Abstract
Aim
The transient receptor potential vanilloid type 1 (TRPV1) is responsible for pain perception in the peripheral nervous system (PNS). TRPV1 is thus considered a versatile target for development of non-opioid analgesics.
Results
Pharmacophore-based clustering of a publicly available data set of TRPV1 antagonists revealed a set of models, which were validated with data sets of inactive compounds, decoys and known drug candidates. The top ranked pharmacophore models were subsequently used for virtual screening. Based on a unique in-house protocol, a set of compounds was selected and biologically tested for modulation of TRPV1 in a voltage-clamp model.
Conclusion
Pharmacophore models extracted from large public data sets are a valuable source for identification of novel scaffolds for TRPV1 receptor modulation.
Transient receptor potential vanilloid type 1 (TRPV1) is a transmembrane non-selective cation channel primarily expressed in the sensory neurons of the PNS in humans. Binding of various endogenous (anandamide, arachidonic acid metabolites) and exogenous (capsaicin (1), resiniferatoxin (RTX), piperine) ligands to TRPV1, as well as exposure to heat (>42°C) and protons (pH <6.0) leads to opening of the channel pore and influx of Ca2+ ions into the cell [1]. Consequent depolarization of the cell membrane activates the neuron signaling pathways and results in the immediate sensation of ‘burning pain’ [2]. Although prolonged Ca2+ influx desensitizes the nociceptive neurons [3], the pain sensation on initial application hinders the clinical development of agonists [4–6]. Therefore, with the discovery of capsazepine [7] (2), the research of both academia [8–11] and pharmaceutical industry [12–15] mainly focused on the development of antagonists as non-opioid analgesics for treatment of various pain conditions.
Key terms.
Capsaicin: Pungent component of chili peppers, an agonist of TRPV1.
Capsazepine: First discovered antagonist of TRPV1 acting competitively to capsaicin.
Pharmacophore: Ensemble of steric and electronic features of a ligand molecule which determines optimal interactions with a specific biological target.
Binding mode: Certain ensemble of interactions between ligand and the receptor at the specific binding site.
LifeChem: Life Chemicals, a vendor database of small organic molecules and molecular fragments.
To date, thousands of chemical compounds are reported in the literature as competitive TRPV1 antagonists, and some of them undergo clinical trials [5,16]. The majority of the compounds have been identified through high-throughput screening experiments [13–15,17]. Some of these compounds were further optimized by isosteric replacements of structural fragments [18–20] and by structure–activity relationship (SAR) studies [9–11,21–30]. Very recently, also a cryo-EM structure of human TRPV1 in a resolution of 3.4 Å has been published. Thus, considering the wealth of data available, in silico virtual screening methods could serve as an efficient and reasonable method for identifying new scaffolds and thus expanding the chemical space of TRPV1 ligands. Considering the type and quality of available public data [31], pharmacophore modeling seems the method of choice [32].
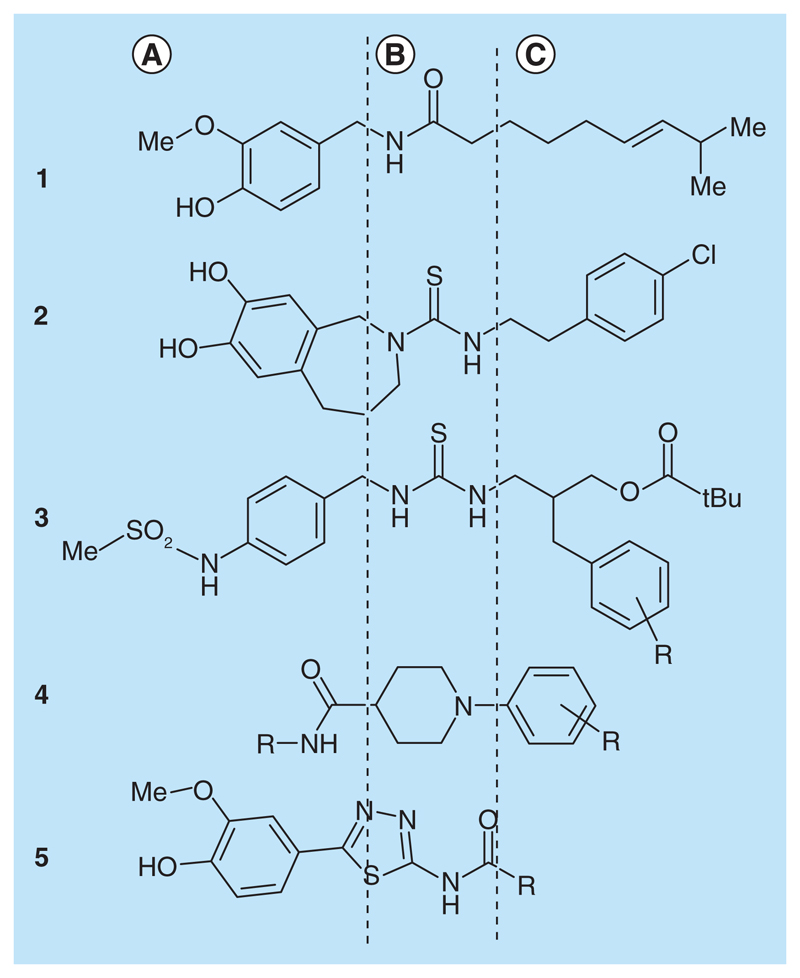
So far, several pharmacophore-like models were proposed for different classes of TRPV1 antagonists. They were derived from a set of unique chemical scaffolds, and highlight three important pharmacophores of TRPV1 ligands: a polar head (A), a linker (B) and a hydrophobic tail (C) (Figure 1) [7,33–36]. In the work of Kim [37], the pharmacophore was derived from a 3D QSAR analysis of substituted capsazepines. The CoMFA maps showed sterically favored substitution in the C region and the possibility of a hydrogen bond interaction between the hydroxyl groups and the amino group of capsazepine derivatives with the receptor. Likewise, Kristam et al. [38] reported a pharmacophore model derived from a 3D QSAR model of imidazole and benzimidazole derivatives. Blumberg and Lee and co-workers [8,10,39–41] as well as Kim [42] further identified important ligand–protein interaction features from docking poses of SAR series of benzylthiourea [10,41] propionamide [39], propanamide [8,40] and phenylbenzyl amide derivatives [42], respectively. Additionally, for 12 compounds being in clinical trials, Kym [16] proposed three essential pharmacophore features in the linker region: a hydrogen-bond acceptor, a hydrogen-bond donor and a ring feature.
Figure 1. Three important pharmacophores of TRPV1 ligands.
Pharmacophoric comparison of (1) capsaicin [7], (2) capsazepine [7], (3) benzylthiourea derivatives [33], (4) piperidine carboxamide derivatives [35], (5) 1,3,4-thiadiazole derivatives [36] according to three structural sections of TRPV1 ligands: (A) polar head, (B) linker and (C) hydrophobic tail.
However, a model abstracting the pharmacophore features of highly active and chemically diverse TRPV1 ligands sharing the same binding site has not been reported to date. Therefore, we aimed to extract and systematize the pharmacophoric patterns from a large set of publicly available TRPV1 ligands independently from the ‘three region’ approach. Pharmacophore-based clustering of this data set led to an array of pharmacophore models, proposing different binding modes of TRPV1 antagonists. These models were computationally validated with data sets of inactive compounds, decoys and known drug candidates [16,43]. Finally, the five best performing models were used for virtual screening of the vendor database Life Chemicals 2012.3 (LifeChem) [61], and 12 compounds with novel scaffolds were selected for biological testing.
Experimental section
Hardware & software specifications
Pharmacophore-based clustering of the data set was performed using Ligandscout 3.1 installed on the local personal computer running Ubuntu 12.04. Generation of conformers for the training set and validation sets, generation of the pharmacophore models and computational validation of the pharmacophore models along with virtual screening were performed using command line packages of Ligandscout 3.1 installed on the local server running Fedora 13: idbgen, espresso and iscreen, respectively. The shell scripts used to execute the calculations are provided in the Supplementary Material.
Preparation of the data sets
Chemical structures and bioactivity data of 2332 compounds reported as TRPV1 antagonists were extracted from ChEMBLdb v.13 [44] and filtered to 408 compounds blocking capsaicin-induced Ca2+ flux into HEK293 cells, as described previously [45]. Compounds were considered as actives if their IC50 value was comparable to the IC50 of reference compound 2 (100 nM) under the same assay conditions [15]. Thereby, a data set of 201 TRPV1 antagonists sharing the same binding site, which are comparable with respect to the assays used, are highly active (IC50 values range from 0.21 to 100 nM), and are chemically diverse, were obtained.
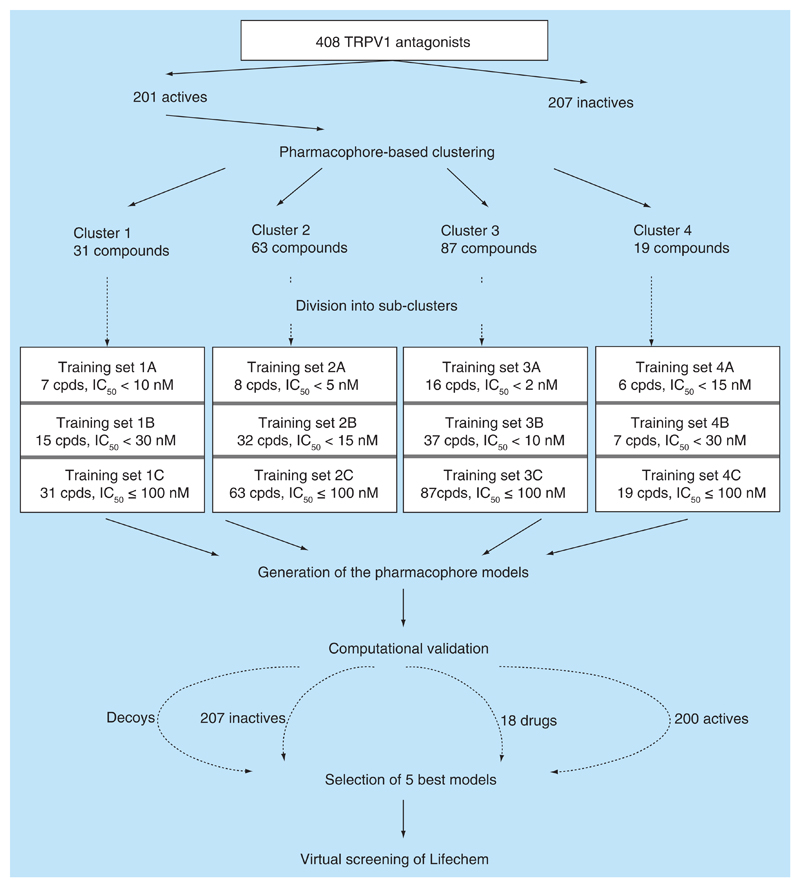
The following data sets were used in the study: a starting data set comprising 201 active TRPV1 antagonists; training sets from 1A to 4C (Figure 2); a decoy set generated for each training set; a validation data set of 207 inactive TRPV1 antagonists; a clinical data set with 17 compounds in clinical trials and a virtual screening data set LifeChem with 305 841 compounds. For each ligand in the data sets 25 conformations were generated with OMEGA [46] algorithm implemented in Ligandscout. Up to 50 decoys were generated for each ligand of the training sets using DUD-E decoy generator developed by Mysinger et al. [47] and openly available [62].
Figure 2. Model generation and computational validation.
Computational studies
Pharmacophore-based clustering was performed for a starting data set using the radial distribution function (RDF) code similarity algorithm of Ligandscout 3.1 with default settings (similarity set to 0.4, conformations set to 3). For each training set ten pharmacophore models were built with the RDF-code similarity algorithm. Shared feature mode was used in order to identify the pharmacophoric pattern present in all the ligands of each training set. Generation of exclusion volumes from the superposed ligands of the training set in the pharmacophore model was turned on. The latter allows definition of the boundary conditions for the size of the ligands. Other settings were set to default. In total, 120 pharmacophore models were built.
Computational validation of the pharmacophore models and virtual screening of LifeChem was performed using the iscreen command line package of Ligandscout 3.1. Checking the match to exclusion volumes was turned on and screening mode ‘stop after first matching conformation’ was used. Other settings were set to default (i.e., omitted features of the pharmacophore models switched off). Each pharmacophore model was screened against the training set it was built on, the corresponding decoy set, the whole cluster to which the training set belonged, the clinical data set, and the inactive data set.
Molecular ACCess System (MACCS) finger-prints [48] were generated for the 12 purchased compounds, reference compound 2 and the most active representatives from clusters 1, 2 and 3 (five per cluster) using the software package MOE v2011.10. Subsequently, pair-wise Tanimoto similarity matrix between the 12 purchased compounds and 16 compounds from the training set was computed in MOE.
Expression of TRPV1 receptors in Xenopus oocytes & two-microelectrode voltage-clamp experiments
Oocytes were surgically removed from anesthetized Xenopus laevis frogs and then carefully shaken for about 60 min in a solution of 82.5 mM NaCl, 2.5 mM KCl, 1 mM MgCl2, 5 mM HEPES and 2 mg/ml collagenase. The Xenopus laevis stage IV-V oocytes were injected with rTRPV1 cRNA in a 1:10 ratio. After injection of cRNA, oocytes were stored at 18°C for 24–48 h in ND96 solution containing penicillin G (10 000 IU/100 ml) and streptomycin (10 mg/100 ml) [49].
Electrophysiological experiments on TRPV1 channels were performed by two-microelectrode voltage-clamp method at a holding potential of -60 mV, making use of a TURBO TEC 01 C amplifier (npi electronic™, Tamm, Germany) and an Axon Digidata 1322A interface (Molecular Devices™, CA, USA). Inward current through TRPV1 receptors (ITRPV1) were measured at a room temperature (21 ± 1°C). Data acquisition was performed using pCLAMP v.9.2 (Molecular Devices). Currents were lowpass-filtered at 1 kHz and sampled at 3 kHz. The bath solution contained 90 mM NaCl, 1 mM KCl, 1 mM MgCl2·6H2O, 1 mM CaCl2 and 5 mM HEPES (pH 7.4). Microelectrodes were filled with 2 M KCl; 2.3 M CsCl, 0.2 M CsOH, 10 mM EGTA and 10 mM HEPES (pH 7.4, nAChR).
The oocyte was penetrated with two microelectrodes, the current electrode for voltage injection and the potential electrode for voltage sensing. The membrane potential measured by the sensing electrode was compared with a command voltage with the difference brought to zero by means of a control amplifier.
Materials
Stock solutions of capsaicin (1), capsazepine (2) and the test compounds (6–17) were prepared in 100% DMSO (100 mM for oocyte experiments, Dimethyl Sulfoxide). All chemicals were purchased from Sigma™, Vienna, Austria except where stated otherwise. The rat TRPV1 clone was kindly donated by Prof David Julius (Department of Cellular and Molecular Pharmacology, University of California, San Francisco, CA, USA).
Data analysis
Compound 1 (1 μM) was applied as control, followed by 5–6 min wash period and subsequent co-application of 1 and a respective test compound (100 μM). Current amplitudes evoked by control 1 were set as 100%. Currents evoked by hit compounds exceeding control suggested agonistic activity while smaller currents revealed TRPV1 inhibition. Equal amounts of DMSO were present in all test solutions. The maximum DMSO concentration in the bath (0.3%) induced no significant effects on TRPV1. Data points represent mean ± SEM from at least three oocytes from ≥2 batches.
Concentration–response curves for modulation of TRPV1 channels were generated by plotting the peak currents evoked by co-application of 1 μM capsaicin (I1, control current) and different concentrations of the hit compound (Icompound) as function of the test compound concentration. Peak currents were normalized to I1 and expressed as percentage of control peak current: Icompound/I1*100.
Half maximum inhibitory concentration (IC50) value and Hill Coefficient (nH) for test compounds were derived from sigmoidal curve fit using Origin 7.0 software(OriginLab®, Northampton, MA, USA).
Results & discussion
As a starting point for this study, we used the data set of TRPV1 antagonists we recently compiled and used for classification studies [45]. The data set has been carefully curated and contains 408 compounds, measured in one protocol. As a threshold for active compounds 100 nM (pIC50 = 7) was defined, which led to a set of 201 actives and 207 inactives. The subsequent workflow for identifying new potential TRPV1 ligands included the following steps: pharmacophore-based clustering of the 201 TRPV1 antagonists, generation and computational validation of the pharmacophore models, virtual screening of a vendor database, prioritization of the hit list, biological testing of selected hits. An overview on the workflow is provided in Figures 2 & 3.
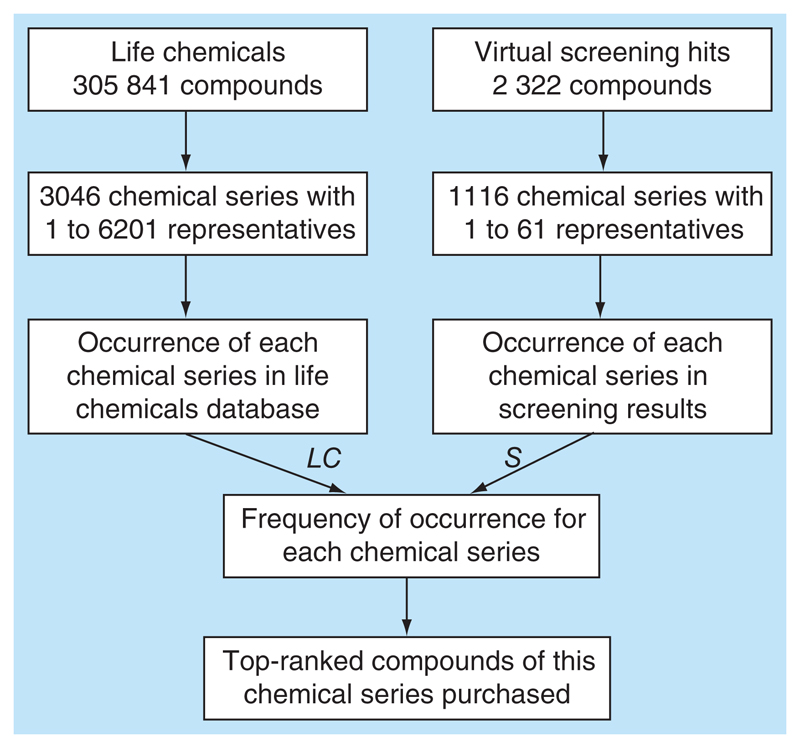
Figure 3. Selection of compounds for biological testing.
LC: LifeChem; S: Hit list.
Pharmacophore-based clustering of the data set of TRPV1 antagonists
The pharmacophore-based clustering of 201 published TRPV1 antagonists, sharing the same binding site, was performed in Ligandscout 3.1 [32] using the RDF-code similarity algorithm [50]. We obtained three large clusters with 31, 63 and 87 compounds, respectively, and nine small clusters with 1–5 compounds each (Table 1). The chemical structures of cluster representatives, their ChEMBL_ID and their activity values are provided in Table 1 of the Supplementary Material.
Table 1. Amount of compounds in each pharmacophore cluster.
| Cluster | Representatives |
|---|---|
| 1 | 31 |
| 2 | 63 |
| 3 | 87 |
| 4 | 5 |
| 5 | 5 |
| 6 | 3 |
| 7 | 2 |
| 8 | 1 |
| 9 | 1 |
| 10 | 1 |
| 11 | 1 |
| 12 | 1 |
Cluster 1 contained 31 thiazolo-pyrimidine derivatives with IC50 values from 1 to 96 nM. Although these compounds belong to the same chemical series [22], we considered them for building pharmacophore models due to their potent activity values and to their diverse substitution pattern around the main scaffold.
Cluster 2 comprised 63 substituted isoquinolin-aralkyl-ureas and -amides [15], hydroxynaphtalen-ureas and -amides [17] and heteroaryl β-tetralin ureas [23] with IC50 values ranging from 2 to 100 nM. A potent clinical candidate, ABT-116 (IC50 7 nM) [51], with an indazole-urea scaffold, was also present in this cluster.
The highest populated and most diverse cluster (cluster 3) contained 87 heterocyclic compounds with IC50 values from 0.21 to 100 nM. It comprised heterocycles [5,12] and various arylurea, biarylamide, aminoquinazoline [18,21], pyrimido-azepine [52], piperazine-benzimidazole [19] and thiazolo-pyrimidine [22] derivatives.
Small clusters contained piperidine carboxamide [35] and azetidone [53] derivatives, as well as benzamide [14] and isoquinoline [15] derivatives with IC50 values from 2.4 to 100 nM. Noteworthy, a potent preclinical candidate SB782443 [54] (IC50 49 nM) was a singleton, though it contains a thiazolo-pyrimidine in its structure (as compounds from cluster 1). Surprisingly, reference compound 2 (IC50 100 nM) represented a unique pharmacophoric pattern in our data set, though the majority of the published studies were based on its structure or used its pharmacophore features as a starting point. In order to obtain a pharmacophoric pattern characteristic for these diverse compounds, we united all the representatives of the small clusters into one additional cluster (cluster 4). However, we excluded iodo-resiniferanotoxin [55] (IC50 = 27 nM), which also appeared as singleton, due to its highly different physicochemical properties compared with other antagonists in the data set (molar refractivity and molecular weight are more than 4 standard deviations away from their mean values for compounds in cluster 4).
As the clusters show a broad variance in the biological activity values (2 orders of magnitude), we further divided each cluster into three subsets based on the IC50 values. This should allow extracting pharmacophoric patterns for highly active representatives of each cluster (see Figure 2 for composition of the clusters).
Generation & computational validation of the pharmacophore models
Consequently, ten ligand-based pharmacophore models were generated for each subset, in other words, training set, using shared feature mode and the RDF-code similarity algorithm of Ligandscout 3.1, thus resulting in 120 pharmacophore models. The ability of the models to differentiate between active and inactive TRPV1 antagonists was assessed using several different data sets. As a positive control, a data set with all compounds from the cluster to which the training set belonged, and a data set of active compounds from other clusters were used. As a negative control served a data set of 207 inactive compounds evaluated in the same assay setup [45] and a data set of decoys generated for each training set [47]. We refer to those data sets as positive and negative validation throughout the manuscript. Finally, a set of 17 compounds that have been in clinical trials [16,43] was established in order to also assess the performance of the pharmacophore models to identify compounds which are structurally different from the initial training sets.
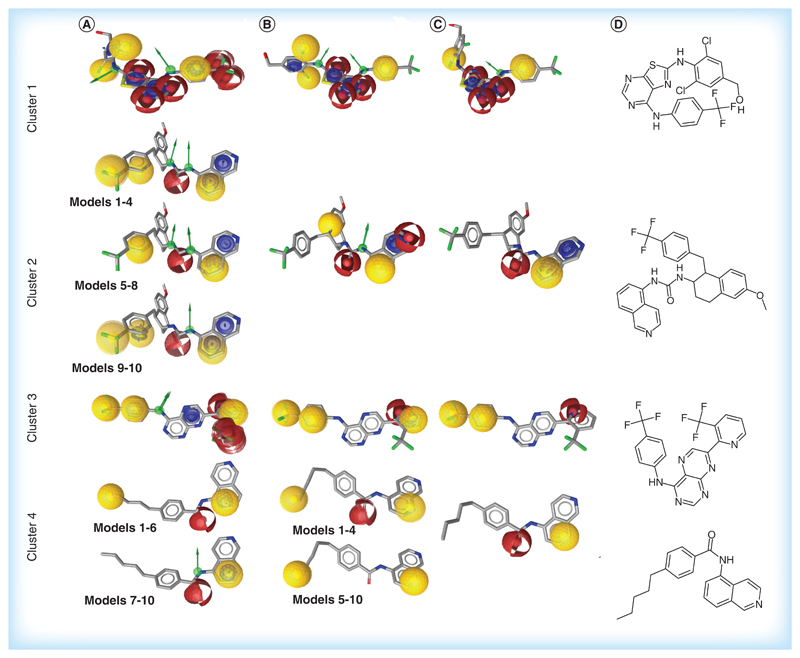
In general, models built on the highly active representatives of the different clusters (Figure 4A) had higher number of features then those derived from less active representatives (Figure 4B) and those built on molecules from the whole cluster (Figure 4C). It is also worth to note that the pharmacophores generated were based on stretched conformations of the ligands, which indicates an elongated binding pocket. This is in agreement with the recently published cryo-electron microscopy structure of the receptor [56].
Figure 4. Pharmacophore models built for 201 TRPV1 antagonists.
Ten models built for each training set are superposed on the chemical structure of the most active representative of the respective cluster. (A) Models built on highly active representatives of the clusters. (B) Models built on highly active and middle active representatives of the cluster. (C) Models built for the whole cluster. (D) The most active representative of each cluster. Pharmacophore features: yellow: lipophilic substitution; blue: Aromatic ring; red: H-bond acceptor; green: H-bond donor.
For colour images see online: www.future-science.com/doi/full/10.4155/FMC.14.168
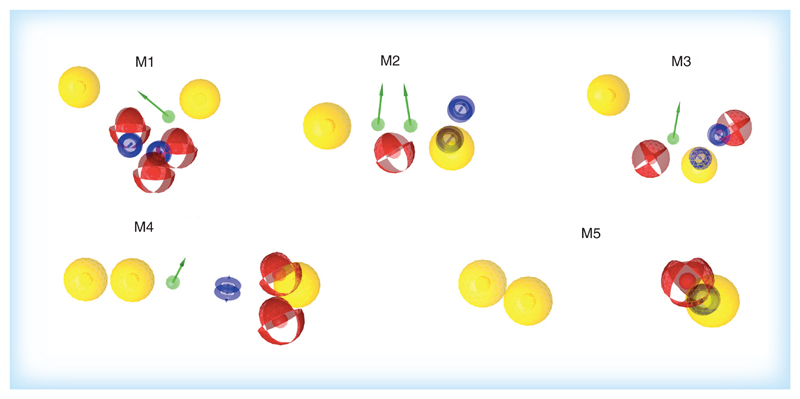
The pharmacophoric patterns of cluster 1 differed only slightly between highly active, middle active and lower active TRPV1 antagonists (Figure 4, row 1). All 30 models performed excellent in positive and negative validation, but failed in retrieving compounds that entered clinical trials (Supplementary Figure 2 row 1). Remarkably, models built on the highly active thiazolo-pyrimidines (training set 1A) showed the same pharmacophoric pattern as reported earlier in the literature [35] for piperidine carboxamide derivatives. Finally, we selected the model based on training set 1C (model M1) for virtual screening experiments, as it contains the lowest number of pharmacophore features (Figure 5).
Figure 5. Pharmacophore models M1-M5 selected for virtual screening of the LifeChem database.
Pharmacophore features: yellow: lipophilic substitution; blue: aromatic ring; red: H-bond acceptor; green: H-bond donor.
For colour images see online: www.future-science.com/doi/full/10.4155/FMC.14.168
The pharmacophore models built on clusters 2 and 3 provided the richest spectrum of information about main features responsible for activity of TRPV1 antagonists. Three types of pharmacophoric patterns were obtained for the highly active isoquinolin-aralkyl-ureas of cluster 2 (training set 2A) (Figure 4A, row 2, models 1–4, 5–8 and 9–10). However, for the middle active representatives (training set 2B), only a single pharmacophoric pattern was identified (Figure 4B, row 2). This pharmacophore mimicked the feature pattern described by Blumberg et al. [33,41] for benzyl-thiourea derivatives, with two lipophilic areas and two H-bond acceptors. Noteworthy, all 20 models built for the training sets 2A and 2B showed excellent performance in both positive and negative validation (Supplementary Figure 2, row 2). We selected model 8 (M2) from the training set 2A and model 9 (M3) from training set 2B for further virtual screening of the vendor database because they retrieved more active representatives of the family cluster as well as several clinical candidates (ABT102 [16], GRC6211 [16] and AMG425619 [43]) (Figure 5 & Table 2).
Table 2. Performance in validation of live models selected for virtual screening.
| Model | TR† | Whole cluster† | Active compounds from other clusters | 207 inactive compounds | 17 compounds in clinical trials | Corresponding decoy set† | EF, 1% | EF, 5% | EF, 10% | AUC, 100% |
|---|---|---|---|---|---|---|---|---|---|---|
| M1 | 31 (31) | 31 (31) | 0 | 12 | 0 | 0 (1600) | 51 | 51 | 51 | 1 |
| M2 | 8 (8) | 57 (63) | 0 | 14 | 3 | 0 (400) | 51 | 51 | 51 | 1 |
| M3 | 32 (32) | 62 (63) | 1 | 17 | 3 | 1 (1650) | 51 | 49,5 | 49,5 | 0,98 |
| M4 | 16 (16) | 63 (87) | 1 | 19 | 1 | 0 (900) | 51 | 51 | 51 | 1 |
| M5 | 37 (37) | 87 (87) | 15 | 78 | 5 | 8 (2050) | 51 | 39,5 | 39,5 | 1 |
Total amount of compounds in a data set is indicated in the brackets.
AUC: Area under the ROC curve; EF: Enrichment factor; TR: Training set.
The 10 models from training set 3A showed one distinct pharmacophoric pattern (Figure 4A, row 3) and performed excellent in positive and negative validation (Supplementary Figure 2, row 3). We selected model 1 (M4) built from the training set 3A for virtual screening, because, when compared with the other nine models from this cluster, M4 identified all compounds from the initial training set 3A and one compound from the clinical data set (NGD8243 [16]) (Figure 5 & Table 2). The pharmacophoric patterns identified for the training set 3B (Figure 4B, row 3) were found in 4 compounds from the clinical data set (AMG425619 [43], A993610, BCTC, NGD8243 [16]) and representatives of other clusters (Supplementary Figure 2, row 3). Finally, model 3 (M5) outperformed other models in the validation with the decoy set therefore we selected this model for virtual screening (Figure 5 & Table 2).
Models obtained for the training sets 2C and 3C contained four and three pharmacophore features, respectively, (Figure 4C, row 2) and thus were quite unspecific. They showed high false positive rates in the validation with the decoy data sets (Supplementary Figure 2, row 2 and row 3) and were thus excluded from further considerations.
Finally, the models built on different training sets of cluster 4 contained from two to four pharmacophore features. Two distinct pharmacophoric patterns were identified for the training set 4A (Figure 4A, row 4). One of them also included three features reported [16] earlier for a data set of TRPV1 antagonists in clinical trials, namely an aromatic ring, an H-bond donor and an H-bond acceptor. However, due to a lower amount of features, the pharmacophore models built on representatives of cluster 4 performed poorly in positive and negative validation (Supplementary Figure 2, row 4). Therefore, none of the models were selected for further virtual screening of the vendor database. Noteworthy, models containing two pharmacophore features (Figure 4B & C, row 4) could not undergo validation screening because the pharmacophore alignment algorithm requires presence of at least three features.
Virtual screening & selection of compounds for biological testing
Virtual screening of LifeChem with the 5 selected models, M1, M2, M3, M4 and M5, led to a hit list of 1909 compounds (Table 3). Each compound mapped all the pharmacophore features of the respective model and did not interfere with the exclusion volume of that model. Finally, 12 compounds were selected for the experimental validation following the protocol described below (Figure 3).
Table 3. Results of virtual screening of a vendor database LifeChem.
| Model | Amount of hits |
|---|---|
| M1 | 46 |
| M2 | 23 |
| M3 | 182 |
| M4 | 32 |
| M5 | 1626 |
The LifeChem compound library used for virtual screening comprises 3046 diverse chemical series, where each chemical series corresponds to a certain compound scaffold with 1 to 6201 derivatives. The 1909 compounds from our hit list clustered in 578 chemical series with 1 to 43 representatives. Consequently, we were interested to pick compounds from those chemical series, which had numerous representatives in the hit list as well as in LifeChem. Therefore, the amount of derivatives for each chemical series in the LifeChem (LC) and in the hit list (S) was retrieved and the frequency (f) of each chemical series in the screening results was calculated according to (Equation 1).
| Equation 1 |
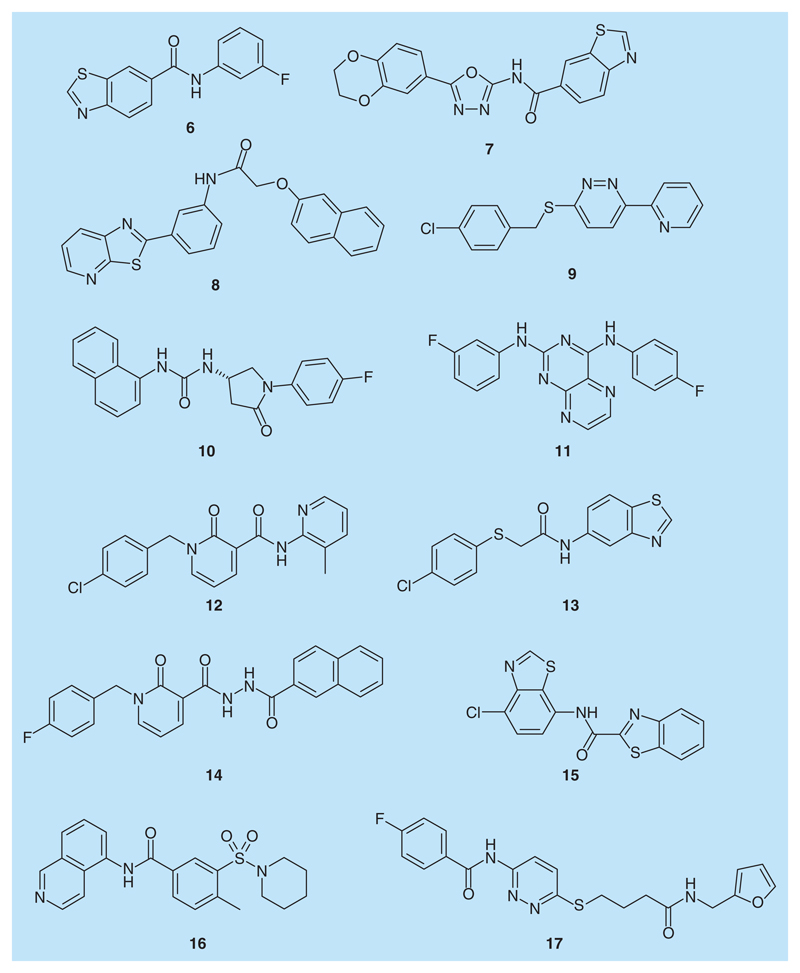
Thus, a higher frequency for a certain chemical scaffold indicates that the number of representatives in the hit list for this scaffold is close to the number of representatives in the LifeChem database. Noteworthy, chemical series with less then 20 representatives in LifeChem were excluded from the analysis. Consequently, we purchased the top-ranked compound of those series showing a frequency greater than 5% (Figure 6). Occurrence of all chemical series in the hit list is provided in Supplementary Table 3.
Figure 6. Twelve compounds selected for biological testing.
Results of biological testing
The biological activity of the 12 selected compounds on a rat TRPV1 clone was analyzed by means of the two-micro-electrode voltage-clamp technique. HPLC and 1H-NMR spectra of the compounds tested are provided in Table 4 and Figures 5-16 of the Supplementary Material.
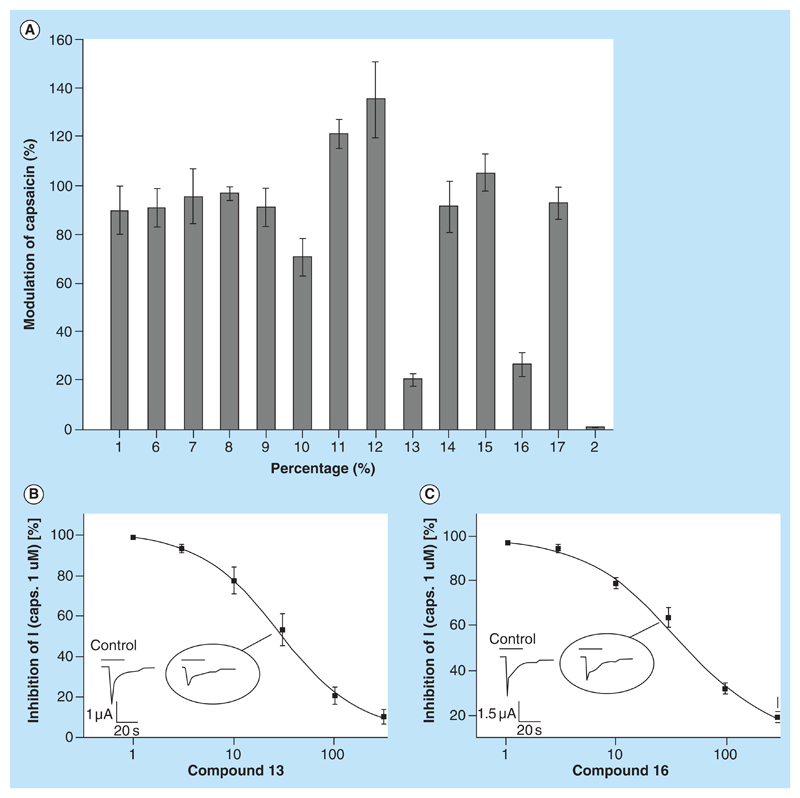
Current amplitudes evoked by 1 μM 1 as control were set as control for reference (100%). When currents evoked by co-application of 1 (1 μM) and tested compounds were smaller than the control, this indicated inhibitory activity. Interestingly, also increase of currents was observed (though statistically not significant), which would indicate either partial agonistic activity or positive allosteric modulation of the receptor. Additionally, we used a TRPV1 antagonist (2) as a reference (Figure 1). In a first screening round all compounds purchased were tested at a concentration of 100 μM. Two compounds (13 and 16) induced significant inhibition of the activated TRPV1 receptor (p < 0.01) while the increase of the current response observed upon co-application of 11 and 12 was not significant (p > 0.05) (Figure 7A). Application of 100 μM 11 and 12 in the absence of 1 induced no inward currents.
Figure 7. Biological testing of 12 selected compounds.
(A) Inhibition of capsaicin (1 μM) induced inward currents trough TRPV1 expressed in Xenopus oocytes, illustrated as mean ±SE residual current in % of control. Test compounds and capsazepine (2) were applied at 100 μM (n = 3–10). (B&C) Concentration-dependent inhibition of TRPV1 currents by test compounds 13 and 16, respectively. The p-value for compound 13 and 16 is less than 0.01 where the p-value for compound 11 and 12 is greater than 0.05.
Analysis of the concentration-dependent inhibition of the inward currents through TRPV1 by 13 and 16 revealed IC50 values of 27.3 and 36.1 μM, respectively (Figure 7B & C). Mean and SE values of IC50 curves are reported in Table S17. Considering the fact that drugs are commonly less potent on ion channels expressed in Xenopus oocytes as compared with channels expressed in either mammalian cells or native tissues [57], these compounds may represent interesting scaffolds for further studies. It is also important to emphasize that having identified TRPV1 inhibitors, though with low μM affinity, we consider this as a proof of concept, that pharmacophore modeling is an efficient method for mining data sets collected from public data sources.
Compounds 13 and 16 were identified by models M5 and M3, respectively. The substances matched all the pharmacophore features of the respective models: compound 13 passed five features of model M5 and compound 16 passed seven features of model M3.
One of the common approaches to prioritize compounds for biological testing includes similarity search to the most active representatives of the training set. However, this results in selection of highly similar compounds. Therefore, in order to confirm that the pharmacophore-based search indeed identified new chemical scaffolds, we calculated the similarity between the 12 purchased compounds and the reference antagonist 2 using MACCS fingerprints [48] and the Tanimoto similarity metrics. Tanimoto coefficients obtained were between 0.29 and 0.46 (Table 4). This confirms that the compounds identified are indeed chemically different from the reference compound. Furthermore, also the similarity of the hits to the highly active representatives of each cluster (Supplementary Table 18) was checked. In this case, Tanimoto coefficients were below 0.6 for 95% of the values. These results once more support the applicability of pharmacophore models for scaffold hopping.
Table 4. Tanimoto similarity between 12 tested compounds and reference compound 2.
| Compound | Similarity |
|---|---|
| 6 | 0.2985 |
| 7 | 0.3415 |
| 8 | 0.3649 |
| 9 | 0.2879 |
| 10 | 0.4583 |
| 11 | 0.2879 |
| 12 | 0.3919 |
| 13 | 0.3571 |
| 14 | 0.3684 |
| 15 | 0.3043 |
| 16 | 0.4024 |
| 17 | 0.4375 |
Conclusion
The public availability of large data sets for development of in silico models provides new opportunities for identification of novel hit compounds. However, due to the heterogeneity of these data, they need careful manual curation. With this manuscript we show that pharmacophore-based clustering of a large set of TRPV1 ligands extracted from the public domain combined with thorough computational and experimental validation allows to identify new TRPV1 inhibitors. This approach could be further projected on the chemical space of other biological targets, which possess abundant pharmacological data but lack structural information.
Future perspective
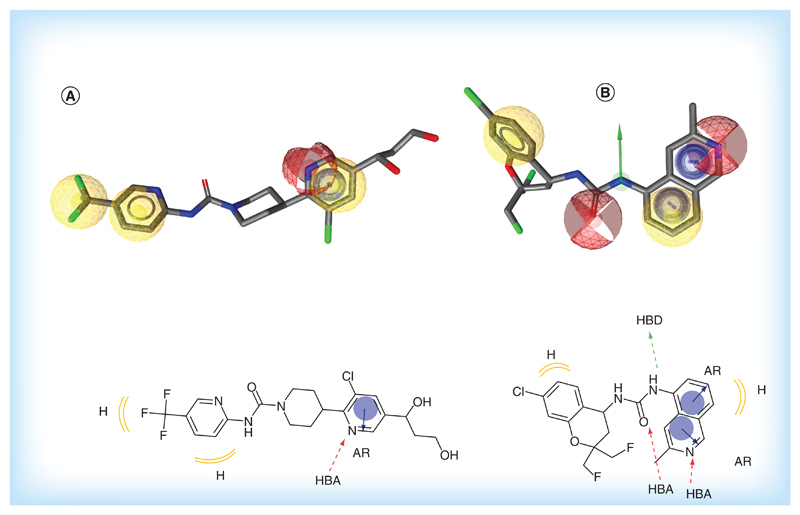
A major side effect of blocking TRPV1 is its implication in thermosensation and thermoregulation pathways. Related side effects, such as impaired heat sensation and hypothermia, often led to withdrawal of compounds from clinical trials [58]. However, very recently two independent groups have reported chemical structures of preclinical candidates, which do not elevate the body temperature [59,60]. These studies confirm the possibility to separate the analgesic effect from hypothermia while modulating TRPV1 with small molecule antagonists. Interestingly, compounds V116517 [59] and A-1165442 [60] fitted models M5 and M3 reported in this work (Figure 8), which further supports the validity of our pharmacophore models.
Figure 8. Preclinical candidates in the validated pharmacophores.
(A) Compound V116517 [59] in pharmacophore M5, 3D and 2D view. (B) Compound A-1165442 [60] in pharmacophore M3, 3D and 2D view. Pharmacophore features: yellow: lipophilic substitution; blue: aromatic ring; red: H-bond acceptor; green: H-bond donor.
For colour images see online: www.future-science.com/doi/full/10.4155/FMC.14.168
Since pharmacophore modeling is already an established approach for efficient identification of novel scaffolds for various biological targets, the approach proposed here could be further optimized and adjusted to the needs of a certain research question. In case of TRPV1, application of pharmacophore-based clustering to potential pyridine-carboxamide [59] and isoquinolin-urea [60] derivatives and subsequent pharmacophore modeling would allow to identify ensemble of pharmacophore features inducing modulation of the receptor by those molecules. Further, one could compare these models with pharmacophore patterns present in compounds failing the body temperature studies and compounds withdrawn from clinic. It would identify pharmacophoric ensembles causing these side effects in candidate molecules.
In addition, recently published structures of TRPV1 allow application of molecular docking. Combined with pharmacophore modeling, docking would suggest protein–ligand interactions at the place in the binding site derived from the pharmacophore features of the small molecules. Therefore, it would be possible to distinguish pharmacophoric interactions between the protein and its ligands with both safe and unsafe profiles. Further, lead optimization of the series with established pharmacophores would focus on development candidates with less undesirable side effects. Taken together, an elaborated pharmacophore approach would allow generation of novel chemical entities with the desired profile.
Supplementary Material
To view the supplementary data that accompany this paper please visit the journal website at: www.future-science.com/doi/full/10.4155/FMC.14.168
Executive summary.
Pharmacophore-based clustering of the data set & computational validation of the pharmacophore models
This study gives a profound systemized classification of the pharmacophoric patterns present in publicly available TRPV1 antagonists sharing the same binding site. Identified pharmacophores reflect the complexity of the binding site in TRPV1, which can accommodate structurally diverse ligands of different size.
Virtual screening & results of biological testing
The applicability of two pharmacophore models for virtual screening was confirmed and led to the identification of novel scaffolds for TRPV1 receptor modulation.
Two out of 12 compounds tested inhibited TRPV1, two other compounds showed modulation of TRPV1 in coapplication with capsaicin.
Pharmacophore modeling is an efficient approach to mine large data sets composed of local structure–activity relationship series
Future perspective
Prosperous models are provided as pml files in the Supplementary Material along with the article and can be freely used by the academic community in the future.
The experimentally validated ligand-based pharmacophore models presented will be further implemented in house in structure-based studies on the recently released structure of the receptor.
Acknowledgements
We thank Inte:Ligand for providing us a free license for Ligandscout 3.1 and Open Eye for an Omega license. We thank M Zehl and E Urban for providing HPLC and 1H-NMR spectral data.
This work was supported by the Austrian Science Fund (FWF doctoral program ‘Molecular drug targets’ W1232). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest; •• of considerable interest
- 1.Jara-Oseguera A, Nieto-Posadas A, Szallasi A, Islas LD, Rosenbaum T. Molecular mechanisms of TRPV1 channel activation. Open Pain J. 2010;3:68–81. [Google Scholar]
- 2.Caterina MJ, Schumacher MA, Tominaga M, Rosen TA, Levine JD, Julius D. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997;389(6653):816–824. doi: 10.1038/39807. [•• The authors report the exact location of transmembrane regions in transient receptor potential vanilloid type 1 (TRPV1) predicted by hydrophilicity analysis (in agreement with structures reported by Cao E. et al., Nature 2013). However, the topology of those transmembrane regions was predicted wrong (β-sheets instead of α-helices). ] [DOI] [PubMed] [Google Scholar]
- 3.Jancso N, Jancso-Gabor A, Szolcsanyi J. Direct evidence for neurogenic inflammation and its prevention by denervation and by pretreatment with capsaicin. Br J Pharmacol Chemother. 1967;31(1):138–151. doi: 10.1111/j.1476-5381.1967.tb01984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Premkumar LS. Targeting TRPV1 as an alternative approach to narcotic analgesics to treat chronic pain conditions. AAPS J. 2010;12(3):361–370. doi: 10.1208/s12248-010-9196-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westaway SM. The potential of transient receptor potential vanilloid type 1 channel modulators for the treatment of pain. J Med Chem. 2007;50(11):2589–2596. doi: 10.1021/jm060637e. [DOI] [PubMed] [Google Scholar]
- 6.Moran MM, Mcalexander MA, Biro T, Szallasi A. Transient receptor potential channels as therapeutic targets. Nat Rev Drug Discov. 2011;10(8):601–620. doi: 10.1038/nrd3456. [•• Authors give an excellent pharmacological overview of TRP channels. Special attention is given to benefits and pitfalls associated with therapeutic targeting each of the channels with small molecules. ] [DOI] [PubMed] [Google Scholar]
- 7.Walpole CS, Bevan S, Bovermann G, et al. The discovery of capsazepine, the first competitive antagonist of the sensory neuron excitants capsaicin and resiniferatoxin. J Med Chem. 1994;37(13):1942–1954. doi: 10.1021/jm00039a006. [DOI] [PubMed] [Google Scholar]
- 8.Kim MS, Ryu H, Kang DW, et al. 2-(3-fluoro-4-methylsulfonylaminophenyl)propanamides as potent transient receptor potential vanilloid 1 (TRPV1) antagonists: structure-activity relationships of 2-amino derivatives in the N-(6-trifluoromethylpyridin-3-ylmethyl) C-region. J Med Chem. 2012;55(19):8392–8408. doi: 10.1021/jm300780p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J, Kang SU, Kil MJ, et al. Analysis of structure-activity relationships for the ‘A-region’ of N-(4-t-butylbenzyl)-N’-[4-(methylsulfonylamino)benzyl]thiourea analogues as TRPV1 antagonists. Bioorg Med Chem Lett. 2005;15(18):4136–4142. doi: 10.1016/j.bmcl.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Lee J, Jin MK, Kang SU, et al. Analysis of structure-activity relationships for the ‘B-region’ of N-(4-t-butylbenzyl)-N’-[4-(methylsulfonylamino)benzyl]-thiourea analogues as TRPV1 antagonists. Bioorg Med Chem Lett. 2005;15(18):4143–4150. doi: 10.1016/j.bmcl.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Appendino G, Daddario N, Minassi A, Moriello AS, De Petrocellis L, Di Marzo V. The taming of capsaicin. Reversal of the vanilloid activity of N-acylvanillamines by aromatic iodination. J Med Chem. 2005;48(14):4663–4669. doi: 10.1021/jm050139q. [DOI] [PubMed] [Google Scholar]
- 12.Blum CA, Caldwell T, Zheng X, et al. Discovery of novel 6,6-heterocycles as transient receptor potential vanilloid (TRPV1) antagonists. J Med Chem. 2010;53(8):3330–3348. doi: 10.1021/jm100051g. [DOI] [PubMed] [Google Scholar]
- 13.Doherty EM, Fotsch C, Bo Y, et al. Discovery of potent, orally available vanilloid receptor-1 antagonists. Structure-activity relationship of N-aryl cinnamides. J Med Chem. 2005;48(1):71–90. doi: 10.1021/jm049485i. [DOI] [PubMed] [Google Scholar]
- 14.Shishido Y, Jinno M, Ikeda T, et al. Synthesis of benzamide derivatives as TRPV1 antagonists. Bioorg Med Chem Lett. 2008;18(3):1072–1078. doi: 10.1016/j.bmcl.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 15.Jetter MC, Youngman MA, Mcnally JJ, et al. N-isoquinolin-5-yl-N’-aralkyl-urea and -amide antagonists of human vanilloid receptor 1. Bioorg Med Chem Lett. 2004;14(12):3053–3056. doi: 10.1016/j.bmcl.2004.04.038. [DOI] [PubMed] [Google Scholar]
- 16.Kym PR, Kort ME, Hutchins CW. Analgesic potential of TRPV1 antagonists. Biochem Pharmacol. 2009;78(3):211–216. doi: 10.1016/j.bcp.2009.02.014. [• Authors propose three pharmacophoric features present in 12 clinical candidates reported to date. Additionally, a homology model of TRPV1 (significantly different from the structures reported by Cao E. et al., Nature 2013) is presented and those 12 clinical candidates are overlaid in the binding pocket. ] [DOI] [PubMed] [Google Scholar]
- 17.Mcdonnell ME, Zhang SP, Nasser N, Dubin AE, Dax SL. 7-Hydroxynaphthalen-1-yl-urea and -amide antagonists of human vanilloid receptor 1. Bioorg Med Chem Lett. 2004;14(2):531–534. doi: 10.1016/j.bmcl.2003.09.090. [DOI] [PubMed] [Google Scholar]
- 18.Zheng X, Hodgetts KJ, Brielmann H, et al. From arylureas to biarylamides to aminoquinazolines: discovery of a novel, potent TRPV1 antagonist. Bioorg Med Chem Lett. 2006;16(19):5217–5221. doi: 10.1016/j.bmcl.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 19.Shao B, Huang J, Sun Q, Valenzano KJ, Schmid L, Nolan S. 4-(2-Pyridyl)piperazine-1-benzimidazoles as potent TRPV1 antagonists. Bioorg Med Chem Lett. 2005;15(3):719–723. doi: 10.1016/j.bmcl.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 20.Li FN, Kim NJ, Chang DJ, et al. Synthesis and structural optimization of multiple H-bonding region of diarylalkyl (thio)amides as novel TRPV1 antagonists. Bioorg Med Chem. 2009;17(24):8149–8160. doi: 10.1016/j.bmc.2009.10.043. [DOI] [PubMed] [Google Scholar]
- 21.Blum CA, Zheng X, Brielmann H, et al. Aminoquinazolines as TRPV1 antagonists: modulation of drug-like properties through the exploration of 2-position substitution. Bioorg Med Chem Lett. 2008;18(16):4573–4577. doi: 10.1016/j.bmcl.2008.07.036. [DOI] [PubMed] [Google Scholar]
- 22.Lebsack AD, Branstetter BJ, Hack MD, et al. Identification and synthesis of 2,7-diamino-thiazolo[5,4-d]pyrimidine derivatives as TRPV1 antagonists. Bioorg Med Chem Lett. 2009;19(1):40–46. doi: 10.1016/j.bmcl.2008.11.024. [DOI] [PubMed] [Google Scholar]
- 23.Jetter MC, Youngman MA, Mcnally JJ, et al. Heteroaryl beta-tetralin ureas as novel antagonists of human TRPV1. Bioorg Med Chem Lett. 2007;17(22):6160–6163. doi: 10.1016/j.bmcl.2007.09.036. [DOI] [PubMed] [Google Scholar]
- 24.Tafesse L, Sun Q, Schmid L, et al. Synthesis and evaluation of pyridazinylpiperazines as vanilloid receptor 1 antagonists. Bioorg Med Chem Lett. 2004;14(22):5513–5519. doi: 10.1016/j.bmcl.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 25.Xi N, Bo Y, Doherty EM, et al. Synthesis and evaluation of thiazole carboxamides as vanilloid receptor 1 (TRPV1) antagonists. Bioorg Med Chem Lett. 2005;15(23):5211–5217. doi: 10.1016/j.bmcl.2005.08.100. [DOI] [PubMed] [Google Scholar]
- 26.Hodgetts KJ, Blum CA, Caldwell T, et al. Pyrido[2,3-b] pyrazines, discovery of TRPV1 antagonists with reduced potential for the formation of reactive metabolites. Bioorg Med Chem Lett. 2010;20(15):4359–4363. doi: 10.1016/j.bmcl.2010.06.069. [DOI] [PubMed] [Google Scholar]
- 27.Li FN, Kim NJ, Paek SM, et al. Design, synthesis, and biological evaluation of novel diarylalkyl amides as TRPV1 antagonists. Bioorg Med Chem. 2009;17(10):3557–3567. doi: 10.1016/j.bmc.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt RG, Bayburt EK, Latshaw SP, et al. Chroman and tetrahydroquinoline ureas as potent TRPV1 antagonists. Bioorg Med Chem Lett. 2011;21(5):1338–1341. doi: 10.1016/j.bmcl.2011.01.056. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi E, Hirano N, Nagahara T, et al. Discovery of potent transient receptor potential vanilloid 1 antagonists: design and synthesis of phenoxyacetamide derivatives. Bioorg Med Chem Lett. 2013;23(11):3154–3156. doi: 10.1016/j.bmcl.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 30.Kim HS, Jin MK, Kang SU, et al. alpha-Methylated simplified resiniferatoxin (sRTX) thiourea analogues as potent and stereospecific TRPV1 antagonists. Bioorg Med Chem Lett. 2014;24(12):2685–2688. doi: 10.1016/j.bmcl.2014.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldmann D, Montanari F, Richter L, Zdrazil B, Ecker GF. Exploiting open data: a new era in pharmacoinformatics. Future Med Chem. 2014;6(5):503–514. doi: 10.4155/fmc.14.13. [DOI] [PubMed] [Google Scholar]
- 32.Wolber G, Langer T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J Chem Inf Model. 2005;45(1):160–169. doi: 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
- 33.Lee J, Kim SY, Lee J, et al. Analysis of structure-activity relationships with the N-(3-acyloxy-2-benzylpropyl)-N’-[4-(methylsulfonylamino)benzyl]thiourea template for vanilloid receptor 1 antagonism. Bioorg Med Chem. 2004;12(13):3411–3420. doi: 10.1016/j.bmc.2004.04.040. [DOI] [PubMed] [Google Scholar]
- 34.Swanson DM, Dubin AE, Shah C, et al. Identification and biological evaluation of 4-(3-trifluoromethylpyridin-2-yl) piperazine-1-carboxylic acid (5-trifluoromethylpyridin-2-yl) amide, a high affinity TRPV1 (VR1) vanilloid receptor antagonist. J Med Chem. 2005;48(6):1857–1872. doi: 10.1021/jm0495071. [DOI] [PubMed] [Google Scholar]
- 35.Cheung WS, Calvo RR, Tounge BA, et al. Discovery of piperidine carboxamide TRPV1 antagonists. Bioorg Med Chem Lett. 2008;18(16):4569–4572. doi: 10.1016/j.bmcl.2008.07.035. [DOI] [PubMed] [Google Scholar]
- 36.Rebolledo CL, Sotelo-Hitschfeld P, Brauchi S, Olavarria MZ. Design and synthesis of conformationally restricted capsaicin analogues based in the 1, 3, 4-thiadiazole heterocycle reveal a novel family of transient receptor potential vanilloid 1 (TRPV1) antagonists. Eur J Med Chem. 2013;66:193–203. doi: 10.1016/j.ejmech.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Kim KH. 3-D-QSAR analysis of N-(3-acyloxy-2-benzylpropyl)-N’-dihydroxytetrahydrobenzazepine and tetrahydroisoquinoline and N-(3-acyloxy-2-benzylpropyl)-N’-(4-hydroxy-3-methoxybenzyl) thioureas analogues as potent vanilloid receptor ligands. Bioorg Med Chem. 2002;10(5):1367–1372. doi: 10.1016/s0968-0896(01)00404-7. [DOI] [PubMed] [Google Scholar]
- 38.Kristam R, Parmar V, Viswanadhan VN. 3D-QSAR analysis of TRPV1 inhibitors reveals a pharmacophore applicable to diverse scaffolds and clinical candidates. J Mol Graph Model. 2013;45:157–172. doi: 10.1016/j.jmgm.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 39.Ryu H, Jin MK, Kim SY, et al. Stereospecific high-affinity TRPV1 antagonists: chiral N-(2-benzyl-3-pivaloyloxypropyl) 2-[4-(methylsulfonylamino)phenyl]propionamide analogues. J Med Chem. 2008;51(1):57–67. doi: 10.1021/jm701049p. [DOI] [PubMed] [Google Scholar]
- 40.Kim YS, Kil MJ, Kang SU, et al. N-4-t-Butylbenzyl 2-(4-methylsulfonylaminophenyl) propanamide TRPV1 antagonists: structure-activity relationships in the A-region. Bioorg Med Chem. 2012;20(1):215–224. doi: 10.1016/j.bmc.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhondwe RS, Kang DW, Kim MS, et al. Structure-activity relationships and molecular modeling of the N-(3-pivaloyloxy-2-benzylpropyl)-N’-[4-(methylsulfonylamino) benzyl] thiourea template for TRPV1 antagonism. Bioorg Med Chem Lett. 2012;22(11):3656–3660. doi: 10.1016/j.bmcl.2012.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim NJ, Li FN, Lee JH, et al. Heterocycle-linked phenylbenzyl amides as novel TRPV1 antagonists and their TRPV1 binding modes: constraint-induced enhancement of in vitro and in vivo activities. Chem Asian J. 2013;8(2):400–409. doi: 10.1002/asia.201200730. [DOI] [PubMed] [Google Scholar]
- 43.Garami A, Shimansky YP, Pakai E, Oliveira DL, Gavva NR, Romanovsky AA. Contributions of different modes of TRPV1 activation to TRPV1 antagonist-induced hyperthermia. J Neurosci. 2010;30(4):1435–1440. doi: 10.1523/JNEUROSCI.5150-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaulton A, Bellis LJ, Bento AP, et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40:d1100–d1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsareva DA, Ecker GF. How far could we go with open data - a case study for TRPV1 antagonists. Mol Inform. 2013;32(5-6):555–562. doi: 10.1002/minf.201300019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hawkins PC, Skillman AG, Warren GL, Ellingson BA, Stahl MT. Conformer generation with OMEGA: algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J Chem Inf Model. 2010;50(4):572–584. doi: 10.1021/ci100031x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mysinger MM, Carchia M, Irwin JJ, Shoichet BK. Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J Med Chem. 2012;55(14):6582–6594. doi: 10.1021/jm300687e. [• An algorithm used for automated decoy generation from ZINC database is validated on numerous examples of biological targets.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Durant JL, Leland BA, Henry DR, Nourse JG. Reoptimization of MDL keys for use in drug discovery. J Chem Inf Model. 2002;42(6):1273–1280. doi: 10.1021/ci010132r. [DOI] [PubMed] [Google Scholar]
- 49.Methfessel C, Witzemann V, Takahashi T, Mishina M, Numa S, Sakmann B. Patch clamp measurements on Xenopus laevis oocytes: currents through endogenous channels and implanted acetylcholine receptor and sodium channels. Pflugers Arch. 1986;407(6):577–588. doi: 10.1007/BF00582635. [DOI] [PubMed] [Google Scholar]
- 50.Wolber G, Dornhofer AA, Langer T. Efficient overlay of small organic molecules using 3D pharmacophores. J Comput Aided Mol Des. 2006;20(12):773–788. doi: 10.1007/s10822-006-9078-7. [•• Authors validate novel pharmacophore alignment algorithm of small molecules on examples of several pharmacologically relevant targets.] [DOI] [PubMed] [Google Scholar]
- 51.Brown BS, Keddy R, Perner RJ, et al. Discovery of TRPV1 antagonist ABT-116. Bioorg Med Chem Lett. 2010;20(11):3291–3294. doi: 10.1016/j.bmcl.2010.04.047. [DOI] [PubMed] [Google Scholar]
- 52.Lebsack AD, Rech JC, Branstetter BJ, et al. 1,2-diamino-ethane-substituted-6,7,8,9-tetrahydro-5H-pyrimido[4,5-d] azepines as TRPV1 antagonists with improved properties. Bioorg Med Chem Lett. 2010;20(23):7142–7146. doi: 10.1016/j.bmcl.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 53.Xiao D, Palani A, Aslanian R, et al. Spiro-piperidine azetidinones as potent TRPV1 antagonists. Bioorg Med Chem Lett. 2009;19(3):783–787. doi: 10.1016/j.bmcl.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 54.Westaway SM, Thompson M, Rami HK, et al. Design and synthesis of 6-phenylnicotinamide derivatives as antagonists of TRPV1. Bioorg Med Chem Lett. 2008;18(20):5609–5613. doi: 10.1016/j.bmcl.2008.08.105. [DOI] [PubMed] [Google Scholar]
- 55.Mcdonnell ME, Zhang SP, Dubin AE, Dax SL. Synthesis and in vitro evaluation of a novel iodinated resiniferatoxin derivative that is an agonist at the human vanilloid VR1 receptor. Bioorg Med Chem Lett. 2002;12(8):1189–1192. doi: 10.1016/s0960-894x(02)00127-0. [DOI] [PubMed] [Google Scholar]
- 56.Cao E, Liao M, Cheng Y, Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 2013;504(7478):113–118. doi: 10.1038/nature12823. [•• Authors present several possible conformation states of TRPV1 based on its single particle electron cryo-microscopy structures of 3.4–4.2 Å resolution.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goldin AL. Expression of ion channels in Xenopus oocytes. In: Clare JJ, Trezise DJ, editors. Expression and Analysis of Recombinant Ion Channels: from Structural Studies to Pharmacological Screening. Wiley-VCH Verlag GmbH & Co. KGaA; Weinheim, Germany: 2006. pp. 1–25. [Google Scholar]
- 58.Holzer P. The pharmacological challenge to tame the transient receptor potential vanilloid-1 (TRPV1) nocisensor. Br J Pharmacol. 2008;155(8):1145–1162. doi: 10.1038/bjp.2008.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tafesse L, Kanemasa T, Kurose N, et al. Structure-activity relationship studies and discovery of a potent transient receptor potential vanilloid (TRPV1) antagonist 4-[3-chloro-5-[(1S)-1,2-dihydroxyethyl]-2-pyridyl]-N-[5-(trifluoromethyl)-2-pyrid yl]-3,6-dihydro-2H-pyridine-1-carboxamide (V116517) as a clinical candidate for pain management. J Med Chem. 2014;57(15):6781–6794. doi: 10.1021/jm500818a. [DOI] [PubMed] [Google Scholar]
- 60.Voight EA, Gomtsyan AR, Daanen JF, et al. Discovery of (R)-1-(7-chloro-2,2-bis(fluoromethyl)chroman-4-yl)-3-(3-methylisoquinolin-5-yl)ur ea (A-1165442): a temperature-neutral transient receptor potential vanilloid-1 (TRPV1) antagonist with analgesic efficacy. J Med Chem. 2014;57(17):7412–7424. doi: 10.1021/jm500916t. [DOI] [PubMed] [Google Scholar]
- 61.Life Chemicals. www.lifechemicals.com.
- 62.DUD-E decoy generator. http://dude.docking.org/generate.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.