. GSK864 was fairly effective against R132Q IDH1, with an IC50 value only 1.4-fold lower than that measured for WT IDH1, yet 34-fold higher than that measured for R132H IDH1 (Figure 5 and Table 3). Thus, R132Q IDH1 also behaves like WT IDH1 in terms of inhibition by this pan-inhibitor. Overall, this suggests that retaining at least moderate WT activity (ICT to αKG conversion) is associated with a loss of inhibition by mutant IDH1 inhibitors unless the compound is also able to inhibit WT IDH1.
Table 3. Biochemical IC50 measurements for the pan-inhibitor GSK864.
Percent activity of an incubation of IDH1, NADPH, and αKG with increasing concentrations of inhibitor were determined and plotted against log[inhibitor] and fit to a sigmoidal function. The 95% confidence intervals are shown as determined from the deviation from these fits.
| IDH1 | GSK864 IC50 (μM) [confidence interval] |
|---|---|
| WT | 0.24 [0.15–0.40] |
| R132H | 0.005 [0.004–0.008] |
| R132Q | 0.17 [0.11–0.25] |
The selective inhibitor AGI-5198 does not effectively inhibit D2HG production in cells expressing R132Q IDH1
To test if R132Q IDH1 was also poorly inhibited in the cellular context, HEK293T cells were transiently transfected with WT, R132H, or R132Q IDH1, and HeLa cells were transiently transfected with R132Q IDH1 (Supplementary Figure S6). Following 48 h treatment with varying concentrations of AGI-5198, GC–MS was used to measure cellular levels of D2HG. While D2HG production was significantly inhibited at inhibitor concentrations as low as 20 nM in cells expressing R132H IDH1, no inhibition was observed in HEK293T cells expressing R132Q IDH1 even at the highest concentration of AGI-5198 tested (3 μM) (Figure 5 and Supplementary Figure S9). Only very modest inhibition was observed in HeLa cells expressing R132Q IDH1, further supporting our biochemical findings (Figure 5 and Supplementary Figure S9). Inhibitor treatment appeared to increase D2HG production in resistant cell lines (i.e. R132Q IDH1), but this increase did not prove to be statistically significant. Of note, basal cellular levels of D2HG were higher in cells expressing R132Q IDH1 compared with R132H IDH1, which has been reported previously [16]. This supports our finding of increased catalytic efficiency of D2HG production by R132Q IDH1 compared with R132H IDH1 (Table 1).
Using MD simulations to probe buried cavity dynamics at the dimer interface
In crystal structures of WT IDH1 complexed with NADP+, Ca2+, and αKG, there are two small pockets between the α9/α10 helices at the inhibitor-binding site that are not solvent accessible in the crystal structures (Supplementary Figure S4; Site 1 and Site 2). The positions of α9 and α10 control the size of this pocket; the latter α helix in the inhibitor-bound R132H IDH1 crystal structures is partially unwound to yield a larger pocket (Supplementary Figure S1D and Tables S2 and S3). The NADP+-bound R132H IDH1 crystal structures are very dynamic, with electron density unresolved (Supplementary Figure S1F). Of interest, Ca2+ is measured to bind competitively with inhibitors, likely due to the coordination of Ca2+ with α10, and Ca2+ is absent from crystal structures of R132H bound only to NADPH or inhibitors (for example, [23,25,53,54]). Additionally, a crystal structure of WT IDH1 without Ca2+ bound has an unraveled α10 [24].
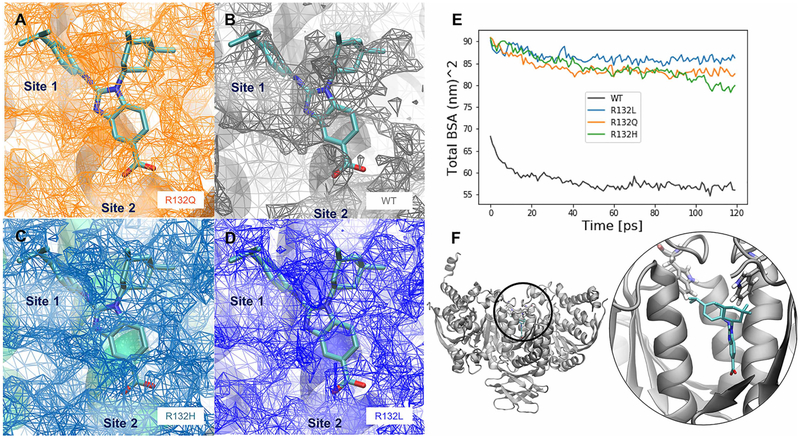
The impact of R132 mutants on the average inhibitor-binding cavity volumes and buried surface area of the dimer interface was calculated in the IDH1 homodimer MD simulations (Figure 6). For all three mutants, the size of the buried cavity was much larger than that calculated in the WT IDH1 simulations in the presence of NADP+ and Ca2+. In simulations of IDH1 in the presence of only NADP+, the inhibitor-binding site was maintained at a small volume in all mutants similar to WT IDH1 (data not shown). This suggests that inhibitor-binding site opening occurs once IDH1 is already in the dynamic open state, since IDH1 simulations in the presence of Ca2+ demonstrate little structural variability between the open/closed states compared with simulations in the absence of Ca2+ (Figure 4B,C).
Figure 6. Inhibitor-binding site differences in R132Q, R132H, and R132L IDH1 simulations.
A BAY1436032 analog, for which a crystal structure in complex with R132H IDH1 has been solved previously (PDB: 5LGE; [53]), was overlaid with our IDH1 models, which are the average structures from each set of MD simulations. The surface of the buried inhibitor-binding pockets averaged across all four simulations are shown in the interface between the monomers where known inhibitors (including the BAY 1 436 032 analog) bind for the (A) R132Q IDH1, (B) WT IDH1, (C) R132H IDH1, and (D) R132L IDH1 simulations. The relative locations of the buried cavity Site 1 and Site 2, as determined from crystal structures (Supplementary Figure S4) are labeled in A–D. (E) The sizes of the average cavity can be compared with the differences in the total buried surface area averaged over all four replicates of simulations. (F) The position of this buried cavity is shown both in zoomed-out and zoomed-in views, along with the positions of residue W124, a possible lid residue for exposing the inhibitor-binding sites. The color scheme for the inhibitor atoms is the same used in Supplementary Figure S1. Note that in A–D, the mesh represents empty space available for inhibitor binding.
In the Ca2+-bound simulations, the buried surface area of all three mutants was greater than the buried surface area of the WT IDH1 cavity. This increase in buried surface area coupled with increased cavity volume seems counter-intuitive. However, this can be explained by the fact that as the cavity expands, there is a corresponding increase in the interfacial surfaces along the perimeter of the buried cavity. Interestingly, all three mutants showed similar sizes in the cavity, although in these models, the α10 helix is intact. In the absence of αKG and Ca2+, R132H IDH1 crystal structures are disordered in this region, resulting in a potentially enlarged inhibitor-binding pocket [23]. Together, these findings suggest that structural changes primarily in the dimer interface in the absence of Ca2+ and in the NADP+/NADPH-binding site may help drive some of the kinetic and inhibitor-binding properties observed in R132Q IDH1 versus R132H and R132L IDH1.
Discussion
Here, we report that the catalytic, structural, and inhibitory properties of R132Q IDH1 vary significantly from all other tumor-relevant mutations described to date. Most mutants show very low and moderate catalytic efficiency for αKG and D2HG production, respectively, while R132L IDH1 has very low and high catalytic efficiency for αKG and D2HG production, respectively. Unusually, R132Q IDH1 maintains moderate activity for αKG production, and has high catalytic efficiency for D2HG production. Previously, cellular models of IDH1-driven tumors have indicated that D2HG levels are higher in HEK293T cells expressing R132Q IDH1 relative to those expressing R132H IDH1 [16], but it is not yet known if these mutations drive differences in tumor phenotypes or patient outcomes. Pusch et al. [55] previously hypothesized in their work on cellular models and patient tissue that lower levels of D2HG production seems most favorable for tumorigenesis based on the frequency of the R132H IDH1 mutation. Our work supports this since we show that the most common IDH1 mutations have only moderate catalytic efficiency for D2HG production, with the lowest catalytic efficiency seen for the most common mutant, R132H IDH1. Since R132Q and R132L IDH1 rapidly consume NADPH and αKG to support high D2HG levels, this catalytic feature is predicted to have an impact on the cellular tumorigenic phenotypes. However, this is likely complicated as IDH mutations are found heterozygously in patients, so WT IDH1 activity is always maintained to some degree, even in non-R132Q IDH1 mutations.
Previously, we reported that, in general, more hydrophobic and smaller amino acid substitutions at residue 132 supported the neomorphic reaction [17], though R132Q IDH1 was shown to be a relative outlier. The robust catalytic efficiency shown by R132L IDH1 supports our previous observations [17]. Interestingly, Pusch et al. [55] show that HEK293T cells stably expressing a series of IDH1 mutants have varying D2HG concentration, with the trend of D2HG levels highest in R132G, followed by R132L, R132C, R132S, and R132H IDH1. Combined with our previously reported findings [17], our kinetic data follow this trend reasonably well except for R132G IDH1, which we show has only moderate efficiency for D2HG production. However, KM in addition to kcat drives catalytic efficiency, and thus, it is not fair to only compare the values of pmol D2HG produced/μg total protein that Pusch et al. report [55] with our kcat/KM values. Indeed, we and Pusch et al. report that KM, ɑKG values for R132G IDH1 are higher than most other mutants except R132H and R132C IDH1 ([55], Table 1), indicating lower efficiency for R132G IDH1. However, this also means that R132L IDH1 does not follow this trend in tumors as well as it appears, since its efficiency is driven by a low KM. The complexities of metabolism in in vivo and in vitro environments clearly blur correlation with the kinetic trends that we can measure in biochemical assays to some degree. However, our assays are able to provide rationale for several major observed trends, such as high D2HG levels in cellular and animal models of R132Q and R132L IDH1-driven tumors.
MD simulations were used to identify the features among the three different mutant types (R132H, R132Q, and R132L IDH1) relative to WT IDH1 to probe differences in catalytic efficiency and inhibitor binding. We noted that both monomers in WT and R132Q IDH1 had lower atomic RMSF values for NADP+ compared with both monomers in R132L and R132H IDH1. There is also variability between monomers of each of the IDH1 mutants. This variability between monomers suggests that there may be an allosteric pathway between the IDH1 active sites in each monomer, and that the mutations not only disrupt WT activity by changing the NADP+-binding site structure, but also negatively impact key allosteric communication between the active sites required for WT activity. Indeed, elegant work by Pietrak et al. [56] showed that ICT to D2HG conversion is coupled between mutant/WT heterodimers. This study describes a mechanism where residue R132 co-ordinates the C3-carboxylate of ICT, the distinguishing feature in ICT versus αKG, to favor binding of ICT over αKG, while mutation of residue 132 allows αKG to more effectively compete with ICT for binding to favor D2HG production over αKG [56]. Residue 132 is also held in place by interactions with D275 which in turn is co-ordinated with Ca2+. Additionally, when this network of inter-residue coordination is displaced, it leads to differences in the openness of the monomers from the closed state on the nanosecond timescale. Due to the differences in NADP+ binding in the WT versus mutant forms of the enzyme, it is possible that residue R132 also plays a role in gating NADP+ versus NADPH to help favor the normal reaction. Such gating may also be affected by inter-subunit allostery. In the future, we will explore catalytic and inhibitory features of WT/mutant heterodimers to explore allostery between the monomers.
Monitoring hydrogen bonding at the ester oxygen of the nicotinamide riboside did not clarify NADP+-binding preference for WT and R132Q IDH1 versus R132L and R132H IDH1. However, when monitoring hydrogen bond formation in the amide oxygen of the nicotinamide, which has the greatest differences in hydrogen bonding patterns when comparing crystal structures of NADP+-bound and NADPH-bound IDH1, we show that R132Q IDH1 can form the largest number of stable hydrogen bonds among the IDH1 mutants. Thus, the finding that R132H and R132L IDH1 have a worse propensity for forming hydrogen bonds at this location compared with R132Q and WT IDH1 may highlight a region for exploring why R132L and R132H IDH1 only catalyze an NADPH-dependent reaction, and not an NADP+-dependent reaction. It will also be interesting to explore the ability of these enzymes to catalyze the reverse normal reaction (αKG to ICT). In general, R132Q IDH1 shows more structural fluctuations between WT-like and mutant-like structures. This is seen in the large fluctuations in distances between several α-helices near the active site (Figure 4B) and in the variability in hydrogen bond formation between the ester oxygen of the nicotinamide riboside (Supplementary Figure S7).
To better understand how the dimer interface cavity accommodates inhibitors, we overlaid results from MD simulations with a crystal structure of R132H IDH1 complexed with an analog of the mutant IDH1 inhibitor BAY1436032 [53]. Both the analog and BAY1436032 are structurally similar to the inhibitors studied here. This was used as a model because IDH1 structures complexed with inhibitors studied here have not yet been solved, as deposited structures [57] do not show density for ML309. The average dimer interface cavity for the holoenzyme mutations and WT IDH1 is overlaid with the BAY1436032 analog in Supplementary Figure S10 [53]. In simulations of apo WT IDH1 (Figure 6) using crystal structures of IDH1 bound only to NADP+ [24], the cavity in the dimeric interface does not overlap with the BAY1436032 analog. Instead, residues from the α10 overlap with this inhibitor. In the mutant NADP+- and Ca2+-bound simulations, the average buried cavity surrounds the BAY1436032 analog except for the phenyl ring of the carboxy-indole. However, this region is likely well accommodated in R132L and R132H IDH1 upon unraveling of α10 helix [23]. This phenyl ring is adjacent to the N-terminus of the α10 helix (shown in gray in the background of Figure 6A–D), the helix for which electron density could not be resolved in the crystal structure of R132H IDH1 [23]. This region also contains D275 that co-ordinates to residue R132 in WT IDH1 structures. Interestingly, the isopropyl group on the phenyl ring of the inhibitor is not well accommodated for either WT or R132Q IDH1, while this region is better accommodated in R132H and R132L IDH1 (Figure 6A–D). Thus, partial unraveling of the α10 N-terminal helix may be possible for the R132Q IDH1 mutant. This may drive some of the differences in affinity of the inhibitors for the various IDH1 mutants observed experimentally. In the future, it will be interesting to expand this work using the recent crystal structure of the pan-inhibitor AG-881, which was reported just prior to publication of this work [58].
The indole moiety in BAY1436032 and the BAY1436032 analog is not novel among IDH1 mutant inhibitors. ML309 similarly contains an indole moiety, whereas AGI-5198 instead contains the smaller imidazole ring. Additionally, the IC50 values for ML309 for IDH1 mutants are generally lower than the IC50 values for AGI-5198, suggesting that this indole moiety contributes to stronger ligand binding in spite of N-terminal helix unfolding. As the IC50 for AGI-5198 is 130-fold higher than that for ML309 for R132Q IDH1, this demonstrates that R132Q IDH1 is better inhibited by the indole moiety-containing inhibitor. Thus, we propose here that accommodation of inhibitors with bulkier indole moieties between the dimeric interface may be dependent upon the unraveling of the N-terminus of the α10 helix. This could enable the binding of the bulkier GSK864 inhibitor and other inhibitors in the dimeric interface as has been observed in many crystal structures (Figure 5 and Supplementary Table S3). Additionally, the BAY1436032 analog binds in a 1-to-2 ratio of analog to IDH1 monomer.
The question remains how the buried cavity becomes solvent accessible to allow ligand binding. From simulations of the R132L and R132V IDH1 bound to NADP+ and Ca2+, torsion angle changes in residue W124 enable opening of the buried inhibitor-binding cavity to solvent (Supplementary Figure S11). These same torsion angle changes were not observed in the R132Q, R132H, or WT IDH1 holo simulations. Concerted phi and psi angle changes in W124 result in a conformational state with a path to the buried cavity. Sliding of α9 and α10 helices provides enough room in simulations for the flipping of this tryptophan. Tryptophan residues have previously been observed to act as gating residues near buried pockets, enabling the transformation from buried to solvent accessible protein cavities on the microsecond timescale [59–62]. This flipping of W124 could be one in a series of concerted changes that enables full access of inhibitors to the buried cavity, as is common in conformational changes that result in solvent exposure of buried cavities [62]. Opening of this buried cavity is one of many steps required for binding, as certainly R132H IDH1 shows excellent affinity for all inhibitors studied here, but cavity opening is not captured within the timeframe of our simulations. These concerted changes will be investigated in future work.
While all-atom explicit-solvent MD simulations revealed interesting differences between the holo mutants and WT IDH1, and apo R132Q IDH1 and WT IDH1, the ~100 ns simulated here for each system is a relatively short timescale and thus cannot reveal the full breadth of conformational changes that are likely within each of these protein systems. Despite these short timescales, significant differences were observed for average distances, NADP+, atomic-level RMSF, torsion changes, and buried cavity volumes across each of the IDH1 protein systems under investigation. Future work will seek to illuminate the microsecond timescale dynamics of these IDH1 protein systems.
Here, we used a combination of kinetic experiments and MD simulations to probe the catalytic, structural, and inhibitor-binding features of a series of IDH1 mutants identified in tumors. We have expanded the established variation [17] in catalytic features seen in IDH1 mutants and show that the ability to catalyze the WT reaction predicts a significant loss of affinity for mutant IDH1 inhibitors. Changes in α-helical regions that shape both substrate and inhibitor-binding pockets, and the open/closed transition of the IDH1 monomer could contribute in tandem to some of these catalytic characteristics. MD simulations suggest that R132Q IDH1 can be viewed as a stochastic structural hybrid of WT and mutant IDH1, consistent with its ability to catalyze both the normal and neomorphic reactions, and suggest that indole, rather than imidazole-containing inhibitors, may be more effective against this mutation type. This work will help inform future drug design strategies and aid in assessing which patients are likely to respond to mutant IDH1 inhibitors based on mutation type.
Supplementary Material
Acknowledgements
The authors thank Yue Xiong for the gift of pcDNA3-Flag-R132H, as well as advice from John Shupe on cell line generation. We also thank T. Matsui and T. Weiss at beamline 4–2 of the Stanford Synchrotron Radiation Lab (SSRL) for technical support in SAXS experiments. We would like to thank Javier Piedrafita (SDSU) for the kind gift of HeLa cells.
Funding
This work was supported by the National Institutes of Health R00 CA187594 (C.D.S.), U54 CA132384 (SDSU) and U54 CA132379 (UC San Diego), and R01 GM110588 (M.A.S), San Diego State University startup funds (C.D.S.), the California Metabolic Research Foundation (C.D.S. and M.A.S), and the SBP NCI Cancer Center Support Grant P30 CA030199 (O.Z. and D.A.S.). Use of the Stanford Synchrotron Radiation Light source, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02–76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- BSA
bovine serum albumin
- D2HG
D-2-hydroxyglutarate
- DMEM
Dulbecco’s Modified Eagle Medium
- DMSO
dimethyl sulfoxide
- GC–MS
gas chromatography–mass spectrometry
- ICT
isocitrate
- IDH1
isocitrate dehydrogenase 1
- IDH2
isocitrate dehydrogenase 2
- MD
molecular dynamics
- NADP+
β-nicotinamide adenine dinucleotide phosphate
- NADPH
β-nicotinamide adenine dinucleotide phosphate reduced
- PEI
polyethylenimine
- RMSD
root-mean-squared deviation
- RMSF
root-mean-squared fluctuation
- SAXS
small-angle X-ray scattering
- VMD
visual molecular dynamics
- WT
wild type
- αKG
α-ketoglutarate
Footnotes
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Warburg O (1925) Über den stoffwechsel der carcinomzelle. Klin Wochenschr. 4, 534–536 10.1007/BF01726151 [DOI] [Google Scholar]
- 2.Warburg O (1956) Origin of cancer cells. Oncologia 9, 75–83 10.1159/000223920 [DOI] [PubMed] [Google Scholar]
- 3.DeBerardinis RJ and Chandel NS (2016) Fundamentals of cancer metabolism. Sci. Adv 2, e1600200 10.1126/sciadv.1600200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P et al. (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321, 1807–1812 10.1126/science.1164382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W et al. (2009) IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med 360, 765–773 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K et al. (2009) Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med 361, 1058–1066 10.1056/NEJMoa0903840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kipp BR, Voss JS, Kerr SE, Barr Fritcher EG Graham RP, Zhang L et al. (2012) Isocitrate dehydrogenase 1 and 2 mutations in cholangiocarcinoma. Hum. Pathol 43, 1552–1558 10.1016/j.humpath.2011.12.007 [DOI] [PubMed] [Google Scholar]
- 8.Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS et al. (2012) Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist 17, 72–79 10.1634/theoncologist.2011-0386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P et al. (2009) Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science 324, 261–265 10.1126/science.1170944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM et al. (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744 10.1038/nature08617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A et al. (2010) Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18, 553–567 10.1016/j.ccr.2010.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR et al. (2011) The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 12, 463–469 10.1038/embor.2011.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dimitrov L, Hong CS, Yang C, Zhuang Z and Heiss JD (2015) New developments in the pathogenesis and therapeutic targeting of the IDH1 mutation in glioma. Int. J. Med. Sci 12, 201–213 10.7150/ijms.11047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirata M, Sasaki M, Cairns RA, Inoue S, Puviindran V, Li WY et al. (2015) Mutant IDH is sufficient to initiate enchondromatosis in mice. Proc. Natl Acad. Sci. U.S.A 112, 2829–2834 10.1073/pnas.1424400112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang B, Zhang J, Xia J, Zhao W, Wu Y, Shi M et al. (2017) IDH1 mutation promotes tumorigenesis by inhibiting JNK activation and apoptosis induced by serum starvation. Cell Rep. 19, 389–400 10.1016/j.celrep.2017.03.053 [DOI] [PubMed] [Google Scholar]
- 17.Avellaneda Matteo D, Grunseth AJ, Gonzalez ER, Anselmo SL, Kennedy MA, Moman P et al. (2017) Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: the role of size and hydrophobicity at residue 132 on catalytic efficiency. J. Biol. Chem 292, 7971–7983 10.1074/jbc.M117.776179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dang L, Yen K and Attar EC (2016) IDH mutations in cancer and progress toward development of targeted therapeutics. Ann. Oncol 27, 599–608 10.1093/annonc/mdw013 [DOI] [PubMed] [Google Scholar]
- 19.Popovici-Muller J, Saunders JO, Salituro FG, Travins JM, Yan S, Zhao F et al. (2012) Discovery of the first potent inhibitors of mutant IDH1 that lower tumor 2-HG in vivo. ACS Med. Chem. Lett 3, 850–855 10.1021/ml300225h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DiNardo CD, Stein EM, de Botton S, Roboz GJ, Altman JK, Mims AS et al. (2018) Durable remissions with Ivosidenib in IDH1-mutated relapsed or refractory AML. N. Engl. J. Med 378, 2386–2398 10.1056/NEJMoa1716984 [DOI] [PubMed] [Google Scholar]
- 21.Wang F, Travins J, DeLaBarre B, Penard-Lacronique V, Schalm S, Hansen E et al. (2013) Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science 340, 622–626 10.1126/science.1234769 [DOI] [PubMed] [Google Scholar]
- 22.Yen K, Travins J, Wang F, David MD, Artin E, Straley K et al. (2017) AG-221, a first-in-class therapy targeting acute myeloid leukemia harboring oncogenic IDH2 mutations. Cancer Discov. 7, 478–493 10.1158/2159-8290.CD-16-1034 [DOI] [PubMed] [Google Scholar]
- 23.Xie X, Baird D, Bowen K, Capka V, Chen J, Chenail G et al. (2017) Allosteric mutant IDH1 inhibitors reveal mechanisms for IDH1 mutant and isoform selectivity. Structure 25, 506–513 10.1016/j.str.2016.12.017 [DOI] [PubMed] [Google Scholar]
- 24.Xu X, Zhao J, Xu Z, Peng B, Huang Q, Arnold E et al. (2004) Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J. Biol. Chem 279, 33946–33957 10.1074/jbc.M404298200 [DOI] [PubMed] [Google Scholar]
- 25.Yang B, Zhong C, Peng Y, Lai Z and Ding J (2010) Molecular mechanisms of ‘off-on switch’ of activities of human IDH1 by tumor-associated mutation R132H. Cell Res. 20, 1188–1200 10.1038/cr.2010.145 [DOI] [PubMed] [Google Scholar]
- 26.Intlekofer AM, Shih AH, Wang B, Nazir A, Rustenburg AS, Albanese SK et al. (2018) Acquired resistance to IDH inhibition through trans or cis dimer-interface mutations. Nature 559, 125–129 10.1038/s41586-018-0251-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schrödinger Release 2018–3, Schrödinger Suite 2018–3 Protein Preparation Wizard, Schrodinger, LLC, New York, NY [Google Scholar]
- 28.(2016) Epik, Schrödinger, LLC, New York, NY [Google Scholar]
- 29.(2016) Impact, Schrödinger, LLC, New York, NY [Google Scholar]
- 30.(2018) Prime, Schrödinger, LLC, New York, NY [Google Scholar]
- 31.Rendina AR, Pietrak B, Smallwood A, Zhao H, Qi H, Quinn C et al. (2013) Mutant IDH1 enhances the production of 2-hydroxyglutarate due to its kinetic mechanism. Biochemistry 52, 4563–4577 10.1021/bi400514k [DOI] [PubMed] [Google Scholar]
- 32.(2018) Schrödinger Release 2018–3: BioLuminate, Schrödinger, LLC, New York, NY [Google Scholar]
- 33.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW and Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys 79, 926–935 10.1063/1.445869 [DOI] [Google Scholar]
- 34.Huang J and MacKerell AD Jr (2013) CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem 34, 2135–2145 10.1002/jcc.23354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beglov D and Roux B (1994) Finite representation of an infinite bulk system: solvent boundary potential for computer simulations. J. Chem. Phys 100, 9050–9063 10.1063/1.466711 [DOI] [Google Scholar]
- 36.Case DA, Cerutti DS, Cheatham TE III, Darden TA, Duke RE, Giese TJ et al. (2017) AMBER 2017, University of California, San Francisco [Google Scholar]
- 37.Salomon-Ferrer R, Case DA and Walker RC (2013) An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci 3, 198–210 10.1002/wcms.1121 [DOI] [Google Scholar]
- 38.Darden T, York D and Pedersen L (1993) Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys 98, 10089–10092 10.1063/1.464397 [DOI] [Google Scholar]
- 39.Perez F and Granger BE (2007) IPython: a system for interactive scientific computing. Comp. Sci. Eng 9, 21–29 10.1109/MCSE.2007.53 [DOI] [Google Scholar]
- 40.Roe DR and Cheatham TE (2013) PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput 9, 3084–3095 10.1021/ct400341p [DOI] [PubMed] [Google Scholar]
- 41.McGibbon RT, Beauchamp KA, Harrigan MP, Klein C, Swails JM, Hernández CX et al. (2015) MDTraj: a modern open library for the analysis of molecular dynamics trajectories. Biophys. J 109, 1528–1532 10.1016/j.bpj.2015.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Humphrey W, Dalke A and Schulten K (1996) VMD: visual molecular dynamics. J. Mol. Graph 14, 33–38; 27–38 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- 43.(2018) Schrödinger Release 2018–3: Maestro, Schrödinger, LLC, New York, NY [Google Scholar]
- 44.(2018) Schrödinger, LLC, New York, NY [Google Scholar]
- 45.Ratnikov B, Aza-Blanc P, Ronai ZA, Smith JW, Osterman AL and Scott DA (2015) Glutamate and asparagine cataplerosis underlie glutamine addiction in melanoma. Oncotarget 6, 7379–7389 10.18632/oncotarget.3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.(2018) SiteMap, Schrödinger, LLC, New York, NY [Google Scholar]
- 47.Halgren TA (2009) Identifying and characterizing binding sites and assessing druggability. J. Chem. Inf. Model 49, 377–389 10.1021/ci800324m [DOI] [PubMed] [Google Scholar]
- 48.Davis MI, Gross S, Shen M, Straley KS, Pragani R, Lea WA et al. (2014) Biochemical, cellular and biophysical characterization of a potent inhibitor of mutant isocitrate dehydrogenase IDH1. J. Biol. Chem 289, 13717–13725 10.1074/jbc.M113.511030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okoye-Okafor UC, Bartholdy B, Cartier J, Gao EN, Pietrak B, Rendina AR et al. (2015) New IDH1 mutant inhibitors for treatment of acute myeloid leukemia. Nat. Chem. Biol 11, 878–886 10.1038/nchembio.1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Popovici-Muller J, Lemieux RM, Artin E, Saunders JO, Salituro FG, Travins J et al. (2018) Discovery of AG-120 (Ivosidenib): a first-in-class mutant IDH1 inhibitor for the treatment of IDH1 mutant cancers. ACS Med. Chem. Lett 9, 300–305 10.1021/acsmedchemlett.7b00421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C et al. (2013) An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science 340, 626–630 10.1126/science.1236062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chaturvedi A, Herbst L, Pusch S, Klett L, Goparaju R, Stichel D et al. (2017) Pan-mutant-IDH1 inhibitor BAY1436032 is highly effective against human IDH1 mutant acute myeloid leukemia in vivo. Leukemia 31, 2020–2028 10.1038/leu.2017.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pusch S, Krausert S, Fischer V, Balss J, Ott M, Schrimpf D et al. (2017) Pan-mutant IDH1 inhibitor BAY 1436032 for effective treatment of IDH1 mutant astrocytoma in vivo. Acta Neuropathol. 133, 629–644 10.1007/s00401-017-1677-y [DOI] [PubMed] [Google Scholar]
- 54.Jones S, Ahmet J, Ayton K, Ball M, Cockerill M, Fairweather E et al. (2016) Discovery and optimization of allosteric inhibitors of mutant isocitrate dehydrogenase 1 (R132H IDH1) displaying activity in human acute myeloid leukemia cells. J. Med. Chem 59, 11120–11137 10.1021/acs.jmedchem.6b01320 [DOI] [PubMed] [Google Scholar]
- 55.Pusch S, Schweizer L, Beck AC, Lehmler JM, Weissert S, Balss J et al. (2014) D-2-Hydroxyglutarate producing neo-enzymatic activity inversely correlates with frequency of the type of isocitrate dehydrogenase 1 mutations found in glioma. Acta Neuropathol. Commun 2, 19 10.1186/2051-5960-2-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pietrak B, Zhao H, Qi H, Quinn C, Gao E, Boyer JG et al. (2011) A tale of two subunits: how the neomorphic R132H IDH1 mutation enhances production of alphaHG. Biochemistry 50, 4804–4812 10.1021/bi200499m [DOI] [PubMed] [Google Scholar]
- 57.Merk A, Bartesaghi A, Banerjee S, Falconieri V, Rao P, Davis MI et al. (2016) Breaking cryo-EM resolution barriers to facilitate drug discovery. Cell 165, 1698–1707 10.1016/j.cell.2016.05.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma R and Yun CH (2018) Crystal structures of pan-IDH inhibitor AG-881 in complex with mutant human IDH1 and IDH2. Biochem. Biophys. Res. Commun 503, 2912–2917 10.1016/j.bbrc.2018.08.068 [DOI] [PubMed] [Google Scholar]
- 59.Mulder FA, Mittermaier A, Hon B, Dahlquist FW and Kay LE (2001) Studying excited states of proteins by NMR spectroscopy. Nat. Struct. Biol 8, 932–935 10.1038/nsb1101-932 [DOI] [PubMed] [Google Scholar]
- 60.Fitzgerald MM, Musah RA, McRee DE and Goodin DB (1996) A ligand-gated, hinged loop rearrangement opens a channel to a buried artificial protein cavity. Nat. Struct. Biol 3, 626–631 10.1038/nsb0796-626 [DOI] [PubMed] [Google Scholar]
- 61.Nucci NV, Fuglestad B, Athanasoula EA and Wand AJ (2014) Role of cavities and hydration in the pressure unfolding of T4 lysozyme. Proc. Natl Acad. Sci. U.S.A 111, 13846–13851 10.1073/pnas.1410655111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schiffer JM, Feher VA, Malmstrom RD, Sida R and Amaro RE (2016) Capturing invisible motions in the transition from ground to rare excited states of T4 lysozyme L99A. Biophys. J 111, 1631–1640 10.1016/j.bpj.2016.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.The PyMOL Molecular Graphics System, Version 1.8.0.4, Schrodinger, LLC [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.