Abstract
Drug-induced liver injury (DILI) is a major issue for both patients and pharmaceutical industry due to insufficient means of prevention/prediction. In the current work we present a 2-class classification model for DILI, generated with Random Forest and 2D molecular descriptors on a dataset of 966 compounds. In addition, predicted transporter inhibition profiles were also included into the models. The initially compiled dataset of 1773 compounds was reduced via a 2-step approach to 966 compounds, resulting in a significant increase (p-value < 0.05) in model performance. The models have been validated via 10-fold cross-validation and against three external test sets of 921, 341 and 96 compounds, respectively. The final model showed an accuracy of 64% (AUC 68%) for 10-fold cross-validation (average of 50 iterations) and comparable values for two test sets (AUC 59%, 71% and 66%, respectively). In the study we also examined whether the predictions of our in-house transporter inhibition models for BSEP, BCRP, P-glycoprotein, and OATP1B1 and 1B3 contributed in improvement of the DILI mode. Finally, the model was implemented with open-source 2D RDKit descriptors in order to be provided to the community as a Python script.
Keywords: Drug-induced liver injury, Random Forest, 2-class classification, Liver transporters, Data curation, Toxicity reports
1. Introduction
Drug-induced liver injury (DILI) is the term used for liver damage that is caused by drugs, herbal agents or nutritional supplements (Ghabril et al., 2010; Watkins and Seeff 2006). DILI has gained increasing attention in recent years (Raschi and De Ponti, 2015), as it is one of the main causes for attrition during clinical and pre-clinical studies and the main reason for drug withdrawal from the market or for labeling with a black box warning (Ballet 1997; Chen et al., 2011; O’Brien et al., 2006; Regev 2014). Thus, great effort has been invested towards elucidating the toxicological processes and mechanisms that result in manifestations of DILI (Vinken, 2015). It is widely accepted that, together with metabolizing enzymes, liver transporters play an important role for maintaining the integrity and proper function of the liver, and also influence the ADMET (absorption, distribution, metabolism, excretion and toxicity) profile of drugs (Faber et al., 2003; Shitara et al., 2013). Actually, there are several recent publications suggesting that inhibition of liver transporters might result in manifestations of DILI. For cholestasis in particular, strong evidence towards the role of the bile salt export pump (BSEP) (Aleo et al., 2014; Dawson et al., 2011; Padda et al., 2011; Qiu et al., 2016; Vinken 2015; Vinken et al., 2013; Welch et al., 2015) has been posed. There is also evidence for the multidrug resistance-associated protein 2 (MRP2) (Padda et al., 2011; Pauli-Magnus and Meier 2006), breast cancer resistance protein (BCRP) (Padda et al., 2011; Pauli-Magnus and Meier 2006), P-glycoprotein (Padda et al., 2011; Pauli-Magnus and Meier 2006) and multidrug resistance-associated protein 3 and 4 (MRP3 and MRP4) (Padda et al., 2011; Pauli-Magnus and Meier 2006; Welch et al., 2015) to be involved. For hyperbilirubinemia, another possible manifestation of hepatotoxicity, involvement of organic anion transporting polypeptides 1B1 and 1B3 (OATP1B1 and OATP1B3) (Chang et al., 2013; Sticova and Jirsa 2013), MRP2 (Sticova and Jirsa, 2013) and to a smaller extent BCRP (Sticova and Jirsa, 2013) is discussed.
Although in vitro predictive methods are efficient for many toxic endpoints, they are time-consuming and expensive (Bowes et al., 2012; Whitebread et al., 2005). In addition, for assessing hepatotoxicity, experimental methods such as in vitro tests and animal models, have been shown to share low concordance (< 50%) with human hepatotoxicity (Chen et al., 2011; Liu et al., 2011; Olson et al., 2000).
This led to the development of predictive computational methods, which are summarized in two recent reviews by (Chen et al., 2014) and (Ekins, 2014). Although all these models generally perform quite well, they sometimes suffer from low statistical performance, imbalanced sensitivity vs specificity, or small data sets (Table 1).
Table 1.
Classification models for DILI reported in literature. Acc stands for accuracy, Sen for sensitivity, Spec for specificity, BA for balanced accuracy, CV for cross validation, EV for external validation and IV for internal validation.
| Reference | Descriptors | Classification algorithm | Data used | Reported performance |
|---|---|---|---|---|
| Cheng and Dixon (2003) | 2D molecular descriptor | Ensemble recursive partitioning | 382 drugs for CV | CV: 76% Acc; 76% Sen; 75% Spec |
| 54 drugs for EV | EV: 81% Acc; 70% Sen; 90% Spec | |||
| Cruz-Monteagudo et al. (2008) | Radial distribution function | Linear discriminant analysis | 74 drugs for CV | CV: 84% Acc; 78% Sen; 90% Spec |
| molecular descriptors | 13 drugs for EV | EV: 82% Acc | ||
| Matthews et al. (2009) | Molecular descriptors | 4 commercial QSAR programs | ~1600 drugs for CV | CV: 39% Sen; 87% Spec |
| 18 drugs for EV | EV: 89% Sen | |||
| Rodgers et al. (2010) | topological indices of molecular structures (MolConnZ) and Dragon molecular descriptors |
k-nearest neighbor | 37 drugs for EV | 84% Acc; 74% Sen; 94% Spec |
| Fourches et al. (2010) | 2D fragments and Dragon | Support vector machine | 531 drugs for CV 18 compounds for EV | CV: 62–68% Accs |
| molecular descriptors | EV: 78% Acc | |||
| Ekins et al. (2010) | extended connectivity functional | Linear discriminant analysis | 295 compound for CV | CV: 59% ACC; 53% Sen; 65% Spec |
| class fingerprints of maximum diameter 6 (ECFC_6) | 237 compounds for EV | EV: 60% Acc; 56% Sen; 67% Spec | ||
| Liew et al. (2011) | PaDEL molecular descriptor | Ensemble of mixed learning | 1087 compounds for CV | CV: 68% Accs; 67% Sen; 70% Spec |
| 120 compounds for EV | EV: 75% Acc; 82% Sen; 65% Spec | |||
| Liu et al. (2011) | functional class | Bayesian models | 888 drugs for training3 data sets with 40–148 drugs for EV | EV: 60–70% Accs |
| fingerprints (FCFP_6) | ||||
| Chen et al. (2013) | Mold2 chemical descriptor | Decision Forest | 197 drugs for CV | CV: 70% Acc |
| Three data sets with 190–348 drugs for EV |
EV: 62–69% Accs | |||
| Liu et al. (2015a) | physicochemical descriptors and fingerprints | Ensemble classifier | 677 compounds for CV | 81% BA; 66% Sen; 95% Spec |
| Muller et al. (2015) | physicochemical descriptors and fingerprints | Ensemble classifier | 677 compounds for CV | 81% BA; 66% Sen; 95% Spec |
| Muller et al. (2015) | ISIDA fragment descriptors | SVM | 424 drugs for CV | 66% BA |
| Xu et al. (2015) | Encoding layers based on SMILES, PaDEL descriptors | Deep Learning | 190, 475 & 1065 compounds for CV | CV: 70–88% Accs; 70–90% Sens; 70–87% Specs |
| 185,320, 236,198 & 119 compounds for EV | EV: 62–87% Accs; 62–83% Sens; 62–93% Specs | |||
| Mulliner et al. (2016) | 2D and 3D physicochemical descriptors | SVM with a genetic algorithm | 3712 compounds for training | IV: 75% Acc; 73% AUC |
| 221 compounds for IV | ||||
| 269 compounds for EV | ||||
| Zhang et al. (2016) | FP4 fingerprints | SVM | 1317 compounds for training | Training set: 66% Acc; 85% Sen; 34% Spec; 55% AUC |
| 88 compounds for EV | EV: 75% Acc; 93% Sen; 38% Spec; 61% AUC |
In this study we generate in silico classification models for DILI by compiling multiple and diverse datasets from literature. We carefully curated these data regarding the chemotypes, as well as the accuracy of the class label. In addition, we are exploring the importance of hepatic transporter inhibition on DILI by using the predictions of a set of in-house in silico classification models as additional descriptors for the DILI model.
2. Methods
2.1. Data compilation
2.1.1. Training set
Searching PubMed, 2017 (http://www.ncbi.nlm.nih.gov/pubmed), Google, 2017 (https://www.google.at) and Scopus, 2017 (https://www.scopus.com/) using the terms: “drug-induced liver injury”, “DILI”, “drug-induced hepatotoxicity” identified 9 unique datasets for human DILI/hepatotoxicity (Table 2).
Table 2.
Description of the sources upon which the training set was built. In number of compounds, “+” denotes the number of DILI-positive compounds and “−” the number of negative compounds. These numbers correspond to the number of compounds remaining after data curation in a source by source basis.
| Source name | Type of data | Number of compounds | Label choice |
|---|---|---|---|
| O’Brien et al. (2006) | In vitro cell-based assay | 132 (100+/32−) | “severely” and “moderately” toxic are considered positives. |
| Rodgers et al. (2010) | FDA reports database | 382 (75+/307−) | Authors classification |
| Fourches et al. (2010) | Text mining | 902 (620+/282−) | Authors classification |
| Greene et al. (2010) | Compilation of published data | 385 (252+/133−) | Authors classification |
| Ekins et al. (2010) | Clinical data for hepatotoxicity | 499 (294+/205−) | Authors classification |
| Chen et al. (2011) | FDA-approved labels | 279 (218+/61−) | “most DILI concern” and “less DILI concern” are considered positives |
| Liu et al. (2011) | SIDER_2 database | 835 (188+/647−) | Authors classification |
| Zhu and Kruhlak (2014) | Post-marketing safety data | 1948 (651+/1297−) | Authors classification, keeping only highest class certainty |
| Liu et al. (2015b) | LiverTox database | 583 (409+/174−) | “hepatotoxic” and “possible hepatotoxic” are considered positives |
For visualizing the data structures and for converting the names into structures Marvin from ChemAxon, 2013 (http://www.chemaxon.com 2013) was used.
2.1.2. External test sets
After compiling the training set and generating the DILI model, we came across one more human DILI dataset that had initially escaped our attention (Liew et al., 2011). Additionally, there were two more datasets published after the model development (Chen et al., 2016; Mulliner et al., 2016) (Table 3).
Table 3.
Description of the sources upon which the test set was built. In number of compounds, “+” denotes the number of DILI-positive compounds and “−” the number of negative compounds. These numbers correspond to the number of compounds remaining after data curation in a source by source basis.
| Source name | Type of data | Number of compounds | Label choice |
|---|---|---|---|
| Liew et al. (2011) | Micromedex reports of adverse reactions | 341 (221+/120−) | Authors classification |
| Mulliner et al. (2016) | Compilation of public data, data from PharmaPendium and Leadscope | 921 (519+/402−) | Authors classification |
| Chen et al. (2016) | Compilation of public data and LiverTox | 96 (50+/46−) | “most DILI concern” and “less DILI concern” are considered positives, “verified no DILI concern” as negatives |
| Merged | The 3 external datasets were merged and the common compounds with contradictory class labels were removed | 996 (541+/455−) | Maintenance of the class labels of the original external test sets |
All datasets (training set, the three external test sets and the merged test set) are provided in the Supplementary material.
2.1.3. Chemical curation
For each dataset we applied the following chemotype curation:
Check for inorganic compounds using MOE 2014.09. (MOE, 2015) and remove any occurring.
Using the Standardiser tool (Atkinson, 2014) created by Francis Atkinson; all salt parts and any compounds containing metals and rare or special atoms are removed from the dataset and the structures are standardized.
Duplicates and permanently charged compounds are removed using MOE 2014.09. (MOE, 2015) Here we must note that stereoisomers, even if biologically can be considered as different compounds, were considered as duplicates in our study, since they give the exactly same vector of descriptors. If two (or more) stereoisomers are of the same class, only one was kept. If they were of different classes, all were removed.
3D structures are generated using CORINA (version 3.4)(Sadowski et al., 1994) and their energy is minimized with MOE 2014.09 (MOE, 2015), using default settings, but changing the gradient to 0.05 RMS kcal/mol/A2. Existing chirality is preserved.
2.1.4. Class-label curation
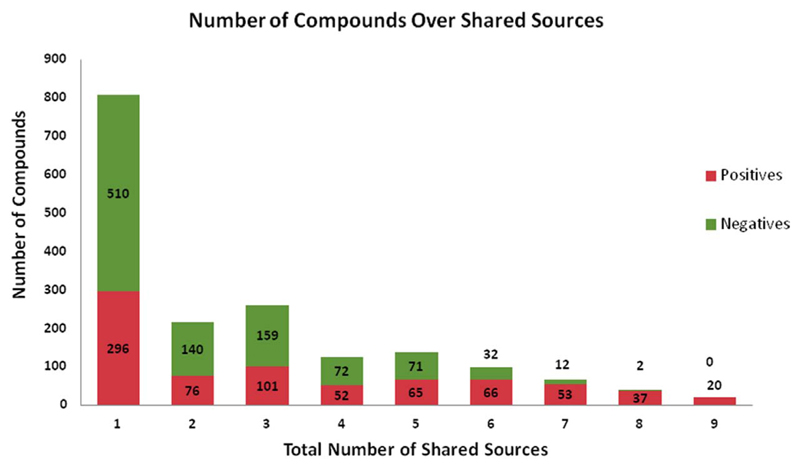
Apart from the chemical curation of the data, we also apply careful curation regarding the class label of the compounds. In particular, after merging all individual datasets in one database, the majority of the compounds are present in more than one dataset. In case of conflicting class labels, the majority label is assigned to the compound. In case the class labels are equally distributed, the compound is considered as “ambiguous” and it is removed from the dataset. This leads to 1773 compounds, 794 positives and 979 negatives. In Chart 1 the overlap of compounds (positives and negatives) across the different amount of sources is depicted. It is notable that for the case of occurrence in all 9 sources, we have only positives for DILI, which is in accordance with the fact that negative results are less often reported.
Chart 1.
Overlap of DILI positives and negatives across the different amount of sources.
However, the first modeling attempt of the dataset gave only moderate results. Re-analyzing the dataset revealed, that for several co-occurring compounds, even labeled as DILI negatives by majority vote, the Fourches source (Fourches et al., 2010) was labeling them as positives. The Fourches dataset was compiled via text mining, a sophisticated but error-prone method (Caporaso et al., 2008; Zhu et al., 2013). Therefore, in order to improve the dataset quality, all compounds that were coming solely from the Fourches dataset (227 compounds) were removed. Subsequently, all compounds coming from only a single source were removed, as they do not allow us to counter check the class label with at least one additional source. Following this concept leads to the removal of additional 584 compounds, which provides the final set of 966 compounds (500 positives and 466 negatives).
The differences in model performance after the class-label curation of the datasets is presented in the Supporting information (Table S1).
2.2. Generation of statistical models
2.2.1. Algorithms used
The 2-class classification models were built using the software package WEKA (version 3.7.12) (Hall et al., 2009). Performance of several base classifiers, such as logistic regression, tree methods (Random Forest and J48), Support Vector Machines (SMO in WEKA with polynomial, RBF and Puk kernels), Naïve Bayes, and k-nearest neighbors, several evaluating methods for attribute selection (AttributeSelectedClassifier), as well as for improving the statistical performance such as Bagging (Breiman, 1996) and Boosting (Freund and Schaphire 1996; Friedman et al., 2000) were evaluated. All in all, Random Forest (Breiman, 2001) with 100 trees was identified as the most promising classifier.
2.2.2. Molecular descriptors
For both datasets, several types of molecular descriptors have been calculated: all 2D MOE descriptors (192 descriptors in total), the 3D Volsurf series of descriptors (MOE 2015), PaDEL descriptors (Yap, 2010) and extended connectivity fingerprints of diameter 6 (ECFP6) using RDKit (Landrum). In general, the 2D MOE descriptors performed best.
In order to investigate the potential influence of transporter inhibition in DILI manifestation, we predicted the transporter inhibition profile of all compounds and used it as additional descriptors (Table S2). In particular, for OATP1B1 and 1B3 inhibition, we use our previously published models based on PaDEL descriptors (Kotsampasakou et al., 2015), as implemented in eTOXlab (Carrio et al., 2015). For BSEP inhibition, we useed the float predictions obtained from the model’s implementation as KNIME workflow (Montanari et al., 2016b). Also for P-glycoprotein (Schwarz et al., 2016) and for BCRP (Montanari et al., 2016a) inhibition, the respective float prediction scores were used.
2.3. DILI model with open-source descriptors
Since MOE is a commercial software package, we also provide a free version of the model using exclusively open-source libraries. For this, the final model set-up (all 2D MOE descriptors and Random Forest with 100 trees) was taken and converted in the following way: descriptors were implemented in RDKit (Landrum, 2016) (196 descriptors in total) and the Random Forest was implemented with the scikit-learn machine learning library for python (Pedregosa et al., 2011). The script for training, cross-validating and using the model is provided as Supplementary material.
2.4. Model validation
For model selection, 10-fold cross validation was used. The performance of each model was examined for accuracy, sensitivity, specificity, area under the curve (AUC) and precision. For the best models obtained, we performed 50 iterations by changing the cross-validation seed (for splitting the data within cross validation) and further performed a Welch (two-sample) t-test in R (http://www.R-project.org/) to assess whether the model performance for the different training data sets (after class label curation) is indeed significantly different. This was also done to compare whether the addition of the predicted transporter interaction profiles significantly improves model performance. The best models are further validated via external testing by using the validation datasets described above.
2.5. Applicability domain of the models
The applicability domain was checked on KNIME with the Enalos nodes (Afantitis et al., 2011; Melagraki et al., 2010) that compute the applicability domain on the basis of the Euclidean distances (Zhang et al., 2006). Additionally, we assessed to which extent the DILI datasets (both training and external test sets) were within the applicability domain of the transporters models, using the same procedure. The number of compounds within the model’s applicability domain for each model and for each DILI dataset is provided in the Supporting information (Table S3).
3. Results and discussion
3.1. Optimizing the training dataset – the importance of curation
Compiling the DILI dataset from the 9 data sources and performing the curation of the chemotypes and class labels according to majority vote initially lead to 1773 compounds. However, the first modeling attempts failed to yield models with acceptable performance. Analyzing the dataset revealed, that one source (Fourches) was compiled from text mining. Although text mining is a powerful approach for collecting data directly from narrative text, it is more prone to errors than manual extraction (Caporaso et al., 2008; Zhu and Kruhlak, 2014). Two examples are tocopherol and carnitine, which were reported as hepatotoxic only by the Fourches source. According to literature, those two compounds rather show a hepatoprotective effect against DILI caused by other drugs (Bohan et al., 2001; Tayal et al., 2007), than being hepatotoxic. Therefore, the compounds coming only from the Fouches dataset were completely removed. This reduction led to a new training set of 1547 compounds and improved the statistical performance of the resulting models (see Table S3 in the Supplementary material). In order to further improve the dataset quality, we also removed all compounds that appear only in one source (581 compounds). In this case, it is not possible to double-check the class label, which definitely adds noise to the data. Indeed, the model trained on this dataset shows additional improvement (Table S1).
In order to evaluate if the difference between the models generated on the three datasets is statistically significant, 50 iterations of 10-fold cross validation were performed by changing the cross-validation seed followed by a two sample t-test (Table S3). As can be seen, all parameters apart from specificity generally increase with higher quality of the data sets. Especially sensitivity, which is of higher importance since we are dealing with a toxicity endpoint, presents a remarkable increase, rising from 46% to 68%.
Remarkably, the analysis also indicates no difference on the model performance whether using the transporters predictions as additional information or not.
3.2. DILI 2-class classification models
For the final training dataset of 966 compounds, the best models are obtained using all 2D MOE descriptors. However, this restricts broader usage, as its application is conditional to a respective license for calculating the descriptors. In order to offer the model to the scientific community in open-source form, we rebuilt it using all 2D RDKit descriptors (196 descriptors in total; Table 4) and provide the respective python script.
Table 4.
Statistical performance of the final Random Forest (100 trees) model A) using all 2D MOE descriptors and transporter predictions (DILI_MOE_transp_RF model) or B) using only the 2D MOE descriptors (DILI_MOE_RF model) and the C) open source model (DILI_RDKit _RF100).
| Accuracy | Sensitivity | Specificity | AUC | Precision | |
|---|---|---|---|---|---|
| A) DILI_MOE_transp _RF100 | |||||
| 10-fold CV (average +/− standard deviation for 50 iterations) | 0.65 ± 0.01 | 0.68 ± 0.01 | 0.61 ± 0.01 | 0.69 ± 0.01 | 0.65 ± 0.01 |
| Mulliner 921 cpds | 0.57 | 0.63 | 0.50 | 0.59 | 0.62 |
| Liew 341 cpds | 0.67 | 0.72 | 0.56 | 0.71 | 0.75 |
| Chen 96 cpds | 0.59 | 0.54 | 0.65 | 0.61 | 0.63 |
| Merged test set 966cpds | 0.59 | 0.68 | 0.50 | 0.62 | 0.62 |
| B) DILI_ MOE _RF100 | |||||
| 10-fold CV (average +/− standard deviation for 50 iterations) | 0.65 ± 0.01 | 0.68 ± 0.01 | 0.61 ± 0.01 | 0.69 ± 0.01 | 0.65 ± 0.01 |
| Mulliner 921 cpds | 0.58 | 0.60 | 0.55 | 0.59 | 0.63 |
| Liew 341 cpds | 0.68 | 0.68 | 0.67 | 0.71 | 0.79 |
| Chen 96 cpds | 0.63 | 0.56 | 0.70 | 0.66 | 0.67 |
| Merged test set 966cpds | 0.60 | 0.64 | 0.56 | 0.62 | 0.63 |
| C) DILI_RDKit_RF100 | |||||
| 10-fold CV (average +/− standard deviation for 50 iterations) | 0.64 ± 0.01 | 0.70 ± 0.01 | 0.57 ± 0.01 | 0.69 ± 0.01 | 0.63 ± 0.01 |
| Mulliner 921 cpds | 0.60 | 0.64 | 0.54 | 0.62 | 0.64 |
| Liew 332 cpds | 0.67 | 0.72 | 0.56 | 0.71 | 0.72 |
| Chen 95 cpds | 0.64 | 0.64 | 0.64 | 0.73 | 0.64 |
| Merged test set 966cpds | 0.60 | 0.67 | 0.52 | 0.64 | 0.63 |
Notes: The number of compounds for the external datasets is slightly different for the predictions on model C because for some compounds (peptides), some descriptor values computed by RDKit were too large to be handled by the machine learning algorithm.
As Table 4 shows, the performance of the models is quite stable and satisfactory for cross validation. As can one see, there is no substantial difference between the model obtained via using transporters predictions as additional descriptors (model A), and the one built with only the 2D MOE descriptors (model B;), which is further confirmed by statistical testing (p-values > 0.05). Furthermore, the open source model and the model built with proprietary descriptors can be considered equivalent, despite some minor changes for 10-fold cross validation and the external validation. The model remains robust also for external validation, with statistics values quite similar to those obtained by cross validation.
However, it has to be taken into account that the DILI dataset is based on toxicity reports. Thus, despite our complex workflow for curating the data, there still might be mislabeled compounds due to the drawbacks of the adverse event reporting system. Among these issues are: 1) under-reporting (Palleria et al., 2013; Rodgers et al., 2010; Zhu and Kruhlak, 2014) due to the voluntary character of the system (Chen et al., 2008; Hauben 2004; Zhu and Kruhlak, 2014), 2) difficulty in finding human toxicity data (often proprietary and post-marketing data difficult to obtain) (Rodgers et al., 2010), 3) non-requirement of causality (Zhu and Kruhlak, 2014). The latter is quite serious in the contemporary era of polypharmacology, where many people, especially the elderly, receive more than one different medication. An indication of these drawbacks is the comparison of the class labels between overlapping compounds of the training and the test sets, as well as between the test sets themselves (formation of the merged external test set), which revealed contradiction of class labels in up to 20% of the compounds
3.3. Association of transporter inhibition profiles and DILI
There is ample evidence in literature for the association of selected liver transporters and DILI. This especially concerns BSEP (Aleo et al., 2014; Dawson et al., 2011; Padda et al., 2011; Qiu et al., 2016; Vinken 2015; Vinken et al., 2013; Welch et al., 2015), BCRP (Padda et al., 2011; Pauli-Magnus and Meier 2006), P-glycoprotein (Padda et al., 2011; Pauli-Magnus and Meier 2006), and OATP1B1/1B3 (Chang et al., 2013; Sticova and Jirsa 2013). This prompted us to introduce predicted inhibition profiles of these transporters into the feature matrix used for predicting DILI. However, we observed the same model performance for the models built with or without the transporter inhibition profile (Table 5, p-values < 0.05).
A possible reason for this might relate to the fact that the transporter inhibition profiles are based on predictions rather than on experimental data. Even though the transport inhibition models are reliable (AUC values in Table S2) and most of the compounds of the DILI training set belong to the respective applicability domains (Table S3), one cannot rule out the possibility of mispredictions, which in turn add noise into the feature matrix. However, when comparing the accuracy of the transporter models with the experience gained in the data curation task, the noise added by wrong predictions is not expected to be far beyond the one present in the DILI class labels.
Furthermore, liver transporters have overlapping substrate and inhibitor profiles (Giacomini et al., 2010; Homolya et al., 2003; König et al., 2013; Shugarts and Benet 2009). Apart from that, the hepatic homeostasis systems have ways to compensate the inhibition of one transporter, by overexpression of another (e.g. OATP1B1/OATP1B3) (Cui et al., 2009; Kalliokoski and Niemi 2009). Thus, inhibition of solely one transporter might not have a great impact in the proper function of the hepatocyte. This is also reflected in the data: the training compounds that are predicted as inhibitors for up to three transporters are not particularly enriched with DILI positives (451 DILI-positives and 443 DILI-negatives), while compounds predicted to inhibit at least four transporters are more likely to be DILI-positives (49 DILI-positives and 23 DILI-negatives, p-value < 0.01). Furthermore, liver transporters other than those included in this study may additionally play a role in DILI: the multidrug resistance-associated protein 2 (MRP2) (Nicolaou et al., 2012; Padda et al., 2011; Pauli-Magnus and Meier 2006), the multidrug resistance protein 3 (MDR3) (Chan and Vandeberg, 2012; Pauli-Magnus and Meier, 2006) and MRP3 and MRP4 (Padda et al., 2011; Pauli-Magnus and Meier 2006; Welch et al., 2015). Unfortunately, due to the lack of experimental data, it was not possible to develop and validate in silico models for these transporters in order to include them in the study.
Finally, it might be that the complexity of the DILI endpoint itself does not allow a strong association between liver transporter inhibition with DILI. Indeed, several other mechanisms produce hepatotoxicity (Vinken, 2015): formation of reactive metabolites by cytochrome P450 (Corsini and Bortolini 2013; Schadt et al., 2015; Utkarsh et al., 2015), formation of glutathione adducts (Schadt et al., 2015) and mitochondrial toxicity (Aleo et al., 2014; Schadt et al., 2015) are examples of mechanisms for causing DILI that are not specifically addressed in this study.
4. Conclusions
Drug-induced liver injury is a major issue for patients and, therefore, also for the process of drug discovery. Within the last decade, several attempts have taken place to predict DILI based on the chemical structure of a compound. In a more mechanistic based approach, one could also think on predicting DILI on basis of i.e. biological fingerprints. As these are usually not available for larger compound sets (at least not in the public domain), we included predicted liver-transporter interaction profiles as additional information. The liver transporter models have been developed in the course of the eTOX project and are available in eTOXsys, the integrated data mining and computational model environment established in the course of the project. Surprisingly, although the role of liver transporter for hepatotoxicity has clearly been demonstrated, this additional piece of information did not significantly improve model performance. Potential reasons for this are outlined above, and most probably the biological fingerprint needs to be substantially broadened by including additional transporter and enzymes to see a significant effect on model performance.
The predictivity of computational models heavily depends on the quality of the respective training data set and the domain it covers. In this work, we compiled datasets for DILI available in literature and carefully curated them both with respect to the chemical structures as well as for their class labels (DILI positive, DILI negative). This reduced the amount of compounds available for classification models from 1773 to 966, and in return remarkably increased the quality of the models developed. While in general bigger datasets are preferred for machine learning approaches, the current work once more stresses out the significance of data quality. However, there might be still an amount of mislabeled compounds, as the conflicting class labels for overlapping compounds in the training and test sets show. This further strengthens the tremendous need for industry-driven collaborative efforts such as the eTOX project to share data and to make them publicly available for mining and exploitation. Only large sets of high quality data will allow deriving predictive in silico models covering a broad chemical space.
Supplementary Material
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.tox.2017.06.003.
Acknowledgements
We are thankful to Dr. Alexander Amberg from Sanofi-Aventis Deutschland GmbH, co-author of Mulliner et al. publication, for providing us with the Supporting information before being available online from the journal.
Funding
The research leading to these results has received financial support from the Innovative Medicines Initiative Joint Undertaking under grant agreements No. 115002 (eTOX) resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007-2013) and EFPIA companies’ in kind contribution; and the Austrian Science Fund, Grant F3502.
Abbreviations
- Acc
Accuracy
- ADMET
absorption, distribution, metabolism, excretion, toxicity
- AUC
area under the curve
- BA
balanced accuracy
- BCRP
breast cancer resistance protein
- cpd(s)
compound(s)
- CV
cross validation
- DILI
drug-induced liver injury
- EV
external validation
- IV
internal validation
- MCC
Matthews correlation coefficient
- MDR3
multidrug resistance protein
- MRP2
multidrug resistance-associated protein 2
- MRP3
multidrug resistance-associated protein 3
- OATP1B1
organic anion transporting polypeptide 1B1
- OATP1B3
organic anion transporting polypeptide 1B3
- P-gp
P-glycoprotein
- RF
Random Forest
- SMO
sequential minimal optimization
- sd
standard deviation
- Sen
sensitivity
- Spec
specificity
- SVM
support vector machines
Footnotes
Conflict of interest
The authors declare no competing financial interest.
Author contributions
Eleni Kotsampasakou performed the data compilation and curation, the classification modeling and developed the final DILI model with 2D MOE descriptors. Floriane Montanari developed the final DILI model with 2D RDKit descriptors, as well as the respective provided python script. Gerhard F. Ecker designed and supervised the study. All authors contributed in the writing and have read and approved the final version of the manuscript.
References
- Afantitis A, Melagraki G, Koutentis PA, Sarimveis H, Kollias G. Ligand-based virtual screening procedure for the prediction and the identification of novel beta-amyloid aggregation inhibitors using Kohonen maps and counterpropagation artificial neural networks. Eur J Med Chem. 2011;46:497–508. doi: 10.1016/j.ejmech.2010.11.029. [DOI] [PubMed] [Google Scholar]
- Aleo MD, Luo Y, Swiss R, Bonin PD, Potter DM, Will Y. Human drug-induced liver injury severity is highly associated with dual inhibition of liver mitochondrial function and bile salt export pump. Hepatology. 2014;60:1015–1022. doi: 10.1002/hep.27206. [DOI] [PubMed] [Google Scholar]
- Atkinson FL. Standardiser. 2014 ( https://github.com/flatkinson/standardiser/tree/1.0.1)
- Ballet F. Hepatotoxicity in drug development: detection, significance and solutions. J Hepatol. 1997;26(Suppl 2):26–36. doi: 10.1016/s0168-8278(97)80494-1. [DOI] [PubMed] [Google Scholar]
- Bohan TP, Helton E, McDonald I, Konig S, Gazitt S, Sugimoto T, Scheffner D, Cusmano L, Li S, Koch G. Effect of L-carnitine treatment for valproate-induced hepatotoxicity. Neurology. 2001;56:1405–1409. doi: 10.1212/wnl.56.10.1405. [DOI] [PubMed] [Google Scholar]
- Bowes J, Brown AJ, Hamon J, Jarolimek W, Sridhar A, Waldron G, Whitebread S. Reducing safety-related drug attrition: the use of in vitro pharmacological profiling. Nat Rev Drug Discov. 2012;11:909–922. doi: 10.1038/nrd3845. [DOI] [PubMed] [Google Scholar]
- Breiman L. Bagging predictors. Mach Learn. 1996;24:123–140. [Google Scholar]
- Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- Caporaso JG, Deshpande N, Fink JL, Bourne PE, Cohen KB, Hunter L. Intrinsic evaluation of text mining tools may not predict performance on realistic tasks. Pac Symp Biocomput. 2008:640–651. [PMC free article] [PubMed] [Google Scholar]
- Carrio P, Lopez O, Sanz F, Pastor M. eTOXlab, an open source modeling framework for implementing predictive models in production environments. J Cheminform. 2015;7:8. doi: 10.1186/s13321-015-0058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Vandeberg JL. Hepatobiliary transport in health and disease. Clin Lipidol. 2012;7:189–202. doi: 10.2217/clp.12.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JH, Plise E, Cheong J, Ho Q, Lin M. Evaluating the in vitro inhibition of UGT1A1, OATP1B1, OATP1B3, MRP2, and BSEP in predicting drug-induced hyperbilirubinemia. Mol Pharm. 2013;10:3067–3075. doi: 10.1021/mp4001348. [DOI] [PubMed] [Google Scholar]
- Marvin. Marvin Suite; 2013. ChemAxon. http://www.chemaxon.com. [Google Scholar]
- Chen Y, Guo JJ, Healy DP, Lin X, Patel NC. Risk of hepatotoxicity associated with the use of telithromycin: a signal detection using data mining algorithms. Ann Pharmacother. 2008;42:1791–1796. doi: 10.1345/aph.1L315. [DOI] [PubMed] [Google Scholar]
- Chen M, Vijay V, Shi Q, Liu Z, Fang H, Tong W. FDA-approved drug labeling for the study of drug-induced liver injury. Drug Discov Today. 2011;16:697–703. doi: 10.1016/j.drudis.2011.05.007. [DOI] [PubMed] [Google Scholar]
- Chen M, Hong H, Fang H, Kelly R, Zhou G, Borlak J, Tong W. Quantitative structure-activity relationship models for predicting drug-induced liver injury based on FDA-approved drug labeling annotation and using a large collection of drugs. Toxicol Sci. 2013;136:242–249. doi: 10.1093/toxsci/kft189. [DOI] [PubMed] [Google Scholar]
- Chen M, Bisgin H, Tong L, Hong H, Fang H, Borlak J, Tong W. Toward predictive models for drug-induced liver injury in humans: are we there yet? Biomark Med. 2014;8:201–213. doi: 10.2217/bmm.13.146. [DOI] [PubMed] [Google Scholar]
- Chen M, Suzuki A, Thakkar S, Yu K, Hu C, Tong W. DILIrank: the largest reference drug list ranked by the risk for developing drug-induced liver injury in humans. Drug Discov Today. 2016;21:648–653. doi: 10.1016/j.drudis.2016.02.015. [DOI] [PubMed] [Google Scholar]
- Cheng A, Dixon SL. In silico models for the prediction of dose-dependent human hepatotoxicity. J Comput Aided Mol Des. 2003;17:811–823. doi: 10.1023/b:jcam.0000021834.50768.c6. [DOI] [PubMed] [Google Scholar]
- Corsini A, Bortolini M. Drug-induced liver injury: the role of drug metabolism and transport. J Clin Pharmacol. 2013;53:463–474. doi: 10.1002/jcph.23. [DOI] [PubMed] [Google Scholar]
- Cruz-Monteagudo M, Cordeiro MN, Borges F. Computational chemistry approach for the early detection of drug-induced idiosyncratic liver toxicity. J Comput Chem. 2008;29:533–549. doi: 10.1002/jcc.20812. [DOI] [PubMed] [Google Scholar]
- Cui YJ, Aleksunes LM, Tanaka Y, Goedken MJ, Klaassen CD. Compensatory induction of liver efflux transporters in response to ANIT-induced liver injury is impaired in FXR-null mice. Toxicol Sci. 2009;110:47–60. doi: 10.1093/toxsci/kfp094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson S, Stahl S, Paul N, Barber J, Kenna JG. In vitro inhibition of the bile salt export pump correlates with risk of cholestatic drug-induced liver injury in humans. Drug Metab Dispos. 2011;40:130–138. doi: 10.1124/dmd.111.040758. [DOI] [PubMed] [Google Scholar]
- Ekins S, Williams AJ, Xu JJ. A predictive ligand-based Bayesian model for human drug-induced liver injury. Drug Metab Dispos. 2010;38:2302–2308. doi: 10.1124/dmd.110.035113. [DOI] [PubMed] [Google Scholar]
- Ekins S. Progress in computational toxicology. J Pharmacol Toxicol Methods. 2014;69:115–140. doi: 10.1016/j.vascn.2013.12.003. [DOI] [PubMed] [Google Scholar]
- Faber KN, Muller M, Jansen PL. Drug transport proteins in the liver. Adv Drug Deliv Rev. 2003;55:107–124. doi: 10.1016/s0169-409x(02)00173-4. [DOI] [PubMed] [Google Scholar]
- Fourches D, Barnes JC, Day NC, Bradley P, Reed JZ, Tropsha A. Cheminformatics analysis of assertions mined from literature that describe drug-induced liver injury in different species. Chem Res Toxicol. 2010;23:171–183. doi: 10.1021/tx900326k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund Y, Schaphire RE. Experiments with a new boosting algorithm. 13th International Conference on Machine Learning; San Francisco. 1996. pp. 148–156. [Google Scholar]
- Friedman J, Hastie T, Tibsharni R. Additive logistic regression: a statistical view of boosting. Annals of Statistics. 2000;95:337–407. [Google Scholar]
- Ghabril M, Chalasani N, Bjornsson E. Drug-induced liver injury: a clinical update. Curr Opin Gastroenterol. 2010;26:222–226. doi: 10.1097/MOG.0b013e3283383c7c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomini KM, Huang SM, Tweedie DJ, Benet LZ, Brouwer KL, Chu X, Dahlin A, Evers R, Fischer V, Hillgren KM, Hoffmaster KA, et al. Membrane transporters in drug development. Nat Rev Drug Discov. 2010;9:215–236. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Google. [last accessed 09/03/2017]; https://www.google.at.
- Greene N, Fisk L, Naven RT, Note RR, Patel ML, Pelletier DJ. Developing structure-activity relationships for the prediction of hepatotoxicity. Chem Res Toxicol. 2010;23:1215–1222. doi: 10.1021/tx1000865. [DOI] [PubMed] [Google Scholar]
- Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: an update. SIGKDD Explor Newsl. 2009;11:10–18. [Google Scholar]
- Hauben M. Early postmarketing drug safety surveillance: data mining points to consider. Ann Pharmacother. 2004;38:1625–1630. doi: 10.1345/aph.1E023. [DOI] [PubMed] [Google Scholar]
- Homolya L, Varadi A, Sarkadi B. Multidrug resistance-associated proteins: export pumps for conjugates with glutathione, glucuronate or sulfate. Biofactors. 2003;17:103–114. doi: 10.1002/biof.5520170111. [DOI] [PubMed] [Google Scholar]
- König J, Muller F, Fromm MF. Transporters and drug–drug interactions: important determinants of drug disposition and effects. Pharmacol Rev. 2013;65:944–966. doi: 10.1124/pr.113.007518. [DOI] [PubMed] [Google Scholar]
- Kalliokoski A, Niemi M. Impact of OATP transporters on pharmacokinetics. Br J Pharmacol. 2009;158:693–705. doi: 10.1111/j.1476-5381.2009.00430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsampasakou E, Brenner S, Jäger W, Ecker GF. Identification of novel inhibitors of organic anion transporting polypeptides 1B1 and 1B3 (OATP1B1 and OATP1B3) using a consensus vote of six classification models. Mol Pharm. 2015;12:4395–4404. doi: 10.1021/acs.molpharmaceut.5b00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum G. RDKit: Open-Source Cheminformatics Software. 2016 http://refhub.elsevier.com/S0300-483X(17)30170-1/sbref0200.
- Liew CY, Lim YC, Yap CW. Mixed learning algorithms and features ensemble in hepatotoxicity prediction. J Comput Aided Mol Des. 2011;25:855–871. doi: 10.1007/s10822-011-9468-3. [DOI] [PubMed] [Google Scholar]
- Liu Z, Shi Q, Ding D, Kelly R, Fang H, Tong W. Translating clinical findings into knowledge in drug safety evaluation–drug induced liver injury prediction system (DILIps) PLoS Comput Biol. 2011;7:e1002310. doi: 10.1371/journal.pcbi.1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Mansouri K, Judson RS, Martin MT, Hong H, Chen M, Xu X, Thomas RS, Shah I. Predicting hepatotoxicity using ToxCast in vitro bioactivity and chemical structure. Chem Res Toxicol. 2015a;28:738–751. doi: 10.1021/tx500501h. [DOI] [PubMed] [Google Scholar]
- Liu R, Yu X, Wallqvist A. Data-driven identification of structural alerts for mitigating the risk of drug-induced human liver injuries. J Cheminform. 2015b;7:4. doi: 10.1186/s13321-015-0053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOE. Molecular Operating Environment (MOE) Chemical Computing Group Inc.; 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H 3A 2R7: 2015. [Google Scholar]
- Matthews EJ, Ursem CJ, Kruhlak NL, Benz RD, Sabate DA, Yang C, Klopman G, Contrera JF. Identification of structure-activity relationships for adverse effects of pharmaceuticals in humans: part B. Use of (Q)SAR systems for early detection of drug-induced hepatobiliary and urinary tract toxicities. Regul Toxicol Pharmacol. 2009;54:23–42. doi: 10.1016/j.yrtph.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Melagraki G, Afantitis A, Sarimveis H, Igglessi-Markopoulou O, Koutentis PA, Kollias G. In silico exploration for identifying structure-activity relationship of MEK inhibition and oral bioavailability for isothiazole derivatives. Chem Biol Drug Des. 2010;76:397–406. doi: 10.1111/j.1747-0285.2010.01029.x. [DOI] [PubMed] [Google Scholar]
- Montanari F, Cseke A, Wlcek K, Ecker GF. Virtual screening of DrugBank reveals two drugs as new BCRP inhibitors. J Biomol Screen. 2016a;22:86–93. doi: 10.1177/1087057116657513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanari F, Pinto M, Khunweeraphong N, Wlcek K, Sohail MI, Noeske T, Boyer S, Chiba P, Stieger B, Kuchler K, Ecker GF. Flagging drugs that inhibit the bile salt export pump. Mol Pharm. 2016b;13:163–171. doi: 10.1021/acs.molpharmaceut.5b00594. [DOI] [PubMed] [Google Scholar]
- Muller C, Pekthong D, Alexandre E, Marcou G, Horvath D, Richert L, Varnek A. Prediction of drug induced liver injury using molecular and biological descriptors. Comb Chem High Throughput Screen. 2015;18:315–322. doi: 10.2174/1386207318666150305144650. [DOI] [PubMed] [Google Scholar]
- Mulliner D, Schmidt F, Stolte M, Spirkl HP, Czich A, Amberg A. Computational models for human and animal hepatotoxicity with a global application scope. Chem Res Toxicol. 2016;29:757–767. doi: 10.1021/acs.chemrestox.5b00465. [DOI] [PubMed] [Google Scholar]
- Nicolaou M, Andress EJ, Zolnerciks JK, Dixon PH, Williamson C, Linton KJ. Canalicular ABC transporters and liver disease. J Pathol. 2012;226:300–315. doi: 10.1002/path.3019. [DOI] [PubMed] [Google Scholar]
- O’Brien PJ, Irwin W, Diaz D, Howard-Cofield E, Krejsa CM, Slaughter MR, Gao B, Kaludercic N, Angeline A, Bernardi P, Brain P, et al. High concordance of drug-induced human hepatotoxicity with in vitro cytotoxicity measured in a novel cell-based model using high content screening. Arch Toxicol. 2006;80:580–604. doi: 10.1007/s00204-006-0091-3. [DOI] [PubMed] [Google Scholar]
- Olson H, Betton G, Robinson D, Thomas K, Monro A, Kolaja G, Lilly P, Sanders J, Sipes G, Bracken W, Dorato M, et al. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul Toxicol Pharmacol. 2000;32:56–67. doi: 10.1006/rtph.2000.1399. [DOI] [PubMed] [Google Scholar]
- Padda MS, Sanchez M, Akhtar AJ, Boyer JL. Drug-induced cholestasis. Hepatology. 2011;53:1377–1387. doi: 10.1002/hep.24229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palleria C, Leporini C, Chimirri S, Marrazzo G, Sacchetta S, Bruno L, Lista RM, Staltari O, Scuteri A, Scicchitano F, Russo E. Limitations and obstacles of the spontaneous adverse drugs reactions reporting: two challenging case reports. J Pharmacol Pharmacother. 2013;4:S66–72. doi: 10.4103/0976-500X.120955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli-Magnus C, Meier PJ. Hepatobiliary transporters and drug-induced cholestasis. Hepatology. 2006;44:778–787. doi: 10.1002/hep.21359. [DOI] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, et al. Scikit-learn: machine learning in python. JMLR. 2011;12:2825–2830. [Google Scholar]
- Home-PubMed-NCBI. [last accessed 09/03/2017]; http://www.ncbi.nlm.nih.gov/pubmed.
- Qiu X, Zhang Y, Liu T, Shen H, Xiao Y, Bourner MJ, Pratt JR, Thompson DC, Marathe P, Humphreys WG, Lai Y. Disruption of BSEP function in HepaRG cells alters bile acid disposition and is a susceptive factor to drug-induced cholestatic injury. Mol Pharm. 2016;13:1206–1216. doi: 10.1021/acs.molpharmaceut.5b00659. [DOI] [PubMed] [Google Scholar]
- R Core Team. A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. (URL). http://www.R-project.org/ [Google Scholar]
- Raschi E, De Ponti F. Drug- and herb-induced liver injury: progress, current challenges and emerging signals of post-marketing risk. World J Hepatol. 2015;7:1761–1771. doi: 10.4254/wjh.v7.i13.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev A. Drug-induced liver injury and drug development: industry perspective. Semin Liver Dis. 2014;34:227–239. doi: 10.1055/s-0034-1375962. [DOI] [PubMed] [Google Scholar]
- Rodgers AD, Zhu H, Fourches D, Rusyn I, Tropsha A. Modeling liver-related adverse effects of drugs using k-nearest neighbor quantitative structure-activity relationship method. Chem Res Toxicol. 2010;23:724–732. doi: 10.1021/tx900451r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski J, Gasteiger J, Klebe G. Comparison of automatic three-dimensional model builders using 639 X-ray structures. J Chem Inf Comput Sci. 1994;34:1000–1008. [Google Scholar]
- Schadt S, Simon S, Kustermann S, Boess F, McGinnis C, Brink A, Lieven R, Fowler S, Youdim K, Ullah M, Marschmann M, et al. Minimizing DILI risk in drug discovery – a screening tool for drug candidates. Toxicol In Vitro. 2015;30:429–437. doi: 10.1016/j.tiv.2015.09.019. [DOI] [PubMed] [Google Scholar]
- Schwarz T, Montanari F, Cseke A, Wlcek K, Visvader L, Palme S, Chiba P, Kuchler K, Urban E, Ecker GF. Subtle structural differences trigger inhibitory activity of propafenone analogues at the two polyspecific ABC transporters: p-glycoprotein (P-gp) and breast cancer resistance protein (BCRP) ChemMedChem. 2016;11:1380–1394. doi: 10.1002/cmdc.201500592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELSEVIER; [last accessed 09/03/2017]. Scopus. https://www.scopus.com/ [Google Scholar]
- Shitara Y, Maeda K, Ikejiri K, Yoshida K, Horie T, Sugiyama Y. Clinical significance of organic anion transporting polypeptides (OATPs) in drug disposition: their roles in hepatic clearance and intestinal absorption. Biopharm Drug Dispos. 2013;34:45–78. doi: 10.1002/bdd.1823. [DOI] [PubMed] [Google Scholar]
- Shugarts S, Benet LZ. The role of transporters in the pharmacokinetics of orally administered drugs. Pharm Res. 2009;26:2039–2054. doi: 10.1007/s11095-009-9924-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticova E, Jirsa M. New insights in bilirubin metabolism and their clinical implications. World J Gastroenterol. 2013;19:6398–6407. doi: 10.3748/wjg.v19.i38.6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayal V, Kalra BS, Agarwal S, Khurana N, Gupta U. Hepatoprotective effect of tocopherol against isoniazid and rifampicin induced hepatotoxicity in albino rabbits. Indian J Exp Biol. 2007;45:1031–1036. [PubMed] [Google Scholar]
- Utkarsh D, Loretz C, Li AP. In vitro evaluation of hepatotoxic drugs in human hepatocytes from multiple donors: identification of P450 activity as a potential risk factor for drug-induced liver injuries. Chem Biol Interact. 2015;255:12–22. doi: 10.1016/j.cbi.2015.12.013. [DOI] [PubMed] [Google Scholar]
- Vinken M, Landesmann B, Goumenou M, Vinken S, Shah I, Jaeschke H, Willett C, Whelan M, Rogiers V. Development of an adverse outcome pathway from drug-mediated bile salt export pump inhibition to cholestatic liver injury. Toxicol Sci. 2013;136:97–106. doi: 10.1093/toxsci/kft177. [DOI] [PubMed] [Google Scholar]
- Vinken M. Adverse outcome pathways and drug-induced liver injury testing. Chem Res Toxicol. 2015;28:1391–1397. doi: 10.1021/acs.chemrestox.5b00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins PB, Seeff LB. Drug-induced liver injury: summary of a single topic clinical research conference. Hepatology. 2006;43:618–631. doi: 10.1002/hep.21095. [DOI] [PubMed] [Google Scholar]
- Welch MA, Kock K, Urban TJ, Brouwer KL, Swaan PW. Toward predicting drug-induced liver injury: parallel computational approaches to identify multidrug resistance protein 4 and bile salt export pump inhibitors. Drug Metab Dispos. 2015;43:725–734. doi: 10.1124/dmd.114.062539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitebread S, Hamon J, Bojanic D, Urban L. Keynote review: in vitro safety pharmacology profiling: an essential tool for successful drug development. Drug Discov Today. 2005;10:1421–1433. doi: 10.1016/S1359-6446(05)03632-9. [DOI] [PubMed] [Google Scholar]
- Xu Y, Dai Z, Chen F, Gao S, Pei J, Lai L. Deep learning for drug-Induced liver injury. J Chem Inf Model. 2015;55:2085–2093. doi: 10.1021/acs.jcim.5b00238. [DOI] [PubMed] [Google Scholar]
- Yap CW. PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J Comput Chem. 2010;32:1466–1474. doi: 10.1002/jcc.21707. [DOI] [PubMed] [Google Scholar]
- Zhang S, Golbraikh A, Oloff S, Kohn H, Tropsha A. A novel automated lazy learning QSAR (ALL-QSAR) approach: method development, applications, and virtual screening of chemical databases using validated ALL-QSAR models. J Chem Inf Model. 2006;46:1984–1995. doi: 10.1021/ci060132x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Cheng F, Li W, Liu G, Lee PW, Tang Y. In silico prediction of drug induced liver toxicity using substructure pattern recognition method. Mol Inform. 2016;35:136–144. doi: 10.1002/minf.201500055. [DOI] [PubMed] [Google Scholar]
- Zhu X, Kruhlak NL. Construction and analysis of a human hepatotoxicity database suitable for QSAR modeling using post-market safety data. Toxicology. 2014;321:62–72. doi: 10.1016/j.tox.2014.03.009. [DOI] [PubMed] [Google Scholar]
- Zhu F, Patumcharoenpol P, Zhang C, Yang Y, Chan J, Meechai A, Vongsangnak W, Shen B. Biomedical text mining and its applications in cancer research. J Biomed Inform. 2013;46:200–211. doi: 10.1016/j.jbi.2012.10.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.