Abstract
The human multispecific drug efflux transporter P-glycoprotein (P-gp) causes drug resistance and modulates the pharmacological profile of systemically administered medicines. It has arisen from a homodimeric ancestor by gene duplication. Crystal structures of mouse MDR1A indicate that P-gp shares the overall architecture with two homodimeric bacterial exporters, Sav1866 and MsbA, which have complete rotational symmetry. For ATP-binding cassette transporters, nucleotide binding occurs in two symmetric positions in the motor domains. Based on the homology with entirely symmetric half-transporters, the present study addressed the key question: can biochemical evidence for the existence of dual drug translocation pathways in the transmembrane domains of P-gp be found? P-gp was photolabeled with propafenone analogs, purified, and digested proteolytically, and peptide fragments were identified by high-resolution mass spectrometry. Labeling was assigned to two regions in the protein by projecting data into homology models. Subsequently, symmetric residue pairs in the putative translocation pathways were identified and replaced by site-directed mutagenesis. Transport assays corroborated the existence of two pseudosymmetric translocation pathways. Although rhodamine123 has a preference to take one path, verapamil, propafenones, and vinblastine preferentially use the other. Two major findings ensued from this study: the existence of two solute translocation pathways in P-gp as a reflection of evolutionary origin from a homodimeric ancestor and selective but not exclusive use of one of these pathways by different P-gp solutes. The pseudosymmetric behavior reconciles earlier kinetic and thermodynamic data, suggesting an alternative concept of drug transport by P-gp that will aid in understanding the off-target quantitative structure activity relationships of P-gp interacting drugs.
Introduction
P-glycoprotein (P-gp, ABCB1) is a primary active multispecific drug efflux transporter that is considered a major determinant of the pharmacokinetic, safety, and efficacy profile of drugs. Recent recommendations of the International Transporter Consortium (Giacomini et al., 2010) have incorporated a decision tree for determination of P-gp solute properties during early drug development. P-glycoprotein is an ATP-binding cassette (ABC) transporter, which uses the energy of nucleotide binding and hydrolysis to prevent xenotoxic compounds from entering the cell. It is composed of two transmembrane domains, which are responsible for translocation of solutes, and two nucleotide binding domains, which provide the energy for outward transport of solutes. Recently published crystal structures of mouse MDR1A (Aller et al., 2009) indicate that P-gp shares the twisted topology of Sav1866, a homodimeric bacterial multidrug transporter (Dawson and Locher, 2006, 2007). A common architecture is also observed with the corrected structures of the bacterial lipid A transporter MsbA (Ward et al., 2007).
Although our understanding of ATP binding and hydrolysis has improved through the availability of a number of structurally resolved ABC importers and isolated nucleotide binding domains (NBDs) of ABC exporters (for review, see Seeger and van Veen, 2009), the mechanism of solute binding and transport remains elusive. Two crystal structures of mouse P-gp cocrystallized in the presence of the cyclic hexapeptide inhibitors QZ59-SSS and QZ59-RRR (PDB IDs 3G60 and 3G61) show these ligands to be located at the interface of the two transmembrane domains (TMDs) in either a central position (QZ59-RRR, between helices 6 and 12) or in two asymmetric positions (QZ59-SSS) termed “upper” and “lower” sites, corroborating the notion that binding of solutes takes place at the interface of the two transmembrane domains (Aller et al., 2009). The wide-open, inward-facing, inverted-V conformation of this structure, in which the NBDs are at a distance of 20 Å, is binding competent but might not correspond to a physiological state, because P-gp cross-linked between residues 175 and 820 in the cytoplasmic portion of transmembrane helices 3 and 9 is able to hydrolyze ATP in the absence of solutes. In addition, basal ATPase activity is stimulated by solutes, indicating that under conditions in which the NBDs cannot disassociate completely, the transporter is still able to bind drugs (Loo et al., 2010).
It is generally believed that the central pore represents an expansive binding pocket for solutes, which accommodates ligands by interaction with a large complement of amino acid residues (Loo and Clarke, 2008; Loo et al., 2009). During exit, solutes take a translocation path, which requires at least the following steps: 1) entry of the solute into the translocation pathway from the inner leaflet; 2) a conformational rearrangement of the transporter (the rate-limiting step, which provides access of the solute to the alternate side of the membrane); and 3) exit of the solute into the cell exterior. This process is powered by binding and hydrolysis of ATP.
Photolabeling with propafenone analogs in the nucleotide free state was previously used to probe the binding sites of P-gp (Pleban et al., 2005). Photolabeling of the benzophenone biradical species proceeds via reaction with any closely located C-H bonds in the protein. In the present study, these data were projected into models of human P-gp in the nucleotide-bound and free states on the basis of crystal structures of Sav1866 (Dawson and Locher, 2006) and mouse P-gp (Aller et al., 2009). Photolabeling is expected to capture the step of carrier reorientation, which may be considered the most highly populated conformational state of the protein. In all models, the C-α distance between the most intensely labeled residues in TM helices 5 and 11 was found to be 23 to 32 Å. Given the size of both the amino acid side chains and the photoactive carbonyl group in the ligand, this distance between the two photolabeled regions is best explained by the existence of two pseudosymmetric rather than one single solute translocation pathway for propafenones.
In further support of this model, we generated transporter mutants in which access of positively charged compounds to one of the two photolabeled regions was prevented by introducing single arginine residues in each of the putative translocation paths in pseudosymmetric positions of the protein. Our results are compatible with a model in which the preferred translocation path for rhodamine123 takes it into vicinity of residue Gln773 in helix 8, whereas verapamil, vinblastine, and propafenone analogs preferentially take a path that brings them close to residue Gln132 in helix 2. In the wild-type transporter, residues Gln773 and Gln132 do not interact with solutes but arginine substitutions cause diagnostic changes in transport or inhibitory potencies of positively charged compounds. Interaction of uncharged compounds with P-gp was not affected by arginine substitutions. This confirmed that mutation of Gln residues at positions 132 and 773 did not disrupt the molecular mechanics of the transporter.
Materials and Methods
Sequence Alignments and Homology Modeling
Multiple sequence alignments of ABCB proteins were performed using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The models of human P-gp were based on the crystal structures of Sav1866 (PDB ID 2HYD; 3.0 Å resolution) and of mouse P-gp (PDB ID 3G5U; 3.8 Å resolution) (Aller et al., 2009) using MODELLER (version 9v2) (Sali and Blundell, 1993; Martí-Renom et al., 2000) in the automodel protocol with the refinement set to “very_slow.” Twenty models were generated with each of the templates. The data-driven models based on Sav1866 (Dawson and Locher, 2006) (PDB ID 2HYD) have been described previously (Stockner et al., 2009) (Protein Model Database ID PM0075213). Model quality was assessed using the discrete optimized protein energy (Shen and Sali, 2006) and GA341 scores (Melo and Sali, 2007) of MODELLER, ProQ (Wallner and Elofsson, 2003), and ProCheck (Laskowski et al., 1993). The models with the lowest MODELLER objective function were selected for visualization. Propafenone labeling data (Pleban et al., 2005) were mapped onto the human P-gp structure by adding labeling intensity on a per-residue basis.
Construction of P-gp Mutants
Site-directed mutagenesis was performed at positions 132 and 773 of hexahistidine-tagged human P-gp cloned into the entry vector pENTR4-MDR1-His6 to generate the Q132A, Q132R, Q773A, Q773R, and the Q132A/Q773A mutants using the following forward and reverse primers: Q132A: forward, 5′-CTG CTT ACA TTG CGG TTT CAT TTT GG-3′; reverse, 5′-CCA AAA TGA AAC CGC AAT GTA AGC AG-3′; Q132R: forward, 5′-GCT GCT TAC ATT CGT GTT TCA TTT TG-3′; reverse, 5′-CAA AAT GAA ACA CGA ATG TAA GCA GC-3′; Q773A: forward, 5′-CAT TTT TCC TTG CGG GTT TCA CAT TTG GC-3′; reverse, 5′-GCC AAA TGT GAA ACC CGC AAG GAA AAA TG-3′; Q773R: forward, 5′-CAT TTT TCC TTC GAG GTT TCA CAT TTG-3′; reverse, 5′-CAT TTT TCC TTC GAG GTT TCA CAT TTG-3′. Double mutant Q132R/Q773R was generated by restriction digestion of pENTR4-Q132R and pENTR4-Q773R with SalI and EcoRI and subsequent ligation of the mutated fragment from pENTR4-Q773R into pENTR4-Q132R using T4 DNA ligase (rapid DNA ligation kit; Fermentas, Vienna, Austria). The wild-type and mutant P-gp were then transferred to the pCEP4 destination vector using gateway cloning (Hartley et al., 2000).
Expression of P-gp Mutants
Wild-type or mutant P-gp was transiently expressed in HEK-293/EBNA cells (obtained from the University of Bielefeld, Germany). When cells reached confluence, they were trypsinized, harvested, and centrifuged at 500g. The cell pellet was incubated on ice water with the mouse monoclonal—P-gp-specific—MRK16 antibody (5 µg/ml; Kamiya Biomedical Company, Seattle, WA) and IgG2A (2.5 µg/ml) as the control antibody for 30 min. Cells were then washed with ice-cold phosphate-buffered saline and centrifuged at 500g followed by a 30-min incubation on ice water with a fluorescein isothiocyanate-labeled goat anti-mouse secondary antibody (12.5 µg/ml). After incubation, cells were again washed with ice-cold PBS and centrifuged. Cells were then resuspended in DMEM and measured in a FACSCalibur flow cytometer (BD Biosciences, Vienna, Austria).
Rhodamine123 Efflux Assays
Transiently transfected HEK-293/EBNA cells were trypsinized, centrifuged at 500g and the cell pellets were resuspended in DMEM containing rhodamine123 (Sigma Chemical., St. Louis, MO) at a final concentration of 0.2 µg/ml (0.53 µM). Cell suspensions were incubated at 37°C for 30 min. Preloading of cells at 37°C was selected because the compound is not taken up to any significant extent on ice water. Tubes were chilled on ice and cells were harvested at 500g in an Eppendorf 5403 centrifuge (Eppendorf AG, Hamburg, Germany). The cell pellet was washed with ice-cold DMEM, pH 7.4, and centrifuged at 500g. Supernatants were removed, and the cell pellet was resuspended in DMEM, pH 7.4, prewarmed to 37°C. After 60, 120, 180, 240, and 300 s, aliquots of the incubation mixture were transferred to tubes containing an equal volume of ice-cold stop solution (DMEM containing 1-(2-(3-(4-(4-fluorophenyl)piperazin-1-yl)-2-hydroxypropoxy)phenyl)-3-phenylpropan-1-one (GPV31) at a final concentration of 5 µM). Aliquots drawn at the respective time points were kept in an ice water bath and measured within 1 h on a FACSCalibur flow cytometer. Viable cells were selected by setting appropriate gates for forward and side scatter. The excitation and emission wavelengths were 488 nm and 534 nm, respectively.
Inhibition Assays
IC50 values for rhodamine123 efflux inhibition were determined as reported previously (Chiba et al., 1996). In brief, cells were sedimented, the supernatant was removed by aspiration, and the cells were resuspended at a density of 106/ml in DMEM containing rhodamine123 (Sigma Chemical, St. Louis, MO) at a final concentration of 0.2 µg/ml (0.53 µM). Cell suspensions were incubated at 37°C for 30 min. Tubes were chilled on ice and centrifuged at 500g in an Eppendorf 5403 centrifuge (Eppendorf AG). Supernatants were removed, and the cell pellet was resuspended in medium prewarmed to 37°C containing either no inhibitor or compounds at various concentrations ranging from 8 nM to 50 µM, depending on the solubility and expected potency of the inhibitor. Eight concentrations (serial 1:2.5 dilution) were tested for each inhibitor. After 60, 120, 180, and 240 s, aliquots of the incubation mixture were transferred to tubes containing an equal volume of ice-cold stop solution (DMEM containing GPV31 at a final concentration of 5 µM). Zero time points were determined by immediately pipetting rhodamine123-preloaded cells into ice-cold stop solution. Samples drawn at the respective time points were kept in an ice water bath and measured within 1 h on a FACSCalibur flow cytometer. Viable cells were selected by setting appropriate gates for forward and side scatter. The excitation and emission wavelengths were 488 and 534 nm, respectively. Five thousand gated events were accumulated for the determination of mean fluorescence values. Exponentials were fitted to the data points taken at different times allowing estimation of the flux rate for the zero time point. The experimental protocol would not require normalization for protein expression, because IC50 values, in contrast to first-order rate constants, are independent of protein expression. First-order rate constants were then used to generate concentration response curves from which the IC50 values were calculated as 50% occupancy values.
Data Analysis
Concentration-response curves for inhibitors [verapamil, GPV31, N-(2-hydroxy-3-(2-(3-phenylpropanoyl)phenoxy)-propyl)-N-propylbenzamide (GPV366)] were fitted to a four-parameter logistic equation (i.e., the Hill equation with a term for background diffusion) using a Marquardt-Levenberg algorithm. In those instances, where the slope factor (Hill coefficient) differed from 1, the data points were also subjected to nonlinear least-squares curve-fitting to equations describing the inhibition via one and two sites. A statistically significant improvement by the two-site model was verified by an F-test based on the extra sum of squares principle. Averages of IC50 values were compared using one-way analysis of variance applying Duncan’s multiple range test in SPSS 10.0 (SPSS Inc., Chicago, IL). A P value of 0.05 was considered significant. Data are represented as mean ± S.E.
Results
Photolabeling of P-gp with Propafenone Analogs
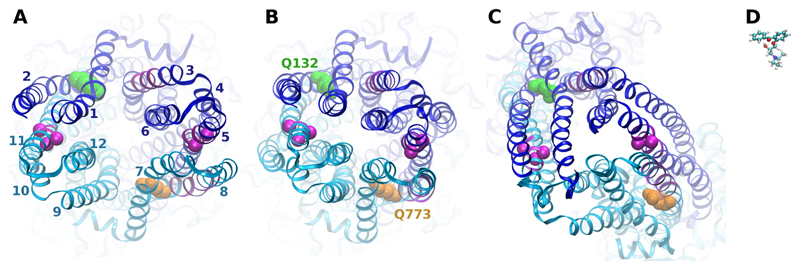
Previously obtained photolabeling data (Pleban et al., 2005) were projected into models of human P-glycoprotein based on the template structures of Sav1866 in the nucleotide-bound state (Fig. 1A) and a previously published data-driven model of P-gp based on the same template, in which the diverging helical bundles in the transmembrane domains were rotated toward each other (Stockner et al., 2009) (Protein Model Database ID PM0075213; Fig. 1B). A third model was based on the recently published mouse MDR1A crystal structure in the nucleotide free state (Fig. 1C) (Aller et al., 2009). Quality parameters for these models are given in Table 1. The quality of all models was high according to the parameters described in the legend for Table 1. C-α atoms of the most intensely photolabeled residues Ala311 and Leu312 (Fig. 1, A–C, right magenta spheres) and Phe951, Ser952, and Tyr953 (Fig. 1, A–C, left magenta spheres) are shown in van der Waals rendering. In all three models, the distances between these residues exceeded the size of the affinity ligands, suggesting that these amino acid residues were unable to react simultaneously with a single propafenone molecule. Models thus indicate that binding of propafenone analogs occurs in two separate pseudosymmetric regions at the interface of the TMDs, involving helix 5 and adjacent helix 8, as well as helix 11. The C-α distance between photolabeled residues is 32 Å in the nucleotide-bound state (Fig. 1A), 24 Å in the data-driven model (Fig. 1B), and 23 Å in the model based on mouse MDR1A (Fig. 1C). For comparison, a propafenone [(2-(3-(diethylamino)-2-hydroxypropoxy)phenyl)-(phenyl)methanone (GPV51)] (Pleban et al., 2005) sized to the same scale as the models is shown in a ball-and-stick representation in Fig. 1D.
Fig. 1.
Projection of photolabeling data into homology models. Top view of P-gp homology models based on the Sav1866 template structure (A), a data-driven P-gp model (B), and mouse MDR1A (C). Photolabeled residues are depicted in magenta. The N-terminal half of the protein is depicted in blue, and the C-terminal half is cyan. C-α atoms of most intensely labeled residues are depicted as magenta spheres. Mutated residues 132 and 773 are depicted in green and orange van der Waals rendering, respectively. D, photoactive propafenone analog GP51 in ball-and-stick representation sized to same scale as the models.
Table 1. Quality of models as assessed by Modeller (Shen et al., 1995; Shen and Sali, 2006; Melo and Sali, 2007), ProQ (Wallner and Elofsson, 2003), and Procheck (Laskowski et al., 1993).
The MODELLER DOPE-score is a statistical potential energy-score trained to recognize correct folding by analyzing nonbonding interactions between atoms in a pair-wise fashion. The more negative the number, the higher the likelihood that a correctly folded model was obtained. The GA341 score is a statistical potential for correct local model geometry. Four parameters contribute to this score: backbone ϕ and ψ angles, atom overlap, atom probability density function, and exposed surface area. The score ranges between 0 and 1, where 1 is indicative of a correctly folded model. A ProQ LG score above 1.5 is indicative of a correct model; above 3, a good model; and above 5, a very good model. A MaxSub score >0.1 indicates a correct model, a score of >0.5 shows a good model, and >0.8 a very good model, whereby the LG score performs better with larger proteins such as P-gp and the MaxSub score is optimized for shorter proteins. ProCheck classifies dihedral angles in favorable, allowed, generously allowed, and disallowed regions of a Ramachandran plot.
| Model |
|||
|---|---|---|---|
| Sav 1866-Based | Data-Driven | MDR1A-Based | |
| Modeller | |||
| DOPE | −140,414 | −149,326 | −130,736 |
| GA341 score | 1.0 | 1.0 | 1.0 |
| ProQ | |||
| LGscore | 4.78 | 5.62 | 3.82 |
| MaxSub | 0.30 | 0.31 | 0.25 |
| ProCheck | |||
| Most favorable | 94.3% | 94.3% | 86.2% |
| Additionally allowed | 5.2% | 5.3% | 10.3% |
| Generously allowed | 0.4% | 0.2% | 2.2% |
| Disallowed | 0.1% | 0.2% | 1.3% |
The irradiation-induced biradical species of the carbonyl group in the benzophenone moiety shows a preference for methionine residues (Dormán and Prestwich, 1994). Helices 5 and 8, as well as their immediate vicinity, are devoid of methionine residues; thus, a reaction preference attributable to the presence of methionines can be ruled out. In contrast, helix 11, which lies in a pseudosymmetric position with respect to helix 5, contains two methionine residues that are accessible to the ligands. The presence of these methionine residues in TM11 might explain that the adjacent helix 2, in contrast to helix 8 in the N terminus, is not strongly labeled. The area between the two photolabeled regions, made up mainly of helices 6 and 12, does not show labeling, indicating that the photoactive group is not oriented toward these helices. Additional labeling was observed for residue Met197 and its vicinity in TM3. This helix is not part of either of the two photolabeled regions discussed above. Met197, however, is predicted by the homology model to be membrane-exposed and close to one of the portals of P-gp in the nucleotide-free crystal structure (Aller et al., 2009). Labeling of this residue is thus indicative of a path—albeit conjectural—along which solutes might access the central cavity of the transporter from the inner leaflet of the membrane.
Analysis of Photolabeling Data and Design of Mutants
Would data discussed in the previous section be indicative of the existence of two pseudosymmetric transport pathways, and could these paths be dissected biochemically? Many of the more than 1000 known P-gp solutes are protonable compounds. We therefore engineered P-gp arginine mutants in which the positive charge of the amino acid residue was expected to block the transport of positively charged solutes when positioned in or close to the respective transport path. Uncharged compounds served as controls for an unaltered functionality of the transporter.
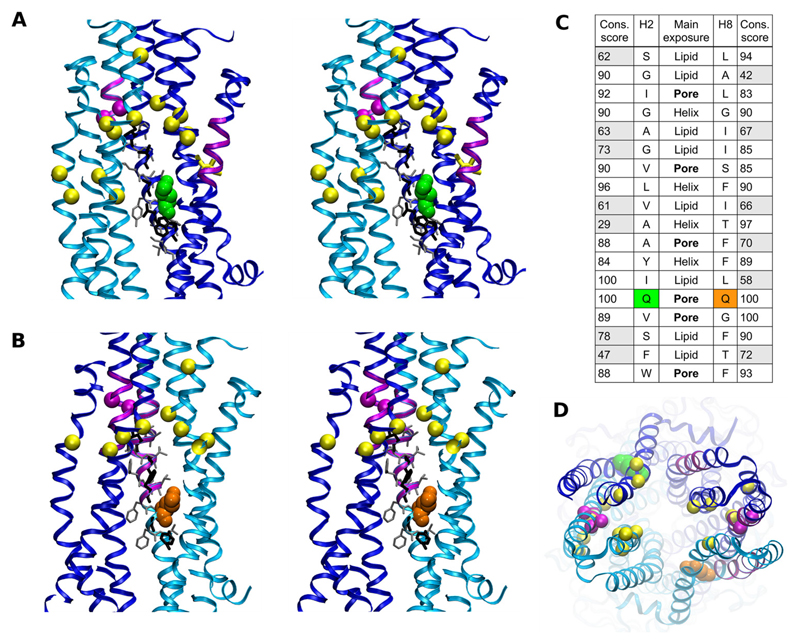
Selection of candidate residues for site directed mutagenesis was based on the following criteria: 1) pore exposure, 2) conservation in symmetric positions, 3) predicted location within the inner leaflet; and 4) a position that lies within the putative transport path and is proximal to the photolabeled regions. Two polar glutamine residue pairs, Gln132 (TM2)/Gln773 (TM8) and Gln195 (TM3)/Gln838 (TM9) were compliant with those criteria. The first of these two pairs was selected because residues are closer to the photolabeled regions in helices 5/8 and 11 and conserved in all annotated P-gp sequences but not in all ABCB family members. Dictyostelium discoideum ABCB subfamily full transporters [GenBank numbers AAL74249 (gi18496816) and AAL74250 (gi18496818)] contain an arginine residue in a position aligning with Gln773 of human ABCB1. In addition, the D. discoideum half-transporters ABCB5 and -6 (Q819W2, Q54RU1) contain an arginine residue in a position aligning with residue Gln132 of the human ABCB1 sequence and Xenopus tropicalis subfamily B member 4 [GenBank number AAH63924 (gi39645391)] contains an arginine residue in a position aligning with Gln773 in human P-gp. Therefore, it was expected that an arginine residue would be tolerated in positions 132 and 773. Figure 2 shows stereo images of the Sav1866-based P-gp model, in which a transverse cross-section along the central axis of the pore allows a view from the pore, centered on the helix 2/11 interface in Fig. 2A and the helix 5/8 interface in Fig. 2B. Figures are rotated 180° with respect to each other. The mutated residues Gln132 (green) and Gln773 (orange) are shown to be pore-exposed. C-α atoms of most intensely photolabeled residues are shown as magenta spheres. The ClustalW conservation score and relative orientation of residues in TM helices 2 and 8 are shown in Fig. 2C. The only completely conserved and aligning residues in annotated P-gp sequences are Gln132 and Gln773. In addition, they constitute the only polar residue pair in helices 2 and 8 for which side chains are predicted to be pore-exposed. It is noteworthy that these residues are located inside the pore and not membrane-exposed. Therefore, they are not located at putative gates of the protein. Figure 2D shows a top view of a Sav1866-based model of P-gp with residues in color identical to that in Fig. 2, A and B.
Fig. 2.
Stereo images of transversal cross sections of P-gp along the central axis of the pore. Images have to be viewed in parallel eye view. A, view on the helix 2/11 interface; residue Gln132 is green; photolabeled residues in magenta (refer to Fig. 1). Yellow spheres represent C-α atoms of residues identified to be involved in verapamil binding (Loo and Clarke, 2008). B, side view on the helix 5/8 interface. Residue Gln773 is depicted in an orange van der Waals rendering. Photolabeled residues are again shown in magenta. Yellow spheres indicate C-α atoms of residues involved in verapamil binding. The N-terminal half of the protein is depicted in blue, and the C-terminal half is cyan. A and B provide views that are at an angle of 180° to each other. C, sequence alignment of helices 2 and 8 with conservation score in multiple sequence alignments of all annotated P-gp sequences as well as exposure to lipid, pore, or toward other helices. D, top view of a Sav1866 based homology model of P-gp. Residues are colored as in Fig. 2, A and B.
Rhodamine123 Efflux in Q132R/A and Q773R/A Mutants
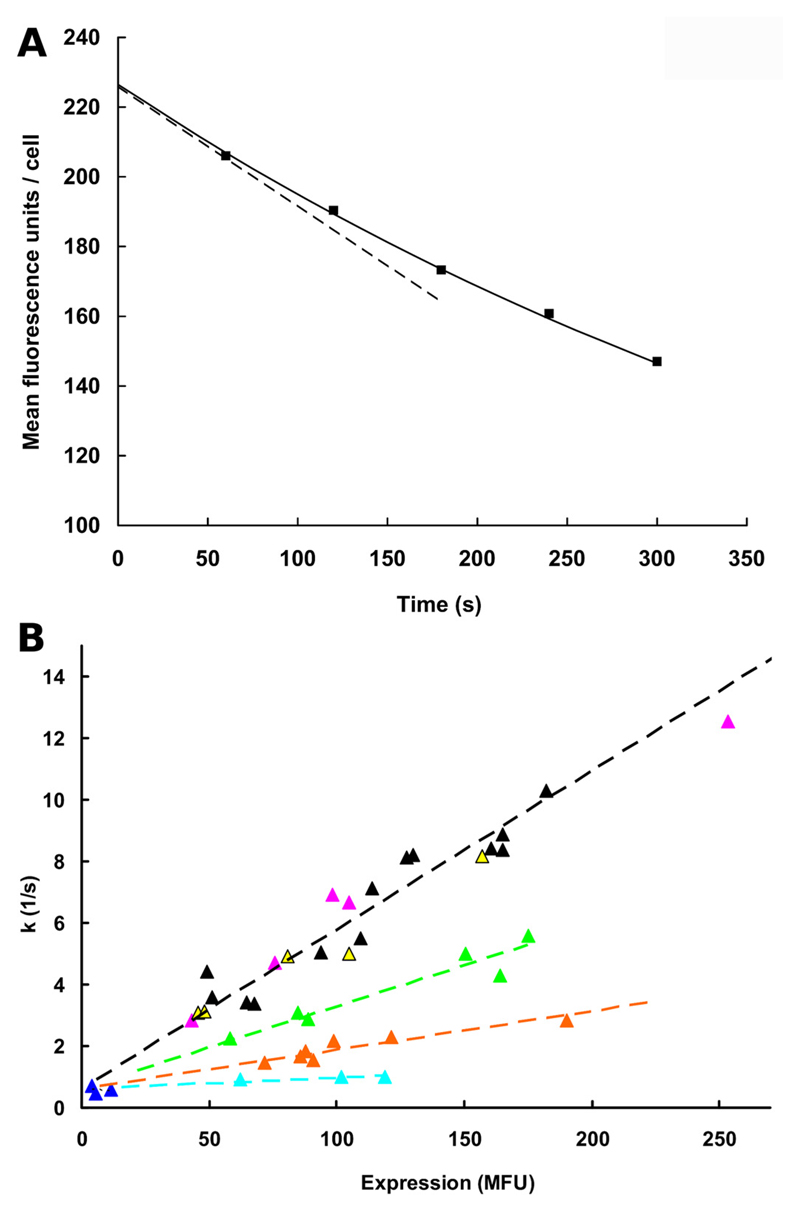
In an initial series of experiments, we monitored rhodamine123 efflux in HEK-293/EBNA cells transiently transfected with either wild-type P-gp or the Q132R and Q773R mutants. The previously described zero trans-efflux protocol was used (Chiba et al., 1996) for the determination of first-order rate constants (FORCs). A representative single experiment for rhodamine123 efflux in the Q773R mutant is shown in Fig. 3A. An exponential curve was fitted to data points taken at 1, 2, 3, 4, and 5 min. Details are given in the legend to Fig. 3A. FORCs depend on protein expression rates, which differ between individual experiments and between wt and mutants. Therefore, FORCs, determined as described in the legend to Fig. 3A, were plotted as a function of expression in Fig. 3B. Data points for cells transfected with wild-type P-gp are shown as black triangles. The slope of the linear regression line (black stippled line) reflects the transport rate for these cells. Introduction of an arginine residue at position 132 (green triangles) decreased transport rates to 52 ± 13% of wild-type (green stippled line), whereas replacement of Gln773 by arginine (orange data points, orange stippled line) reduced transport rates to 24 ± 6%. A comparable decrease in transport activity was also observed at a rhodamine123 concentration of 5 µM, indicating the reaction conditions to be first order (data not shown). A mutation of both glutamine residues to arginine resulted in complete loss of transport activity (Fig. 3B, cyan data points, cyan stippled line), even though the protein was expressed at the cell surface. The residual rate is similar for all data points irrespective of expression rate (0.0007–0.0008/s) and reflects outward diffusion of rhodamine123 from cells. As a control, mock-transfected cells were used (blue data points). These show a low flux rate corresponding to diffusion. Replacement of glutamine residues in either position 132 (yellow triangles) or 773 (magenta triangles) with alanine residues did not affect rhodamine123 transport, thereby ruling out a direct interaction of these glutamine residues with the solutes (Fig. 3B).
Fig. 3.
Transport activity of wild-type and mutants as measured by rhodamine123 efflux. A, Rhodamine123 efflux is shown for the Q773R mutant as a representative example. An exponential curve was fitted to the data points taken at 1, 2, 3, 4, and 5 min according to the equation y(MFU/cell) = a × e−k · t + c, where a is initial loading, e is Euler’s number, k is the rate constant, t is the time in minutes, MFU is mean fluorescence units, and c is background fluorescence of the cells. The rate constant corresponds to the slope in the origin of the curve (−k · a, stippled black line) normalized to 1. Therefore, this rate constant is independent of initial loading (a), which is different for wild type and mutants and also between different experiments. The rate constant was measured in first order and thus represents a first-order rate constant, which for each data point was determined in duplicate and plotted on the ordinate in Fig. 3B. B, first-order rate constants were plotted as a function of P-gp surface expression, determined by MRK16 staining. For this figure, a total number of 42 efflux experiments were performed in duplicate. In parallel, protein expression was determined. The slope of each linear regression line is a measure of relative transport activity (stippled lines). A linear least-squares fit of wild-type P-gp (black) is shown by the stippled black line. Negative controls (blue triangles) lie close to that line with a y-offset corresponding to diffusion and an x-offset corresponding to background fluorescence of nonexpressing cells. Mutants Q132A (yellow) and Q773A (magenta) lie close to the stippled black line and therefore show transport rates that are comparable with wild-type. Q132R (green) and Q773R (orange) show transport rates of 52 ± 13 and 24 ± 6% of wild type, respectively (error propagation accounted for). The double mutant Q132R/Q773R (cyan) shows flux rates that are equal to simple diffusion (0.7– 0.8/s).
Effect of Propafenone Analogs on Rhodamine123 Efflux in Q132R/A and Q773R/A Mutants
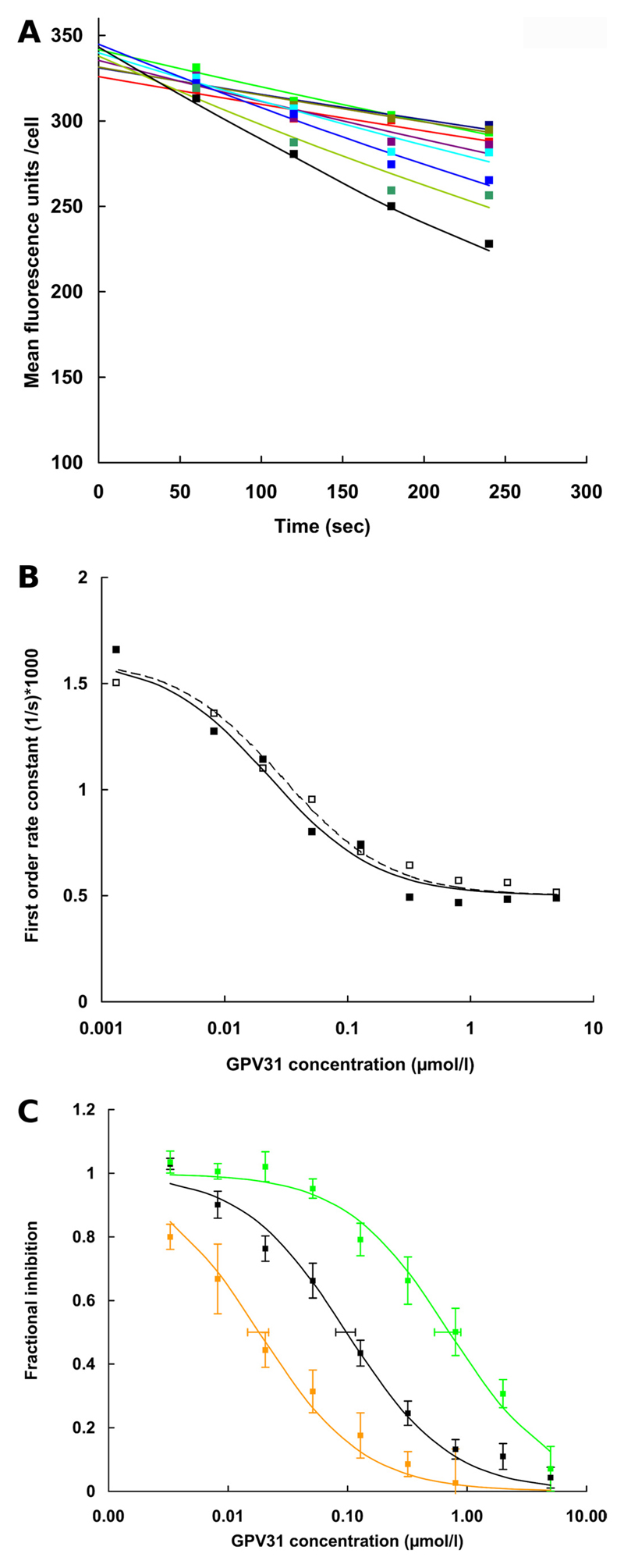
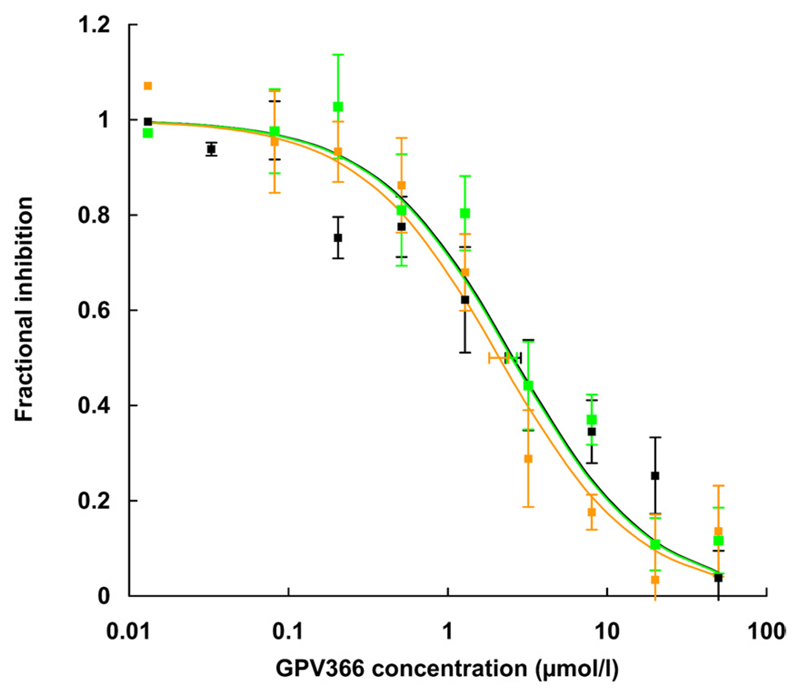
Propafenone analogs are solutes and inhibitors of P-gp. These were evaluated based on experimentally determined IC50 values for the inhibition of rhodamine123 transport, because a direct measurement of P-gp mediated propafenone flux rates is not possible as a result of rapid membrane permeation by diffusion (Schmid et al., 1999). Most propafenone analogs contain a tertiary nitrogen atom that becomes positively charged upon protonation. One of these compounds, GPV31, a p-fluorophenylpiperazine analog of propafenone (Chiba et al., 1996) was used first. For each concentration of GPV31, a time-dependent decrease in mean cellular fluorescence was monitored over 4 min. A representative experiment for the Q773R mutant is shown in Fig. 4A. Details are described in the figure legend. FORCs for controls and different GPV31 concentrations were then plotted as a function of the inhibitor concentration. A representative concentration-response curve determined in duplicate is shown in Fig. 4B. A composite graph of the results of six independent experiments performed in duplicate is shown in Fig. 4C. For each data point, the mean ± S.E. is given. Data are represented as fractional inhibition values to enable comparison of values from different experiments. The IC50 value for wild-type P-gp was found to be 0.11 ± 0.06 µM (black), whereas mutant Q132R showed an almost 7-fold higher IC50 value of 0.71 ± 0.46 µM (green). In contrast, the Q773R mutant showed an IC50 value of 0.02 ± 0.01 µM (orange). A 35-fold difference in IC50 values was thus observed between the two mutants. To show that charge repulsion was indeed the underlying principle for the observed effects, the acid amide analog of propafenone, GPV366 (Ecker et al., 1999), which is not protonable and thus permanently uncharged, was used in subsequent control experiments. Again, six independent experiments were performed in duplicate. IC50 values for wild type (black) and mutants Q132R (green) and Q773R (orange) (Fig. 5, Table 2) were indistinguishable, demonstrating that uncharged P-gp solutes were not affected by introduction of a positive charge in either position 132 or 773.
Fig. 4.
Inhibition of rhodamine123 transport by propafenone analog GPV31 in wild-type and mutant P-gp. Cells were loaded with rhodamine123 at 37°C for 30 min. Subsequently different concentrations of GPV31 were added to inhibit rhodamine123 efflux. A, for each concentration of GPV31 (none, black; 0.008 µM, dark green; 0.02 µM, light blue; 0.05 µM, cyan; 0.13 µM, lavender; 0.32 µM, olive; 0.80 µM, dark blue; 2.00 µM, light green; and 5.00 µM, red) a time-dependent decrease in mean cellular fluorescence was monitored over 4 min. A representative experiment for Q773R P-gp is shown. An exponential curve was fitted by the method of least-squares to data points taken at 60, 120, 180, and 240 s. First-order rate constants were determined as described in the legend to Fig. 3A. These were then plotted as a function of GPV31 concentration as described previously (Ecker et al., 1999). In theory, the estimate for the zero time point (initial loading) should be identical for all curves. Values between 326 and 345 reflect experimental scatter. B shows the two concentration response curves for one representative experiment performed with the Q773R mutant. Filled squares correspond to the time dependencies shown in A, open squares are data points for the duplicate determination. The GPV31 concentration is shown on a logarithmic scale on the abscissa, and first-order rate constants of rhodamine123 transport are given on the ordinate. For this representative experiment, IC50 values were determined to be 39 and 40 nM. Attention is drawn to the fact that the experimental protocol would not require normalization for protein expression, because IC50 values, in contrast to first-order rate constants, are independent of protein expression. C, fractional inhibition of rhodamine123 transport is given as a percentage of first-order rate constants in the absence of GPV31 for six independent experiments performed in duplicate. Note that IC50 values are independent of expression rates. The S.E.E. for IC50 values is represented by horizontal error bars. Wild-type P-gp, black curve; Q132R, green curve; Q773R, orange curve. For IC50 values, see Table 2.
Fig. 5.
Inhibition of rhodamine123 transport in wild-type and mutant P-gp by the nonprotonable propafenone analog GPV366. The GPV366 concentration (0.08 –50 µM) is shown on a logarithmic scale on the abscissa and fractional inhibition of rhodamine123 transport is on the ordinate. Fractional inhibition of rhodamine123 transport is given as a percentage of fluorescence in the absence of GPV366 to allow a composite graph of all experiments. Solid lines represent hyperbolic dose response curves, which were fitted to the data points by the method of least-squares. IC50 values were calculated from these curves as 50% occupancy values and are represented as mean ± S.E (six independent experiments; S.E.E. represented as horizontal error bars). Wild-type P-gp, black curve; Q132R, green curve; Q773R, orange curve.
Table 2. IC50 values for rhodamine123 efflux inhibition by verapamil, vinblastine, and propafenone analogs.
IC50 values were calculated from concentration response curves (shown in Figs 4–7). Results are represented as mean ± S.D. of at least three individual experiments performed in duplicate.
| Mutation | IC50 |
|||
|---|---|---|---|---|
| Verapamil | Vinblastine | GPV31 | GPV366 | |
| µM | ||||
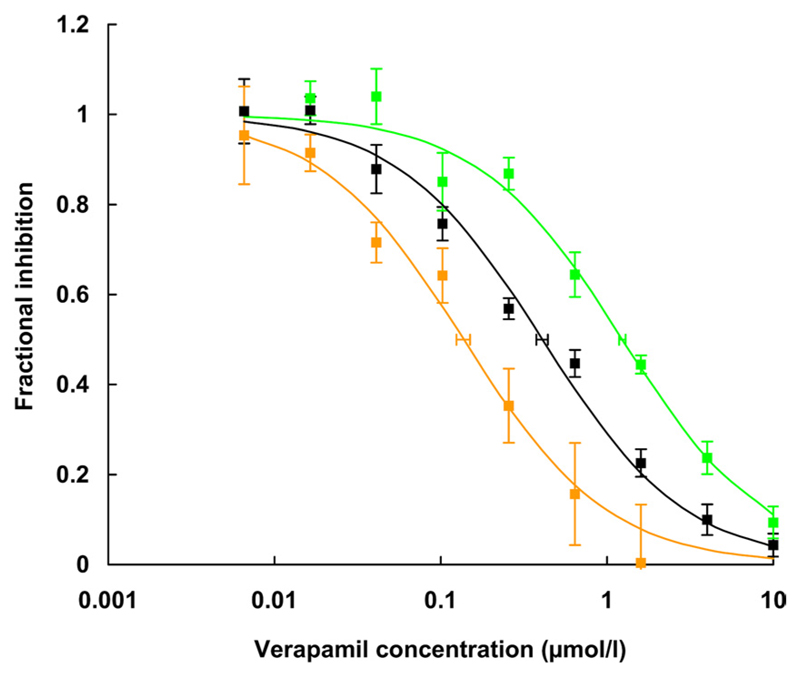
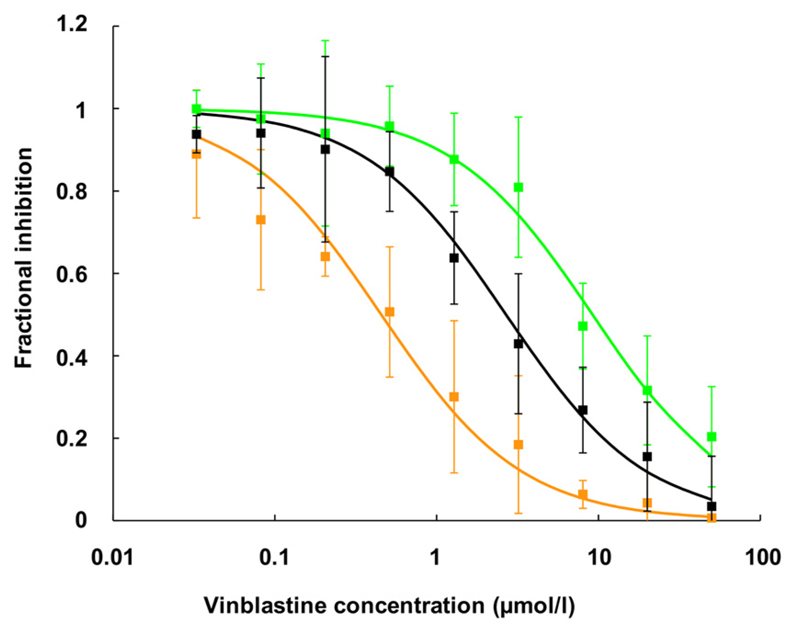
| Wild-type P-gp | 0.43 ± 0.10 | 2.68 ± 0.93 | 0.11 ± 0.06 | 2.60 ± 0.69 |
| Q132A | 0.30 ± 0.07 | 2.58 ± 0.31 | 0.10 ± 0.03 | 2.57 ± 0.69 |
| Q773A | 0.42 ± 0.15 | 2.80 ± 0.12 | 0.11 ± 0.05 | 2.40 ± 1.47 |
| Q132A/Q773A | 0.44 ± 0.16 | 2.24 ± 0.43 | 0.12 ± 0.01 | 2.87 ± 0.63 |
| Q132R | 1.24 ± 0.13* | 9.19 ± 1.20* | 0.71 ± 0.46* | 2.64 ± 0.59 |
| Q773R | 0.14 ± 0.04* | 0.45 ± 0.16* | 0.02 ± 0.01* | 2.09 ± 0.90 |
P < 0.05, significantly different from wild-type values.
Additional experiments were performed with chalcone analogs, which are potent inhibitors of P-gp. These are uncharged compounds. Similar to GPV366, these compounds showed indistinguishable IC50 values in wild type and mutants (data not shown).
Effect of Verapamil and Vinblastine on Rhodamine123 Efflux in Q132R/A and Q773R/A Mutants
In a third series of experiments, we evaluated the effect of verapamil and vinblastine on rhodamine123 transport in wild type and mutants Q132R and Q773R. Similar to propafenones, verapamil is a solute and inhibitor of P-gp that has been shown to bind to the Hoechst (H) site of the protein (Spoelstra et al., 1994; Shapiro and Ling, 1997; Al-Shawi et al., 2003; Al-Shawi and Omote, 2005). First-order rate constants of rhodamine123 efflux were determined at different verapamil concentration. A composite graph of five independent experiments performed in duplicate is shown in Fig. 6. Verapamil inhibited rhodamine123 transport of wild-type P-gp with an IC50 value of 0.43 ± 0.10 µM. In contrast, the IC50 values for the Q132R and Q773R mutants were 1.24 ± 0.13 and 0.14 ± 0.04 µM, respectively.
Fig. 6.
Inhibition of rhodamine123 transport by verapamil in wild-type and mutant P-gp. The composite graph shows fractional inhibition of rhodamine123 transport as a function of verapamil concentration of five independent experiments. Data points are given as mean ± S.E. of fractional inhibition values. IC50 values were calculated from concentration response curves as 50% occupancy values (S.E.E. represented as horizontal error bars) as described in the legend to Fig. 4. Black curve, wild-type P-gp; green curve, Q132R; orange curve, Q773R. One thousand and eighty individual data points were compiled for this graph. For IC50 values, see Table 2.
For vinblastine, a composite graph of three independent experiments performed in duplicate is presented in Fig. 7. The respective IC50 values were 2.68 ± 0.93 µM for wild type, 0.45 ± 0.16 µM for mutant Q773R, and 9.19 ± 1.2 µM for the Q132R mutant. Mutation of glutamine residues to alanine did not change IC50 values for GPV31, GPV366, verapamil, or vinblastine (in line with the results observed for rhodamine123 efflux), suggesting that molecular mechanics and function are not altered by the mutations. Table 2 gives a synopsis of IC50 values for the inhibition of rhodamine123 efflux in wild type and mutants.
Fig. 7.
Inhibition of rhodamine123 transport by vinblastine in wild-type and mutant P-gp. The composite graph shows fractional inhibition of rhodamine123 transport as a function of vinblastine concentration of three independent experiments. Data points are given as mean ± S.E. of fractional inhibition values. IC50 values were calculated from concentration response curves as 50% occupancy values. Black curve, wild-type P-gp; green curve, Q132R; orange curve, Q773R. For IC50 values, see Table 2.
Biphasic Characteristics of Concentration Response Curves for GPV31 and Verapamil in Wild-Type P-gp
The concentration response curves for wild type should be a composite of the concentration response curves obtained for the two mutants. In theory, two sites ought to be resolved in the wild-type transporter with affinity differences of the magnitude observed above. Assuming the presence of two translocation pathways, the two sites ought to be present at a 1:1 ratio; under these conditions, an affinity difference of 10-fold ought to suffice to discriminate between one- and two-site models, provided that the error is sufficiently small (De Lean et al., 1982). However, these theoretical insights are based on Monte Carlo simulations, which do not necessarily recapitulate the nature of the variability in the current experiments. In addition, the fact that rhodamine123 prefers one of the two sites also affects the ability to accurately discriminate between one and two site models (Nanoff et al., 1987). In fact, convergence was achieved in most instances, when the data for inhibition of wild-type P-gp by GPV31 and by verapamil were fitted to a two-site model. In addition, in several instances, the fit to the biphasic curve resulted in a significant improvement over a monophasic inhibition curve, but this was not uniformly seen in all experiments. We therefore conclude that the current data are consistent with the presence of two pathways for verapamil and GPV31 in wild-type P-gp but that the experimental error is too large to unequivocally resolve these in concentration response curves for wild-type P-gp.
Discussion
Despite the availability of several ABC-exporter structures the molecular details of P-gp-dependent solute transport remain unresolved. The availability of the mouse MDR1A structure in the nucleotide free state in complex with the cyclic hexapeptide inhibitors QZ59-SSS and QZ59-RRR (PDB IDs 3G5U, 3G60, and 3G61) (Aller et al., 2009) corroborated the notion that ligands are bound at the interface of the transmembrane domains as suggested by earlier experimental evidence (Loo and Clarke, 1999; Pleban et al., 2005). The translocation pathway taken by the solutes however remained undefined.
P-gp is thought to have arisen from a half-transporter by gene duplication (Gottesman and Pastan, 1993) and has retained an overlapping solute specificity with several bacterial efflux half-transporters (Reuter et al., 2003; Velamakanni et al., 2008). These half-transporters require homodimerization for function and show complete axial rotational symmetry, implying that every structural element in the protein is found twice. Therefore, solutes would choose to bind to either of two sites formed by an identical complement of amino acid residues and identical 3D structure. Ensuing ligand poses would therefore be required to represent mirror images of each other. Indeed an example of a two site cooperative behavior was found for vinblastine binding of the bacterial half-transporter LmrA (van Veen et al., 2000). In contrast to these half transporters, P-gp is a full transporter in which gene duplication has led to a fusion of both monomers into a single polypeptide chain. Figure 1 shows photolabeling of P-gp in two distinct pseudosymmetric regions, providing evidence that ligands are still recognized by two—initially identical—regions of the protein. Evolutionary diversification changed properties of these two regions in a way that make them biochemically dissectible. Results from photolabeling experiments indicate positioning of the photoactive benzophenone group in either of the two regions at the time of photoactivation and -labeling. It is noteworthy that photolabeling is unable to discriminate between binding of solutes in either position in different P-gp molecules or simultaneous binding of two substrates to the same P-gp molecule. In addition, parts of alternatively bound solute molecules might extend into and interact within an overlapping and more central region of the transporter.
In this study, mutant P-gp was generated to address the question of whether solutes reach the two photolabeled regions via two translocation pathways that reflect the 2-fold rotational (pseudo)symmetry of the transmembrane domains. Positively charged arginine residues were placed in symmetric positions of the transporter to prevent positively charged ligands from reaching either of the photolabeled regions. Both the Q132R and the Q773R mutant showed reduced transport activity for rhodamine123, which, however, was more pronounced in the Q773R mutant. In contrast, verapamil, vinblastine, and propafenone analogs were more strongly affected by mutation of residue 132 to arginine. IC50 values for rhodamine123 efflux inhibition in the two Arg mutants differed by a factor of 8.9 for verapamil, 20 for vinblastine, and 36 for the protonable propafenone analog GPV31. Indistinguishable IC50 values were found for either compound in the Q132A, the Q773A, and the Q132A/Q773A mutants. In addition, the uncharged propafenone analog GPV366 showed identical IC50 values for wild type and mutants. This was also the case for an uncharged chalcone derivative. Three conclusions can be drawn from these experiments: 1) charge repulsion is the underlying principle for selectively blocking one of the solute translocation pathways; 2) glutamine residues are not directly interacting with solutes; and 3) molecular mechanics of the transporter are not altered by the mutations. Finally, these data demonstrate that a biochemical dissection of solute translocation pathways of P-gp is possible by site-directed mutagenesis and that these pathways are both used by rhodamine123, propafenone analogs, verapamil, and vinblastine. Although rhodamine123 shows preference for one of them, propafenones, verapamil, and vinblastine are preferentially using the other.
How do these results compare to data from the literature? As early as 1997, Shapiro and Ling reported positively cooperative sites for drug transport by P-glycoprotein with distinct drug specificities. These sites were then named the R- and H-sites because of their preferential binding of rhodamine123 and Hoechst33342, respectively. Verapamil was reported to preferentially interact with the H-site (Shapiro and Ling, 1997). Earlier work by Spoelstra et al. (1994) indicated daunorubicin, an R-site compound, to be inhibited by verapamil in a noncompetitive fashion at low daunorubicin concentration and in a competitive manner at higher concentrations. This was interpreted to indicate that daunorubicin bound to two sites with different affinities and that verapamil bound to the lower affinity site. In addition, Guiral et al. (1994) reported that daunorubicin was transported by P-gp in a positively cooperative manner. Our findings are in accordance with a two-site model and for the first time, to our knowledge, relate these sites to (pseudo)symmetric positions in the transporter. Data also do not contradict the model of multiple binding sites (Martin et al., 2000), because mutations introduced in this study block access of compounds to pseudosymmetric interaction regions, which might have partially overlapping binding properties for different chemical entities. In addition, evidence for the existence of allosteric (nonsubstrate) solute-binding sites has been presented (Martin et al., 2000).
Evolutionary pressure seems to have led to conservation of amino acid ensembles in the TMDs of P-gp (Chiba et al., 2006). This might enable binding pockets to procure redundant binding interactions with solutes. A review on the nature of polyspecificity has appeared recently (Gutmann et al., 2010). Highest conservation is observed for residues that are pore-exposed, and these are candidate residues for interaction with solutes. Although aromatic residues have been found to provide approximately half of the residues interacting with cyclic hexapeptide inhibitors (Aller et al., 2009), only 2 of 16 residues that interact with verapamil are aromatic (Loo and Clarke, 2008). Therefore, in addition to aromatic residues, aliphatic and other polar residues do contribute to solute interaction. Interaction redundancy, as well as the ability of the transporter to accommodate more than one solute molecule at a time, do not imply, however, that the process of drug interaction is proceeding in a random fashion and does not contradict a 2-fold (pseudo)symmetric interaction of solutes with the transporter as found in this study.
In two reports (Loo et al., 2008, 2009), arginine scanning was employed for identification of residues in the solute translocation pathway of P-gp. For this purpose, all residues in membrane-spanning portions of the TMDs were mutated to arginine in a trafficking-deficient background, and rescue was used as the measure. A subset of mutants in which arginines were pore-oriented were able to enhance trafficking. The 132Arg mutant did not increase trafficking of the G251V background, whereas mutant 773Arg showed a complete trafficking deficiency. Therefore, Loo et al. (2009) did not study these mutations further.
In Fig. 2D, the Sav1866-based P-gp model, which is considered to represent the outward-facing (low-affinity, solute-releasing) conformation of the transporter, either of the two mutated residues Gln132 (green) and Gln773 (orange) is located in one of the wing-like extensions of the structure. One wing, shown on the left, contains residue 132 and is formed by helices 1 and 2 of the N-terminal TMD and helices 9 through 12 of the C-terminal TMD. The most intensely photolabeled residues 951 to 953 in helix 11 are shown as magenta spheres. Additional labeling is observed for adjacent C-terminal residues in TM11 (magenta ribbon). Of 16 residues that have been identified to be important for verapamil binding (Loo and Clarke, 2008), 11 lie in the left wing. C-α atoms of these residues are depicted as yellow spheres and lie at a distance of 7 to 15 Å from the photolabeled residues 951 to 953 in TM helix 11.
The wing shown on the right (Fig. 2D) is composed of helices 7 and 8 of the C-terminal and helices 3 through 6 of the N-terminal half of P-gp. The most strongly photolabeled residues, 311 and 312 in helix 5, are again shown as magenta spheres with adjacent regions of high labeling in helices 5 and 8 shown as magenta ribbons. Five residues located in this wing have been identified to be involved in verapamil binding. These are located at a C-α distance of 8 to 13 Å from photolabeled residues. Residues 132 and 773 adopt a position in a region of the protein that is in contact with the inner leaflet of the membrane and closer to the cell interior than any other of the highlighted residues (magenta and yellow). This can be appreciated from the side views in Fig. 2, A and B. Our data suggest that a positive charge in either of these positions selectively prevents positively charged solutes from reaching the respective interaction region within the same wing. In addition, the majority of verapamil residues (11 of 16) are located in close proximity to residue 132, which is in agreement with our findings that this part of the protein contains the preferred interaction region for verapamil.
In conclusion, two major findings emerged from this study: proof of the existence of two solute translocation pathways in P-gp as a reflection of evolutionary origin from a homodimeric ancestor and selective but not exclusive use of one of these pathways by different P-gp solutes. Geometric considerations suggest that these paths might partially overlap. This has important implications for the process of drug development and a molecular understanding of the interaction of P-gp with drugs.
Acknowledgments
This study was supported by the Austrian Science Fund [Grant SFB3509]. Z.P. is the recipient of a scholarship from the Higher Education Commission Pakistan.
Abbreviations
- P-gp
P-glycoprotein
- ABC
ATP-binding cassette
- MDR
multidrug resistance
- NBD
nucleotide binding domain
- PDB
Protein Data Bank
- TMD
transmembrane domain
- DMEM
Dulbecco’s modified Eagle’s medium
- HEK
human embryonic kidney
- FORC
first-order rate constant
- TM
transmembrane
- EBNA
Epstein-Barr virus nuclear antigen
- GPV31
1-(2-(3-(4-(4-fluorophenyl)piperazin-1-yl)-2-hydroxypropoxy)-phenyl)-3-phenylpropan-1-one
- GPV366
N-(2-hydroxy-3-(2-(3-phenylpropanoyl)phenoxy)propyl)-N-propylbenzamide
- GPV51
(2-(3-(diethylamino)-2-hydroxypropoxy)phenyl)(phenyl)methanone
Footnotes
Authorship Contributions
Participated in research design: Parveen, Stockner, and Chiba.
Conducted experiments: Parveen, Stockner, Bentele, and Pferschy.
Contributed new reagents or analytic tools: Stockner.
Performed data analysis: Parveen, Bentele, Freissmuth, Ecker, and Chiba.
Wrote or contributed to the writing of the manuscript: Parveen, Stockner, Freissmuth, and Chiba.
Other: Chiba acquired funding for the research.
Contributor Information
Zahida Parveen, Institute of Medical Chemistry, Medical University of Vienna, Vienna, Austria.
Thomas Stockner, Institute of Medical Chemistry, and Institute of Pharmacology, Medical University of Vienna, Vienna, Austria.
Martin Kraupp, Institute of Medical Chemistry, Medical University of Vienna, Vienna, Austria.
Michael Freissmuth, Institute of Pharmacology, Medical University of Vienna, Vienna, Austria.
Gerhard F. Ecker, Emerging Field Pharmacoinformatics, Department of Medicinal Chemistry, University of Vienna, Vienna, Austria
Peter Chiba, Institute of Medical Chemistry, Medical University of Vienna, Vienna, Austria.
References
- Al-Shawi MK, Omote H. The remarkable transport mechanism of P-glycoprotein: a multidrug transporter. J Bioenerg Biomembr. 2005;37:489–496. doi: 10.1007/s10863-005-9497-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shawi MK, Polar MK, Omote H, Figler RA. Transition state analysis of the coupling of drug transport to ATP hydrolysis by P-glycoprotein. J Biol Chem. 2003;278:52629–52640. doi: 10.1074/jbc.M308175200. [DOI] [PubMed] [Google Scholar]
- Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, et al. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba P, Ecker G, Schmid D, Drach J, Tell B, Goldenberg S, Gekeler V. Structural requirements for activity of propafenone-type modulators in P-glycoprotein-mediated multidrug resistance. Mol Pharmacol. 1996;49:1122–1130. [PubMed] [Google Scholar]
- Chiba P, Mihalek I, Ecker GF, Kopp S, Lichtarge O. Role of transmembrane domain/transmembrane domain interfaces of P-glycoprotein (ABCB1) in solute transport. Convergent information from photoaffinity labeling, site directed mutagenesis and in silico importance prediction. Curr Med Chem. 2006;13:793–805. doi: 10.2174/092986706776055607. [DOI] [PubMed] [Google Scholar]
- Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- Dawson RJ, Locher KP. Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett. 2007;581:935–938. doi: 10.1016/j.febslet.2007.01.073. [DOI] [PubMed] [Google Scholar]
- De Lean A, Hancock AA, Lefkowitz RJ. Validation and statistical analysis of a computer modeling method for quantitative analysis of radioligand binding data for mixtures of pharmacological receptor subtypes. Mol Pharmacol. 1982;21:5–16. [PubMed] [Google Scholar]
- Dormán G, Prestwich GD. Benzophenone photophores in biochemistry. Biochemistry. 1994;33:5661–5673. doi: 10.1021/bi00185a001. [DOI] [PubMed] [Google Scholar]
- Ecker G, Huber M, Schmid D, Chiba P. The importance of a nitrogen atom in modulators of multidrug resistance. Mol Pharmacol. 1999;56:791–796. [PubMed] [Google Scholar]
- Giacomini KM, Huang SM, Tweedie DJ, Benet LZ, Brouwer KL, Chu X, Dahlin A, Evers R, Fischer V, Hillgren KM, et al. Membrane transporters in drug development. Nat Rev Drug Discov. 2010;9:215–236. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman MM, Pastan I. Biochemistry of multidrug resistance mediated by the multidrug transporter. Annu Rev Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- Guiral M, Viratelle O, Westerhoff HV, Lankelma J. Cooperative P-glycoprotein mediated daunorubicin transport into DNA-loaded plasma membrane vesicles. FEBS Lett. 1994;346:141–145. doi: 10.1016/0014-5793(94)00447-1. [DOI] [PubMed] [Google Scholar]
- Gutmann DA, Ward A, Urbatsch IL, Chang G, van Veen HW. Understanding polyspecificity of multidrug ABC transporters: closing in on the gaps in ABCB1. Trends Biochem Sci. 2010;35:36–42. doi: 10.1016/j.tibs.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- Loo TW, Bartlett MC, Clarke DM. Arginines in the first transmembrane segment promote maturation of a P-glycoprotein processing mutant by hydrogen bond interactions with tyrosines in transmembrane segment 11. J Biol Chem. 2008;283:24860–24870. doi: 10.1074/jbc.M803351200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo TW, Bartlett MC, Clarke DM. Identification of residues in the drug translocation pathway of the human multidrug resistance P-glycoprotein by arginine mutagenesis. J Biol Chem. 2009;284:24074–24087. doi: 10.1074/jbc.M109.023267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo TW, Bartlett MC, Clarke DM. Human P-glycoprotein is active when the two halves are clamped together in the closed conformation. Biochem Biophys Res Commun. 2010 doi: 10.1016/j.bbrc.2010.04.057. [DOI] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. The transmembrane domains of the human multi-drug resistance P-glycoprotein are sufficient to mediate drug binding and trafficking to the cell surface. J Biol Chem. 1999;274:24759–24765. doi: 10.1074/jbc.274.35.24759. [DOI] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Mutational analysis of ABC proteins. Arch Biochem Biophys. 2008 doi: 10.1016/j.abb.2008.02.025. [DOI] [PubMed] [Google Scholar]
- Martí-Renom MA, Stuart AC, Fiser A, Sánchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- Martin C, Berridge G, Higgins CF, Mistry P, Charlton P, Callaghan R. Communication between multiple drug binding sites on P-glycoprotein. Mol Pharmacol. 2000;58:624–632. doi: 10.1124/mol.58.3.624. [DOI] [PubMed] [Google Scholar]
- Melo F, Sali A. Fold assessment for comparative protein structure modeling. Protein Sci. 2007;16:2412–2426. doi: 10.1110/ps.072895107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanoff C, Freissmuth M, Schütz W. The role of a low beta 1-adrenoceptor selectivity of [3H]CGP-12177 for resolving subtype-selectivity of competitive ligands. Naunyn Schmiedebergs Arch Pharmacol. 1987;336:519–525. doi: 10.1007/BF00169308. [DOI] [PubMed] [Google Scholar]
- Pleban K, Kopp S, Csaszar E, Peer M, Hrebicek T, Rizzi A, Ecker GF, Chiba P. P-glycoprotein substrate binding domains are located at the transmembrane domain/transmembrane domain interfaces: a combined photoaffinity labeling-protein homology modeling approach. Mol Pharmacol. 2005;67:365–374. doi: 10.1124/mol.104.006973. [DOI] [PubMed] [Google Scholar]
- Reuter G, Janvilisri T, Venter H, Shahi S, Balakrishnan L, van Veen HW. The ATP binding cassette multidrug transporter LmrA and lipid transporter MsbA have overlapping substrate specificities. J Biol Chem. 2003;278:35193–35198. doi: 10.1074/jbc.M306226200. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Schmid D, Ecker G, Kopp S, Hitzler M, Chiba P. Structure-activity relationship studies of propafenone analogs based on P-glycoprotein ATPase activity measurements. Biochem Pharmacol. 1999;58:1447–1456. doi: 10.1016/s0006-2952(99)00229-4. [DOI] [PubMed] [Google Scholar]
- Seeger MA, van Veen HW. Molecular basis of multidrug transport by ABC transporters. Biochim Biophys Acta. 2009;1794:725–737. doi: 10.1016/j.bbapap.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Shapiro AB, Ling V. Positively cooperative sites for drug transport by P-glycoprotein with distinct drug specificities. Eur J Biochem. 1997;250:130–137. doi: 10.1111/j.1432-1033.1997.00130.x. [DOI] [PubMed] [Google Scholar]
- Shen DW, Akiyama S, Schoenlein P, Pastan I, Gottesman MM. Characterisation of high-level cisplatin-resistant cell lines established from a human hepatoma cell line and human KB adenocarcinoma cells: cross-resistance and protein changes. Br J Cancer. 1995;71:676–683. doi: 10.1038/bjc.1995.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoelstra EC, Westerhoff HV, Pinedo HM, Dekker H, Lankelma J. The multidrug-resistance-reverser verapamil interferes with cellular P-glycoprotein-mediated pumping of daunorubicin as a non-competing substrate. Eur J Biochem. 1994;221:363–373. doi: 10.1111/j.1432-1033.1994.tb18748.x. [DOI] [PubMed] [Google Scholar]
- Stockner T, de Vries SJ, Bonvin AM, Ecker GF, Chiba P. Data-driven homology modelling of P-glycoprotein in the ATP-bound state indicates flexibility of the transmembrane domains. Febs J. 2009;276:964–972. doi: 10.1111/j.1742-4658.2008.06832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veen HW, Margolles A, Müller M, Higgins CF, Konings WN. The homodimeric ATP-binding cassette transporter LmrA mediates multidrug transport by an alternating two-site (two-cylinder engine) mechanism. EMBO J. 2000;19:2503–2514. doi: 10.1093/emboj/19.11.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velamakanni S, Yao Y, Gutmann DA, van Veen HW. Multidrug transport by the ABC transporter Sav1866 from Staphylococcus aureus. Biochemistry. 2008;47:9300–9308. doi: 10.1021/bi8006737. [DOI] [PubMed] [Google Scholar]
- Wallner B, Elofsson A. Can correct protein models be identified? Protein Sci. 2003;12:1073–1086. doi: 10.1110/ps.0236803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc Natl Acad Sci USA. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]