Abstract
Modifications of histone proteins are fundamental to the regulation of epigenetic phenotypes. Dysregulations of histone modifications have been linked to the pathogenesis of diverse human diseases. However, identifying differential histone modifications in patients with immune-mediated diseases has been challenging, in part due to the lack of a powerful analytic platform to study histone modifications in the complex human immune system. We recently developed a highly multiplexed platform, Epigenetic landscape profiling using cytometry by Time-Of-Flight (EpiTOF), to analyze the global levels of a broad array of histone modifications in single cells using mass cytometry. In this review, we summarize the development of EpiTOF and discuss its potential applications in biomedical research. We anticipate that this platform will provide new insights into the roles of epigenetic regulation in hematopoiesis, immune cell functions and immune system aging, and reveal aberrant epigenetic patterns associated with immune-mediated diseases.
Keywords: Epigenetics, Mass cytometry, EpiTOF, Chromatin, Histone post-translational modifications, Immune System
Introduction
The field of epigenetics, which studies the links between genotypes and phenotypes, has emerged to be key to our understanding of cellular development and functions1. With the exception of antigen receptor loci in lymphocytes, all cells in the human immune system, from hematopoietic stem cells to terminally differentiated immune cells, share identical genomic content. Nevertheless, the diverse immune cell subtypes (e.g. B, T, NK cells and monocytes) and their distinct functional subgroups (e.g. naïve and memory, helper and cytotoxic T cells) best exemplify the degree of phenotypic variations that can be derived from the same genome. At the individual level, rich data from twin-based studies have found a large collection of human traits, including susceptibility to autoimmune diseases 2, discordant between genetically identical twins 3, suggesting the importance of molecular mechanisms other than genetics underlying the heterogeneity of these human traits.
In recent years, epigenetics often refers to research on chromatin-based events that regulate DNA-templated biological processes4. Chromatin, the nuclear structure containing nucleic acids wrapped around core histone proteins, is the physiologically relevant template atop of which DNA-based biological processes, such as transcription, replication, and recombination, occur. Compaction of chromatin into higher-order structure precludes DNA-binding of transcription factors and other trans-acting regulators to loci that are rendered inaccessible. The extent to which chromatin is packaged and the accessibility of the underlying DNA can thus influence transcription programs and other biological functions, leading to diverse phenotypes without altering DNA sequence. Post-translational modifications of histone proteins (chromatin marks) are fundamental to the regulation of chromatin dynamics5. For instance, covalent modifications of histone proteins such as acetylation and phosphorylation modulate chromatin packaging through disruption of DNA-histone electrostatic interactions 5. Further, these chromatin marks create unique docking sites for modification-dependent protein-protein interactions at chromatin 6, which stabilize chromatin binding of protein complexes mediating chromatin remodeling or other unique DNA-templated biological functions. Finally, distinct sequences and modification patterns of histone variants relative to their canonical counterparts alter chromatin structure, nucleosome stability and interactions with chromatin-binding proteins 7. Together, histone modifications act in concert with other molecular mechanisms such as cytosine methylation and noncoding RNAs to regulate chromatin dynamics and, therefore, epigenetic phenotypes.
Until recently, studying the regulatory roles of chromatin marks in hematopoiesis and immune functions has been hindered by technical difficulties associated with conventional experimental methods. Chromatin immunoprecipitation (ChIP) and Western blotting require a large number of cells and only generate averaged snapshots of cell populations that are potentially heterogeneous. The results fall short of demonstrating the individuality of immune cells and taking into account the heterogeneity within a population of cells. On the other hand, although immunofluorescence and immunohistochemistry allow single-cell measurements of chromatin marks, the low throughput and labor-intensive protocols prevent the assays from being widely applied to immunology research. To overcome these limitations, we leveraged the multiplexing capacity and single-cell resolution of mass cytometry to develop a new analytical platform, Epigenetic landscape profiling using cytometry by Time-Of-Flight (EpiTOF), for profiling the cellular levels of a diverse array of chromatin marks in single immune cells. In this review, we first summarize the design of EpiTOF and the methods to construct antibody panels for analyzing chromatin marks. We describe the selection and validation of antibodies, sample processing, and data pre-processing, normalization, analysis and visualization. We discuss how EpiTOF data will enrich our understanding of chromatin-based regulation of epigenetic phenotypes, in the contexts of hematopoietic development, immune responses against pathogens and/or external stimuli and the pathobiology of immune-mediated diseases.
EpiTOF assay design
The landmark invention of mass cytometry (i.e. Cytometry by Time-Of-Flight or CyTOF) has revolutionized the fields of cytometry and single-cell phenotypic and functional analyses 8. Staining cells with lanthanide-labeled antibodies followed by detection using mass spectrometry greatly increases the number of parameters that can be extracted from individual cells 9,10. This is a significant advancement in comparison with the conventional fluorescence-based flow cytometry, which is inevitably limited by spectral overlap between fluorochromes. The latest generation of mass cytometer is tuned to detect atomic mass between 75 and 209. Thirty-five lanthanides are readily available commercially (Fluidigm, CA) to construct customized antibody panels. To establish antibody panels for EpiTOF, 35 lanthanide channels are assigned to conjugate to three categories of antibodies: i) antibodies targeting chromatin marks; ii) antibodies recognizing total histones in a modification-independent fashion; iii) antibodies against immunophenotypic markers to reveal immune cell identity.
The sensitivity of mass cytometry to detect signals from different lanthanide channels is not uniformly distributed across the mass window. Thulium (Tm)-169 yields the greatest signal, and the sensitivity declines for atomic mass with increasing distance from Tm-169. For this reason, a recommendation has been to assign channels surrounding Tm-169 for the detection of low-abundance markers 11. We generally place chromatin marks in the center of the mass window where maximal sensitivity can be achieved. However, chromatin marks known to be present at high levels in cells, for instance, di-methylation of H4 at lysine 20 (H4K20me2) that has been shown to mark up to 85% of all H4 molecules 12, may be detected using channels with less-than-optimal sensitivity. Similarly, well-characterized immunophenotypic markers such as lineage-specific CD3, CD14, CD19 and CD56 may be detected using atomic mass distant from Tm-169. It is important to note that methanol-based permeabilization is used in EpiTOF to achieve maximal permeabilization in order to reveal epitopes in nuclei. However, epitopes of many cell surface markers are sensitive to methanol, and hence the selection of antibodies against immunophenotypic markers needs to be carefully validated.
Further, we incorporate several methods in EpiTOF to ensure that high-quality data are collected and that accurate comparisons are made between biological samples. First, we employ pulse treatment with the DNA-intercalating drug cisplatin to distinguish live cells from dead cells with compromised plasma membranes 13. This viability staining is especially crucial for cryopreserved clinical samples, where the viability of cells can vary greatly due to sample processing and long-term storage. Second, we perform intracellular staining of chromatin marks in a single tube where cells from independent samples are labeled with palladium barcodes 14,15. Sample barcoding is essential for EpiTOF in order to minimize technical variability and to eliminate doublet events during data collection. Third, we integrate two antibodies recognizing bulk histone proteins independent of modifications in EpiTOF panels to serve as controls for single-cell variations in total histone content, nuclear epitope accessibility and background signals proportional to proteome size and complexity. Lastly, we preferably select monoclonal antibodies (over polyclonal antibodies) for EpiTOF to enhance long-term assay reproducibility.
Selection and validation of affinity reagents targeting chromatin marks
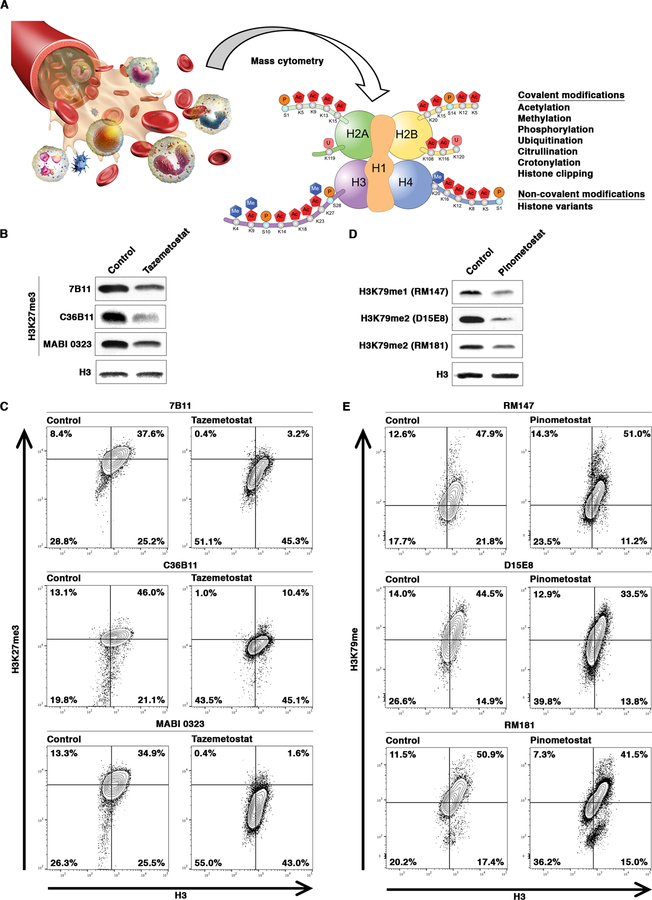
Post-translational modifications (PTMs) of histone proteins have been extensively studied, with greater than 130 modifications detected at over 60 sites on histones 16. Nucleosomes containing non-canonical histone variants with unique PTM patterns further expand the diversity of chromatin modifications 17. We prioritize and select chromatin marks for EpiTOF based on the following criteria: i) involved in the regulation of pivotal cellular and/or physiological functions; ii) known to be dysregulated or associated with human diseases; iii) generated and removed by known chromatin-modifying enzymes; iv) that can be pharmacologically manipulated by approved or investigational drugs; and v) against which highly specific affinity reagents are available (Figure 1A).
Figure 1. Selection and Validation of Affinity Reagents for EpiTOF.
(A) Overview of EpiTOF platform.
(B) Validation of H3K27me3 antibodies under denaturing condition. Western blotting analysis of whole-cell extract from Jurkat cells cultured in the absence or presence of EZH2 inhibitor Tazemetostat using the indicated monoclonal antibodies. Control, DMSO-treated cells.
(C) Validation of H3K27me3 antibodies under native condition. Mass cytometry analysis of the cells as in (B) using the indicated monoclonal antibodies. Contour plots: x-axis, antibodies against total histone H3; y-axis, antibodies against H3K27me3. Percentage of cells in each quadrant is shown.
(D) Validation of H3K79me antibodies under denaturing condition. Western blotting analysis of whole-cell extract from Jurkat cells cultured in the absence or presence of DOT1L inhibitor Pinometostat using the indicated monoclonal antibodies. Control, DMSO-treated cells.
(E) Validation of H3K79me antibodies under native condition. Mass cytometry analysis of the cells as in (D) using the indicated monoclonal antibodies. Contour plots: x-axis, antibodies against total histone H3; y-axis, antibodies against H3K79me. Percentage of cells in each quadrant is shown.
The next critical steps in early EpiTOF development include the identification and validation of chromatin mark-specific antibodies. We adopt several strategies to test the specificity of commercially available antibodies against chromatin marks and their compatibility with cytometry-based assays. Only antibodies capable of detecting known changes in chromatin marks in flow and/or mass cytometry assays are selected.
Pharmacological inhibition of chromatin-modifying enzymes
The highly druggable characteristics of chromatin-modifying enzymes and the reversible nature of chromatin-based epigenetic phenomena led to the development of many FDA-approved drugs and drug candidates at various stages of clinical trials 18. For instance, a potent lysine methyltransferase EZH2 inhibitor, Tazemetostat (EPZ-6438), has demonstrated promising efficacy in treating various forms of hematological malignancies and solid tumors 19,20. Inhibition of EZH2 by Tazemetostat reduces the levels of all three degrees of methylation on histone H3 at lysine 27 (H3K27me1/2/3). While several clones of monoclonal antibodies can detect Tazemetostat-induced H3K27me reduction using western blotting (Figure 1B), we found that clone MABI 0323 not only accurately detects repressed H3K27me3 levels in Tazemetostat-treated cells but also demonstrates the best signal-to-noise dynamic range in CyTOF (Figure 1C). Based on the results, MABI 0323 was chosen for EpiTOF.
Manipulation of the expression of chromatin-modifying enzymes
During antibody validation, we altered the levels of enzymes that catalyze the generation or removal of chromatin marks to create cells with differential chromatin marks. Transient transfection of the deubiquitinases USP21 or USP49 reduces ubiquitination of H2A at lysine 119 (H2AK119ub) or H2B at lysine 120 (H2BK120ub), respectively 21,22. Ectopic overexpression of the lysine demethylase KDM4A/JMJD2A removes di- and tri-methylation of H3 at lysines 9 and 36 (H3K9me2/3 and H3K36me2/3), resulting in the accumulation of the mono-methylated species at these residues (H3K9me1 and H3K36me1) 23. Similarly, overexpression of KDM4D/JMJD2D leads to decreased H3K9me2 and H3K9me3 in cells 23. Lysine methyltransferases SUV420H1 and SUV420H2 catalyze di- and tri-methylation of H4 at lysine 20 (H4K20me2/3) using mono-methylated H4K20 residues as a substrate 12. Cells overexpressing SUV420H1 and SUV420H2 show elevated H4K20me2/3 and decreased mono-methylation (H4K20me1) levels. Using these cellular systems where the expression of chromatin-modifying enzymes is experimentally increased, we selected highly specific monoclonal antibodies for EpiTOF.
Additionally, RNAi- or CRISPR-mediated depletion of key regulators of chromatin marks may be employed to manipulate global levels of chromatin marks. For instance, knocking down the expression of WDR5, an essential component of several protein complexes that catalyze H3 methylation at lysine 4 (H3K4me), by RNAi reduces the bulk of di- and tri-methylation of H3K4. Similarly, RNAi-mediated depletion of lysine methyltransferase SETD2 specifically reduces tri-methylation of H3 at lysine 36 (H3K36me3) 24. CRISPR-mediated deletion of arginine methyltransferase PRMT1, which generates over 90% of asymmetric di-methylarginine (ADMA) in cells 25, results in accumulation of mono-methylarginine (MMA). Deletion of the allele of lysine methyltransferase NSD2/MMSET/WHSC1 overexpressed in multiple myeloma cells carrying t(4;14) chromosomal translocation reduces di-methylation of H3 at lysine 36 (H3K36me2). Using these experimental systems, we identified additional monoclonal antibodies suitable for EpiTOF.
Induction of differential chromatin marks by external stimuli
Many chromatin marks are known to be induced in response to external stimuli or under specific growth conditions. Genotoxic stress such as neocarzinostatin (NCS) treatment induces phosphorylation of H2AX at serine 139 (γ-H2AX), which then recruits DNA damage repair machinery to double-strand break sites 26. The chemotherapy agent etoposide induces an apoptosis marker, phosphorylation of H2B at serine 14 (H2BS14ph), in acute promyelocytic leukemia cells 27. We identified monoclonal antibodies recognizing γ-H2AX or H2BS14ph for EpiTOF using NCS- or etoposide-treated cells, respectively. To find affinity reagents recognizing crotonylation marks, we cultured cells in media containing high concentrations of crotonate, increasing intracellular crotonyl-CoA concentrations and promoting protein crotonylation 28. This system allowed the identification of an anti-crotonylated lysine antibody for EpiTOF.
Cell cycle-dependent changes in chromatin marks
Elevated phosphorylation of histones, in particular at several residues of H3, is a signature of mitosis 29. Co-staining of cycling cells with antibodies recognizing phosphorylated histones and reagents measuring DNA content allows us to test if antibodies specifically mark mitotic cells with duplicated DNA content. We adopted this strategy to identify antibodies against H3 phosphorylated at serine 10 (H3S10ph) 30 and H3.3 phosphorylated at serine 31 (H3.3S31ph) 31. Tozasertib, which inhibits H3S10ph-catalyzing Aurora kinases, abolishes the positive staining 32. The same experimental system may be used to validate other mitotic markers such as H3 phosphorylation at threonine 3 (H3T3ph) 33 and serine 28 (H3S28ph) 34. Additionally, histones that are newly-synthesized during S phase are marked with H4 acetylation at lysine 5 (H4K5ac) 35. We selected the antibody clone that can optimally detect this enrichment in cells undergoing DNA replication for EpiTOF.
Despite an extensive search for affinity reagents against several physiologically and pathologically important chromatin marks for EpiTOF, we were unable to identify antibodies that can accurately detect differential levels of several chromatin marks by CyTOF. For instance, inhibition of lysine methyltransferase DOT1L by Pinometostat reduces the bulk of H3 methylation at lysine 79 (H3K79me) 36. Several monoclonal antibodies can identify reduced H3K79me in Pinometostat-treated cells by western blotting under denaturing conditions (Figure 1D). However, none is able to detect similar changes in mass cytometry assays, where chromatin structure is maintained in the native state (Figure 1E). Similarly, we were unable to find affinity reagents, either monoclonal antibodies or modification-recognizing protein modules (e.g. methyl-CpG-binding domains (MBDs)) 37, to detect azacytidine (DNA methyltransferase inhibitor)-induced reduction in DNA methylation by CyTOF 38. Given the critical functions of these chromatin marks in normal physiological functions and in human diseases, and that potent inhibitors modulating these modifications are available and have been used in treating human patients, it is important in future studies to identify or develop monoclonal antibodies against these chromatin marks with high specificity and are compatible with CyTOF.
Data pre-processing and normalization
For peripheral blood mononuclear cells (PBMCs) samples, approximately 1e05 cells are required to obtain sufficient events for rare immune cells such as circulating dendritic cells and hematopoietic progenitors. After identifying cells from individual samples using palladium barcodes, the immune cell subtype of each event is determined using immunophenotypic markers, generating a dataset containing the levels of a broad spectrum of chromatin marks in millions of individual single cells.
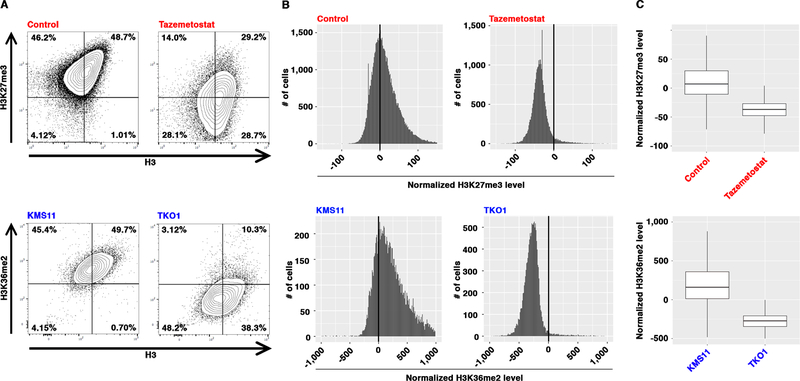
A regression model has been successfully employed to correct for morphological variations between biological samples of flow cytometry data 39. Analysis of EpiTOF datasets comparing doublet with singlet events shows approximately two-fold signals in doublets across all chromatin marks and total histones, indicating the linear nature of these measurements. We therefore applied a linear regression model utilizing signals from total histones as predictor variables to obtain normalized chromatin mark levels independent of the variations in total histones. Using cells with differential levels of chromatin marks, including Tezametostat-treated cells with reduced H3K27me3 and multiple myeloma cells carrying a t(4;14) chromosomal translocation with elevated H3K36me2 (Figure 2A), the residuals after correction by linear model (normalized mark levels) are near normally distributed (Figure 2B). This suggests that our data analysis fully corrects the signals captured by antibodies against total histones. This approach allows us to compare the levels of chromatin marks between cells and between samples, while taking into account the variability in histones expression and non-specific signals shared by all antibodies (Figure 2C).
Figure 2. Normalization of EpiTOF Data.
(A) Raw EpiTOF data. Mass cytometry analysis of control (DMSO) and Tazemetostat-treated Jurkat cells (top) or KMS11 and TKO1 cells (bottom) using the indicated antibodies. Contour plots: x-axis, antibodies against total histone H3; y-axis, antibodies against H3K27me3 (top) or H3K36me2 (bottom). Percentage of cells in each quadrant is shown.
(B) Distributions of linear regression residuals of EpiTOF data. Histograms of linear regression residuals of the data in (A). x-axis, residual levels; y-axis, numbers of cells with given residual levels; top, H3K27me3 in DMSO (left)- or Tazemetostat (right)-treated Jurkat cells; bottom, H3K36me2 in KMS11 (left) or TKO1 (right) cells.
(C) Normalized EpiTOF data. Box plots show the normalized H3K27me3 (top) or H3K36me2 (bottom) levels of the cells in (B). y-axis, normalized chromatin mark levels (residuals of a linear model using total H3 as predictor variable).
EpiTOF data analysis and visualization
Typically, an EpiTOF dataset comprises billions of data points derived from over 100,000 immune cells from each barcoded biological sample. Over 70 parameters, including chromatin marks, immunophenotypic markers, viability staining and sample barcodes, are collected from individual cells. The complexity of the dataset requires extensive bioinformatic and biostatistical analyses.
An overview of chromatin modification profiles of various immune cell populations
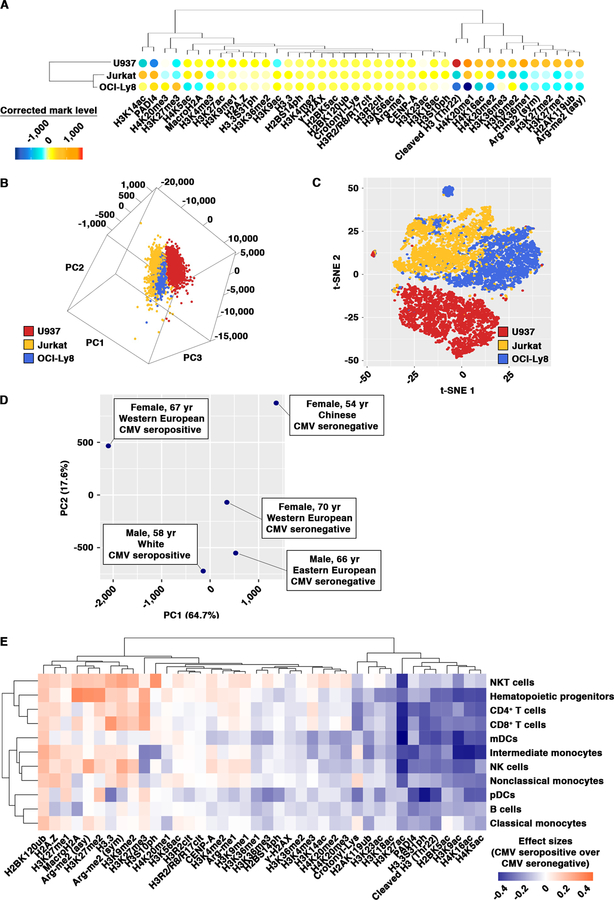
We utilize multi-dimension heatmaps to present a global view of a typical EpiTOF dataset. Using EpiTOF analysis of three leukemia and lymphoma cell lines, U937, Jurkat and OCI-Ly8, as an example (Figure 3A), averages of normalized chromatin mark levels (residuals after correction by linear model) are computed for individual chromatin marks in each cell line and are represented by the color in the heatmap. Signals from each chromatin mark are rescaled, with 0 representing the average across all three cell lines. Warm colors indicate higher-than-average levels of chromatin marks, whereas cold colors show levels lower than the average. Chromatin marks and the three cell lines are shown at the x- and y-axes, respectively, which are both ordered based on unsupervised clustering. Site-specific proteolytic cleavage of histone H3 at threonine 22 (Thr22) is observed at a higher level in U937 cells in comparison with Jurkat and OCI-Ly8 cells. The differential levels of cleaved H3 Thr22 are validated by western blotting on whole cell extracts from these three cell lines (unpublished data). When multiple biological samples are analyzed and the heatmap color represents the averages of chromatin marks across several donors or patients, we use circle size to demonstrate the biological heterogeneity using Simpson’s diversity index. This data visualization method allows us to gain a rapid and high-level view on the chromatin modification profiles of individual immune cell subtypes and to compare differential levels of chromatin marks between immune cell subsets.
Figure 3. Data analysis and visualization of EpiTOF Data.
(A) Distinct chromatin modification profiles of immune cell subtypes. Heatmap representation of the normalized levels of 40 chromatin marks (x-axis) in three leukemia and lymphoma cell lines (y-axis). Color, normalized chromatin mark levels. Means of individual samples are used for plotting and are centered around the mean across all three samples. Warm colors, higher than the mean of all samples; cold color, lower than the means of all samples. Dendrograms, unsupervised clustering.
(B and C) Segregations of single cells based on chromatin modification profiles. PCA (B) or t-SNE (C) analysis of EpiTOF data as in (A). Each dot represents a single cell. Color represents the sample to which individual cell belongs (red, U937; yellow, Jurkat; blue, OCI-Ly8). The first three principal components (B) or two t-SNE axes (C) computed from 20 chromatin marks are used to visualize the data.
(D) Segregation of blood donors based on chromatin modification profiles. PCA of EpiTOF data collected from 5 healthy subjects. Each dot represents a single donor with label depicting the demographics. Principal components are calculated based on the levels of 40 chromatin marks in 11 immune cell subtypes (440 data points) from individual donors. The percentage of variance each principal component explains is shown.
(D) Differential chromatin marks between CMV-seropositive and CMV-seronegative blood donors. Heatmap representation of the effect sizes of the levels of the indicated chromatin mark (x-axis) and immune cell subtype (y-axis) pairs in CMV-seropositive over CMV-seronegative donors. Dendrograms, unsupervised clusterings.
Single-cell chromatin modification profiles
At the single-cell level, dimensionality reduction algorithms such as principal component analysis (PCA) 40 are employed to visualize the variance in chromatin modification profiles between and within immune cell subtypes. Each principal component is a linear combination of several chromatin marks that account for the maximal variance in chromatin modification profile in the dataset. We can then plot single cells in a reduced-dimension space (2D or 3D) where the first two to three principal components explain as much variance in the dataset as possible. Each data point represents a single cell, which is color-coded based on the immune cell subtype. For instance, using the dataset described in Figure 3A, single cells from the three cell lines, painted with distinct colors, are situated in a three-dimension space where each dimension (principal component) is calculated based on different combinations of chromatin marks (Figure 3B). Despite the fact that all cells are immortal, single cells from the three cell lines are clustered by cell lines, suggesting that they remain distinct at the epigenetic level. Other visualization tools can also be employed. For example, t-Distributed Stochastic Neighbor Embedding (t-SNE) is a powerful dimensionality reduction technique for identifying non-linear relationships between single cells 41, which can be overlooked in PCA due to the linear transformation of variables. t-SNE analysis of single cells from the three cell lines described above reveals closer relationships between B cell lymphoma OCI-Ly8 and T cell leukemia Jurkat cells, relative to their distances from the myeloid lymphoma cells U937 (Figure 3C). We have used both PCA and t-SNE to visualize the segregations of single cells between immune cell subtypes or biological samples based on their chromatin modification profiles. Importantly, this analysis has the potential to find previously uncharacterized immune cell subsets defined by chromatin marks.
Clustering and segregation of biological samples based on epigenetic markers
We used PCA to visualize the variance in chromatin modification profiles between samples from different subjects or patients. For instance, Figure 3D shows the PCA of an EpiTOF dataset containing the levels of 40 chromatin marks in 11 immune cell subtypes (440 data points) from five healthy donors. The first principal component, which explains 64.7% of the variance of the dataset, separates donors with cytomegalovirus (CMV) infection from others who are CMV-seronegative. Investigating the variables selected by PCA from the 440 parameters allows us to identify specific chromatin marks that best contribute to the separation of subjects by the first principal component. For EpiTOF datasets containing different groups of biological samples, such as immune cells purified from healthy donors or patients with rheumatic diseases, or clinical trial samples from placebo-controlled or treatment arms, we performed PCA to find epigenetic signatures shared by individual groups of samples and to identify chromatin marks segregating one group from another. Markers representing individual samples on PCA plots can be labeled with different colors, shapes and sizes to present additional layers of clinical information available for the samples, such as disease severity scores, treatment groups, sex and disease subtypes. Notably, batch effect should be carefully addressed when samples from independent experiments are compared in PCA. In experiments where combined processing and staining of all samples are not possible due to large sample numbers, we employed an empirical Bayes framework to correct for batch effects prior to PCA 42.
Comparative analysis between biological samples
For comparative analysis, we compute effect sizes between different groups of samples as a robust and reliable measurement of mean differences between groups 43–45. In Figure 3E, we used a heatmap to visualize the effect sizes of the 440 data points computed between CMV-seropositive and -seronegative blood donors shown in Figure 3D. Warm colors represents chromatin marks elevated in specific cell types in the experimental group (CMV-seropositive) versus the control group (CMV-seronegative), and the cold colors indicate the repressed chromatin marks in the experimental group. Both x and y-axes are ordered based on unsupervised hierarchical clustering to facilitate data interpretation. The heatmap provides a useful summary of the comparative analysis results and allows us to find the following: i) Chromatin marks altered in multiple or unique immune cell subsets. The immune cell subsets where differential levels of chromatin marks are observed provide insights into the hematopoietic stage at which alterations of chromatin marks may have occurred. For instance, changes across multiple immune cell subsets are likely due to reprogramming of chromatin marks upstream in hematopoietic stem cells, whereas some changes may be restricted to specific hematopoietic lineages or cell subtypes. ii) Concurrent change of functionally related chromatin marks. An open chromatin state is generally associated with acetylated histones and methylation of H3 at lysine 4 (H3K4me) and 36 (H3K36me), whereas condensed and inaccessible chromatin is marked with tri-methylation of H3 at lysines 9 (H3K9me3) or 27 (H3K27me3) or H4 at lysine 20 (H4K20me3). Systematic alterations of several of these chromatin marks indicate global changes in chromatin accessibility, which may be experimentally tested utilizing Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) 46. iii) Chromatin marks regulated by the same groups of chromatin-modifying enzymes. The homeostasis of chromatin marks is dynamically maintained by enzymes that generate or remove chromatin marks. For instance, differential levels of H3K27 di- and tri-methylation may suggest altered activities of lysine methyltransferases EZH1 and/or EZH2, the enzymatic subunits of polycomb repressive complex 2 (PRC2). In contrast, changes in mono-methylation and/or acetylation of H3K27 suggest altered activities of KDM6A/UTX and/or KDM6B/JMJD3, two lysine demethylases that catalyze the removal of H3K27me3 and conversion to a lower degree of methylation. Ideally, studies where EpiTOF and transcription profiling datasets are available from the same sample will allow us to identify differential chromatin marks in addition to the altered expression of the corresponding chromatin-modifying enzymes. Experimental manipulations of chromatin-modifying enzymes can provide direct evidence for the involvement of the corresponding chromatin marks in the phenotypes of interest.
Applications of EpiTOF to biomedical research
EpiTOF is a powerful platform that allows highly multiplexed analysis of chromatin modifications in single cells and facilitates the identification of global changes of chromatin marks between biological samples. We envision that it may be broadly employed to help us better understand the roles of epigenetic mechanisms in the regulation of hematopoietic differentiation and immune cell physiological functions. Changes of chromatin marks can be studied in the context of lineage commitment (e.g. lymphoid versus myeloid), maintenance of cell identity (e.g. FOXP3+ regulatory T cells), activation in response to stimulations such as cytokines and pathogens (e.g. changes at chromatin along with gene expression changes in these settings), aging of the immune system 47, and other fundamental areas of immunology research. Importantly, EpiTOF can be employed to identify chromatin marks dysregulated in immune-mediated diseases, as the etiology of many autoimmune and autoinflammatory disorders remain largely unknown, and the discordance of disease incidence between genetically identical monozygotic twins suggests a key role for epigenetic mechanisms in their pathogenesis and disease progression 2. Additionally, the use of EpiTOF to study the impacts of pathogens on the host epigenome in infectious disease patients, the involvement of epigenetic mechanisms in asthma and allergic diseases 48, epigenetic changes associated with treatment response, and single-cell heterogeneity in chromatin modification profiles in hematological malignancies will provide an unprecedented opportunity to discover new therapeutic targets and biomarkers.
Conclusions
Epigenetics has emerged as a fast-moving area on many fronts of biomedical research in recent years. However, understanding the physiological roles of chromatin-related mechanisms in the human immune system is still in its infancy. The development of new single-cell analytic platforms, such as bisulfite sequencing to measure DNA methylation 49,50, ATAC-seq to measure chromatin accessibility 51,52, chromosome conformation capture (3C) to measure chromatin spatial organization 53, and now EpiTOF to measure global levels of chromatin marks 47, allow interrogation of epigenetic mechanisms that regulate immune functions and identification of the dysregulation of these mechanism in the context of human diseases. Between genetically unrelated individuals, over 99% of the genetic code is identical 54. It is almost certain that all morphological and phenotypic heterogeneity is at least in part explained by variations in epigenetics or chromatin-based regulation. The same concept also applies to the variability in aging-dependent deteriorations of immune functions, disease susceptibility, and responses to treatments and vaccines. We anticipate that in the near future, an increasing number of epigenetic markers will be identified to explain functionally-important variations in the human immune system 55.
Acknowledgements
We thank the Stanford Blood Center for recruiting subjects and collecting blood samples and the Stanford Shared FACS Facility (SSFF) where EpiTOF was performed. CyTOF at SSFF was obtained using NIH S10 Shared Instrument Grant (S10OD016318–01). The development of EpiTOF was supported in part by grants from Autoimmunity Center of Excellence (5U19AI110491–04 and 5UM1A110498–04), a research consortium supported by the NIAID/NIH (to P.J.U.), the Bill & Melinda Gates Foundation (to P.K.), the Donald E. and Delia B. Baxter Foundation (to P.J.U.), Elizabeth F. Adler (to P.J.U.), the Henry Gustav Floren Trust (to P.J.U.), NIH (5R01AI125197–02) (to P.J.U. and P.K.), NIH (5U19AI057229–15) (to P.K.) and NIH (5U19AI109662–05) (to P.K.) (listed alphabetically). P.C. was supported by NIH T32 training grant 2T32AR050942–06A1 and Novo Nordisk Senior Postdoctoral Fellowship. F.V. was supported by NIH K12 Award 5K12HL120001–02. A.J.K. was supported by NIH T32 training grant 5T32AI007290–29 and Novo Nordisk Senior Postdoctoral Fellowship.
Footnotes
Conflict of Interest
The authors declare no competing interests.
References
- 1.Goldberg AD, Allis CD & Bernstein E Epigenetics: a landscape takes shape. Cell 128, 635–638, doi: 10.1016/j.cell.2007.02.006 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Bogdanos DP et al. Twin studies in autoimmune disease: genetics, gender and environment. J Autoimmun 38, J156–169, doi: 10.1016/j.jaut.2011.11.003 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Polderman TJ et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet 47, 702–709, doi: 10.1038/ng.3285 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Allis CD & Jenuwein T The molecular hallmarks of epigenetic control. Nat Rev Genet 17, 487–500, doi: 10.1038/nrg.2016.59 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Kouzarides T Chromatin modifications and their function. Cell 128, 693–705, doi: 10.1016/j.cell.2007.02.005 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Patel DJ & Wang Z Readout of epigenetic modifications. Annu Rev Biochem 82, 81–118, doi: 10.1146/annurev-biochem-072711-165700 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Talbert PB & Henikoff S Histone variants--ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol 11, 264–275, doi: 10.1038/nrm2861 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Bendall SC et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696, doi: 10.1126/science.1198704 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bendall SC, Nolan GP, Roederer M & Chattopadhyay PK A deep profiler’s guide to cytometry. Trends Immunol 33, 323–332, doi: 10.1016/j.it.2012.02.010 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spitzer MH & Nolan GP Mass Cytometry: Single Cells, Many Features. Cell 165, 780–791, doi: 10.1016/j.cell.2016.04.019 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leipold MD, Newell EW & Maecker HT Multiparameter Phenotyping of Human PBMCs Using Mass Cytometry. Methods Mol Biol 1343, 81–95, doi: 10.1007/978-1-4939-2963-4_7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schotta G et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev 22, 2048–2061, doi: 10.1101/gad.476008 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fienberg HG, Simonds EF, Fantl WJ, Nolan GP & Bodenmiller B A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry A 81, 467–475, doi: 10.1002/cyto.a.22067 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zunder ER et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc 10, 316–333, doi: 10.1038/nprot.2015.020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bodenmiller B et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol 30, 858–867, doi: 10.1038/nbt.2317 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan M et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028, doi: 10.1016/j.cell.2011.08.008 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maze I, Noh KM, Soshnev AA & Allis CD Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nat Rev Genet 15, 259–271, doi: 10.1038/nrg3673 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dawson MA & Kouzarides T Cancer epigenetics: from mechanism to therapy. Cell 150, 12–27, doi: 10.1016/j.cell.2012.06.013 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Knutson SK et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci U S A 110, 7922–7927, doi: 10.1073/pnas.1303800110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Italiano A et al. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet Oncol, doi: 10.1016/S1470-2045(18)30145-1 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Yuan G et al. Histone H2A ubiquitination inhibits the enzymatic activity of H3 lysine 36 methyltransferases. J Biol Chem 288, 30832–30842, doi: 10.1074/jbc.M113.475996 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z et al. USP49 deubiquitinates histone H2B and regulates cotranscriptional pre-mRNA splicing. Genes Dev 27, 1581–1595, doi: 10.1101/gad.211037.112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin S & Janknecht R Diversity within the JMJD2 histone demethylase family. Biochem Biophys Res Commun 353, 973–977, doi: 10.1016/j.bbrc.2006.12.147 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Kuo AJ et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell 44, 609–620, doi: 10.1016/j.molcel.2011.08.042 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhar S et al. Loss of the major Type I arginine methyltransferase PRMT1 causes substrate scavenging by other PRMTs. Sci Rep 3, 1311, doi: 10.1038/srep01311 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fillingham J, Keogh MC & Krogan NJ GammaH2AX and its role in DNA double-strand break repair. Biochem Cell Biol 84, 568–577, doi: 10.1139/o06-072 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Cheung WL et al. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell 113, 507–517 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Sabari BR et al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol Cell 58, 203–215, doi: 10.1016/j.molcel.2015.02.029 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawicka A & Seiser C Histone H3 phosphorylation - a versatile chromatin modification for different occasions. Biochimie 94, 2193–2201, doi: 10.1016/j.biochi.2012.04.018 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hendzel MJ et al. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106, 348–360 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Hake SB et al. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc Natl Acad Sci U S A 102, 6344–6349, doi: 10.1073/pnas.0502413102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao B et al. Modulation of kinase-inhibitor interactions by auxiliary protein binding: crystallography studies on Aurora A interactions with VX-680 and with TPX2. Protein Sci 17, 1791–1797, doi: 10.1110/ps.036590.108 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polioudaki H et al. Mitotic phosphorylation of histone H3 at threonine 3. FEBS Lett 560, 39–44, doi: 10.1016/S0014-5793(04)00060-2 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Goto H et al. Identification of a novel phosphorylation site on histone H3 coupled with mitotic chromosome condensation. J Biol Chem 274, 25543–25549 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Sobel RE, Cook RG, Perry CA, Annunziato AT & Allis CD Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci U S A 92, 1237–1241 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stein EM et al. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood, doi: 10.1182/blood-2017-12-818948 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du Q, Luu PL, Stirzaker C & Clark SJ Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics 7, 1051–1073, doi: 10.2217/epi.15.39 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Issa JP, Kantarjian HM & Kirkpatrick P Azacitidine. Nat Rev Drug Discov 4, 275–276, doi: 10.1038/nrd1698 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Knijnenburg TA et al. A regression model approach to enable cell morphology correction in high-throughput flow cytometry. Mol Syst Biol 7, 531, doi: 10.1038/msb.2011.64 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hotelling H Analysis of a complex of statistical variables into principal components (1933). [Google Scholar]
- 41.van der Maaten L & Hinton G Visualizing Data using t-SNE. Journal of Machine Learning Research 9, 2579–2605 (2008). [Google Scholar]
- 42.Johnson WE, Li C & Rabinovic A Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127, doi: 10.1093/biostatistics/kxj037 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Andres-Terre M et al. Integrated, Multi-cohort Analysis Identifies Conserved Transcriptional Signatures across Multiple Respiratory Viruses. Immunity 43, 1199–1211, doi: 10.1016/j.immuni.2015.11.003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lofgren S et al. Integrated, multicohort analysis of systemic sclerosis identifies robust transcriptional signature of disease severity. JCI Insight 1, e89073, doi: 10.1172/jci.insight.89073 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Azad TD et al. Inflammatory macrophage-associated 3-gene signature predicts subclinical allograft injury and graft survival. JCI Insight 3, doi: 10.1172/jci.insight.95659 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–1218, doi: 10.1038/nmeth.2688 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheung P et al. Single-Cell Chromatin Modification Profiling Reveals Increased Epigenetic Variations with Aging. Cell, doi: 10.1016/j.cell.2018.03.079 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Begin P & Nadeau KC Epigenetic regulation of asthma and allergic disease. Allergy Asthma Clin Immunol 10, 27, doi: 10.1186/1710-1492-10-27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo H et al. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res 23, 2126–2135, doi: 10.1101/gr.161679.113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smallwood SA et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods 11, 817–820, doi: 10.1038/nmeth.3035 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buenrostro JD et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490, doi: 10.1038/nature14590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cusanovich DA et al. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348, 910–914, doi: 10.1126/science.aab1601 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagano T et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502, 59–64, doi: 10.1038/nature12593 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Venter JC et al. The sequence of the human genome. Science 291, 1304–1351, doi: 10.1126/science.1058040 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Brodin P et al. Variation in the human immune system is largely driven by non-heritable influences. Cell 160, 37–47, doi: 10.1016/j.cell.2014.12.020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]