Abstract
Nontyphoidal Salmonella is a major contributor to the global burden of foodborne disease, with invasive infections contributing substantially to illnesses and deaths. We analyzed notifiable disease surveillance data for invasive nontyphoidal Salmonella disease (iNTS) in Queensland, Australia. We used Poisson regression to estimate incidence rate ratios by gender, age group, and geographical area over 2007–2016. There were 995 iNTS cases, with 945 (92%) confirmed by blood culture. Salmonella Virchow accounted for 254 (25%) of 1,001 unique iNTS isolates. Invasive NTS disease notification rates peaked among infants, during the summer months, and in outback Queensland where the notification rate (95% CI) was 17.3 (14.5–20.1) cases per 100,000 population. Overall, there was a 6,5% annual increase (p<0.001) in iNTS disease incidence. In conclusion, high iNTS rates among males, infants, and the elderly require investigation of household level risk factors for NTS infection. Controlling Salmonella Virchow infections is a public health priority.
Author summary
We identified increasing incidence of invasive infections due to nontyphoidal Salmonella in Queensland with particularly high rates of disease among males, infants, elderly people, and cases infected with Salmonella serotype Virchow. Salmonella serotypes Choleraesuis, Dublin, and Panama had the highest proportion of invasive isolates.
Introduction
Nontyphoidal Salmonella (NTS) infections are a serious public health concern globally. In high-income settings, NTS predominantly causes a self-limiting diarrhoeal illness with low case fatality risk [1]. NTS may also invade beyond the gastrointestinal tract and cause severe disease, including bacteremia and infection of other normally sterile sites referred to as invasive nontyphoidal Salmonella (iNTS) disease. Children under the age of five years, the elderly, those with comorbid illnesses, and the immunocompromised are at particular risk of developing iNTS disease [2,3]. Invasive disease caused by NTS is particularly common in low-resource settings with high prevalence of HIV, malaria, anemia, and malnutrition [4].
The case fatality risk of NTS bacteremia is as high as 19% in sub-Saharan Africa and 26% in Vietnam [2,5]. Due to its severity and health impact, there have been several attempts to estimate the global burden of iNTS disease since 2010. The study of Ao et al. estimated 3.4 million cases of iNTS disease resulting in 681,316 deaths annually [6]. Another study, which unlike Ao et al. excluded HIV-associated iNTS disease, estimated 596,824 cases of iNTS accounting for 63,312 deaths in 2010 [7]. However, both estimates are based on very few studies, therefore, additional data are needed to improve current estimates and obtain a more complete picture of the burden of disease.
In Australia, salmonellosis is one of the most important causes of foodborne diarrheal disease due to its increasing incidence and propensity to cause large outbreaks. Almost all states and territories have experienced increasing rates of NTS infection since 2000 [8]. Tropical areas of Australia experience rates amongst the highest in the industrialised world [9]. In 2016, 18,059 notifications of Salmonella infection were reported across Australia at a rate of 74.6 cases per 100,000 population, a 25% increase compared with the mean rate for the previous 5 years. In Queensland, the NTS notification rate was 99.2 cases per 100,000 population in 2016—one of the highest rates in Australia [10]. Recent studies of NTS in the Australian Capital Territory and Victoria found that iNTS comprised 1.6% [11] and 2.5% [12] of all NTS notifications with Salmonella Typhimurium being the most commonly reported serotype.
The epidemiology of iNTS disease in Queensland is poorly understood in terms of severity, risk factors, and the distribution of infecting serotypes. Understanding disease incidence and the geographical distribution of iNTS disease is important for effective control of foodborne diseases. In this study, we investigated the descriptive epidemiology of iNTS in Queensland to identify serotypes and high-risk populations to prioritise control measures.
Materials and methods
We used non-identifiable human illness data on Salmonella enterica notifications in Queensland from 2007 through 2016 to analyze trends in disease incidence. Queensland is one of eight Australian states and territories. We chose to analyze iNTS in Queensland due to high rates of salmonellosis overall, varied climate encompassing tropical, subtropical, hot arid, and warm temperate zones, and to include populations living in lower socio-economic areas who might be at greater risk for invasive bacterial infections. Area-level socio-economic indices identify remote parts of Queensland, Townsville, and Cairns as among the most disadvantaged in Australia [13].
Surveillance system
All Australian states and territories have public health legislation requiring doctors and pathology laboratories to notify any laboratory-confirmed case of salmonellosis [14]. State and territory health departments record details of notified cases on surveillance databases. It is estimated that that for each nontyphoidal Salmonella infection, there are 7 undiagnosed cases in the community [15] confirming that notification rates to the surveillance system underestimate disease incidence.
In Queensland, notifiable conditions are reported to Queensland Health and are held in the Notifiable Conditions System (NOCS). Data from NOCS and other databases are aggregated into a national database, the National Notifiable Diseases Surveillance System, operating since 1991 [10].
Data sources
We obtained non-identified Salmonella enterica notifications from NOCS including: ‘notification id,’ ‘person id,’ ‘age at onset,’ ‘onset year,’ ‘onset month,’ ‘diagnosis date,’ ‘collection date,’ ‘hospital and health service,’ ‘postcode,’ ‘locality,’ ‘Statistical Local Area (SLA),’ ‘sex,’ ‘test id,’ ‘specimen,’ ‘serotype,’ ‘subtype,’ and ‘site of infection.’
SLAs have been in use since 1984 as general purpose spatial units covering Australia without gaps or overlaps. There were 475 SLAs in Queensland, defined under The Australian Standard Geographical Classification [16]. In 2011, this system was replaced by The Australian Statistical Geography Standard (ASGS), which has seven hierarchical levels comprising in ascending order: Mesh Block, Statistical Areas Levels 1–4 (SA1-4), State and Territory, and Australia. Each level directly aggregates to the level above [17].
For the purposes of our analyses, SLAs were converted into 19 SA4s (Fig 1), the largest sub-State spatial units in the main structure of the ASGS. The conversion was done in several steps. Firstly, SLA was converted into SA2, a medium-sized area built up from whole SA1, and consequently into SA4 using the conversion file from the Australian Bureau of Statistics (ABS) which develops standard statistical geographies and frameworks [18]. Secondly, SLA codes, postcodes, and locations from NOCS were compared and basic checks were done. Lastly, for selected non-matching locations, manual mapping of SLAs and SA4s was performed using the ABS map [19].
Fig 1. Map of Queensland divided into statistical areas level 4.
We defined an NTS case as a notified infection in a resident of Queensland with a culture-confirmed NTS isolated from any source. If an isolate came from blood, cerebrospinal fluid, peritoneal fluid, pleural fluid, synovial fluid, bone, or other normally sterile site, it was considered as an iNTS case. An individual could meet the case definition more than once if a subsequent infection occurred >30 days after an episode of culture-confirmed NTS infection of a normally sterile site, hereafter referred to as a recurrent iNTS case. If an isolate came from stool, urine, vomitus, sputum, skin, soft tissue abscesses, and wounds, it was defined as a non-invasive NTS case. If an individual had Salmonella isolated from the gallbladder only, it was also considered as non-invasive infection due to possible persistence of carriage at that site [20]. Due to possible long-term Salmonella shedding in stool [21], we considered recurrent non-invasive NTS cases as those where the period between culture-confirmed episodes of salmonellosis was more than 6 months if caused by the same serotype, and 3 months if serotypes differed.
Cases of Salmonella enterica serotype Typhi and serotypes Paratyphi A, B, and C were excluded except for Salmonella Paratyphi B biovar Java which predominantly causes enterocolitis rather than paratyphoid fever. Salmonella Paratyphi B biovar Java infections were grouped with other NTS serotypes [22,23]. NTS cases residing or having been diagnosed in other states or territories of Australia or abroad and cases with missing information on age, gender, or collection site were excluded from the analysis. Specimens collected post-mortem were excluded from the analysis due to the possibility of bacterial translocation into normally sterile sites after death potentially resulting in misclassification of invasive disease.
We used the date a specimen was collected for all analyses as it was the closest available date to a person’s onset of illness. Phage type data were analyzed only for the two most common iNTS serotypes. In specimen analysis, all normally sterile sites from which iNTS specimens were collected from the same case were analyzed to demonstrate the most common infection site. Otherwise, for the purposes of all analyses, each case was registered once per episode of iNTS infection, regardless of the number of specimens or serotypes per iNTS case.
Data analyses
We calculated an invasiveness index as the proportion of invasive isolates to the total number of isolates recovered for each serotype. For serotype analysis and invasiveness index calculations, we used the number of unique iNTS isolates as some cases were infected by multiple different serotypes. To determine the effect of age group, gender, geographical area, time, and serotype on invasiveness, we calculated crude odds ratios (OR) and adjusted odds ratios (aOR) comparing invasive and non-invasive NTS infections. The final model was constructed using variables significantly associated with the outcome in the univariable analysis.
In addition, we calculated crude and adjusted incidence rate ratios (IRR) with associated 95% confidence intervals (CI) and p-values by age group, gender, geographical area, and time. Results with p ≤ 0.05 were considered significant. Chi squared tests were used to test for heterogeneity and trends in proportions, two sample t-tests to compare means, and Pearson rank correlations to determine the correlation between the variables and crude incidence rate. For the multivariable analysis of incidence rates, we used a Poisson model, confirming that there was very little overdispersion in the data. Rates of iNTS disease per 100,000 population were estimated using the data on the population residing in Queensland as of the June quarter for each year between 2007 and 2016, obtained from the ABS [24] as the offset in the model.
In the descriptive analysis, invasiveness analysis, and for IRR calculations, age was categorized into 11 age groups: 3 age groups (<1, 1–4, 5–9) until the age of 10 years and then into 10-year age groups until 80 years and over. In the multivariable analysis of incidence rates, ten age groups were included as the first two age groups were combined into one age group (0–4). This combination was necessary due to the unavailability of population data by age, gender, and location from 2007–2010. Age group 30 to 39 years of age was used as a reference category to highlight high-risk age categories. All analyses were performed using Stata version 14 (StataCorp, USA). ArcGIS v10.5 (ESRI, USA) was used to create a map of Australia and Queensland statistical divisions.
Research ethics
Ethics approval was obtained from the Australian National University Human Research Ethics Committee prior to the conduct of the study [protocol 2017/545]. The Queensland Health data custodian provided approval to access the notifiable disease data.
Results
There were 32,117 NTS cases reported over the 10-year period in Queensland. Of these, 153 (0.5%) cases were excluded due to following reasons: 136 (88.9%) cases resided or were diagnosed outside Queensland, seven (4.6%) had incomplete information on age and gender, five (3.3%) had isolates collected post-mortem, four (2.6%) had a non-specified specimen source, and one (0.7%) was incorrectly categorized as NTS. Of 31,964 NTS cases, 995 (3.1%) were identified as iNTS and further analyzed.
Among 995 iNTS cases, 13 were infected with two different serotypes per episode of salmonellosis, giving a total of 1008 unique iNTS isolates. Among 1,001 (99.3%) iNTS isolates serotyped, 254 (25.4%) were serotype Virchow, 203 (20.3%) serotype Typhimurium, and 74 (7.4%) serotype Aberdeen. The invasiveness index was 9.06% (254/2804) for serotype Virchow, 2.04% (203/9942) for serotype Typhimurium, and 6.11% (74/1212) for serotype Aberdeen (Table 1). The proportion of iNTS serotypes differed among statistical areas (S1 Table). The most commonly isolated invasive strain of Salmonella serotypes Virchow and Typhimurium were phage types 8 and 135A, respectively (S2 and S3 Tables). When compared to serotype Typhimurium, Salmonella serotypes Virchow and Aberdeen were associated with invasive disease (aOR, 4.7; 95% CI, 3.9–5.8; and aOR, 2.2; 95% CI, 2.2–3.9, respectively) when adjusted for age group and gender (Table 1). Time, geographical location, and season were not significant predictors for invasive disease. Overall, the odds ratios for invasive disease were highest among males, infants, the elderly (S4 Table), and those infected with Salmonella serotypes Choleraesuis, Dublin, and Panama. However, these serotypes represented a small proportion of all iNTS cases (Table 1).
Table 1. Distribution and invasiveness of most common iNTS serotypes in Queensland, 2007–2016.
| Serotype | No. (%) of iNTS isolates | No. (%) of non-iNTS isolates | Invasive-ness index* | aOR† | 95% CI | p-value | |
|---|---|---|---|---|---|---|---|
| VIRCHOW | 254 (25.37) | 2550 (8.76) | 9.06 | 4.74 | 3.90 | 5.76 | <0.001 |
| TYPHIMURIUM | 203 (20.28) | 9739 (33.44) | 2.04 | reference | |||
| ABERDEEN | 74 (7.39) | 1138 (3.91) | 6.11 | 2.90 | 2.19 | 3.85 | <0.001 |
| ENTERITIDIS | 45 (4.5) | 1279 (4.39) | 3.40 | 1.63 | 1.17 | 2.27 | <0.001 |
| SAINTPAUL | 44 (4.4) | 2183 (7.50) | 1.98 | 0.97 | 0.69 | 1.34 | 0.84 |
| CHESTER | 36 (3.6) | 569 (1.95) | 5.95 | 3.08 | 2.13 | 4.45 | <0.001 |
| BIRKENHEAD | 33 (3.3) | 1054 (3.62) | 3.04 | 1.34 | 0.92 | 1.96 | 0.13 |
| WAYCROSS | 31 (3.1) | 677 (2.32) | 4.38 | 2.11 | 1.42 | 3.13 | <0.001 |
| MGULANI | 19 (1.9) | 221 (0.76) | 7.92 | 4.25 | 2.58 | 7.00 | <0.001 |
| JAVIANA | 18 (1.8) | 236 (0.81) | 7.09 | 3.08 | 1.86 | 5.11 | <0.001 |
| CORVALLIS | 16 (1.6) | 266 (0.91) | 5.67 | 2.50 | 1.47 | 4.25 | 0.001 |
| HEIDELBERG | 15 (1.5) | 143 (0.49) | 9.49 | 5.24 | 3.00 | 9.16 | <0.001 |
| SUBSPECIES 1 | 13 (1.3) | 431 (1.48) | 2.93 | 1.32 | 0.74 | 2.34 | 0.35 |
| ZANZIBAR | 11(1.1) | 332 (1.14) | 3.21 | 1.33 | 0.72 | 2.48 | 0.37 |
| HVITTINGFOSS | 10(1) | 871 (2.99) | 1.14 | 0.51 | 0.27 | 0.97 | 0.04 |
| STANLEY | 10(1) | 220 (0.76) | 4.35 | 2.25 | 1.17 | 4.31 | 0.02 |
| AGONA | 9 (0.9) | 419 (1.44) | 2.10 | 0.93 | 0.47 | 1.83 | 0.84 |
| MONTEVIDEO | 9 (0.9) | 162 (0.56) | 5.26 | 2.55 | 1.28 | 5.09 | 0.01 |
| PARATYPHI B JAVA | 9 (0.9) | 346 (1.19) | 2.54 | 1.33 | 0.67 | 2.61 | 0.41 |
| DUBLIN | 8 (0.8) | 13 (0.04) | 38.1 | 26.93 | 10.66 | 68.02 | <0.001 |
| READING | 8 (0.8) | 329 (1.13) | 2.37 | 1.17 | 0.57 | 2.40 | 0.67 |
| ANATUM | 7 (0.7) | 299 (1.03) | 2.29 | 0.98 | 0.45 | 2.10 | 0.95 |
| INFANTIS | 6 (0.6) | 248 (0.85) | 2.36 | 1.05 | 0.46 | 2.40 | 0.90 |
| MISSISSIPPI | 6 (0.6) | 75 (0.26) | 7.41 | 3.21 | 1.36 | 7.53 | 0.01 |
| NEWPORT | 6 (0.6) | 154 (0.53) | 3.73 | 1.92 | 0.84 | 4.41 | 0.12 |
| BOVISMORBIFICANS | 5 (0.5) | 224 (0.77) | 2.18 | 0.95 | 0.39 | 2.34 | 0.91 |
| BREDENEY | 5 (0.5) | 36 (0.12) | 12.2 | 6.48 | 2.49 | 16.88 | <0.001 |
| ORANIENBURG | 5 (0.5) | 83 (0.28) | 5.68 | 2.81 | 1.12 | 7.04 | 0.03 |
| PANAMA | 5 (0.5) | 16 (0.05) | 23.81 | 15.26 | 5.43 | 42.86 | <0.001 |
| SINGAPORE | 5 (0.5) | 108 (0.37) | 4.42 | 2.11 | 0.85 | 5.25 | 0.11 |
| CHOLERAESUIS | 4 (0.4) | 2 (0.01) | 66.67 | 77.78 | 13.21 | 457.97 | <0.001 |
| ORIENTALIS | 4 (0.4) | 171 (0.59) | 2.29 | 0.91 | 0.33 | 2.49 | 0.86 |
| POTSDAM | 4 (0.4) | 292 (1.00) | 1.35 | 0.56 | 0.21 | 1.52 | 0.25 |
| SUBSPECIES 4 | 4 (0.4) | 55 (0.19) | 6.78 | 3.09 | 1.10 | 8.69 | 0.03 |
| THOMPSON | 4 (0.4) | 154 (0.53) | 2.53 | 1.19 | 0.43 | 3.25 | 0.74 |
| WELIKADE | 4 (0.4) | 151 (0.52) | 2.58 | 1.18 | 0.43 | 3.24 | 0.75 |
| Other‡ | 50 (4.99) | 3874 (13.30) | |||||
| Total§ | 1001 | 29120 | |||||
*Proportion of invasive isolates to the total number of isolates recovered.
†Odds ratio of invasiveness compared to serotype Typhimurium adjusted for gender and age groups.
§Includes 13 iNTS cases which were infected with two different serotypes, 369 non-iNTS cases infected with 2 different serotypes and 4 non-iNTS cases infected with 3 different serotypes. Seven iNTS and 2221 non-iNTS single isolates had missing information on serotype.
‡Includes 30 remaining iNTS and 125 non-iNTS serotypes
In the specimen analysis, 945 (91.7%) of 1,031 isolates came from blood with the proportions of remaining specimens depicted in S5 Table. The characteristics of iNTS cases with recurrent infection and of those infected by multiple serotypes are summarized in S6 and S7 Tables.
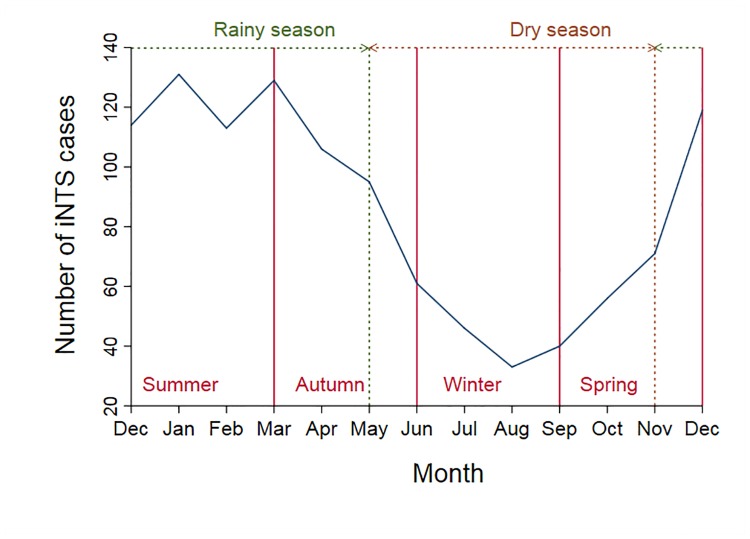
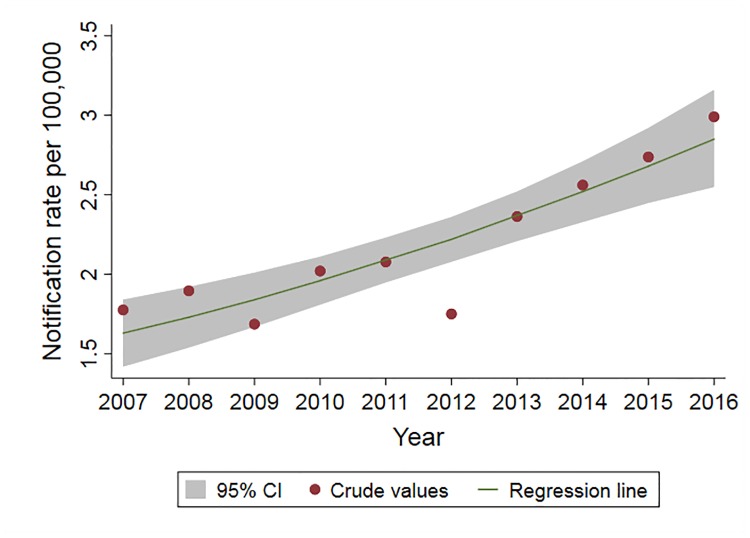
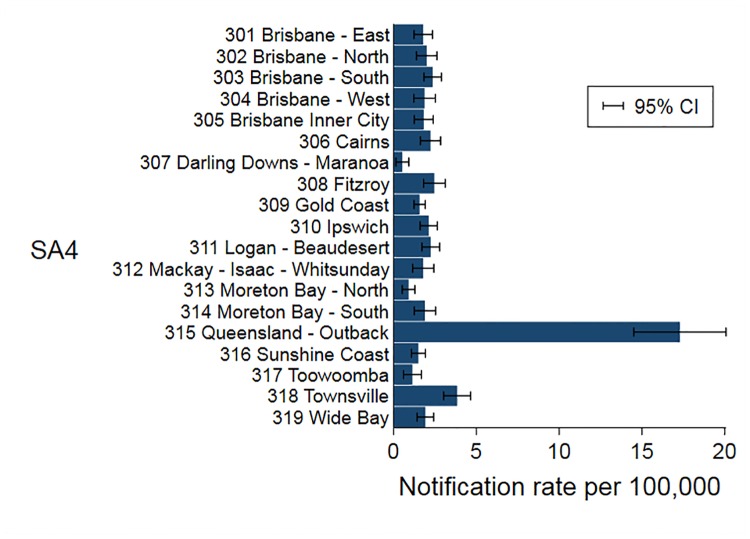
Of 995 iNTS cases included in the IRR analysis, 567 (57%) were males. The mean (range) age was 34 (0–95) years, with a significant difference in age between females and males (31. vs. 35 years, respectively; p<0.04). Of iNTS cases, 363 (36%) of 995 occurred in summer and 329 (33%) in autumn (Fig 2). The crude annual notification rate was lowest in 2012 at 1.8 per 100,000 and increased to 3.0 per 100,000 in 2016 (Fig 3). When adjusted for age group, gender, and statistical area, there was an annual 6.5% increase in the number of iNTS cases (95% CI 1.04–1.09; p<0.001) (Table 2). Crude IRR of iNTS peaked among infants (age group <1 years) for both males and females with values as high as 32 and 29 cases per 100,000 persons, respectively. The IRR was also high among young children (1–4 years old) and older adults (≥70 years old), although notification rates among older adults were much lower than in infants (Fig 4). Geographically, notifications rates ranged between 0.6 (Darling Downs—Maranoa; SA4 307) and 17.6 (Queensland—Outback; SA4 315) cases per 100,000 persons (Fig 5).
Fig 2. Number of iNTS cases in Queensland by month, 2007–2016.
Fig 3. Crude and adjusted notification rates of iNTS disease in Queensland, 2007–2016.
Table 2. Adjusted incidence rate ratios calculated using Poisson regression of iNTS disease in Queensland by sex, age, location, and year, 2007–2016.
| IRR* | 95% CI† | P-value | ||
|---|---|---|---|---|
| Gender (reference = female) | ||||
| Male | 1.314778 | 1.16 | 1.49 | <0.001 |
| Age groups (reference = 30–39 years) | ||||
| 0–4 | 10.46591* | 7.96 | 13.76 | <0.001 |
| 5–9 | 1.357503 | 0.91 | 2.02 | 0.13 |
| 10–19 | 1.155499 | 0.82 | 1.63 | 0.41 |
| 20–29 | 1.579273 | 1.15 | 2.17 | 0.01 |
| 40–49 | 0.8737139 | 0.60 | 1.26 | 0.47 |
| 50–59 | 1.26938 | 0.90 | 1.79 | 0.18 |
| 60–69 | 2.433061 | 1.77 | 3.35 | <0.001 |
| 70–79 | 5.172689 | 3.79 | 7.05 | <0.001 |
| 80+ | 4.844231 | 3.40 | 6.90 | <0.001 |
| SA4 (reference = 301 Brisbane—East) | ||||
| 302 Brisbane—North | 1.118061 | 0.72 | 1.74 | 0.62 |
| 303 Brisbane South | 1.316775 | 0.89 | 1.94 | 0.16 |
| 304 Brisbane—West | 1.044321 | 0.65 | 1.67 | 0.86 |
| 305 Brisbane Inner City | 1.017825 | 0.65 | 1.59 | 0.94 |
| 306 Cairns | 1.24689 | 0.82 | 1.89 | 0.30 |
| 307 Darling Downs—Maranoa | 0.2956672 | 0.13 | 0.66 | <0.01 |
| 308 Fitzroy | 1.375837 | 0.91 | 2.08 | 0.13 |
| 309 Gold Coast | 0.8774511 | 0.60 | 1.28 | 0.50 |
| 310 Ipswich | 1.185018 | 0.80 | 1.76 | 0.40 |
| 311 Logan—Beaudesert | 1.252945 | 0.84 | 1.86 | 0.26 |
| 312 Mackay—Isaac—Whitsunday | 1.001692 | 0.62 | 1.61 | 0.99 |
| 313 Moreton Bay—North | 0.5057448 | 0.30 | 0.86 | 0.01 |
| 314 Moreton Bay—South | 1.057549 | 0.67 | 1.68 | 0.81 |
| 315 Queensland—Outback | 9.660672 | 6.79 | 13.75 | <0.001 |
| 316 Sunshine Coast | 0.8387001 | 0.55 | 1.28 | 0.41 |
| 317 Toowoomba | 0.6356508 | 0.36 | 1.12 | 0.12 |
| 318 Townsville | 2.143418* | 1.47 | 3.13 | <0.001 |
| 319 Wide Bay | 1.071832 | 0.71 | 1.61 | 0.74 |
| Year (2007–2016) | 1.064551 | 1.04 | 1.09 | <0.001 |
*Adjusted incidence rate ratio
†Confidence interval
Fig 4. Adjusted notification rates of iNTS disease in Queensland by gender and age group, 2007–2016.
Fig 5. Adjusted notification rates of iNTS disease in Queensland by Statistical Area level 4, 2007–2016.
Of 65 different serotypes identified in Queensland, 39 (60%) were found in outback Queensland. Among 151 isolates in outback Queensland, 33 (22.2%) were Salmonella serotypes Virchow, 28 (18.5%) Typhimurium, and 13 (8.6%) Aberdeen (S1 Table). Gender and age distributions of iNTS in outback Queensland were similar to other locations, with most iNTS cases being male (56%) and infants representing the most common age group (20.8%). Crude and predicted notification rates by year, gender, age group, and statistical area are presented in S8–S10 Tables.
Discussion
We analysed 10 years of Queensland passive surveillance data to identify trends in the reported incidence and distribution of infecting Salmonella serotypes causing invasive disease. Males, people living in remote areas, and children had elevated incidence of iNTS disease. Higher rates of iNTS disease in remote areas were driven by an overall high number of NTS infections. However, high rates in males, infants, and elderly remained significant predictors for iNTS disease.
There are over 2,500 NTS serotypes worldwide with the great majority of infections caused by Salmonella Enteritidis [25]. In Australia, Salmonella Typhimurium is the most common serotype, as Salmonella Enteritidis is not endemic in egg layer flocks and is mostly acquired abroad [26]. In our study, Salmonella Virchow followed by Salmonella Typhimurium were the most common serotypes causing invasive disease in Queensland. Similarly, a study that reviewed laboratory records in a northern Queensland town over 1978–1988 also showed high proportions of Salmonella Virchow, accounting for 46% of all Salmonella bacteremias [27]. In another Australian states and territories, Salmonella Virchow was second to Salmonella Typhimurium as the most common iNTS pathogen comprising 11% and 13% of all iNTS isolates [11,12]. Salmonella Virchow is rare in the United States [28] but has emerged as an important pathogen in Europe [29], and Israel [30–32]. In Australia, Salmonella Virchow has been found in chickens, comprising 3.8% of all chicken NTS isolates; whereas bovine, porcine, and raw meat do not seem to be a common source of this serotype [33]. Surveys of vertebrates and reptiles in Queensland have found Salmonella Virchow in wallabies, kangaroos, and Asian house geckos [34,35].
The majority of iNTS cases occurred in summer and autumn which mostly corresponds to the wet season (November—April) in Queensland. As evident from our analyses, the seasonality of iNTS disease was driven by an overall increase in NTS cases. This is consistent with the findings of a study evaluating the effect of climate variation on Salmonella infection in subtropical and tropical regions in Queensland where increases in both maximum and minimum temperatures were associated with an increase in Salmonella infections [36].
Disease forms differ substantially by serotype, with serotypes Dublin and Choleraesuis having the highest invasiveness index [3,37,38]. This could be due to these serotypes being either more invasive or less diarrheagenic. Our results are consistent with other studies [12,37,38], although we identified very few Salmonella Choleraesuis and Salmonella Dublin infections. As previously described [2,3], infants, young children, and the elderly were most likely to develop invasive disease when compared to a referent category of 30–39 year olds. When looking at serotype distribution in specific age categories, Salmonella Virchow was more prevalent in infants and young children, whereas Salmonella Typhimurium was more common in older adults. Similar results were reported by a study from Greece that identified Salmonella Virchow as the most common serotype in children under one year of age [39]. As previously reported [12], being male was a significant predictor for invasive disease. Higher iNTS incidence in males was also found in another multi-national population-based cohort study that speculated that higher incidence in males might be a consequence of agricultural exposure or diet [40]. In contrast, studies investigating all NTS isolates in Australia found higher incidence in women [8].
Our study demonstrated high incidence rates of iNTS in outback Queensland. The outback is a very remote area of 1.18 million km2, which makes up about 68% of the land area of the state [41], and is largely (88%) agricultural [42]. Outback Queensland has a population of 79,700 with a density of 0.1 person per km2, the lowest of all SA4s in Queensland [43]. Our finding is in line with previous studies which identified considerably higher incidence of iNTS in rural areas when compared to urban areas [44,45]. However, when investigating the proportion of invasive and non-invasive NTS cases by statistical area, no significant difference was observed for outback Queensland (OR, 1.01; 95% CI, 0.71–1.43) suggesting that high incidence in this area is driven by an overall higher number of NTS cases.
There are several potential reasons for substantially higher rates of NTS in outback Queensland. Higher rates of NTS infection in remote areas may be linked to socio-economic status and poverty, which may mediate risk through malnutrition and comorbidities. We did not have Indigenous status in our dataset. However, outback Queensland has a higher proportion of people who identify as Aboriginal and/or Torres Strait Islander (26,560 persons) which represents 33.3% of the region’s total population [43]. Aboriginal and Torres Strait Islander people living in remote areas often live in conditions that may pose risk factors for iNTS, including poor housing conditions, food insecurity, chronic diseases, and co-infections [46–49].
While this is a large population-based study with a clearly defined denominator, there are several limitations. As we were relying only on passively collected data for surveillance, incidence of NTS infections is almost certainly underestimated, especially in remote and rural areas where access to health care is very limited [50]. As iNTS disease is more severe than non-invasive NTS and requires urgent medical attention to prevent death, selective reporting bias could be present. In addition to not having information on Aboriginal and Torres Strait Islander status, we also lacked information on comorbidities, and immunocompromising conditions that would enable us to analyze associations with high disease incidence in specific ethnic groups and statistical areas. We were not able to analyze our data by socio-economic status as Socio-Economic Indexes for Areas were unavailable at the same geographical level in which we conducted our analyses (SA4). Finally, we did not have information on travel abroad or interstate. However, a study estimating foodborne illness in Australia circa 2010 found that approximately 15% of Salmonella notifications were travel-associated [51].
Other investigators have used whole-genome sequencing and PCR to distinguish recrudescence from reinfection [52]. However, these methods are not included in routinely collected data in Australia. One study conducted in sub-Saharan Africa concluded that 78% of recurrences were caused by recrudescence with similar or identical Salmonella isolates, and reinfection accounted only for 22% of recurrences [52]. In our study, only two (0.2%) of 995 cases with iNTS disease had recurrent infection caused by the same serotype (Salmonella Typhimurium and Salmonella Chester). As we did not have molecular subtyping information on the isolates, our division was based on the time difference between two episodes of salmonellosis. As both individuals had two episodes of iNTS disease more than 30 days apart (6 months and 10 months), we considered them as reinfection and as such we counted them as separate cases of iNTS disease. However, due to such a small number of recurrent infections, this does not represent a major limitation to our study.
Our study provides an important insight into the epidemiology of iNTS disease in Australia. High rates of iNTS among males, infants, and elderly require further investigation of spatial clusters of disease, and clarification of patient and household risk factors through the conduct of case-control studies. In addition, there is a particular need to investigate and control food, animal, and environmental sources of Salmonella Virchow, and conduct further molecular and genomic analysis of invasive Salmonella Virchow isolates. Lastly, more studies are needed to update information on the global burden of iNTS disease including risk factors, predominant sources of infection, modes of transmission, and antimicrobial resistance profiles.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We would like to thank Ms. Nicole Burt, a data specialist at Queensland Health, for her help with data extraction and transfer of SLAs into SA4 codes.
Data Availability
The data that support the findings of this study are available upon request from Dr Sonya Bennett, Communicable Diseases Branch, Prevention Division, Queensland Health (email: Sonya.Bennett@health.qld.gov.au). Data deposition or provision in supporting information is not possible as it involves human research participants data.
Funding Statement
AP is supported by the Endeavour Postgraduate Scholarship. MDK is supported by the National Health and Medical Research Council career development fellowship (GNT1136112). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Majowicz SE, Musto J, Scallan E, et al. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 2010; 50(6): 882–9. 10.1086/650733 [DOI] [PubMed] [Google Scholar]
- 2.Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. Invasive non-typhoidal Salmonella disease: an emerging and neglected tropical disease in Africa. Lancet 2012; 379(9835): 2489–99. 10.1016/S0140-6736(11)61752-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uche IV, MacLennan CA, Saul A. A Systematic review of the incidence, risk factors and case fatality rates of invasive nontyphoidal Salmonella (iNTS) disease in Africa (1966 to 2014). PLoS Negl Trop Dis 2017; 11(1): e0005118 10.1371/journal.pntd.0005118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vugia DJ, Samuel M, Farley MM, et al. Invasive Salmonella infections in the United States, FoodNet, 1996–1999: incidence, serotype distribution, and outcome. Clin Infect Dis 2004; 38 Suppl 3: S149–56. [DOI] [PubMed] [Google Scholar]
- 5.Phu Huong Lan N, Le Thi Phuong T, Nguyen Huu H, et al. Invasive Non-typhoidal Salmonella infections in Asia: Clinical observations, disease outcome and dominant serovars from an infectious disease hospital in Vietnam. PLoS Negl Trop Dis 2016; 10(8): e0004857 10.1371/journal.pntd.0004857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ao TT, Feasey NA, Gordon MA, Keddy KH, Angulo FJ, Crump JA. Global burden of invasive nontyphoidal Salmonella disease, 2010(1). Emerg Infect Dis 2015; 21(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirk MD, Pires SM, Black RE, et al. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: A Data synthesis. PLoS Med 2015; 12(12): e1001921 10.1371/journal.pmed.1001921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ford L, Glass K, Veitch M, et al. Increasing incidence of Salmonella in Australia, 2000–2013. PLoS One 2016; 11(10): e0163989 10.1371/journal.pone.0163989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The Department of Health. National Notifiable Diseases Surveillance System data 1991–2017 [cited 2018 April 28]. http://www9.health.gov.au/cda/source/rpt_4.cfm
- 10.The Department of Health. National Notifiable Diseases Surveillance System. Canberra (ACT): Commonwealth of Australia; [cited 2018 April 29]. http://www9.health.gov.au/cda/source/rpt_4_sel.cfm
- 11.Wilson HL, Kennedy KJ, Moffatt CRM. Epidemiology of non-typhoid Salmonella infection in the Australian Capital Territory over a 10-year period. Intern Med J. 2018. March;48(3):316–23. 10.1111/imj.13625 [DOI] [PubMed] [Google Scholar]
- 12.Williamson DA, Lane CR, Easton M, Valcanis M, Strachan J, Veitch MG, et al. Increasing Antimicrobial Resistance in Nontyphoidal Salmonella Isolates in Australia from 1979 to 2015. Antimicrob Agents and Chemother. 2018. February;62(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Australian Bureau of Statistics. Census of Population and Housing: Socio-Economic Indexes for Areas (SEIFA), Australia, 2016 [cited 2018 May 28]. http://www.abs.gov.au/ausstats/abs@.nsf/mf/2033.0.55.001
- 14.The Department of Health. Links to State and Territory Public Health Legislation, the Quarantine Act, and the National Health Security Act 2007. Canberra (ACT): Commonwealth of Australia [cited 2018 May 28]. http://www.health.gov.au/internet/main/publishing.nsf/Content/cda-state-legislation-links.htm.
- 15.Hall G, Yohannes K, Raupach J, Becker N, Kirk M. Estimating community incidence of Salmonella, Campylobacter, and Shiga toxin—producing Escherichia coli infections, Australia. Emerging Infectious Diseases. 2008. 14(10):1601–9. 10.3201/eid1410.071042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Australian Bureau of Statistics. Queensland Statistical Local Areas (SLA), 2011 [cited 2018 April 28]. http://www.qgso.qld.gov.au/products/maps/qld-sla-asgc-2011/index.php
- 17.Australian Bureau of Statistics. Australian Statistical Geography Standard: Volume 1—Main Structure and Greater Capital City Statistical Areas, 2016 [cited 2018 April 28]. http://www.abs.gov.au/ausstats/abs@.nsf/Lookup/by%20Subject/1270.0.55.001~July%202016~Main%20Features~Statistical%20Area%20Level%204%20(SA4)~10016
- 18.Australian Bureau of Statistics. Queensland Concordance—SLA (ASGC 2011) to SA2 (ASGS 2011) [cited 2018 April 28]. http://www.qgso.qld.gov.au/products/maps/qld-conc-sla-2011-sa2-2011/index.php
- 19.Australian Bureau of Statistics. Queensland statistical areas, level 4 (SA4), 2016 [cited 2018 April 28]. http://stat.abs.gov.au/itt/r.jsp?ABSMaps
- 20.Gunn JS, Marshall JM, Baker S, Dongol S, Charles RC, Ryan ET. Salmonella chronic carriage: epidemiology, diagnosis, and gallbladder persistence. Trends Microbiol 2014; 22(11): 648–55. 10.1016/j.tim.2014.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Im J, Nichols C, Bjerregaard-Andersen M, Sow AG, Løfberg S, Tall A, et al. Prevalence of Salmonella Excretion in Stool: A Community Survey in 2 Sites, Guinea-Bissau and Senegal. Clin Infect Dis 2016. 03/01;62(Suppl 1):S50–S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barker RM, Kearney GM, Nicholson P, Blair AL, Porter RC, Crichton PB. Types of Salmonella Paratyphi B and their phylogenetic significance. J Med Microbiol 1988; 26(4): 285–93. 10.1099/00222615-26-4-285 [DOI] [PubMed] [Google Scholar]
- 23.Gupta SK, Medalla F, Omondi MW, et al. Laboratory-based surveillance of paratyphoid fever in the United States: travel and antimicrobial resistance. Clin Infect Dis 2008; 46(11): 1656–63. 10.1086/587894 [DOI] [PubMed] [Google Scholar]
- 24.Australian Bureau of Statistics. Population by age and sex, regions of Australia, 2007–2016 [cited 2018 April 28]. http://www.abs.gov.au/AUSSTATS
- 25.Galanis E, Wong D, Patrick ME, et al. Web-based surveillance and global Salmonella distribution, 2000–2002. Emerg Infect Dis 2006; 12(3): 381–8. 10.3201/eid1203.050854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The OzFoodNet Working Group. Monitoring the incidence and causes of diseases potentially transmitted by food in Australia: Annual report of the OzFoodNet network, 2011. Commun Dis Intell Q Rep 2015; 39(2): E236–64. [PubMed] [Google Scholar]
- 27.Ashdown LR, Ryan PJ. Invasive disease due to Salmonella Virchow: a north Queensland problem. Med J Aus 1990; 153(6): 330–2, 4–5. [DOI] [PubMed] [Google Scholar]
- 28.Centers for Disease Control and Prevention. An Atlas of Salmonella in the United States, 1968–2011 [cited 2018 May 28]. https://www.cdc.gov/salmonella/reportspubs/salmonella-atlas/index.html
- 29.Snow LC, Davies RH, Christiansen KH, Carrique-Mas JJ, Wales AD, O’Connor JL, et al. Survey of the prevalence of Salmonella species on commercial laying farms in the United Kingdom. Vet Rec 2007;161(14):471–6. [DOI] [PubMed] [Google Scholar]
- 30.Solnik-Isaac H, Weinberger M, Tabak M, Ben-David A, Shachar D, Yaron S. Quinolone resistance of Salmonella enterica serovar Virchow isolates from humans and poultry in Israel: evidence for clonal expansion. J Clin Microbiol 2007;45(8):2575–9. 10.1128/JCM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinberger M, Solnik-Isaac H, Shachar D, Reisfeld A, Valinsky L, Andorn N, et al. Salmonella enterica serotype Virchow: epidemiology, resistance patterns and molecular characterisation of an invasive Salmonella serotype in Israel. Clin Microbiol Infect 2006;12(10):999–1005. 10.1111/j.1469-0691.2006.01466.x [DOI] [PubMed] [Google Scholar]
- 32.Marzel A, Desai PT, Nissan I, et al. Integrative analysis of Salmonellosis in Israel reveals association of Salmonella enterica Serovar 9,12:l,v:- with extraintestinal infections, dissemination of endemic S. enterica Serovar Typhimurium DT104 biotypes, and severe underreporting of outbreaks. J Clin Microbiol 2014; 52(6): 2078–88 10.1128/JCM.00399-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heuzenroeder MW, Murray CJ, Davos D, Ross IL. Salmonella typing and colonization of chickens by characterized Salmonella Sofia. Canberra: Rural Industries Research and Development Corporation, 2004. [cited 2018 May 28]. http://www.agrifutures.com.au/wp-content/uploads/publications/04-138.pdf [Google Scholar]
- 34.Callaway Z, Thomas A, Melrose W, Buttner P, Speare R. Salmonella Virchow and Salmonella Weltevreden in a random survey of the Asian house gecko, Hemidactylus frenatus, in houses in northern Australia. Vector Borne Zoonotic Dis 2011;11(6):621–5. 10.1089/vbz.2010.0015 [DOI] [PubMed] [Google Scholar]
- 35.Speare R, Thomas AD. Orphaned kangaroo and wallaby joeys as a potential zoonotic source of Salmonella spp. Med J Aust 1988;148(12):619, 22–3. [PubMed] [Google Scholar]
- 36.Zhang Y, Bi P, Hiller JE. Climate variations and Salmonella infection in Australian subtropical and tropical regions. Sci Total Environ 2010; 408(3): 524–30. 10.1016/j.scitotenv.2009.10.068 [DOI] [PubMed] [Google Scholar]
- 37.Harvey RR, Friedman CR, Crim SM, et al. Epidemiology of Salmonella enterica serotype Dublin infections among humans, United States, 1968–2013. Emerg Infect Dis 2017; 23(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones TF, Ingram LA, Cieslak PR, et al. Salmonellosis outcomes differ substantially by serotype. J Infect Dis 2008; 198(1): 109–14. 10.1086/588823 [DOI] [PubMed] [Google Scholar]
- 39.Galanakis E, Bitsori M, Maraki S, Giannakopoulou C, Samonis G, Tselentis Y. Invasive non-typhoidal salmonellosis in immunocompetent infants and children. Int J Infect Dis 2007; 11(1): 36–9. 10.1016/j.ijid.2005.09.004 [DOI] [PubMed] [Google Scholar]
- 40.Laupland KB, Schonheyder HC, Kennedy KJ, et al. Salmonella enterica bacteraemia: a multi-national population-based cohort study. BMC Infect Dis 2010; 10: 95 10.1186/1471-2334-10-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Australian Bureau of Statistics. Queensland Outback: region data summary, 2016 [cited 2018 April 28]. http://stat.abs.gov.au/itt/r.jsp?RegionSummary®ion=315&dataset=ABS_REGIONAL_ASGS2016&geoconcept=ASGS_2016&measure=MEASURE&datasetASGS=ABS_REGIONAL_ASGS2016&datasetLGA=ABS_REGIONAL_LGA2016®ionLGA=LGA_2016®ionASGS=ASGS_2016
- 42.Department of Agriculture and Water Resources. About my region—Queensland—Outback. 2017 [cited 2018 April 28]. http://www.agriculture.gov.au/abares/research-topics/aboutmyregion/qld-outback#agricultural-sector
- 43.Queensland Government Statistician’s Office. Queensland regions compared, Census 2016. [cited 2018 April 28]. http://www.qgso.qld.gov.au/products/reports/qld-regions-compared-c16/qld-regions-compared-c16.pdf
- 44.Cruz Espinoza LM, Nichols C, Adu-Sarkodie Y, et al. Variations of invasive Salmonella infections by population size in Asante Akim north municipal, Ghana. Clin Infect Dis 2016; 62 Suppl 1: S17–22. [DOI] [PubMed] [Google Scholar]
- 45.Muthumbi E, Morpeth SC, Ooko M, et al. Invasive salmonellosis in Kilifi, Kenya. Clin Infect Dis 2015; 61 Suppl 4: S290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andersen MJ, Skinner A, Williamson AB, Fernando P, Wright D. Housing conditions associated with recurrent gastrointestinal infection in urban Aboriginal children in NSW, Australia: findings from SEARCH. Aust N Z J Public Health 2018; 42(3):247–253 10.1111/1753-6405.12786 [DOI] [PubMed] [Google Scholar]
- 47.The Kirby Institute. Australian NSP Survey National Data Report 2013–2017 [cited 2018 July 5]. https://kirby.unsw.edu.au/sites/default/files/kirby/report/ANSPS_National-Data-Report-2013-2017.pdf
- 48.Australian Institute of Health and Welfare. Chronic kidney disease in Aboriginal and Torres Strait Islander people: 2011 [cited 2018 april 28]. https://www.aihw.gov.au/reports/chronic-kidney-disease/chronic-kidney-disease-indigenous-australians/contents/table-of-contents
- 49.Australian Institute of Health and Welfare. The health and welfare of Australia’s Aboriginal and Torres Strait Islander peoples: 2015 [cited 2018 April 28]. https://www.aihw.gov.au/getmedia/584073f7-041e-4818-9419-39f5a060b1aa/18175.pdf.aspx?inline=true
- 50.Australian Institute of Health and Welfare. Australia’s health: 2016 [cited 2018 April 28]. https://www.aihw.gov.au/getmedia/9844cefb-7745-4dd8-9ee2-f4d1c3d6a727/19787-AH16.pdf.aspx?inline=true
- 51.Kirk M, Ford L, Glass K, Hall G. Foodborne illness, Australia, circa 2000 and circa 2010. Emerg Infect Dis 2014; 20(11): 1857–64. 10.3201/eid2011.131315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okoro CK, Kingsley RA, Quail MA, et al. High-resolution single nucleotide polymorphism analysis distinguishes recrudescence and reinfection in recurrent invasive nontyphoidal Salmonella typhimurium disease. Clin Infect Dis 2012; 54(7): 955–63. 10.1093/cid/cir1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
The data that support the findings of this study are available upon request from Dr Sonya Bennett, Communicable Diseases Branch, Prevention Division, Queensland Health (email: Sonya.Bennett@health.qld.gov.au). Data deposition or provision in supporting information is not possible as it involves human research participants data.