ABSTRACT
To develop a practically applicable method for T-cell receptor (TCR)-engineered T cell immunotherapy targeting neoantigens, we have been attempting to identify neoantigen-specific T cell receptors (TCRs) and establish TCR-engineered T cells in a 3–4-month period. In this study, we report the characterization of T cell repertoires in tumor microenvironment (TME) and identification of neoantigen-specific TCRs after stimulation of patient-derived T cells. We screened 15 potential neoantigen peptides and successfully identified two CD8+HLA-dextramer+ T cells, which recognized MAGOHBG17A and ZCCHC14P368L. All three dominant TCR clonotypes from MAGOHBG17A-HLA dextramer-sorted CD8+ T cells were also found in T cells in TME, while none of dominant TCR clonotypes from ZCCHC14P368L-HLA dextramer-sorted CD8+ T cells was found in the corresponding TME. The most dominant TCRA/TCRB pairs for these two neoantigens were cloned into HLA-matched healthy donors’ T lymphocytes to generate TCR-engineered T cells. The functional assay showed MAGOHBG17A TCR-engineered T cells could be significantly activated in a mutation-specific, HLA-restricted and peptide-dose-dependent manner while ZCCHC14P368L TCR-engineered T cells could not. Our data showed neoantigen-reactive T cell clonotypes that were identified in the patient’s peripheral blood could be present in the corresponding TME and might be good TCRs targeting neoantigens.
KEYWORDS: Head and neck squamous cell carcinoma (HNSCC), T cell receptor (TCR), adoptive T cell therapy, neoantigen, cytotoxic T lymphocyte (CTL), engineered T cells
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer type and has a very poor prognosis.1 Despite advances in diagnosis and therapeutic modalities, the 5-year survival rate is still ~50% and improved very slightly in the last few decades.2,3 Development of novel treatments with higher efficacy is therefore urgently required. There are evidences that HNSCC can be targeted by immunotherapy: HNSCCs associated with HPV infection carry virus-related antigens that might be detected by T cells4,5 and HNSCC tumors carry a relatively high number of somatic mutations which might lead to a substantial number of immunogenic neoantigens.6,7 However, HNSCCs often cause dysfunction of the immune system, particularly in their tumor microenvironment (TME).8-10 Hence, the development of adoptive immunotherapies using T cells engineered with neoantigen-specific T cell receptors (TCRs) might be a new therapeutic option for this type of cancer.
Adoptive cell therapies (ACT) using tumor-infiltrating lymphocytes (TILs) achieve complete and durable regression in a subset of melanoma patients with thousands of mutations.11-13 However, ACT with TILs showed very limited efficacy in other types of solid tumors, which may correlate with the lower mutational burden in these cancers and, potentially, a lower number of neoantigen-specific T cells in the TIL populations.14-16 Based on the success of ACT using T cells engineered with chimeric antigen receptors (CARs),17-21 ACT with T cells engineered with neoantigen-specific TCRs is considered as a promising immunotherapeutic strategy.22-24 To facilitate engineering of T cells with neoantigen-specific TCRs for ACT, we previously developed a six-step protocol including: (1) prediction of neoantigen epitopes through exome and transcriptome analyses, (2) isolation of neoantigen-specific T cells after stimulation of CD8+ peripheral blood lymphocytes (PBLs) with neoantigen-loaded dendritic cells (DCs), (3) identification of a neoantigen-specific TCR sequences, (4) production of viral vectors for transfer of neoantigen-specific TCR genes, (5) engineering of T cells with neoantigen-specific TCRs and (6) functional evaluation TCR-modified T cells.25,26
While engineering of T cells has become a well-established technique, identification of cancer neoantigens as therapeutic targets and isolation of neoantigen-specific TCRs remain challenging. Our screening system allows to identify neoantigen-specific TCRs after one stimulation of CD8+ T cells with neoantigen peptides.25,26 In our previous reports, we focused on the isolation of neoantigen-specific T cells from peripheral blood mononuclear cells (PBMCs) derived from healthy donors, because (1) advanced cancer patients often suffer from myelosuppression caused by multiple regimens of chemotherapy27 and (2) such cancer patients might also have a smaller T cell diversity when compared to the T cell repertoire of healthy donors.27 In this study, we attempted to examine whether our screening approach is similarly applicable to identify neoantigen-reactive T cells in peripheral blood of HNSCC patients by stimulating patient-derived PBMCs with autologous neoantigen-loaded DCs. We further investigated whether neoantigen-reactive T cell clonotypes that were identified in the patient’s peripheral blood could be present in the corresponding TME.
Results
HNSCCS harbor a varying number of somatic mutations and are infiltrated by T cells
To analyze neoantigen-specific T cell responses in HNSCC TME, we performed whole exome sequencing (WES) and TCR repertoire analysis using tumor samples isolated from 10 HNSCC patients (Table 1), eight HPV-positive and two HPV-negative tumors (nine stage IV and one stage III tumors); these tumors were located at tongue, tonsils, mouth or larynx. WES analysis of these samples (normal and tumor DNAs) had an average sequencing depth of 78.9 × per base and identified a total number of 750 non-synonymous mutations (11–175 mutations per sample, Supplementary Table 1, Figure 1(a)). We then used available algorithms to predict neoepitopes based on the binding affinity of mutant peptides to the HLA-A alleles (estimated binding affinity of ≤500 nM, Figure 1(a), Table 2). Possible neoantigen epitopes that were predicted to bind to all HLA class I molecules are summarized in Supplementary Table 2 (the estimated binding affinity of ≤500 nM).
Table 1.
Clinicopathological characteristics of 10 HNSCC patients.
| Patient | Gender | Age | Smoking status (py) |
Stage | TNM | HPV status |
Anatomic site | Adjuvant chemoradiotherapy |
Prognosis | |
|---|---|---|---|---|---|---|---|---|---|---|
| A1 | Female | 27 | 0–10 | IVA | T4N1 | Positive | Left lateral tongue | FHX-based* chemoradiotherapy | No recurrence | |
| A2 | Male | 67 | >10 | IVA | T1N2b-c | Positive | Right tonsil | FHX-based* chemoradiotherapy | No recurrence | |
| A3 | Male | 58 | >10 | IVA | T4N2b-c | Positive | Base of tongue | FHX-based* chemoradiotherapy | No recurrence | |
| A4 | Male | 59 | 0–10 | IVA | T3N2b-c | Positive | Base of tongue | FHX-based* chemoradiotherapy | No recurrence | |
| A5 | Male | 57 | 0–10 | IVA | T2N2b-c | Positive | Base of tongue | FHX-based* chemoradiotherapy | No recurrence | |
| A6 | Male | 69 | >10 | IVA | T4N2b-c | Positive | Floor of mouth | FHX-based* chemoradiotherapy | Lung metastasis | |
| A7 | Male | 66 | 0–10 | IVA | T3N2b-c | Positive | Left tonsil | FHX-based* chemoradiotherapy | No recurrence | |
| A8 | Male | 59 | 0–10 | IVA | T2N2b-c | Positive | Left tonsil | FHX-based* chemoradiotherapy | No recurrence | |
| A9 | Male | 74 | 0–10 | IVA | T1N2b-c | Negative | Base of tongue | FHX-based* chemoradiotherapy | No recurrence | |
| A10 | Male | 70 | >10 | III | T3N0 | Negative | Larynx (true vocal cords) | FHX-based* chemoradiotherapy | No recurrence | |
py: packs/year.
*FHX; F: 5-fluorouracil, H: hydroxyurea, X: radiotherapy.
Figure 1.
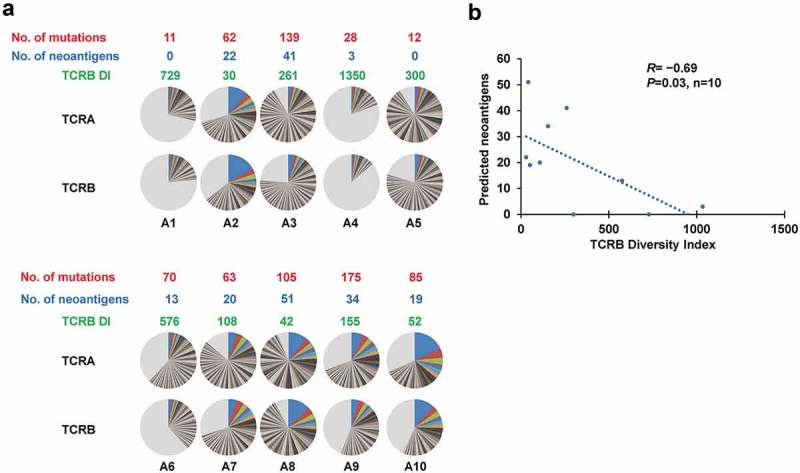
TCR repertoire analysis in TME.
(a) Distribution of both TCRA and TCRB CDR3 unique clonotypes, the numbers of mutations, the numbers of neoantigens restricted to HLA-A (the estimated binding affinity of ≤500nM) in individual tumors as well as TCRB DIs of TILs in HNSCC tumors. Pie charts depict clonotypes with the frequency of 0.1% or higher in each patient. Common colors among different pie charts do not represent identical clonotypes. The light gray portion of each pie chart contains clonotypes with the frequency of less than 0.1%. (b) Correlation analysis between the numbers of predicted neoantigen candidates and TCRB diversity index (DI). (R= −0.69, P= 0.03, n = 10).
Table 2.
Summary of patients’ predicted HLA A-restricted neoepitopes.
| Number of predicted neoepitopes |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient | Nonsynonymous mutation | Affinity (IC50) | ≦10 nM | ≦50 nM | ≦100 nM | ≦500 nM | Affinity (IC50) | ≦10 nM | ≦50 nM | ≦100 nM | ≦500 nM | Total number |
| A1 | 11 | A*32:01 | 0 | 0 | 0 | 0 | A*32:01 homozygous | 0 | ||||
| A2 | 62 | A*02:01 | 2 | 0 | 1 | 4 | A*68:01 | 0 | 5 | 3 | 7 | 22 |
| A3 | 139 | A*11:01 | 0 | 10 | 8 | 12 | A*24:02 | 0 | 1 | 3 | 7 | 41 |
| A4 | 28 | A*01:01 | 0 | 0 | 0 | 0 | A*30:01 | 1 | 1 | 0 | 1 | 3 |
| A5 | 12 | A*24:02 | 0 | 0 | 0 | 0 | A*24:02 homozygous | 0 | ||||
| A6 | 70 | A*02:01 | 1 | 3 | 3 | 3 | A*24:02 | 0 | 1 | 1 | 1 | 13 |
| A7 | 63 | A*02:02 | 2 | 2 | 1 | 10 | A*23:01 | 0 | 2 | 0 | 3 | 20 |
| A8 | 105 | A*02:01 | 1 | 3 | 4 | 10 | A*68:01 | 2 | 19 | 4 | 8 | 51 |
| A9 | 175 | A*01:01 | 0 | 1 | 0 | 3 | A*03:01 | 0 | 5 | 6 | 19 | 34 |
| A10 | 85 | A*01:01 | 0 | 1 | 0 | 2 | A*02:01 | 1 | 4 | 2 | 9 | 19 |
The TCR sequence analysis of TILs in the TME yielded total sequence reads of 395,399 ± 187,399 (mean ± one standard deviation (SD)) for TCRα and 593,294 ± 186,696 for TCRβ. These sequences included 15,944 ± 10,980 unique TCRα and 31,213 ± 13,751 unique TCRβ complementarity determining region 3 (CDR3) sequences (Supplementary Table 1). The individual number and frequency of T cell clonotypes (Figure 1(a), pie charts) were used to determine the diversity index (DI) of TILs (Figure 1(a)), which represents the clonal expansion status of T cells in the TME (higher clonal expansion results in lower DI). Interestingly, the DI calculated for TCRβ clonotypes negatively correlated with the number of predicted neoantigens (R= −0.69, P= 0.03, Figure 1(b)), implying that dominance of certain T clones in the TME of HNSCC might be indicative for T cell responses against neoantigens.
Based on previous studies demonstrating the importance of high peptide-HLA-A affinity28 and high neoantigen expression levels for the success of adoptive T cell therapy,29,30 we selected neoantigen peptides that were predicted to have higher binding affinity to HLA-A molecules and revealed high RNA expression (RNA sequencing: ≥10 reads among nearly 20,000,000 sequence reads). To this end, we selected 15 candidate mutant peptides to induce neoantigen-specific T cells from PBMCs of the HNSCC patients (1–3 peptides per patient, Table 3).
Table 3.
List of predicted neoepitopes tested with patients’ PBMC.
| Mutant peptide |
Wild-type peptide |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient | Gene | Amino acid substitution | Sequence | Affinity to HLA-A(IC50 nM) | Sequence | Affinity to HLA-A (IC50 nM) | Tumor_var (RNA) | Mutation ratio | HLA alleles |
| A2 | BRE | S170L | FLARFLLKL | 9 | FSARFLLKL | 1711 | 8 | 16% | A*02:01 |
| KRT18 | D238H | LTVEVHAPK | 21 | LTVEVDAPK | 22 | 145 | 17% | A*68:01 | |
| A3 | FASN | E2113K | MVLSSFVLAK | 15 | MVLSSFVLAE | 10204 | 42 | 15% | A*11:01 |
| EXOC3 | A111T | ATAVENLK | 24 | AAAVENLK | 81 | 10 | 24% | A*11:01 | |
| A6 | DHRS7 | I194V | SILGVISVPL | 85 | SILGIISVPL | 74 | 47 | 15% | A*02:01 |
| MAGOHB | G17A | RYYVGHKAKF | 53 | RYYVGHKGKF | 119 | 13 | 20% | A*24:02 | |
| A7 | ATP2C1 | T806S | TMSFTCFVF | 39 | TTMTFTCFV | 18 | 132 | 30% | A*23:01 |
| IFITM3 | P70T | LFMNTCCLGF | 42 | FMNPCCLGFI | 5 | 7 | 22% | A*23:01 | |
Neoantigen-specific T cells can be captured from peripheral blood of HNSCC patients
We applied our previously established protocol to stimulate neoantigen-specific T cells using CD8+ cells from the patients as starting materials. Among the 15 selected peptides, 2 induced the expansion of neoantigen-specific CD8+ T cell after in vitro stimulation as measured by staining with peptide-loaded HLA-dextramers (Figure 2(a–b)). The peptides MAGOHBG17A and ZCCHC14P368L were identified in patient A6 (predicted IC50: 53 nM, presented by HLA-A*24:02) and patient A10 (predicted IC50: 44 nM, HLA-A*02:01), respectively.
Figure 2.

Induction of neoantigen-specific CTLs and identification of TCRA and TCRB sequences of sorted CD8+/Dextramer+ T cells.
(a) Peptide-HLA dextramer assay for CD8+ T cells co-cultured with autologous DCs with/without MAGOHBG17A (left); the pie-chart showed the frequencies of unique TCRA and TCRB CDR3 sequences of sorted CD8+/Dextramer+ T cells (Middle); the line-chart showed the rank and frequency of TCRA/TCRB of HLA-dextramer-sorted cells in their corresponding TME (right). Antigen peptide of CMV pp65 for HLA-A*24:02 was used as a positive control. (b) Peptide-HLA dextramer assay for CD8+ T cells co-cultured with autologous DCs with/without ZCCHC14P368L (left); the pie-chart showed the frequencies of unique TCRA and TCRB CDR3 sequences of sorted CD8+/Dextramer+ T cells (right). Antigen peptide of CMV pp65 for HLA-A*02:01 was used as a positive control.
In the in vitro-stimulated T cells of patient A6 with autologous dendritic cells (DCs) pulsed with MAGOHBG17A peptide, the proportion of CD8+HLA-dextramer+ T cells was 0.23% (Figure 2(a) (left)). A total of 489 cells were captured and used for TCRα and TCRβ sequencing as previously described.31 The TCR sequencing revealed an oligoclonal T cell population with three dominant clones (Figure 2(a) (middle)). The clonotype with the highest TCRα frequency (33%) in the CD8+HLA-dextramer+ T cell population was identified as the most abundant clonotype in the patient’s TME (1.8%). Similarly, the most dominant TCRβ sequence (40%) after in vitro stimulation was detected to be most abundant in the TME (1.83%). The two other clonally expanded TCRα (19.9% and 19.4%) and TCRβ sequences (16.9% and 16.8%) were also found in the TME with the lower frequency (0.25–0.60%) as shown in Figure 2(a) (right). The simultaneous analysis of T cells after neoantigen-specific expansion in vitro and those in the TME provides evidence that the tumor has some levels of T cell response against the MAGOHBG17A peptide and that the predicted neoepitope is very likely to be processed and presented by cells in the TME. We selected the dominant TCR alpha and beta pair for generating TCR-encoding vectors and further performed functional analysis using TCR-engineered T cells.
After stimulation with a neoepitope ZCCHC14P368L, we sorted 626 CD8+HLA-dextramer+ T cells (0.026% of the cultured lymphocytes, Figure 2(b) (left)). TCR sequencing revealed a single dominant TCRα clonotype (93.0%) and oligoclonal TCRβ clonotypes with the most abundant one of 44% frequency (Figure 2(b) right). In contrast to the in vitro-stimulation with the MAGOHBG17A peptide, these TCR sequences were not found in the TME of patient A10. However, we selected the most abundant TCRα and TCRβ sequences in the CD8+HLA-dextramer+ T cell population for generation of TCR-engineered T cells.
Specificity of TCRs obtained from HNSCC patients can be analyzed using TCR-engineered T cells
Retroviral vectors encoding either the MAGOHBG17A or the ZCCHC14P368L-specific TCR cDNA sequences were used to engineer T cells isolated from blood of healthy donors. TCR expression was verified using antibodies directed against the TCRβ mouse constant region of the optimized TCR sequences. Transgenic TCR expression in the CD8+ T cell population was 20.9% and 10.9% for the MAGOHBG17A- or the ZCCHC14P368L-specific TCR, respectively (Figure 3(a)). We then examined whether the TCR-engineered T cells specifically bind to the neoantigen-loaded HLA-dextramers. MAGOHBG17A-specific TCR-engineered T cells were only stained when HLA-dextramers were loaded with the mutant peptide but not when loaded with the wild-type peptide (Figure 3(b) (upper)). T cells engineered with the TCR raised against ZCCHC14P368L, on the other hand, were not stained by either the mutant or the wild-type peptide-loaded HLA dextramer (Figure 3(b)(lower)).
Figure 3.

Peptide-HLA-dextramer staining for TCR-engineered T cells.
(a) The proportion of T cells expressing MAGOHBG17A- (upper panels) and ZCCHC14P368L- (lower panels) specific TCRs that were stained with an antibody against the mouse constant region of TCRβ. (b) MAGOHBG17A- (upper panels) and ZCCHC14P368L- (lower panels) TCR-engineered T cells stained with an HLA-dextramer loaded with the corresponding wild-type or mutant peptides.
To obtain further proof for neoantigen-specific reactivity of the TCR-engineered T cells, we used C1R cells expressing either HLA-A*24:02 (A24) or HLA-A*02:01 (A2) as antigen-presenting cells (APCs). C1R cells were loaded with high concentrations of either the mutant or wild-type peptide (10−5 M) and incubated with the TCR-engineered T cells. T cell activation was measured by an IFN-γ ELISPOT assay. Comparable to the HLA dextramer-binding assay, MAGOHBG17A-specific TCR-engineered T cells secreted IFN-γ only when incubated with HLA-matched C1R-A24 cells loaded with the mutant peptide. No obvious IFN-γ secretion was detected when the T cells were incubated with HLA-mismatched C1R-A2 cells or with C1R-A24 cells loaded with the wild-type MAGOHB peptide. Incubation of the C1R cell panel with T cells engineered with the TCR raised against ZCCHC14P368L confirmed that the isolated TCR was probably not specific or the establishment of TCR-engineered T cells was not functional (Supplementary Figure 1).
MAGOHBG17A-specific TCR-engineered T cells recognize low concentrations of neoantigen
To determine the functional activity of TCR-engineered T cells targeting the MAGOHBG17A neoantigen, we performed sensitivity assays and analyzed dose-dependent cytokine secretion, T cell activation, and cytotoxicity. C1R-A24 cells were loaded with different concentrations of the MAGOHBG17A peptide (ranging from 10−6 M to 10−11 M). The concentration of 10−8 M seemed to be sufficient to induce IFN-γ secretion as measured by an ELISPOT assay (Figure 4(a)). This sensitivity was confirmed when determining quantitative amounts of the TH1 cytokines IFN-γ (Figure 4(b)), IL-2 (Figure 4(c)), and TNF-α (Figure 4(d)). Cytokine levels were at background levels when TCR-engineered T cells were incubated with C1R-A24 cells loaded with the wild-type MAGOHB peptide (Figure 4(a–d)). We also evaluated the activation of TCR-engineered T cells after incubation with MAGOHBG17A peptide-loaded C1R-A24 cells by staining for the surface molecule CD137, which is upregulated upon T cell activation. CD137 upregulation was dependent on the presence of the MAGOHBG17A peptide and 10−9–10−10 M concentrations were sufficient to induce T cell activation (Figure 4(e)). To validate the cytotoxic activity of the MAGOHBG17A-specific TCR-engineered T cells, we explored peptide-dependent target cell killing. C1R-A24 cells were loaded with 10−6 M of either the mutant or the corresponding wild-type peptide, and co-cultures with TCR-engineered T cells were performed at different effector/target cell ratios. Cytotoxic activity was restricted to C1R-A24 cells loaded with the mutant peptide and could be titrated when decreasing the effector/target cell ratio (Figure 4(f)). Very modest cytotoxicity was observed when TCR-engineered T cells were incubated with C1R-A24 cells (E/T ratio of 50:1) pulsed with the wild-type peptide (Figure 4(f)).
Figure 4.
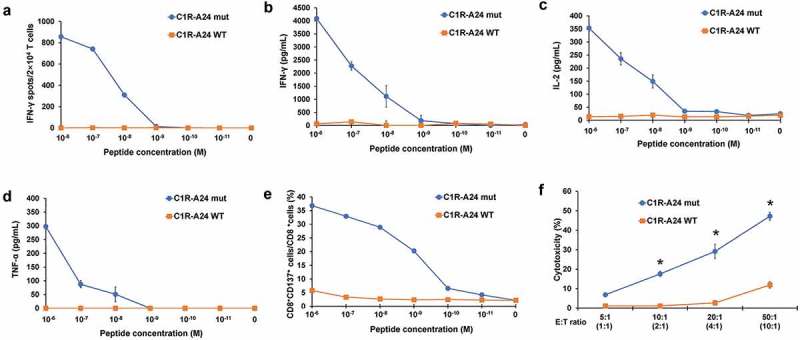
Functional assay of MAGOHBG17A-TCR-engineered T cells.
(a) IFN-γ ELISPOT assay of MAGOHBG17A TCR-engineered T cells stimulated by C1R-A24 cells pulsed with different concentrations of mutant or wild-type peptide. (b) IFN-γ ELISA assay of MAGOHBG17A TCR-engineered T cells stimulated by C1R-A24 cells loaded with different concentrations of the mutant or wild-type peptide. (c) IL-2 ELISA assay of MAGOHBG17A TCR-engineered T cells stimulated by C1R-A24 cells loaded with different concentrations of the mutant or wild-type peptide. (d) TNF-α ELISA assay of MAGOHBG17A TCR-engineered T cells stimulated by C1R-A24 cells loaded with different concentrations of the mutant or wild-type peptide. (e) CD137 staining of MAGOHBG17A TCR-engineered T cells stimulated by C1R-A24 cells pulsed with different concentrations of mutant or wild-type peptide. (f) Cytotoxic activity of MAGOHBG17A TCR-engineered T cells with different effector cell/target cell ratios. Four different ratios (5:1, 10:1, 20:1 and 50:1) were tested and the MAGOHBG17A TCR+CD8+ cell/target cell ratios were calculated to be approx. 1:1, 2:1, 4:1, 10:1 based on the peptide-dextramer staining. The asterisks indicate the statistically significant difference (p < 0.05) between two groups.
Discussion
HNSCC, which is classified to a highly immunosuppressive type, is poorly infiltrated by lymphocytes (including neoantigen-specific T cells).8 The transfer of ex vivo-activated TCR-engineered T cells that target cancer-specific neoantigens would be a promising option to overcome the suppression of endogenous immune responses. To facilitate neoantigen-specific ACT, our group established an effective and rapid method to identify neoantigen-specific TCRs and to generate TCR-engineered T cells for the clinical application. This strategy of in vitro-induction of neoantigen-specific T cells was based on whole exome and RNA sequencing of a tumor sample followed by a selection of predicted neoantigens. In a previous study using PBMCs derived from healthy donors,25 we showed that the approach is time-efficient as only two weeks were required from the T cell stimulation with neoantigen peptides to the identification of a neoantigen-specific TCR. These results were corroborated with neoantigen candidates from seven ovarian tumors, for which we identified neoantigen-specific TCRs, generated TCR-engineered T cells and confirmed their neoantigen-specific function.26 The study focusing on ovarian cancer showed that our approach would also be applicable for tumors with a relatively low mutational load.26
In our previous studies, we used T cells of HLA-matched healthy donors that were considered to be more efficient for the isolation of neoantigen-specific T cells because advanced cancer patients often suffer from myelosuppression by multiple regimens of chemotherapy. However, previous studies also showed that neoantigen-reactive TCRs could be isolated from the peripheral T cell pool of melanoma patients by tetramer staining32 or by using PD-1 as a selective marker.33 In the present study, we, therefore, addressed whether neoantigen-reactive T cells are present in peripheral blood and/or TME of HNSCC patients and whether these cells can be activated and expanded from PBMCs using cognate neoantigen peptides for stimulation.22,34,35 We further examined whether TCRs of these in vitro-stimulated T cells were included in TILs of corresponding patients.
Among the 15 peptides that we selected for in vitro-stimulation of neoantigen-specific T cells, only one led to the isolation of a functional TCR targeting a mutation identified in the HNSCC patient A6 (MAGOHBG17A). T cells engineered with this TCR recognized the mutant peptide, but not its corresponding wild-type peptide. When comparing these results to our previous screening efficiency (15–25%) using PBMCs from healthy donors,26 the efficiency of isolating neoantigen-specific TCRs from patients’ PBMCs was lower. As the MAGOHBG17A-specific TCR sequences were also the most frequent in the TME, a paralleled TCR repertoire analysis of in vitro-stimulated T cells and tumor samples might be advantageous for selecting functional TCRs. Along these lines, the TCR sequences identified after stimulating peripheral CD8+ T cells of patient A10 with the ZCCHC14P368L neoantigen peptide were not functional and not detected in the TME. And ZCCHC14P368L-TCR-engineered T cells could not recognize the ZCCHC14P368L-dextramer and were not activated by the exposure to ZCCHC14P368L-pulsed APCs although the TCR was identified from CTLs sorted out by ZCCHC14P368L-dextramer. One of the possibilities for this unexpected result could be the use of the mouse TCR constant regions for cloning of TCR. There are 38 (21%) and 50 (36%) amino-acid differences in the C regions of TCRα and TCRβ, respectively, between human and mouse. By replacing the human TCRα and TCRβ constant regions with their murine counterparts, the structure of the antigen-recognition site of TCR may be slightly influenced and TCR possibly reduced the binding affinity to the HLA-neoantigen complex.
In the present study, the screening for neoantigen-specific TCRs was limited to the patients’ HLA-A alleles due to the availability of dextramers that are needed for T cell sorting. The probability to detect neoantigen-reactive T cells in the patient’s blood would further improve, when considering all potential six MHC Class I alleles of individual patients.
In summary, the experiments have provided the first evidence that neoantigen-reactive T cells are present in the peripheral blood of HNSCC patients. Our screening approach facilitated the identification of neoantigen-specific TCR genes targeting a mutation in the cellular gene MAGOHB. Functional and specificity testing was done by generating TCR-engineered T cells using PBMCs of healthy donors. As the TCR proved to be mutation-specific and was isolated from the autologous host, its clinical application would bear virtually no toxicity risk associated with the chosen TCR and target antigen.36 Our strategy will also support the identification of targetable neoantigens for immunotherapy of HNSCC.
Methods and materials
Patient
Ten patients with HNSCC who were previously treated with or without platinum-based chemotherapy were enrolled from October 2016 till April 2017 at the University of Chicago. The clinicopathological characteristics of these patients are summarized in Table 1. Frozen tumor samples were collected either during diagnostic biopsies or during definitive surgery from 10 patients (A1-A10) and were used for extraction of DNA and RNA. DNA of peripheral blood was also extracted as a normal control. PBMCs from 50 mL of peripheral blood were separated for induction of neoantigen-specific CTLs. The study protocol was approved by the Institutional Review Board of the University of Chicago (approval number 8980, 13-0797, and 13-0526). All patients provided written informed consent for research use. This study’s involvement with human subjects complies with the Declaration of Helsinki.
Cell line
C1R cells stably expressing HLA-A*02:01 (C1R-A02) or HLA-A*24:02 (C1R-A24) were kindly provided by OncoTherapy Science, Inc (Kawasaki, Japan). Briefly, the cDNA encoding an open reading frame of HLA-A*02:01 or HLA-A*24:02 gene was amplified by PCR and inserted into an expression vector, pCAGGSn3FC. The C1R cells were transfected with these HLA-A expression vectors by electroporation with Neon Transfection System (Invitrogen) and then cultured in the presence of 1.0 mg/mL of G418 (Invitrogen) for 14 days. The expression of transfected HLA class I on the C1R cells was confirmed by flow cytometry analysis with BD FACSCanto II (BD Biosciences).
Whole-exome and transcriptome analysis
Genomic DNAs and total RNAs were extracted from frozen tumors using the AllPrep DNA/RNA mini kit (Qiagen, Catalog number 80204) according to the manufacturer’s instructions. Control genomic DNAs were extracted from peripheral blood samples using QIAamp DNA Blood Midi Kit (Qiagen, Catalog number 51183). Whole-exome libraries were built up as described previously37 and sequenced by 100-bp paired-end reads on HiSeq2500 Sequencer (Illumina, San Diego, CA, USA). The obtained sequence data were analyzed by an in-house pipeline as described previously.38 Somatic variants (single nucleotide variations (SNVs) and indels) were called using the following parameters, (i) base quality of ≥15, (ii) sequence depth of ≥10, (iii) variant depth of ≥4, (iv) variant frequency in tumor of ≥10%, (v) variant frequency in normal of <2%, and (vi) Fisher P value of <0.05.39 SNVs and indels were annotated based on RefGene using ANNOVAR.40
Identification of potential neoantigens
HLA class I genotypes of these patients were determined by OptiType algorithm41 using whole-exome data of normal samples. We then examined the binding affinities of all possible 8- to 11-mer peptides harboring each amino acid substitution to HLA class I molecules and filtered out with the predicted binding affinity to HLA molecules lower than 500 nM, using NetMHCv3.4 software.42-44 In addition, we used the transcriptome data to further select non-synonymously mutant peptides with a defined level (at least 10 reads among ~20,000,000 sequence reads) of gene expressions in tumor cells for induction of neoantigen-specific T cells. The neoantigen candidate peptides were synthesized by Innopep Inc. (San Diego, CA, USA).
Induction of neoantigen-specific cytotoxic T lymphocytes (CTLs)
Induction of neoantigen-specific T cells was performed by the protocol we developed previously.25,26 Antigen peptides of CMV pp65 for HLA-A*24:02 or HLA-A*02:01 were used as a positive control. Briefly, on day 1, 1 × 108 patient-derived PBMCs were collected using Vacutainer CPT Cell Preparation Tube (BD Biosciences, Catalog number 362761). Dynabeads CD8 Positive Isolation Kit (Thermo Fisher Scientific, Catalog number 11333D) were used to isolate CD8+ T cells from PBMCs and the remaining CD8− cells were used to generate monocyte-derived dendritic cells (DCs) using a plastic-adherence method. CD8− cells were cultured in CellGro DC (Cellgenix, Catalog number 20801-0500) medium containing 1% human AB serum (ABS, Thermo Fisher Scientific, Catalog number 34005-100), 500 U/mL of IL-4 (Thermo Fisher Scientific, Catalog number 11846-HNAE-25) and 1,000 U/mL of GM-CSF (Thermo Fisher Scientific, Catalog number PHC2015) for 72 h in 6-well plate (Thermo Fisher Scientific, Catalog number 353046) at 37°C, 5% CO2.
On day 4, 100 U/mL of IFN-γ (Thermo Fisher Scientific, Catalog number PHC4031) and 10 ng/mL of Lipopolysaccharides (LPS, Millipore-Sigma, Catalog number L4391-1MG) were added into the culture medium to induce the maturation of DCs. A total of 5.0 × 105 matured DCs were seeded into one well of 24-well plate and respective peptides with 10 µM final concentration were added to the culture medium to pulse the DCs for 16 h at 37°C. On day 5, pulsed DCs were treated with 30 µg/mL of mitomycin C (Millipore-Sigma, Catalog number M4287-5X2MG) at 37°C for 30 min, then co-cultured with autologous CD8+ T cells/expanded TILs in CellGro DC/5% ABS with 30 ng/mL of IL-21 (Thermo Fisher Scientific, Catalog number PHC0214) (each well in a 48-well plate contained 1.0 × 105 peptide-pulsed DCs and 5 × 105 CD8+ T cells). On day 8, day 10, and day 12, IL15 (Novoprotein, Catalog number C016) and IL7 (R&D Systems, Catalog number 207-IL-200) (final concentration 5 ng/mL) were newly added into the respective wells.
On day 15, neoantigen-specific T cells were stained using peptide-HLA dextramers (Immudex, Copenhagen, Denmark) for each neoantigen peptide and analyzed by flow cytometry technology. CD8+/Dextramer+ T cells were sorted out and used for the following TCR sequencing analysis.
TCR sequencing analysis of original tumor and sorted neoantigen-specific T cells
TCR sequencing was performed using the methods described previously.31,43,45 In brief, total RNAs were extracted from tumors with AllPrep DNA/RNA mini kit (Qiagen, Catalog number 80207) or sorted CD8+/Dextramer+ T cells with PicoPure RNA Isolation Kit (Life Technologies, Catalog number KIT0204). The cDNAs with common 5ʹ-RACE adapters were synthesized from total RNA using SMART library construction kit (Clontech, Catalog number 634901). The TCRA and TCRB cDNAs were amplified by PCR using a forward primer for the SMART adapter and reverse primers corresponding to the constant regions of each of TCRA and TCRB. After adding the Illumina index sequences with barcodes using the Nextera XT Index kit (Illumina, Catalog number FC-131-2004), the prepared libraries were sequenced by 300-bp paired-end reads on Illumina MiSeq platform, using MiSeq Reagent v3 600-cycles kit (Illumina, Catalog number MS-102-3003). Obtained sequence reads were analyzed using Tcrip software.31
Engineered-TCR T cells
Both TCRA and TCRB sequences were codon-optimized, synthesized by GeneArt (ThermoFisher Scientific, Waltham, MA) and cloned into pMP71-PRE vector as described previously.46 To increase TCR surface expression, we used TCRs with mouse constant regions.47 Transient retroviral supernatants were generated and PBMCs from healthy donors were transduced as described previously.48
Enzyme-linked immunospot (ELISPOT) and enzyme-linked immunosorbent assay (ELISA) assay
Interferon (IFN)-γ secretion of T cells were detected by ELISPOT using Human IFN-γ ELISpotPRO kit (MABTECH, Catalog number 3420-2APW-10) according to the manufacturer’s instruction. Briefly, APCs (C1R-A24 or C1R-A02 cells) were pulsed with each respective peptide before co-culture for 16 h at 37°C, 5% CO2. T cells were pre-treated with IL-2 (35 U/mL) for 16 h and then co-cultured with the peptide-pulsed APCs (2 × 104 APCs and 5 × 104 engineered T cells/well) at 37°C for 20 h in 96-well plate. Spots were captured and analyzed by an automated ELISPOT reader, ImmunoSPOT S4 (Cellular Technology Ltd, Shaker Heights, OH) and the ImmunoSpot Professional Software package, Version 5.1 (Cellular Technology Ltd).
OptEIA Human IFN-γ ELISA set, OptEIA Human IL2 ELISA set, OptEIA Human TNFα ELISA set (BD Biosciences, Catalog number 555142, 555190, 555212) were used to measure the secreted IFN-γ, IL2, and TNFα levels in the supernatant of co-cultured engineered T cells and APCs. Briefly, APCs were pulsed with respective peptides at 37°C for 16 h and 5% CO2. T cells were pre-treated with IL-2 (35 U/mL) for 16 h and then cocultured with the peptide-pulsed APCs (2 × 104 cells/well) at 37°C for 20 h in a 96-well plate. The supernatant was transferred into another 96-well plate, and the concentration of each protein was measured according to the manufacturer’s instruction.
Cytotoxic assay
The cytotoxic assay was performed using CytoTox 96 Non-Radioactive Cytotoxicity Assay kit (Promega, Catalog number G1780) according to the manufacturer’s instruction. Briefly, C1R-A24 cells were pulsed with each of respective peptides (10−6 M) at 37°C for 16 h and used as target cells. Effector cells and target cells were incubated in 96-well plate at 5:1, 10:1, 20:1 and 50:1 ratios for 4 h at 37°C, 5% CO2. Experiments were conducted in triplicate. Maximum lactate dehydrogenase (LDH) release from target cells was measured by the addition of lysis solution. The spontaneous LDH release of effector and target cells was measured by separate incubation of the respective population. After 4-h incubation, the plate was centrifuged at 250 × g for 4 min. The supernatant was transferred to another 96-well plate. The substrate was added to each well, and the plate was incubated for 30 min in the dark at room temperature. Stop solution was added to terminate the reaction and absorbance at 490 nm was recorded. The percentage of cytotoxic activity was calculated according to the following formula: % Cytotoxicity = [(Experimental-Effector Spontaneous-Target Spontaneous)/(Target Maximum-Target Spontaneous)] × 100.
Statistical analysis
Correlation analysis and data description were done using GraphPad Prism version 7.01 (GraphPad software, La Jolla, CA). P value of <0.05 was considered to be statistically significant.
Funding Statement
This work was supported partly by a research grant from OncoTherapy Science, Inc.
Acknowledgments
We thank Drs. Rui Yamaguchi, Seiya Imoto, and Satoru Miyano at The University of Tokyo for developing the algorithm of TCR repertoire analysis and helpful support in data management. The super-computing resource (http://sc.hgc.jp/shirokane.html) was provided by Human Genome Center, Institute of Medical Science, The University of Tokyo.
Disclosure of potential conflicts of interest
Y. N. is a stockholder and a scientific advisor of OncoTherapy Science, Inc.; J-H. Park is a scientific advisor of OncoTherapy Science, Inc. No potential conflicts of interest were disclosed by the other authors.
Authors’ contributions
Y.N. designed, supervised the project and edited the manuscript; L.R. performed CTL induction, evaluation of TCR-engineered T cells and wrote the manuscript; M.L. built the engineered T cells and edited the manuscript; B.D. and T.M. assisted experiments; K.K. analyzed data and interpreted data; M.H. performed the TCR-sequencing of sorted cells; T.K. and J.P. directed and supervised the techniques involved; V.S., T.S., E.V., and N.A. provided the samples, clinical information, and advice to the project.
Supplemental material
Supplemental data for this article can be accessed on the publisher’s website.
Abbreviations
- HNSCC
Head and neck squamous cell carcinoma
- CDR3
Complementarity determining region 3
- DI
Diversity index
- TCR
T-cell receptor
- CTLs
Cytotoxic T lymphocytes
- TILs
Tumor-infiltrating lymphocytes
- TME
Tumor microenvironment
- ACT
Adoptive cell therapies
References
- 1.Global Burden of Disease Cancer C, Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, Dandona L, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3:524–548. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaturvedi AK, Anderson WF, Lortet-Tieulent J, Curado MP, Ferlay J, Franceschi S, Rosenberg PS, Bray F, Gillison ML.. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol. 2013;31:4550–4559. doi: 10.1200/JCO.2013.50.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Razzaghi H, Saraiya M, Thompson TD, Henley SJ, Viens L, Wilson R. Five-year relative survival for human papillomavirus-associated cancer sites. Cancer. 2018;124:203–211. doi: 10.1002/cncr.30947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lorenz FKM, Ellinger C, Kieback E, Wilde S, Lietz M, Schendel DJ, Uckert W. Unbiased identification of T-Cell receptors targeting immunodominant peptide-MHC complexes for T-cell receptor immunotherapy. Hum Gene Ther. 2017;28:1158–1168. doi: 10.1089/hum.2017.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scholten KB, Kramer D, Kueter EW, Graf M, Schoedl T, Meijer CJ, Schreurs MWJ, Hooijberg E. Codon modification of T cell receptors allows enhanced functional expression in transgenic human T cells. Clin Immunol. 2006;119:135–145. doi: 10.1016/j.clim.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexandroff AB, Nicholson S, Patel PM, Jackson AM. Recent advances in bacillus Calmette-Guerin immunotherapy in bladder cancer. Immunotherapy. 2010;2:551–560. doi: 10.2217/imt.10.32. [DOI] [PubMed] [Google Scholar]
- 8.Moskovitz JM, Ferris RL. Tumor immunology and immunotherapy for head and neck squamous cell carcinoma. J Dent Res. 2018;97:622–626. doi: 10.1177/0022034518759464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saleh K, Eid R, Haddad FG, Khalife-Saleh N, Kourie HR. New developments in the management of head and neck cancer - impact of pembrolizumab. Ther Clin Risk Manag. 2018;14:295–303. doi: 10.2147/TCRM.S125059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothschild U, Muller L, Lechner A, Schlosser HA, Beutner D, Laubli H, Zippelius A, Rothschild SI, et al. Immunotherapy in head and neck cancer - scientific rationale, current treatment options and future directions. Swiss Med Wkly. 2018;148:w14625. doi: 10.4414/smw.2018.14575. [DOI] [PubMed] [Google Scholar]
- 11.Saint-Jean M, Knol AC, Volteau C, Quereux G, Peuvrel L, Brocard A, Pandolfino M-C, Saiagh S, Nguyen J-M, Bedane C, et al. Adoptive cell therapy with tumor-infiltrating lymphocytes in advanced melanoma patients. J Immunol Res. 2018;2018:3530148. doi: 10.1155/2018/3530148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersen R, Donia M, Ellebaek E, Borch TH, Kongsted P, Iversen TZ, Hölmich LR, Hendel HW, Met Ö, Andersen MH, et al. Long-lasting complete responses in patients with metastatic melanoma after adoptive cell therapy with tumor-infiltrating lymphocytes and an attenuated IL2 regimen. Clin Cancer Res. 2016;22:3734–3745. doi: 10.1158/1078-0432.CCR-15-1879. [DOI] [PubMed] [Google Scholar]
- 13.Cipponi A, Wieers G, van Baren N, Coulie PG. Tumor-infiltrating lymphocytes: apparently good for melanoma patients. But why? Cancer Immunol Immunother. 2011;60:1153–1160. doi: 10.1007/s00262-011-1026-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayor P, Starbuck K, Zsiros E. Adoptive cell transfer using autologous tumor infiltrating lymphocytes in gynecologic malignancies. Gynecol Oncol. 2018;150:361–369. doi: 10.1016/j.ygyno.2018.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butt SU, Malik L. Role of immunotherapy in bladder cancer: past, present and future. Cancer Chemother Pharmacol. 2018;81:629–645. doi: 10.1007/s00280-018-3518-7. [DOI] [PubMed] [Google Scholar]
- 16.Inarrairaegui M, Melero I, Sangro B. Immunotherapy of hepatocellular carcinoma: facts and hopes. Clin Cancer Res. 2018;24:1518–1524. doi: 10.1158/1078-0432.CCR-17-0289. [DOI] [PubMed] [Google Scholar]
- 17.Prasad V. Immunotherapy: tisagenlecleucel - the first approved CAR-T-cell therapy: implications for payers and policy makers. Nat Rev Clin Oncol. 2018;15:11–12. doi: 10.1038/nrclinonc.2017.156. [DOI] [PubMed] [Google Scholar]
- 18.Androulla MN, Lefkothea PC. CAR T-cell therapy: a new era in cancer immunotherapy. Curr Pharm Biotechnol. 2018;19:5–18. doi: 10.2174/1389201019666180418095526. [DOI] [PubMed] [Google Scholar]
- 19.Lulla PD, Hill LC, Ramos CA, Heslop HE. The use of chimeric antigen receptor T cells in patients with non-Hodgkin lymphoma. Clin Adv Hematol Oncol. 2018;16:375–386. [PMC free article] [PubMed] [Google Scholar]
- 20.Arabi F, Torabi-Rahvar M, Shariati A, Ahmadbeigi N, Naderi M. Antigenic targets of CAR T cell therapy. A retrospective view on clinical trials. Exp Cell Res. 2018;369:1–10. doi: 10.1016/j.yexcr.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 21.June CH, O’Connor RS, Kawalekar OU, Ghassemi S, Milone MC. CAR T cell immunotherapy for human cancer. Science. 2018;359:1361–1365. doi: 10.1126/science.aar6711. [DOI] [PubMed] [Google Scholar]
- 22.Ye B, Smerin D, Gao Q, Kang C, Xiong X. High-throughput sequencing of the immune repertoire in oncology: applications for clinical diagnosis, monitoring, and immunotherapies. Cancer Lett. 2018;416:42–56. doi: 10.1016/j.canlet.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 23.Schmitt TM, Stromnes IM, Chapuis AG, Greenberg PD. New strategies in engineering T-cell receptor gene-modified T cells to more effectively target malignancies. Clin Cancer Res. 2015;21:5191–5197. doi: 10.1158/1078-0432.CCR-15-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blankenstein T, Leisegang M, Uckert W, Schreiber H. Targeting cancer-specific mutations by T cell receptor gene therapy. Curr Opin Immunol. 2015;33:112–119. doi: 10.1016/j.coi.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato T, Matsuda T, Ikeda Y, Park JH, Leisegang M, Yoshimura S, Hikichi T, Harada M, Zewde M, Sato S, et al. Effective screening of T cells recognizing neoantigens and construction of T-cell receptor-engineered T cells. Oncotarget. 2018;9:11009–11019. doi: 10.18632/oncotarget.24232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuda T, Leisegang M, Park JH, Ren L, Kato T, Ikeda Y, Harada M, Kiyotani K, Lengyel E, Fleming GF, et al. Induction of neoantigen-specific cytotoxic T cells and construction of T-cell receptor-engineered T cells for ovarian cancer. Clin Cancer Res. 2018;24:5357–5367. doi: 10.1158/1078-0432.CCR-18-0142. [DOI] [PubMed] [Google Scholar]
- 27.Stronen E, Toebes M, Kelderman S, van Buuren MM, Yang W, van Rooij N, Donia M, Böschen M-L, Lund-Johansen F, Olweus J, et al. Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science. 2016;352:1337–1341. doi: 10.1126/science.aaf2288. [DOI] [PubMed] [Google Scholar]
- 28.Engels B, Engelhard VH, Sidney J, Sette A, Binder DC, Liu RB, Kranz DM, Meredith SC, Rowley DA, Schreiber H. Relapse or eradication of cancer is predicted by peptide-major histocompatibility complex affinity. Cancer Cell. 2013;23:516–526. doi: 10.1016/j.ccr.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leisegang M, Engels B, Schreiber K, Yew PY, Kiyotani K, Idel C, Arina A, Duraiswamy J, Weichselbaum RR, Uckert W, et al. Eradication of large solid tumors by gene therapy with a T-cell receptor targeting a single cancer-specific point mutation. Clin Cancer Res. 2016;22:2734–2743. doi: 10.1158/1078-0432.CCR-15-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leisegang M, Kammertoens T, Uckert W, Blankenstein T. Targeting human melanoma neoantigens by T cell receptor gene therapy. J Clin Invest. 2016;126:854–858. doi: 10.1172/JCI83465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang H, Yamaguchi R, Liu X, Daigo Y, Yew PY, Tanikawa C, Matsuda K, Imoto S, Miyano S, Nakamura Y. Quantitative T cell repertoire analysis by deep cDNA sequencing of T cell receptor alpha and beta chains using next-generation sequencing (NGS). Oncoimmunology. 2014;3:e968467. doi: 10.4161/21624011.2014.968467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen CJ, Gartner JJ, Horovitz-Fried M, Shamalov K, Trebska-McGowan K, Bliskovsky VV, Parkhurst MR, Ankri C, Prickett TD, Crystal JS, et al. Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes. J Clin Invest. 2015;125:3981–3991. doi: 10.1172/JCI82416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gros A, Parkhurst MR, Tran E, Pasetto A, Robbins PF, Ilyas S, Prickett TD, Gartner JJ, Crystal JS, Roberts IM, et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med. 2016;22:433–438. doi: 10.1038/nm.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han Y, Li H, Guan Y, Huang J. Immune repertoire: a potential biomarker and therapeutic for hepatocellular carcinoma. Cancer Lett. 2016;379:206–212. doi: 10.1016/j.canlet.2015.06.022. [DOI] [PubMed] [Google Scholar]
- 35.Wang CY, Yu PF, He XB, Fang YX, Cheng WY, Jing ZZ. alphabeta T-cell receptor bias in disease and therapy (Review). Int J Oncol. 2016;48:2247–2256. doi: 10.3892/ijo.2016.3492. [DOI] [PubMed] [Google Scholar]
- 36.Kunert A, Obenaus M, Lamers CHJ, Blankenstein T, Debets R. T-cell receptors for clinical therapy: in vitro assessment of toxicity risk. Clin Cancer Res. 2017;23:6012–6020. doi: 10.1158/1078-0432.CCR-17-1012. [DOI] [PubMed] [Google Scholar]
- 37.Ren L, Matsuda T, Deng B, Kiyotani K, Kato T, Park JH, Seiwert TY, Vokes EE, Agrawal N, Nakamura Y. Similarity and difference in tumor-infiltrating lymphocytes in original tumor tissues and those of in vitro expanded populations in head and neck cancer. Oncotarget. 2018;9:3805–3814. doi: 10.18632/oncotarget.23454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 40.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szolek A, Schubert B, Mohr C, Sturm M, Feldhahn M, Kohlbacher O. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics. 2014;30:3310–3316. doi: 10.1093/bioinformatics/btu548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kiyotani K, Park JH, Inoue H, Husain A, Olugbile S, Zewde M, Nakamura Y, Vigneswaran WT. Integrated analysis of somatic mutations and immune microenvironment in malignant pleural mesothelioma. Oncoimmunology. 2017;6:e1278330. doi: 10.1080/2162402X.2016.1278330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choudhury NJ, Kiyotani K, Yap KL, Campanile A, Antic T, Yew PY, Steinberg G, Park JH, Nakamura Y, O’Donnell PH. Low T-cell receptor diversity, high somatic mutation burden, and high neoantigen load as predictors of clinical outcome in muscle-invasive bladder cancer. Eur Urol Focus. 2016;2:445–452. doi: 10.1016/j.euf.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 44.Lundegaard C, Lund O, Nielsen M. Accurate approximation method for prediction of class I MHC affinities for peptides of length 8, 10 and 11 using prediction tools trained on 9mers. Bioinformatics. 2008;24:1397–1398. doi: 10.1093/bioinformatics/btn128. [DOI] [PubMed] [Google Scholar]
- 45.Kato T, Iwasaki T, Uemura M, Nagahara A, Higashihara H, Osuga K, Ikeda Y, Kiyotani K, Park J-H, Nonomura N, et al. Characterization of the cryoablation-induced immune response in kidney cancer patients. Oncoimmunology. 2017;6:e1326441. doi: 10.1080/2162402X.2017.1326441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leisegang M, Turqueti-Neves A, Engels B, Blankenstein T, Schendel DJ, Uckert W, Noessner E. T-cell receptor gene-modified T cells with shared renal cell carcinoma specificity for adoptive T-cell therapy. Clin Cancer Res. 2010;16:2333–2343. doi: 10.1158/1078-0432.CCR-09-2897. [DOI] [PubMed] [Google Scholar]
- 47.Cohen CJ, Zhao Y, Zheng Z, Rosenberg SA, Morgan RA. Enhanced antitumor activity of murine-human hybrid T-cell receptor (TCR) in human lymphocytes is associated with improved pairing and TCR/CD3 stability. Cancer Res. 2006;66:8878–8886. doi: 10.1158/0008-5472.CAN-06-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leisegang M, Engels B, Meyerhuber P, Kieback E, Sommermeyer D, Xue SA, Reuss S, Stauss H, Uckert W. Enhanced functionality of T cell receptor-redirected T cells is defined by the transgene cassette. J Mol Med (Berl). 2008;86:573–583. doi: 10.1007/s00109-008-0317-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


