Abstract
Understanding the genetic basis of heat and drought stress tolerance in wheat is prerequisite for wheat breeding program. In the present study, a wheat panel comprising of 192 elite bread wheat genotypes was phenotyped in eight environments for yield and related traits in field conditions. Four stress environments were created by implying four different treatments differing in sowing date and water availability, panel was evaluated for two years in field conditions. The panel was genotyped with 15K Illumina chip and 9236 polymorphic markers concentrated on B genome were employed in GWAS analysis. Consistent, fast LD decay was observed on D genome and structure analysis germplasm divided panel into three major populations. GWAS was performed using BLUEs values of combined environment data in R package GAPIT using log10(P) = 3.96 as significance threshold. The significance of association was further checked using FDR<0.05 threshold. The GWAS identified 487 loci associated with the traits and were significant at log10(p) threshold out of these 350 loci were significant at FDR threshold. For two stress indices 108 associations were significant at FDR threshold. Nine genomic regions were shared among all treatment, while multiple pleiotropic regions were present on chromosome 7D followed by unmapped chromosome. The present study validated many marker trait associations for yield and other traits, MTAs significant under combined drought and heat stress were novel. These regions are important and can be used for fine mapping and marker assisted selection to discover new genes responsible for heat and drought tolerance in wheat.
Introduction
Bread wheat is world’s 3rd most cultivated cereal crop planted over more than 20% area and provide 20% calories and 20% plant derived proteins to global population [1,2]. Global wheat demand is increasing with continuous increase in population and it is estimated that there is a need to increase global wheat production by 70% in 2050 [3]. Due to uncertainty in climatic conditions, it is estimated that wheat yield will reduce by 50% in South Asia in 2050 which is 7% of total global crop reduction [4]. This vulnerability in wheat yield is due to changes in patterns of rainfall, increase in temperature and occurrence of simultaneous drought and heat stress during the grain filling period. Increase in air temperature, radiation stress, high levels of CO2 and increase in the amount of greenhouse gasses further increase the intensity of drought and heat stress [5–7]. In coming few decade scenario of climate change will worsen, it is predicted that global temperature will increase by 3–5°C and annual precipitation will decrease by 4–27% in different parts of world [8]. The major adverse effects of heat stress on wheat include reduction in crop cycle, increase in soil temperature and rate of evaporation while drought stress mainly effects sink and source strength. Interactive effects of drought and heat stress may come from increased vapour pressure deficit. Many attempts have been made by using conventional breeding strategies to improve grain yield and quality of bread wheat, but these approaches altogether increased yield by less than one percent per year [9–12]. Future wheat breeding program is based on dissecting molecular and genetic basis of heat and drought stress tolerance through complementary approaches of association mapping and QTL mapping [13–15]. Till many years QTL mapping was considered as a powerful tool for genetic dissection of complex traits in plants but now QTL mapping is replaced by association mapping. Association mapping is based on linkage disequilibrium (LD) and is a powerful approach with higher resolution due to presence of higher genetic diversity and historic recombination of alleles among association mapping populations [16]. Association mapping is used to identify genomic regions associated with heat and drought tolerance in many association mapping populations [17–20] but these studies focus only either on heat or drought stress. Only few studies focused on combined drought and heat stress [21–24]. In the present study a diverse panel of bread wheat genotype was phenotyped under optimum [C], drought [D], heat [H] and combined heat and drought stressed [HD] conditions for two cropping years. In addition to assessing genetic diversity of the panel significant markers traits associations were identified for each stress treatment. Furthermore, common association among stress treatment and pleiotropic regions shared by multiple traits were also assessed.
Materials and methods
Plant material
The plant material comprised of 192 diverse bread wheat lines from International Wheat and Maize Research Center (CIMMYT) heat and drought nurseries for South Asia. Ten local high yielding varieties were also used in the present study to compare yield and to estimate effects of combination of both stresses on their yield. Detailed information about selection history and entry number is given in S1 Table. Seed of whole germplasm was obtained from Crop Physiology, Laboratory National Agriculture Research Center (NARC), Islamabad. Field was ploughed three weeks prior to seed sowing and fertilizer was added with standard rate. All standard agronomic practices were performed in all plots.
Experimental setup
The experiments were planted in a lattice design with four replications and net plot size of 3 × 2m. Three rows per genotype with a row length of 2m were sown using a small plot grain drill machine with row to row distance of 22 cm for two cropping seasons. The averaged value of minimum, maximum and average percent humidity and temperature for both cropping seasons are given in Fig 1A and Fig 1B. Following stress treatments were imposed on entire germplasm for two years. The Control [C] or non-stress treatment genotypes were sown at 22 November and grown at 100 percent moisture content throughout cropping cycle. For drought [D] treatment genotypes were sown at 20 November and grown at 35% field capacity after heading. For Heat [H] stress the planting date was 01 January and germplasm was grown at 100% field capacity after heading. For a combination of heat and drought stress [HD] treatment germplasm was sown at 01 January and grown at 35% field capacity after heading. The germplasm in all plots was grown with normal agronomic practices and 100% field capacity till 50% heading S1 Fig. The treatments were started after heading and last till maturity after which all the plots were rehydrated. The [HD] and [D] treatment were protected from the rain after heading by covering tunnel with polythene sheaths and to avoid percolation of water one-meter-deep ditch was dug around the boundary and polythene sheet was inserted in it to avoid rain water seepage inside the tunnel.
Fig 1.
a: Averaged two-year maximum minimum and average temperature of experimental location. b. Averaged two-year maximum, minimum and average percent humidity of experimental location.
Data recording
The yield traits were recorded from all three plants per genotype while physiological data was recorded from three randomly selected plants. Following yield traits were recorded during study; awn length (AL) (from tip of spike), days to anthesis (DTA) (number of days taken from emergence to appearance of anthers), day to maturity (DTM) (number of days taken from emergence to maturity), grains per spike (GPS) (number of grains in spike was counted), grain yield (GY) (weighing grains form all harvested plants), harvest index (HI) (percent ratio of grain yield and above ground plant dry weight (DW), peduncle extrusion (PEXT) (from tip of flag leaf to base of spike), peduncle length (PL) (from first node to base of spike), plant height (PH) (from ground level to spike tip excluding awns), spikelet number (SLP) (counting number of fertile tillers), spike length (SL) (manually with ruler in cm). Stress tolerance index (STI) Tolerance index (TOL)
where Ys and Yp represent yield under stress and non-stress treatment and Yp2 is the mean yield of wheat lines evaluated under non-stress conditions [25].
Statistical analysis
Summary statistics, Pearson’s correlation analyses, broad sense heritability estimates and association mapping were estimated from best linear unbiased estimated values calculated from mean value of each trait keeping genotype as fixed factor in Genestat v 18. The germplasm was genotyped with 15K Illumina chip and filtering of polymorphic markers was done following [26]. Map position of SNPs was indicated by using wheat genetic map published by [27], after filtering 9236 polymorphic markers were used in GWAS analyses. Genetic diversity, polymorphic information content and major allele frequency was estimated by using Powermarker v3.25 (http://statgen.ncsu.edu/powermarker) software. GenStat v18 was used to calculate chromosome-wise linkage disequilibrium (LD) decay by calculating pairwise marker allele squared correlation (r2) and plotting the r2 values against the genetic distances (cM). Population structure was estimated using STRUCTURE v 2.3.4 [28], as described by [29]. All the polymorphic markers were used to determine population structure by inferring K from 2 to 10 using 100,000 burn-in iterations followed by 100,000 MCMC (Markov-Chain Monte Carlo) iterations and 5 replications for each K. The obtained results were analyzed using STRUCTURE harvester (http://taylor0.biology.ucla.edu/structureHarvester) to get appropriate K value.
Association mapping
Best linear unbiased estimate values of each trait were used to perform GWAS using mixed linear model approach [30,31] in R package GAPIT (Genome Association and Prediction Integrated Tool) [32]. The correction for population stratification and cryptic relatedness was performed by employing coefficient of co-ancestry kinship and first three principal components as random effects in the linear mixed-effect model [33]. Appropriateness of model used for association analysis in present study was checked by drawing QQ plots between expected and observed log10(P) values. Significance threshold was set by following [34] which was equal or greater than log10 (P) ≥3.98 and after application of Bonferroni correction the log10 (P) threshold rose to 5.26 [35].
Results
Phenotypic results
Grain yield varied greatly across the environments, high grain yield was recorded under non stress treatment [C] (Average = 387.24, H2 = 0.86) followed by Heat [H] (Average = 281.40, H2 = 0.86), drought [D] (Average = 290.80, H2 = 0.97) and combined heat and drought treatment [HD] (Average = 187.16, H2 = 0.83). These results showed that independent heat had more pronounced effects on grain yield than independent drought stress while combination of both stresses had more severe effects than independent drought or heat stress. The same trend was observed for other traits and FLL, FLW HI, LA and TILL were more effected by drought stress than heat and regarded as drought sensitive traits while DW, DTM, PH, SL and SPLS were more effected by heat stress and regarded as heat sensitive traits. The heritability values were higher under all stress treatment with DW and GY having highest H2 value under [C] treatment, FLL and LA had highest H2 value under [D] treatment, FLL had highest H2 value under [H] treatment and DTM had highest H2 value under [HD] treatment. The range, mean and heritability values for yield and related traits in the normal environment [C], drought [D], heat [H] and combined heat and drought [HD] stressed environments are summarized in Table 1.
Table 1. Averages and standard deviations for eleven traits evaluated in the GWAS in four stressed environments under control [C], drought [D], heat [H] and combined drought and heat [HD] stress for two cropping seasons.
| CONTROL | DROUGHT | HEAT | HEAT*DROUGHT | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Min | Max | Mean | H2 | Min | Max | Mean | H2 | Min | Max | Mean | H2 | Min | Max | Mean | H2 |
| DW | 246.5 | 878.49 | 542.94 | 0.96 | 186.72 | 715 | 459.43 | 0.83 | 218 | 653.14 | 419.5 | 0.76 | 150.5 | 618 | 372.99 | 0.96 |
| DTM | 132 | 160 | 145.5 | 0.88 | 121 | 147 | 133.23 | 0.64 | 109 | 138.5 | 121.91 | 0.9 | 104.9 | 124.02 | 117.09 | 0.97 |
| FLL | 23.03 | 44.4 | 32.95 | 0.96 | 16.89 | 39.09 | 26.62 | 0.98 | 20 | 40.7 | 30.77 | 0.99 | 14.77 | 32.1 | 22.37 | 0.92 |
| FLW | 1.46 | 3.35 | 2.41 | 0.92 | 1.29 | 2.64 | 1.9 | 0.8 | 1.47 | 3.16 | 2.17 | 0.93 | 1.02 | 2.65 | 1.69 | 0.77 |
| GY | 216.5 | 565.5 | 387.24 | 0.86 | 108.5 | 490.5 | 290.8 | 0.97 | 212.3 | 360 | 281.4 | 0.86 | 102 | 286.5 | 187.16 | 0.83 |
| HI | 40.08 | 88.49 | 59.51 | 0.95 | 30.17 | 70.92 | 48.62 | 0.96 | 33.77 | 71.94 | 52.94 | 0.92 | 16.62 | 61.32 | 38.5 | 0.59 |
| LA | 31.57 | 73.03 | 50.8 | 0.94 | 22 | 65.67 | 40.09 | 0.98 | 32.16 | 65.85 | 46.09 | 0.86 | 20 | 44.46 | 30.87 | 0.69 |
| PH | 66.83 | 109.67 | 86.84 | 0.61 | 61.13 | 104.13 | 80.34 | 0.5 | 59.33 | 85.64 | 71.31 | 0.79 | 44 | 75.77 | 60.63 | 0.89 |
| SL | 11.17 | 20.63 | 16.48 | 0.91 | 9 | 18.67 | 14.26 | 0.85 | 8.67 | 19.37 | 13.67 | 0.79 | 6.68 | 14 | 10.2 | 0.79 |
| SPLS | 13.33 | 24.67 | 19.56 | 0.94 | 12.01 | 22.96 | 17.86 | 0.97 | 9.5 | 19.83 | 14.46 | 0.92 | 8.17 | 15.96 | 11.36 | 0.96 |
| TILL | 4 | 11 | 7.27 | 0.39 | 4 | 9.47 | 6.27 | 0.52 | 3.88 | 9.14 | 6.43 | 0.77 | 1.33 | 7.3 | 4.82 | 0.53 |
DW, Plant Above Ground Dry Weight; DTM, Days to Maturity; FLL, Flag Leaf Length; FLW, Flag Leaf Width; GY, Grain Yield; HI, Harvest Index; LA, Leaf Area; PH, Plant Height; SL, Spike Length; SPLS, Spikelets per spike; TILL, Tillers per plant
Min, Minimum value; Max, Maximum value, H2, Broad sense heritability
Grain yield showed consistent negative correlation with day to maturity under all stress treatments, suggesting that early maturing plants had higher yield than the late maturing plants under stressed environment. Under non stress treatment [C] grain yield had highest positive correlation with plant above ground dry weight (r = 0.58**) while under drought stress [D] it had highest positive correlation with Spikelets per spike (r = 0.88**), under heat stress it had highest correlation with harvest index (r = 0.92**) and under combined heat and drought stress it had highest correlation with plant above ground dry weight (r = 0.83**). Stress tolerance index had positive correlation with grain yield under all stress treatments while tolerance index had positive correlation under individual drought (r = 0.14*) and individual heat stress (r = 0.18*) but it had negative correlation with grain yield under combined drought and heat stress (r = -0.33**). The detailed information about correlations among all studied traits is given in Fig 2. The results from analysis of variance showed that genotype, treatment, cropping years and their interaction had significant effects on studied traits.
Fig 2. Pearson correlation between grain yield and all studied traits and stress indices.
a: Correlation under non stress treatment b: correlation under drought stress c: correlation under heat stress d: correlation under combined heat and drought stress.
Genotypic results
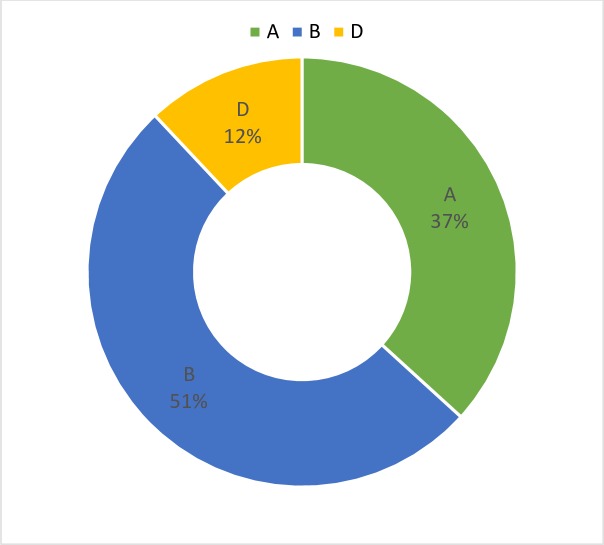
The association mapping panel was mapped with 15k ilumina chip from Gatersleben, Germany. After filtering (removing SNPs with MAF <5%), 9236 polymorphic markers were employed in GWAS analysis. Most of these polymorphic SNPs were concentrated on B genome (51%) followed by A (37%) and D (12%) genome (Fig 3). Chromosome 2B had highest number (903) of polymorphic SNPs while chromosome 4D had least number (42) of these SNPs. The average polymorphic information content was 0.27 with maximum PIC value (0.35) for chromosome 6B and least (0.14) for chromosome 5D (S2 Fig). Based on R2 model highest LD decay (r2 = 0.64) was seen on chromosome 4D while least LD (r2 = 0.38) was seen on chromosome 5A with an average Pearson's correlation (r2) value of 0.45. The LD decay was faster for A and B genome resulting smaller genetic distance when compared to D genome. The rate of LD decay was ~8 cM; ~5 cM ~10 cM for A, B and D genome, respectively with mean genetic length of 8.1 cM for all groups showing that one marker is enough within ~8.1 cM region for each chromosome (S1 Appendix).
Fig 3. Distribution of polymorphic SNP markers on different wheat genomes.
Population structure
Population structure analysis divided the whole population in three sub-populations, population_01 containing 36 genotypes while population_02 and population_03 contained 51 and 105 genotypes respectively. The members of population _01 contained genotypes with VORB, PASTOR and PWB65 as one of their parent while genotypes of population_02 contained CHIBIA, KACHU #, ND643 and PRL as one of their parents. The population_03 contained maximum 54% genotypes of whole studied panel and most of these genotypes contained ATTILA, KIRITATI, PASTOR, SERI and WAXWING as one of their parents (Fig 4A and 4B, S1 Table).
Fig 4. STRUCTURE analysis used to define genetic relationships in the AM panel.
a) Each horizontal entry represents one AM panel entry. The existence of three sub-populations was inferred. b) STRUCTURE analysis used to define genetic relationships in the AM panel. Δk plot, with k ranging from1-10.
Association mapping
Genome wide association mapping was performed for independent drought and independent heat stress and for combination of both heat and drought stress using R package GAPIT with a significance threshold of log 10(P)>3.98 and FDR<0.05. Total 148, 95, 151 and 93 significant associations were detected for studied traits under non stress [C], drought [D], Heat [H] and for combined heat and drought [HD] treatment respectively. Out of these, 105, 63, 122 and 60 were significant at FDR threshold under non stress [C], drought [D], Heat [H] and for combined heat and drought [HD] treatment respectively (Fig 5). Manhattan and QQ plots for each trait under each stress treatment are given in (S2 Appendix).
Fig 5.
Number of loci significantly associated with studied trait under [C], [D], [H] and [HD] treatments.
Marker trait association under non-stress treatment [C]
Under optimum growing conditions 148 marker trait associations (MTAs) were significant at Log(P) value threshold while 105 were significant at FDR threshold. Maximum number of significant associations (36) were reported for plant above ground biomass (DW) followed by 23 significant associations for grain yield (GY). Only one marker significant at Log(P) threshold was associated with leaf area under [C] treatment while no significant association was recorded at FDR threshold for FLL, HI, LA and PH. On average all significant associations under [C] treatment explained 20.25% phenotypic variation. Maximum number of associations (18) were recorded on chromosome 3D followed by 17 associations on 5B and 6A while chromosome 2D and 3A had only one significant association. Many genomic regions on chromosome 3D were pleiotropic and were associated with more than one trait. The most potent MTAs for grain yield under optimum conditions [C] were recorded on chromosome 3D (GENE-1752_162 at 101.24 and RFL_Contig2471_119 at 23.41 cM) and explained 28.65 and 23.61% phenotypic variation. Grain yield had common QTL with DW, DTM, FLL, FLW, HI, SPLS and TILL and most consistent regions controlling these traits were present on 3D, 5B and 6A (Table 2, S2 Table).
Table 2. Centi-Morgan position of significant marker traits associations detected for yield and related traits from BLUEs value under non stress [C] treatment.
| Chromosome | DW | DTM | FLL | FLW | GY | HI | PH | SPL | SPLS | TILL |
|---|---|---|---|---|---|---|---|---|---|---|
| 1A | 51.09, 71.05 | - | - | - | - | - | 93.61 | - | - | - |
| 1B | 30.68, 43.86, 62.58, 63.8 | - | - | 158.59 | 158.59 | 158.59 | - | - | 158.59 | 158.59 |
| 1D | 87.36, 88.85, 71.47 | - | - | - | - | - | - | - | - | - |
| 2B | - | - | 48.54 | - | 96.14 | - | 108.04 | - | 96.14 | 96.14 |
| 2D | 97.14 | - | - | - | - | - | - | - | - | - |
| 3A | 26.01,87.78, 86.16,97.14 | - | - | - | - | - | - | - | - | - |
| 3B | 37.29, 67.67, 67.78, 137.84 | - | - | - | - | - | - | - | - | - |
| 3D | 101.24 | 23.41,101.24 | 23.41, 25.11 | 142.32, 143.01 | 23.41, 101.24 | - | - | - | 23.41,101.24 | 23.41, 101.24, 82.56, 96.33, 97.66 |
| 4A | - | - | - | 48.84 | - | - | - | - | - | - |
| 4B | 5.99 | - | - | - | - | - | 34.15 | - | - | - |
| 5A | - | - | - | - | 53.47 | - | - | - | - | - |
| 5B | 11.23 | 32.73, 49.01, 100.64, 141.91 | 32.73 | 141.91 | 32.73, 49.01, 100.64, 141.91 | 141.91 | - | 109.53, 115.69 | 32.73 | 32.73 |
| 5D | - | 69.96 | - | - | 69.96 | - | - | - | - | - |
| 6A | 85.07 | 133.74 | - | 133.74 | 133.74 | 133.74 | 140.7 | 25.53, 27.64, 37.03, 40.6, 43.1 | - | - |
| 6B | - | - | - | - | - | - | - | 67.24, 71.76 | - | - |
| 6D | 153.08 | - | - | - | 153.08 | - | - | - | - | - |
| 7A | - | 63.21 | - | - | 63.21 | - | - | - | - | - |
| 7B | 49.38,61.14 | - | - | 89.82, 90.42, 92.52, 143.23, 144.8, 148.65, 150.6 | 163.3 | 148.65 | - | - | 163.3 | 163.3 |
| 7D | 197.58, 203.58 | - | - | - | 208.1 | - | - | - | 208.1 | 208.1 |
DW, Plant Above Ground Dry Weight; DTM, Days to Maturity; FLL, Flag Leaf Length; FLW, Flag Leaf Width; GY, Grain Yield; HI, Harvest Index; LA, Leaf Area; PH, Plant Height; SPL, Spike Length; SPLS, Spikelets per spike; TILL, Tillers per plant
Marker trait association under drought stress treatment [D]
Under Drought stress treatment [D], 95 associations were significant at Log(P) threshold while 63 associations were significant at FDR threshold. The maximum number of associations (22) were recorded for TILL followed by STI (19) and only a single marker was significantly associated with leaf area. Tillers per plant and stress tolerance index had also highest number of significant associations at FDR threshold each with 10 and 19 associations respectively. Harvest index, leaf area and spike length had no significant association beyond FDR threshold. Under [D] treatment, all the significant associations explained 19.33% phenotypic variation with maximum phenotypic variation (28.93% each) explained by markers IACX203 (67.24 cM) and wsnp_Ex_c18372_27196625 (71.76 cM) on chromosome 5D and associated with STI. Chromosome wise highest number of significant associations (24) were detected on chromosome 7B followed by 17 associations on chromosome 2B.
Significant associations for GY under [D] stress were detected on chromosome 3D, 6A, 6D, 7B and on unmapped chromosome, CAP7_c1274_206 on unmapped chromosome was most significantly associated with GY and explained 23.49% phenotypic variance. Pleiotropic regions under [D] stress were concentrated on chromosome 7B followed by chromosome 4B, chromosomal region significant for GY on 7B was ~15 cM apart from region associated with DW, FLW, HI, LA, PH and TILL. The chromosomal region on 3D, 6A and 6D were specifically associated with grain yield (Table 3, S2 Table).
Table 3. Marker Centi-Morgan position of significant marker traits associations detected for yield and related traits from BLUEs value under drought stress [D] treatment.
| Chromosome | DW | DTM | FLL | FLW | GY | HI | LA | PH | SPL | SPLS | TILL |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1B | - | - | - | - | - | - | - | - | - | - | 56.88 |
| 2B | - | - | - | - | - | - | - | - | - | 109.53 | 109.53 |
| 3D | - | - | - | - | 66.57 | - | - | - | - | - | - |
| 4B | - | 55.55, 55.96 | - | 55.55, 55.96 | - | - | - | - | 55.96 | 55.55, 55.96 | 55.55, 55.96, 62.92 |
| 6A | - | - | - | 28.46, 29.53, 31.89 | 6.98 | - | - | - | - | - | - |
| 6B | - | - | - | - | - | - | - | - | 0.37 | - | - |
| 6D | - | - | - | - | 9.47 | - | - | - | - | - | - |
| 7A | - | 178.42 | - | - | - | - | 135.81 | - | - | - | - |
| 7B | 163.3 | - | 163.3 | - | 134.06, 148.65, 150.6 | 163.3 | 163.3 | 163.3 | - | - | 76.31, 73.79, 71.66, 163.3, 171.11 |
| 7D | 208.1 | - | 208.1 | - | - | 208.1 | 208.1 | 208.1 | - | - | - |
Marker trait association under heat stress treatment [H]
Under [H], 151 associations were significant at Log(P) threshold while 122 of these were significant at FDR. TOL had maximum number (21) of significant associations followed by STI (20) and FLL (12) and all these MTAs were also significant at FDR. Only DW had no significant association at FDR threshold among all the studied traits. Under [H] stress the potent associations for grain yield were present on two chromosomes viz. 7B and unmapped chromosome and IACX5767 on 7B explained 21.62% phenotypic variation. All significant associations under [H] stress explained 20% phenotypic variation with maximum value of phenotypic variation of 24.17% for leaf area. Grain yield under [H] stress shared genomic region with all studied traits except FLL, PH, SL and TILL and these common genomic regions were present between 134.06–150.6 cM on chromosome 7B (Table 4, S2 Table).
Table 4. Centi-Morgan position of significant marker traits associations detected for yield and related traits from BLUEs value under heat stress [H] treatment.
| Chromosome | Biomass | DTM | FLL | FLW | GY | HI | LA | PH | SPL | SPLS | TILL |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2A | - | - | 52.74 | - | - | - | - | 52.74, 53.18 | - | - | - |
| 3A | - | - | 89.04, 89.48 | - | - | - | - | 89.48 | - | - | - |
| 3D | 66.57 | - | - | - | - | - | 66.57, 142.32 | - | - | - | - |
| 5B | - | - | - | - | - | - | 129.83 | - | - | - | - |
| 6A | - | 6.98 | - | 6.98 | - | - | - | - | - | 6.98 | - |
| 6D | - | 9.47 | - | 9.47 | - | - | - | - | - | 9.47 | - |
| 7A | - | 178.42 | - | 178.42 | - | - | - | - | - | 178.42 | - |
| 7B | 134.06, 148.65, 150.6 | 134.06, 148.65, 150.6 | - | 134.06, 148.65, 150.6 | 134.06, 148.65, 150.6 | 134.06, 143.23, 144.8, 148.65, 150.6 | 134.06, 148.65, 150.6 | - | - | 134.06, 148.65, 150.6 | - |
| 7D | - | - | 149.97, 197.58, 202.54, 206.75 | - | - | - | - | 149.97, 197.58, 202.54, 206.75 | 161.13, 197.58, 202.54, 203.58 | - | 161.13, 197.58, 202.54, 203.58 |
Marker trait association under combined heat and drought stress treatment [HD]
Under [HD] treatment, 93 associations were significant at Log(P) threshold while 60 of these were significant at FDR. Maximum numbers of significant associations were recorded for STI (29) and TOL (13) index and both these traits have higher number of significant associations on FDR threshold. DTM, FLW, GY and TILL were only agronomic traits which had association significant at FDR threshold. Chromosome wise, highest associations were recorded on chromosome 4B (12) followed by 2B, 7B and 7D each carrying 10 significant MTAs. MTAs for grain yield were present on 1A, 2D, 2B, 4A, 7B, 7D and on unmapped chromosome, the potent marker Kukri_rep_c68068_95 was present on chromosome 2D and explained 22.55% phenotypic variation. All significant associations significant for all studied traits on average explained 19% phenotypic variation with maximum contribution (23.67%) by RFL_Contig854_2253 present on 3A significantly associated with TOL. Grain yield share many pleotropic regions with other traits and most of these pleiotropic regions were concentrated on chromosome 7B and 7D where shares the chromosomal region with FLL, FLW, HI, PH, SL, SPLS and TILL (Table 5, S2 Table).
Table 5. Centi-Morgan position of significant marker traits associations detected for yield and related traits from BLUEs value under Combined drought and heat stress [HD] treatment.
| Chromosome | Biomass | DTM | FLL | FLW | GY | HI | LA | PH | SPL | SPLS | TILL |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1A | - | - | - | 108.06 | 108.06 | - | - | - | - | - | - |
| 2B | - | - | - | 16.88 | 16.88 | - | - | - | - | - | - |
| 2D | - | - | - | 100.19 | 100.19 | - | - | - | - | - | - |
| 4A | - | - | - | 48.52 | 48.52 | - | - | - | - | - | - |
| 4B | - | 61.31, 61.84, 62.56, 62.92 | - | - | - | - | - | 55.55, 55.96 | - | 55.55, 55.96 | - |
| 5A | 15.61 | - | - | - | - | - | - | - | - | - | - |
| 5B | 60.31 | - | - | - | - | - | 40.55, 43.56, 45.36, 45.4, 46.47 |
- | - | - | - |
| 5D | - | 69.13 | - | - | - | - | - | 135.81 | - | - | - |
| 7A | - | - | - | - | - | - | - | - | - | - | - |
| 7B | - | - | 163.3 | 98.3, 163.3 | 98.3, 163.3 | 163.3 | - | 163.3 | 163.3 | 163.3 | 163.3 |
| 7D | - | - | 208.1 | 208.1 | 208.1 | 208.1 | - | 208.1 | 208.1 | 208.1 | 208.1 |
Marker trait association for drought stress indices [D]
Marker trait associations for STI under [D] stress were recorded on chromosome 1A, 1B, 1D, 2B, 3A, 3D and 5D. BS00077789_51 on chromosome 2B at 37.03 was most potent loci associated with STI under drought stress and explained 29.08% phenotypic variation. Many genomic regions were shared between STI and other stress indices for example, genomic region on chromosome 2B and 5D were shared by STI and TOL under [D] treatment. Genomic regions significant for TOL under [D] stress were detected on chromosome 2B, 4D and 5D with the highest number of associations on chromosome 5D. The genomic region significantly associated for TOL under drought stress was common for both STI and TOL under all stress treatments. Locus BS00077789_51 on 2B at 37.03 cM was most significantly associated with TOL and explained 26.69% phenotypic variation (Table 6, S2 Table).
Table 6. Centi-Morgan position of significant marker traits associations detected for stress Indices for grain yield under all studied stress treatments.
| Chromosome | STI_D | STI_H | STI_HD | TOL_D | TOL_H | TOL_HD |
|---|---|---|---|---|---|---|
| 1A | 60.25, 80.13 | 51.07, 60.25, 61.41 | 60.25, 61.89, 69.53 | - | 25.09, 76.6, 80.13 | - |
| 1B | 31.10 | - | - | - | 29.6, 31.1 | 28.1, 29.6, 31.1, 37, 37.7 |
| 1D | 132.74 | - | - | - | - | 134.75 |
| 2B | 25.53, 27.64, 37.03, 40.6, 43.1 | 37.03, 40.6 | 37.03, 40.6, 85.07 | 25.53, 27.64, 37.03, 37.13, 40.6, 43.1 | 25.53, 27.64, 37.03, 37.13, 40.6, 43.1 | 25.53, 27.64, 37.13, 43.1 |
| 3A | 81.91, 209.25 | - | 51.36 | - | 81.91, 209.25 | 81.91 |
| 3D | 11.23, 176.18 | 11.23, 94.89, 96.7, 97.28, 127.96, 141.91, 176.18 | 11.23, 96.7, 97.28, 110.2, 141.91, 176.18 | - | 176.18 | - |
| 4A | - | - | 67.72,71.47 | - | - | - |
| 4B | - | 42.91 | 99.79 | - | - | - |
| 4D | - | 97.71 | 31.04, 40.94,97.71 | 158.59 | - | - |
| 5A | - | - | 87.78 | - | - | - |
| 5D | 25.45, 25.82, 67.24,71.76 | 67.24, 71.76 | 39.24, 67.24, 71.76 | 67.24, 71.76 | 25.45, 25.82, 67.24, 71.76 | 69.56 |
| 6A | - | 4.56 | - | - | 146.51 | - |
| 6B | - | 121.6 | 10.06 | - | - | - |
STI_D: Stress Tolerance index for GY under drought stress
STI_H: Stress Tolerance index for GY under heat stress
STI_HD: Stress Tolerance index for GY under combined drought and heat stress
TOL_D: Tolerance index for GY under drought stress
TOL_H: Tolerance index for GY under heat stress
TOL_HD: Tolerance index for GY under combined drought and heat stress
Marker trait association for heat stress indices [H]
Twenty significant MTAs were recorded for STI under [H] stress and were concentrated on chromosome 3D. The most significant association was present on 3D at 11.23 cM and explained 22.56% phenotypic variation. The region on 3D at 176.18 cM was also significant for TOL index. The TOL index had 21 significant associations under [H] stress most of which were concentrated on chromosome 2B and on average explained 21% phenotypic variation. The most potent association on 2B was present at 37.03 cM and explained 28.8% phenotypic variation. TOL index shared regions on chromosome 2B with stress indices under studied [D], [H] and [HD], similarly genomic region significant at chromosome 5D was also shared among all stress indices in all three treatments (Table 6, S2 Table).
Marker trait association for combined heat and drought stress indices [HD]
Twenty-nine significant associations were detected for STI under [HD] treatment concentrated on chromosome 3D and explaining average 19.62% phenotypic variation. IACX203 (67.24 cM) and wsnp_Ex_c18372_27196625 (71.76 cM) were most potent loci associated with STI under [HD] treatment and explained 21.22% phenotypic variation. Thirteen significant associations were detected for TOL under [HD] treatment concentrated on 2B and average phenotypic variation value of 20.05%. RFL_Contig854_2253 on 3A at 81.91 cM was most significant loci for TOL and explained 23.67% phenotypic variation (Table 6, S2 Table).
Comparison between different stress treatments
Many significant associations were shared between two or more stress treatments. [C] and [H] stress share four loci, [D] and [H] share 13 loci, [C] and [HD] share 3 loci, [H] and [HD] had two common loci while no common locus was recorded between [C] and [D] treatment. Some loci were common among the three treatments were also recorded [C], [D] and [H] had six loci in common [C], [H] and [HD] had three loci common among them [C], [D] and [HD] had two loci in common and [D], [H] and [HD] had three loci in common. Nine loci were common among all four treatments and were recorded as consistent loci (Fig 6).
Fig 6. Stress specific and consistent associations shared among more than one stress treatments.
Traits having common associations
A total 69 pleiotropic SNPs were associated with multiple traits under all stress treatments, [H] with maximum (31) of these followed by [C], [HD] and [D] each having 20, 10 and 8 associations respectively (Fig 7A). The maximum number of pleiotropic associations were recorded on chromosome 7D followed by unmapped chromosome and 7B (Fig 7B). Locus BS00022775_51 was associated with eight studied traits, i.e. Biomass, DTM, FLW, GY, HI, LA, SLPS and SL, similarly locus BobWhite_c28058_232 and wsnp_Ex_c8400_14157060 on chromosome 7B at 134.06 cM were associated with seven out of eleven studied traits. Under [C] treatment locus Tdurum_contig52086_129 on chromosome 1B at 158.59 cM was associated with six multiple traits while BobWhite_s64797_152 and Tdurum_contig33737_157 on 4B at 55 cM were associated with five studied traits under [D] stress. Locus BobWhite_c28058_232 and wsnp_Ex_c8400_14157060 on chromosome 7B at 134.06 cM were associated with seven traits under [H] stress while locus RAC875_rep_c110526_324 on 7B at 163.3 cM was associated with 8 studied traits under [HD] treatment (Table 7).
Fig 7.
a) Number of peliotropic SNPs associated with studied traits under all stress treatments i.e. [C], [D], [H] and [HD]. b) Chromosome wise distribution of pleiotropic SNPs associated with traits under all stress treatments.
Table 7. Chromosome, centi Morgan position and associated traits of pleiotropic SNPs recoded from association analysis of BLUEs values.
| Significant SNPs | Chr. | Position | Control [C] | Drought [D] | Heat [H] | Combination [HD] |
|---|---|---|---|---|---|---|
| wsnp_Ex_c33831_42253707, wsnp_Ku_c17726_26872129 | 1A | 70.1–71.05 | Biomass, DTM | |||
| Excalibur_c26688_138 | 1A | 108.06 | FLW, GY | |||
| Tdurum_contig52086_129 | 1B | 158.59 | DTM, FLL, GY, HI, SPLS, TILL | |||
| RAC875_c62936_139 | 2A | 52.74 | FLL, PH | |||
| Excalibur_c2656_3198 | 2B | 16.88 | FLW, GY | |||
| BS00077789_51, tplb0060j17_879 | 2B | 37.03–40.6 | STI, TOL | |||
| IAAV1903 | 2B | 96.14 | GY, SPLS, TILL | |||
| Excalibur_c6922_1393-Kukri_s115194_71 | 2B | 109.53 | SPLSS, TILL | |||
| Kukri_rep_c68068_95 | 2D | 100.19 | FLW, GY | |||
| wsnp_Ex_c14340_22315611, wsnp_Ex_rep_c66907_65324299 | 3A | 89.48 | FLL, PH | |||
| Tdurum_contig100787_79 | 3B | 137.84 | Biomass, SPL | |||
| RFL_Contig2471_119 | 3D | 23.41 | DTM, FLL, GY, SPLS, TILL | |||
| BS00021930_51 | 3D | 66.57 | Biomass, LA | |||
| BS00095640_51 | 3D | 82.56 | SPLS, TILL | |||
| GENE-1752_162 | 3D | 101.24 | Biomass, DTM, GY, SPLS, TILL | |||
| BS00029720_51 | 3D | 176.18 | STI, TOL | |||
| Ex_c17894_1159 | 4A | 48.52 | FLW, GY | |||
| BobWhite_s64797_152, Tdurum_contig33737_157 | 4B | 55.55–55.96 | DTM, FLW, SPL, SPLSS, TILL | PH, SPLS | ||
| Tdurum_contig49841_618 | 5B | 32.73 | DTM, FLL, GY, SPLS, TILL | |||
| Ra_c22730_460 | 5B | 100.64 | DTM, GY | |||
| BS00029540_51 | 5B | 141.91 | DTM, FLW, GY, HI | |||
| IACX203, wsnp_Ex_c65985_64188864, wsnp_Ex_c18372_27196625 | 5D | 67.24–71.76 | DTM, GY | STI, TOL | ||
| Tdurum_contig27939_357 | 6A | 6.98 | DTM, FLW, SLPS | |||
| BobWhite_rep_c63956_254 | 6A | 133.74 | DTM, FLW, GY, HI | |||
| Excalibur_c4789_2748 | 6D | 9.47 | DTM, FLW, SLPS | |||
| RFL_Contig2615_982 | 6D | 153.08 | Biomass, GY | |||
| Kukri_c34887_734 | 7A | 63.21 | DTM, GY | |||
| wsnp_Ex_c53442_56678505 | 7A | 178.42 | DTM, FLW, SLPS |
Discussion
In last few decades lots of efforts had been made to study the effects of a single stress on plants under control and field conditions [36]. However, in open field conditions, plant have to face multiple biotic and abiotic stresses at same time and one cannot really understand the effects of these combined stresses by applying one stress separately in controlled lab conditions. The combination of two or more stresses alters plant proteomics, metabolism and genetics in a unique way which is totally different from changes imposed by individual stress [23,37,38]. To address this scenario, we aimed to determine independent and combined effect of drought and heat stress on wheat yield and related traits and finding out genomic regions associated with heat and drought stress tolerance.
A diverse panel of 192 advance wheat lines was planted at National Agriculture Research Center Islamabad for two cropping seasons in four treatments, i.e. [C], [D], [H] and [HD]. The GY was severely reduced by [HD] stress (51.67% reduction) followed by [H] (27.33% reduction) and [D] treatment (24.9% reduction). Similar results were reported by [15,39,40] for bread wheat genotypes grown under heat and drought stressed environments. Heritability estimates were high for many traits showing uniformity in the performance of genotypes across the years, these values are similar as reported by [26,41]. Consistent negative association of GY with DTM under stressed conditions suggests early maturity is beneficial in stressed environments [42]. Plant height had a consistent positive association with grain yield this might be due to positive correlation of PH with DW [43–45]. This suggests that under stressed environment tall genotypes can accumulate and mobilize more reserve to grain and thus had higher yield than short stature genotypes [43].
Association mapping is a powerful approach to elucidate genetic basis of complex traits including grain yield [41,46–48], in spite of its high-resolution there are higher chances to detect false positive associations due to population structure [49,50]. In our study mixed linear model was used to detect significant MTAs and significance was further tested to avoid false positive associations using FDR threshold. The MTAs for GY were detected on almost all chromosomes with maximum number of significant associations under [C] treatment. QTLs for grain yield and its related traits are reported on almost all wheat chromosome [51–54]. The maximum number of associations for grain yield were detected on chromosome 7B under both stress and non-stress conditions. The association on 7B at 134.06 cM was approximately 5 cM apart from genomic region (129.77 cM) reported by [41]for plant height and the genomic region reported for tolerance index by [15]. A region significant for grain yield on chromosome 4A at 48.52 cM was close to locus Kukri_c12563_52 at 66.28 cM reported significant for effective number of spikes per square meter by [41]. The same study reported a genomic region on 5B at 39.64 cM for GY was 7 cM away from significant region for grain yield on 5B at 32.73 cM reported in the present study. Locus RAC875_c29431_1849 on 5B at 49.01 was close to genomic regions Tdurum_contig52439_196 at 40.56 cM [41], wsnp_CAP7_c2086_1018815 at 43.42 cM [55]and wsp_Ex_rep_c66651_64962429 at 49.01 cM [56] significantly associated with GY, normalized differential vegetative index at vegetative stage and thousand-kernel weight respectively. Locus BobWhite_c28058_232 and wsnp_Ex_c8400_14157060 at 134.06 cM and BS00076622_51 at on 148.65 cM on 7B were associated with yield in two out of four environments and similar regions were previously reported by [57] for SPN (134.5 cM) and KNL (149.5 cM).
Germplasm evaluation in multiple environmental conditions helps us to detect and compare QTLs for desired traits common among more than one condition. Genotype × environment interaction is the main problem in breeding for diverse adaptation in bread wheat. Occurrence of common genomic region among more than one stress shows common adaptation and tolerance mechanism of accessions under these stress treatments. In our study nine common regions were identified which were associated with traits under all four treatments i.e. [C], [D], [H] and [HD] treatment. In addition, two different stress indices were also used to find out stable regions across the environments. Multiple consistent regions for both stress indices were recorded on chromosome 2B and 5D. Many regions were shared among grain yield and stress indices for instance the genomic region on 5D at 67.24–71.76 cM was shared by DTM, GY, STI and TOL. A set of MTAs on chromosome 2B at 85.07 cM was near to IWB2285 locus at 70.4 cM significant for grain yield and grains per spike in collection of tetraploid wheat [54]. The genomic regions on 2B were present at 25.53, 27.64 and 37.03 cM, genomic regions in the same range (24.7 cM and 36.9 cM) as were reported for heat susceptibility index for kernel number, flag-leaf length and grain yield by [58]. The associations common among two or more treatments and associations significant for stress indices are of vital importance and can be used in marker-assisted selection to improve wheat tolerance to combined heat and drought stress.
Makers shared among more than one trait are very useful in marker-assisted selection as they may play a vital role in increasing QTL pyramiding efficiency. Multi trait markers were detected on all chromosomes in present study except 1D, 4D, 5A and 6B. These markers share two or more than two traits and maximum number of these multi trait markers was detected on 7B. The locus RAC875_rep_c110526_324 on 7B at 163.3 cM and chromosomal region on chromosome 7D at 208.1 cM were shared among 8 studied traits including GY. This implies that there are genetic bases for the high and consistent phenotypic correlation recorded among grain yield and these traits. The region on chromosome 7B was approximately 39 cM and 52 cM apart from two genomic regions M1441 and M7175 respectively identified by [59] for GY and associated traits in CIMMYT wheat accessions. The distal regions of chromosome 7A and 7B are reported to contain QTLs for grain yellow pigment content which is controlled by Phytoene synthase 1 (PSY-1) gene, presence of this gene might be responsible for pleiotropic associations on 7B [60]. Pleiotropic regions identified on chromosome 7D are may be due to pleiotropic effects of 7DL.7Ag translocation in wheat [61]. In addition to yield many genomic regions were shared between leaf characters (FLL, FLW, LA) and spike characters under stress treatments for example marker Excalibur_c26688_138 and BS00022775_51 on 1A and unmapped chromosome respectively were shared by FLW and GY. Another region on chromosome 7D at 202.54 cM was shared by FLL, PH, SL, TILL and BS00076622_51 on 7B at 148.65 cM was shared by FLW and HI. Sharing of these regions may be attributed to effective translocation of leaf photosynthates to growing spike and kernels during stress [62].
Conclusion
The identification and introgression of major-effect QTLs are one of the best and proven approaches to improving the stress tolerance of wheat varieties. The accuracy and consistency of QTLs have clearly shown that structured association mapping with genome-wide molecular markers is an attractive option to identify major-effect QTLs for GY under different stress treatments. Association mapping results revealed many novel regions associated with combined drought and heat stress tolerance, along with nine consistent regions common among all four treatments and many pleiotropic regions associated with more than one traits. Further exploration of common regions among treatments through marker assisted selection can help in understanding complex mechanisms of abiotic stress tolerance and fine mapping of these regions can lead to new gene discoveries. Intensity of climate change will increase in next few years and will decrease wheat production, thus it is need of time to improve germplasm by arranging crosses between diverse parents and improve genomic technology and couple these technologies with marker assisted selection. The present study will help in understanding irrigation use efficiency and tolerance of exotic lines to rain fed conditions. It will help in identification and inclusion of tolerant genotypes in breeding program for further strengthening.
Supporting information
(XLSX)
(XLSX)
(TIF)
(TIF)
(PDF)
(PDF)
Acknowledgments
The authors are grateful for Higher Education Commission (HEC) Pakistan for International Research Support Initiative Support Program (IRSIP), Dr Jalal-ud-Din head Crop Physiology group for providing space for field experiments and lab work at National Agriculture Research Center Islamabad Pakistan and Dr Marion S Roeder group leader at Gene and Genome mapping group IPK, Gatersleben, Germany for facilitating data analysis and genotyping.
Abbreviations
- DW/Biomass
Plant above ground dry weight
- DTM
Days to maturity
- FDR
False discovery rate
- FLL
Flag leaf length
- FLW
Flag leaf width
- GWAS
Genome wide association studies
- GY
grain yield, HI: Harvest index
- LA
Leaf area
- LD
Linkage disequilibrium
- MAF
Minor allele frequency
- MTAs
Marker trait associations
- PH
Plant height
- SL
Spike length
- SPLS
Spikelets per spike
- TILL
Tillers per plant
- STI
Stress tolerance index
- TOL
Tolerance index
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Acevedo E, Silva P, Silva H. Wheat growth and physiology. Bread Wheat, Improvement and Production. 2002:30. [Google Scholar]
- 2.Godfray HCJ, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, et al. Food security: the challenge of feeding 9 billion people. science. 2010:1185383. [DOI] [PubMed] [Google Scholar]
- 3.Ray DK, Ramankutty N, Mueller ND, West PC, Foley JA. Recent patterns of crop yield growth and stagnation. Nature communications. 2012;3:1293 10.1038/ncomms2296 [DOI] [PubMed] [Google Scholar]
- 4.Ali S, Liu Y, Ishaq M, Shah T, Ilyas A, Din IU. Climate change and its impact on the yield of major food crops: Evidence from Pakistan. Foods. 2017;6(6):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen N. Global climate changes and rice food security. Rome: FAO; 2002. [Google Scholar]
- 6.Atlin GN, Cairns JE, Das B. Rapid breeding and varietal replacement are critical to adaptation of cropping systems in the developing world to climate change. Global food security. 2017;12:31–7. 10.1016/j.gfs.2017.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassia R, Nocioni M, Correa-Aragunde N, Lamattina L. Climate Change and the Impact of Greenhouse Gasses: CO2 and NO, Friends and Foes of Plant Oxidative Stress. Frontiers in plant science. 2018;9:273 10.3389/fpls.2018.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flato G, Marotzke J, Abiodun B, Braconnot P, Chou SC, Collins W, et al. Evaluation of climate models. Cambridge University Press; 2013. [Google Scholar]
- 9.Crespo-Herrera LA, Crossa J, Huerta-Espino J, Autrique E, Mondal S, Velu G, et al. Genetic yield gains in CIMMYT’s international elite spring wheat yield trials by modeling the genotype× environment interaction. Crop Science. 2017;57(2):789–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sukumaran S, Lopes M, Dreisigacker S, Reynolds M. Genetic analysis of multi-environmental spring wheat trials identifies genomic regions for locus-specific trade-offs for grain weight and grain number. Theoretical and Applied Genetics. 2018;131(4):985–98. 10.1007/s00122-017-3037-7 [DOI] [PubMed] [Google Scholar]
- 11.Aisawi K, Reynolds M, Singh R, Foulkes M. The physiological basis of the genetic progress in yield potential of CIMMYT spring wheat cultivars from 1966 to 2009. Crop Science. 2015;55(4):1749–64. [Google Scholar]
- 12.Sharma R, Crossa J, Velu G, Huerta-Espino J, Vargas M, Payne T, et al. Genetic gains for grain yield in CIMMYT spring bread wheat across international environments. Crop Science. 2012;52(4):1522–33. [Google Scholar]
- 13.Graziani M, Maccaferri M, Royo C, Salvatorelli F, Tuberosa R. QTL dissection of yield components and morpho-physiological traits in a durum wheat elite population tested in contrasting thermo-pluviometric conditions. Crop and Pasture Science. 2014;65(1):80–95. [Google Scholar]
- 14.Zhu C, Gore M, Buckler ES, Yu J. Status and prospects of association mapping in plants. The plant genome. 2008;1(1):5–20. [Google Scholar]
- 15.Sukumaran S, Reynolds MP, Sansaloni C. Genome-Wide Association Analyses Identify QTL Hotspots for Yield and Component Traits in Durum Wheat Grown under Yield Potential, Drought, and Heat Stress Environments. Frontiers in Plant Science. 2018;9(81). 10.3389/fpls.2018.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorkheh K, Malysheva-Otto LV, Wirthensohn MG, Tarkesh-Esfahani S, Martínez-Gómez P. Linkage disequilibrium, genetic association mapping and gene localization in crop plants. Genetics and Molecular Biology. 2008;31(4):805–14. [Google Scholar]
- 17.Sonah H, O'Donoughue L, Cober E, Rajcan I, Belzile F. Identification of loci governing eight agronomic traits using a GBS‐GWAS approach and validation by QTL mapping in soya bean. Plant biotechnology journal. 2015;13(2):211–21. 10.1111/pbi.12249 [DOI] [PubMed] [Google Scholar]
- 18.Maccaferri M, El-Feki W, Nazemi G, Salvi S, Canè MA, Colalongo MC, et al. Prioritizing quantitative trait loci for root system architecture in tetraploid wheat. Journal of Experimental Botany. 2016;67(4):1161–78. 10.1093/jxb/erw039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valluru R, Reynolds MP, Davies WJ, Sukumaran S. Phenotypic and genome‐wide association analysis of spike ethylene in diverse wheat genotypes under heat stress. New Phytologist. 2017;214(1):271–83. 10.1111/nph.14367 [DOI] [PubMed] [Google Scholar]
- 20.Zanke CD, Ling J, Plieske J, Kollers S, Ebmeyer E, Korzun V, et al. Analysis of main effect QTL for thousand grain weight in European winter wheat (Triticum aestivum L.) by genome-wide association mapping. Frontiers in plant science. 2015;6:644 10.3389/fpls.2015.00644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qaseem MF, Qureshi R, Muqaddasi QH, Shaheen H, Kousar R, Röder MS. Genome-wide association mapping in bread wheat subjected to independent and combined high temperature and drought stress. PloS one. 2018;13(6):e0199121 10.1371/journal.pone.0199121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thoen MP, Davila Olivas NH, Kloth KJ, Coolen S, Huang PP, Aarts MG, et al. Genetic architecture of plant stress resistance: multi‐trait genome‐wide association mapping. New Phytologist. 2017;213(3):1346–62. 10.1111/nph.14220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z, Qin J, Tian X, Xu S, Wang Y, Li H, et al. Global profiling of alternative splicing landscape responsive to drought, heat and their combination in wheat (Triticum aestivum L.). Plant biotechnology journal. 2018;16(3):714–26. 10.1111/pbi.12822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tricker PJ, ElHabti A, Schmidt J, Fleury D. The physiological and genetic basis of combined drought and heat tolerance in wheat. Journal of experimental botany. 2018;69(13):3195–210. 10.1093/jxb/ery081 [DOI] [PubMed] [Google Scholar]
- 25.Tahmasebi S, Heidari B, Pakniyat H, McIntyre CL. Mapping QTLs associated with agronomic and physiological traits under terminal drought and heat stress conditions in wheat (Triticum aestivum L.). Genome. 2016;60(1):26–45. 10.1139/gen-2016-0017 [DOI] [PubMed] [Google Scholar]
- 26.Qaseem MF, Qureshi R, Muqaddasi QH, Shaheen H, Kousar R, Röder MSJPo. Genome-wide association mapping in bread wheat subjected to independent and combined high temperature and drought stress. 2018;13(6):e0199121 10.1371/journal.pone.0199121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, et al. Characterization of polyploid wheat genomic diversity using a high‐density 90 000 single nucleotide polymorphism array. Plant biotechnology journal. 2014;12(6):787–96. 10.1111/pbi.12183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maccaferri M, Zhang J, Bulli P, Abate Z, Chao S, Cantu D, et al. A Genome-Wide Association Study of Resistance to Stripe Rust (Puccinia striiformis f. sp. tritici) in a Worldwide Collection of Hexaploid Spring Wheat (Triticum aestivum L.). G3: Genes|Genomes|Genetics. 2015;5(3):449–65. 10.1534/g3.114.014563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alomari DZ, Eggert K, Von Wirén N, Pillen K, Röder MS. Genome-Wide Association Study of Calcium Accumulation in Grains of European Wheat Cultivars. Frontiers in plant science. 2017;8:1797 10.3389/fpls.2017.01797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu J, Pressoir G, Briggs WH, Bi IV, Yamasaki M, Doebley JF, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nature genetics. 2006;38(2):203–8. 10.1038/ng1702 [DOI] [PubMed] [Google Scholar]
- 32.Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, et al. GAPIT: genome association and prediction integrated tool. Bioinformatics. 2012;28(18):2397–9. 10.1093/bioinformatics/bts444 [DOI] [PubMed] [Google Scholar]
- 33.Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, et al. Efficient control of population structure in model organism association mapping. Genetics. 2008;178(3):1709–23. 10.1534/genetics.107.080101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Peng Z, Yang X, Wang W, Fu J, Wang J, et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nature genetics. 2013;45(1):43–50. 10.1038/ng.2484 [DOI] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society Series B (Methodological). 1995:289–300. [Google Scholar]
- 36.Mittler R, Blumwald E. Genetic engineering for modern agriculture: challenges and perspectives. Annual review of plant biology. 2010;61:443–62. 10.1146/annurev-arplant-042809-112116 [DOI] [PubMed] [Google Scholar]
- 37.Rizhsky L, Liang H, Mittler R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant physiology. 2002;130(3):1143–51. 10.1104/pp.006858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zandalinas SI, Rivero RM, Martínez V, Gómez-Cadenas A, Arbona V. Tolerance of citrus plants to the combination of high temperatures and drought is associated to the increase in transpiration modulated by a reduction in abscisic acid levels. BMC plant biology. 2016;16(1):105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinto RS, Reynolds MP, Mathews KL, McIntyre CL, Olivares-Villegas J-J, Chapman SC. Heat and drought adaptive QTL in a wheat population designed to minimize confounding agronomic effects. Theor Appl Genet. 2010;121(6):1001–21. 10.1007/s00122-010-1351-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sukumaran S, Reynolds MP, Sansaloni C. Genome-wide association analyses identify QTL hotspots for yield and component traits in durum wheat grown under yield potential, drought, and heat stress environments. Frontiers in Plant Science. 2018;9:81 10.3389/fpls.2018.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S-X, Zhu Y-L, Zhang D-X, Shao H, Liu P, Hu J-B, et al. Genome-wide association study for grain yield and related traits in elite wheat varieties and advanced lines using SNP markers. PloS one. 2017;12(11):e0188662 10.1371/journal.pone.0188662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mondal S, Joshi AK, Huerta-Espino J, Singh RP, editors. Early Maturity in Wheat for Adaptation to High Temperature Stress2015; Tokyo: Springer Japan. [Google Scholar]
- 43.Zarei L, Cheghamirza K, Farshadfar E. Evaluation of grain yield and some agronomic characters in durum wheat ('Triticum turgidum'L.) Under rainfed conditions. Australian Journal of Crop Science. 2013;7(5):609. [Google Scholar]
- 44.Sadeghzadeh D, Alizadeh K. Relationship between grain yield and some agronomic characters in durum wheat under cold dryland conditions of Iran. Pak J Biol Sci. 2005;8:959–62. [Google Scholar]
- 45.Qaseem MF, Qureshi R, Illyas N, Jalal-Ud-Din, Shabbir G. Multivariate statistical analysis for yield and yield components in bread wheat planted under rainfed conditions. Pakistan Journal of Botany. 2017;(49):6. [Google Scholar]
- 46.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273(5281):1516–7. [DOI] [PubMed] [Google Scholar]
- 47.Sun C, Zhang F, Yan X, Zhang X, Dong Z, Cui D, et al. Genome‐wide association study for 13 agronomic traits reveals distribution of superior alleles in bread wheat from the Yellow and Huai Valley of China. Plant biotechnology journal. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morris GP, Ramu P, Deshpande SP, Hash CT, Shah T, Upadhyaya HD, et al. Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proceedings of the National Academy of Sciences. 2013;110(2):453–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang LY, Liu DC, Guo XL, Yang WL, Sun JZ, Wang DW, et al. Genomic distribution of quantitative trait loci for yield and yield‐related traits in common wheat. Journal of integrative plant biology. 2010;52(11):996–1007. 10.1111/j.1744-7909.2010.00967.x [DOI] [PubMed] [Google Scholar]
- 50.Yu J, Pressoir G, Briggs WH, Bi IV, Yamasaki M, Doebley JF, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nature genetics. 2006;38(2):203 10.1038/ng1702 [DOI] [PubMed] [Google Scholar]
- 51.Cui F, Zhao C, Ding A, Li J, Wang L, Li X, et al. Construction of an integrative linkage map and QTL mapping of grain yield-related traits using three related wheat RIL populations. Theoretical and applied genetics. 2014;127(3):659–75. 10.1007/s00122-013-2249-8 [DOI] [PubMed] [Google Scholar]
- 52.Araus JL, Slafer GA, Royo C, Serret MD. Breeding for yield potential and stress adaptation in cereals. Critical Reviews in Plant Science. 2008;27(6):377–412. [Google Scholar]
- 53.Gupta PK, Balyan HS, Gahlaut V. QTL analysis for drought tolerance in wheat: present status and future possibilities. Agronomy. 2017;7(1):5. [Google Scholar]
- 54.Mangini G, Gadaleta A, Colasuonno P, Marcotuli I, Signorile AM, Simeone R, et al. Genetic dissection of the relationships between grain yield components by genome-wide association mapping in a collection of tetraploid wheats. PloS one. 2018;13(1):e0190162 10.1371/journal.pone.0190162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sukumaran S, Dreisigacker S, Lopes M, Chavez P, Reynolds MP. Genome-wide association study for grain yield and related traits in an elite spring wheat population grown in temperate irrigated environments. Theoretical and applied genetics. 2015;128(2):353–63. 10.1007/s00122-014-2435-3 [DOI] [PubMed] [Google Scholar]
- 56.Ain Q-u, Rasheed A, Anwar A, Mahmood T, Imtiaz M, Mahmood T, et al. Genome-wide association for grain yield under rainfed conditions in historical wheat cultivars from Pakistan. Frontiers in plant science. 2015;6 10.3389/fpls.2015.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edae EA, Byrne PF, Haley SD, Lopes MS, Reynolds MP. Genome-wide association mapping of yield and yield components of spring wheat under contrasting moisture regimes. Theoretical and applied genetics. 2014;127(4):791–807. 10.1007/s00122-013-2257-8 [DOI] [PubMed] [Google Scholar]
- 58.Mason RE, Mondal S, Beecher FW, Pacheco A, Jampala B, Ibrahim AM, et al. QTL associated with heat susceptibility index in wheat (Triticum aestivum L.) under short-term reproductive stage heat stress. Euphytica. 2010;174(3):423–36. [Google Scholar]
- 59.Sehgal D, Autrique E, Singh R, Ellis M, Singh S, Dreisigacker S. Identification of genomic regions for grain yield and yield stability and their epistatic interactions. Scientific Reports. 2017;7:41578 10.1038/srep41578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang W, Dubcovsky J. Association between allelic variation at the Phytoene synthase 1 gene and yellow pigment content in the wheat grain. Theoretical and Applied Genetics. 2008;116(5):635–45. 10.1007/s00122-007-0697-8 [DOI] [PubMed] [Google Scholar]
- 61.Monneveux P, Reynolds M, Aguilar JG, Singh R, Weber W. Effects of the 7DL. 7Ag translocation from Lophopyrum elongatum on wheat yield and related morphophysiological traits under different environments. Plant breeding. 2003;122(5):379–84. [Google Scholar]
- 62.Lupton F. Translocation of photosynthetic assimilates in wheat. Ann Appl Biol. 1966;57(3):355–64. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(TIF)
(TIF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.