Abstract
Characterizing glycans and glycoconjugates in the context of three-dimensional structures is important in understanding their biological roles and developing efficient therapeutic agents. Computational modeling and molecular simulation have become an essential tool complementary to experimental methods. Here, we present a computational tool, Glycan Modeler for in silico N-/O-glycosylation of the target protein and generation of carbohydrate-only systems. In our previous study, we developed Glycan Reader, a web-based tool for detecting carbohydrate molecules from a PDB structure and generation of simulation system and input files. As integrated into Glycan Reader in CHARMM-GUI, Glycan Modeler (Glycan Reader & Modeler) enables to generate the structures of glycans and glycoconjugates for given glycan sequences and glycosylation sites using PDB glycan template structures from Glycan Fragment Database (http://glycanstructure.org/fragment-db). Our benchmark tests demonstrate the universal applicability of Glycan Reader & Modeler to various glycan sequences and target proteins. We also investigated the structural properties of modeled glycan structures by running 2-μs molecular dynamics simulations of HIV envelope protein. The simulations show that the modeled glycan structures built by Glycan Reader & Modeler have the similar structural features compared to the ones solved by X-ray crystallography. We also describe the representative examples of glycoconjugate modeling with video demos to illustrate the practical applications of Glycan Reader & Modeler. Glycan Reader & Modeler is freely available at http://charmm-gui.org/input/glycan.
Keywords: Glycan Reader, molecular dynamics simulation, N-linked glycosylation, O-linked glycosylation, protein–glycan interaction, template-based glycan modeling
Introduction
Carbohydrate moieties, also referred to as glycans, are one of the most abundant cell components (Ohtsubo and Marth 2006). They can be highly branched and covalently attached to protein (glycoprotein) or lipid (glycolipid), and can exist as free ligands (Dwek 1996). The covalent attachment of glycans to proteins (protein glycosylation) is one of the most important and abundant post-translational protein modifications. More than half of all proteins are expected to be glycosylated based on SWISS-PROT database (Apweiler et al. 1999). Glycolipids and unbranched polysaccharides such as glycosaminoglycans (GAGs) have been found in many different bacterial species and their structural forms have the enormous diversity (Curatolo 1987; Muthana et al. 2012). Therefore, it is not surprising that glycans play a critical role in living organisms. Indeed, numerous studies have shown that glycans are associated with a variety of important biological processes, such as immune recognition and response (Rabinovich and Toscano 2009), tumor growth and metastasis (El Ghazal et al. 2016), protein quality control and trafficking (Trombetta 2003), and cell–cell communications (Collins and Paulson 2004).
To understand how glycosylation is functionally involved in these physiological and pathological processes, knowledge of the three-dimensional structures of glycans, their dynamic properties, and their interactions with proteins is essential (Malik and Ahmad 2007; Nagae and Yamaguchi 2012; Malik et al. 2014; Jo et al. 2016; Qi et al. 2016; Dong et al. 2018). Experimental methods such as X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy have been widely used to provide the atomic details of biomolecular structural information (Wormald et al. 2002). During last decades, X-ray crystallography has been improved substantially with recent advances such as the synchrotron radiation (Perez and de Sanctis 2017). However, despite these technical advances, solving the complex carbohydrate structures remains nontrivial. For example, during the crystallization of glycoproteins, heterogeneity of glycans on the protein surface hinders the crystal packing, and the electron density maps of these glycans are not often fully resolved because of their high flexibility (Perez and de Sanctis 2017). NMR spectroscopy also has been advanced in terms of the labeling strategies such as labeled glycans with stable NMR-active isotopes. For instance, the use of 13C-labeled glycans provides the large signal dispersion and high sensitivity and also makes it possible to observe the fast exchanges of ligand complexes in the NMR chemical shift timescales (Arda and Jimenez-Barbero 2018). Nevertheless, some limitations are still remained in terms of the relatively low intrinsic sensitivity and the dependency on the availability of isotopic labeling or specific chemical tags required in the case of large biomolecules (Marchetti et al. 2016).
In order to alleviate these difficulties, combination of experimental techniques with molecular modeling and simulation methodologies such as Monte Carlo and molecular dynamics (MD) simulations has emerged as an indispensable tool to investigate glycan conformations in a biological environment (Imberty and Perez 2000; Marchetti et al. 2016; Hamark et al. 2017). Over the years, several carbohydrate force fields such as GLYCAM06 (Kirschner et al. 2008), CHARMM36 (Guvench et al. 2009), GROMOS 53A6GLYC (Pol-Fachin et al. 2012), OPLS-AA-SEI (Kony et al. 2002), and MM4 (Allinger et al. 2003) have been developed. However, building the simulation systems containing glycans and glycoconjugates still remains nontrivial due to the structural complexity of glycans (i.e., branches and various types of glycosidic linkages (Campbell et al. 2015)). To facilitate the computational modeling of glycans, standalone programs such as POLYS (Engelsen et al. 2014), doGlycans (Danne et al. 2017), CarbBuilder (Kuttel et al. 2016), and RosettaCarbohydrate (Labonte et al. 2017) have been developed. Several web-based tools, such as GlyProt (Bohne-Lang and von der Lieth 2005) and SWEET-II (Bohne et al. 1999) at GLYCOSCIENCES.de (Lutteke et al. 2006), GLYCAM Carbohydrate Builder (Woods-Group 2005–2018) at GLYCAM-web, Glycan Fragment Database (GFDB) (Jo and Im 2013) and GS-align (glycan structure alignment) (Lee and Jo et al. 2015) at GlycanStructure.ORG (Im-Group 2011-2018), Glycan Reader in CHARMM-GUI (Jo et al. 2011), and PROCARB (Malik et al. 2010) provide glycan-related applications and databases, allowing users to model glycan structures with experimentally determined glycan structures as a template without any installation of program in user-side. However, except CHARMM-GUI and GLYCAM-web, these tools require significant post-processing efforts to prepare simulation systems with the generated glycan structure. Currently, CHARMM-GUI is the only available tool for the valid input setup of major MD simulation programs (Lee et al. 2016) such as CHARMM (Brooks et al. 2009), NAMD (Phillips et al. 2005), GROMACS (Hess et al. 2008), AMBER (Case et al. 2005), GENESIS (Jung et al. 2015), LAMMPS (Plimpton 1995), Desmond (Bowers et al. 2006), OpenMM (Eastman et al. 2013) and CHARMM/OpenMM (Arthur and Brooks 2016) (using the CHARMM force field).
CHARMM-GUI, http://www.charmm-gui.org, is a web-based graphical user interface and provides various functional modules to prepare complex biomolecular systems and input files for molecular simulations. During the 13 years of successful services, CHARMM-GUI has contributed to facilitate molecular modeling and simulation of glycans and glycoconjugates by developing Glycolipid Modeler (Lee et al. 2019), LPS Modeler (Lee et al. 2019) and Glycan Reader (Jo et al. 2011). In particular, Glycan Reader has greatly simplified the reading of PDB and PDBx/mmCIF structure files containing glycans by automatic detection of carbohydrate molecules and glycosidic linkage information. In the recent update (Park et al. 2017), Glycan Reader not only detects most sugar types and chemical modifications in the PDB, but also allows users to edit the glycan sequences through addition/deletion/change of sugar types, chemical modifications, glycosidic linkages, and anomeric states. However, CHARMM-GUI Glycan Reader did not support in silico glycosylation and addition of a sugar at the reducing end of an existing glycan chain.
In this work, we present Glycan Modeler, a new CHARMM-GUI module for in silico N-/O-glycosylation and generation of carbohydrate-only systems. CHARMM-GUI Glycan Modeler can generate the most appropriate representative glycan structures through GFDB database searches and determines proper orientations relative to target protein. Although GFDB stores all available complex carbohydrate structures in the PDB after the validation processes such as reannotation of carbohydrate residue names based on their Cartesian coordinates using Glycan Reader, not every possible carbohydrate sequence is available in GFDB. In the absence of target glycan sequence in GFDB, our module generates the structures by using the valid internal coordinate information in the CHARMM force field (see Materials and methods for details). Glycan Reader & Modeler (as Glycan Modeler is integrated into Glycan Reader) also allows addition of a sugar at the reducing end of a pre-existing glycan chain. We performed benchmark analysis and MD simulations to validate the reliability of modeled N-glycan structures. In addition, we also describe the representative examples of glycoconjugate modeling with video demos to illustrate the practical applications of Glycan Reader & Modeler (http://www.charmm-gui.org/demo).
Results and discussion
Glycan Modeler can build N-/O-glycans at multiple glycosylation sites with or without pre-existing glycan(s) in a protein structure. The inputs for N-/O-glycan modeling are a protein structure, glycosylation site(s), and glycan sequence for each glycosylation site (see Materials and methods for details). In addition to N-/O-glycans, Glycan Modeler can build a carbohydrate-only structure with a user-specified glycan sequence.
Benchmark test of Glycan Modeler against PDB non-redundant N-glycan set
To evaluate the quality of glycan structure models generated by Glycan Modeler, we measured the structural similarities of glycan models to their native PDB structures for a set of target N-glycans. We first prepared a set of target proteins and their N-glycan structures by collecting non-redundant (unique) N-glycan sequences containing at least five carbohydrates from the PDB and randomly selecting one PDB entry for each unique sequence. We used the PDB glycan sequence and glycosylation site information to generate N-glycans structure models onto their parent protein structure (see Materials and methods for how we set up the benchmark set in detail).
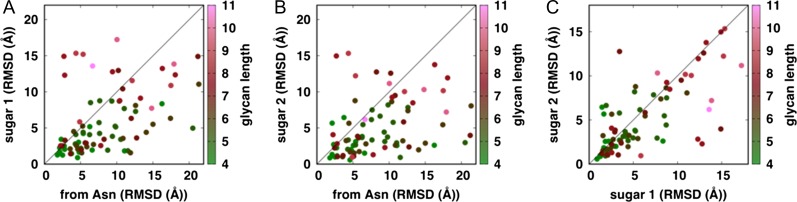
Similarities between the N-glycan models and their target PDB glycan structures were measured by the root-mean-squared deviation (RMSD) of glycans without superposition between the modeled and PDB glycan structures; i.e., the measured RMSD represents structural deviation of modeled glycan from its native one, resulting from differences in their conformation and orientation with respect to the target protein. We also examined if pre-existing glycan can improve the quality of the modeled glycan structures and orientations, as a small orientational difference of first one or two N-glycan residues could lead a big difference of the entire N-glycan orientation. We tested three approaches by building glycan models from (1) Asn residue (“from Asn”), (2) first sugar unit from pre-existing glycan in the PDB file (“sugar 1”) or (3) first two sugar units from pre-existing glycan in the PDB file (“sugar 2”). Figure 1 shows RMSD comparisons of the three approaches. The RMSD values of some N-glycan models are as high as 20 Å due to their orientational differences with respect to proteins, while the RMSD values after superposition between modeled and crystal N-glycan structure (i.e., only for glycan conformational differences) are mainly within 1−3 Å (Figure S1). Most RMSD values of “sugar 1” and “sugar 2” are much smaller than those of “from Asn”, indicating that the use of a pre-existing partial glycan can provide better orientation in the modeled glycans. This is also in agreement with our previous PDB glycan structure survey study (Jo et al. 2013) that the difficulties in finding the global orientation of the glycan with respect to the protein, even when the homologous N-glycan templates are present, can be significantly alleviated when a partial glycan structure is available.
Fig. 1.
Benchmark result of Glycan Modeler. For 82 non-redundant N-glycan structures, glycan RMSDs were measured against actual crystal N-glycan structures. The length of each target N-glycan is represented by color. Note that the RMSD was calculated without superposition between crystal and modeled N-glycan structures, so the RMSD value includes the glycan conformational and orientational differences on the protein surface.
Benchmark test of Glycan Modeler against PDB deglycosylated protein set
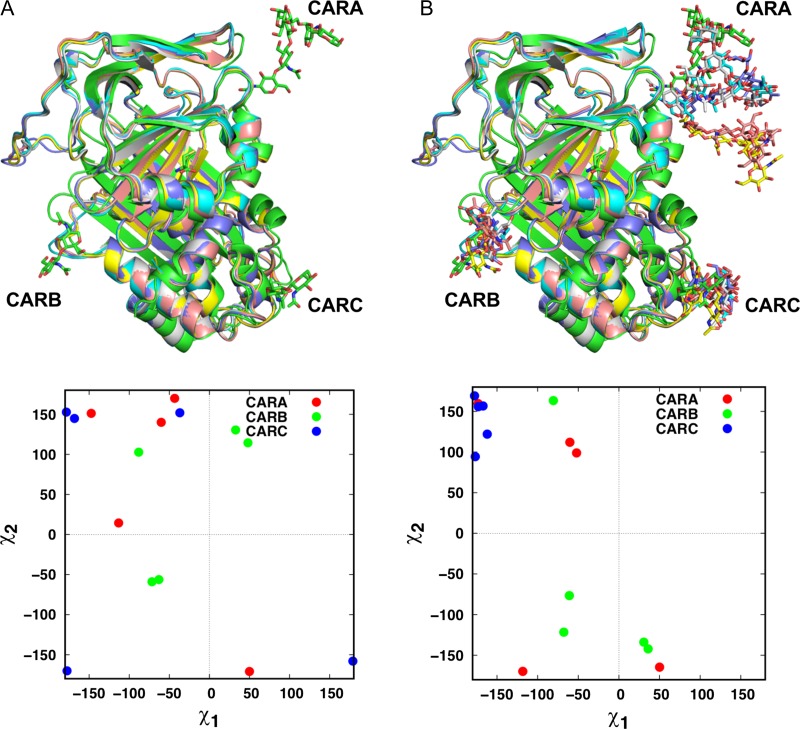
In our previous work, Lee et al. measured the local and global structural similarities between glycosylated and deglycosylated forms of identical proteins (GP/P pairs) in the PDB to investigate the impact of N-glycosylation on protein structure (Lee and Qi et al. 2015). To examine if Glycan Modeler is applicable to in silico glycosylation on a deglycosylated form of proteins, we performed another benchmark test using the same GP/P pairs. We discarded the pairs containing only monosaccharide, resulting in a total of 1,543 GP/P pairs. Using Glycan Modeler, we generated N-glycan models on the deglycosylated proteins based on the glycan sequence and glycosylation site information from the paired glycoproteins. Since the pairs often did not have the exactly same sequences, the glycosylation sites were determined based on the aligned structures of GP/P pairs by TM-align (Zhang and Skolnick 2005). Figure 2 shows the examples of predicted N-glycans on the crystal structures of a target deglycosylated protein (human alpha 1-antitrypsin (Engh et al. 1989)). In the deglycosylated protein structures, the side chains of Asn residues at the glycosylation sites have different orientations even though the structures come from an identical protein (Figure 2A). Because Glycan Modeler determines the orientations by randomly searching the torsional angle space of Asn side chain with an energy cutoff, predicted χ1 and χ2 of target Asn residues can be diverse, as shown in Figure 2B. Nonetheless, for the 1,543 GP/P pairs, all N-glycan chains were successfully generated without structural/chirality errors and bad contacts with their parent proteins, demonstrating the universal applicability of Glycan Modeler in generating glycan structures regardless of pre-existing glycans.
Fig. 2.
An example of in silico glycosylation on deglycosylated protein targets. (A) Green is a glycosylated protein (PDB: 7API) and others are deglycosylated proteins (PDB: 1QLP, 2QUG, 3CWL, 3CWM, 3NE4) in the PDB. On the left bottom, the scatter plot shows the distribution of χ1 and χ2 of target Asn residues of deglycosylated proteins. (B) The N-glycan models were generated on deglycosylated proteins with the identical glycan sequences and glycosylation sites. The χ1 and χ2 of target Asn residues after the modeling are displayed as scatter plots.
Simulation study of HIV envelope protein with glycans from PDB and Glycan Modeler
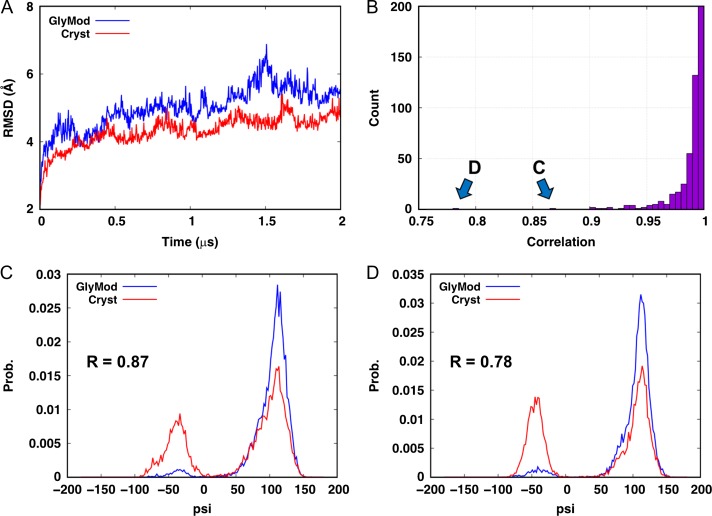
To further examine the reliability of modeled glycan structures, we performed MD simulations of two fully glycosylated HIV envelope trimer systems. One system (Scryst) was built based on all 66 glycan structures directly taken from the crystal structure (PDB:5FYL (Stewart-Jones et al. 2016)) (see the next section for system building in detail). In the other system (Sglymod), all glycan structures were removed and modeled by Glycan Modeler using the same sequence as those in the crystal structure. For a sufficient sampling of glycan conformations, each system was simulated for 2 μs using Anton 2 (Shaw et al. 2014) (see Materials and methods for simulation details; see also Supporting Movies, HIVenv_cryst.mpg and HIVenv_glymod.mpg). As shown in Figure 3A, the RMSDs of the envelope trimers reached a comparable value after 2 μs (i.e., the average RMSDs during the last 1 μs are 4.59 ± 0.24 Å (Scryst) and 5.42 ± 0.40 Å (Sglymod)), indicating the protein is stable in both systems.
Fig. 3.
Simulations of the HIV envelope trimer. (A) Backbone RMSDs of the protein. (B) Distribution of the linear correlation between the glycosidic dihedrals in the two MD systems. (C, D) Distributions of the ψ dihedrals that have a correlation of less than 0.9.
To compare the conformations of the glycans, we calculated the φ, ψ dihedrals of all 243 glycosidic linkages in each system. The distributions of these dihedrals for the same glycosidic linkage in both systems were compared using the Pearson correlation coefficient, i.e., if the two systems sample very similar dihedral distributions, the coefficient would be close to 1. Only two out of the 486 dihedrals have correlations less than 0.9 (Figure 3B), and both are the ψ dihedral in the Manβ(1→4)GlcNAc linkage. The distributions of these two dihedrals show that the deviations mostly come from minor populations at –60° (Figure 3C, D). To examine the preference of this dihedral, we searched for all Manβ(1→4)GlcNAc linkages in the PDB using GFDB (Jo and Im 2013) and found that ψ indeed has a minor population (~2.5%) around -60° (Figure S2), which may require much longer simulations to reach sampling convergence in the context of glycoprotein. Taken together, these results demonstrate that there is no significant difference in the torsion angle distributions of glycosidic linkages between crystal and modeled glycans.
Examples of practical applications of Glycan Modeler
In this section, we describe the representative examples (Figure 4) to illustrate practical applications of Glycan Reader & Modeler for glycoconjugate modeling and simulation with the following four video demos in CHARMM-GUI (http://www.charmm-gui.org/demo): (1) solution system building of the HIV envelope protein, (2) modeling N-glycans on immunoglobulin G1 Fc, (3) membrane system building of a cholera toxin B and ganglioside GM1 complex, and (4) solution system building of a heparin molecule. It should be noted that Glycan Reader & Modeler is embedded in PDB Reader, which makes Glycan Reader & Modeler available in most other CHARMM-GUI modules such as Solution Builder, Membrane Builder (Jo et al. 2007; 2009; Wu et al. 2014; Lee et al. 2019), HMMM Builder (Qi et al. 2015), Nanodisc Builder (Qi et al. 2019), Micelle Builder (Cheng et al. 2013), and PBEQ-Solver (Im et al. 1998; Jo et al. 2008).
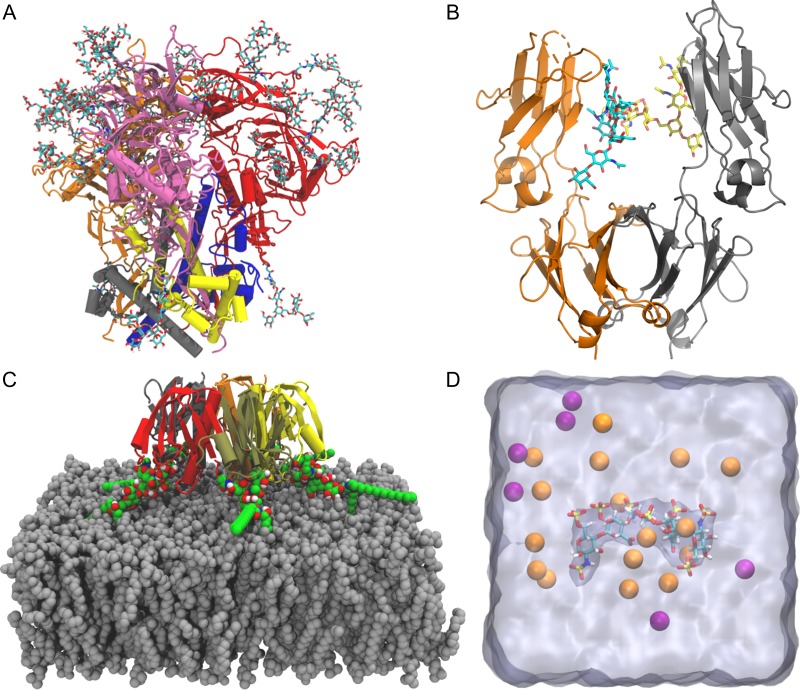
Fig. 4.
Practical applications of Glycan Reader & Modeler. (A) HIV envelope protein trimer of heterodimer with N-glycans. Each heterodimer is composed of gp120 (gray, blue, and yellow) paired with gp41 (orange, red, and pink, respectively). (B) Immunoglobulin G Fc dimer (orange and gray) with N-glycans. (C) Cholera toxin B and ganglioside GM1 pentameric complex shown with each protein segment colored differently. GM1 glycolipids are shown as green sphere. A membrane bilayer (gray) is composed of DMPC. (D) Heparan sulfate pentasaccharide surrounded by K+ (orange) and Cl– ions (purple). Water box is shown as a translucent surface.
Solution system building of the HIV envelope protein: To build a solution system of a glycoprotein like the HIV envelope protein (PDB:5FYL (Stewart-Jones et al. 2016)), users can use Solution Builder. The PDB contains a closed prefusion structure of the HIV envelope protein (Figure 4A). The envelope protein is synthesized as a gp160 precursor and processed into a trimer of heterodimer consisting of glycoproteins gp120 and gp41 that are heavily glycosylated and mediate receptor binding and membrane fusion. Note that the gp41 transmembrane domain is truncated in this structure and thus not included in this example. In addition to the envelope protein, this PDB file contains antibodies PGT122 and 35O22 that interact with the envelope protein. In this system building example, these antibodies are excluded for convenience. PDB:5FYL contains only one copy of the trimeric envelope protein, so users need to generate the remaining two copies during the PDB manipulation step. One can follow the step-by-step procedure in Section 1 of Supplementary Data to build a solution system of the HIV envelope protein.
Modeling N-glycans immunoglobulin G1 Fc Immunoglobulin G1 (IgG1) is a subclass of human serum antibodies and is the most widely used platform for developing therapeutic monoclonal antibodies (Wei et al. 2017). The antibody-mediated effector functions require binding of the crystallizable fragment (Fc) of IgG1 to Fc γ receptors that are expressed on the surface of recruited cells. IgG1 Fc is a symmetric homodimer and an N-glycan is attached to Asn297 in each chain. Our recent simulation study reports that changes in N-glycan composition alter the conformational ensembles of C’E loop and Cγ2- Cγ3 orientation in Fc chains, eventually affecting the binding affinity to its Fc receptor (Lee and Im 2017). The results also suggest that computational optimization of Fc N-glycans can guide engineering of IgG1-based antibodies for their better efficiency. In this example, a modified PDB file (see Section 2 of Supplementary Data) that contains only the first N-acetylglucosamine (GlcNAc) on chain B is used to build the original N-glycans in both chains using Glycan Reader & Modeler (Figure 4B).
Membrane system of a cholera toxin B and ganglioside GM1 complex: Cholera toxin (CT) produced by Vibrio cholerae is an AB5 toxin with a single enzymatic A subunit and a pentameric B subunit (CTB). CTB can bind to ganglioside GM1 receptors in the plasma membrane for CT’s entry to the cell. PDB:3CHB (Merritt et al. 1998) has five CTB subunits and associated GM1 carbohydrates with no ceramide lipid part. In addition, two GM1 glycan structures do not have β-glucose (Glc) at the reducing end. This example illustrates how to build a complete CTB:GM1 pentameric complex using Glycan Reader & Modeler and to insert the ceramide lipid portions into a membrane bilayer using Membrane Builder (Jo et al. 2009; Wu et al. 2014) (Figure 4C); see Section 3 of Supplementary Data for the detailed steps. Note that it is relatively easy to move one acyl chain of GM1 initially located outside the membrane into the bilayer during the equilibration.
Solution system of a heparin molecule: PDB:1AXM (DiGabriele et al. 1998) contains a heparin molecule complexed with a fibroblast growth factor. Heparin is a linear polysaccharide sulphated on alternating IdoA (l-iduronic acid) and GlcNS (6-O, N-sulfonated glucosamine), which can modulate endothelial cell proliferation and migration (Giraux et al. 1998). One can build a solution system only with this heparin molecule using Solution Builder. However, this example illustrates how to use Glycan Reader & Modeler to build a carbohydrate-only solution system (Figure 4D); see Section 4 of Supplementary Data for the detailed steps.
Conclusions
We have described the development of a new CHARMM-GUI module, Glycan Modeler that provides the functionality of in silico N-/O-glycosylation onto the target protein and carbohydrate-only system building. Our benchmark tests show that the Glycan Modeler framework can be efficiently used to model multiple glycan structures onto the target protein with or without pre-existing glycan structures. We also carried out the 2-μs MD simulations of a fully glycosylated HIV envelope protein to compare the structural properties of glycan structures modeled by Glycan Modeler to those from the crystal structure. The simulation results show no significant difference in glycan conformations, demonstrating the reliability of the glycan structure models generated by Glycan Modeler for molecular simulation studies. In addition to the benchmark evaluations, we described practical representative examples with video demos (http://www.charmm-gui.org/demo) to illustrate how to build complex glycoconjugate systems using Glycan Reader & Modeler.
Glycan Reader & Modeler can handle the glycan modeling with all kinds of glycan residues described in Symbol Nomenclature for Glycans (SNFG) (Varki et al. 2015) through addition/deletion/changes of sugar types, chemical modifications, glycosidic linkages, and anomeric states. Furthermore, the method allows the generation of simulation systems and inputs of user-defined glycoconjugate systems as it is integrated into other CHARMM-GUI modules such as Solution Builder or Membrane Builder. We hope that Glycan Reader & Modeler can become a useful tool enabling researchers to easily carry out innovative glycan modeling and simulation studies to acquire novel insight into structures, dynamics, and underlying mechanisms of complex glycan-related biological systems.
Materials and methods
N-/O-glycan modeling
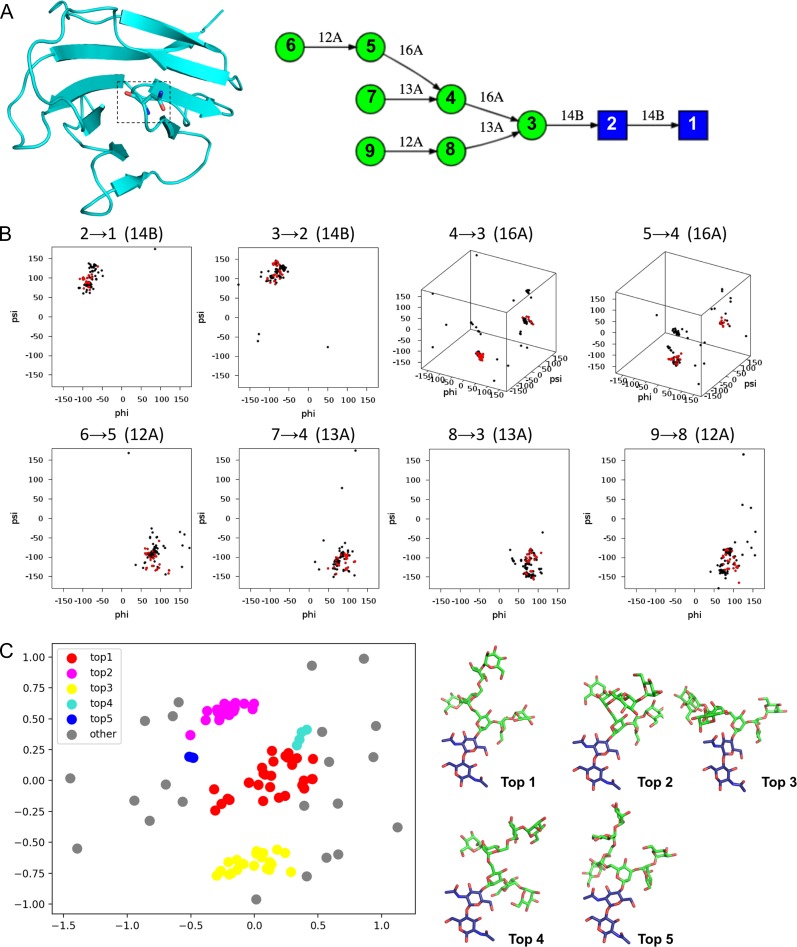
Glycan Modeler uses PDB glycan structures as templates for glycan structure modeling. To identify the relevant template structures, Glycan Modeler starts with Glycan Fragment Database (GFDB) searches between a query glycan sequence (for each glycosylation site, Figure 5A) and the PDB glycan structures in the database (Jo and Im 2013). PDB glycan structures containing the query glycan sequence are all searched in this step (Figure 5B). Glycosidic torsion angle based clustering (Jo and Im 2013) is performed to generate five representative glycan structures among the searched glycans (Figure 5C). Briefly, the clustering algorithm is composed of the following steps: (1) construction of the pairwise distance matrix where a distance is defined as the torsion angle difference between the same glycosidic linkages for all-to-all template structure pairs, (2) identification of the first cluster determined by the maximum number of neighbors within a cutoff radius of 30°, (3) identification of the second cluster after excluding the first cluster members, and (4) repeat of the clustering analysis for third, fourth and fifth clusters. After the clustering analysis, representative glycan models are generated by CHARMM using average glycosidic torsion angles from each cluster (Figure 5C). If an uploaded protein structure contains pre-existing glycan structure(s) and the pre-existing glycan is the part of a query glycan sequence, Glycan Modeler maps the pre-existing glycan onto the glycan models to conserve the coordinates of the pre-existing glycan structures. If there is no match in GFDB searches for a given glycan sequence, its structure is generated based on the internal coordinate (IC) information in the CHARMM carbohydrate force field (Guvench et al. 2009).
Fig. 5.
Illustration of glycan modeling in Glycan Modeler. (A) An example of a query glycan sequence and uploaded protein structure (PDB ID: 1CDB). Target glycosylation site (stick) and query glycan sequence were obtained from the identical protein with a glycan structure, PDB ID:1GYA (human CD2). (B) GFDB database search for glycan fragments that contain the query glycan sequence. For the glycan sequence in (A), 112 glycan fragments were identified. Torsion angles of all glycosidic linkages (ϕ, ψ, and ω) are used for the cluster determination. Red dots belong to the largest cluster based on a clustering analysis that is briefly described in Method. (C) Visualization of top five clusters of glycan fragments through a multi-dimensional scaling algorithm in a scikit-learn python module (Pedregosa et al. 2011).
Glycan conformation sampling algorithm
Glycan Modeler aims to build glycan models onto glycosylation sites of a target protein within a reasonable computational time without structural errors and bad contacts between glycan models and the protein. To achieve this goal, Glycan Modeler conducts conformational sampling by rigid-body rotations of each glycan chain and evaluates the protein–glycan interaction energy using the CHARMM force field.
Figure S3 shows the overall procedure of the conformation sampling. (1) The protein structure is fixed except glycosylated Asn residue(s). The all missing atoms in the protein structures are generated using the IC information in the CHARMM force field and remains to be flexible during the orientation search. (2) The glycan structure from the first GFDB cluster is attached to each glycosylation site (or onto pre-existing glycans) and is restrained by BESTFIT harmonic restraints (to maintain the overall glycan structure) and improper restraints (to maintain proper chirality of each sugar). (3) 500 iterations of orientation search are performed for each glycan chain to find an initial orientation. The orientation search is accomplished by changing torsional angles (ϕ and ψ angles) of a rotatable glycosidic linkage; the selection of a rotatable glycosidic linkage is elaborated below. (4) The number of bad contacts between the glycan and the protein is counted using a heavy atom distance cutoff of 2.5 Å. If there are less than five bad contacts, a short energy-minimization (10 steepest descent steps) is performed to further avoid bad contacts. Otherwise, the orientation trial is rejected and proceeds to the next iteration. Note that, to reduce the computational time, a bad contact checkup is performed before the minimization. (5) After the energy minimization in (4), if the glycan–protein interaction energy is lower than the previous interaction energy, the glycan orientation is accepted, and IC information of the accepted glycan orientation is stored. (6) After the initial orientation search in (3)–(5), another iteration of orientation search is performed to find lower energy orientation with the same methods described in (4) and (5). In any step, if the glycan–protein interaction energy is lower than a cutoff value (60 kcal/mol per glycan chain, which was empirically determined in this study), the orientation search is terminated and Glycan Modeler provides the accepted structure. Otherwise, the procedure goes to (6). The maximum number of cycles for a given glycosylation site is 500. If the procedure fails to find a glycan orientation that satisfies the given criteria, the procedure goes to (6) with the glycan structure from the second GFDB cluster. If the procedure fails to find a glycan orientation that satisfies the given criteria even with all five glycan cluster structures, the minimum-energy glycan orientation among all the searched glycan orientations is provided. (7) The accepted (or minimum-energy) glycan structure(s) in all glycosylation sites are further minimized only with the improper dihedral restraints. In this way, inherent protein–glycan or glycan–glycan bad contacts due to the GFDB average structure are removed without BESTFIT restraints.
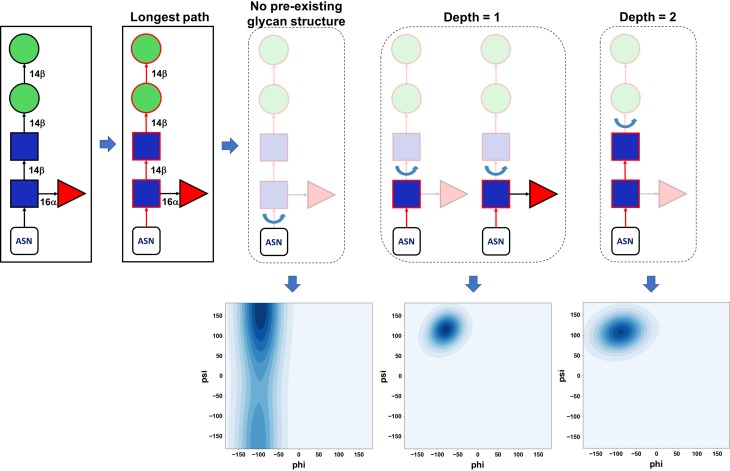
In (3) and (6), Glycan Modeler only changes ϕ and ψ torsion angles of a selected glycosidic linkage (even for 1–6 linkages) with preservation of other glycosidic linkages to maintain the glycan structures from GFDB. Figure 6 illustrates our selection scheme using an example of a query glycan sequence, Manβ4Manβ4GlcNAcβ4(Fucα6)GlcNAcβ-Asn. When the query glycan has branches, the longest path on the glycan sequence is first determined (red in Figure 6) and then a rotatable glycosidic linkage is defined as a linkage between the residues with and without coordinates on the longest path. For the glycosylation site with no pre-existing glycan structure, the glycosidic linkage to protein (e.g., CG–ND2–C1–O5 (ϕ) and CB–CG–ND2–C1 (ψ) for N-linked glycan) is selected for torsion angle variations. In the case where the glycosylation site contains a pre-existing glycan structure, the rotatable linkage is defined as the one between the existing glycan and model glycan, as shown in Figure 6. If the depth is greater than two in this exemplary case, Glycan Modeler generates the structure only using IC information in the CHARMM carbohydrate force field. Note that all atomic coordinates in the pre-existing glycan structures are conserved. For each (3) and (6) step, the torsion angles are randomly selected within the ϕ/ψ distribution in the PDB glycan structures of the selected glycosidic linkage type (Figure 6). We use a kernel density estimation algorithm (Pedregosa et al. 2011) to fit the PDB ϕ/ψ distribution and randomly generate the torsional values within the distribution.
Fig. 6.
Rotatable glycosidic linkage. Graphical illustration of rotatable glycosidic linkage determination depending on the extent of a pre-existing glycan structure in a protein. After the longest path on the glycan sequence at each glycosylation site is identified (red), a rotatable glycosidic linkage is defined as a linkage between residues with and without coordinates on the longest path. If there are no coordinates only for the last glycans (i.e., the depth more than two in this case), they are generated simply by IC information in the CHARMM force field and has no rotation during the modeling step. Note that all glycosidic linkages in the pre-existing glycan structure are fixed. Random glycosidic torsion angles are generated based on glycosidic torsion angle populations from GFDB by using the kernel density estimation algorithm in a scikit-learn python module (Pedregosa et al. 2011).
In some cases where a protein structure does not contain a pre-existing glycan for a certain glycosylation site, the glycosylation site can be buried inside the protein and thus does not have proper side chain orientation for glycan modeling. To handle such cases, Glycan Modeler randomly changes the χ angle of the glycosylation site. For an N-linked glycan, the χ2 (CA-CB-CG-OD1) angle is changed, while the χ1 (N-CA-CB-OG/OG1) angle is changed for O-linked glycans.
Web implementation in CHARMM-GUI: glycoprotein modeling
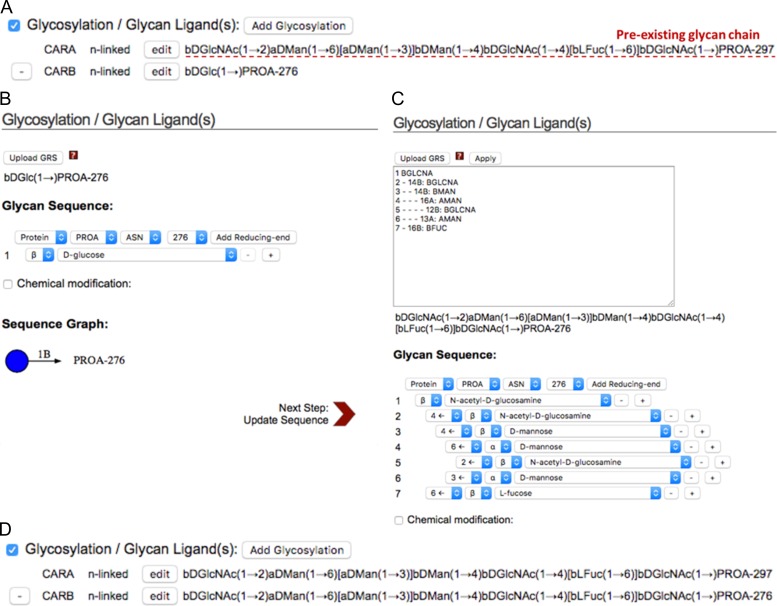
Glycan Modeler is integrated into Glycan Reader in CHARMM-GUI (Glycan Reader & Modeler). Glycan Reader & Modeler displays the detected carbohydrate chains in a PDB entry or in an uploaded PDB file and allows users to select the glycan chains of interest (Park et al. 2017). The selected glycans are listed and their sequences are shown in CASPER sequence representation (Lundborg and Widmalm 2011) under the “Glycosylation/Glycan Ligand(s)” category on the PDB manipulation page (Figure 7A). As described in our previous work (Park et al. 2017), an edit button next to each glycan chain enables users to edit a specific glycan chain on the web interface. Specifically, CHARMM-GUI displays a new popup window that allows users to add, remove, or change sugar types and anomeric states, glycosidic linkages, glycosylation types, and chemical modifications (Figure 7B). The new features of Glycan Reader & Modeler are (1) availability for in silico glycosylation on any Asn/Ser/Thr residues on the protein (with or without pre-existing glycans), (2) glycan modeling using GRS (Glycan Reader sequence) format (Figure 7C), and (3) addition of a sugar at the reducing end of glycan chain. For example, when users click the “Add Glycosylation” button, a new glycan chain is listed at the bottom of the Glycosylation/Glycan Ligand(s) category (Figure 7A). On the new popup window displayed after clicking the “edit” button, users can specify its sequence and target glycosylation site (Figure 7B) or GRS format (Figure 7C) can be used to specify a glycan sequence. The selected glycan is updated under the Glycosylation/Glycan Ligand(s) category (Figure 7D).
Fig. 7.
Snapshots of Glycan Reader & Modeler. (A) The glycans that exist in a PDB entry or in an uploaded PDB file are listed and their sequences are shown in CASPER sequence representation (Lundborg and Widmalm 2011) under the Glycosylation/Glycan Ligand(s) category on the PDB manipulation page. In silico glycosylation can be applied using the “Add Glycosylation” button. After clicking this, a new glycan chain is listed at the bottom of existing glycan structures. (B) A new popup window is displayed after clicking the “edit” button, so users can build their glycan model by selection of the glycosylation site, as well as additions, deletions, and changes of sugar residues. (C) Users can also use GRS (Glycan Reader sequence) format for a glycan sequence. (D) The CARB glycan chain in (A) is then updated based on the input GRS format in (C).
Web implementation in CHARMM-GUI: carbohydrate-only structure modeling
The carbohydrate-only solution system can be generated by selecting “Glycan Only System” option in the main page of Glycan Reader & Modeler. In the next page, a new web interface is provided, allowing users to build a glycan sequence of their interest. Using the sequence information provided by users, Glycan Reader & Modeler generates CHARMM input files to model the glycan structure, and the glycan model structure is generated using the IC information in the CHARMM carbohydrate force field. The generated glycan model is then solvated with ions (upon user’s selection) following the standard building procedures in CHARMM-GUI Solution Builder. The glycan system, topology and parameter files, simulation inputs for various MD simulation programs (based upon user’s selection) (Lee et al. 2016), as well as all the CHARMM input files used to generate the glycan system are provided as a downloadable single tar file (“download.tgz”).
Video demos of Glycan Reader & Modeler
In CHARMM-GUI Video Demo (http://www.charmm-gui.org/demo), we created the following four Glycan Reader & Modeler demos: (1) solution system building of the HIV envelop protein using PDB:5FYL (Stewart-Jones et al. 2016), (2) modeling N-glycans on a Fc heterodimer of immunoglobulin G1 using PDB:5TPS (Wei et al. 2017), (3) membrane system building of a cholera toxin B and ganglioside GM1 complex using PDB:3CHB (Merritt et al. 1998), and (4) solution system building of a heparin molecule. These examples are further elaborated in Results and discussion.
Preparation of non-redundant N-glycan structure set from PDB
PDB structures having resolution better than 3 Å were obtained from the RCSB Protein Data Bank (as of December 2017). Glycan Reader (Jo et al. 2011) was used to identify PDB entries with at least one N-glycan chain, resulting in 11,844 PDB entries having 28,364 N-linked glycan structures in total. All the N-glycan structures were categorized based on the glycan sequences to prepare the unique glycan sequences. For each unique glycan sequence having more than 5 PDB entries, we randomly selected one PDB entry as a target. The unique glycan sequences with glycan length smaller than four were discarded to avoid small glycans. Consequently, the benchmark set was composed of 82 target N-glycans and parent glycoproteins. Each target glycoprotein has one specific target glycan to measure the performance, although target glycoproteins can have multiple glycans.
MD simulations
MD simulations of a fully glycosylated HIV envelope trimer (from PDB:5FYL or using Glycan Modeler) were performed using the CHARMM36m force field (Huang et al. 2017) and NAMD 2.12 (Phillips et al. 2005) with a time step of 2 fs. Proteins were solvated in TIP3P cubic boxes (~160 Å in each side) with 150 mM KCl, yielding 375,305 atoms (a system based on PDB:5FYL) and 380,451 atoms (a system based on Glycan Modeler). The non-bonded interactions were smoothly switched off at 10–12 Å with a force-switching function (Steinbach and Brooks 1994). Long-range electrostatic forces were calculated with the particle mesh Ewald algorithm (Essmann et al. 1995). Temperature was maintained at 303.15 K using Langevin dynamics with a friction coefficient of 1 ps–1. Pressure was controlled at 1 bar using the Langevin-piston method with a piston period of 50 fs and a piston decay of 25 fs (Feller et al. 1995). After equilibration with NAMD, the MD systems were transferred to Anton2 (Shaw et al. 2014) for 2-μs production runs with a time step of 2 fs. The non-bonded interactions were cut off at 10 Å. Long-range electrostatic interactions were calculated with the u-series approach (Shaw et al. 2014). Pressure and temperature were controlled at 1 bar and 303.15 K using the semi-isotropic MTK barostats and Nosé–Hoover thermostats under the Multigrator framework (Lippert et al. 2013). Trajectory analysis was performed with CHARMM (Brooks et al. 2009) and VMD (Humphrey et al. 1996). Trajectories for all 2 μs simulation were used to calculate the backbone RMSD and torsion angle distributions.
Supplementary Material
Acknowledgements
The Anton machine at PSC was generously made available by D.E. Shaw Research. We thank Seonghoon Kim for preforming Anton simulations.
Funding
This work was supported by the National Science Foundation (DBI-1707207), XSEDE Resources (MCB070009) and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (NRF-2017R1E1A1A01077717). Anton computer time was provided by the National Center for Multiscale Modeling of Biological Systems (MMBioS) through Grant P41GM103712-S1 from the National Institutes of Health and the Pittsburgh Supercomputing Center (PSC).
Conflict of interest statement
None declared.
References
- Allinger NL, Chen KH, Lii JH, Durkin KA. 2003. Alcohols, ethers, carbohydrates, and related compounds. I. The MM4 force field for simple compounds. J Comput Chem. 24:1447–1472. [DOI] [PubMed] [Google Scholar]
- Apweiler R, Hermjakob H, Sharon N. 1999. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1473:4–8. [DOI] [PubMed] [Google Scholar]
- Arda A, Jimenez-Barbero J. 2018. The recognition of glycans by protein receptors. Insights from NMR spectroscopy. Chem Commun (Camb). 54:4761–4769. [DOI] [PubMed] [Google Scholar]
- Arthur EJ, Brooks CL. 2016. Parallelization and improvements of the generalized born model with a simple sWitching function for modern graphics processors. J Comput Chem. 37:927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohne A, Lang E, von der Lieth CW. 1999. SWEET – WWW-based rapid 3D construction of oligo- and polysaccharides. Bioinformatics. 15:767–768. [DOI] [PubMed] [Google Scholar]
- Bohne-Lang A, von der Lieth CW. 2005. GlyProt: In silico glycosylation of proteins. Nucleic Acids Res. 33:W214–W219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers KJ, Chow DE, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti FD et al. 2006. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. SC 2006 Conference, Proceedings of the ACM/IEEE. p. 43-43.
- Brooks BR, Brooks CL 3rd, Mackerell AD Jr., Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S et al. 2009. CHARMM: The biomolecular simulation program. J Comput Chem. 30:1545–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MP, Royle L, Rudd PM. 2015. GlycoBase and autoGU: Resources for interpreting HPLC-glycan data. Methods Mol Biol. 1273:17–28. [DOI] [PubMed] [Google Scholar]
- Case DA, Cheatham TE 3rd, Darden T, Gohlke H, Luo R, Merz KM Jr., Onufriev A, Simmerling C, Wang B, Woods RJ. 2005. The Amber biomolecular simulation programs. J Comput Chem. 26:1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Jo S, Lee HS, Klauda JB, Im W. 2013. CHARMM-GUI micelle builder for pure/mixed micelle and protein/micelle complex systems. J Chem Inf Model. 53:2171–2180. [DOI] [PubMed] [Google Scholar]
- Collins BE, Paulson JC. 2004. Cell surface biology mediated by low affinity multivalent protein-glycan interactions. Curr Opin Chem Biol. 8:617–625. [DOI] [PubMed] [Google Scholar]
- Curatolo W. 1987. Glycolipid function. Biochim Biophys Acta. 906:137–160. [DOI] [PubMed] [Google Scholar]
- Danne R, Poojari C, Martinez-Seara H, Rissanen S, Lolicato F, Rog T, Vattulainen I. 2017. doGlycans-tools for preparing carbohydrate structures for atomistic simulations of glycoproteins, glycolipids, and carbohydrate polymers for GROMACS. J Chem Inf Model. 57:2401–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGabriele AD, Lax I, Chen DI, Svahn CM, Jaye M, Schlessinger J, Hendrickson WA. 1998. Structure of a heparin-linked biologically active dimer of fibroblast growth factor. Nature. 393:812–817. [DOI] [PubMed] [Google Scholar]
- Dong C, Lee J, Kim S, Lai W, Webb EB 3rd, Oztekin A, Zhang XF, Im W. 2018. Long-ranged protein-glycan interactions stabilize von willebrand factor A2 domain from mechanical unfolding. Sci Rep. 8:16017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwek RA. 1996. Glycobiology: Toward understanding the function of sugars. Chem Rev. 96:683–720. [DOI] [PubMed] [Google Scholar]
- Eastman P, Friedrichs MS, Chodera JD, Radmer RJ, Bruns CM, Ku JP, Beauchamp KA, Lane TJ, Wang L-P, Shukla D et al. 2013. OpenMM 4: A reusable, extensible, hardware independent library for high performance molecular simulation. J Chem Theory Comput. 9:461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Ghazal R, Yin X, Johns SC, Swanson L, Macal M, Ghosh P, Zuniga EI, Fuster MM. 2016. Glycan sulfation modulates dendritic cell biology and tumor growth. Neoplasia. 18:294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelsen SB, Hansen PI, Perez S. 2014. POLYS 2.0: An open source software package for building three-dimensional structures of polysaccharides. Biopolymers. 101:733–743. [DOI] [PubMed] [Google Scholar]
- Engh R, Lobermann H, Schneider M, Wiegand G, Huber R, Laurell CB. 1989. The S variant of human alpha 1-antitrypsin, structure and implications for function and metabolism. Protein Eng. 2:407–415. [DOI] [PubMed] [Google Scholar]
- Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. 1995. A smooth particle mesh Ewald method. J Chem Phys. 103:8577–8593. [Google Scholar]
- Feller SE, Zhang YH, Pastor RW, Brooks BR. 1995. Constant-pressure molecular-dynamics simulation – The Langevin Piston Method. J Chem Phys. 103:4613–4621. [Google Scholar]
- Giraux JL, Matou S, Bros A, Tapon-Bretaudiere J, Letourneur D, Fischer AM. 1998. Modulation of human endothelial cell proliferation and migration by fucoidan and heparin. Eur J Cell Biol. 77:352–359. [DOI] [PubMed] [Google Scholar]
- Guvench O, Hatcher ER, Venable RM, Pastor RW, Mackerell AD. 2009. CHARMM additive all-atom force field for glycosidic linkages between hexopyranoses. J Chem Theory Comput. 5:2353–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamark C, Berntsson RP, Masuyer G, Henriksson LM, Gustafsson R, Stenmark P, Widmalm G. 2017. Glycans confer specificity to the recognition of ganglioside receptors by botulinum neurotoxin A. J Am Chem Soc. 139:218–230. [DOI] [PubMed] [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Lindahl E. 2008. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 4:435–447. [DOI] [PubMed] [Google Scholar]
- Huang J, Rauscher S, Nawrocki G, Ran T, Feig M, de Groot BL, Grubmuller H, MacKerell AD Jr. 2017. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat Methods. 14:71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. 1996. VMD: Visual molecular dynamics. J Mol Graph. 14:33–38, 27-38. [DOI] [PubMed] [Google Scholar]
- Im W, Beglov D, Roux B. 1998. Continuum Solvation Model: Computation of electrostatic forces from numerical solutions to the Poisson-Boltzmann equation. Comput Phys Commun. 111:59–75. [Google Scholar]
- Im-Group 2011-2018. GlycanStructure.ORG.
- Imberty A, Perez S. 2000. Structure, conformation, and dynamics of bioactive oligosaccharides: Theoretical approaches and experimental validations. Chem Rev. 100:4567–4588. [DOI] [PubMed] [Google Scholar]
- Jo S, Im W. 2013. Glycan fragment database: A database of PDB-based glycan 3D structures. Nucleic Acids Res. 41:D470–D474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Kim T, Im W. 2007. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One. 2:e880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Lee HS, Skolnick J, Im W. 2013. Restricted N-glycan conformational space in the PDB and its implication in glycan structure modeling. PLoS Comput Biol. 9:e1002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Lim JB, Klauda JB, Im W. 2009. CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys J. 97:50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Qi Y, Im W. 2016. Preferred conformations of N-glycan core pentasaccharide in solution and in glycoproteins. Glycobiology. 26:19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Song KC, Desaire H, MacKerell AD Jr, Im W. 2011. Glycan Reader: Automated sugar identification and simulation preparation for carbohydrates and glycoproteins. J Comput Chem. 32:3135–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Vargyas M, Vasko-Szedlar J, Roux B, Im W. 2008. PBEQ-Solver for online visualization of electrostatic potential of biomolecules. Nucleic Acids Res. 36:W270–W275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Mori T, Kobayashi C, Matsunaga Y, Yoda T, Feig M, Sugita Y. 2015. GENESIS: A hybrid-parallel and multi-scale molecular dynamics simulator with enhanced sampling algorithms for biomolecular and cellular simulations. Wiley Interdiscip Rev Comput Mol Sci. 5:310–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner KN, Yongye AB, Tschampel SM, Gonzalez-Outeirino J, Daniels CR, Foley BL, Woods RJ. 2008. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J Comput Chem. 29:622–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kony D, Damm W, Stoll S, Van Gunsteren WF. 2002. An improved OPLS-AA force field for carbohydrates. J Comput Chem. 23:1416–1429. [DOI] [PubMed] [Google Scholar]
- Kuttel MM, Stahle J, Widmalm G. 2016. CarbBuilder: Software for building molecular models of complex oligo- and polysaccharide structures. J Comput Chem. 37:2098–2105. [DOI] [PubMed] [Google Scholar]
- Labonte JW, Adolf-Bryfogle J, Schief WR, Gray JJ. 2017. Residue-centric modeling and design of saccharide and glycoconjugate structures. J Comput Chem. 38:276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, Wei S, Buckner J, Jeong JC, Qi Y et al. 2016. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comput. 12:405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Im W. 2017. Effects of N-glycan composition on structure and dynamics of IgG1 Fc and their implications for antibody engineering. Sci Rep-Uk. 7:12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Jo S, Mukherjee S, Park SJ, Skolnick J, Lee J, Im W. 2015. GS-align for glycan structure alignment and similarity measurement. Bioinformatics. 31:2653–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Patel DS, Stahle J, Park SJ, Kern NR, Kim SH, Lee J, Cheng X, Valvano MA, Holst O et al. 2019. CHARMM-GUI membrane builder for complex biological membrane simulations with glycolipids and lipoglycans. J Chem Theory Comput 15:775–786. [DOI] [PubMed] [Google Scholar]
- Lee HS, Qi Y, Im W. 2015. Effects of N-glycosylation on protein conformation and dynamics: Protein Data Bank analysis and molecular dynamics simulation study. Sci Rep. 5:8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert RA, Predescu C, Ierardi DJ, Mackenzie KM, Eastwood MP, Dror RO, Shaw DE. 2013. Accurate and efficient integration for molecular dynamics simulations at constant temperature and pressure. J Chem Phys. 139:164106. [DOI] [PubMed] [Google Scholar]
- Lundborg M, Widmalm G. 2011. Structural analysis of glycans by NMR chemical shift prediction. Anal Chem. 83:1514–1517. [DOI] [PubMed] [Google Scholar]
- Lutteke T, Bohne-Lang A, Loss A, Goetz T, Frank M, von der Lieth CW. 2006. GLYCOSCIENCES.de: An Internet portal to support glycomics and glycobiology research. Glycobiology. 16:71R–81R. [DOI] [PubMed] [Google Scholar]
- Malik A, Ahmad S. 2007. Sequence and structural features of carbohydrate binding in proteins and assessment of predictability using a neural network. BMC Struct Biol. 7:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik A, Firoz A, Jha V, Ahmad S. 2010. PROCARB: A database of known and modelled carbohydrate-binding protein structures with sequence-based prediction tools. Adv Bioinformatics. 2010:436036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik A, Lee J, Lee J. 2014. Community-based network study of protein-carbohydrate interactions in plant lectins using glycan array data. PLoS One. 9:e95480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti R, Perez S, Arda A, Imberty A, Jimenez-Barbero J, Silipo A, Molinaro A. 2016. “Rules of Engagement” of protein-glycoconjugate interactions: A molecular view achievable by using nmr spectroscopy and molecular modeling. ChemistryOpen. 5:274–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt EA, Kuhn P, Sarfaty S, Erbe JL, Holmes RK, Hol WG. 1998. The 1.25 A resolution refinement of the cholera toxin B-pentamer: Evidence of peptide backbone strain at the receptor-binding site. J Mol Biol. 282:1043–1059. [DOI] [PubMed] [Google Scholar]
- Muthana SM, Campbell CT, Gildersleeve JC. 2012. Modifications of glycans: Biological significance and therapeutic opportunities. ACS Chem Biol. 7:31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagae M, Yamaguchi Y. 2012. Function and 3D structure of the N-glycans on glycoproteins. Int J Mol Sci. 13:8398–8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo K, Marth JD. 2006. Glycosylation in cellular mechanisms of health and disease. Cell. 126:855–867. [DOI] [PubMed] [Google Scholar]
- Park SJ, Lee J, Patel DS, Ma H, Lee HS, Jo S, Im W. 2017. Glycan Reader is improved to recognize most sugar types and chemical modifications in the Protein Data Bank. Bioinformatics. 33:3051–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V et al. 2011. Scikit-learn: Machine learning in Python. J Mach Learn Res. 12:2825–2830. [Google Scholar]
- Perez S, de Sanctis D. 2017. Glycoscience@Synchrotron: Synchrotron radiation applied to structural glycoscience. Beilstein J Org Chem. 13:1145–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. 2005. Scalable molecular dynamics with NAMD. J Comput Chem. 26:1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plimpton S. 1995. Fast parallel algorithms for short-range molecular-dynamics. J Comput Phys. 117:1–19. [Google Scholar]
- Pol-Fachin L, Rusu VH, Verli H, Lins RD. 2012. GROMOS 53A6GLYC, an improved GROMOS force field for hexopyranose-based carbohydrates. J Chem Theory Comput. 8:4681–4690. [DOI] [PubMed] [Google Scholar]
- Qi Y, Cheng X, Lee J, Vermaas JV, Pogorelov TV, Tajkhorshid E, Park S, Klauda JB, Im W. 2015. CHARMM-GUI HMMM builder for membrane simulations with the highly mobile membrane-mimetic model. Biophys J. 109:2012–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Jo S, Im W. 2016. Roles of glycans in interactions between gp120 and HIV broadly neutralizing antibodies. Glycobiology. 26:251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Lee J, Klauda JB, Im W. 2019. CHARMM-GUI Nanodisc builder for modeling and simulation of various nanodisc systems. J Comput Chem. 40:893–899. [DOI] [PubMed] [Google Scholar]
- Rabinovich GA, Toscano MA. 2009. Turning ‘sweet’ on immunity: Galectin-glycan interactions in immune tolerance and inflammation. Nat Rev Immunol. 9:338–352. [DOI] [PubMed] [Google Scholar]
- Shaw DE, Grossman JP, Bank JA, Batson B, Butts JA, Chao JC, Deneroff MM, Dror RO, Even A, Fenton CH et al. 2014. Anton 2: Raising the bar for performance and programmability in a special-purpose molecular dynamics supercomputer. Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis. New Orleans, Louisana: IEEE Press. p. 41–53.
- Steinbach PJ, Brooks BR. 1994. New spherical-cutoff methods for long-range forces in macromolecular simulation. J Comput Chem. 15:667–683. [Google Scholar]
- Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens AJ et al. 2016. Trimeric HIV-1-Env structures define glycan shields from Clades A, B, and G. Cell. 165:813–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta ES. 2003. The contribution of N-glycans and their processing in the endoplasmic reticulum to glycoprotein biosynthesis. Glycobiology. 13:77R–91R. [DOI] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Aebi M, Packer NH, Seeberger PH, Esko JD, Stanley P, Hart G, Darvill A, Kinoshita T et al. 2015. Symbol nomenclature for graphical representations of glycans. Glycobiology. 25:1323–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Cai H, Jin Y, Wang P, Zhang Q, Lin Y, Wang W, Cheng J, Zeng N, Xu T et al. 2017. Structural basis of a novel heterodimeric Fc for bispecific antibody production. Oncotarget. 8:51037–51049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods-Group 2005. –2018. GLYCAM Web.
- Wormald MR, Petrescu AJ, Pao YL, Glithero A, Elliott T, Dwek RA. 2002. Conformational studies of oligosaccharides and glycopeptides: Complementarity of NMR, X-ray crystallography, and molecular modelling. Chem Rev. 102:371–386. [DOI] [PubMed] [Google Scholar]
- Wu EL, Cheng X, Jo S, Rui H, Song KC, Davila-Contreras EM, Qi Y, Lee J, Monje-Galvan V, Venable RM et al. 2014. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J Comput Chem. 35:1997–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. 2005. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33:2302–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.