Abstract
Human P-glycoprotein is an ATP-binding cassette transporter that plays an important role in the defence against potentially harmful molecules from the environment. It is involved in conferring resistance against cancer therapeutics and plays an important role for the pharmacokinetics of drugs. The lack of a high resolution structure of P-glycoprotein has hindered its functional understanding and represents an obstacle for structure based drug development. The homologous bacterial exporter Sav1866 has been shown to share a common architecture and overlapping substrate specificity with P-glycoprotein. The structure of Sav1866 suggests that helices in the transmembrane domains diverge at the extracytoplasmic face, whereas cross-link information and a combination of small angle X-ray scattering and cryo-electron crystallography data indicate that helices 6 and 12 of P-glycoprotein are closer in P-glycoprotein than in the crystal structure of Sav1866. Using homology modelling, we present evidence that the protein possesses intrinsic structural flexibility to allow cross-links to occur between helices 6 and 12 of P-glycoprotein, thereby reconciling crystallographic models with available experimental data from cross-linking.
Keywords: ABC transporter, ATP-bound P-gp model, cysteine cross-links, data-driven homology modelling, P-glycoprotein
Multidrug transporters are active efflux pumps that can prevent the influx of potentially harmful compounds by expelling them from cells in an energy-dependent manner [1,2]. Drug resistance of cancer cells and microbial organisms has been related to the expression of multidrug efflux transporters [3,4]. An important hallmark of ATP-binding cassette (ABC) transporters [5] is the presence of nucleotide binding domains (NBD), which use the energy of ATP binding and hydrolysis to fuel the transport process. The human ABC transporter P-glycoprotein (P-gp) plays an important role in xeno-protection because it is expressed in cells lining the intestinal tract, as well as in the liver and kidney, and at the blood–brain barrier [6]. Its expression profile has important implications for the pharmaceutical industry because it has a strong influence on the pharmacokinetics (absorption, distribution, metabolism, excretion and toxicity) of drugs. At present, no high resolution structure of P-gp is available, but structures of the homologous bacterial efflux transporters Sav1866 and MsbA have been solved recently. These structures represent Sav1866 from Staphylococcus aureus in complex with nucleotides [AMP-PNP, ADP; Protein Data Bank (PDB) codes 2ONJ and 2HYD, respectively] [7,8] and MsbA in different conformational states (open apo, closed apo, AMP-PMP-bound, ADP/Vi-bound; PDB codes 3B5W, 3B5X, 3B5Y, 3B5Z, 3B60) [9]. The structures indicate a common architecture of Sav1866 and MsbA. P-gp has sufficient sequence similarity with these two transporters to assume that they share a common fold. Additional evidence for structural homology stem from cross-linking experiments indicating that helices 10 and 11 of transmembrane domain (TMD) 2 reach over to the other side of the protein to form contacts with NBD1 [10] and from electron density maps obtained by a combination of small angle X-ray scattering and cryo-electron crystallography [11]. Furthermore, Sav1866 has at least partially overlapping substrate specificity with P-gp because Sav1866 has been found to transport the known P-gp substrates Hoechst 33324, verapamil and vinblastine [12]. Several recent homology models of P-gp have been based on the Sav1866 structure [13–15]. Globisch et al. [13] and Ravna et al. [15] used the structure of Sav1866 as a template to investigate pharmacological aspects of P-gp. O’Mara and Tieleman [14] created P-gp models in different conformational states relying on the different structures of the nucleotide binding domain (MalK) of the MalFGK2 bacterial maltose importer. These modelling attempts provide structural insights into drug binding and possible conformational transitions.
In the present study, we used the crystal structure of the ATP-bound state of Sav1866 (PDB code 2HYD, 3.0 Å resolution) [7] as a template for the generation of an outward facing P-gp model and compared its compliance with data obtained by cysteine cross-linking. Large deviations of the pore spanning H6–H12 cross-links were apparent. We developed a model of the cross-linked conformation of P-gp that would explain the spatial proximity of transmembrane helices 6 and 12 [16–21] as suggested by cysteine cross-linking. Our models indicate that the wing-like extensions can approach each other to a point to allow formation of cross-links while retaining structural integrity. The model is discussed in light of the currently available experimental evidence for the location of the substrate-binding region.
Results
The crystal structures of the bacterial exporters Sav1866 and MsbA in presence of the nonhydrolysable ATP-analogue AMP-PNP are very similar. These crystal structures show two symmetrically bound nucleotides sandwiched between the nucleotide binding domains at the NBD1 and NBD2 compound sites. The structures of Sav1866 obtained in complex with either AMP-PNP [8] or ADP [7] (PDB code 2ONJ and 2HYD, respectively) show an rmsd of < 0.1 Å, whereas the rmsd between Sav1866 (2HYD) and MsbA from Salmonella typhimurium (PDB code 3B60 resolved at 3.7 Å) [9] in complex with AMP-PNP is 2.0 Å. An additional structure of MsbA in the AMP-PNP bound state at a resolution of 4.5 Å (PDB code 3B5Y) is a Cα-trace only. We therefore used the ADP-bound state of Sav1866 (PDB code 2HYD) resolved at 3.0 Å [7] as a template for model building. Small differences between the two Sav1866 and the MsbA structures are observed towards the extracytoplasmic face of the transporters. In the Sav1866 crystal structure, helices 1 and 2 are extended by one helical turn at the extracellular side compared to MsbA. A similar helical length for P-gp compared to Sav1866 can be inferred for P-gp from the sequence alignment for helices 7 and 8. Modelling of the extracytoplasmic loop 1 has not been attempted due to lack of a suitable template.
Model of the open outward facing conformation
The Sav1866 template structure has been solved in an outward facing conformation and we built a model of P-gp based on this template. Our best outward facing model was compared with the experimental evidence provided by cysteine cross-linking data for P-gp in either the ATP or the AMP-PNP bound state (Table 1). Sixteen unique cross-link distances between pairs of cysteine residues were available [16–19], of which 14 corresponded to distances between helices in the N- and C-terminal TMDs, whereas two were available for the NBDs. All distances given in Table 1 are Cα–Cα distances. The majority of cross-links were obtained with Cu-phenanthroline, a zero length cross-linker, which oxidizes two cysteine residues to form a disulfide bond; two were obtained with 3,6,9,12-tetraoxatetradecane-1,14-diyl bismethanethiosulfonate (M14M); and one was obtained with tris-(2-maleimidoethyl)amine. Seven of the cross-links were found in the presence of ATP as well as AMP-PNP, indicating that a hydrolysis event is not required for the formation of these cross-links. The five thiol cross-links between helices 1 and 11 showed an average deviation per cross-link of 3.0 ± 1.5 Å for the outward facing model. The experimentally determined distances between the two cysteine pairs in helices 2 and 11 were satisfied. The same residues were also cross-linked in the non-energized state, indicating that ATP-binding does not change the relative orientation of helices 2 and 11. Residues L339 (TM6) and F728 (TM7), as well as F343 (TM6) and F728 (TM7), were cross-linked with M14M (spacer length 20.8 Å), corresponding to a Cα distance of 29.2 Å. This distance between cross-linked residues is a maximal distance in which the spacer is present in an extended conformation. Lower bounds for the cross-linkers are not available experimentally and the reported values therefore represent an upper limit. Distances in our model were 9.7 and 12.3 Å, respectively, and are thus consistent with the experimental data. In addition, two cross-links between the NBDs were measured. The Walker A motif of each NBD could be cross-linked by Cu oxidation with the signature sequence of the other NBD [20]. Table 1 shows that experimentally determined cross-link distances between residues in helices 1/11, 2/11 and 6/7 deviate on average by only 3 Å.
Table 1.
Residue pair distances determined by cysteine cross-linking compared with distances in the initial and final model. aThe reported cross-linker distances correspond to the maximum possible distance obtainable based on the structure of the linker used. bNegative deviations indicate that the distances in the models are shorter than the maximally possible distances obtained with the respective linker. cA1, Walker A motif of NBD1; A2, Walker A motif of NBD2; C1, signature sequence of NBD1; C2, signature sequence of NBD2.
| Residue 1 | TM | Residue 2 | TM | Cross-link distancea | Initial model | Deviationb | Final model | Deviationb | Reference |
|---|---|---|---|---|---|---|---|---|---|
| M68 | 1 | Y950 | 11 | 7.0 | 11.3 | 4.3 | 12.1 | 5.1 | [18] |
| M68 | 1 | Y953 | 11 | 7.0 | 11.0 | 4.0 | 10.9 | 3.9 | [18] |
| M68 | 1 | A954 | 11 | 7.0 | 10.8 | 3.8 | 10.5 | 3.5 | [18] |
| M69 | 1 | A954 | 11 | 7.0 | 8.2 | 1.2 | 8.1 | 1.1 | [18] |
| M69 | 1 | F957 | 11 | 7.0 | 9.2 | 2.2 | 8.6 | 1.6 | [18] |
| V133 | 2 | G939 | 11 | 7.0 | 5.5 | 0.0 (−1.5) | 5.6 | 0.0 (−1.4) | [17] |
| C137 | 2 | A935 | 11 | 7.0 | 6.6 | 0.0 (−0.4) | 6.6 | 0.0 (−0.4) | [17] |
| L339 | 6 | F728 | 7 | 29.2 | 9.7 | 0.0 (−19.5) | 9.6 | 0.0 (−19.6) | [19] |
| F343 | 6 | F728 | 7 | 29.2 | 12.3 | 0.0 (−16.9) | 12.3 | 0.0 (−16.9) | [19] |
| L332 | 6 | L975 | 12 | 7.0 | 20.4 | 13.4 | 7.3 | 0.3 | [19] |
| F343 | 6 | M986 | 12 | 7.0 | 16.1 | 9.1 | 11.3 | 4.3 | [19] |
| F343 | 6 | V982 | 12 | 18.7 | 22.5 | 3.8 | 10.9 | 0.0 (−7.8) | [19] |
| G346 | 6 | G989 | 12 | 7.0 | 19.2 | 12.2 | 16.9 | 9.9 | [16] |
| P350 | 6 | S993 | 12 | 7.0 | 17.2 | 10.2 | 16.3 | 9.3 | [16] |
| C431 | A1c | L1176 | C2c | 7.0 | 10.9 | 3.9 | 10.9 | 3.9 | [20] |
| L531 | C1c | C1074 | A2c | 7.0 | 10.9 | 3.9 | 10.9 | 3.9 | [20] |
Model of the cross-linked conformation
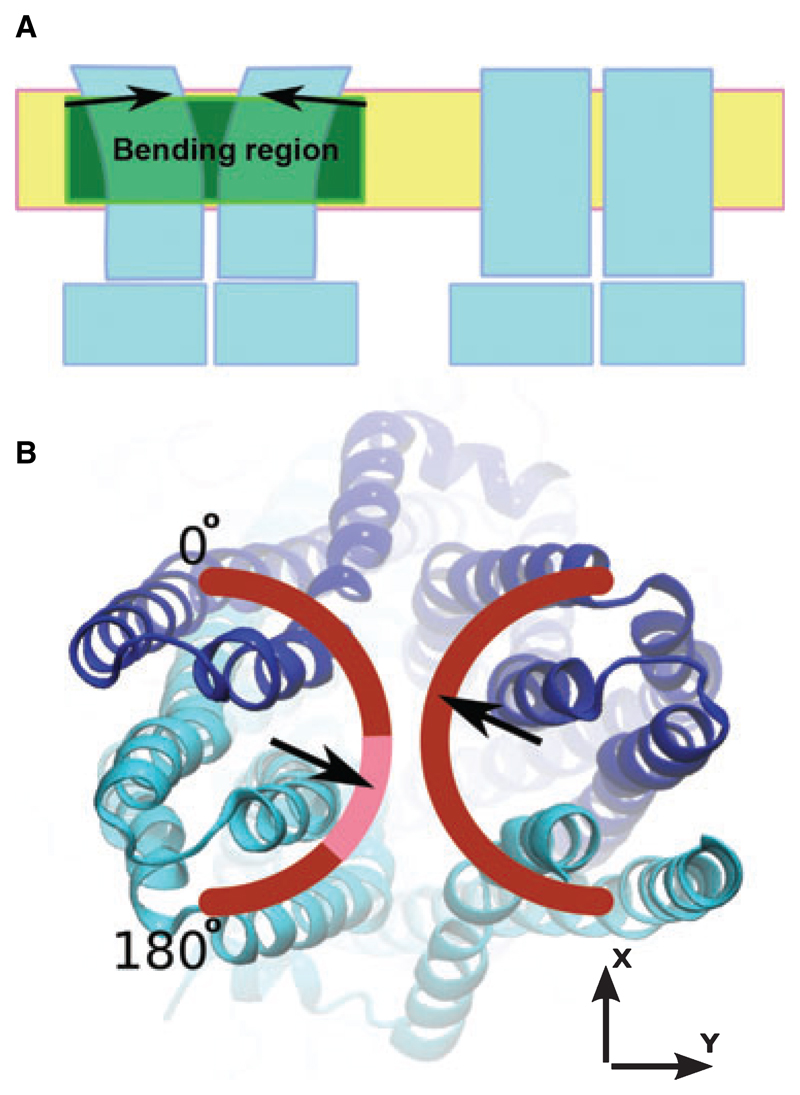
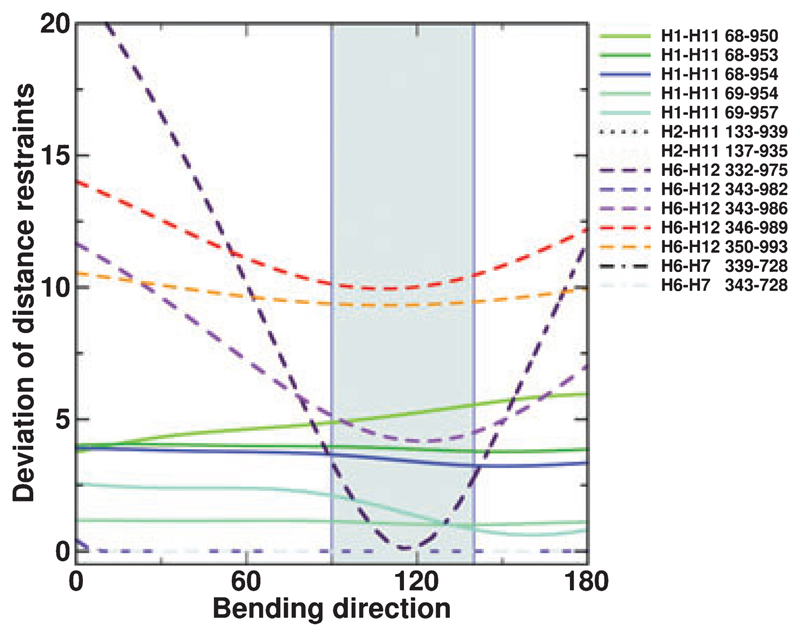
Although the model of P-gp in the open outward facing conformation is in accordance with many cysteine cross-links and the overall electron microscopy (EM) density maps, additional density in the centre of the transmembrane region has been observed and we find that distances between helices 6 and 12 are much larger in the outward facing model than suggested by cysteine cross-links (average deviation of 9.7 ± 3.7Å). Cross-links that were consistent with our model structure were all found within one of the wing-like extensions, whereas those deviating reached over to the other wing. The question therefore arose as to whether it would be feasible to explain this experimental information by flexibility in the TMD region. The concept illustrated schematically in Fig. 1 was developed to build a P-gp model that would satisfy experimental data. Parts of the TMDs were defined as mobile units and symmetrically bent towards each other while holding the intracellular portion of the transporter (intracellular domains and NBDs) in place. A smooth rotation gradient was applied in a bending region corresponding to that portion of the TMDs predicted to be in contact with the hydrophobic core of the plasma membrane. The axis of rotation was oriented at a 90° angle relative to the Z-axis (the Z-axis is parallel to the axis of symmetry of the molecule and thus perpendicular to the membrane plane) and the bending direction was sampled in 5° increments in the XY-plane. This is illustrated in Fig. 1B. Thirty-seven models were generated and assessed for compliance with experimental cysteine cross-linking data. For each cross-link, the deviation between the distances measured in the model and the experimentally determined distances was plotted as a function of the bending direction in the XY-plane (Figs 1B and 2). A consistent minimum of individual traces in the range 90–140° was observed, indicating that a simultaneous optimization of distance constraints was possible. The initial direction and amplitude of bending was determined based only on Cα–Cα distances of the cross-linked residues and does not take into account side-chain orientation, size and atom overlap. Therefore, in a subsequent step, the bending direction and amplitude were adjusted simultaneously to obtain optimal helix packing. The model of the cross-linked conformation was obtained for a bending direction of 110° and a bending amplitude of 10° (Fig. 3). In Table 1, the cross-linking distances for this model are compared with those found for the outward facing model. The overall deviation from the experimental data is reduced by 30% in the closed model; in particular, the strong distance deviations between residues in helices 6 and 12 were decreased by 50%. A summary of the quality of the energy minimized closed TMD model is presented in Table 2. The structural and geometric parameters are very good and indicate that the bending procedure did not compromise the structural integrity of our model.
Fig. 1.
(A) Principle of the bending procedure applied to the TMDs of the initial P-gp homology model. The coordinates of the intra-cellular part remained unchanged. The bending region, placed in a section of the protein predicted to be in contact with the hydrophobic core of the membrane, is shown in green. As indicated by the arrows, the extracellular portion of the transporter was rigidly rotated by 10° (referred to as the ‘bending angle’ in the text). (B) The range of tested bending directions, which were sampled in 5° increments in the range 0–180°. The highlighted part of the arch indicates the bending directions that resulted in models that complied best with experimental cross-linking data. The two halves were bent symmetrically.
Fig. 2.
Generation of a model of the cross-linked conformation: the deviations between measured distances in the models and experimentally determined cross-link distances are shown as a function of the bending direction. The area shaded in grey indicates the most favourable range of bending directions. Distances between H2 and H11, as well as between H6 and H7, always fulfil the experimental constraints.
Fig. 3.
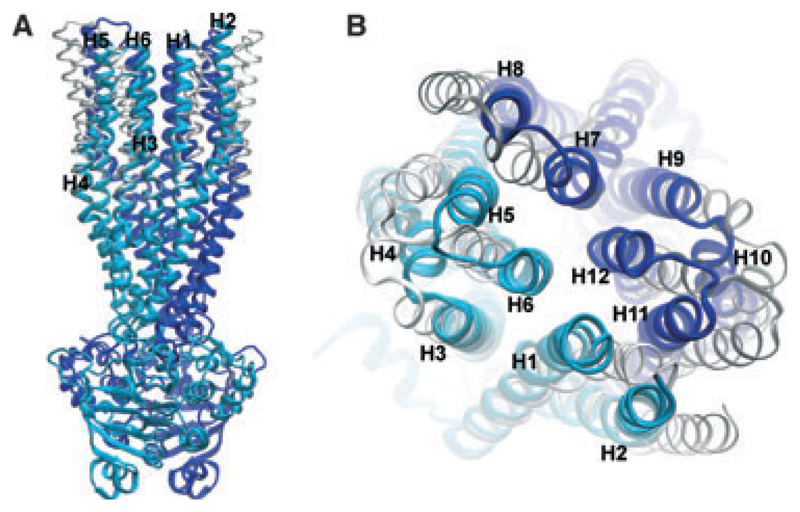
Comparison of the models representing the outward facing and the cross-linked conformations in (A) side and (B) top view. The model representing the cross-linked conformation is shown in cyan (N-terminus) and blue (C-terminus). The outward facing model is superimposed in white (N-terminus) and grey (C-terminus).
Table 2.
Quality assessment of the final energy minimized model.
| Total potential energy | −6.5e + 04 kJ⋅mol−1 |
|---|---|
| proq | |
| LGscore | 5.621 (extremely good model) |
| MaxSub | 0.312 (very good model) |
| prosaii | |
| Z-score | −12.38 |
| procheck | |
| Ramachandran (%) | |
| Most favourable | 94.3 |
| Additionally allowed | 5.3 |
| Generously allowed | 0.2 |
| Disallowed | 0.2 |
| G-factor | |
| Dihedral | −0.04 |
| Covalent | 0.19 |
| Overall | 0.06 |
| whatcheck | |
| Structure Z-scores | |
| 1st generation packing quality | −0.137 |
| 2nd generation packing quality | −0.836 |
| Ramachandran plot appearance | 0.152 |
| chi-1/chi-2 rotamer normality | −1.644 |
| Backbone conformation | −0.652 |
| Rms Z-scores | |
| Bond lengths | 0.706 |
| Bond angles | 1.073 |
| Omega angle restraints | 0.168 (tight) |
| Side-chain planarity | 1.718 |
| Improper dihedral distribution | 2.508 (loose) |
| Inside/outside distribution | 1.141 |
Discussion
P-gp models
In the present study, we probed the compatibility of available experimental data from cross-linking and small angle X-ray scattering (SAXS)/EM with models of P-gp in the ATP-bound state by combining homology modelling with data-driven structural modification. The structure of Sav1866 (2HYD) in the ATP-bound state was used as a template for model generation. The selection of the template was motivated by the highest crystallographic resolution and the recently documented similarity in the TMD/NBD interface [10]. In the template structure, a canonical NBD/NBD interface is present [7–9] and the transmembrane segments of helix pairs 2/11 and 5/8 are in contact. The latter has also been demonstrated for P-gp [17,22]. A clear helical periodicity of the formation of cross-links was observed for residues in these helix pairs and the residues forming cross-links were oriented towards each other, whereas those that did not pointed in opposite directions.
Overall, the outward facing P-gp model conforms to its EM density map because the template Sav1866 structure had previously been shown to fit into this electron density map [11]. Our outward facing homology model was validated by comparison with the available cross-linking information. The compliance of the model with experimental evidence is good, with average distance deviations of 3 Å for residues in helix pairs 1/11, 2/11 and 6/7. These distance deviations lie in the range of thermal fluctuations of proteins that typically are 1–2 Å [23–25].
By contrast, on average, helices 6 and 12 were 10 Å further apart in the outward facing model than as determined by measurements in cross-linking experiments. Furthermore, SAXS/EM density maps of P-gp indicate that helices 6 and 12 are positioned closer to each other than in the Sav1866 crystal structure. This suggested the existence of a P-gp conformational state differing from that observed in the crystal structure of the template Sav1866. We thus tested the hypothesis that a structurally intact P-gp model can be generated, which is in agreement with all available cross-linking information. We thereby observed a simultaneous decrease of helix 6 helix 12 distance deviations for a narrow range of bending directions (Fig. 2). This finding suggests that these cysteine cross-links can occur simultaneously in one conformational state of the protein. The finding that the L332/L975 cross-link only occurs under conditions in which ATP hydrolysis can take place indicates that movements of helices 6 and 12 are more intricate than reflected by simply bending the model structure in the way described in the present study. This is also suggested by two studies demonstrating considerable conformation changes in the segment V331 to F343 of TM6 [26,27].
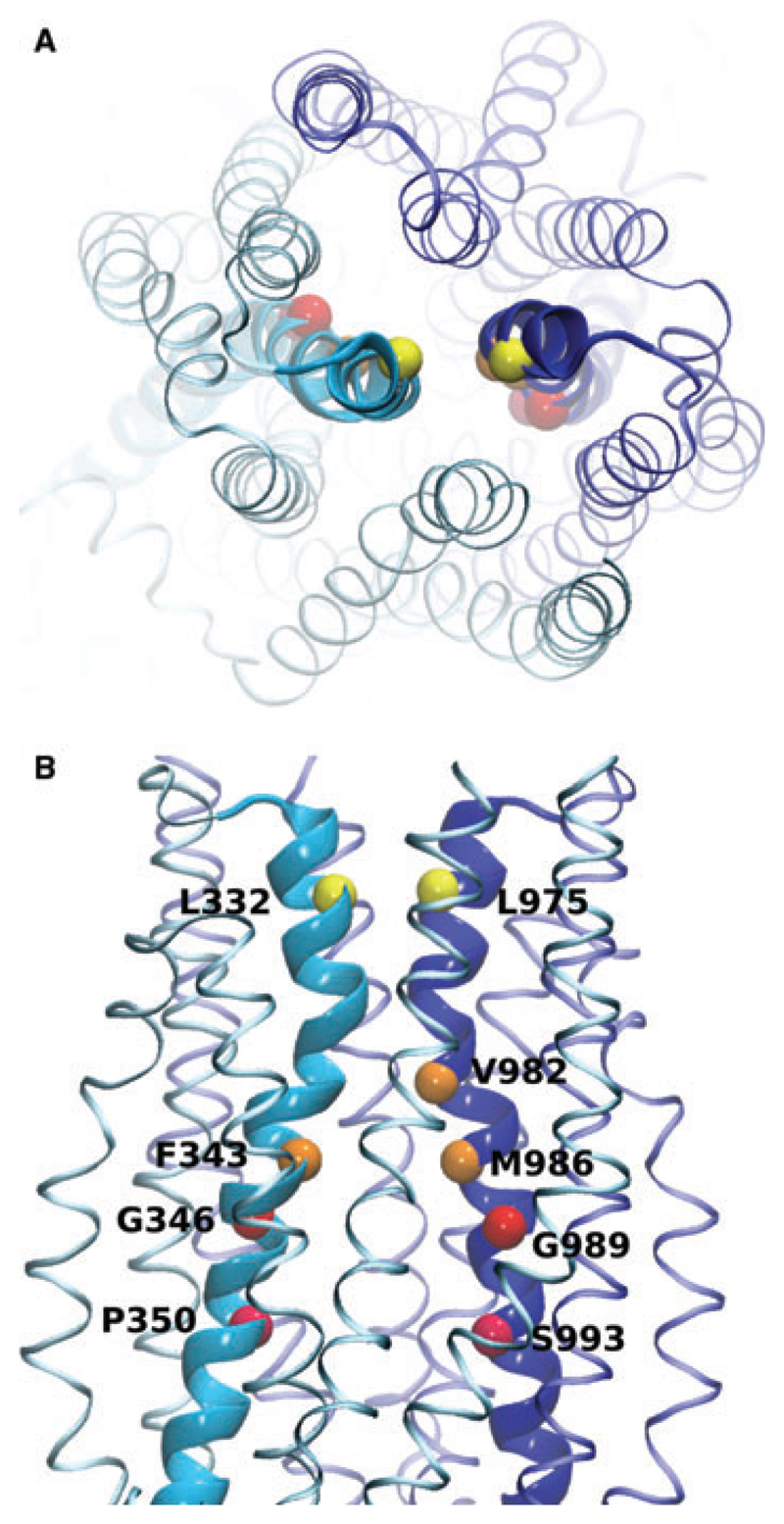
The residues in transmembrane helices 6 and 12 that were cross-linked with Cu-phenanthroline are highlighted in Fig. 4. These residues face the central cavity, are oriented towards each other, and come into close proximity in the cross-linked model. We therefore suggest that P-gp can fluctuate between an outward facing and a cross-linked conformation. Figure 3 shows an overlay of the outward facing and the cross-linked conformation. These two model structures suggest that a transition between these two conformations is possible without encountering any obvious barriers and without compromising the structural integrity of the transmembrane domains.
Fig. 4.
Model of the cross-linked conformation of P-gp showing residues in helices H6 and H12 that were cross-linked. (A) Top view. (B) Side view. The Cα-atoms of the cross-linked residues are highlighted. Helices 6 and 12 are shown in bold. Identical colours indicate cross-linked residues.
Experimental conditions of crystallography, SAXS/EM and cysteine cross-linking studies may select for different thermodynamically accessible conformations. The techniques used to obtain crystals of transmembrane proteins and protein preparations for EM studies might influence the conformational equilibrium of P-gp because the transporter has to be removed from its native environment and detergent solubilized [28]. Forces experienced by the transporter in the lipid bilayer might thus be lacking in the crystal environment. The lateral pressure profile of a phospholipid bilayer indicates that considerable lateral forces can act on membrane inserted proteins [29,30]. On the other hand, cysteine cross-linking is a non-equilibrium method and might not reflect the most populated conformation of P-gp inserted in a physiological membrane, but certainly represents a thermodynamically accessible state. EPR studies on the multidrug resistance transporter MsbA from Escherichia coli suggest that the central helices 6 and 12 are neither water nor lipid accessible [31]. In the outward facing model, a large portion (~ 4400 Å2) of the TMD surfaces facing the central cavity are hydrophobic, but water exposed. Residues that form cross-links between TM6 and TM12 are located in regions with higher hydrophobicity. In the cross-linked model, a hydrophobic surface area of approximately 2000 Å2 is buried, which contributes approximately 24 kJ⋅mol)−1 of stabilization energy [32]. Hydrophobic water-exposed surfaces of similar size are commonly interpreted as surface patches that drive protein–protein interactions [33]. We therefore suggest that P-gp can fluctuate between an outward facing and a cross-linked conformation. This would reconcile the apparently contradictory lines of evidence from X-ray crystallography, SAXS/EM studies and cross-linking experiments.
Possible implications for substrate binding
The experimental evidence indicates that the transporter conformation changes upon substrate binding [19]. In the absence of substrate, residues P350 and S993 of helices 6 and 12 can be cross-linked by Cu-phenanthroline in all stages of the catalytic cycle. Substrate binding induces a conformational change that reduces the formation of this cross-link, but a new cross-link from residue P350 to residue V991 at the opposite side of helix 12 is favoured, indicating a translational and/or rotational movement of the central helices 6 and 12.
Site-directed mutagenesis studies have identified residues F728 in helix 7, F343 in helix 6 and I306 in helix 5 as being involved in the binding of rhodamine 123 (F343) or verapamil (I306, F728), respectively, and increasing the rate of ATP-hydrolysis [34,35]. Reaction of these residues with 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium-coupled substrates led to a permanent stimulation of P-gp ATPase activity, indicating the importance of these residues for the transmission of conformational changes from the TMDs to the motor domains. I306, F343 and F728 in helices 5, 6 and 7 are predicted by our model to lie close to each other within a hydrophobic patch. Interestingly, a cross-link between V982 in TM12 and F343 in TM6 has been identified and V982 has been shown to be located close to the substrate binding region [19]. In our closed TMD model, V982 is juxtaposed to I306, F343 and F728. Hydrophobic substrates might be able to insert into this putative hydrophobic contact region and trigger the observed changes in cross-links upon substrate binding.
Experimental procedures
Multiple sequence alignments
Sequences of P-gp N and C-terminal halves, Sav1866 and MsbA from E. coli, Vibrio cholerae and S. typhimurium were aligned with clustalw [36] using a gap penalty of 10 and a gap extension penalty of 0.05. The alignment was subsequently adjusted manually in helix 1 and in the vicinity of the extracytoplasmic loop 1 (see Fig. S1). Validity of the sequence alignment was provided by the orientation of residues at the TMD contact helices 5/8 and 2/11 of the models.
Model generation
The crystal structure of Sav1866 (PDB code 2HYD) was used as a template for model building. Two hundred and fifty models were generated with the modeller 9v2 software [37] (refinement settings: ‘very_slow’, automodel procedure). The extracytoplasmic loop 1 and the linker region were excluded.
Optimization of the model
The best scoring model was further refined in a data driven optimization procedure. For this, a c++ program was written to allow bending of transmembrane domain helices. The program divides the protein into three parts: a static core region, a flexible bending region and a rigidly rotated region. The borders of the bending region are defined by their positions along the Z-axis. The rotation axis is set normal to the Z-axis. During the bending procedure, the core region remains in place and the rigid region is rotated around the rotational axis by a specified angle, whereas the atoms in the bending region are rotated depending on their Z-position within the bending region. This procedure allows bending parts of a protein while avoiding chain breaks.
The protein’s main axis (corresponding to the axis of rotational symmetry) was aligned with the Z-axis, normal to the membrane plane. The bending region was defined to coincide with those parts of the protein predicted to be in contact with the hydrophobic core of the plasma membrane (Fig. 1A). The axis of rotation was positioned parallel to the membrane plane in an orientation that makes it pass through the centre of the cavity. Its orientation in the XY-plane was sampled in 5° increments over a 180° range, whereby the bending moved the rotated parts of the protein towards each other (Fig. 1B). The optimal bending direction was determined by comparing the residue–residue distances in the model with experimental cross-linking distances. The bending amplitude was optimized to achieve good helix packing. The final structure was energy minimized with Gromacs [38] using the vacuum force field Gromos96 G43B1 [39]. The quality of the final model was assessed with procheck [40], prosaii [41], proq [42] and whatcheck [43].
Supplementary Material
The following supplementary material is available:
This supplementary material can be found in the online version of this article.
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Acknowledgements
This work was supported by a grant from the Austrian Science Fund (SFB-35, F3509).
Abbreviations
- ABC
ATP-binding cassette
- EM
electron microscopy
- M14M
3,6,9,12-tetraoxatetradecane-1,14-diyl bismethanethiosulfonate
- NBD
nucleotide binding domain
- P-gp
P-glycoprotein
- SAXS
small angle X-ray scattering
- TMD
transmembrane domain
References
- 1.Gottesman MM, Pastan I. Biochemistry of multidrug resistance mediated by the multidrug transporter. Annu Rev Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- 2.Lomovskaya O, Zgurskaya HI, Totrov M, Watkins WJ. Waltzing transporters and ‘the dance macabre’ between humans and bacteria. Nat Rev Drug Discov. 2007;6:56–65. doi: 10.1038/nrd2200. [DOI] [PubMed] [Google Scholar]
- 3.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 4.Lomovskaya O, Bostian KA. Practical applications and feasibility of efflux pump inhibitors in the clinic – a vision for applied use. Biochem Pharmacol. 2006;71:910–918. doi: 10.1016/j.bcp.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 6.Sarkadi B, Homolya L, Szakacs G, Varadi A. Human multidrug resistance ABCB and ABCG transporters: participation in a chemoimmunity defense system. Physiol Rev. 2006;86:1179–1236. doi: 10.1152/physrev.00037.2005. [DOI] [PubMed] [Google Scholar]
- 7.Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 8.Dawson RJ, Locher KP. Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett. 2007;581:935–938. doi: 10.1016/j.febslet.2007.01.073. [DOI] [PubMed] [Google Scholar]
- 9.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc Natl Acad Sci USA. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zolnerciks JK, Wooding C, Linton KJ. Evidence for a Sav1866-like architecture for the human multidrug transporter P-glycoprotein. FASEB J. 2007;21:3937–3948. doi: 10.1096/fj.07-8610com. [DOI] [PubMed] [Google Scholar]
- 11.McDevitt CA, Shintre CA, Grossmann JG, Pollock NL, Prince SM, Callaghan R, Ford RC. Structural insights into P-glycoprotein (ABCB1) by small angle X-ray scattering and electron crystallography. FEBS Lett. 2008;582:2950–2956. doi: 10.1016/j.febslet.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 12.Velamakanni S, Yao Y, Gutmann DA, van Veen HW. Multidrug transport by the ABC transporter Sav1866 from Staphylococcus aureus. Biochemistry. 2008;47:9300–9308. doi: 10.1021/bi8006737. [DOI] [PubMed] [Google Scholar]
- 13.Globisch C, Pajeva IK, Wiese M. Identification of putative binding sites of P-glycoprotein based on its homology model. ChemMedChem. 2008;3:280–295. doi: 10.1002/cmdc.200700249. [DOI] [PubMed] [Google Scholar]
- 14.O’Mara ML, Tieleman DP. P-glycoprotein models of the apo and ATP-bound states based on homology with Sav1866 and MalK. FEBS Lett. 2007;581:4217–4222. doi: 10.1016/j.febslet.2007.07.069. [DOI] [PubMed] [Google Scholar]
- 15.Ravna AW, Sylte I, Sager G. Molecular model of the outward facing state of the human P-glycoprotein (ABCB1), and comparison to a model of the human MRP5 (ABCC5) Theor Biol Med Model. 2007;4:33. doi: 10.1186/1742-4682-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loo TW, Clarke DM. Drug-stimulated ATPase activity of human P-glycoprotein requires movement between transmembrane segments 6 and 12. J Biol Chem. 1997;272:20986–20989. doi: 10.1074/jbc.272.34.20986. [DOI] [PubMed] [Google Scholar]
- 17.Loo TW, Bartlett MC, Clarke DM. Val133 and Cys137 in transmembrane segment 2 are close to Arg935 and Gly939 in transmembrane segment 11 of human P-glycoprotein. J Biol Chem. 2004;279:18232–18238. doi: 10.1074/jbc.M400229200. [DOI] [PubMed] [Google Scholar]
- 18.Loo TW, Bartlett MC, Clarke DM. ATP hydrolysis promotes interactions between the extracellular ends of transmembrane segments 1 and 11 of human multidrug resistance P-glycoprotein. Biochemistry. 2005;44:10250–10258. doi: 10.1021/bi050705j. [DOI] [PubMed] [Google Scholar]
- 19.Loo TW, Bartlett MC, Clarke DM. Nucleotide binding, ATP hydrolysis, and mutation of the catalytic carboxylates of human P-glycoprotein cause distinct conformational changes in the transmembrane segments. Biochemistry. 2007;46:9328–9336. doi: 10.1021/bi700837y. [DOI] [PubMed] [Google Scholar]
- 20.Loo TW, Bartlett MC, Clarke DM. Drug binding in human P-glycoprotein causes conformational changes in both nucleotide-binding domains. J Biol Chem. 2003;278:1575–1578. doi: 10.1074/jbc.M211307200. [DOI] [PubMed] [Google Scholar]
- 21.Loo TW, Bartlett MC, Clarke DM. The ‘LSGGQ’ motif in each nucleotide-binding domain of human P-glycoprotein is adjacent to the opposing walker A sequence. J Biol Chem. 2002;277:41303–41306. doi: 10.1074/jbc.C200484200. [DOI] [PubMed] [Google Scholar]
- 22.Loo TW, Bartlett MC, Clarke DM. Disulfide cross-linking analysis shows that transmembrane segments 5 and 8 of human P-glycoprotein are close together on the cytoplasmic side of the membrane. J Biol Chem. 2004;279:7692–7697. doi: 10.1074/jbc.M311825200. [DOI] [PubMed] [Google Scholar]
- 23.Vanderkooi JM. The protein state of matter. Biochim Biophys Acta. 1998;1386:241–253. doi: 10.1016/s0167-4838(98)00097-1. [DOI] [PubMed] [Google Scholar]
- 24.Bond PJ, Sansom MS. Membrane protein dynamics versus environment: simulations of OmpA in a micelle and in a bilayer. J Mol Biol. 2003;329:1035–1053. doi: 10.1016/s0022-2836(03)00408-x. [DOI] [PubMed] [Google Scholar]
- 25.Daniel RM, Dunn RV, Finney JL, Smith JC. The role of dynamics in enzyme activity. Annu Rev Biophys Biomol Struct. 2003;32:69–92. doi: 10.1146/annurev.biophys.32.110601.142445. [DOI] [PubMed] [Google Scholar]
- 26.Rothnie A, Storm J, Campbell J, Linton KJ, Kerr ID, Callaghan R. The topography of transmembrane segment six is altered during the catalytic cycle of P-glycoprotein. J Biol Chem. 2004;279:34913–34921. doi: 10.1074/jbc.M405336200. [DOI] [PubMed] [Google Scholar]
- 27.Storm J, Modok S, O’Mara ML, Tieleman DP, Kerr ID, Callaghan R. Cytosolic region of TM6 in P-glycoprotein: topographical analysis and functional perturbation by site directed labeling. Biochemistry. 2008;47:3615–3624. doi: 10.1021/bi7023089. [DOI] [PubMed] [Google Scholar]
- 28.Hollenstein K, Dawson RJ, Locher KP. Structure and mechanism of ABC transporter proteins. Curr Opin Struct Biol. 2007;17:412–418. doi: 10.1016/j.sbi.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Cantor RS. The lateral pressure profile in membranes: a physical mechanism of general anesthesia. Biochemistry. 1997;36:2339–2344. doi: 10.1021/bi9627323. [DOI] [PubMed] [Google Scholar]
- 30.Lindahl E, Edholm O. Mesoscopic undulations and thickness fluctuations in lipid bilayers from molecular dynamics simulations. Biophys J. 2000;79:426–433. doi: 10.1016/S0006-3495(00)76304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong J, Yang G, McHaourab HS. Structural basis of energy transduction in the transport cycle of MsbA. Science. 2005;308:1023–1028. doi: 10.1126/science.1106592. [DOI] [PubMed] [Google Scholar]
- 32.Eisenberg D, McLachlan AD. Solvation energy in protein folding and binding. Nature. 1986;319:199–203. doi: 10.1038/319199a0. [DOI] [PubMed] [Google Scholar]
- 33.Nooren IM, Thornton JM. Diversity of protein-protein interactions. EMBO J. 2003;22:3486–3492. doi: 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loo TW, Bartlett MC, Clarke DM. Permanent activation of the human P-glycoprotein by covalent modification of a residue in the drug-binding site. J Biol Chem. 2003;278:20449–20452. doi: 10.1074/jbc.C300154200. [DOI] [PubMed] [Google Scholar]
- 35.Loo TW, Bartlett MC, Clarke DM. Transmembrane segment 7 of human P-glycoprotein forms part of the drug-binding pocket. Biochem J. 2006;399:351–359. doi: 10.1042/BJ20060715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 38.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 39.van Gunsteren WF, Billeter SR, Eising AA, Hünenberger PH, Krüger PH, Mark AE, Scott WRP, Tironi IG. Biomolecular Simulation: The GROMOS96 Manual and User Guide. Hochschulverlag AG an der ETH Zürich; Zürich: 1996. [Google Scholar]
- 40.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 41.Sippl MJ. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- 42.Wallner B, Elofsson A. Can correct protein models be identified? Protein Sci. 2003;12:1073–1086. doi: 10.1110/ps.0236803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hooft RWWVG. Whatcheck: A Structure Validation System. EMBL; Heidelberg: 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.