Abstract
With the discovery of P-glycoprotein (P-gp), it became evident that ABC-transporters play a vital role in bioavailability and toxicity of drugs. They prevent intracellular accumulation of toxic compounds, which renders them a major defense mechanism against xenotoxic compounds. Their expression in cells of all major barriers (intestine, blood–brain barrier, blood–placenta barrier) as well as in metabolic organs (liver, kidney) also explains their influence on the ADMET properties of drugs and drug candidates. Thus, in silico models for the prediction of the probability of a compound to interact with P-gp or analogous transporters are of high value in the early phase of the drug discovery process. Within this review, we highlight recent developments in the area, with a special focus on the molecular basis of drug–transporter interaction. In addition, with the recent availability of X-ray structures of several ABC-transporters, also structure-based design methods have been applied and will be addressed.
Keywords: ABC transporters, Computational models, Bioassays, Machine learning, Pharmacophore modeling, Transport inhibition
1. Introduction
ATP-binding cassette transporters (ABC-transporters) form a large superfamily of membrane proteins. Members of the ABC-transporters can be found in all living organisms from prokaryotes to mammals. Generally speaking, these transporters participate in active transport, i.e. they hydrolyze ATP and use its energy to transport their substrates. In humans, 49 ABC-transporters are recognized to date and belong to 7 distinct subfamilies [1], ABCA to ABCG. The usual “transport unit” consists of two intracellular nucleotide binding domains and two transmembrane domains. The nucleotide binding domains (NBDs), usually well conserved across subfamilies, bind and hydrolyze ATP. The transmembrane domains create the translocation chamber across which the substrates diffuse. These regions are usually little conserved and are responsible for the substrate specificity of the different transporters. Members of the ABCBA subfamily transport cholesterol and lipids [2]. Members of the B, C and G subfamilies are multi-drug resistance-associated transporters or associated with diseases.
It was in 1976 when Juliano and Ling [3] linked the phenomenon of anticancer multiple drug resistance to a single glycoprotein expressed in the membranes of Chinese hamster ovary cells. As multiple drug resistance was characterized by a decreased accumulation of the anticancer agents in the tumor cells, they named the protein P-glycoprotein (P for permeability). Soon after, it became evident that P-glycoprotein (P-gp) functions as an ATP-driven, transmembrane efflux pump with an extremely broad substrate specificity (polyspecificity or promiscuity). Obviously, there were immediate attempts to develop compounds which would block the P-gp mediated efflux of anticancer drugs and thus resensitize multidrug resistant tumor cells. The first representative of this new class of so-called modulators of multidrug resistance (MDR-modulators) was the calcium channel blocker verapamil [4,5]. As for substrates, also in case of inhibitors P-gp is characterized by an extremely broad ligand profile. Thus, there are currently more than 5000 compounds retrieved when you search the Open PHACTS Discovery Platform [6] for compounds interacting with P-glycoprotein. Several compounds were subject to clinical studies, but none was approved so far. This raised the question of the druggability of P-gp, and the research focus shifted towards its potential role as antitarget [7].
1.1. ABC-transporters and ADMET
With the increasing knowledge on the tissue expression and function of P-glycoprotein, its important role in absorption of drugs and drug candidates became evident. This is now broadly accepted and has been also picked up by regulatory authorities. Based on a proposal from the International Transporter Consortium, the FDA now recommends a standardized set of experiments to assess the likelihood of a compound to interact with P-glycoprotein and the Breast Cancer Resistance Protein (BCRP/ABCG2) [8], another member of this super-family of ABC-transporters. According to the multiple roles of P-gp and analogs, both substrate and inhibitor properties of compounds need to be explored. The latter especially is important for drug–drug interactions. There are numerous cases reported where co-administration of a P-gp inhibitor with a P-gp substrate considerably increased the blood levels of the latter, leading to serious side effects. Classical examples are drug–drug interactions with digoxin (dronedarone, quinidine, ranolazine), loperamide (tipranavir, ritonavir), saquinavir (tipranavir, ritonavir) for P-gp and interactions with topotecan (GF120918) for BCRP.
Compounds inducing expression of P-gp will lead to analogous results. However, as clearly exemplified in the Biopharmaceutics Classification System (BCS) [9], the solubility of the compounds plays also an indispensable role for assessing the final risk for transporter-related low bioavailability. As P-gp is an ATP-driven transporter, its transport capacity has limits and it can be saturated. P-gp does not play any role in the bioavailability of highly soluble compounds, irrespective of whether they are substrates or not. P-gp becomes the limiting step only for substrates with low solubility. This of course increases the complexity and renders the task of predicting bioavailability of a compound by in silico models quite a challenge.
The blood–brain barrier (BBB) has been recognized as a tissue/barrier where P-gp and BCRP play a major role in controlling the transcellular flux of small molecules. The BBB is characterized by tight junctions, which force all solutes to take the transcellular route. Both P-gp and BCRP are highly expressed at the BBB and thus are the major functional constituents of this barrier. This has implications for the development of CNS-active drugs, as these need to cross the BBB, and thus should be devoid of P-gp and BCRP substrate properties. Especially for the therapy of brain tumours, this is of major relevance, as most anticancer agents are substrates of P-gp and BCRP [10,11]. Very recent examples are provided by the group of Schinkel, who demonstrate that brain accumulation of the PARP inhibitor rucaparib and the JAK1/2 inhibitor CYT387 in mice is restricted by Abcg2 and Abcb1a/1b [12,13]. In contrast, for compounds supposed not to interact with CNS-targets, favoring P-gp substrate properties would be a versatile approach for preventing them from entering the brain. A classical example in this respect is the class of antihistaminic agents: the first generation of compounds (e.g. diphenhydramine) showed remarkable CNS-related side effects, such as dizziness, whereas the 3rd generation of drugs, such as fexofenadine, is devoid of CNS side effects due to their P-gp substrate properties [14]. In addition, as already outlined previously, drug–drug interactions mediated by P-gp and BCRP are an important issue also at the BBB [15].
1.2. ABC-transporters and liver toxicity
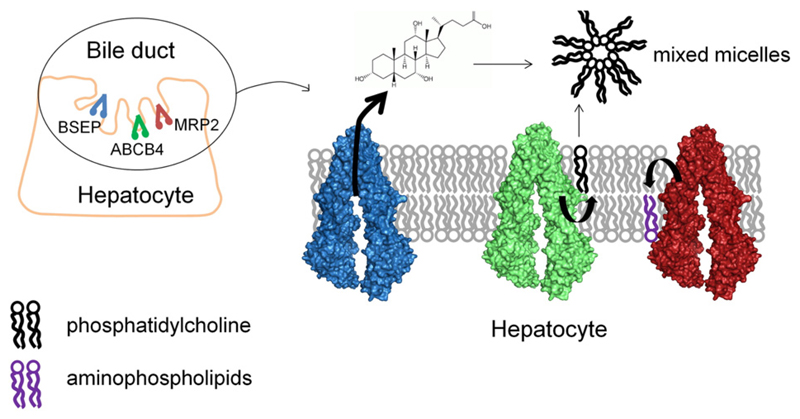
Canalicular ABC-transporters, which mediate the excretion of individual bile constituents, play a key role in bile formation and cholestasis. Some of these constituents, such as bile acids, cause serious damage to hepatocytes and bile duct cells, which might lead to inflammation, fibrosis, cirrhosis, sitosterolemia, hyperbilirubinemia, cholestasis, and potentially also cancer [16,17]. Especially, the proper interplay (see Fig. 1) of the bile salt export pump (BSEP, gene ABCB11) with MDR3 (gene ABCB4) is critical for the formation of bile salt micelles, and inhibition of BSEP has been clearly linked to drug-induced liver injury (DILI) [18]. However, besides BSEP and MDR3, MRP2 (gene ABCC2) as well as P-gp and BCRP are involved. Thus, there are multiple possibilities for drugs and nutrients to interfere with the liver transportome, and we are just beginning to understand how this is linked to hepatotoxicity. One possible starting point are diseases linked to ABC-transporter mutations. For example, homozygous-null MDR3 mutations cause progressive familial intrahepatic cholestasis [19]. MDR3 flops phosphatidylcholine into the bile canaliculus to protect the biliary tree from the detergent activity of bile salts. Thus, a misbalance of BSEP and MDR3 activity leads to toxic concentrations of bile salts either in the hepatocyte or in the bile duct.
Fig. 1.
Cooperation of BSEP, ABCB4 and MRP2 in the canalicular membrane of hepatocytes. BSEP (blue) exports the bile salts, ABCB4 (green) flips phosphatidylcholine to the outer leaflet of the membrane, where it is recruited by bile salts to form mixed micelles. MRP2 (red) maintains the asymmetry in lipid composition by flipping aminophospholipids to the inner leaflet of the membrane.
1.3. Diseases related to malfunction of ABC-transporters
On a more general level, there are numerous diseases which have been linked to improper functioning ABC-transporters. The paradigm example is cystic fibrosis, which is caused by mutations of the CFTR chloride channel [20]. The CFTR chloride channel is encoded by the ABCC7 gene, which is mutated in patients with cystic fibrosis. ATP-driven conformational changes open and close a gate to allow transmembrane flow of chloride anions down their electrochemical gradient. Very recently, Vertex launched a drug which potentiates the function of deltaF508 mutated CFTR and thus compensates for the impaired function. Another compound developed by Vertex acts as pharmacochaperone, thus increasing the concentration of CFTR in the membrane.
Other examples are the link of MRP2 to Dubin–Johnson syndrome [21], of ABCA1 to Tangier disease [22], and of BCRP to gout [23].
Finally, there is increasing evidence that P-gp, BCRP, MRP1 (gene ABCC1) and the cholesterol transporter ABCA1 may contribute to the pathogenesis of Alzheimer's Disease (for a review see [24]). Thus, modulation of their activity might be a new concept for the treatment of Alzheimer.
1.4. Drug–drug and drug–nutrient interactions
In an aging society, drug–drug interactions become an extremely important issue. Elderly patients are quite often subject to complex medications, and the risk of severe drug–drug interactions increases with the number of drugs. Most often, these interactions are linked to cytochrome P450-related metabolism of compounds, i.e. compound A blocks the metabolism of compound B, which increases the concentration of compound B beyond the toxic level. However, there are numerous reports in the literature pointing towards drug–transporter interaction as additional contributor to severe drug–drug interactions. It could be that compound A blocks a transporter which is transporting compound B, thus influencing the distribution of compound B. Another scenario is that compound A induces the expression of a certain transporter, which then influences the distribution of all substrates of this transporter. A selected example is the interaction of rifampin with the P-gp substrate digoxin, where patients treated with rifampin and digoxin show considerably increased digoxin levels. As the renal clearance and half-life of digoxin was not altered by rifampin, this is most probably due to an increase of the intestinal P-gp content due to an induction of P-gp expression [25].
Another well documented example is the influence of P-gp inhibitors on the distribution of HIV-1 protease inhibitors into brain and testes [26]. However, a very recent study based on a detailed analysis of clinical drug–drug interaction studies revealed that the risk for drug–drug interactions caused by P-gp inhibition is quite limited. A significant risk could only be detected when both P-gp and CYP3A are inhibited [27].
One of the major functions of ABC-transporters is the transport of natural toxins. Therefore, they are definitely also linked to drug–nutrient interactions. One prominent example are flavonoids, which have been shown in numerous studies to interact with P-gp [28–30] and BCRP [31]. Of course, induction of protein expression — like it has been shown for St. Johns wort (Hypericum perforatum) — and cytochromes might also be a major issue, especially when considering that nuclear receptors are involved for both cytochromes and ABC-transporters [32].
Considering the multiple involvement of ABC-transporters in ADMET properties of drugs as well as their potential role as targets for treatment of multidrug resistant tumours, it is evident that numerous computational studies have been performed with the aim to predict potential compound–transporter interaction and to explore the molecular basis of the polyspecificity of these transporters. These started with ligand-based approaches, which extended to structure-based studies when the first X-ray structures became available.
2. Ligand-based models
2.1. Machine learning approaches for predicting inhibitors of ABC-transporters
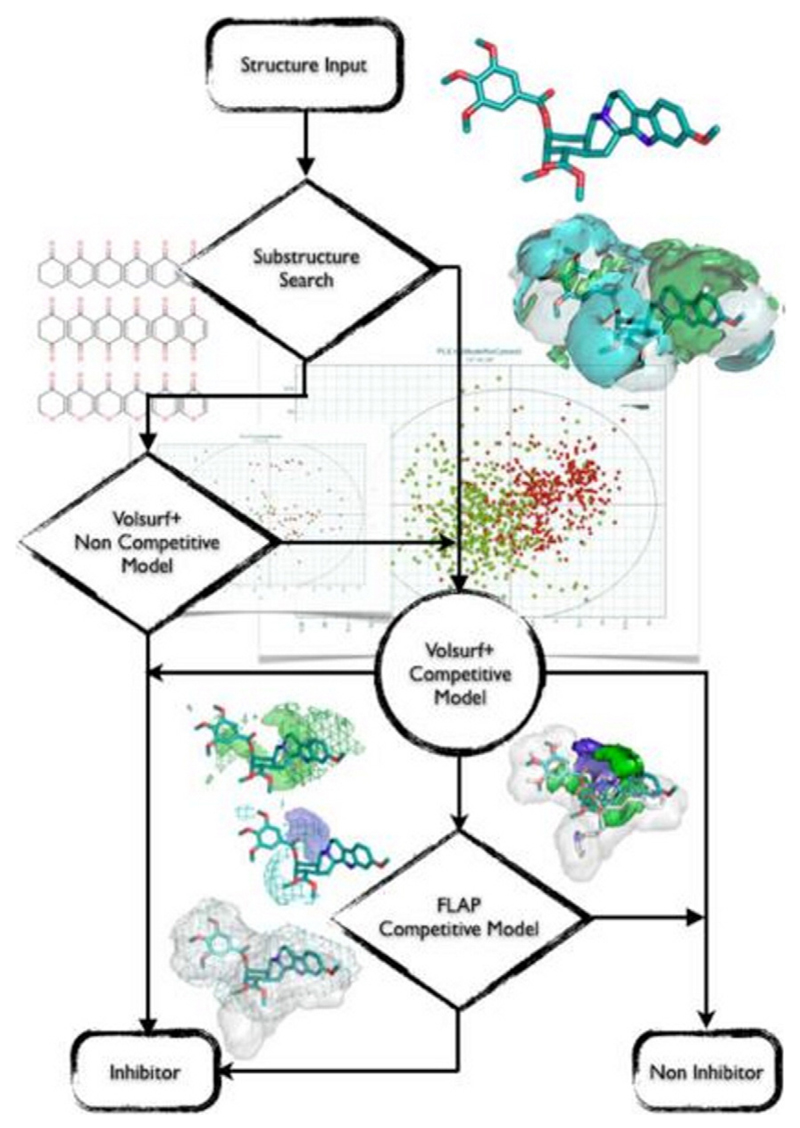
P-glycoprotein is definitely the paradigm protein for the whole family of ABC-transporters. Thus, basically all methods available for ligand-based design have been applied. These include conventional Hansch analysis, linear and non-linear classification algorithms, pharmacophore modeling, as well as supervised and unsupervised artificial neural networks. There are numerous reviews published which summarize these studies, and the reader is referred to a small selection for further reading [33–38]. However, the challenges in the field of ABC-transporter modeling are manifold, and the main question — what is the molecular basis for the polyspecificity — is still not solved. A large number of chemical scaffolds for inhibitors of P-gp have been published, and, for basically all of them, structure–activity relationships could be derived. This indicates that there are local effects (binding sites?) which translate to a distinct structure–activity relationship (SAR). For each scaffold investigated, clear determinants for high and low inhibitory activity could be established. They most often relate to quite basic physicochemical parameters, such as lipophilicity, H-bonding, aromatic rings, and may be also charge. However, after more than 30 years of intense research, there is still no clear understanding of the molecular basis of compound–transporter interaction which would translate to a set of general rules for medicinal chemists that could help them to enhance or to avoid P-gp inhibitor properties in a lead optimization program. Interestingly, also the concepts of ligand efficiency and lipophilic efficiency have to be applied in a different way than for conventional targets [39]. In recent years, the focus shifted to classification models for large data sets in order to allow in silico profiling of compound libraries. Also in this area a number of publications appeared in the literature, and we will just summarize a few recent ones to outline the main strategies followed. One of the groundbreaking contributions is the work of Broccatelli and colleagues [40], who used a combination of molecular field analysis, pharmacophore-based representation of the compounds, as well as physicochemical descriptors to develop both global and local models for P-gp inhibitors. Based on a data set of 1275 compounds derived from 61 references, the authors established a workflow which combines specific (pharmacophore) and nonspecific (general physicochemical) descriptors (Fig. 2). The final model points towards flexibility, hydrophobic surface area, and logP as main discriminating physicochemical parameters for inhibitors/non-inhibitors. Furthermore, shape also emerged as a crucial factor, indicating the importance of the 3D description of the molecules. The authors reported an accuracy of 0.86, specificity of 0.8, sensitivity of 0.9 and Cohen's kappa of 0.7 on a true external set.
Fig. 2.
Flow chart of the Composite Model. Reprinted with permission from [40]. Copyright 2011 American Chemical Society.
Chen and colleagues compiled a large data set from literature, comprising in total 1273 compounds [41]. Their classification approach is based on recursive partitioning and naive Bayes categorization using a set of physicochemical descriptors and various fingerprints. Also in their models, logP is an important contributor to distinguish inhibitors from non-inhibitors. The introduction of fingerprints remarkably improved the prediction accuracy of the models (from a sensitivity of 0.69 and specificity of 0.70 with only physicochemical descriptors on an external test set to a maximum of 0.84 in sensitivity and 0.87 in specificity on the same set but with fingerprints) and furthermore allowed to identify molecular fragments which are favorable or unfavorable for P-gp inhibition. However, one should bear in mind that all methods linking substructures/fragments to biological activity of course heavily depend on the presence/absence of these fragments in the data set. For example, a data set which includes a series of propafenone analogs will of course point towards the importance of an aryloxypropanolamine moiety for P-gp inhibitory potency. However, detailed structure–activity relationship studies showed that the hydroxy-group of the propanolamine seems not to be involved in compound–transporter interaction [42].
While in the case of P-gp datasets of considerable size are available in the literature, for most of the other ABC-transporters there is still a lack of data for establishing in silico models. Thus, for MDR3, which is a phospholipid transporter expressed in the liver, only 5 compounds are retrieved by the Open PHACTS Discovery Platform [6], including taxol, vinblastine and verapamil. Considering the fact that MDR3 is the closest homolog to P-glycoprotein (sequence identity 75%), it seems quite unlikely that the protein is inhibited only by five compounds. In case of BCRP, BSEP, MRP1, and MRP2, considerable progress has been made within the past few years, allowing developing in silico models.
Very recently, the hitherto largest data set for BCRP has been compiled by Montanari and Ecker, and includes 978 unique compounds extracted from 47 studies [43]. Subsequently, the data set was used to derive a Bayesian classification model using ECFP_6 fingerprints. This allowed extracting important substructures, which are mostly in line with currently published SAR studies around BCRP inhibition. Basically, the number of nitrogen atoms, the aromaticity and the presence of fused aromatic heterocycles seem to favor inhibition, while the presence of sulfur atom, five-membered rings, or amide linkers seems to favor inactivity. The authors report an accuracy of 0.92 and an area under the ROC curve of 0.85 in cross-validation for this naive Bayes model.
In case of the human bile salt export pump (BSEP), Warner et al. [44] used a recently described in vitro membrane vesicle BSEP inhibition assay to quantify transporter inhibition for a set of 624 compounds. Relating a set of physicochemical properties of the compounds to BSEP inhibition, they showed that lipophilicity and molecular size are significantly correlated with BSEP inhibition. BSEP inhibitor classification by a support vector machine model leads to a total accuracy of 0.87. The model could be further used to minimize the propensity of drug candidates to inhibit BSEP.
In case of MRP2, an ABC-transporter which also might be involved in drug–drug interactions in the liver, Pedersen et al. [45] measured a set of 191 structurally diverse drugs and drug-like compounds for inhibition of MRP2-mediated transport of estradiol-17-d-glucuronide (E17G) in inside-out membrane vesicles from Sf9 cells overexpressing human MRP2. Based on these data, a multivariate orthogonal partial least squares discriminant analysis (OPLS-DA) model that distinguishes between MRP2 inhibitors and non-inhibitors was built. The model was capable of correctly classifying 72% of the inhibitors and 71% of the non-inhibitors in the test set. The coefficients in the final model show that a combination of increased lipophilicity, aromaticity, and size is a major determinant for the MRP2 inhibitory effect. Interestingly, the authors also performed an analysis to examine whether inhibitors that have also been reported to be substrates, and which are thus likely to compete with E17G binding at the transport site, were structurally different from other inhibitors. They were indeed on average less lipophilic than other inhibitors and also had a higher molecular weight and a larger polar surface area.
Interestingly, there are also early attempts to perform selectivity profiling studies over several ABC-transporters. For example, Matsson and colleagues [55] used a set of 122 structurally diverse drugs to study the inhibition patterns of P-gp, BCRP, and MRP2. The inhibitor specificities of P-gp, BCRP and MRP2 were shown to be highly overlapping, and a computational model based on multivariate statistics correctly classified 80% of general ABC transporter inhibitors and non-inhibitors in an external test set.
2.2. Pharmacophore models for ABC-transporter inhibitors
Of course, in addition to classical QSAR of local compound series and inhibitor/non-inhibitor classification models, numerous pharmacophore models have been derived. This is driven by the aim to understand pharmacophoric and pharmacophobic features which determine ligand–transporter interactions. However, due to the high structural diversity of the ligands, also pharmacophore modeling so far did not lead to a better molecular understanding of the molecular basis of polyspecificity. However, most of the pharmacophore models derived show good capabilities in identifying new ligands with new chemical scaffolds, thus proving their utility. Briefly, Palmeira et al. [46] created a pharmacophore model based on 26 known P-gp inhibitors from the flavonoid family, which was then used to screen DrugBank. 167 structures were found to comply with the pharmacophore model with an RMSD of < 1 Å. Out of these 167 structures, 91 fulfilled the Lipinski rules of 5. Finally, 21 compounds were selected for biological testing, whereby 12 were found to significantly increase the intracellular accumulation of Rhodamine-123, a P-gp substrate. Analogously, Pan et al. [47] created a pharmacophore model based on 25 BCRP inhibitors and screened the Collaborative Drug Discovery Database, which comprises 2815 FDA-approved drugs selected from all medications on the market since 1938. 33 drugs were tested in vitro for their inhibitory effects on BCRP-mediated transport of [3H]-mitoxantrone in MCF-7/AdrVp cells, and 19 compounds were identified with significant inhibitory effect on BCRP transport function. For BSEP, a small set of 5 compounds served as basis for a pharmacophore model, which was validated against a set of 59 compounds, including registered drugs. The model recognized 9 out of 12 inhibitors, which could not be identified based on general parameters (such as molecular weight or SlogP) alone. Finally, the model was used to screen a virtual compound database of commercially available compounds. A number of compounds found via virtual screening were tested and displayed statistically significant BSEP inhibition, ranging from 13 ± 1% to 67 ± 7% of control (P < 0.05) [48].
A pharmacophore for MRP1 was recently built on five diverse and potent inhibitors [36]. It is composed of 3 aromatic rings and 3 H-bond donor features and was able to retrieve 3 known inhibitors of MRP1 among a large database of 500 drugs. For MRP2, Zhang and colleagues [49] reported a pharmacophore built on nine potent and diverse inhibitors. It contains two H-bond acceptor features and one hydrophobic feature. The model gave a sensitivity of 78% and a specificity of 70%, with an overall accuracy of 74%.
2.3. Models for predicting substrates
While for inhibitors a set of assays is available which leads to precise IC50 values, the case of substrates is much more complicated. Most commonly a polarized transport assay across a monolayer of cells overexpressing a distinct transporter is used. Thus, the in silico models derived on basis of these data mostly use binary classification algorithms (substrate versus non substrate). To our knowledge the largest data set for P-gp substrates/non-substrates in the public domain was compiled by Li et al. [50] (423 substrates, 300 non-substrates). Analyzing the distributions of eight basic physicochemical properties for the substrates and non-substrates showed that molecular weight and solubility are the main factors differentiating P-gp substrates from non-substrates. When comparing the 423 substrates with a set of 735 P-gp inhibitors, inhibitors proved to be significantly more hydrophobic than substrates while substrates tend to have more H-bond donors than inhibitors. Applying a naive Bayes classifier using a set of simple molecular properties, topological descriptors, and molecular fingerprints, a classification model with very good performance was retrieved (Matthews correlation coefficient (MCC) = 0.824, prediction accuracy = 91.2% for leave 20% out cross-validation, prediction accuracy of 83.5% for a test set of 200 molecules). The most important structural fragments provided by the Bayesian classifier indicate that H-bond acceptors arranged in distinct spatial patterns as well as flexibility are quite essential for P-gp substrate-likeness.
In another setting, Wang et al. [51] used a set of 332 compounds to develop a classification model using support vector machine. The best model (MCC = 0.73) shows a prediction accuracy of 0.88 on a test set. Examination of the model based on ECFP_4 fingerprints revealed substructures such as nitrile and sulfoxide, which have a higher frequency in non-substrates than in substrates. However, as already previously stated, this should be taken with caution, as substructure analysis depends on the occurrence of the respective fragments in the training set. Also for BCRP, a larger set of 263 substrates and non-substrates has been collated from literature and classified via a support vector machine [52]. The final SVM model had an overall prediction accuracy of 73% for an independent test set of 40 compounds and was integrated to a free web server (http://bcrp.althotas.com).
While finding a large dataset for MRP2 substrates and non-substrates is not an easy task to date, Pinto and colleagues [53] made use of a fuzzy dataset correlating transporter expression in cancer cell lines with the substrate capability of the tested compounds [54] to build classification models predicting MRP2 substrates. The authors reached a sensitivity of 0.77 and specificity of 0.72 in their best settings where 16 physicochemical descriptors were used to build a cost sensitive Random Forest. This technique allowed taking into account the imbalance of the data by penalizing predictions errors made by the model on the minority class.
In summary, there are numerous models published which are able to predict inhibitors and/or substrates of the most important ABC-transporters. All the models described here are summarized in Table 1. However, a set of general rules with respect to the main driving factors for ligand–transporter interaction, which go beyond lipophilicity, size, and H-bonding, is still missing. Furthermore, most of the models lack proper applicability domain assessment, which renders it difficult to judge their performance in a broader chemical space. Finally, when checking the original publications where the data were coming from, it becomes evident that numerous different assays are used to measure ABC-transporter inhibition. Thus, an upfront careful curation of the data seems mandatory before using them for large scale models.
Table 1. Summary of all ligand-based models described in Section 2.
| Transporter | Type of model | Dataseta | Predictivity | Publication |
|---|---|---|---|---|
| P-gp | Combined | 1275 inhibitors | Accuracy: 0.86 | Broccatelli et al. [40] |
| P-gp | Naive Bayes | 1273 inhibitors | Sensitivity: 0.835 Specificity: 0.866 |
Chen et al. [41] |
| P-gp | Pharmacophore | 26 inhibitors | 12/21 tested were active | Palmeira et al. [46] |
| P-gp | Naive Bayes | 723 substrates | Accuracy: 0.84 | Li et al. [50] |
| P-gp | SVMb | 332 substrates | Accuracy: 0.88 | Wang et al. [51] |
| BCRP | Naive Bayes | 978 inhibitors | Accuracy: 0.92 | Montanari and Ecker [43] |
| BCRP | Pharmacophore | 25 inhibitors | 19/33 tested were active | Pan et al. [47] |
| BCRP | SVMb | 263 substrates | Accuracy: 0.73 | Hazai et al. [52] |
| BSEP | SVMb | 624 inhibitors | Accuracy: 0.87 | Warner et al. [44] |
| BSEP | Pharmacophore | 5 inhibitors | Sensitivity: 0.75 | Ritschel et al. [48] |
| MRP2 | OPLS-DAc | 191 inhibitors | Sensitivity: 0.72 Specificity: 0.71 |
Pedersen et al. [45] |
| MRP2 | Pharmacophore | 9 inhibitors | Accuracy: 0.74 | Zhang et al. [49] |
| MRP2 | Random Forest | 1204 substrates | Sensitivity: 0.77 Specificity: 0.72 |
Pinto et al. [53] |
| MRP1 | Pharmacophore | 5 inhibitors | Not clear | Chang et al. [36] |
| P-gp, BCRP, MRP2 | PLS-DAd | 122 inhibitors | Accuracy: 0.8 | Matsson et al. [55] |
Size and type of data (for models that are not pharmacophores, both active and inactive are present).
Support vector machine.
Orthogonal partial least squares discriminant analysis.
Partial least squares discriminant analysis.
3. Data curation
Working with large datasets seems to be the way to build high quality models and derive general trends for compounds interacting with ABC-transporters. Data, however, is scarce, at least for some less studied transporters. Typical medicinal chemistry studies report bioactivities for a small set of chemically related compounds. Scientists wanting to build large datasets must collect and merge together such data, using databases like ChEMBL [56] and Pubchem [57], but also manual search through MEDLINE. Now, what if the groups measuring ABC-transporters substrate or inhibition activities each use their own assay design? Then merging together data becomes a challenging task. Zdrazil et al. [58], studied all bioassays from ChEMBL for P-gp inhibition and transport, when these assays reported IC50, EC50 or Ki values. Subsequently, they annotated assays according to their potential for being combined together in a large QSAR dataset. The results show the importance of overlapping binding sites for the different substrates used in the bioassays, as well as the cell line in which the transporter is expressed.
In another recent study [59], the authors compared IC50 values obtained across several laboratories for P-gp inhibition, each using several assay methodologies. The variability range was over 20 fold for all compounds tested, and the study concluded that the most important actor was inter-laboratory variability rather than inter-assay variability. Beyond the assay diversity and inter-laboratory variabilities, Balimane and colleagues [60] have pointed out yet another source of variability, namely the calculation of inhibition given one raw set of data measured on one assay by one laboratory. It seems that, depending on the calculation method used, one can draw entirely different conclusions regarding the inhibition capability of compounds.
For other less studied transporters like BCRP, the picture gets worse: most of the assays use different cell lines, and little is known about the binding sites of the different substrates used in these assays [61]. The resulting problem is that a compound may show activity in an assay with a given substrate, but no activity in the presence of another substrate. One solution is to compare activities reported across distinct assays, exclusively use the data for building classification models, apply a threshold for activity assay by assay and remove problematic compounds [43].
To alleviate the aforementioned problems related with bioactivity data in the field of transporters, one could propose some simple measures to apply and change the current habits in the field. While agreeing on a specific assay methodology seems a bit irrealistic, a set of reference compounds for each transporter could be defined (both inactives and actives) and recommend that each laboratory willing to publish new bioactivity data must also report the bioactivities obtained on their assay for this group of reference compounds. That way, inter-laboratory and inter-assay differences would be immediately spotted and taken care of appropriately when merging data from different sources.
4. Structure-based models
Due to the tremendous progress in the field of structural biology, structures of transmembrane transporters, including several ABC-transporters, became available. Most of them were from prokaryotes, and only very recently structures from eukaryotic organisms were also resolved in a resolution which allows starting structure-based approaches. However, the only human ABC-transporter crystallized so far is ABCB10 [62]. Nevertheless, the whole field of ABC-transporter research was inspired by the first structures being deposited in the Protein Data Bank [63], and protein homology models of P-gp immediately became available.
More recent templates available for homology modeling are provided in Table 2.
Table 2. Existing 3D structures of ABC transporters in the Protein Data Bank (PDB).
| Year | Publication | PDB IDs | Species | Protein | Res.a | State |
|---|---|---|---|---|---|---|
| 2012 | Jin et al. [64] | 4F4C | C. elegans | Pgp-1 (Uniprot: P34712) | 3.4 Å | Open-in |
| 2012 | Shintre et al. [62] |
3ZDQ, 4AYT, 4AYX, 4AYW |
H. sapiens | ABC transporter 10 protein (Uniprot: Q9NRK6) | 2.85 Å | Open-in |
| 2009 | Aller et al. [65] |
3G5U 3G60 3G61 |
M. musculus | MDR1A (Uniprot: P21447) | 3.8 Å | Open-in |
| 2013 | Ward et al. [66] |
4KSB 4KSC 4KSD 4LSG |
M. musculus | MDR1A (Uniprot: P21447) | 3.8 Å | Open-in |
| 2014 | Li et al. [67] |
4M1M, 4M2S, 4M2T |
M. musculus | MDR1A (Uniprot: P21447) | 3.8 Å | Open-in |
| 2007 | Dawson and Locher [68] | 2ONJ | S. aureus | SAV1866 (Uniprot: Q99T13) | 3.4 Å | Open-out |
| 2006 | Dawson and Locher [69] | 2HYD | S. aureus | SAV1866 (Uniprot: Q99T13) | 3.0 Å | Open-out |
| 2007 | Ward et al. [70] |
3B5Y, 3B5Z, 3B60 |
S. typhimurium | Permease protein msbA (Uniprot: P63359) | 3.7 Å | Open-out |
| 2012 | Hohl et al. [71] | 3QF4 | T. maritima | Uncharacterized ABC transporter (Uniprot: Q9WYC4) | 2.9 Å | Open-in |
Resolution. When more than one PDB ID is given, the lowest resolution is reported.
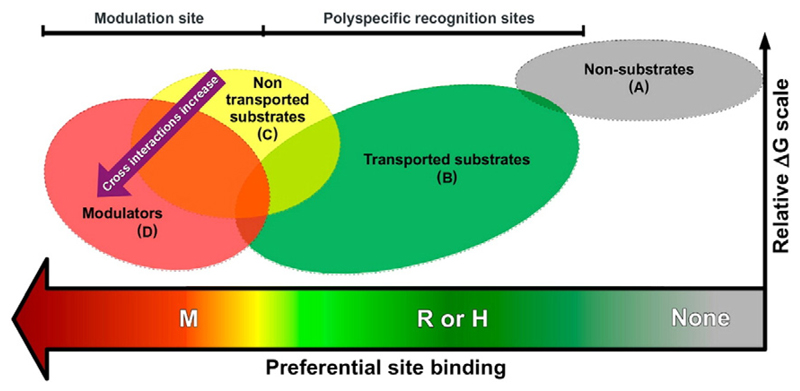
Although most of these structures are in sufficient resolution to serve as templates for structure-based studies, one needs to bear in mind that there is still no protein structure cocrystallized with a classical substrate/inhibitor, such as verapamil or cyclosporin. The only structure which includes a small molecule is the one from mouse P-gp [65]. Furthermore, the transporters undergo a substantial conformational change when progressing through the transport cycle, which renders all structures available only snapshots of a very distinct point in the whole conformational space. Nevertheless, especially for P-gp, numerous docking studies of selected ligands into homology models were performed with the aim to understand the molecular determinants of binding (for reviews, see e.g. [33,34,35,72,37]). However, experimental validation, especially with respect to prospective validation of the binding hypotheses retrieved, is mostly missing. In this section we will thus focus on recent advances where structure-based studies were used more in the sense of virtual screening rather than providing distinct poses for selected compounds. Dolghih et al. [73] implemented a flexible receptor docking protocol for docking a set of 26 drugs known to interact with P-gp. 102 endogenous metabolites, assuming that they will not interact with the transporter, served as negative control. As a template, mouse P-gp bound to the cyclic peptide QZ59-RRR was used. Subsequently, the dataset from Doan et al. [74] of FDA-approved drugs that included results of the monolayer efflux and CAM inhibition assays, was used for docking. The results suggest that many P-gp substrates bind deeper in the cavity than the cyclic peptide in the crystal structure, and that specificity in P-gp is better understood in terms of physicochemical properties of the ligands (and the binding site), rather than being defined by specific sub-sites. Klepsch et al. [75] also built on the mouse P-gp complexed with QZ59-RRR and implemented a docking protocol which exhaustively samples the pose space of a small set of ligands which show a distinct SAR pattern followed by common scaffold clustering. The SAR information is then used to priorities the pose cluster in order to retrieve an experimental-data-guided binding hypothesis. Subsequently, the docking protocol was used to classify a large set of 1608 inhibitors and non-inhibitors of P-gp. Although the performance of the structure-based classification was considerably lower (61% for the external test set) than those obtained by Random Forest or SVM classification (73% and 75%, respectively), it shows that structure-based classification of ligands of ABC-transporter is within reach [76]. A very comprehensive approach was used by Ferreira et al. [77]. Using a previously refined structure of murine P-gp, they characterized the M-, H- and R-site by means of molecular docking. The drug-binding pockets were defined as substrate- or modulator-binding sites according to the molecules that preferentially docked in each location. For the authors, “modulator” refers to compounds that appear to block the efflux of substrates. The substrate-binding sites H and R refer to Hoechst 33342 and rhodamine-123, respectively. Analogously, the modulator-binding (M) site was linked to the main interaction site for verapamil. Subsequently, they carried out further docking studies with molecules classified as substrates or modulators in order to retrieve a structure-based classification model with the ability to discriminate substrates from modulators. The classification scheme contains four main categories: (i) non-substrates, (ii) transported substrates, (iii) non-transported substrates, and (iv) modulators (Fig. 3). Their model properly predicted 14 modulators out of 19 (74%), 20 substrates out of 32 (63%) and 2 out of 3 non-substrates. However, the authors rightly conclude that “…. the substrate-binding sites may present different characteristics at different steps of the efflux mechanism, possibly interconverting the H-site and the R-site in one another, partially explaining the induced-fit and polyspecificity models proposed for Pgp substrate recognition”, and point towards the importance of molecular dynamics simulations for further insights into the dynamics of the protein.
Fig. 3.
Classification scheme for P-gp substrates. Reprinted with permission from [78]. Copyright 2013 American Chemical Society.
In a different publication [78], the authors indeed performed 100 ns molecular dynamics simulations in order to refine their homology model. Subsequent 20 ns production runs with a small set of ligands indicated that the number of interactions established between several ligands and the drug binding pocket might allow distinguishing inhibitors from substrates. Indeed, the modulators studied consistently established a higher number of nonbonded interactions, mainly aromatic ones, when compared with substrates. In the particular case of verapamil, the increased nonbonded interactions established, which is also shown by the modulator tariquidar, classifies the molecule as a modulator. This is well in accordance with a previously developed pharmacophore [79], where the ability to establish a greater number of hydrophobic interactions within the pocket is one of the major features that allows a molecule to block competitively the substrate binding. These studies convincingly demonstrate that structure-based modeling in the field of ABC-transporter has become a valuable tool for a deeper understanding of the molecular features driving ligand–transporter interaction. However, almost all studies focus on P-gp. This is mainly due to the fact that both mammalian structures available are from transporters belonging to the B-family (P-gp and ABCB10). The C-family has the so-called TMD 0, which consists of 5 transmembrane helices (thus having 5 + 12 transmembrane helices) where no suitable template exists. Even worse is the case of members of the G family (BCRP), which show a reversed order of nucleotide binding domain and transmembrane domain. Although there is a considerable substrate and inhibitor overlap between P-gp and BCRP, there is no suitable template for modeling the whole transporter. Thus, new structures are heavily awaited by the community. These might well use also other methods than X-ray crystallography, such as the one of TmrAB published by the Tampe group in November 2014 [80].
5. Future challenges
Challenges in the field of ABC-transporter are manifold. With respect to the prediction of drug transporter interaction, there are on our point of view several immediate issues which should be mentioned. The most obvious one is the availability of an atomic resolution structure of human P-glycoprotein in complex with a prototype ligand such as verapamil. This would allow benchmarking all docking studies on this structure, which definitely would increase the validity of the binding hypotheses retrieved. Nevertheless, in early drug discovery, in silico models based on machine learning will still be the main tools for prioritization of large compound libraries. In silico classification models will only show high predictivity if the underlying data are of high quality and of a considerable size. In case of P-glycoprotein, the size of the datasets available in the public domain is sufficient, but the use of almost 50 different assays currently does not allow combining all the data and to compile a large, high quality dataset for training the models. For ABC-transporters other than P-glycoprotein, the situation is even worse, as already the available datasets are small. For solving the issue of different assays, a transporter assay ontology combined with the definition of a set of standard reference compounds would be highly recommended.
Another important issue is the flexibility inherent to P-gp and most probably to all transmembrane transporters [81]. Even if there would be a set of crystal structures, they still would only cover a small portion of the conformational space of the transporter. With the ever increasing computing power, also pushed by GPU clusters, large grids and special computer hardware, molecular dynamics simulations of transmembrane proteins in the ms range are possible. Simulating an ABC-transporter through the whole transport cycle and validating/constraining the simulation by respective biophysical and biochemical experiments seems already feasible. Once the system is established, this would also allow including small molecules, be it substrates, inhibitors, or modulators. However, also the composition of the membrane, its cholesterol content, as well as the behavior of the ligand in the membrane needs to be considered [82]. This opens another layer of complexity for full atomic simulations. Another issue linked to dynamics is the on- and off-kinetics of the ligands. There is increasing evidence that the dissociation kinetics of a given drug from its target (its residence time) may be more relevant for the in vivo efficacy than its in vitro equilibrium binding constant. Recent examples demonstrate that receptor subtype selectivity might be driven by differences in dissociation kinetics rather than by affinity differences [83]. We have first evidence in our lab that this is also the case for propafenone-type inhibitors of P-gp, and also other groups already speculated on this [84].
The importance of ligand–transporter interaction for prediction of toxicity and safety needs much more attention. As outlined in the introduction, ABC-transporters play a major role in toxicity related to drug–drug interaction. However, due to the multiple interplay of several transporters, this requires more complex strategies. In might well be that there are a sort of redundant backup systems, where one transporter can compensate for the functional failure of another one. A well established example is the interplay of P-gp and BCRP at the blood–brain barrier. But also in the liver there are numerous ABC-transporters, which have to be considered. There are already a few publications available which attempt to simultaneously predict the interaction of a compound of interest with several transporters (not necessarily only ABC-transporter; see e.g. [85]). Selectivity profiling, of course, is strongly linked to the availability of proper data, as one would need a matrix of a set of compounds measured at all transporters of interest. These models could then be further used for linking in vitro interaction profiles to in vivo effects, as recently has been shown for a set of antidepressant drugs and their side effects observed in clinical studies [86]. This would allow including ligand–transporter interaction profiles into very early safety considerations and help to reduce late stage failures in clinical studies.
6. Conclusions
ABC-transporters represent an integral and important part of the human transportome. Although there are only 49 genes described in humans, they fulfill important roles and are strongly linked to drug absorption, distribution, and elimination. Furthermore, besides cytochromes, they are also involved in drug–drug interactions and thus also toxicity of drugs. With the increasing accessibility of biological data and the tremendous progress of structural biology, our understanding of the molecular basis of ligand–transporter interaction is progressing. In this review, we have outlined recent ligand-based models built on large datasets rather than on congeneric series. While these models allow screening rapidly large databases of molecules to predict their substrate or inhibition properties, their interpretation remains at the level of substructures or general physico-chemical properties. On the structure-based side, the presence of crystal structures of the B subfamily allowed building high quality homology models for P-gp and a mapping of different binding sites has started. Such advances have not yet been noted for other ABC-transporters, but we believe that new structures will appear in the PDB that will allow similar studies to be performed.
However, the influence of on- and off-kinetics of ligands on their efficacy as well as the multiple interplay of ABC-transporters under in vivo conditions pose additional challenges which the community will face in the near future.
Funding
The research leading to this publication has received support from the Austrian Science Fund (FWF), Grant F03502 and from the University of Vienna, doctoral programme Biopromotion.
None of the funding sources took part in the study design, collection, analysis or interpretation of the data, nor in the writing and decision to submit the article for publication.
References
- [1].Vasiliou V, Vasiliou K, Nebert DW. Human ATP-binding cassette (ABC) transporter family. Hum Genet. 2009;3:281–290. doi: 10.1186/1479-7364-3-3-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Tarling EJ, de Aguiar Vallim TQ, Edwards PA. Role of ABC transporters in lipid transport and human disease. Trends Endocrinol Metab. 2013;24:342–350. doi: 10.1016/j.tem.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Juliano RL, Ling V. A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochim Biophys Acta Biomembr. 1976;455:152–162. doi: 10.1016/0005-2736(76)90160-7. [DOI] [PubMed] [Google Scholar]
- [4].Chatterjee M, Robson CN, Harris AL. Reversal of multidrug resistance by verapamil and modulation by alpha 1-acid glycoprotein in wild-type and multidrug-resistant Chinese hamster ovary cell lines. Cancer Res. 1990;50:2818–2822. [PubMed] [Google Scholar]
- [5].Lehnert M, Dalton WS, Roe D, Emerson S, Salmon SE. Synergistic inhibition by verapamil and quinine of P-glycoprotein-mediated multidrug resistance in a human myeloma cell line model. Blood. 1991;77:348–354. [PubMed] [Google Scholar]
- [6].Gray AJG, Groth P, Loizou A, Askjaer S, Brenninkmeijer C, Burger K, et al. Applying linked data approaches to pharmacology: architectural decisions and implementation. Semantic Web. 2014;5:101–113. doi: 10.3233/SW-2012-0088. [DOI] [Google Scholar]
- [7].Kalvass JC, Polli JW, Bourdet DL, Feng B, Huang S-M, Liu X, et al. Why clinical modulation of efflux transport at the human blood–brain barrier is unlikely: the ITC evidence-based position. Clin Pharmacol Ther. 2013;94:80–94. doi: 10.1038/clpt.2013.34. [DOI] [PubMed] [Google Scholar]
- [8].Food and Drug Administration. Guidance for Industry Drug Interaction Studies — Study Design, Data Analysis, Implications for Dosing, and Labeling Recommendations. U.S.Department of Health and Human Services Food and Drug Administration Center for Drug Evaluation and Research (CDER); 2012. [accessed January 20, 2014]. http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm292362.pdf. [Google Scholar]
- [9].Wu C-Y, Benet LZ. Predicting drug disposition via application of BCS: transport/absorption/elimination interplay and development of a Biopharmaceutics Drug Disposition Classification System. Pharm Res. 2005;22:11–23. doi: 10.1007/s11095-004-9004-4. [DOI] [PubMed] [Google Scholar]
- [10].Demeule M, Régina A, Jodoin J, Laplante A, Dagenais C, Berthelet F, et al. Drug transport to the brain: key roles for the efflux pump P-glycoprotein in the blood–brain barrier. Vasc Pharmacol. 2002;38:339–348. doi: 10.1016/s1537-1891(02)00201-x. [DOI] [PubMed] [Google Scholar]
- [11].Laquintana V, Trapani A, Denora N, Wang F, Gallo JM, Trapani G. New strategies to deliver anticancer drugs to brain tumors. Expert Opin Drug Deliv. 2009;6:1017–1032. doi: 10.1517/17425240903167942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Durmus S, Sparidans RW, van Esch A, Wagenaar E, Beijnen JH, Schinkel AH. Breast cancer resistance protein (BCRP/ABCG2) and P-glycoprotein (P-GP/ABCB1) restrict oral availability and brain accumulation of the PARP inhibitor rucaparib (AG-014699) Pharm Res. 2014:1–10. doi: 10.1007/s11095-014-1442-z. [DOI] [PubMed] [Google Scholar]
- [13].Durmus S, Xu N, Sparidans RW, Wagenaar E, Beijnen JH, Schinkel AH. P-glycoprotein (MDR1/ABCB1) and breast cancer resistance protein (BCRP/ABCG2) restrict brain accumulation of the JAK1/2 inhibitor, CYT387. Pharmacol Res. 2013;76:9–16. doi: 10.1016/j.phrs.2013.06.009. [DOI] [PubMed] [Google Scholar]
- [14].Bagal SK, Bungay PJ. Minimizing drug exposure in the CNS while maintaining good oral absorption. ACS Med Chem Lett. 2012;3:948–950. doi: 10.1021/ml300378n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sanchez-Covarrubias L, Slosky LM, Thompson BJ, Zhang Y, Laracuente M-L, DeMarco KM, et al. P-glycoprotein modulates morphine uptake into the CNS: a role for the non-steroidal anti-inflammatory drug diclofenac. PLoS ONE. 2014;9:e88516. doi: 10.1371/journal.pone.0088516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cuperus FJC, Claudel T, Gautherot J, Halilbasic E, Trauner M. The role of canalicular ABC transporters in cholestasis. Drug Metab Dispos. 2014;42:546–560. doi: 10.1124/dmd.113.056358. [DOI] [PubMed] [Google Scholar]
- [17].Wlcek K, Stieger B. ATP-binding cassette transporters in liver. BioFactors. 2014;40:188–198. doi: 10.1002/biof.1136. [DOI] [PubMed] [Google Scholar]
- [18].Dawson S, Stahl S, Paul N, Barber J, Kenna JG. In vitro inhibition of the bile salt export pump correlates with risk of cholestatic drug-induced liver injury in humans. Drug Metab Dispos. 2012;40:130–138. doi: 10.1124/dmd.111.040758. [DOI] [PubMed] [Google Scholar]
- [19].Andress EJ, Nicolaou M, Romero MR, Naik S, Dixon PH, Williamson C, et al. Molecular mechanistic explanation for the spectrum of cholestatic disease caused by the S320F variant of ABCB4. Hepatology. 2014;59:1921–1931. doi: 10.1002/hep.26970. [DOI] [PubMed] [Google Scholar]
- [20].Gadsby DC, Vergani P, Csanady L. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature. 2006;440:477–483. doi: 10.1038/nature04712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Machida I, Wakusawa S, Sanae F, Hayashi H, Kusakabe A, Ninomiya H, et al. Mutational analysis of the MRP2 gene and long-term follow-up of Dubin–Johnson syndrome in Japan. J Gastroenterol. 2005;40:366–370. doi: 10.1007/s00535-004-1555-y. [DOI] [PubMed] [Google Scholar]
- [22].Cameron J, Ranheim T, Halvorsen B, Kulseth MA, Leren TP, Berge KE. Tangier disease caused by compound heterozygosity for ABCA1 mutations R282X and Y1532C. Atherosclerosis. 2010;209:163–166. doi: 10.1016/j.atherosclerosis.2009.08.039. [DOI] [PubMed] [Google Scholar]
- [23].Matsuo H, Takada T, Ichida K, Nakamura T, Nakayama A, Suzuki H, et al. ABCG2/BCRP dysfunction as a major cause of gout. Nucleosides Nucleotides Nucleic Acids. 2011;30:1117–1128. doi: 10.1080/15257770.2011.633954. [DOI] [PubMed] [Google Scholar]
- [24].Abuznait AH, Kaddoumi A. Role of ABC transporters in the pathogenesis of Alzheimer's disease. ACS Chem Neurosci. 2012;3:820–831. doi: 10.1021/cn300077c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Greiner B, Eichelbaum M, Fritz P, Kreichgauer H-P, von Richter O, Zundler J, et al. The role of intestinal P-glycoprotein in the interaction of digoxin and rifampin. J Clin Invest. 1999;104:147–153. doi: 10.1172/JCI6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Choo EF, Leake B, Wandel C, Imamura H, Wood AJJ, Wilkinson GR, et al. Pharmacological inhibition of P-glycoprotein transport enhances the distribution of HIV-1 protease inhibitors into brain and testes. Drug Metab Dispos. 2000;28:655–660. [PubMed] [Google Scholar]
- [27].Umeyama Y, Fujioka Y, Okuda T. Clarification of P-glycoprotein inhibition-related drug–drug interaction risks based on a literature search of the clinical information. Xenobiotica. 2014;44:1135–1144. doi: 10.3109/00498254.2014.928958. [DOI] [PubMed] [Google Scholar]
- [28].Conseil G, Baubichon-Cortay H, Dayan G, Jault JM, Barron D, Di Pietro A. Flavonoids: a class of modulators with bifunctional interactions at vicinal ATP- and steroid-binding sites on mouse P-glycoprotein. Proc Natl Acad Sci USA. 1998;95:9831–9836. doi: 10.1073/pnas.95.17.9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Comte G, Daskiewicz JB, Bayet C, Conseil G, Viornery-Vanier A, Dumontet C, et al. C-isoprenylation of flavonoids enhances binding affinity toward P-glycoprotein and modulation of cancer cell chemoresistance. J Med Chem. 2001;44:763–768. doi: 10.1021/jm991128y. [DOI] [PubMed] [Google Scholar]
- [30].Wesołowska O, Wiśniewski J, Środa K, Krawczenko A, Bielawska-Pohl A, Paprocka M, et al. 8-Prenylnaringenin is an inhibitor of multidrug resistanceassociated transporters, P-glycoprotein and MRP1. Eur J Pharmacol. 2010;644:32–40. doi: 10.1016/j.ejphar.2010.06.069. [DOI] [PubMed] [Google Scholar]
- [31].Tan KW, Li Y, Paxton JW, Birch NP, Scheepens A. Identification of novel dietary phytochemicals inhibiting the efflux transporter breast cancer resistance protein (BCRP/ABCG2) Food Chem. 2013;138:2267–2274. doi: 10.1016/j.foodchem.2012.12.021. [DOI] [PubMed] [Google Scholar]
- [32].Gurley BJ, Swain A, Williams DK, Barone G, Battu SK. Gauging the clinical significance of P-glycoprotein-mediated herb–drug interactions: comparative effects of St. John's wort, Echinacea, clarithromycin, and rifampin on digoxin pharmacokinetics. Mol Nutr Food Res. 2008;52:772–779. doi: 10.1002/mnfr.200700081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chang C, Swaan PW. Computational approaches to modeling drug transporters. Eur J Pharm Sci. 2006;27:411–424. doi: 10.1016/j.ejps.2005.09.013. [DOI] [PubMed] [Google Scholar]
- [34].Chen L, Li Y, Yu H, Zhang L, Hou T. Computational models for predicting substrates or inhibitors of P-glycoprotein. Drug Discov Today. 2012;17:343–351. doi: 10.1016/j.drudis.2011.11.003. [DOI] [PubMed] [Google Scholar]
- [35].Ecker GF, Stockner T, Chiba P. Computational models for prediction of interactions with ABC-transporters. Drug Discov Today. 2008;13:311–317. doi: 10.1016/j.drudis.2007.12.012. [DOI] [PubMed] [Google Scholar]
- [36].Chang C, Ekins S, Bahadduri P, Swaan PW. Pharmacophore-based discovery of ligands for drug transporters. Adv Drug Deliv Rev. 2006;58:1431–1450. doi: 10.1016/j.addr.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Demel MA, Krämer O, Ettmayer P, Haaksma EEJ, Ecker GF. Predicting ligand interactions with ABC transporters in ADME. Chem Biodivers. 2009;6:1960–1969. doi: 10.1002/cbdv.200900138. [DOI] [PubMed] [Google Scholar]
- [38].Gandhi YA, Morris ME. Structure–activity relationships and quantitative structure–activity relationships for breast cancer resistance protein (ABCG2) AAPS J. 2009;11:541–552. doi: 10.1208/s12248-009-9132-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Jabeen I, Pleban K, Rinner U, Chiba P, Ecker GF. Structure–activity relationships, ligand efficiency, and lipophilic efficiency profiles of benzophenone-type inhibitors of the multidrug transporter P-glycoprotein. J Med Chem. 2012;55:3261–3273. doi: 10.1021/jm201705f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Broccatelli F, Carosati E, Neri A, Frosini M, Goracci L, Oprea TI, et al. A novel approach for predicting P-glycoprotein (ABCB1) inhibition using molecular interaction fields. J Med Chem. 2011;54:1740–1751. doi: 10.1021/jm101421d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chen L, Li Y, Zhao Q, Peng H, Hou T. adme evaluation in drug discovery. 10. Predictions of P-glycoprotein inhibitors using recursive partitioning and naive bayesian classification techniques. Mol Pharm. 2011;8:889–900. doi: 10.1021/mp100465q. [DOI] [PubMed] [Google Scholar]
- [42].Chiba P, Annibali D, Hitzler M, Richter E, Ecker G. Studies on propafenone-type modulators of multidrug resistance VI. Synthesis and pharmacological activity of compounds with varied spacer length between the central aromatic ring and the nitrogen atom. Il Farm. 1998;53:357–364. doi: 10.1016/S0014-827X(98)00035-4. [DOI] [Google Scholar]
- [43].Montanari F, Ecker GF. BCRP inhibition: from data collection to ligand-based modeling. Mol Inform. 2014;33:322–331. doi: 10.1002/minf.201400012. [DOI] [PubMed] [Google Scholar]
- [44].Warner DJ, Chen H, Cantin L-D, Kenna JG, Stahl S, Walker CL, et al. Mitigating the inhibition of human bile salt export pump by drugs: opportunities provided by physicochemical property modulation, in silico modeling, and structural modification. Drug Metab Dispos. 2012;40:2332–2341. doi: 10.1124/dmd.112.047068. [DOI] [PubMed] [Google Scholar]
- [45].Pedersen JM, Matsson P, Bergström CAS, Norinder U, Hoogstraate J, Artursson P. Prediction and identification of drug interactions with the human ATP-binding cassette transporter multidrug-resistance associated protein 2 (MRP2; ABCC2) J Med Chem. 2008;51:3275–3287. doi: 10.1021/jm7015683. [DOI] [PubMed] [Google Scholar]
- [46].Palmeira A, Rodrigues F, Sousa E, Pinto M, Vasconcelos MH, Fernandes MX. New uses for old drugs: pharmacophore-based screening for the discovery of P-glycoprotein inhibitors. Chem Biol Drug Des. 2011;78:57–72. doi: 10.1111/j.1747-0285.2011.01089.x. [DOI] [PubMed] [Google Scholar]
- [47].Pan Y, Chothe PP, Swaan PW. Identification of novel breast cancer resistance protein (BCRP) inhibitors by virtual screening. Mol Pharm. 2013;10:1236–1248. doi: 10.1021/mp300547h. [DOI] [PubMed] [Google Scholar]
- [48].Ritschel T, Hermans SMA, Schreurs M, van den Heuvel JJMW, Koenderink JB, Greupink R, et al. In silico identification and in vitro validation of potential cholestatic compounds through 3D ligand-based pharmacophore modeling of BSEP inhibitors. Chem Res Toxicol. 2014;27:873–881. doi: 10.1021/tx5000393. [DOI] [PubMed] [Google Scholar]
- [49].Zhang H, Xiang M-L, Zhao Y-L, Wei Y-Q, Yang S-Y. Support vector machine and pharmacophore-based prediction models of multidrug-resistance protein 2 (MRP2) inhibitors. Eur J Pharm Sci. 2009;36:451–457. doi: 10.1016/j.ejps.2008.11.014. [DOI] [PubMed] [Google Scholar]
- [50].Li D, Chen L, Li Y, Tian S, Sun H, Hou T. ADMET evaluation in drug discovery. 13. Development of in silico prediction models for P-glycoprotein substrates. Mol Pharm. 2014;11:716–726. doi: 10.1021/mp400450m. [DOI] [PubMed] [Google Scholar]
- [51].Wang Z, Chen Y, Liang H, Bender A, Glen RC, Yan A. P-glycoprotein substrate models using support vector machines based on a comprehensive data set. J Chem Inf Model. 2011;51:1447–1456. doi: 10.1021/ci2001583. [DOI] [PubMed] [Google Scholar]
- [52].Hazai E, Hazai I, Ragueneau-Majlessi I, Chung SP, Bikadi Z, Mao Q. Predicting substrates of the human breast cancer resistance protein using a support vector machine method. BMC Bioinformatics. 2013;14:130. doi: 10.1186/1471-2105-14-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Pinto M, Trauner M, Ecker GF. An in silico classification model for putative ABCC2 substrates. Mol Inform. 2012;31:547–553. doi: 10.1002/minf.201200049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Szakács G, Annereau J-P, Lababidi S, Shankavaram U, Arciello A, Bussey KJ, et al. Predicting drug sensitivity and resistance: profiling ABC transporter genes in cancer cells. Cancer Cell. 2004;6:129–137. doi: 10.1016/j.ccr.2004.06.026. [DOI] [PubMed] [Google Scholar]
- [55].Matsson P, Pedersen JM, Norinder U, Bergström CAS, Artursson P. Identification of novel specific and general inhibitors of the three major human ATP-binding cassette transporters P-gp, BCRP and MRP2 among registered drugs. Pharm Res. 2009;26:1816–1831. doi: 10.1007/s11095-009-9896-0. [DOI] [PubMed] [Google Scholar]
- [56].Bento AP, Gaulton A, Hersey A, Bellis LJ, Chambers J, Davies M, et al. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 2014;42:D1083–D1090. doi: 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Zhou Z, et al. PubChem's bioassay database. Nucleic Acids Res. 2012;40:D400–D412. doi: 10.1093/nar/gkr1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zdrazil B, Pinto M, Vasanthanathan P, Williams AJ, Balderud LZ, Engkvist O, et al. Annotating human P-glycoprotein bioassay data. Mol Inform. 2012;31:599–609. doi: 10.1002/minf.201200059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bentz J, O'Connor MP, Bednarczyk D, Coleman J, Lee C, Palm J, et al. Variability in P-glycoprotein inhibitory potency (IC50) using various in vitro experimental systems: implications for universal digoxin drug–drug interaction risk assessment decision criteria. Drug Metab Dispos. 2013;41:1347–1366. doi: 10.1124/dmd.112.050500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Balimane PV, Marino A, Chong S. P-gp inhibition potential in cell-based models: which “calculation” method is the most accurate? AAPS J. 2008;10:577–586. doi: 10.1208/s12248-008-9068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Giri N, Agarwal S, Shaik N, Pan G, Chen Y, Elmquist WF. Substrate-dependent breast cancer resistance protein (Bcrp1/Abcg2)-mediated interactions: consideration of multiple binding sites in in vitro assay design. Drug Metab Dispos. 2009;37:560–570. doi: 10.1124/dmd.108.022046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Shintre CA, Pike ACW, Li Q, Kim J-I, Barr AJ, Goubin S, et al. Structures of ABCB10, a human ATP-binding cassette transporter in apo- and nucleotide-bound states. Proc Natl Acad Sci. 2013;110:9710–9715. doi: 10.1073/pnas.1217042110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Brice MD, Rodgers JR, et al. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- [64].Jin MS, Oldham ML, Zhang Q, Chen J. Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature. 2012;490:566–569. doi: 10.1038/nature11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, et al. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ward AB, Szewczyk P, Grimard V, Lee C-W, Martinez L, Doshi R, et al. Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain. Proc Natl Acad Sci. 2013;110:13386–13391. doi: 10.1073/pnas.1309275110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Li J, Jaimes KF, Aller SG. Refined structures of mouse P-glycoprotein. Protein Sci. 2014;23:34–46. doi: 10.1002/pro.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Dawson RJP, Locher KP. Structure of the multidrug ABC transporter Sav 1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett. 2007;581:935–938. doi: 10.1016/j.febslet.2007.01.073. [DOI] [PubMed] [Google Scholar]
- [69].Dawson RJP, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- [70].Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc Natl Acad Sci. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Hohl M, Briand C, Grütter MG, Seeger MA. Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation. Nat Struct Mol Biol. 2012;19:395–402. doi: 10.1038/nsmb.2267. [DOI] [PubMed] [Google Scholar]
- [72].McDevitt CA, Callaghan R. How can we best use structural information on P-glycoprotein to design inhibitors? Pharmacol Ther. 2007;113:429–441. doi: 10.1016/j.pharmthera.2006.10.003. [DOI] [PubMed] [Google Scholar]
- [73].Dolghih E, Bryant C, Renslo AR, Jacobson MP. Predicting binding to P-glycoprotein by flexible receptor docking. PLoS Comput Biol. 2011;7:e1002083. doi: 10.1371/journal.pcbi.1002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Mahar Doan KM, Humphreys JE, Webster LO, Wring SA, Shampine LJ, Serabjit-Singh CJ, et al. Passive permeability and P-glycoprotein-mediated efflux differentiate central nervous system (CNS) and non-CNS marketed drugs. J Pharmacol Exp Ther. 2002;303:1029–1037. doi: 10.1124/jpet.102.039255. [DOI] [PubMed] [Google Scholar]
- [75].Klepsch F, Chiba P, Ecker GF. Exhaustive sampling of docking poses reveals binding hypotheses for propafenone type inhibitors of P-glycoprotein. PLoS Comput Biol. 2011;7:e1002036. doi: 10.1371/journal.pcbi.1002036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Klepsch F, Vasanthanathan P, Ecker GF. Ligand and structure-based classification models for prediction of P-glycoprotein inhibitors. J Chem Inf Model. 2014;54:218–229. doi: 10.1021/ci400289j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ferreira RJ, Ferreira M-JU, dos Santos DJVA. Molecular docking characterizes substrate-binding sites and efflux modulation mechanisms within P-glycoprotein. J Chem Inf Model. 2013;53:1747–1760. doi: 10.1021/ci400195v. [DOI] [PubMed] [Google Scholar]
- [78].Ferreira RJ, Ferreira M-JU, dos Santos DJVA. Insights on P-glycoprotein's efflux mechanism obtained by molecular dynamics simulations. J Chem Theory Comput. 2012;8:1853–1864. doi: 10.1021/ct300083m. [DOI] [PubMed] [Google Scholar]
- [79].Ferreira RJ, dos Santos DJVA, Ferreira M-JU, Guedes RC. Toward a better pharmacophore description of P-glycoprotein modulators, based on macrocyclic diterpenes from Euphorbia species. J Chem Inf Model. 2011;51:1315–1324. doi: 10.1021/ci200145p. [DOI] [PubMed] [Google Scholar]
- [80].Kim J, Wu S, Tomasiak TM, Mergel C, Winter MB, Stiller SB, et al. Subnanometre-resolution electron cryomicroscopy structure of a heterodimeric ABC exporter. Nature. 2014;517:396–400. doi: 10.1038/nature13872. (advance online, publication) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Wen P-C, Verhalen B, Wilkens S, Mchaourab HS, Tajkhorshid E. On the origin of large flexibility of P-glycoprotein in the inward-facing state. J Biol Chem. 2013;288:19211–19220. doi: 10.1074/jbc.M113.450114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Rauch C, Paine SW, Littlewood P. Can long range mechanical interaction between drugs and membrane proteins define the notion of molecular promiscuity? Application to P-glycoprotein-mediated multidrug resistance (MDR) Biochim Biophys Acta Gen Subj. 2013;1830:5112–5118. doi: 10.1016/j.bbagen.2013.06.038. [DOI] [PubMed] [Google Scholar]
- [83].Copeland RA, Pompliano DL, Meek TD. Drug–target residence time and its implications for lead optimization. Nat Rev Drug Discov. 2006;5:730–739. doi: 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- [84].Clay AT, Sharom FJ. Lipid bilayer properties control membrane partitioning, binding, and transport of p-glycoprotein substrates. Biochemistry (Mosc) 2013;52:343–354. doi: 10.1021/bi301532c. [DOI] [PubMed] [Google Scholar]
- [85].Sedykh A, Fourches D, Duan J, Hucke O, Garneau M, Zhu H, et al. Human intestinal transporter database: QSAR modeling and virtual profiling of drug uptake, efflux and interactions. Pharm Res. 2013;30:996–1007. doi: 10.1007/s11095-012-0935-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Michl J, Scharinger C, Zauner M, Kasper S, Freissmuth M, Sitte HH, et al. A multivariate approach linking reported side effects of clinical antidepressant and antipsychotic trials to in vitro binding affinities. Eur Neuropsychopharmacol. 2014;24:1463–1474. doi: 10.1016/j.euroneuro.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]